Abstract
Francisella tularensis is a highly infectious gram-negative bacterium with potential for use as a bioweapon. Analysis of the F. tularensis live vaccine strain (LVS) ultrastructure by electron microscopy revealed the presence of long, thin fibers, similar in appearance to type 4 pili. The highly virulent F. tularensis Schu S4 strain was found to contain type 4 pilus genes, and we confirmed that these genes are present and expressed in the LVS.
Francisella tularensis, the causative agent of tularemia, is a zoonotic pathogen found in various animal hosts and can also be recovered from the environment. F. tularensis infects humans through the skin, mucous membranes, gastrointestinal tract, and lungs and through the bite of infected arthropods. The primary pneumonic form of tularemia is the most severe, with mortality rates as high as 60% if not treated (7, 8). Aerosolized F. tularensis is extremely infectious, and a number of naturally occurring airborne outbreaks have been documented (8). The high infectivity and lethality of the aerosolized form have led to classification of F. tularensis as a category A agent of bioterrorism (7, 27).
F. tularensis is a small, nonmotile, pleomorphic, gram-negative coccobacillus. It is a facultative intracellular pathogen, able to survive and replicate inside macrophages (30). There are two main subspecies of F. tularensis associated with human disease. Francisella tularensis subsp. tularensis is highly virulent and predominant in North America. Francisella tularensis subsp. holarctica is predominant in Eurasia and produces a milder form of the disease in humans but is still highly virulent in mice. A live, attenuated vaccine (the live vaccine strain [LVS]) was developed from an F. tularensis subsp. holarctica strain (7).
The molecular mechanisms that account for the high virulence of F. tularensis are largely unknown. F. tularensis makes an unusual lipopolysaccharide that has a low toxicity and produces a low proinflammatory response compared to that of lipopolysaccharide from other bacteria (9, 29). F. tularensis expresses a capsule that protects it against serum-mediated lysis and appears to be necessary for full virulence (28). Several genes associated with growth inside macrophages have been identified (2, 15, 16). However, apart from the capsule, toxins or other secreted virulence factors have not been identified to date in F. tularensis. We describe here the presence of pilus-like structures on the surface of the F. tularensis LVS.
F. tularensis LVS ultrastructure.
The F. tularensis LVS (ATCC 29684; American Type Culture Collection, Manassas, Va.) and a capsule-deficient LVS mutant (28) were cultured in supplemented Mueller-Hinton broth (MH; BD Microbiology Systems, Sparks, Md.) at 37°C and 5% CO2 as described previously (19). Frozen aliquots were streaked onto Mueller-Hinton chocolate agar (MHC; BD Microbiology Systems), and single bacterial colonies were inoculated into MH or Chamberlain's defined medium (CDM), prepared as described previously (3, 4). These cultures were incubated at 37°C in 5% CO2 with shaking at 100 rpm for 16 to 40 h. For transmission electron microscopy (TEM), bacteria were washed once with phosphate-buffered saline, adsorbed onto polyvinyl formal-carbon-coated grids (Ernest F. Fullam, Latham, N.Y.) for 2 min, and fixed with 1% glutaraldehyde (Sigma-Aldrich, St. Louis, Mo.) for 1 min. The grids were washed twice with phosphate-buffered saline and twice with water and then negatively stained with 0.5% phosphotungstic acid (Ted Pella Inc., Redding, Calif.) for 25 s. The grids were viewed in a JEOL 1200EXII transmission electron microscope (JEOL USA, Peabody, Mass.) at 80-kV accelerating voltage.
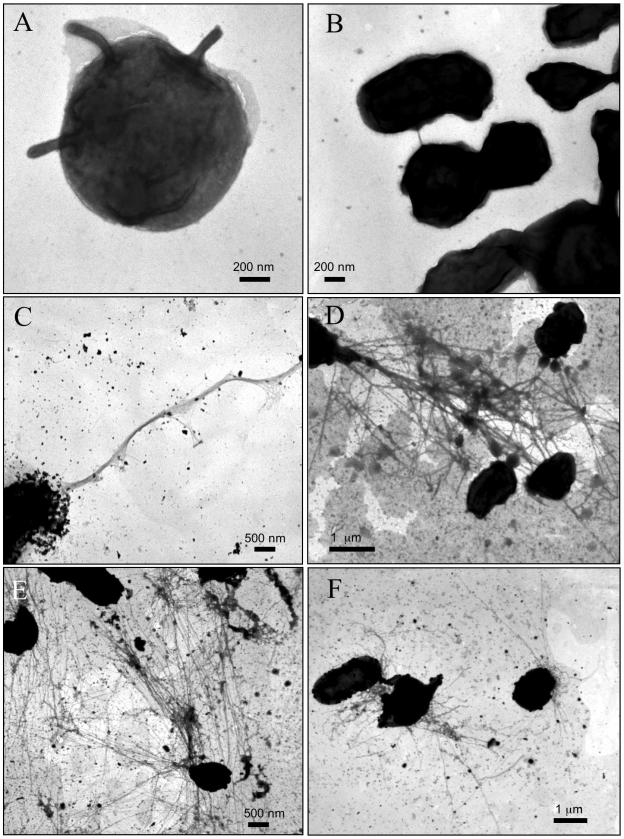
We first examined the ultrastructure of the LVS grown on solid medium (MHC). The majority of bacteria appeared as small, round or oval cells, often surrounded by lighter-staining material that likely represents the capsule. One or more short, thick, horn-like structures could be seen protruding from the surface of some bacteria (Fig. 1A). These thick projections, roughly 60 nm in diameter, could be related to large membrane protrusions reported in a previous electron microscopy study (12). Bacterial aggregates were common, and short bridges of various sizes could be seen joining closely spaced bacteria (Fig. 1B). We next examined bacteria grown in a rich liquid medium (MH) to early (16-h) and late (40-h) log phases of growth. The short, thick protrusions were not detected in the broth-grown bacteria. However, long, thin fibers could be seen emanating from an occasional grouping of cells. These pilus-like fibers often had a polar location (Fig. 1C). In some cases, the fibers were highly expressed and formed tangled networks that connected multiple bacteria (Fig. 1D). Bacterial expression of the fibers was more abundant at 16 than at 40 h of growth. The LVS grown in a defined medium (CDM) exhibited the same features as did bacteria grown in MH, but a greater proportion of the cells expressed the long, thin fibers. Polar bundles of fibers were again observed, and the nets of highly intertwined fibers were especially prominent (Fig. 1E and F). Expression of the fibers was highest at 16 h of growth. We found that a capsule-deficient LVS mutant (28) produced the same long, thin fibers as did the parent strain, indicating that these fibers are not related to or dependent on the capsule (data not shown).
FIG. 1.
Whole-bacterium, negative-stain TEM of F. tularensis LVS grown on different media. (A and B) Bacteria grown on MHC. (A) Bacterium with thick, horn-like protrusions. (B) Cluster of bacteria, with two or more bacteria directly connected or joined by a short bridge. (C and D) Bacteria grown in MH. Bacteria expressed long, polar bundles (C) or tangled networks (D) of long, thin fibers. (E and F) Bacteria grown in CDM. Both panels show bacteria joined by a tangled network of the long, thin fibers.
F. tularensis Schu S4 contains type 4 pilus genes.
The long, thin fibers observed in MH and CDM were similar in appearance to type 4 pili (T4P), which are expressed by a number of gram-negative pathogens (19, 22, 25, 32). T4P function in adhesion to host tissues (32), DNA uptake (11), biofilm formation, and twitching motility (21, 23, 31). The LVS genome (http://bbrp.llnl.gov/bbrp/html/microbe.html) was not available at the time of these studies. Therefore, we examined a draft sequence of the highly virulent F. tularensis subsp. tularensis Schu S4 strain (26) for the presence of T4P genes. BLAST analysis using the server available at the F. tularensis Genome Sequencing Consortium's website (http://artedi.ebc.uu.se/Projects/Francisella/blast) identified orthologs to T4P genes present in Neisseria spp. and Pseudomonas aeruginosa.
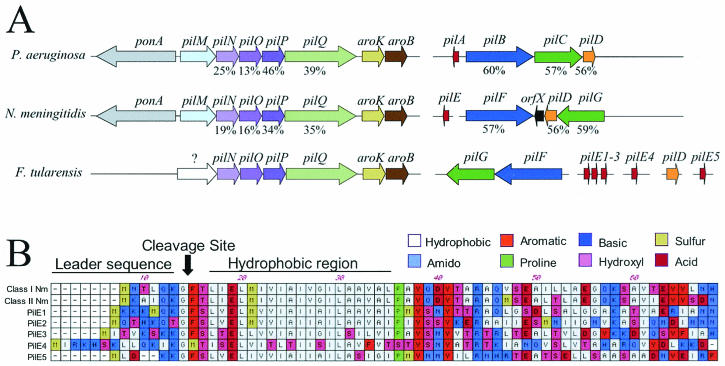
The Schu S4 T4P genes are distributed in clusters or individually at different locations in the genome (Fig. 2A). This fractionated distribution is also found in other bacteria, including Neisseria spp. and P. aeruginosa (10, 22, 33). According to the Neisseria nomenclature, one F. tularensis cluster contains the genes pilNOPQ. The PilQ secretin forms the outer membrane channel for pilus secretion to the cell surface, and PilP is a lipoprotein that functions as a “pilot” to stabilize the secretin in the outer membrane (5, 22). The functions of PilN and PilO are not known (10, 22). The structure of this cluster is conserved in other bacteria (Fig. 2A) (10, 22). Downstream of pilQ in F. tularensis are the aroK and aroB genes, which are involved in the metabolism of aromatic amino acids (26). These genes are found at the same position in other bacteria (Fig. 2A). In contrast, the upstream region of the F. tularensis cluster lacks pilM, which is usually found preceding pilN (Fig. 2A) (10, 22). Instead, F. tularensis has a gene without significant homology to other genes at this position. The function of PilM in T4P biogenesis is not known (10, 22). F. tularensis also lacks the gene ponA, involved in peptidoglycan biosynthesis, which is typically found upstream of pilM (10, 22). A second F. tularensis cluster contains the genes pilFG, which also occur together in other bacteria (Fig. 2A) (10, 22, 33). PilF is the cytoplasmic ATPase responsible for pilus assembly, and PilG is an integral inner membrane protein (10, 22, 33). The genes pilT and pilD occur at separate locations in the Schu S4 genome. PilT is the ATPase responsible for retraction of the pilus (23, 31) and is required for twitching motility and DNA uptake (6, 21). PilD is the inner membrane peptidase responsible for processing of the prepilin signal sequence and methylation of the mature pilin (20).
FIG. 2.
(A) Distribution of the T4P genes in P. aeruginosa, Neisseria meningitidis, and F. tularensis Schu S4. Homologous genes have the same color. Homology values (identical plus similar residues, percentages of total protein) from pairwise sequence alignments of the Neisseria and P. aeruginosa T4P proteins with the Schu S4 gene products are shown below the arrows. The alignments were done with ClustalW using the MacVector software program (Oxford Molecular Ltd., Madison, Wis.). Arrows point in the direction of the open reading frame, and interrupted lines represent different regions of the genome. (B) Alignment of the conserved N-terminal regions of the Schu S4 pilin candidates with those of two N. meningitidis pilins (class I, GenBank accession no. AY077727; class II, GenBank accession no. U8155). Characteristic features of pilin proteins are indicated, including the leader sequence, cleavage site, and hydrophobic region.
We identified at least five different genes in the Schu S4 genome that could code for pilus subunit proteins or pilins (pilE1 to pilE5). Three of these genes (pilE1 to pilE3) occurred together in a cluster (Fig. 2A). Each of the candidate pilins has a characteristic, short N-terminal leader sequence containing positively charged amino acids (Fig. 2B) (18). A typical prepilin peptidase cleavage site is found following a conserved glycine, and there is a conserved glutamate at position +5 of the mature protein (Fig. 2B) (32). As with pilin subunits of other bacteria, the conserved glutamate is followed by a hydrophobic region (Fig. 2B) (32). Each of the Schu S4 pilins also has a conserved pair of cysteines in its C-terminal region (32). Four of the pilE candidates (pilE1 to pilE3 and pilE5) belong to the type A pilin subfamily, which has a short leader sequence and for which the first amino acid of the mature protein is phenylalanine (32). The pilE4 gene belongs to the type B subfamily, which has a longer leader sequence and for which the first mature amino acid is not phenylalanine. Type B pili often form long, polar bundles and are associated with enteric pathogens such as Escherichia coli and Vibrio cholerae (13, 14, 17). In addition to the five candidate pilE genes, we identified several genes in the Schu S4 genome coding for proteins with more limited pilin homology (data not shown). A number of such proteins, termed pseudopilins, have been found in T4P biogenesis systems and also in the closely related type II secretion pathway (1, 24, 32).
T4P genes are present and expressed in the LVS.
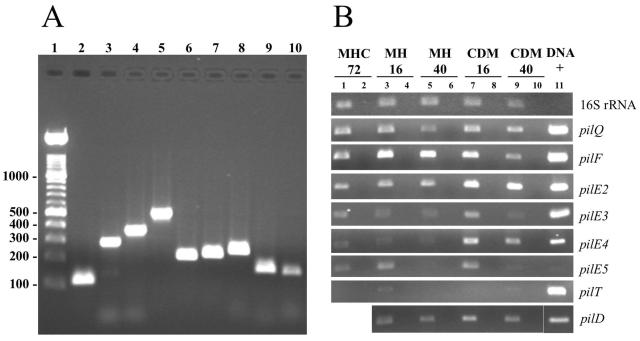
To confirm that the T4P genes identified in the Schu S4 genome were present in the LVS, PCR primers were designed based on the Schu S4 sequence to amplify a portion of one gene from each cluster (pilQ, pilF, pilD, and pilT) as well as the five candidate pilin genes (pilE1 to pilE5) (Table 1). Whole-bacterium PCR was performed using 5 μl of an LVS colony boiled for 10 min in 100 μl of water. PCR products of the expected sizes were obtained for each of the pilus genes tested (Fig. 3A), except pilE1 (data not shown). To verify the identity of the LVS genes, the PCR products were purified with a Wizard PCR Preps DNA purification kit (Promega, Madison, Wis.) and sequenced using the BigDye Terminator V3.1 cycle sequencing kit (Applied Biosystems, Foster City, Calif.) and the Stony Brook University DNA Sequencing Facility. The pilQ, pilF, pilD, pilE4, and pilE5 PCR products were essentially identical to the Schu S4 sequences. However, the sequence of the pilT, pilE2, and pilE3 PCR products revealed the presence of mutations that introduced stop codons into their open reading frames (a frameshift in pilE3 and point mutations in pilT and pilE2). Despite the introduction of stop codons into these genes, the LVS should still be able to assemble pili. The pilT ATPase is not required for pilus assembly (6, 21), and although candidate pilin genes pilE1 to pilE3 are not likely to be functional, pilE4 and pilE5 appear to be intact. The differences in the pil genes between the Schu S4 strain (F. tularensis subsp. tularensis) and the LVS (F. tularensis subsp. holarctica) could relate to the different virulence characteristics of these two subspecies. An alternate possibility is that the mutations could be specific to the LVS and contribute to its attenuation.
TABLE 1.
Primers used for PCR and RT-PCR
| Gene | Gene size (bp) | Primer | Sequence | Amplicon sizea (bp) |
|---|---|---|---|---|
| 16S rRNA | 1,523 | 16S-724F | 5′-TGG ACC GAT ACT GAC ACT GAG G | 115 |
| 16S-838R | 5′-AGA GCC TTT ACA CCG ACT CC | |||
| pilQ | 1,785 | PQ-1259F | 5′-GGC TTG AGA TGG AAG TCA GAG | 289 |
| PQ-1547R | 5′-AAA TCA CCT CCT TGT GGG G | |||
| pilF | 1,779 | PF-667F | 5′-AAA AAC TTT CGC ATC CGC | 364 |
| PF-1030R | 5′-CAG AAC CTG TTG GTC CTG TTA C | |||
| pilT | 1,029 | PT-186F | 5′-GCT GAT AGA AAC CTA CGA ATG TG | 497 |
| PT-682R | 5′-TCC CTA AAA CCA AAT GCC C | |||
| pilE1 | 408 | PE1-232F | 5′-GCA AAT GGT CTT CCA AGT GG | 123 |
| PE1-354R | 5′-ACA AGT CCA AGT AAT AGC ACC G | |||
| PE1-78F | 5′-TGT AGC GAT CCC GAT GTA CTC | 169 | ||
| PE1-247R | 5′-TTG GAA GAC CAT TTG CAG C | |||
| pilE2 | 441 | PE2-81F | 5′-AGC AAT ACC GAT TTA CTC AAG C | 214 |
| PE2-294R | 5′-GTG TTG TGG TGA AGT TTG GC | |||
| pilE3 | 597 | PE3-88F | 5′-ATC CCA GCG TAT TCA AAC TAT G | 226 |
| PE3-313R | 5′-TGC TTA CAC CAA CTA TTC TAC CG | |||
| pilE4 | 594 | PE4-91F | 5′-GCG GTG TTT GTA ACG TCT ACC | 248 |
| PE4-339R | 5′-AGG ATC TAC TGC TTC TGG ACG | |||
| pilE5 | 408 | PE5-119F | 5′-CTG AAG CAA CTT CTG AGC TG | 164 |
| PE5-283R | 5′-TAG GGC ACT GAT GTT CTC C | |||
| pilD | 819 | PD-38F | 5′-TTC TAT TTG GTG CTG CTA TTG G | 161 |
| PD-198R | 5′-TGG GCA TTT TGA TGG TGT C |
Expected size of the PCR product with the primer pair for each gene.
FIG. 3.
(A) Presence of T4P genes in the LVS. Shown are results of PCR analysis of the LVS with primers based on the Schu S4 pilus genes (Table 1). Lane 1, DNA ladder; lane 2, 16S rRNA (expected PCR product size, 115 bp); lane 3, pilQ (289 bp); lane 4, pilF (364 bp); lane 5, pilT (497 bp); lane 6, pilE2 (214 bp); lane 7, pilE3 (226 bp); lane 8, pilE4 (248 bp); lane 9, pilE5 (164 bp); lane 10, pilD (161 bp). Samples were run in a 1% agarose gel and stained with ethidium bromide. Gels were visualized with a Gel Doc 2000 gel documentation system (Bio-Rad Laboratories, Hercules, Calif.). (B) Expression of the LVS T4P genes. The LVS was grown in different conditions and analyzed for expression of the indicated genes by RT-PCR. Lanes 1 and 2, MHC at 72 h of growth; lanes 3 and 4, MH at 16 h of growth; lanes 5 and 6, MH at 40 h of growth; lanes 7 and 8, CDM at 16 h of growth; lanes 9 and 10, CDM at 40 h of growth. Even-numbered lanes represent control samples in which the RT step was omitted to check for DNA contamination. PCR was performed for 30 cycles, except for 13 cycles for the 16S rRNA and 35 cycles for pilT. Lane 11, whole-bacterium DNA of F. tularensis as a positive control for the PCR.
Reverse transcription-PCR (RT-PCR) was used to ascertain expression of the T4P genes in the LVS under the growth conditions used for the TEM analysis. RNA was extracted using TRI Reagent-LS (Molecular Research Center, Cincinnati, Ohio) from an MHC colony or from a pellet obtained by centrifugation of a 1-ml CDM or MH culture at 16 or 40 h of growth. Contaminating DNA was removed by treatment with RNase-free DNase I (Roche) at 37°C for 1 h. For the RT reaction, 15 μl of RNA was incubated with 3 μg of random primers (Invitrogen) and 50 U of avian myeloblastosis virus reverse transcriptase (Roche). The obtained cDNA was used at a 1:10 dilution for PCR. Each of the T4P genes tested was expressed (Fig. 3B), although expression of pilT was very low. Expression of the genes was highest in CDM at 16 h of growth, particularly for pilE3 to pilE5 (Fig. 3B). Growth in CDM at 16 h was the condition under which the surface fibers were most highly expressed as visualized by TEM (Fig. 1E and F).
In summary, we have shown that F. tularensis, a facultative intracellular pathogen and potential bioterrorism agent, expresses pilus-like surface fibers. Whole-bacterium TEM examination of the LVS revealed the presence of surface fibers with the characteristics of T4P: long, thin, polarly localized structures that may form bundles or interlinked networks (19, 22, 24, 25). In agreement with this, analysis of a draft sequence of the highly virulent Schu S4 strain (26) revealed the presence of orthologs to T4P genes found in Neisseria spp. and P. aeruginosa. PCR and sequence analysis confirmed that the T4P genes were present in the LVS. Furthermore, RT-PCR demonstrated that the LVS genes are expressed. We propose to name the F. tularensis genes pil and designate the genes according to the Neisseria nomenclature (pilE, major pilin; pilQ, secretin, etc.). Given that surface structures are critical to the virulence of pathogenic bacteria, we also propose that the F. tularensis pili represent virulence factors for this organism.
Addendum in Proof
A preliminary draft of the LVS genome was made available by the BBRP Sequencing Group at Lawrence Livermore National Laboratory (http://bbrp.llnl.gov/bbrp/html/microbe.html) after submission of this paper. The genome sequence matches our findings from analysis of the PCR products. That only a small fragment of the pilE1 gene is present in the LVS genome (the first 50 nucleotides) explains our inability to amplify this gene by PCR.
Acknowledgments
We thank K. L. Elkins for providing the LVS and U. Eriksson for providing the capsule-deficient mutant of the LVS. We thank M. B. Furie for helpful discussions and members of the Furie laboratory for assistance with growth and handling of the LVS, K. Studholme for assistance with electron microscopy, and M. Ivanov for translation of Russian manuscripts.
This work was supported by National Institutes of Health grant AI-48492. Preliminary sequence data were obtained from the F. tularensis strain Schu S4 Genome Sequencing Consortium, a joint effort supported by the U.S. Department of the Army, UK Ministry of Defence, Swedish Ministry of Defense, and the U.S. Defense Advanced Research Projects Agency.
Editor: V. J. DiRita
REFERENCES
- 1.Alm, R. A., and J. S. Mattick. 1995. Identification of a gene, pilV, required for type 4 fimbrial biogenesis in Pseudomonas aeruginosa, whose product possesses a pre-pilin leader sequence. Mol. Microbiol. 16:485-496. [DOI] [PubMed] [Google Scholar]
- 2.Baron, G. S., and F. E. Nano. 1998. MglA and MglB are required for the intramacrophage growth of Francisella novicida. Mol. Microbiol. 29:247-259. [DOI] [PubMed] [Google Scholar]
- 3.Chamberlain, R. E. 1965. Evaluation of live tularemia vaccine prepared in a chemically defined medium. Appl. Microbiol. 13:232-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherwonogrodzky, J. W., M. H. Knodel, and M. R. Spence. 1994. Increased encapsulation and virulence of Francisella tularensis live vaccine strain (LVS) by subculturing on synthetic medium. Vaccine 12:773-775. [DOI] [PubMed] [Google Scholar]
- 5.Collins, R. F., R. C. Ford, A. Kitmitto, R. O. Olsen, T. Tonjum, and J. P. Derrick. 2003. Three-dimensional structure of the Neisseria meningitidis secretin PilQ determined from negative-stain transmission electron microscopy. J. Bacteriol. 185:2611-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comolli, J. C., A. R. Hauser, L. Waite, C. B. Whitchurch, J. S. Mattick, and J. N. Engel. 1999. Pseudomonas aeruginosa gene products PilT and PilU are required for cytotoxicity in vitro and virulence in a mouse model of acute pneumonia. Infect. Immun. 67:3625-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dennis, D. T., T. V. Inglesby, D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. D. Fine, A. M. Friedlander, J. Hauer, M. Layton, S. R. Lillibridge, J. E. McDade, M. T. Osterholm, T. O'Toole, G. Parker, T. M. Perl, P. K. Russell, and K. Tonat. 2001. Tularemia as a biological weapon: medical and public health management. JAMA 285:2763-2773. [DOI] [PubMed] [Google Scholar]
- 8.Feldman, K. A., D. Stiles-Enos, K. Julian, B. T. Matyas, S. R. Telford III, M. C. Chu, L. R. Petersen, and E. B. Hayes. 2003. Tularemia on Martha's Vineyard: seroprevalence and occupational risk. Emerg. Infect. Dis. 9:350-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forestal, C. A., J. L. Benach, C. Carbonara, J. K. Italo, T. J. Lisinski, and M. B. Furie. 2003. Francisella tularensis selectively induces proinflammatory changes in endothelial cells. J. Immunol. 171:2563-2570. [DOI] [PubMed] [Google Scholar]
- 10.Friedrich, A., C. Prust, T. Hartsch, A. Henne, and B. Averhoff. 2002. Molecular analyses of the natural transformation machinery and identification of pilus structures in the extremely thermophilic bacterium Thermus thermophilus strain HB27. Appl. Environ. Microbiol. 68:745-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fussenegger, M., T. Rudel, R. Barten, R. Ryll, and T. F. Meyer. 1997. Transformation competence and type-4 pilus biogenesis in Neisseria gonorrhoeae—a review. Gene 192:125-134. [DOI] [PubMed] [Google Scholar]
- 12.Gerasimov, V. N., V. Y. Dolotov, A. V. Stepanov, and N. N. Urakov. 1997. The morphology, ultrastructure, and populational features of the bacteria Francisella. Vestn. Ross. Akad. Med. Nauk. 6:24-30. [PubMed] [Google Scholar]
- 13.Giron, J. A., A. S. Ho, and G. K. Schoolnik. 1991. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science 254:710-713. [DOI] [PubMed] [Google Scholar]
- 14.Giron, J. A., M. M. Levine, and J. B. Kaper. 1994. Longus: a long pilus ultrastructure produced by human enterotoxigenic Escherichia coli. Mol. Microbiol. 12:71-82. [DOI] [PubMed] [Google Scholar]
- 15.Golovliov, I., M. Ericsson, G. Sandstrom, A. Tarnvik, and A. Sjostedt. 1997. Identification of proteins of Francisella tularensis induced during growth in macrophages and cloning of the gene encoding a prominently induced 23-kilodalton protein. Infect. Immun. 65:2183-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray, C. G., S. C. Cowley, K. K. Cheung, and F. E. Nano. 2002. The identification of five genetic loci of Francisella novicida associated with intracellular growth. FEMS Microbiol. Lett. 215:53-56. [DOI] [PubMed] [Google Scholar]
- 17.Herrington, D. A., R. H. Hall, G. Losonsky, J. J. Mekalanos, R. K. Taylor, and M. M. Levine. 1988. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in human. J. Exp. Med. 168:1487-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hobbs, M., and J. S. Mattick. 1993. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complex. Mol. Microbiol. 10:233-243. [DOI] [PubMed] [Google Scholar]
- 19.Kennan, R. M., O. P. Dhungyel, R. J. Whittington, J. R. Egerton, and J. I. Rood. 2001. The type IV fimbrial subunit gene (fimA) of Dichelobacter nodosus is essential for virulence, protease secretion, and natural competence. J. Bacteriol. 183:4451-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lory, S., and M. S. Strom. 1997. Structure-function relationship of type-IV prepilin peptidase of Pseudomonas aeruginosa—a review. Gene 192:117-121. [DOI] [PubMed] [Google Scholar]
- 21.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 22.Mattick, J. S., C. B. Whitchurch, and R. A. Alm. 1996. The molecular genetics of type-4 fimbriae in Pseudomonas aeruginosa—a review. Gene 179:147-155. [DOI] [PubMed] [Google Scholar]
- 23.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98-102. [DOI] [PubMed] [Google Scholar]
- 24.Nunn, D. 1999. Bacterial type II protein export and pilus biogenesis: more than just homologies? Trends Cell Biol. 9:402-408. [DOI] [PubMed] [Google Scholar]
- 25.Park, H. S., M. Wolfgang, J. P. van Putten, D. Dorward, S. F. Hayes, and M. Koomey. 2001. Structural alterations in a type IV pilus subunit protein result in concurrent defects in multicellular behaviour and adherence to host tissue. Mol. Microbiol. 42:293-307. [DOI] [PubMed] [Google Scholar]
- 26.Prior, R. G., L. Klasson, P. Larsson, K. Williams, L. Lindler, A. Sjostedt, T. Svensson, I. Tamas, B. W. Wren, P. C. Oyston, S. G. Andersson, and R. W. Titball. 2001. Preliminary analysis and annotation of the partial genome sequence of Francisella tularensis strain Schu 4. J. Appl. Microbiol. 91:614-620. [DOI] [PubMed] [Google Scholar]
- 27.Rotz, L. D., A. L. Khan, S. R. Lillibridge, S. M. Ostroff, and J. M. Hughes. 2002. Public health assessment of potential biological terrorism agents. Emerg. Infect. Dis. 8:225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandstrom, G., S. Lofgren, and A. Tarnvik. 1988. A capsule-deficient mutant of Francisella tularensis LVS exhibits enhanced sensitivity to killing by serum but diminished sensitivity to killing by polymorphonuclear leukocytes. Infect. Immun. 56:1194-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandstrom, G., A. Sjostedt, T. Johansson, K. Kuoppa, and J. C. Williams. 1992. Immunogenicity and toxicity of lipopolysaccharide from Francisella tularensis LVS. FEMS Microbiol. Immunol. 5:201-210. [DOI] [PubMed] [Google Scholar]
- 30.Sjostedt, A. 2003. Family XVII. Francisellaceae, genus I. Francisella, p. 111-135. In D. J. Brenner (ed.), Bergey's manual of systematic bacteriology. Springer, New York, N.Y.
- 31.Skerker, J. M., and H. C. Berg. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. USA 98:6901-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strom, M. S., and S. Lory. 1993. Structure-function and biogenesis of the type IV pili. Annu. Rev. Microbiol. 47:565-596. [DOI] [PubMed] [Google Scholar]
- 33.Tonjum, T., and M. Koomey. 1997. The pilus colonization factor of pathogenic neisserial species: organelle biogenesis and structure function relationships—a review. Gene 192:155-163. [DOI] [PubMed] [Google Scholar]