Abstract
Legionella pneumophila, the causative agent of Legionnaires' disease, is an intracellular pathogen of amoebae, macrophages, and epithelial cells. The pathology of Legionella infections involves alveolar cell destruction, and several proteins of L. pneumophila are known to contribute to this ability. By screening a genomic library of L. pneumophila, we found an additional L. pneumophila gene, plaB, which coded for a hemolytic activity and contained a lipase consensus motif in its deduced protein sequence. Moreover, Escherichia coli harboring the L. pneumophila plaB gene showed increased activity in releasing fatty acids predominantly from diacylphospho- and lysophospholipids, demonstrating that it encodes a phospholipase A. It has been reported that culture supernatants and cell lysates of L. pneumophila possess phospholipase A activity; however, only the major secreted lysophospholipase A PlaA has been investigated on the molecular level. We therefore generated isogenic L. pneumophila plaB mutants and tested those for hemolysis, lipolytic activities, and intracellular survival in amoebae and macrophages. Compared to wild-type L. pneumophila, the plaB mutant showed reduced hemolysis of human red blood cells and almost completely lost its cell-associated lipolytic activity. We conclude that L. pneumophila plaB is the gene encoding the major cell-associated phospholipase A, possibly contributing to bacterial cytotoxicity due to its hemolytic activity. On the other hand, in view of the fact that the plaB mutant multiplied like the wild type both in U937 macrophages and in Acanthamoeba castellanii amoebae, plaB is not essential for intracellular survival of the pathogen.
Legionella pneumophila is an inhabitant of fresh water, where it intracellularly colonizes protozoa (20). When bacteria-laden aerosols are inhaled by humans, L. pneumophila exploits alveolar macrophages and epithelial cells for its multiplication, leading to a severe pneumonia characterized by destruction of alveolar cells (60). The cytopathology of Legionnaires' disease involves several cytotoxic or hemolytic factors produced by L. pneumophila, for example, the zinc metalloprotease ProA, the legiolysin Lly, and several pore-forming toxins, one of which is an RTX (repeats in structural toxin) protein (12, 26, 31, 35, 36, 47, 61). The zinc metalloprotease ProA is the major extracellular protease of L. pneumophila and its export depends on the L. pneumophila type II protein secretion system (28, 37). The enzyme hydrolyzes a broad spectrum of protein substrates and confers hemolytic as well as cytolytic activities (47, 53). Additionally, ProA has been shown to contribute to bacterial pathogenesis in a guinea pig model of pneumonia (38). Another hemolytic, but not cytotoxic, protein is the L. pneumophila legiolysin Lly, which is also responsible for color production and fluorescence of the bacterium (61). Pore-forming activities of L. pneumophila confer contact-dependent hemolytic and cytotoxic activities toward a variety of cells, especially at high bacterial numbers (12, 31, 36). The L. pneumophila RTX toxin RtxA has been shown to contribute to cytotoxicity, to pore formation, and to the initial steps of infection, namely, adherence and entry of the bacterium into host cells (12, 13). Pore formation is furthermore an indispensable step in bacterial egress after bacterial intracellular multiplication in both amoebae and macrophages (1, 40). Pore formation in L. pneumophila depends on the integrity of several genes of the icm/dot locus (e.g., dotA, icmQ, icmR, and icmT), responsible for the expression of a type IV protein secretion machinery involved in pathogenesis (14, 36, 40).
In addition to proteases and pore-forming toxins, bacterial lipolytic enzymes are known to confer hemolytic and cytotoxic activities by the destruction of cell membrane lipids and/or generation of membrane-perturbing reaction products, as shown for Clostridium perfringens alpha-toxin, a phospholipase C, or Campylobacter coli PldA, a phospholipase A (27, 54, 59). L. pneumophila also possesses a variety of lipolytic activities, including lipase, phospholipase A, and lysophospholipase A activities, secreted into the culture medium as well as associated with the bacterium (3, 4, 22, 23, 25). So far, research has focused primarily on activities secreted into the extrabacterial environment. The lipase LipA and the putative lipase/esterase LipB hydrolyze esters of palmitic or caprylic acid, respectively, but are not essential for the cytopathogenicity and intracellular survival of L. pneumophila in U937 macrophages and Hartmannella amoebae (4). The lysophospholipase A PlaA, specific for lysophospholipid hydrolysis, detoxifies lysophosphatidylcholine, a pore-forming agent which can be generated, for example, by the action of Legionella phospholipase A on lung surfactant (24, 25). plaA knockout mutants are not impaired in their intracellular infection of macrophages and amoebae (25). The secreted phospholipase A activities hydrolyzing diacylphospholipids, as well as the cell-associated phospholipase A activity, still remain to be characterized on the molecular level. Here, we identify the gene for the major cell-associated phospholipase A, which was found by screening a genomic L. pneumophila library for new hemolysis genes. Furthermore, we examine its contribution to the hemolytic activity of the bacterium and its significance in intracellular infection.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
L. pneumophila sg1 strain Corby (33) was used to construct an expression library in Escherichia coli (30) for mutagenesis of the Legionella plaB gene and later served as a wild-type control. Legionella strains used for Southern blot analysis were L. pneumophila sg1 strain Philadelphia I (ATCC 33152), L. pneumophila sg1 strain Msp19 (7), L. pneumophila sg1 strain 685 (7), L. pneumophila sg3 strain Bloomington (ATCC 33155), L. pneumophila sg3 strain U22 (7), L. pneumophila sg4 strain Los Angeles (ATCC 33156), L. pneumophila sg6 strain Chicago-2 (ATCC 33215), L. pneumophila 664 sg6 (patient isolate) (7), L. pneumophila type strains (sg7, sg10, sg12, and sg13) (kindly provided by P. C. Lück, Dresden, Germany), L. anisa (21), L. dumoffii (ATCC 33279), L. erythra (51), L. gormanii (ATCC 33297), L. hackeliae sg1 and sg2 (ATCC 33250 and ATCC 35999, respectively), L. israelensis (ATCC 43119), L. longbeachae sg1 and sg2 (ATCC 33462 and ATCC 33484, respectively), L. micdadei (ATCC 33218), L. oakridgensis (ATCC 33761), and Sarcobium (Legionella) lyticum (PCM 2298; from the Polish Culture of Microorganisms).
L. pneumophila was routinely grown on buffered charcoal-yeast extract (BCYE) agar for 2 days at 37°C (19). For extracellular growth, L. pneumophila was cultured in buffered yeast extract (BYE) broth at 37°C with shaking at 350 rpm. Bacterial growth was monitored by determining the optical density at 660 nm (OD660) of the culture with a Beckman spectrophotometer DU520 (Beckman Coulter, Unterschleißheim, Germany), following inoculation to an OD660 of 0.2 to 0.3. E. coli strain DH5α, the host for new recombinant plasmids, was grown in Luria-Bertani (LB) broth or agar (6). An E. coli (EIEC 12860, a clinical isolate provided by Helge Karch; serotype O:124) cylA knockout mutant was kindly provided by Christian Hüttinger (University of Würzburg, Würzburg, Germany). When appropriate, media were supplemented with antibiotics at final concentrations suitable for L. pneumophila or E. coli: kanamycin at 25 or 50 μg/ml, respectively; chloramphenicol at 6 or 30 μg/ml, respectively; and ampicillin at 100 μg/ml.
Preparation of culture supernatants and cell lysates.
Culture supernatants for assessment of hydrolytic activities were obtained at the end of exponential growth (OD660, 2.2 to 2.3) by centrifugation for 5 min at 5,000 × g. For the generation of cell lysates, bacteria from the late-exponential phase were pelleted by centrifugation as described above and then lysed as described previously, except that the lysate was repeatedly passaged through a 22-gauge needle (25). Culture supernatants and cell lysates were either tested immediately for enzymatic activities or stored overnight at 4°C.
DNA techniques and sequence analysis.
An expression library of L. pneumophila Corby was constructed as described previously (30). E. coli DH5α was used for the propagation of recombinant plasmid DNA. The following vectors were used: pUC18 (backbone in plasmids pKHL102, pCL102-1, pCL102-2, pCL102-3, pKH190, pKH194, and phlyCABD) or pUC19 (Amersham Biosciences, Freiburg, Germany), pBCKS+ (backbone in plasmid pKH192; Stratagene, Heidelberg, Germany), and pBOC20 (backbone in plasmid pKH195) (44). Genomic and plasmid DNAs were prepared according to standard protocols (49). PCR was carried out using a TRIO-Thermoblock or a T-Gradient thermocycler (Biometra, Göttingen, Germany) and AmpliTaq polymerase (Perkin-Elmer, Weiterstadt, Germany) or Taq DNA polymerase (New England Biolabs, Frankfurt am Main, Germany). Foreign DNA was introduced into bacterial strains by electroporation with a Bio-Rad (Munich, Germany) gene pulser according to the manufacturer's specifications. E. coli or L. pneumophila strains were electroporated at 2.0 or 2.3 kV, respectively, 200 or 100 Ω, respectively, and 25 mF. Both strands of plasmid DNA were sequenced with infrared dye-labeled primers by using an automated DNA sequencer (LI-COR-DNA 4000; MWG-Biotech, Ebersberg, Germany). Nucleotide and translated protein sequences were analyzed by using the Genetics Computer Group package, the PEDANT website (http://pedant.gsf.de/), SMART (http://smart.embl-heidelberg.de), the SignalP program (42), and the L. pneumophila genome project web page (http://genome3.cpmc.columbia.edu/∼legion/). Sequence database searches as well as protein alignments were performed by the BLAST algorithm (2).
Southern hybridization.
Chromosomal DNA from various Legionella strains was digested with SacII and HindIII and then subjected to electrophoresis, and fragments were transferred to a nylon membrane (Pall, Dreieich, Germany) by capillary blotting. The DNA probe, generated from primers pla1 (5′-GAATTCAGTATAAAATAATCTAATATC-3′; located 70 bp in front of the start codon) and pla2 (5′-TCTAGATTAATCTATCTTTTTCCCAGTTG-3′; ending with the stop codon of plaB), containing the complete L. pneumophila plaB gene, was used as a plaB-specific probe. DNA probes were labeled and detected by using the nonradioactive enhanced chemiluminescence detection kit (ECL; Amersham Biosciences). Hybridization was performed at 42°C overnight. Membranes were washed twice, for 10 min each time, at 42°C with wash buffer I (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]-0.1% sodium dodecyl sulfate [SDS]) and twice, for 5 min each time, at room temperature with wash buffer II (2× SSC). Detection was carried out as described previously (30).
Gene cloning and Legionella mutant construction.
Two different strategies were used for L. pneumophila plaB mutant generation. For both, a plasmid containing the disrupted plaB gene and allelic exchange were used to introduce a Kmr insertion mutation into the chromosome of strain Corby. The complete plaB gene was amplified by primers pla1 and pla2, cloned into pUC18, yielding pKH190, and sequenced. A Kmr cassette was then inserted into the SacII restriction site of the plaB gene of pKH190, resulting in pKH194 (see Fig. 1). This construct was subcloned into pBOC20, yielding pKH195. L. pneumophila plaB knockout mutants were generated by electroporation of pKH195 into L. pneumophila Corby and allelic exchange. Screening for mutants obtained by double crossover was performed as described recently (17). Integration of the Kmr cassette into the chromosomal plaB gene was verified by Southern blot analysis (data not shown). The second method for mutant generation involved the introduction of plasmid pKH194 into L. pneumophila Corby and of the Kmr cassette into the chromosomal plaB gene by natural transformation and homolog recombination (25). PCR and Southern blot analysis were used to examine Kmr legionellae for the presence of the plaB mutation (49). For complementation studies, the complete plaB gene of pKH190 was subcloned into vector pBCKS+, resulting in plasmid pKH192 (see Fig. 1). pBCKS+ and pKH192 were introduced into wild-type and mutant L. pneumophila by electroporation.
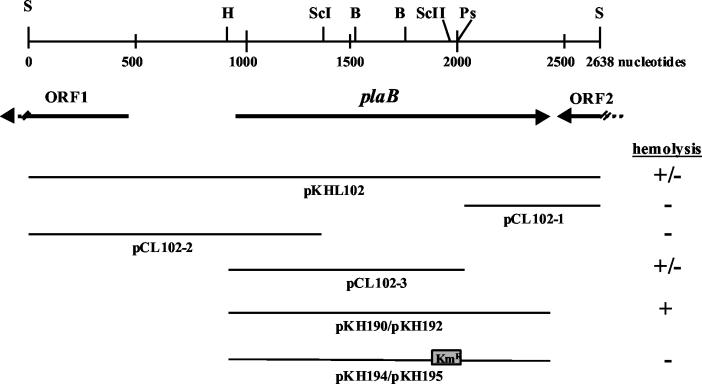
FIG. 1.
plaB locus in L. pneumophila and recombinant E. coli. (Top) Diagram represents the L. pneumophila chromosome region that contains the phospholipase A gene plaB, along with the locations of the relevant restriction enzyme sites (B, BamHI; H, HindIII; Ps, PstI; S, Sau3A1; ScI, SacI; ScII, SacII). (Center) Horizontal arrows depict the relative location, size, and orientation of plaB and neighboring ORFs. (Bottom) Lines represent the segments of Legionella DNA that were cloned into plasmid vectors. Plasmids pKH194 and pKH195 contained a Kmr gene cassette. Plus and minus signs indicate whether recombinant E. coli exhibited hemolytic activity on human red blood agar plates.
Hemolysis assays.
For detection of hemolytic activity of L. pneumophila or E. coli strains on blood agar media, 30 ml of blood/liter was added to BCYE− (without charcoal) or LB agar medium, respectively. Bacterial strains were inoculated onto human or sheep blood agar plates and incubated for 48 h at 37°C. Recombinant E. coli and wild-type, mutant, or complemented L. pneumophila strains were prepared for quantitative liquid hemolysis assays in the following manner. E. coli logarithmic-growth-phase cultures were set to an OD600 of 1, 2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added, and bacteria were incubated for 2 h. L. pneumophila strains were grown on BCYE agar at 37°C for 1 day. Subsequently, bacteria were resuspended in phosphate-buffered saline (PBS) and adjusted to an OD600 of 1. A 200-μl volume of E. coli or L. pneumophila bacteria was mixed with 800 μl of a suspension of 0.5 ml of human red blood cells in 40 ml of PBS. To test for the hemolytic activity of L. pneumophila, sample mixtures were centrifuged (at 800 × g for 2 min) prior to incubation. After incubation for 20 h (E. coli) or 7 h (Legionella) at 37°C, samples were centrifuged (at 800 × g for 2 min), and the OD415 of the supernatant was determined.
Enzymatic assay for lipolytic activities.
Enzymatic activities were detected as described previously (22, 25) with minor modifications. Briefly, different phospholipids or lipids were incubated with bacterial culture supernatants or cell lysates in a mixture containing 6.7 mM lipid substrate (1-monopalmitoyllysophosphatidylcholine [MPLPC], 1-monopalmitoyllysophosphatidylglycerol [MPLPG], 1-monopalmitoylglycerol [1-MPG], 1,2-dipalmitoylphosphatidylglycerol [DPPG], 1,2-dipalmitoylphosphatidylcholine [DPPC], 1,2-dipalmitoylglycerol [1,2-DG], or tripalmitoylglycerol [TG]), 3 mM NaN3, 0.5% (vol/vol) Triton X-100, and 20 mM Tris-HCl (pH 7.2). All lipids, including standards for thin-layer chromatography (TLC), were obtained from Sigma Chemicals (Munich, Germany) or Avanti Polar Lipids, Inc. (Alabaster, Ala.). Prior to incubation, the lipid substrates were vortexed for 15 min at 37°C and then exposed to ultrasonication (Sonoplus; Bandelin, Berlin, Germany) three times, for 15 s each time, at cycle 4 × 10%, with the power set to 65%. Incubations with bacterial products were performed at 37°C with continuous agitation at 100 rpm for overnight incubations and at 170 rpm for various shorter times, which are given in descriptions of specific experiments. Levels of free fatty acids (FFA) were determined by use of the NEFA-C kit (WAKO Chemicals, Neuss, Germany) according to the manufacturer's instructions. Depending on the nature of the experiment, BYE broth, LB broth, or 40 mM Tris-HCl (pH 7.2) (25°C) was incubated, treated like the cultures, and subsequently used as a negative control.
Lipid extraction and TLC.
For detection of distinct polar and apolar lipids, reaction mixtures of lipids with cell lysates or corresponding negative controls were subjected to lipid extraction (9, 22). The lower chloroform phase was subsequently used for separation of lipids by TLC. For detection of polar lipids, silica gel plates (Merck, Darmstadt, Germany) were developed in tanks containing a solvent mixture of chloroform-methanol-water in a 65:25:4 (vol/vol/vol) ratio (22, 55). A mixture of n-hexane-diethyl ether-glacial acetic acid in a 70:30:4 (vol/vol/vol) ratio was used for separation of apolar lipids (22). For visualization, silica plates were then stained with naphthol blue black (Aldrich, Milwaukee, Wis.) (25, 46).
Intracellular infection of U937 cells and Acanthamoeba castellanii amoebae.
A. castellanii amoebae and U937 (CRL-1593.2; American Type Culture Collection, Manassas, Va.), a human cell line that differentiates into macrophage-like cells upon treatment with phorbol esters (incubation for 36 to 48 h with 80 nM phorbol-12-myristate-13-acetate [P-8139; Sigma Chemicals]), were used as hosts for in vitro infection by L. pneumophila (10, 39). Amoebae and U937 cells were maintained and infected as previously described (10, 37, 39). To assess the intracellular growth of L. pneumophila, wells containing amoebae or U937 cells at a concentration of 105 or 106/ml, respectively, were infected with wild-type bacteria or isogenic mutants at a multiplicity of infection of 0.01 for amoebal and 1 for U937 cell infections (time point, 0 h). U937 macrophages were incubated for 2 h with the added bacteria in plain RPMI; then monolayers were washed three times with plain RPMI to remove unbound bacteria and were subsequently incubated with RPMI containing 10% (vol/vol) fetal calf serum (PAA, Linz, Austria). At various time points, coincubations of U937 cells and legionellae were treated with 10% (wt/vol) saponin (Sigma Chemicals) for lysis of the host cells, and serial dilutions were plated on BCYE agar. The number of bacteria within the amoebal coculture was determined by plating serial dilutions on BCYE agar.
Nucleotide sequence accession number.
The L. pneumophila Corby plaB sequence has been deposited in GenBank at the National Center for Biotechnology Information (NCBI) under accession no. AJ565849.
RESULTS
Identification of a new L. pneumophila gene encoding a hemolytic protein.
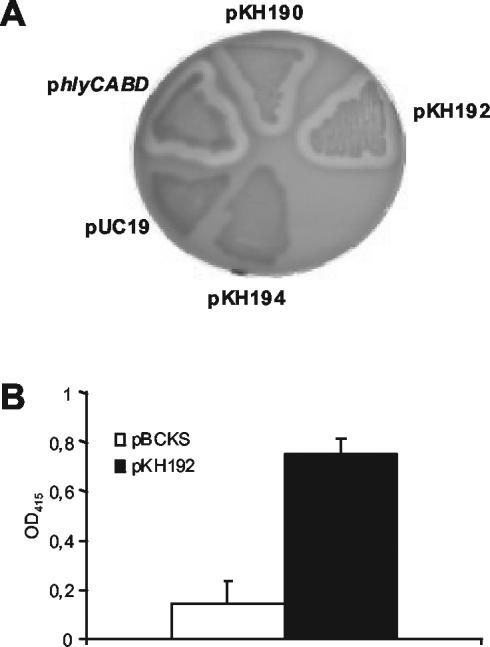
To isolate new L. pneumophila genes responsible for hemolytic activity, a total of 8,000 E. coli clones of an L. pneumophila expression library were screened for hemolytic activity on human red blood agar plates. Three clones showing hemolytic activity were obtained. After retransformation, plasmids pKHL82 and pKHL102, continuously displaying hemolytic activity, were found to be identical by restriction analysis. Subclones (pCL102-1, pCL102-2, pCL102-3, pKH190, and pKH192) containing different segments of the Legionella DNA present in pKHL102 were generated and tested for hemolytic activity (Fig. 1). Hemolysis of human blood agar was mediated by E. coli clones with plasmid pKHL102, pCL102-3, pKH190, or pKH192 but not by clones containing pCL102-1 or pCL102-2 (Fig. 1 and 2A). This suggests that the genetic region coding for the hemolysin is located downstream from the HindIII cleavage site of the L. pneumophila sequence present in pKHL102 (Fig. 1). In pKHL102, only one complete open reading frame (ORF) (insert nucleotides 918 to 2342) was identified and designated plaB. It was completely (pKH190 and pKH192) or approximately two-thirds (pCL102-3) present in all clones showing hemolytic activity. Hemolysis of E. coli containing pKH190 was not caused by induction of the latent E. coli hemolysin CylA (eqivalent to SheA) (16), because an E. coli cylA knockout mutant harboring pKH190 was still hemolytic (data not shown). Inserting a Kmr cassette into the SacII restriction site (nucleotide position 1080 within the ORF) of plaB present in pKH190 abolished the hemolytic activity of the corresponding recombinant E. coli clone, confirming that the ORF identified was responsible for the hemolytic phenotype (Fig. 1 and 2A). It was also noted that clones harboring plasmids with L. pneumophila plaB did not show hemolytic activity on sheep blood agar and were also not hemolytic when human blood agar was incubated at 30 instead of 37°C (data not shown). Independently of blood cell origin and incubation temperature, E. coli containing the E. coli hemolysin locus hlyCABD on plasmid pUC18 was able to cause hemolysis under all conditions used (data not shown). To quantitatively confirm the results from the hemolysis plate assays, E. coli harboring pBCKS+ or pKH192 was tested for hemolysis of human red blood cells in a liquid hemolysis assay. As shown in Fig. 2B, recombinant E. coli containing pKH192 was more hemolytic than E. coli harboring pBCKS+.
FIG. 2.
Hemolysis of human red blood cells by recombinant E. coli containing L. pneumophila plaB. (A) Human blood agar was inoculated with recombinant E. coli harboring different plasmids and then incubated for 48 h at 37°C. Plasmids contained the following inserts: pKH190 and pKH192, intact plaB; pKH194, disrupted plaB; pUC19, empty vector; phlyCABD, hemolysin-encoding operon of E. coli 536 in pUC18 (compare Fig. 1). (B) Recombinant E. coli containing either plain pBCKS+ or pBCKS+ with L. pneumophila plaB (pKH192) was grown to the logarithmic-growth phase and subsequently treated with IPTG. Afterwards, human red blood cells were incubated with the bacteria for 20 h and then quantitatively assessed for hemolysis. Results are means and standard deviations from triplicate cultures and are representative of three independent experiments.
The L. pneumophila plaB gene sequence was predicted to encode a protein of 474 amino acids. As examined by the BlastP algorithm, the deduced protein sequence showed significant homology to three proteins: a hypothetical protein from a Nostoc sp. (gi17230632), a hypothetical protein from Pseudomonas aeruginosa (gi15598123), and the putative lipase LipB from L. pneumophila (gi21666984) (2). All three proteins showed homology to the N-terminal half of L. pneumophila PlaB. LipB of L. pneumophila contains a lipase consensus sequence [(L/I/V)-X-(L/I/V/F/Y)-(L/I/V/M/S/T)-G-(H/Y/W/V)-S-X-G-(G/S/T/A/C)] (PROSITE accession no. PS00120) (4), which we found was substantially conserved in the Legionella hemolysis protein (78-FACITHSTGG-89). Neither LipB nor the hemolysin protein shares the first conserved glycine residue with the lipase consensus sequence; both contain a tyrosine residue instead, suggesting that both proteins may possess biochemical properties distinct from those of other lipases. Indeed, since an L. pneumophila lipB mutant released smaller amounts of fatty acids from tricaprylin but not from lipase substrates such as 1-MPG or 1,2-DG (4), there is still some ambiguity as to whether LipB should be considered a lipase. Using the conserved-domain search tool from the NCBI BLAST page, we found that the N-terminal ∼270 amino acids mainly revealed regions homologous to hydrolytic enzymes, such as the LipA domain of predicted acetyltransferases and hydrolases with the α/β-hydrolase fold (COG1075), the PldB domains of lysophospholipase A (COG2267), and the lipase class 2 domain (pfam01674). Interestingly, the homologous Nostoc sp. protein, but not the P. aeruginosa protein or Legionella LipB, also shared homology with the C-terminal region of Legionella PlaB.
The protein sequence of L. pneumophila PlaB indicated similarities to lipolytic enzymes. Since the majority of the secreted lipid-hydrolyzing activity is exported depending on the type II protein secretion machinery (23, 48), we examined the protein sequence of the L. pneumophila hemolysis protein for a signal sequence. The protein was not predicted to have an N-terminal signal peptide, as tested with SignalP and the PSORT server, but was calculated to be located in the bacterial cytoplasm (41, 42).
Two uncharacterized genes, orf1 and orf2, flanked the L. pneumophila hemolysis gene (Fig. 1). The closest homolog of the orf1 protein product was the DlrA protein of Dictyostelium discoideum, and the closest homologs of the orf2 protein product were several hypothetical proteins from Mesorhizobium loti, Brucella melitensis, and Agrobacterium tumefaciens. Both the upstream orf1 gene and the downstream orf2 gene were oriented in the opposite direction from the hemolysis gene, suggesting that the hemolysis gene message is monocistronic. Farther upstream, two genes encoding proteins with homology to enzymes of the fatty acid oxidation complex were found.
Analysis of different Legionella strains for the presence of plaB genes.
Since the species L. pneumophila has been most frequently associated with the development of Legionnaires' disease, researchers often seek to identify virulence factors which are present only in L. pneumophila and not in less pathogenic species. Therefore, we were also interested in whether the gene coding for PlaB was present in Legionella species other than L. pneumophila. To investigate this, genomic DNAs of 16 L. pneumophila strains and 15 non-L. pneumophila strains were treated with restriction enzymes and examined by Southern blot analysis. We found that under low-stringency conditions, the plaB gene probe did hybridize exclusively to DNA from the L. pneumophila strains tested, and not to DNA from non-L. pneumophila species (data not shown). This indicates that the plaB gene may be restricted to L. pneumophila. However, even though a growing number of L. pneumophila-specific genes have been identified, in some cases the failure of cross-hybridization may arise from the high DNA diversity found in different Legionella strains.
Enzymatic activities of E. coli clones containing the L. pneumophila plaB gene.
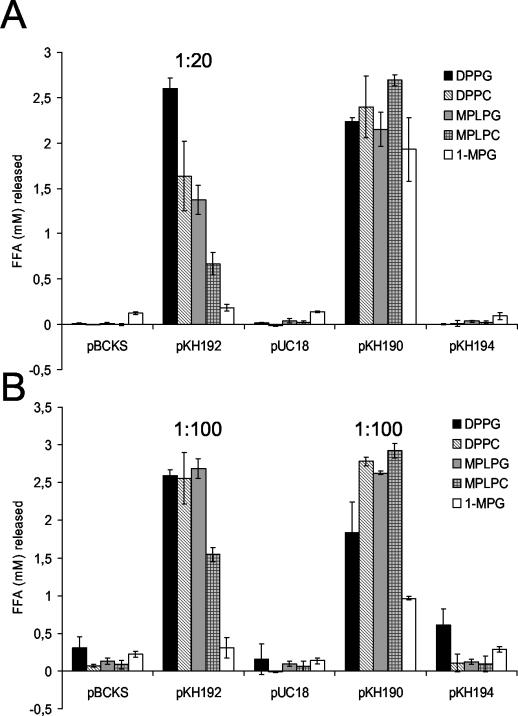
Since the protein sequence of the newly characterized L. pneumophila plaB gene showed sequence homology to lipolytic hydrolases, we tested both culture supernatants and cell lysates of recombinant E. coli clones containing the plaB gene for fatty acid release from various lipid substrates, in particular from diacylglycerophospholipids (DPPG and DPPC) to check for phospholipase A activity, from 1-monoacylphospholipids(lysophospholipids) (MPLPG and MPLPC) to examine for lysophospholipase A activity, and from monoacylglycerol (1-MPG) to test for lipase activity. Both culture supernatants and cell lysates of the hemolytic E. coli clones containing either pKH190 or pKH192 released significantly more FFA from all of the tested lipid substrates than E. coli harboring the corresponding control vectors (Fig. 3). Clones containing plaB hydrolyzed predominantly the diacyl- and lysophospholipid substrates, showing that the respective protein product is both a phospholipase A and a lysophopholipase A. Moreover, the clone harboring pKH194 and its inactivated plaB gene did not liberate increased amounts of FFA from the different lipids (Fig. 3). Because the Legionella plaB gene cloned into E. coli predominantly possessed phospholipase A and lysophospholipase A activity, we designated the gene plaB, for phospholipase A gene B.
FIG. 3.
Lipolytic activities of recombinant E. coli containing L. pneumophila plaB. Culture supernatants (A) and cell lysates (B) of E. coli containing pBCKS or its derivative pKH192, or pUC18 or its derivative pKH190 or pKH194, were mixed with DPPG, DPPC, MPLPG, MPLPC, or 1-MPG. In some cases, culture supernatants or cell lysates were diluted prior to incubation with lipids as indicated. After a 2-h (A) or 50-min (B) incubation at 37°C, the release of FFA was quantified. Data are expressed as differences between the amount of FFA released by the culture supernatant or cell lysate and the amount released by uninoculated LB broth or Tris-HCl buffer, respectively. Results are means and standard deviations of duplicate cultures and are representative of three independent experiments.
Isolation of L. pneumophila plaB mutants.
In order to determine the degree to which plaB is responsible for phospholipase A-lysophospholipase A activity in Legionella and whether PlaB is a protein associated with the bacterial cell or transported into the culture supernatant, we constructed two sets of L. pneumophila plaB mutants. The first mutants were generated by cloning the interrupted plaB gene of pKH194 into pBOC20 and by subsequent allelic exchange forced by the counterselectable sacB gene present in pBOC20 (11, 43). Two plaB mutants (plaB37 and plaB60) were obtained and confirmed by PCR and Southern blot analysis (data not shown). The second set of mutants was generated by using pKH194 without further cloning steps and with allelic exchange to introduce a Kmr cassette into the chromosomal plaB gene of strain Corby. Two plaB mutants (plaB1 and plaB4) were obtained following two separate DNA transformations and allelic exchange selections. PCR and Southern blot analysis confirmed the mutations in plaB (data not shown). All of the subsequent experiments involving enzymatic and hemolytic activities, except genetic complementation, were performed with plaB37, plaB60, plaB1, and plaB4 with comparable results, although in most cases data for plaB60 and in some cases also for plaB1 are presented. In host cell infection assays, all four mutants were tested in amoebae, whereupon mutants plaB1 and plaB4 behaved similarly, but mutants plaB37 and plaB60 were found to contain a second-site mutation in a uncharacterized gene (data not shown). The plaB1, plaB37, and plaB60 mutants were examined for intracellular growth in U937 macrophages and yielded comparable results. Furthermore, a plaB mutant was generated in L. pneumophila strain 130b (ATCC strain BAA-74, also known as Wadsworth or AA100) by the second mutagenesis method. This mutant behaved similarly to L. pneumophila Corby mutants plaB1 and plaB4 in the enzymatic as well as the host cell infection assays (data not shown).
To assess the importance of plaB for the extracellular growth of L. pneumophila, we compared the growth of strain Corby and the plaB mutants in BYE broth, the standard medium for culturing legionellae. As measured by the OD660 of the cultures, the plaB mutants grew comparably to the wild type throughout the logarithmic- and stationary-growth phases when incubated at 37°C with shaking. Furthermore, the mutants grew normally on BCYE agar, the standard Legionella medium. Thus, these observations indicate that plaB is not required for normal extracellular growth in liquid or solid bacteriological media (data not shown).
Lipolytic activities of an L. pneumophila plaB mutant.
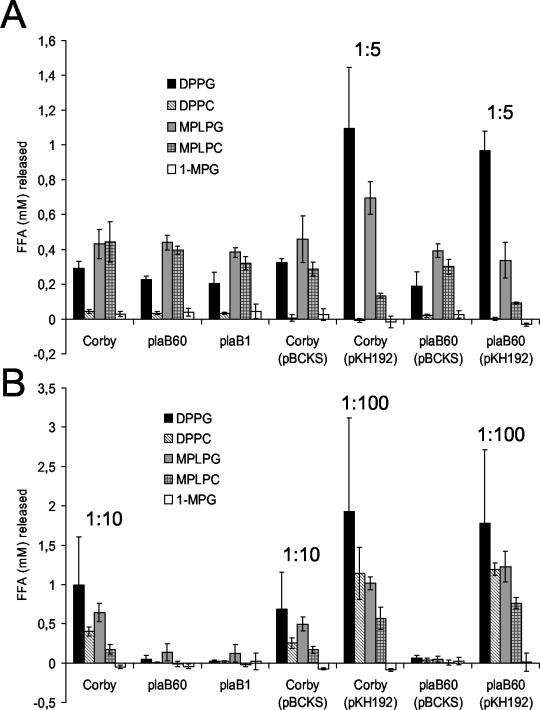
L. pneumophila has been shown to secrete several lipolytic activities into the culture supernatant as well as to possess lipolytic activities associated with the bacterial cell (3, 4, 22, 23, 25, 40). To assess the plaB mutant with respect to lipid hydrolysis, we tested both culture supernatants and bacterial-cell lysates for their abilities to release FFA from DPPG, DPPC, MPLPG, MPLPC, and 1-MPG. The supernatants of wild-type L. pneumophila as well as those of mutants plaB60 and plaB1 showed comparable hydrolysis of DPPG, DPPC, MPLPG, MPLPC, and 1-MPG, suggesting that PlaB is not secreted into the culture supernatant of L. pneumophila (Fig. 4A). Next, we examined whether plaB contributes to the cell-associated lipolytic activities of L. pneumophila. Indeed, the cell-associated activities hydrolyzing DPPG, DPPC, MPLPG, and MPLPC were dramatically reduced in the plaB mutants (Fig. 4B). These data show that plaB is the gene for the major cell-associated phospholipase A-lysophospholipase A activity of L. pneumophila. The ability of plaB60 to fully release FFA from DPPG, DPPC, MPLPG, and MPLPC was restored after trans-complementation with plaB on plasmid pKH192 (Fig. 4). The activities of both the complemented wild type and complemented plaB60 against DPPG, DPPC, MPLPG, and MPLPC were more than 10-fold higher than wild-type activities, a result that is likely due to multiple copies of plaB.
FIG. 4.
Lipolytic activities of wild type, plaB mutant, and genetically complemented L. pneumophila strains. Culture supernatants (A) and cell lysates (B) of wild type, plaB mutant, and genetically complemented L. pneumophila strains were incubated with DPPG, DPPC, MPLPG, MPLPC, or 1-MPG for 2 h (A) or 30 min (B) at 37°C, and then the release of FFA was quantified. In some cases, culture supernatants or cell lysates were diluted with lipids as indicated prior to incubation. Data are expressed as differences between the amount of FFA released by the culture supernatant or cell lysate and the amount released by uninoculated BYE broth or Tris-HCl buffer, respectively. Results are means and standard deviations from duplicate cultures and are representative of three independent experiments.
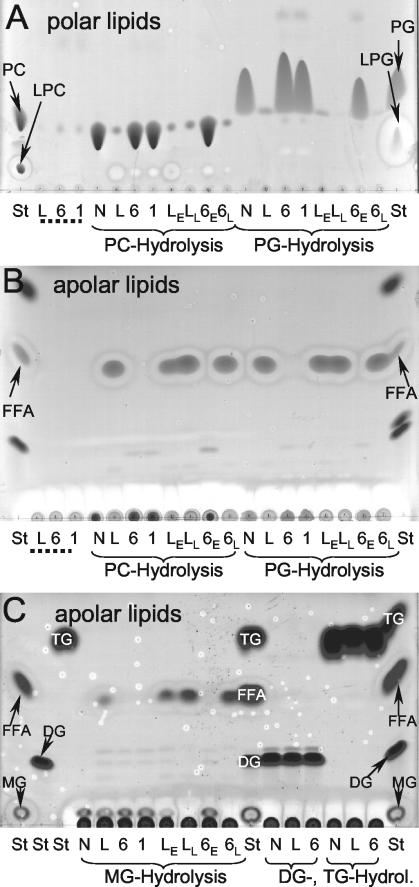
To confirm that the plaB mutant had altered lipolytic activities, we used TLC to examine the pattern of phospholipid cleavage caused by cell lysates. In agreement with our FFA release data from the colorimetric assay, strain Corby cell lysates almost completely hydrolyzed the DPPG and DPPC substrates and liberated large amounts of FFA (Fig. 5A and B). Only minor amounts of lysophospholipids were detected, corroborating that the cell-associated lipolytic activity of L. pneumophila exhibits both phospholipase A and lysophospholipase A activity. Smaller amounts of FFA were also observed for 1-MPG incubations with L. pneumophila wild-type cell lysates; however, no detectable amounts of FFA were released from 1,2-DG or TG, indicating that the cell-associated lipolytic activity of L. pneumophila preferentially hydrolyzes phospholipids (Fig. 5C). On the other hand, cell lysates of plaB mutants did not release detectable amounts of FFA from any of the substrates examined (Fig. 5), showing that plaB is responsible for the cell-associated lipolytic activities of L. pneumophila. The phenotype of the L. pneumophila plaB mutant was fully complemented by a plasmid copy of plaB (Fig. 5). In summary, L. pneumophila PlaB is a cell-associated phospholipase A and lysophospholipase A that preferentially cleaves phospholipids (both diacylphospholipids and lysophospholipids) and, to a lesser extent, also hydrolyzes the nonphospholipid 1-MPG.
FIG. 5.
TLC analysis of lipid hydrolysis by cell lysates of wild-type, plaB mutant, and genetically complemented L. pneumophila strains. Cell lysates of wild-type L. pneumophila (L), plaB mutants plaB60 (6) and plaB1 (1), or genetically complemented L. pneumophila (wild type and plaB60 mutant harboring the empty vector [LE and 6E, respectively] and wild type and plaB60 mutant harboring pKH192, containing intact plaB [LL and 6L, respectively]) were incubated with DPPC (PC) or DPPG (PG) (A and B) or with 1-MPG (MG), 1,2-DG (DG), or TG (C) for 24 h at 37°C, and then lipids were extracted and subjected to TLC. A polar or apolar solvent mixture was used for the separation of the polar (A) or apolar (B and C) lipids, respectively. A mixture of Tris-HCl buffer and the lipids was also incubated and served as a negative control (N). Furthermore, bacterial-cell lysates not incubated with any lipid were extracted and served as the bacterial lipid background control (designated by dotted lines). In all incubations, samples were examined for degradation of the lipid substrate (for DPPC and DPPG, polar TLC [A]; for 1-MPG, 1,2-DG, and TG, apolar TLC [C]), enrichment of FFA (apolar TLC [B and C]), and in the case of DPPC and DPPG, incubations for enrichment of the respective lysophospholipid (polar TLC [A]). For qualitative identification of the lipid spots, lanes containing lipid standards (St) were included. The observations depicted here were made on one more occasion.
Hemolytic activities of an L. pneumophila plaB mutant.
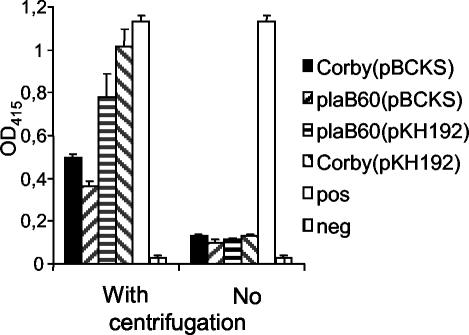
Next, we examined whether the plaB gene contributes to the hemolytic activity of L. pneumophila. L. pneumophila harboring pKH192, which contains the plaB gene, caused faster and more prominent hemolysis on human blood agar plates than the wild-type strain (data not shown). To confirm and quantitatively examine this observation, we additionally used a hemolysis assay, where the different bacterial strains were added to human red blood cells and, after 7 h of incubation, hemolysis was determined by OD415 readings. When the bacteria were centrifuged onto red blood cells, wild-type L. pneumophila harboring pBCKS+ lysed approximately 50% of the cells (Fig. 6). A plaB mutant containing pBCKS+ lysed less than 40% of the red blood cells, demonstrating that L. pneumophila PlaB contributes to the hemolytic activity of the bacterium but is, as expected, not the only L. pneumophila protein mediating hemolysis. Overexpression of plaB in L. pneumophila led to twofold-higher hemolysis of red blood cells relative to that by the wild type strain. Additionally, an intact plaB gene reintroduced on a plasmid into an L. pneumophila plaB mutant compensated for the loss of hemolytic activity. The prominent hemolytic activity of L. pneumophila has been shown to be contact dependent (36). In agreement with this finding, we observed that centrifugation of the bacteria onto the red blood cells potentiated hemolysis and made the hemolysis defect of the plaB mutant detectable (Fig. 6). In conclusion, we have shown that plaB is partially responsible for the hemolytic phenotype of L. pneumophila.
FIG. 6.
Hemolysis of human red blood cells by recombinant wild-type, plaB mutant, and genetically complemented L. pneumophila strains. Wild-type (Corby), plaB mutant (plaB60), and genetically complemented L. pneumophila strains were grown for 24 h at 37°C on BCYE agar plates. Afterward, the bacteria were added to human red blood cells, centrifuged onto the cells or not, incubated for 7 h at 37°C, and then quantitatively assessed for hemolysis. SDS served as a positive control, which was able to lyse 100% of the red blood cells. As a negative control, PBS was added to the cells instead of bacteria. Results are means and standard deviations from triplicate cultures and are representative of three independent experiments.
Intracellular infection by L. pneumophila plaB mutants.
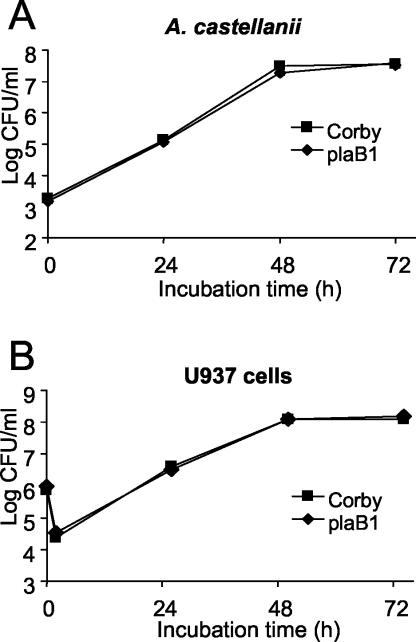
After enzymatically profiling the plaB knockout mutation in L. pneumophila, we investigated the importance of the hemolytic phospholipase A PlaB for the intracellular infection of A. castellanii amoebae and U937 macrophages by L. pneumophila. To that end, we compared the abilities of L. pneumophila Corby and plaB mutants to grow in amoebae. Whereas strain Corby and mutants plaB1 and plaB4 revealed the typical pattern of intracellular growth, in which bacterial numbers increased about 10,000- to 100,000-fold by 48 h postinoculation, the numbers of mutants plaB37 and plaB60 increased only 100- to 1,000-fold (Fig. 7A and data not shown). Complementation experiments with mutants plaB37 and plaB60 clarified that the defect in intracellular infection was caused by a mutation other than that in plaB (data not shown). The nature of this mutation was not further studied here. To determine the role of PlaB in U937 cell infection, macrophage cultures were inoculated with wild-type or plaB mutant L. pneumophila and the numbers of bacteria were recorded at different times. Comparable numbers of wild-type and mutant bacteria were recovered from the macrophage cocultures (Fig. 7B). Taken together, our data indicate that plaB is not required for intracellular infection by L. pneumophila.
FIG. 7.
Intracellular infection by wild-type and plaB mutant L. pneumophila. Strains Corby and plaB1 were used to infect monolayers of A. castellanii amoebae (A) or cultures of U937 macrophages (B) at a multiplicity of infection of 0.01 or 1, respectively. At various time points postinoculation, bacteria were quantitated by plating aliquots on BCYE agar. Results are means and standard deviations from triplicate samples and are representative of three independent experiments.
DISCUSSION
In this investigation, we identified the gene for a new hemolytic protein of L. pneumophila, the major cell-associated phospholipase A PlaB. L. pneumophila PlaB releases fatty acids from a variety of phospholipids and lysophospholipids, and also, to a lesser extent, from the nonphospholipid 1-MPG, but not in detectable amounts from the respective diacyl- and triacylglycerols, showing that it is a phospholipase A with only a low affinity for typical lipase substrates. The finding that enzymes hydrolyzing predominantly phospholipids can also degrade nonphospholipids has been reported for other enzymes. For example, the L. pneumophila lysophospholipase A PlaA primarily hydrolyzes lysophospholipids but can also release FFA to a lesser extent from 1-MPG (25). Furthermore, the Staphylococcus hyicus lipase turned out to preferentially degrade phospholipids over nonphospholipids (57). L. pneumophila PlaB contains a short region of similarity to a lipase consensus motif which is also conserved in lipase family I, including lipases from P. aeruginosa (accession no. D50587) and Bacillus subtilis (M74010) and the phospholipase A from S. hyicus (X02844) (5). Enzymes of this family possess a catalytic triad characterized by the nucleophilic agent serine, embedded in the consensus sequence GXSXG, aspartate, and histidine (5). Interestingly, the first glycine residue of the putative nucleophile-containing motif is not conserved in L. pneumophila PlaB. It is also not conserved in L. pneumophila LipB; both of these proteins contain a tyrosine residue instead (4). The lipid substrates hydrolyzed by LipB and its location in the bacterial cell still need to be elucidated, but since PlaB and LipB show sequence homology in one of their putative catalytic domains, it seems reasonable to test LipB for phospholipase A activity. This is also supported by the observation that outer membrane phospholipases of several gram-negative bacteria, such as E. coli, Helicobacter pylori, and Salmonella enterica serovar Typhimurium, also comprise a primary sequence motif with the catalytically active serine where the first glycine is replaced by a histidine residue, which serves as one of the other members of the catalytic triad (15). The two other important catalytic domains of lipase family I, containing the residues aspartate (motif NDGLV) and histidine [motif (N/D)HLD], were not found to be recognizably conserved in PlaB. Interestingly, the motif HPT, in a region where the catalytically active histidine would be expected, was found in PlaB. This motif is conserved in lipolytic enzymes of the GDSL family, for example, in L. pneumophila lysophospholipase PlaA (5, 25). The amino acid environment of the catalytically active histidine, in particular, has been shown to be important for expression of either lipase or phospholipase A activity. For example, S. hyicus phospholipase shares high conservation of the catalytic domains with other lipases of family I, with one important exception. The amino acid adjacent to the catalytically active histidine is a serine, a more polar amino acid, instead of the hydrophobic residues leucine, valine, etc., present in most other lipases. Replacement of serine 356 by valine decreased phospholipase activity more than 10-fold (56); additionally, the lipase of Bacillus thermocatenulatus was converted into a phospholipase A by replacement of the leucine adjacent to the active-site histidine with serine (34). The question of which sequential or structural determinant defines the phospholipase activity of L. pneumophila PlaB needs further elucidation, for example, by site-directed mutagenesis and by solution of the three-dimensional structure.
In contrast to L. pneumophila LipB, PlaB contains a C-terminal extension of about 200 amino acids adjacent to the potential catalytic active domain. At the moment, it is difficult to even speculate on the possible functions of this domain. However, it is noteworthy that the L. pneumophila plaB mutants constructed in this study were defective in cell-associated phospholipase A activity although the Kmr cassette disrupting the plaB gene was placed after the putative catalytic domains, suggesting that the C terminus might be important for activation, stability, or proper transport of PlaB. Indeed, the cytotoxic activity of the type III secreted P. aeruginosa cytotoxin ExoU, recently found to be a phospholipase A (45, 50), has been suggested to depend on the C-terminal region adjacent to the catalytic domain, since a mutant containing a transposon insertion 88 nucleotides from the exoU stop codon secretes a stable protein but is defective in cell killing (29). Furthermore, several type I secreted bacterial proteins, such as the E. coli hemolysin HlyA or the Erwinia chrysanthemi metalloprotease PrtG, contain their signal for transport via an ABC transporter in their C termini (8). A putative type I secretion system has recently been identified in L. pneumophila (32). Since an L. pneumophila plaB mutant showed reduced red blood cell lysis activity upon bacterial contact with the target cell, we believe that PlaB is located in the outer bacterial membrane and is presented to the external environment. Interestingly, PlaB seems not to contain a recognizable signal sequence, suggesting that it is not sec-dependently transported to the periplasm. Further experiments are necessary to explore the mechanisms involved in PlaB export and activation.
L. pneumophila PlaB shares several properties with the outer membrane phospholipase A (OMPLA) of several enteropathogenic bacteria such as E. coli or Salmonella serovar Typhimurium and other gram-negative bacteria such as H. pylori or Neisseria meningitidis. As shown here for PlaB, OMPLA also hydrolyzes phospholipids, lysophospholipids, and nonphospholipids; under laboratory conditions, the enzyme is not essential for the growth rate; and in C. coli and H. pylori, OMPLA has been found to be hemolytic (15, 18, 27). Whether other characteristics—(i) involvement of a nucleophilic serine in catalysis, (ii) activation of OMPLA upon bacterial disintegration, (iii) regulation of OMPLA activity by dimerization, (iv) requirement of calcium ions for activity, or (v) contribution to bacterial pathogenesis-are true for PlaB, as they are for H. pylori OMPLA, remains to be determined (15, 18).
We have shown that L. pneumophila PlaB is a phospholipase A contributing to the total cytotoxic activity of the pathogen. Other bacterial factors involved in the destruction of eukaryotic cells, such as the RtxA toxin (12) or the icm/dot gene products (1, 36, 40), have already been characterized. However, especially in the case of the icm/dot genes, which code for a type IV protein secretion system (52, 58), it is possible that these determinants do not represent the structural genes actually accounting for the cell-lysing agent. The genes might rather code for proteins important for toxin translocation.
In conclusion, we have shown that L. pneumophila plaB is the gene for the major cell-associated phospholipase A exhibiting hemolytic activity on human red blood cells. The hemolytic activity of PlaB could be one factor among others contributing to bacterial cytotoxicity, but it is not essential for intracellular infection of A. castellanii amoebae or U937 macrophages. Future experiments will be designed to examine the catalytic active residues necessary for phospholipase and hemolytic activities as well as the impact of the different protein domains of PlaB on its activation and transport.
Acknowledgments
We thank Claudia Loske and Elisabeth Bogusch for technical assistance.
This work was supported by grants from the Deutsche Forschungsgesellschaft (GRK587/1-01 and HE284/2-1).
Editor: J. T. Barbieri
REFERENCES
- 1.Alli, O. A., L. Y. Gao, L. L. Pedersen, S. Zink, M. Radulic, M. Doric, and Y. Abu Kwaik. 2000. Temporal pore formation-mediated egress from macrophages and alveolar epithelial cells by Legionella pneumophila. Infect. Immun. 68:6431-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aragon, V., S. Kurtz, A. Flieger, B. Neumeister, and N. P. Cianciotto. 2000. Secreted enzymatic activities of wild-type and pilD-deficient Legionella pneumophila. Infect. Immun. 68:1855-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aragon, V., O. Rossier, and N. P. Cianciotto. 2002. Legionella pneumophila genes that encode lipase and phospholipase C activities. Microbiology 148:2223-2231. [DOI] [PubMed] [Google Scholar]
- 5.Arpigny, J. L., and K. E. Jaeger. 1999. Bacterial lipolytic enzymes: classification and properties. Biochem. J. 343:177-183. [PMC free article] [PubMed] [Google Scholar]
- 6.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1989. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 7.Bender, L., M. Ott, A. Debes, U. Rdest, J. Heesemann, and J. Hacker. 1991. Distribution, expression, and long-range mapping of legiolysin gene (lly)-specific DNA sequences in legionellae. Infect. Immun. 59:3333-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Binet, R., S. Letoffe, J. M. Ghigo, P. Delepelaire, and C. Wandersman. 1997. Protein secretion by Gram-negative bacterial ABC exporters—a review. Gene 192:7-11. [DOI] [PubMed] [Google Scholar]
- 9.Bligh, E. G., and W. J. Dyer. 2003. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 10.Cianciotto, N. P., and B. S. Fields. 1992. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc. Natl. Acad. Sci. USA 89:5188-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cianciotto, N. P., R. M. Long, B. I. Eisenstein, and N. C. Engleberg. 1988. Site-specific mutagenesis in Legionella pneumophila by allelic exchange using counterselectable ColE1 vectors. FEMS Microbiol. Lett. 56:203-208. [Google Scholar]
- 12.Cirillo, S. L., L. E. Bermudez, S. H. El-Etr, G. E. Duhamel, and J. D. Cirillo. 2001. Legionella pneumophila entry gene rtxA is involved in virulence. Infect. Immun. 69:508-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cirillo, S. L., J. Lum, and J. D. Cirillo. 2000. Identification of novel loci involved in entry by Legionella pneumophila. Microbiology 146:1345-1359. [DOI] [PubMed] [Google Scholar]
- 14.Coers, J., J. C. Kagan, M. Matthews, H. Nagai, D. M. Zuckman, and C. R. Roy. 2000. Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol. Microbiol. 38:719-736. [DOI] [PubMed] [Google Scholar]
- 15.Dekker, N. 2000. Outer-membrane phospholipase A: known structure, unknown biological function. Mol. Microbiol. 35:711-717. [DOI] [PubMed] [Google Scholar]
- 16.del Castillo, F. J., S. C. Leal, F. Moreno, and I. del Castillo. 1997. The Escherichia coli K-12 sheA gene encodes a 34-kDa secreted haemolysin. Mol. Microbiol. 25:107-115. [DOI] [PubMed] [Google Scholar]
- 17.Dietrich, C., K. Heuner, B. C. Brand, J. Hacker, and M. Steinert. 2001. Flagellum of Legionella pneumophila positively affects the early phase of infection of eukaryotic host cells. Infect. Immun. 69:2116-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorrell, N., M. C. Martino, R. A. Stabler, S. J. Ward, Z. W. Zhang, A. A. McColm, M. J. Farthing, and B. W. Wren. 1999. Characterization of Helicobacter pylori PldA, a phospholipase with a role in colonization of the gastric mucosa. Gastroenterology 117:1098-1104. [DOI] [PubMed] [Google Scholar]
- 19.Edelstein, P. H. 1981. Improved semiselective medium for isolation of Legionella pneumophila from contaminated clinical and environmental specimens. J. Clin. Microbiol. 14:298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fields, B. S. 1996. The molecular ecology of legionellae. Trends Microbiol. 4:286-290. [DOI] [PubMed] [Google Scholar]
- 21.Fields, B. S., J. M. Barbaree, G. N. Sanden, and W. E. Morrill. 1990. Virulence of a Legionella anisa strain associated with Pontiac fever: an evaluation using protozoan, cell culture, and guinea pig models. Infect. Immun. 58:3139-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flieger, A., S. Gong, M. Faigle, M. Deeg, P. Bartmann, and B. Neumeister. 2000. Novel phospholipase A activity secreted by Legionella species. J. Bacteriol. 182:1321-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flieger, A., S. Gong, M. Faigle, S. Stevanovic, N. P. Cianciotto, and B. Neumeister. 2001. Novel lysophospholipase A secreted by Legionella pneumophila. J. Bacteriol. 183:2121-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flieger, A., S. Gongab, M. Faigle, H. A. Mayer, U. Kehrer, J. Mussotter, P. Bartmann, and B. Neumeister. 2000. Phospholipase A secreted by Legionella pneumophila destroys alveolar surfactant phospholipids. FEMS Microbiol. Lett. 188:129-133. [DOI] [PubMed] [Google Scholar]
- 25.Flieger, A., B. Neumeister, and N. P. Cianciotto. 2002. Characterization of the gene encoding the major secreted lysophospholipase A of Legionella pneumophila and its role in detoxification of lysophosphatidylcholine. Infect. Immun. 70:6094-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedman, R. L., B. H. Iglewski, and R. D. Miller. 1980. Identification of a cytotoxin produced by Legionella pneumophila. Infect. Immun. 29:271-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grant, K. A., I. U. Belandia, N. Dekker, P. T. Richardson, and S. F. Park. 1997. Molecular characterization of pldA, the structural gene for a phospholipase A from Campylobacter coli, and its contribution to cell-associated hemolysis. Infect. Immun. 65:1172-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hales, L. M., and H. A. Shuman. 1999. Legionella pneumophila contains a type II general secretion pathway required for growth in amoebae as well as for secretion of the Msp protease. Infect. Immun. 67:3662-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hauser, A. R., P. J. Kang, and J. N. Engel. 1998. PepA, a secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol. Microbiol. 27:807-818. [DOI] [PubMed] [Google Scholar]
- 30.Heuner, K., L. Bender-Beck, B. C. Brand, P. C. Luck, K. H. Mann, R. Marre, M. Ott, and J. Hacker. 1995. Cloning and genetic characterization of the flagellum subunit gene (flaA) of Legionella pneumophila serogroup 1. Infect. Immun. 63:2499-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Husmann, L. K., and W. Johnson. 1994. Cytotoxicity of extracellular Legionella pneumophila. Infect. Immun. 62:2111-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacobi, S., and K. Heuner. 2003. Description of a putative type I secretion system in Legionella pneumophila. Int. J. Med. Microbiol. 293:349-358. [DOI] [PubMed] [Google Scholar]
- 33.Jepras, R. I., R. B. Fitzgeorge, and A. Baskerville. 1985. A comparison of virulence of two strains of Legionella pneumophila based on experimental aerosol infection of guinea-pigs. J. Hyg. (London) 95:29-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kauffmann, I., and C. Schmidt-Dannert. 2001. Conversion of Bacillus thermocatenulatus lipase into an efficient phospholipase with increased activity towards long-chain fatty acyl substrates by directed evolution and rational design. Protein Eng. 14:919-928. [DOI] [PubMed] [Google Scholar]
- 35.Keen, M. G., and P. S. Hoffman. 1989. Characterization of a Legionella pneumophila extracellular protease exhibiting hemolytic and cytotoxic activities. Infect. Immun. 57:732-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirby, J. E., J. P. Vogel, H. L. Andrews, and R. R. Isberg. 1998. Evidence for pore-forming ability by Legionella pneumophila. Mol. Microbiol. 27:323-336. [DOI] [PubMed] [Google Scholar]
- 37.Liles, M. R., P. H. Edelstein, and N. P. Cianciotto. 1999. The prepilin peptidase is required for protein secretion by and the virulence of the intracellular pathogen Legionella pneumophila. Mol. Microbiol. 31:959-970. [DOI] [PubMed] [Google Scholar]
- 38.Moffat, J. F., P. H. Edelstein, D. P. J. Regula, J. D. Cirillo, and L. S. Tompkins. 1994. Effects of an isogenic Zn-metalloprotease-deficient mutant of Legionella pneumophila in a guinea-pig pneumonia model. Mol. Microbiol. 12:693-705. [DOI] [PubMed] [Google Scholar]
- 39.Moffat, J. F., and L. S. Tompkins. 1992. A quantitative model of intracellular growth of Legionella pneumophila in Acanthamoeba castellanii. Infect. Immun. 60:296-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molmeret, M., O. A. Alli, S. Zink, A. Flieger, N. P. Cianciotto, and Y. Abu Kwaik. 2002. icmT is essential for pore formation-mediated egress of Legionella pneumophila from mammalian and protozoan cells. Infect. Immun. 70:69-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakai, K., and M. Kanehisa. 1991. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins 11:95-110. [DOI] [PubMed] [Google Scholar]
- 42.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 43.O'Connell, W. A., J. M. Bangsborg, and N. P. Cianciotto. 1995. Characterization of a Legionella micdadei mip mutant. Infect. Immun. 63:2840-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Connell, W. A., E. K. Hickey, and N. P. Cianciotto. 1996. A Legionella pneumophila gene that promotes hemin binding. Infect. Immun. 64:842-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phillips, R. M., D. A. Six, E. A. Dennis, and P. Ghosh. 2003. In vivo phospholipase activity of the Pseudomonas aeruginosa cytotoxin ExoU and protection of mammalian cells with phospholipase A2 inhibitors. J. Biol. Chem. 278:41326-41332. [DOI] [PubMed] [Google Scholar]
- 46.Plekhanov, A. Y. 1999. Rapid staining of lipids on thin-layer chromatograms with amido black 10B and other water-soluble stains. Anal. Biochem. 271:186-187. [DOI] [PubMed] [Google Scholar]
- 47.Quinn, F. D., and L. S. Tompkins. 1989. Analysis of a cloned sequence of Legionella pneumophila encoding a 38 kD metalloprotease possessing haemolytic and cytotoxic activities. Mol. Microbiol. 3:797-805. [DOI] [PubMed] [Google Scholar]
- 48.Rossier, O., and N. P. Cianciotto. 2001. Type II protein secretion is a subset of the PilD-dependent processes that facilitate intracellular infection by Legionella pneumophila. Infect. Immun. 69:2092-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Sato, H., D. W. Frank, C. J. Hillard, J. B. Feix, R. R. Pankhaniya, K. Moriyama, V. Finck-Barbancon, A. Buchaklian, M. Lei, R. M. Long, J. Wiener-Kronish, and T. Sawa. 2003. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 22:2959-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saunders, N. A., N. Doshi, and T. G. Harrison. 1992. A second serogroup of Legionella erythra serologically indistinguishable from Legionella rubrilucens. J. Appl. Bacteriol. 72:262-265. [DOI] [PubMed] [Google Scholar]
- 52.Segal, G., J. J. Russo, and H. A. Shuman. 1999. Relationships between a new type IV secretion system and the icm/dot virulence system of Legionella pneumophila. Mol. Microbiol. 34:799-809. [DOI] [PubMed] [Google Scholar]
- 53.Thompson, M. R., R. D. Miller, and B. H. Iglewski. 1981. In vitro production of an extracellular protease by Legionella pneumophila. Infect. Immun. 34:299-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Titball, R. W. 1993. Bacterial phospholipases C. Microbiol. Rev. 57:347-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Touchstone, J. C., S. S. Levin, M. F. Dobbins, L. Matthews, P. C. Beers, and S. G. Gabbe. 1983. (3-sn-Phosphatidyl)cholines (lecithins) in amniotic fluid. Clin. Chem. 29:1951-1954. [PubMed] [Google Scholar]
- 56.van Kampen, M., J. W. Simons, N. Dekker, M. R. Egmond, and H. M. Verheij. 1998. The phospholipase activity of Staphylococcus hyicus lipase strongly depends on a single Ser to Val mutation. Chem. Phys. Lipids 93:39-45. [DOI] [PubMed] [Google Scholar]
- 57.van Oort, M. G., A. M. Deveer, R. Dijkman, M. L. Tjeenk, H. M. Verheij, G. H. de Haas, E. Wenzig, and F. Gotz. 1989. Purification and substrate specificity of Staphylococcus hyicus lipase. Biochemistry 28:9278-9285. [DOI] [PubMed] [Google Scholar]
- 58.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 59.Weltzien, H. U. 1979. Cytolytic and membrane-perturbing properties of lysophosphatidylcholine. Biochim. Biophys. Acta 559:259-287. [DOI] [PubMed] [Google Scholar]
- 60.Winn, W. C. J., and R. L. Myerowitz. 1981. The pathology of the Legionella pneumonias. A review of 74 cases and the literature. Hum. Pathol. 12:401-422. [DOI] [PubMed] [Google Scholar]
- 61.Wintermeyer, E., U. Rdest, B. Ludwig, A. Debes, and J. Hacker. 1991. Characterization of legiolysin (lly), responsible for haemolytic activity, colour production and fluorescence of Legionella pneumophila. Mol. Microbiol. 5:1135-1143. [DOI] [PubMed] [Google Scholar]