Abstract
Rates and mechanisms of intron gain and loss have traditionally been inferred from alignments of highly conserved genes sampled from phylogenetically distant taxa. We report a population-genomic approach that detected 24 discordant intron/exon boundaries between the whole-genome sequences of two Daphnia pulex isolates. Sequencing of presence/absence loci across a collection of D. pulex isolates and outgroup Daphnia species shows that most polymorphisms are a consequence of recent gains, with parallel gains often occurring at the same locations in independent allelic lineages. More than half of the recent gains are associated with short sequence repeats, suggesting an origin via repair of staggered double-strand breaks. By comparing the allele-frequency spectrum of intron-gain alleles with that for derived single-base substitutions, we also provide evidence that newly arisen introns are intrinsically deleterious and tend to accumulate in population-genetic settings where random genetic drift is a relatively strong force.
Introns are noncoding sequences that interrupt eukaryotic exons and are removed from premature mRNAs by the spliceosomal machinery before translation (1–3). Intron colonization affects the evolution of gene structure and is a factor in the emergence of genomic and organismal complexity, as newly arisen introns are thought to be intrinsically deleterious owing to the increased mutational target that they impose on their host genes (4, 5). The number of introns in a genome is determined by the relative rates of intron gain and loss over evolutionary time, which differ among lineages. Across eukaryotes, intron numbers range from >100,000 per vertebrate genome to only two in Giardia lamblia (6, 7). The fundamental causes of this variation remain controversial (8, 9), partly because of a lack of population-level analyses with the power to infer the properties of recent gain or loss alleles.
The early eukaryotic progenitor has been assumed to be intron-rich on the basis of the presence of introns in homologous positions of orthologous genes of widely divergent eukaryotes (10–12) and the likely presence of a complex spliceosome in the eukaryotic ancestor (13). In this context, intron-poor lineages are assumed to reflect a long-term history of intron loss (14). Alternatively, moderate ancestral intron density followed by lineage-specific gains (15) may have occurred, even at orthologous positions in divergent taxa (16). However, most comparative studies of introns have examined only a small subset of highly conserved genes between deeply divergent lineages, and although some studies have documented unambiguous examples of intron gain (17–19) and some statistical procedures allow an indirect inference of parallel gains and/or losses (20, 21), comparative studies of taxa with extreme sequence divergence have essentially no possibility of directly inferring parallel intron gains.
Because they potentially retain the molecular signatures of the process of intron origin, intron presence or absence alleles segregating in natural populations provide material to infer gain or loss mechanisms and to estimate taxon-specific turnover rates. Such polymorphisms do exist. A standing intron presence/absence polymorphism was found at a locus in natural isolates of Drosophila teisseri (22), and two intron-gain alleles segregate with an intron-free version at a locus in the microcrustacean Daphnia pulex (23). The latter study, in particular, inspired us to look more deeply for evidence of recent intron gain or loss in D. pulex.
By artificially removing intron sequences from all predicted gene sequences of the annotated D. pulex genome [clone TCO (24)] and querying the exon-exon boundaries (n = 110,021) against another D. pulex genome sequence (TRO), we detected putative intron-free alleles. After filtering for paralogy and false positives, such as processed pseudogenes, we sequenced the genomic regions surrounding 24 intron presence/absence positions across 84 natural isolates of North American D. pulex species as well as in eight Daphnia outgroup species. Gene trees constructed from flanking-exon sequence for each presence/absence polymorphism revealed the phylogenetic relationships of the polymorphic alleles, and from these data we inferred that 87.5% (21/24) of the intron polymorphisms reflect recent intron gains, with three reflecting intron losses (figs. S1 to S24). Most of the gains (15/28) were exclusive to Oregon populations, a genetically isolated subclade of North American D. pulex (25, 26) with a historically low effective population size (27). Active splicing of all polymorphic introns was confirmed with reverse transcription polymerase chain reaction sequencing.
The features of newly arisen introns in D. pulex are inconsistent with most hypothesized mechanisms of intron origin (7). We found no support for intron gains resulting from tandem duplications of fragments of coding DNA or insertions of transposable elements. Furthermore, the polymorphic intron sequences identified seem to be evolutionary novelties absent from well-characterized eukaryotic and prokaryotic genomes. Except for gains at one locus (Dappu-42116_2, fig. S20), Blastn searches using recently gained intron sequences against the D. pulex genome assembly (http://wfleabase.org/blast), D. pulex genome trace files, and the full GenBank repository did not retrieve any homologous sequence hits.
We observed that short direct repeats, ranging in size from 5 to 12 base pairs, flank many (12/28) of the intron gains, with one repeat positioned within the end of an adjacent exon and the other repeat near the opposite end of the intron sequence (figs. S1 to S3, S7 to S11, S15 to S18, and S21). These sequences suggest that intron gains in D. pulex result from recent repair of staggered double-strand breaks (DSBs) accompanied by small segmental insertions (either preexisting fragments or newly synthesized). In other systems, DSBs and subsequent nonhomologous end joining (NHEJ) repair are known to be associated with small insertions of exogenous DNA (28), including mitochondrial DNAs (29). Consistent with this model, one recent intron gain identified in our study was homologous to the 16S ribosomal subunit of the D. pulex mitochondrial genome, although, as noted above, the source of other gained introns remains unresolved. We also noted that the AT content of recently gained introns (80.9 ± 1.3%) was significantly higher than that of other, nonpolymorphic introns in the same genes (70.8 ± 0.5%), which are themselves high in AT relative to surrounding exons (54.1 ± 1.0%). This suggests that AT-rich insertion sequences are particularly prone to intronization.
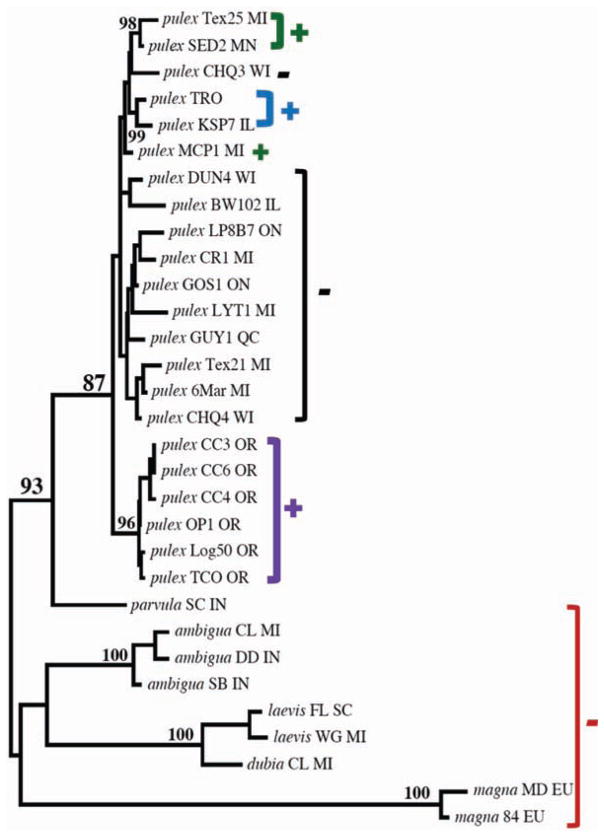
In four of the loci, multiple intron origins have arisen at the exact same site (Fig. 1, Fig. 2, and figs. S15, S19, S20, and S21). If these observations were each due to a single intron-gain event followed by subsequent divergence of intronic sequence, we would expect an overall high rate of allelic divergence in the surrounding exons, but the latter exhibit typical levels of sequence divergence (no more than 3 to 6%), as do other introns in the same gene. Among the parallel intron gains identified, two involve a phase difference (figs. S15 and S19), where the intron interrupts a different position of the same codon. Thus, we conclude that the divergent intron alleles observed at homologous sites are independently derived. Because 4 of 21 total sites of intron gain harbor such independent sequences, this further implies that intron-insertion hot spots may exist in the D. pulex genome.
Fig. 1.
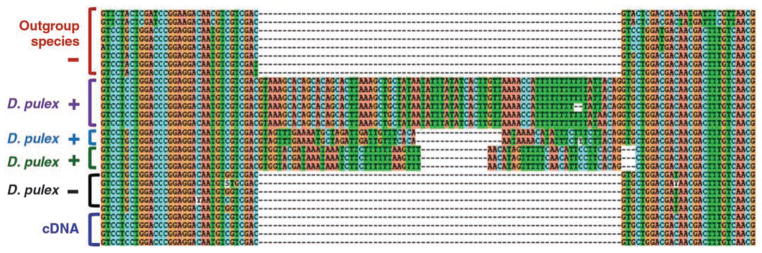
Sequence alignment showing three parallel intron gains in D. pulex at locus Dappu-42116_2. Homologous intron sequences are indicated by color-coded brackets; different gains are indicated by different colors; intron-containing and intron-absent alleles are indicated by plus and minus signs, respectively.
Fig. 2.
Neighbor-joining gene tree with bootstrap values greater than 85% showing three parallel intron gains in D. pulex at locus Dappu-42116_2. Color codes and symbols are as in Fig. 1.
A motif search of exon sequences immediately surrounding the polymorphic introns did not yield any motif common to the insertion sites. Whereas nearly half (45%) of established D. pulex introns reside in between-codon positions (phase 0), most of the gained introns in this study were inserted after the first positions within codons (phase 1) (24). Because introns that split codons are more prone to deleterious consequences during faulty splicing and/or intron sliding (4), they may be less likely to be fixed, which suggests that some percentage of intron gains are transient and not typical of established introns.
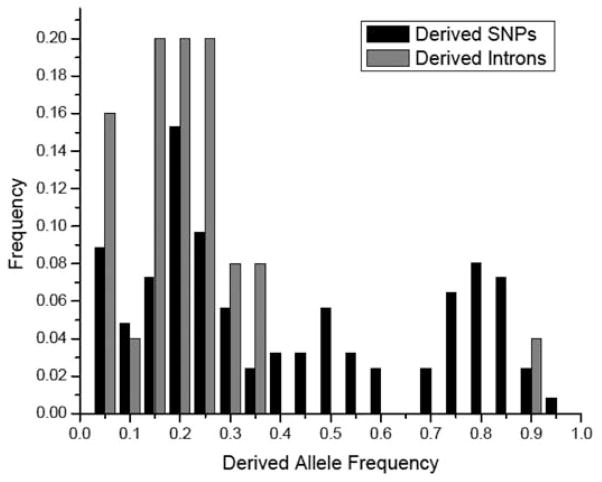
Purifying selection on derived introns relative to other base-substitution polymorphisms can be inferred by comparing the allele-frequency spectrum of derived single-nucleotide polymorphisms (SNPs) and gained introns (Fig. 3). We found that the allele-frequency distribution of recently gained introns is skewed to low levels relative to derived SNPs (average frequency of derived introns is 19.5 ± 2.6% versus 39.4 ± 3.4% for derived SNPs).
Fig. 3.
Allele-frequency spectra of derived SNPs and introns in D. pulex. Gained introns are skewed to lower frequency than derived SNPs.
The presence of upstream and downstream introns argues against presence/absence polymorphisms being artifacts of processed pseudo-genes, where adjacent introns are expected to be missing because of the incorporation of a fully spliced cDNA transcript. Moreover, this additional sequencing also yielded another intron presence/absence polymorphism within D. pulex that was not previously detected with our bio-informatic survey of TCO and TRO, which suggests that there may be many more allelic variants yet to be discovered. Such sequencing also uncovered intron gains in two outgroup species, D. obtusa and D. parvula, indicating that intron gains may not be exclusive to D. pulex. In addition, we detected a parallel intron gain at locus Dappu-323635_3 in D. laevis (fig. S16), consistent with the view that the existence of intron colonization hot spots is not restricted to D. pulex. Our study has an ascertainment bias, making identification of intron presence preferential in TCO-related populations, because the exon boundaries used to detect intron absences were generated from the predicted annotations of the TCO genome. However, any polarizations as a gain or loss are not affected by this bias.
Intron gain has been argued to be a rare event, with a rate on the order of <4 × 10−6 per coding site per million years, which is orders of magnitude lower than estimated rates of loss (5, 30). Massive losses of ancestral introns have been postulated to have occurred in select lineages (14, 31, 32), and it has been suggested that rates of intron gain and loss have been declining in the past 1.3 billion years in most eukaryotes, with a greater decline of gains than losses (32). Regardless of whether these are accurate inferences, they are not consistent with our observations of Daphnia intron turnover, where the rate of gain is higher than the rate of loss and is minimally 1.2 × 10−5 per coding site per million years (24).
Our data suggest that rates of intron turnover, particularly intron gain, are higher than previously appreciated. Furthermore, the documentation of parallel gains occurring at many of the same sites is contrary to the assumptions in many prior analyses. If similar processes occur in other taxa, analyses of intron turnover rates that fail to account for rampant parallel gain may lead to underestimates of the rates of gain and overestimates of the rates of loss. In addition, our identification of the short direct repeats found in association with recently originated intron-containing alleles suggests that introns may be gained fortuitously as a consequence of DNA damage, with repair of staggered DSBs being occasionally accompanied by insertions that by chance harbor the sequences necessary to elicit a splicing reaction.
Supplementary Material
Footnotes
References and Notes
- 1.Berget SM, Moore C, Sharp PA. Proc Natl Acad Sci USA. 1977;74:3171. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chow LT, et al. Cell. 1977;12:1. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- 3.Gilbert W. Nature. 1978;271:501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- 4.Lynch M. Proc Natl Acad Sci USA. 2002;99:6118. doi: 10.1073/pnas.092595699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lynch M. The Origins of Genome Architecture. Sinauer; Sunderland, MA: 2007. [Google Scholar]
- 6.Nixon JEJ, et al. Proc Natl Acad Sci USA. 2002;99:3701. [Google Scholar]
- 7.Roy SW, Gilbert W. Nat Rev Genet. 2006;7:211. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- 8.Belshaw R, Bensasson D. Heredity. 2006;96:208. doi: 10.1038/sj.hdy.6800791. [DOI] [PubMed] [Google Scholar]
- 9.Jeffares DC, Mourier T, Penny D. Trends Genet. 2006;22:16. doi: 10.1016/j.tig.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Fedorov A, Merican AF, Gilbert W. Proc Natl Acad Sci USA. 2002;99:16128. doi: 10.1073/pnas.242624899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogozin IB, Wolf YI, Sorokin AV, Mirkin BG, Koonin EV. Curr Biol. 2003;13:1512. doi: 10.1016/s0960-9822(03)00558-x. [DOI] [PubMed] [Google Scholar]
- 12.Stajich JE, Dietrich FS, Roy SW. Genome Biol. 2007;8:R223. doi: 10.1186/gb-2007-8-10-r223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins L, Penny D. Mol Biol Evol. 2005;22:1053. doi: 10.1093/molbev/msi091. [DOI] [PubMed] [Google Scholar]
- 14.Roy SW, Penny D. Mol Biol Evol. 2007;24:1926. doi: 10.1093/molbev/msm102. [DOI] [PubMed] [Google Scholar]
- 15.Roy SW, Penny D. Mol Biol Evol. 2007;24:1447. doi: 10.1093/molbev/msm048. [DOI] [PubMed] [Google Scholar]
- 16.Tarrío R, Rodríguez-Trelles F, Ayala FJ. Proc Natl Acad Sci USA. 2003;100:6580. doi: 10.1073/pnas.0731952100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coghlan A, Wolfe KH. Proc Natl Acad Sci USA. 2004;101:11362. doi: 10.1073/pnas.0308192101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hankeln T, Friedl H, Ebersberger I, Martin J, Schmidt ER. Gene. 1997;205:151. doi: 10.1016/s0378-1119(97)00518-0. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen CB, et al. PLoS Biol. 2004;2:e422. doi: 10.1371/journal.pbio.0020422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carmel L, Rogozin IB, Wolf YI, Koonin EV. BMC Evol Biol. 2007;7:192. doi: 10.1186/1471-2148-7-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen HD, Yoshihama M, Kenmochi N. PLOS Comput Biol. 2005;1:e79. doi: 10.1371/journal.pcbi.0010079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Llopart A, Comeron JM, Brunet FG, Lachaise D, Long M. Proc Natl Acad Sci USA. 2002;99:8121. doi: 10.1073/pnas.122570299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Omilian AR, Scofield DG, Lynch M. Mol Biol Evol. 2008;25:2129. doi: 10.1093/molbev/msn164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.See supporting material on Science Online.
- 25.Colbourne JK, et al. Biol J Linn Soc. 1998;65:347. [Google Scholar]
- 26.Paland S, Colbourne JK, Lynch M. Evolution. 2005;59:800. [PubMed] [Google Scholar]
- 27.Lynch M, et al. Evolution. 1999;53:100. [Google Scholar]
- 28.Yu X, Gabriel A. Mol Cell. 1999;4:873. doi: 10.1016/s1097-2765(00)80397-4. [DOI] [PubMed] [Google Scholar]
- 29.Hazkani-Covo E, Covo S. PLoS Genet. 2008;4:e1000237. doi: 10.1371/journal.pgen.1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy SW, Gilbert W. Proc Natl Acad Sci USA. 2005;102:5773. doi: 10.1073/pnas.0500383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho S, Jin S, Cohen A, Ellis RE. Genome Res. 2004;14:1207. doi: 10.1101/gr.2639304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carmel L, Wolf YI, Rogozin IB, Koonin EV. Genome Res. 2007;17:1034. doi: 10.1101/gr.6438607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Supported by NSF grants MCB-0342431, EF-082741, and EF-0328516 (M.L.). We thank F. Catania, T. Doak, J. Colbourne, and K. Montooth for discussions, and A. Seyfert, S. Schaack, A. Omilian, and E. Williams for early contributions to this project. Genome sequences and annotation are available through the collaboration of JGI and the Daphnia Genomics Consortium at http://genome.jgi-psf.org/Dappu1/Dappu1.home.html. Intron data are provided in the online supplement. Sequences have been deposited in GenBank, accession numbers GQ984366 to GQ985204 (for details see tables S6 and S7).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.