Abstract
Apical membrane antigen 1 (AMA-1) is a micronemal protein secreted to the surface of merozoites of Plasmodium species and Toxoplasma gondii tachyzoites in order to fulfill an essential but noncharacterized function in host cell invasion. Here we describe cloning and characterization of a Babesia bovis AMA-1 homologue designated BbAMA-1. The overall level of similarity of BbAMA-1 to P. falciparum AMA-1 was low (18%), but characteristic features like a transmembrane domain near the C terminus, a predicted short cytoplasmic C-terminal sequence with conserved sequence properties, and an extracellular domain containing 14 conserved cysteine residues putatively involved in disulfide bridge formation are typical of AMA-1. Rabbit polyclonal antisera were raised against three synthetic peptides derived from the N-terminal region and domains II and III of the putative extracellular domain and were shown to recognize specifically recombinant BbAMA-1 expressed in Escherichia coli. Immunofluorescence microscopy showed that there was labeling of the apical half of merozoites with these antisera. Preincubation of free merozoites with all three antisera reduced the efficiency of invasion of erythrocytes by a maximum of 65%. Antisera raised against the N-terminal peptide detected a 82-kDa protein on Western blots and a 69-kDa protein in the supernatant that was harvested after in vitro invasion, suggesting that proteolytic processing and secretion take place during or shortly after invasion. A combination of two-dimensional Western blotting and metabolic labeling allowing direct identification of spots reacting with the BbAMA-1 peptide antisera together with the very low silver staining intensity of these spots indicated that very low levels of BbAMA-1 are present in Babesia merozoites.
Babesia bovis is an obligatory intraerythrocytic bovine parasite that belongs to the phylum Apicomplexa. Although members of the Apicomplexa infect different host and cell types, they have similar host cell invasion processes. When an extracellular merozoite enters an erythrocyte, it forms an initial reversible attachment, which leads to reorientation of the merozoite that brings the anterior apical pole in contact with the plasma membrane of the erythrocyte (9, 36). A tight junction is formed, through which the parasite invades the red blood cell. The process is completed when the parasite is inside a parasitophorous vacuole of the red blood cell. From the first attachment until completion of the invasion process the parasite secretes proteins from apical organelles into the merozoite membrane and into the environment. Proteins secreted from micronemes, rhoptries, and dense granules are thought to play a central role in invasion and in the establishment of infection by apicomplexan parasites (4, 36). This supposed critical function and the exposure to the immune system, localized on the surface of the merozoites, have marked merozoites as potential vaccine candidates (2). One of these candidates is apical membrane antigen 1 (AMA-1) (8, 15, 20, 25, 31, 37), which is expressed in the late schizont stage of the asexual life cycle of the Plasmodium falciparum parasite (31). AMA-1 is a type I integral membrane protein with three characteristic structures: (i) an N-terminal, cysteine-rich ectodomain, (ii) a single transmembrane domain, and (iii) a C-terminal cytoplasmic tail. The ectodomain is organized into domains I, II, and III by the formation of disulfide bridges between conserved cysteine residues. Full-length P. falciparum AMA-1 (83 kDa) is a micronemal protein (19) that is transported to the merozoite surface membrane as a 66-kDa protein upon proteolytic cleavage in the N-terminal ectodomain (19). During invasion of merozoites, P. falciparum AMA-1 is further processed to 44- and 48-kDa soluble fragments (19, 20). Although the biological function of AMA-1 is unknown, the subcellular localization, stage-specific expression, and secretion during host cell invasion suggest that it is involved in merozoite invasion. A strong correlation was found between protection and P. falciparum AMA-1 antibodies that were generated against different peptide sequences (34). Furthermore, passive transfer of rabbit AMA-1 antibodies protected mice against Plasmodium chabaudi infection (1), and antibodies against Plasmodium reichenowi (24) and Plasmodium vivax AMA-1 (23) were shown to inhibit red blood cell invasion. Recently, eight peptides of the P. falciparum AMA-1 protein that have specific erythrocyte binding activities were mapped (13, 38). An AMA-1 homologue is present in all Plasmodium species studied and Toxoplasma gondii, which supports the suggestion that this protein is involved in an essential function (5, 8, 11, 24, 27).
Here we report the complete sequence of the B. bovis AMA-1 (BbAMA-1) cDNA. We studied the protein on one-dimensional (1D) and two-dimensional (2D) Western blots and by immunofluorescence microscopy, and we analyzed inhibition of in vitro invasion by antisera directed against specific regions.
MATERIALS AND METHODS
B. bovis in vitro culture.
The B. bovis Israel isolate (clonal line C61411) was cultured in vitro in bovine erythrocytes as previously described (12). Briefly, B. bovis cultures were maintained in 24-well plates (total volume, 1.2 ml) or in 25-cm2 bottles (total volume, 15 ml) containing medium M199 with 40% bovine serum, 25 mM sodium bicarbonate, and bovine erythrocytes at a packed cell volume (PCV) of 5%. Cultures were incubated at 37°C in 5% CO2 in air, and the level of parasitemia was kept between 1 and 12% by daily dilution.
For metabolic labeling, a B. bovis culture (8 to 10% parasitemia) was centrifuged (2,000 × g, 10 min, 15°C), washed once with phosphate-buffered saline (PBS), and resuspended in RPMI 1640 medium without methionine and cysteine containing 20 mM 3-(N-tris[hydroxymethyl]methylamino)-2-hydroy-propanesulfonic acid (Sigma) and 1% glutamine (Sigma). 35S-labeled methionine and cysteine (167 μCi/ml) were added, and this was followed by incubation for 18 h at 37°C in CO2 in air.
B. bovis in vitro cell invasion.
The4 cell invasion procedure was performed as described previously (12), with slight modifications. B. bovis-infected red blood cells (6 to 8% parasitemia) were centrifuged (2,000 × g, 10 min, 15°C) and resuspended in an equal volume of VyMs buffer (4°C). Samples (800 μl) were subjected to five intermittent (10 s with incubation at 0°C in between pulses) high-voltage pulses (2.5 kV, 200 Ω, 25 μF) in 4-mm cuvettes (Bio-Rad) by using a Bio-Rad Gene Pulser with a pulse controller. The lysed samples (800 μl) were washed with 8 ml of PBS containing 25 mM sodium bicarbonate (PBSbc) (pH 8.0) at 20°C, and this was followed by centrifugation at 1,800 × g for 10 min at 15°C. A second similar wash was performed, except that the centrifugation speed was lower (1,300 × g). The final merozoite pellet was resuspended in 800 μl of PBSbc. Invasion was initiated by addition of 1 volume of resuspended merozoites to 9 volumes of suspended bovine erythrocytes (5.5% PCV in PBS [pH 8.0] containing PBSbc, preincubated for 30 min at 37°C in 5% CO2 in air) in 24-well plates (final volume, 1.2 ml), in 25-cm2 flasks (15 ml), or in 80-cm2 flasks (50 ml) at 37°C in 5% CO2 in air. Giemsa-stained slides were prepared after 1 h, and the number of parasitized erythrocytes out of a total of 5,000 erythrocytes was determined.
In vitro inhibition of invasion by rabbit antisera.
B. bovis merozoites (200 μl) that were liberated by high-voltage pulsing and resuspended in PBSbc as described above were incubated with 40 μl of rabbit antiserum for 1 h at 20°C. After 1 h, 960 μl of suspended bovine erythrocytes (6.25% PCV in PBSbc, preincubated for 30 min at 37°C in 5% CO2 in air) was added, and this was followed by 1 h of incubation, after which Giemsa-stained slides were prepared and examined to determine the level of invasion. The rabbit antisera used were raised against synthetic peptides derived from the BbAMA-1 sequence, and a control serum was raised against an unrelated peptide (YAGRLFSKRTAATAYKLQ), designated peptide C. Peptides were linked to maleimide-activated keyhole limpet hemocyanin (KLH) (Pierce) prior to immunization. Preimmune sera were also included in the test.
Preparation of total merozoite protein extracts and proteins solubilized upon invasion.
Samples of merozoites (800 μl), prepared as described above for in vitro invasion, were partially separated from erythrocyte ghosts by filtration with 1.2-μm-pore-size polypropylene prefilters (Millipore). Filtered merozoites were pooled and washed twice in 20 volumes of PBSbc, and this was followed by centrifugation at 2,000 × g for 20 min at 4°C. After the second wash the pellet was resuspended in an equal volume of PBSbc, the suspension was divided into 200-μl aliquots that were centrifuged (10,000 × g, 5 min at 4°C), and 100-μl cell pellets (2 × 109 merozoites) were stored at −20°C after removal of the supernatants. Frozen merozoite pellets were thawed just before use and then lysed, reduced, and alkylated by using a total protein extraction kit (Proteoprep; Sigma) according to the manufacturer's instructions; finally, they were suspended in 1.7 ml of buffer compatible with direct application to sodium dodecyl sulfate (SDS)-polyacrylamide gels or isoelectrofocusing strips. Insoluble material was removed by centrifugation at 16,000 × g for 3 min at 4°C. As the extracts contained considerable amounts of erythrocyte proteins, control extracts were prepared in the same way except that we started with a culture of noninfected erythrocytes.
Proteins that were solubilized upon invasion were obtained by gently removing the overlaying buffer after 1 h of in vitro invasion as described above. The samples were centrifuged (2,000 × g, 10 min, 4°C), after which the supernatant was centrifuged again at a high speed for removal of membrane fragments (20 min, 12,000 × g, 4°C). The final supernatant was dialyzed (Pierce; Snakeskin pleated dialysis tubing) overnight against 10 mM Tris—NaCl (pH 7.5).
Construction and screening of a B. bovis cDNA library.
A cDNA library was constructed from 5 μg of B. bovis mRNA by using a λZAP-cDNA synthesis kit (Stratagene) according to the manufacturer's instructions. cDNA fragments (0.5 to 4 kb) were collected by gel filtration on a Sepharose CL-4B column and ligated into the EcoRI/XhoI site of the λ uniZAP-XR Express vector. Giga Pack III Gold was used for packaging into phage particles, and this was followed by transformation of Escherichia coli XL-1 Blue MRF′ cells. A total of 3 × 105 plaques were obtained, and an amplified library was constructed from these plaques.
The cDNA library was screened with probe F1. Oligonucleotides p1 (5′-CCACGGCTCTGGAATCTATGTC-3′) at position 329 and p2 (5′-CAAAAGGATACCTATATTTGGTAC-3′) at position 703 (derived from an expressed sequence tag available at www.sanger.ac.uk; numbering according to the numbering of the sequence deposited under GenBank accession number AY486101) were used to amplify probe F1 by PCR performed with a 50-μl mixture containing each deoxynucleoside triphosphate at a concentration of 0.2 mM, 20 pmol of each primer μl−1, 100 ng of B. bovis genomic DNA, and 0.5 U of Taq DNA polymerase in standard buffer (Promega). Amplification was performed for 30 cycles (92°C for 30 s, 58°C for 30 s, 72°C for 30 s) with initial denaturation for 3 min at 95°C and final elongation at 72°C for 10 min. The fragment was purified from an agarose gel and was labeled with 50 μCi of [α-32P]dATP (3,000 Ci mmol−1) by using a Random Primer labeling kit (Roche). A total of 1.2 × 106 plaques were screened by standard procedures (35) to obtain the AMA-1 sequence (35). After two cycles of plaque purification, two clones were excised in vivo for isolation of the phagemid inserts as described in the manufacturer's instructions (Stratagene) and were sequenced on both strands by performing automated cycle sequencing by the dye terminator method (ABI PRISM dye terminator kit; Pharmacia). A full-length AMA-1 cDNA including the noncoding 5′ end was obtained with a GeneRacer kit by using a specific primer (5′-GATGAAATGGGATCGAGGAAGTCG-3′ [Invitrogen]) according to the manufacturer's instructions, and the clone obtained was sequenced on both strands.
Expression of recombinant BbAMA-1 in E. coli.
A cDNA clone of BbAMA-1 was amplified by PCR by using primers that introduced a BamHI site prior to base 1 (numbering from the first base of the initiation codon) and a HindIII site after base 1504. Primers p3 (5′-CCCGGATCCATGCAGTTACATAACAAA-3′) and p4 (5′-GGGAAGCTTCTGAGCAAAGGAAATAGG-3′) with BamHI and HindIII sites were used to clone the AMA-1 PCR products in vector pET-32a (Novagen), which allowed their expression as fusion products with an N-terminal thioredoxin domain and an internal six-histidine tag. After PCR (1 min at 94°C, 1 min at 55°C, and 1 min at 72°C for 30 cycles) of cDNA clones, the fragment was gel purified, ligated in the pET-32a vector, and used for transformation of the E. coli NovaBlue strain. Plasmids containing the appropriate insert were used to transform expression host strain BL21(DE3). Fusion proteins with thioredoxin were obtained after induction with 1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) for 4 h at 37°C, as shown by analysis of total cell samples at zero time and 4 h after induction (see Fig. 2). Bacterial pellets were heated at 95°C in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer containing 2% (vol/vol) β-mercaptoethanol, electrophoresed on SDS—10% PAGE minigels, and stained with Coomassie blue to confirm expression.
FIG. 2.
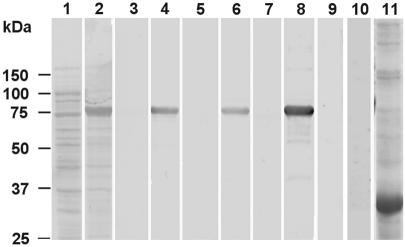
Western blots of recombinant BbAMA-1 probed with polyclonal rabbit antisera against synthetic peptides: Direct Blue-stained strips of polyvinylidene difluoride membrane obtained after blotting of display E. coli lysates of uninduced cells (lane 1) and induced cells (lanes 2 and 11) expressing BbAMA-1 (lane 2) and B. bovis rab5 (lane 11). Immunoblots of recombinant BbAMA-1 (lanes 3 to 9) were incubated with preimmune serum (lanes 3, 5, and 7), with immune serum against peptide 1 (lane 4), peptide 2 (lane 6), or peptide 3 (lane 8), or with B. bovis rab5 antiserum (lane 9). Lane 10 is an immunoblot of B. bovis recombinant protein rab5 with AMA-1 antisera. Molecular masses are indicated on the left.
Peptide selection and immunization.
Synthetic peptides for immunization were derived from the full-length BbAMA-1 sequence by selection of amphiphatic alpha-helices having a high probability for surface localization and several charged residues by using the Protean software package (Lasergene) for protein sequence analysis. Peptides were selected from the N-terminal region (amino acids 46 to 60; cysteine-AFHKEPNNRRLTKRS; peptide 1), domain II (amino acids 395 to 409; cysteine-RGVGMNWATYDKDSG; peptide 2), and domain III (amino acids 453 to 467; cysteine-YVEPRAKNTNKYLDV; peptide 3). The peptides were synthesized and coupled to KLH as a carrier protein by following the manufacturer's recommendations (Pierce). The peptide-carrier conjugates were used to generate rabbit polyclonal antisera.
Three groups of New Zealand White rabbits (each group containing two rabbits) were immunized five times subcutaneously at 3-week intervals for 8 months. Before the first immunization blood serum was collected from each rabbit, which was used as a negative control. Each rabbit was inoculated with 250 μg of peptide that was coupled to 250 μg of KLH and an equal volume of the adjuvant Stimune (ID-DLO, Lelystad, The Netherlands) in a 1,000-μl (total volume) mixture. Sera were tested periodically for reactivity by an enzyme-linked immunosorbent assay (ELISA). Plasmaphoresis was performed 1 week after the immunization starting at month 4.
ELISA.
Production of rabbit antibodies against the peptides was monitored by an ELISA. Ninety-six-well microtiter plates were coated with 150 ng of peptide 1, peptide 2, or peptide 3 per well in 0.1 M Tris-HCl (pH 8.0), incubated for 30 min at 37°C, and blocked for 1 h with PBS containing 0.25% bovine serum albumin at 37°C. Dilutions (1:50 to 1:50,000) of individual rabbit sera were incubated for 1 h at 37°C. The plates were washed, and swine anti-rabbit-immunoglobulin G-horseradish peroxidase-conjugated (DAKO) (1:2,000) secondary antibody was incubated for 1 h. The plates were washed and developed for 45 min with 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) (ABTS)-peroxidase substrate (Roche Biochemicals). The optical density at 405 nm was recorded, and comparative ELISA titers were calculated.
SDS-PAGE and Western blotting.
Total merozoite extracts or proteins that were solubilized upon invasion were separated by SDS—10% PAGE and transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore). The blot was blocked with 5% skim milk diluted in PBS containing 0.05% Tween (PBST) for 1 h at 37°C. The rabbit antisera were diluted (1:500) in PBST containing 2% skim milk and incubated overnight at 4°C. The blot was washed with PBST and then incubated with anti-rabbit-immunoglobulin—horseradish peroxidase (1:10,000) (DAKO) for 1 h at 37°C. After the blot was washed with PBST, it was developed either with a TMB MB substrate kit (Lucron Bioproducts b.v.) or with an enhanced chemiluminescence ECL Plus kit (Amersham).
2D electrophoresis.
Total merozoite extract (300 μg) and invasion supernatant (100 μg) were dissolved in a rehydration solution containing 7 M urea, 2 M thiourea, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 2% carrier ampholyte mixture (pH 4 to 7 in Immobiline Drystrip gels), and 20 mM dithiothreitol and loaded on 13-cm IPG strips (pH 4 to 7). The isoelectric focusing and subsequent 2D SDS-PAGE procedures were performed as described previously (39). Silver staining was used to visualize proteins after 2D SDS-PAGE. Images of the gels and blots were acquired by using LabScan v3.0 software with a Umax flatbed scanner and were digitally analyzed by using the ImageMaster 2D v4.01 software (Amersham Biosciences).
Confocal immunofluorescence microscopy.
Recognition of B. bovis merozoites by anti-AMA-1 antibodies was tested by indirect immunofluorescence by using a confocal microscope. Thin blood smears were fixed in acetone for 10 min and air dried. Primary incubation with polyclonal rabbit serum directed against anti-AMA-1 peptide 1, 2, or 3 (1:20) for 35 min was followed by three 5-min washes with PBS. The slides were then incubated with goat anti-rabbit immunoglobulin G conjugated with Alexa 488 (20 μg/ml; Molecular Probes Inc., Eugene, Oreg.) for 30 min and washed with PBS. Subsequently, for dual labeling, the slides were incubated with 4′,6′-diamidino-2-phenylindole (DAPI) (0.5 μM; Molecular Probes Inc.) for 20 min and washed. FluorSave solution was applied, and the slides were left overnight at room temperature, covered, in a horizontal position. Fluorescent signals were visualized by using a Bio-Rad Radiance 2100MP confocal and multiphoton system equipped with a Nikon TE300 inverted microscope. The DAPI probes were excited by multiphoton excitation at 780 nm by using a mode-locked titanium-sapphire laser (Tsunami; Spectra-Physics) pumped by a 10-W solid-state laser (Milennia Xs; Spectra-Physics), while the Alexa 488 probe was excited by an argon laser at 488 nm.
Nucleotide sequence accession number.
The nucleotide sequence described in this paper has been deposited in the GenBank database under accession no. AY486101.
RESULTS
Identification and cloning of a full-length cDNA encoding BbAMA-1. A B. bovis expressed sequence tag displaying 33.1% identity to domain I of P. falciparum AMA-1 over a stretch of 145 amino acids was identified by BLAST analysis. Probing a B. bovis cDNA library with a 350-bp PCR product derived from this expressed sequence tag resulted in cloning and sequencing of a 2,036-bp cDNA containing a 1,818-bp open reading frame and a 189-bp 3′ noncoding region terminating in a poly(A) tail. To determine the 5′ capped end of the full-length mRNA, total mRNA was dephosphorylates, after which the 5′ caps, which were left intact, were removed by tobacco acid pyrophosphatase; this was followed by ligation of a specific RNA oligonucleotide. Subsequently, nested PCR of first-strand cDNA allowed cloning and sequencing of a 755-bp fragment derived from the 5′ end of the B. bovis sequence, which revealed a 246-bp 5′ untranslated region preceding the first methionine codon of the 1,818-bp open reading frame.
Comparison of BbAMA-1 with the T. gondii and Plasmodium AMA-1 proteins.
Conceptual translation of the 1,818-bp open reading frame resulted in prediction of a 67.2-kDa protein with a pI of 6.35, which was aligned with the full-length sequences of the P. falciparum, P. vivax, and T. gondii AMA-1 proteins (Fig. 1). Reminiscent of the previously described AMA-1 proteins, the hydrophobic N-terminal 39 amino acids of BbAMA-1 were predicted to form a signal peptide (SignalP2.0), whereas the hydrophobic stretch from Ile-523 to Trp-541 is likely to form a transmembrane region with a predicted topology of a type Ia membrane protein (TMHMM2.0), leaving a 64-amino-acid cytoplasmic C terminus. The signal peptide cleavage sites and terminal residues of the transmembrane segments as predicted by using SignalP2.0 and TMHMM2.0 for all four AMA-1 proteins are precisely aligned in Fig. 1. The predicted signal peptide of BbAMA-1 is nearly twice as long as those of the other proteins. However, the possibility that translation initiation does not start at the first ATG codon could not be eliminated. The transmembrane segment of BbAMA-1 is two to four amino acids shorter than the other transmembrane segments, whereas the boundaries of the four segments are remarkably conserved. The lengths of the cytoplasmic domains of the AMA-1 molecules examined are similar (53 to 64 amino acids), but only the 30 C-terminal residues exhibit considerable similarity (e.g., 43.3% identity between the B. bovis and P. falciparum sequences). The extracellular domain of P. falciparum AMA-1 has been shown to be organized in three structural domains (domains I to III) that are stabilized by eight intradomain disulfide bridges (as shown in Fig. 1) between 16 cysteine residues (12 of which are conserved in T. gondii). Fourteen of these cysteine residues are easily aligned with cysteine residues in the B. bovis sequence, and only the third and sixth cysteine residues of domain III, forming disulfide bridge IIIc, are absent. Whereas domain I (Pro96 to Pro303) and domain II (Met-304 to Glu-438) constitute the best-conserved regions of AMA-1 (40.3 and 29.3% identity, respectively, in P. falciparum and B. bovis, excluding gap positions), the remaining sequences, including domain III, show very little conservation. P. falciparum AMA-1 is special when it is compared with the AMA-1 proteins of other Plasmodium species and T. gondii because it has an N-terminal prodomain that is cleaved off during merozoite maturation. BbAMA-1 is the first AMA-1 that also has an extended N-terminal sequence, although this sequence is shorter than its P. falciparum homologue. If gap regions are disregarded, the full-length AMA-1 sequences of B. bovis and P. falciparum are only slightly more similar to each other (28% identity) than they are to the T. gondii sequence (25% identity in both cases), but alignment of the T. gondii AMA-1 sequence with the P. falciparum and B. bovis sequences requires introduction of 24 gap regions, whereas the P. falciparum and B. bovis proteins align with 14 gaps.
FIG. 1.
Multiple-sequence alignment of AMA-1 proteins of B. bovis (Bb), T. gondii (Tg), P. vivax (Pv), and P. falciparum (Pf). Similar and identical residues are shaded. Black shading indicates similarity in all four species, and gray shading indicates similarity in three species. Synthetic peptides 1, 2, and 3 are indicated by black bars. The signal peptide cleavage site is indicated by an arrow, and the transmembrane region is indicated by a grey bar. Cysteine residues that form disulfide bonds in P. falciparum AMA-1 are indicated by domain (I, II, and III) and bond (a, b, and c) designations.
Recognition of recombinant BbAMA-1 by antisera against short, BbAMA-1-derived peptides.
To enable further studies of the BbAMA-1 protein, rabbits were immunized with KLH-linked synthetic peptides (Fig. 1) that were derived from the N-terminal domain (peptide 1, Ala-46 to Ser-60), domain II (peptide 2, Arg-395 to Gly-409), and domain III (peptide 3, Tyr-453 to Val-467). All three antisera specifically recognized a recombinant fusion product of thioredoxin and the extracellular domain of BbAMA-1 (Met-1 to Ser-501) that was expressed in E. coli BL21 cells (Fig. 2). PAGE of total cell lysates obtained before (Fig. 2, lane 1) and after (lane 2) induction with IPTG resulted in identification of the recombinant fusion product as an 80-kDa product (calculated size, 65 kDa) that was recognized by all three immune sera (lanes 4, 6, and 8) and not by preimmune sera (lanes 3, 5, and 7) on immunoblots. Immune recognition was specific for the BbAMA-1 part of the fusion product, as a recombinant fusion product of B. bovis rab5 (lane 11) expressed in PET32a (amino acids 298 to 1801; GenBank accession no. AY324137) was not recognized (10). Also, immune recognition was peptide specific and not due to antibodies induced by the KLH carrier protein used for immunization, as antiserum raised against a KLH-linked synthetic peptide unrelated to AMA-1 did not recognize the BbAMA-1 recombinant fusion product (lane 9).
Confocal immunofluorescence microscopy.
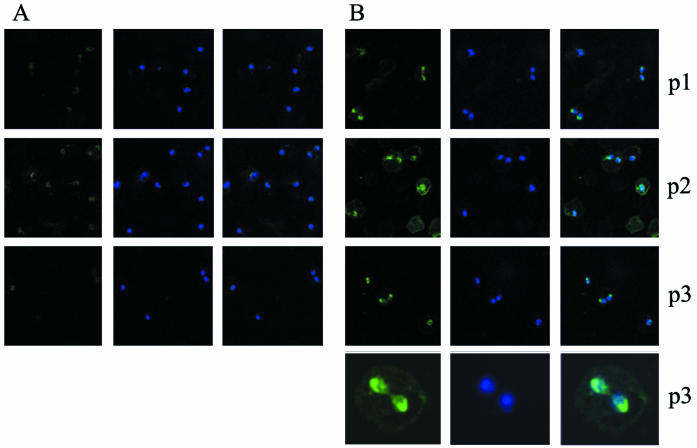
To localize BbAMA-1, rabbit antisera against the three KLH-linked peptides were incubated with B. bovis in vitro cultures attached to glass slides. Staining of DNA with DAPI allowed identification of B. bovis parasites due to their fluorescent nuclei (Fig. 3, middle columns). Fluorescent parasites were clearly visible after incubation with the three immune sera (Fig. 3B, left column), and overlay images with fluorescence derived from DAPI-stained nuclei (Fig. 3B, right column) indicated that all parasites in any microscope field were recognized by anti-AMA-1 antisera. The duplicated, double-pear-shaped parasites usually showed more intense staining than the nonduplicated forms. Nuclei identified by DAPI fluorescence were located posterior to the intense apical staining with the anti-AMA-1 antisera. This was most clearly observed in erythrocytes harboring duplicated parasites whose posterior ends remain linked by a residual body, which allowed easy determination of the apical end (Fig. 3B). Preimmune sera were included as a control and displayed only a faint background fluorescent signal (Fig. 3A).
FIG. 3.
Immunofluorescence reactivity of antiserum against BbAMA-1 incubated with acetone-fixed B. bovis-infected bovine erythrocytes. (A) Incubation with preimmune sera. (B) Incubation with immune sera against peptide 1 (p1), peptide 2 (p2), and peptide 3 (p3), as indicated on the right. Panels in the left column show anti-AMA-1 staining; panels in the middle column show DAPI staining; and panels in the right column are overlay images. The images in the bottom row of panel B are enlargements of duplicated B. bovis merozoites reacting with anti-peptide 3.
Inhibition of in vitro invasion by peptide-specific antisera.
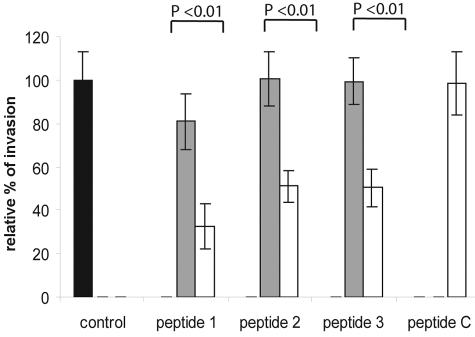
The AMA-1 proteins of P. falciparum and T. gondii are secreted from apically located micronemes and are thought to be involved in host cell invasion (13, 16). A B. bovis in vitro invasion assay that allowed us to study the invasion of erythrocytes by free merozoites in a protein-free buffer within a time span of 1 h was used to assess the effect of antisera directed against the three peptides derived from different domains of BbAMA-1. Free merozoites were preincubated for 1 h at 20°C with the three anti-BbAMA-1 sera and the control serum directed against a nonrelated peptide, after which invasion was started by addition of erythrocytes. All three antisera against the specific BbAMA-1 peptides gave rise to significant inhibition of invasion, whereas preimmune sera and a control antiserum did not have a significant effect on invasion efficiency (Fig. 4). The strongest effect (65% ± 13% inhibition) was observed with the antiserum directed against the N-terminally located peptide, whereas antisera directed against the domain II and III peptides showed less inhibition. Preincubation of merozoites with a combination of antisera directed against peptide 1 and either peptide 2 or peptide 3 did not result in increased inhibition compared to the inhibition observed with antiserum against peptide 1 alone (results not shown).
FIG. 4.
Inhibition of erythrocyte invasion by B. bovis merozoites with antisera raised against four different synthetic peptides. The black bar represents the value for invasion of B. bovis merozoites that were directly added to medium after liberation, and this value was considered the 100% value with which all incubation data were compared. The open bars indicate the values for invasion of erythrocytes by B. bovis merozoites after preincubation with immune sera against peptide 1, peptide 2, peptide 3, and peptide C, whereas the grey bars indicate the values for invasion of B. bovis merozoites after preincubation with preimmune serum. Each bar indicates the average value for six individual experiments, and the error bars indicate standard deviations. Data were examined by using the Kruskal-Wallis nonparametric test. Pairwise comparisons of the groups were performed by post hoc analysis as advised by Kruskall-Wallis, and P values are indicated at the top.
Detection of BbAMA-1 secreted into the surroundings during erythrocyte invasion.
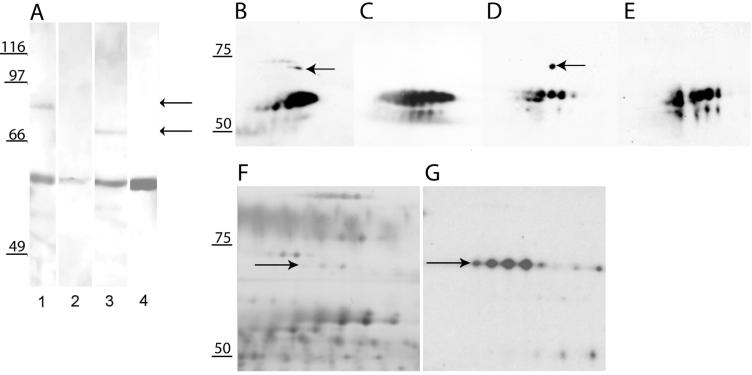
Despite the easy recognition of recombinant BbAMA-1 on Western blots by antisera against all three BbAMA-1-derived peptides (Fig. 2), only minor amounts could be detected directly in B. bovis extracts by sensitive chemoluminescence staining methods. The best antiserum for this purpose appeared to be the antiserum raised against peptide 1 (N-terminal peptide), which detected a B. bovis-specific 82-kDa band in total merozoite extracts (Fig. 5A, lane 1) and a smaller 69-kDa band in the protein pool secreted after invasion of fresh erythrocytes by liberated merozoites for 1 h (lane 3). Antisera against the other two peptides occasionally recognized bands of comparable sizes, but the signals were very weak. In addition, variation in repeated experiments also included the occasional detection of other minor bands (results not shown) by sera against peptides 2 and 3, making the results obtained by Western blotting with these two antisera inconclusive. A reaction of anti-peptide 1 serum with 55- to 60-kDa proteins was observed in lanes containing the merozoite extract and invasion supernatant (Fig. 5, lanes 1 and 3), as well as in control lanes containing only erythrocyte proteins (Fig. 5, lanes 2 and 4). Additional controls showed that several other unrelated peptides, when linked to KLH, induced rabbit antisera that reacted with a group of erythrocyte cytosolic proteins at this position (results not shown). Merozoite extracts contain erythrocyte cytosolic proteins derived from a fraction of erythrocytes not lysed during isolation of merozoites, whereas proteins secreted during invasion have been shown to be contaminated with erythrocyte cytosolic proteins (12), as a small number of erythrocytes are lysed during invasion.
FIG. 5.
Western blot analysis of total merozoites and invasion supernatant incubated with sera raised against AMA-1 peptides after 1D SDS-PAGE (A) and 2D SDS-PAGE (B to G). (A) Total merozoites (lane 1) and invasion supernatant (lane 3) were incubated with anti-peptide 1 antiserum. The arrows indicate bands at 82 kDa (lane 1) and 69 kDa (lane 3). Lanes 2 and 4 contained an erythrocyte control incubated with anti-peptide 1 antiserum. (B and D) Western blots of 2D gels loaded with invasion material and incubated with anti-peptide 1 and anti-peptide 3 antisera, respectively. (C and E) Erythrocyte controls incubated with anti-peptide 1 and anti-peptide 3 antisera, respectively. (F and G) Silver-stained image of a 2D gel loaded with invasion supernatant obtained from metabolically labeled B. bovis run in parallel (F) and subsequently exposed to film (G). The arrows indicate BbAMA-1-specific spots in total merozoite and invasion supernatants. Molecular masses are indicated on the left.
To obtain additional proof of the erythrocytic nature of the bands and the B. bovis-specific nature of the 69-kDa band secreted upon invasion, 2D gel electrophoresis was employed. Western blots of 2D gels were probed with anti-peptide 1 serum (Fig. 5B) and anti-peptide 3 serum (Fig. 5D) and matched with a silver-stained image of another gel (Fig. 5F). Subsequent metabolic labeling of parasites with 35S prior to invasion allowed localization of B. bovis-specific spots (Fig. 5G). Erythrocytic proteins at 55 to 60 kDa (nonlabeled) were recognized by both antisera and were also present in controls carrying only erythrocyte cytosolic proteins (Fig. 5C and E). In addition, spots that specifically reacted with anti-peptide 1 serum (Fig. 5B) and anti-peptide 2 serum (Fig. 5D) were detected on the 2D Western blots, matching the most abundant spots in a row of four metabolically labeled 69-kDa spots that in turn matched a row of weak spots on the silver-stained gel. We concluded on basis of this set of data that AMA-1 is a low-abundance protein and that a 69-kDa form is secreted upon invasion of erythrocytes by B. bovis merozoites.
DISCUSSION
Apicomplexan organisms are defined by a common set of apically located secretory organelles required for host cell invasion by a mechanism having many conserved features. Despite the conserved nature of the invasion process, only very few proteins which are involved exhibit significant similarity when T. gondii is compared with Plasmodium species. AMA-1 is one such protein conserved in T. gondii and P. falciparum, and here we describe cloning of the B. bovis homologue, which we designated BbAMA-1. Overall, BbAMA-1 is only slightly more similar to P. falciparum AMA-1 than to T. gondii AMA-1 (28 and 25%, respectively). Most notably, these proteins have 10 conserved cysteine residues that have been shown to form disulfide bridges and have been suggested to stabilize the proposed domains I and II of the P. falciparum AMA-1 ectodomain (17, 30). However, a few conserved residues, like the four cysteine residues that form two of the three disulfide bridges present in domain III of P. falciparum AMA-1, may represent a functional feature conserved in B. bovis and Plasmodium species that is not present in T. gondii. Also, the number of insertions or deletions is much smaller in a pairwise comparison of B. bovis and P. falciparum than in comparisons with T. gondii. So far, molecular phylogenetic analyses have not unambiguously resolved the evolutionary relationship among the genera Toxoplasma, Babesia, and Plasmodium, and it is tempting to hypothesize that some features of the AMA-1 ectodomain may be slightly more conserved in Babesia and Plasmodium because of their identical host cell target, the erythrocyte.
Like other micronemal proteins, AMA-1 has a short cytoplasmic domain whose length varies between 64 amino acids (B. bovis) and 53 amino acids (P. falciparum), of which the 30 C-terminal residues are remarkably similar, indicating a conserved function. The cytoplasmic tail of micronemal proteins has been suggested to function as an organellar targeting signal (7, 18), but a more recent study has shown that after removal of this domain there is still correct localization of P. falciparum micronemal proteins like EBA-175 and TRAP (14). The cytoplasmic tail may be involved in binding to other intracellular proteins that may govern the timing of secretion, which is not synchronous for different micronemal proteins (15). Alternatively, it may be involved in transmitting a signal upon contact of the ectodomain with a host cell ligand, or it may be involved in linkage to cytoskeletal structures like those recently described for members of the TRAP family of micronemal proteins (21).
P. falciparum AMA-1 is proteolytically processed at several positions, giving rise to a complicated pattern of bands recognized by polyclonal and monoclonal antisera (3). An 83-kDa type I transmembrane protein with aberrant mobility (predicted Mw, 64,000) is created after cleavage of the signal peptide. Upon translocation to the apical region a 72-amino-acid propeptide is removed, leaving a protein that migrates at 66 kDa on polyacrylamide gels (19). T. gondii AMA-1 does not possess such a propeptide, whereas BbAMA-1 was shown here to contain an N-terminal region whose size is intermediate between the sizes of the regions in T. gondii AMA-1 and P. falciparum AMA-1. Peptide 1-directed antisera recognized a 82-kDa band in B. bovis total merozoite extracts, suggesting that there was aberrant mobility (the predicted Mw for the region from the signal peptide cleavage site to the C terminus is 62,700), as observed for P. falciparum AMA-1. Whether a propeptide is cleaved remains to be determined as Western blots probed with anti-peptide 2 and anti-peptide 3 sera yielded inconclusive results, potentially due to the small amounts of BbAMA-1 present.
Several forms of soluble P. falciparum AMA-1 are continuously released from the merozoite surface into the extracellular milieu. A 48-kDa form results from cleavage at a position 29 residue N-terminal to the transmembrane region (19), whereas a 52-kDa form probably results from cleavage just beside or within the transmembrane region (19). Further processing gives rise to a 44-kDa protein (19). Here we demonstrated the release of a 69-kDa form of BbAMA-1 that is recognized by anti-peptide 1 and anti-peptide 3 sera upon invasion of erythrocytes, suggesting that there is cleavage N terminal of the transmembrane region for BbAMA-1 as well. Analysis on 2D gels allowed mapping of this band to a row of spots of low abundance and provided definite proof of the Babesia origin of these spots by making use of metabolic labeling. Whereas on 1D gels only anti-peptide 1 serum consistently recognized this band, peptide 3-directed antisera also recognized these spots on 2D gels, probably due to the larger amount of protein loaded onto a 2D gel, arguing against cleavage of a propeptide.
Immunofluorescence showed that BbAMA-1 was localized at the merozoite surface and was specifically distributed around the apical region, like the N-terminal processed form of P. falciparum AMA-1. P. falciparum AMA-1 has been shown to be localized near the periphery of micronemes in developing schizonts, most likely as a transmembrane protein (3). Upon release of mature merozoites, secretion from micronemes results in spreading of P. falciparum AMA-1 over the outside of the plasma membrane. Immunofluorescence studies indicated that P. falciparum AMA-1 remains mainly confined to the apical half of the merozoite. Upon erythrocyte invasion traces of P. falciparum AMA-1 were shown to be carried into the host cell, but immunofluorescence soon faded away (10). Immunofluorescence studies with BbAMA-1 anti-peptide sera resulted in staining of the apical part of B. bovis parasites present in asynchronous in vitro cultures. Staining revealed a punctuate pattern which could represent BbAMA-1 located in the plasma membrane, as well as in apical organelles. This staining pattern and the fact that the more intense staining was detected mainly on apparently mature and duplicated intraerythrocytic forms suggest localization and temporal expression similar to those observed for P. falciparum AMA-1.
The inability to make knockout mutants suggests that AMA-1 has a critical function in T. gondii (16) and P. falciparum (37). Several lines of evidence suggest that AMA-1 has a role in host cell invasion. Monoclonal and polyclonal antisera inhibit host cell invasion (1, 16, 17, 24), as do peptides binding specifically to AMA-1 (22, 26). Also, repacement of the P. falciparum AMA-1 gene with the P. chabaudi AMA-1 gene results in P. falciparum parasites that invade mouse erythrocytes better (37). Antisera directed against BbAMA-1-derived peptides specifically reduced the in vitro invasion efficiency of B. bovis, indicating that AMA-1 is indeed located on the surface of merozoites and is accessible to antibodies. Invasion inhibition may result in blocking of some specific function of AMA-1 or may involve inhibition of proteolytic processing, as recently shown for P. falciparum AMA-1 (10). Alternatively, antibodies may just cross-link merozoites, although the low dilutions required to obtain an immunofluorescent signal argue against this option. Peptides derived from loop 1 of domain III of P. falciparum AMA-1 have been shown to induce antibodies that inhibit P. falciparum growth (29). The immunodominant epitopes in these peptides are located in the region that is aligned with peptide 3 of BbAMA-1 in Fig. 1.
The epitopes recognized by other invasion-blocking antisera or peptides have not been mapped yet. Antisera directed against peptide 1 (from the N terminus) and peptide 2 (from domain II) also gave rise to inhibition of B. bovis invasion, indicating that inhibitory antibodies can be directed against epitopes over the full length of the ectodomain. The peptides were selected on the basis of predictions that they form amphiphatic α-helices with a high surface probability and a considerable number of charged residues. Studies of the population genetics of the AMA-1 gene in endemic areas have provided evidence that there is selective pressure operating on domains I and III, indicating that they are targets for protective immunity (6, 33). Remarkably, the AMA-1 sequences in domain I are most conserved in genera (Fig. 1), implying that there are strong functional constraints, whereas the population studies mentioned above indicated that this region of P. falciparum AMA-1 is the most variable region.
P. falciparum AMA-1 is considered one of the prime candidates for incorporation into a recombinant vaccine (10) for reasons briefly discussed above. Soluble parasitic antigens of B. bovis secreted into the environment during in vitro culture have been shown to confer protection against B. bovis infection in cattle (28, 32), although no individual antigens contributing to this effect have been isolated. As shown by the results presented here, we have identified BbAMA-1 as such a secreted protein, and the inhibition of invasion by rabbit antibodies directed against this protein indicates that it might be one of the protective components of soluble parasitic antigens. Further proof obviously requires immunization experiments with cattle in which native or recombinant BbAMA-1 is used. Genetic studies on BbAMA-1 diversity in the field should help in determining the potential of this protein as a vaccine component.
Acknowledgments
We thank A. W. C. A. Cornelissen for critical reading of the manuscript. Anko de Graaf is gratefully acknowledged for advice and assistance with confocal imaging. Omar Taoufik is thanked for technical support. B. bovis clonal line C61411 was provided by E. Pipano (Kimron Veterinary Institute, Bet Dagan, Israel).
This study was supported in part by The Netherlands Foundation for Advancement of Tropical Research (NWO-WOTRO) and by Intervet International B.V.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Anders, R. F., P. E. Crewther, S. Edwards, M. Margetts, M. L. Matthew, B. Pollock, and D. Pye. 1998. Immunisation with recombinant AMA-1 protects mice against infection with Plasmodium chabaudi. Vaccine 16:240-247. [DOI] [PubMed] [Google Scholar]
- 2.Anders, R. F., D. J. McColl, and R. L. Coppel. 1993. Molecular variation in Plasmodium falciparum: polymorphic antigens of asexual erythrocytic stages. Acta Trop. 53:239-253. [DOI] [PubMed] [Google Scholar]
- 3.Bannister, L. H., J. M. Hopkins, A. R. Dluzewski, G. Margos, I. T. Williams, M. J. Blackman, C. H. Kocken, A. W. Thomas, and G. H. Mitchell. 2003. Plasmodium falciparum apical membrane antigen 1 (PfAMA-1) is translocated within micronemes along subpellicular microtubules during merozoite development. J. Cell Sci. 116:3825-3834. [DOI] [PubMed] [Google Scholar]
- 4.Carruthers, V. B. 1999. Armed and dangerous: Toxoplasma gondii uses an arsenal of secretory proteins to infect host cells. Parasitol. Int. 48:1-10. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, Q., and A. Saul. 1994. Sequence analysis of the apical membrane antigen I (AMA-1) of Plasmodium vivax. Mol. Biochem. Parasitol. 65:183-187. [DOI] [PubMed] [Google Scholar]
- 6.Cortes, A., M. Mellombo, I. Mueller, A. Benet, J. C. Reeder, and R. F. Anders. 2003. Geographical structure of diversity and differences between symptomatic and asymptomatic infections for Plasmodium falciparum vaccine candidate AMA1. Infect. Immun. 71:1416-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Cristina, M., R. Spaccapelo, D. Soldati, F. Bistoni, and A. Crisanti. 2000. Two conserved amino acid motifs mediate protein targeting to the micronemes of the apicomplexan parasite Toxoplasma gondii. Mol. Cell. Biol. 20:7332-7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donahue, C. G., V. B. Carruthers, S. D. Gilk, and G. E. Ward. 2000. The Toxoplasma homolog of Plasmodium apical membrane antigen-1 (AMA-1) is a microneme protein secreted in response to elevated intracellular calcium levels. Mol. Biochem. Parasitol. 111:15-30. [DOI] [PubMed] [Google Scholar]
- 9.Dubremetz, J. F., N. Garcia-Reguet, V. Conseil, and M. N. Fourmaux. 1998. Apical organelles and host-cell invasion by Apicomplexa. Int. J. Parasitol. 28:1007-1013. [DOI] [PubMed] [Google Scholar]
- 10.Dutta, S., J. D. Haynes, J. K. Moch, A. Barbosa, and D. E. Lanar. 2003. Invasion-inhibitory antibodies inhibit proteolytic processing of apical membrane antigen 1 of Plasmodium falciparum merozoites. Proc. Natl. Acad. Sci. 100:12295-12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dutta, S., P. Malhotra, and V. S. Chauhan. 1995. Sequence analysis of apical membrane antigen 1 (AMA-1) of Plasmodium cynomolgi bastianelli. Mol. Biochem. Parasitol. 73:267-270. [DOI] [PubMed] [Google Scholar]
- 12.Franssen, F. F., F. R. Gaffar, A. P. Yatsuda, and E. de Vries. 2003. Characterisation of erythrocyte invasion by Babesia bovis merozoites efficiently released from their host cell after high-voltage pulsing. Microbes Infect. 5:365-372. [DOI] [PubMed] [Google Scholar]
- 13.Fraser, T. S., S. H. Kappe, D. L. Narum, K. M. VanBuskirk, and J. H. Adams. 2001. Erythrocyte-binding activity of Plasmodium yoelii apical membrane antigen-1 expressed on the surface of transfected COS-7 cells. Mol. Biochem. Parasitol. 117:49-59. [DOI] [PubMed] [Google Scholar]
- 14.Gilberger, T. W., J. K. Thompson, M. B. Reed, R. T. Good, and A. F. Cowman. 2003. The cytoplasmic domain of the Plasmodium falciparum ligand EBA-175 is essential for invasion but not protein trafficking. J. Cell Biol. 162:317-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Healer, J., S. Crawford, S. Ralph, G. McFadden, and A. F. Cowman. 2002. Independent translocation of two micronemal proteins in developing Plasmodium falciparum merozoites. Infect. Immun. 70:5751-5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hehl, A. B., C. Lekutis, M. E. Grigg, P. J. Bradley, J. F. Dubremetz, E. Ortega-Barria, and J. C. Boothroyd. 2000. Toxoplasma gondii homologue of Plasmodium apical membrane antigen 1 is involved in invasion of host cells. Infect. Immun. 68:7078-7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hodder, A. N., P. E. Crewther, M. L. Matthew, G. E. Reid, R. L. Moritz, R. J. Simpson, and R. F. Anders. 1996. The disulfide bond structure of Plasmodium apical membrane antigen-1. J. Biol. Chem. 271:29446-29452. [DOI] [PubMed] [Google Scholar]
- 18.Hoppe, H. C., H. M. Ngo, M. Yang, and K. A. Joiner. 2000. Targeting to rhoptry organelles of Toxoplasma gondii involves evolutionarily conserved mechanisms. Nat. Cell Biol. 2:449-456. [DOI] [PubMed] [Google Scholar]
- 19.Howell, S. A., I. Well, S. L. Fleck, C. Kettleborough, C. R. Collins, and M. J. Blackman. 2003. A single malaria merozoite serine protease mediates shedding of multiple surface proteins by juxtamembrane cleavage. J. Biol. Chem. 278:23890-23898. [DOI] [PubMed] [Google Scholar]
- 20.Howell, S. A., C. Withers-Martinez, C. H. Kocken, A. W. Thomas, and M. J. Blackman. 2001. Proteolytic processing and primary structure of Plasmodium falciparum apical membrane antigen-1. J. Biol. Chem. 276:31311-31320. [DOI] [PubMed] [Google Scholar]
- 21.Jewett, T. J., and L. D. Sibley. 2003. Aldolase forms a bridge between cell surface adhesins and the actin cytoskeleton in apicomplexan parasites. Mol. Cell 11:885-894. [DOI] [PubMed] [Google Scholar]
- 22.Keizer, D. W., L. A. Miles, F. Li, M. Nair, R. F. Anders, A. M. Coley, M. Foley, and R. S. Norton. 2003. Structures of phage-display peptides that bind to the malarial surface protein, apical membrane antigen 1, and block erythrocyte invasion. Biochemistry 42:9915-9923. [DOI] [PubMed] [Google Scholar]
- 23.Kocken, C. H., M. A. Dubbeld, A. Van Der Wel, J. T. Pronk, A. P. Waters, J. A. Langermans, and A. W. Thomas. 1999. High-level expression of Plasmodium vivax apical membrane antigen 1 (AMA-1) in Pichia pastoris: strong immunogenicity in Macaca mulatta immunized with Plasmodium vivax AMA-1 and adjuvant SBAS2. Infect. Immun. 67:43-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kocken, C. H., D. L. Narum, A. Massougbodji, B. Ayivi, M. A. Dubbeld, A. van der Wel, D. J. Conway, A. Sanni, and A. W. Thomas. 2000. Molecular characterisation of Plasmodium reichenowi apical membrane antigen-1 (AMA-1), comparison with Plasmodium falciparum AMA-1, and antibody-mediated inhibition of red cell invasion. Mol. Biochem. Parasitol. 109:147-156. [DOI] [PubMed] [Google Scholar]
- 25.Kocken, C. H., C. Withers-Martinez, M. A. Dubbeld, A. van der Wel, F. Hackett, A. Valderrama, M. J. Blackman, and A. W. Thomas. 2002. High-level expression of the malaria blood-stage vaccine candidate Plasmodium falciparum apical membrane antigen 1 and induction of antibodies that inhibit erythrocyte invasion. Infect. Immun. 70:4471-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, F., A. Dluzewski, A. M. Coley, A. Thomas, L. Tilley, R. F. Anders, and M. Foley. 2002. Phage-displayed peptides bind to the malarial protein apical membrane antigen-1 and inhibit the merozoite invasion of host erythrocytes. J. Biol. Chem. 277:50303-50310. [DOI] [PubMed] [Google Scholar]
- 27.Marshall, V. M., M. G. Peterson, A. M. Lew, and D. J. Kemp. 1989. Structure of the apical membrane antigen I (AMA-1) of Plasmodium chabaudi. Mol. Biochem. Parasitol. 37:281-283. [DOI] [PubMed] [Google Scholar]
- 28.Montenegro-James, S., M. Ristic, M. Toro Benitez, E. Leon, and R. Lopez R. 1985. Heterologous strain immunity in bovine babesiosis using a culture-derived soluble Babesia bovis immunogen. Vet. Parasitol. 18:321-337. [DOI] [PubMed] [Google Scholar]
- 29.Mueller, M. S., A. Renard, F. Boato, D. Vogel, M. Naegeli, R. Zurbriggen, J. A. Robinson, and G. Pluschke. 2003. Induction of parasite growth-inhibitory antibodies by a virosomal formulation of a peptidomimetic of loop I from domain III of Plasmodium falciparum apical membrane antigen 1. Infect. Immun. 71:4749-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nair, M., M. G. Hinds, A. M. Coley, A. N. Hodder, M. Foley, R. F. Anders, and R. S. Norton. 2002. Structure of domain III of the blood-stage malaria vaccine candidate, Plasmodium falciparum apical membrane antigen 1 (AMA1). J. Mol. Biol. 322:741-753. [DOI] [PubMed] [Google Scholar]
- 31.Narum, D. L., and A. W. Thomas. 1994. Differential localization of full-length and processed forms of PF83/AMA-1, an apical membrane antigen of Plasmodium falciparum merozoites. Mol. Biochem. Parasitol. 67:59-68. [DOI] [PubMed] [Google Scholar]
- 32.Patarroyo, J. H., A. A. Prates, C. A. Tavares, C. L. Mafra, and M. I. Vargas. 1995. Exoantigens of an attenuated strain of Babesia bovis used as a vaccine against bovine babesiosis. Vet. Parasitol. 59:189-199. [DOI] [PubMed] [Google Scholar]
- 33.Polley, S. D., and D. J. Conway. 1994. Strong diversifying selection on domains of the Plasmodium falciparum apical membrane antigen 1 gene. Genetics 158:1505-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salazar, L. M., M. P. Alba, M. H. Torres, M. Pinto, X. Cortes, L. Torres, and M. E. Patarroyo. 2002. Protection against experimental malaria associated with AMA-1 peptide analogue structures. FEBS Lett. 527:95-100. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual: 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Soldati, D., J. F. Dubremetz, and M. Lebrun. 2001. Microneme proteins: structural and functional requirements to promote adhesion and invasion by the apicomplexan parasite Toxoplasma gondii. Int. J. Parasitol. 31:1293-1302. [DOI] [PubMed] [Google Scholar]
- 37.Triglia, T., J. Healer, S. R. Caruana, A. N. Hodder, R. F. Anders, B. S. Crabb, and A. F. Cowman. 2000. Apical membrane antigen 1 plays a central role in erythrocyte invasion by Plasmodium species. Mol. Microbiol. 38:706-718. [DOI] [PubMed] [Google Scholar]
- 38.Urquiza, M., J. E. Suarez, C. Cardenas, R. Lopez, A. Puentes, F. Chavez, J. C. Calvo, and M. E. Patarroyo. 2000. Plasmodium falciparum AMA-1 erythrocyte binding peptides implicate AMA-1 as erythrocyte binding protein. Vaccine 19:508-513. [DOI] [PubMed] [Google Scholar]
- 39.Yatsuda, A. P., J. Krijgsveld, A. W. Cornelissen, A. J. Heck, and E. de Vries. 2003. Comprehensive analysis of the secreted proteins of the parasite Haemonchus contortus reveals extensive sequence variation and differential immune recognition. J. Biol. Chem. 278:16941-16951. [DOI] [PubMed] [Google Scholar]