Summary
Recognition of the mRNA 5′ m7GTP cap is key to translation initiation for most eukaryotic mRNAs. The cap is bound by the eIF4F complex, consisting of a cap binding protein (eIF4E), a “scaffold” protein (eIF4G), and an RNA helicase (eIF4A). As a central early step in initiation, regulation of eIF4F is crucial for cellular viability. Although the structure and function of eIF4E have been defined, a dynamic mechanistic picture of its activity at the molecular level in the eIF4F•mRNA complex is still unavailable. Here, using single-molecule fluorescence, we measured the effects of Saccharomyces cerevisiae eIF4F factors, mRNA secondary structure, and the poly(A)-binding protein Pab1p on eIF4E-mRNA binding dynamics. Our data provide an integrated picture of how eIF4G and mRNA structure modulate eIF4E-mRNA interaction, and uncover an eIF4G- and poly(A)-independent activity of poly(A)-binding protein that prolongs the eIFE•mRNA complex lifetime.
Introduction
Four decades of study have identified eukaryotic translation initiation factors and an outline of when and how they act to coordinate initiation. However, the dynamic contributions of factor and mRNA composition and conformation to the initiation process, and how these dynamics are perturbed by regulatory mechanisms, are still poorly understood (Aitken and Lorsch, 2012). Eukaryotic initiation is an intricate sequence of events involving an mRNA, initiator tRNA, ribosomal subunits, and at least eleven protein factors that guide assembly of the 80S ribosome positioned correctly at the start codon to establish reading frame.
The earliest phase of canonical initiation involves binding of a complex of protein factors to the mRNA 5′ 7-methylguanosine m7G(5′)ppp(5′)N cap. This complex, called eIF4F, consists of the cap binding protein (eIF4E), a multi-domain, multifunctional “scaffold” protein (eIF4G), and an ATP-dependent RNA helicase (eIF4A) that is thought to unwind secondary structures close to the cap (von der Haar et al, 2004, Rajagopal et al., 2012). Interactions between eIF4G and downstream components of the initiation machinery result in recruitment of the eIF4F-bound mRNA to the small ribosomal subunit (Walker et al., 2012). The Saccharomyces cerevisiae genome encodes two paralogs of eIF4G, eIF4G1 and eIF4G2, that exhibit extensive functional overlap (Clarkson et al., 2010).
mRNA binding by eIF4F is a highly dynamic process; the eIF4E cap-binding, eIF4G ssRNA-binding, and the eIF4A helicase activities must engage the RNA with the appropriate timing and conformation to prime the RNA efficiently for recognition by the factor-associated 40S subunit. However, detailed investigation of the dynamics – the time-evolution of composition and conformation in the eIF4F-mRNA interaction – and the modulation of these dynamics by initiation factors and RNA structure is difficult using traditional bulk biochemical or static structural methods. This is a key challenge to understanding translation initiation and translational control in eukaryotes.
Initiation is heavily regulated, since it represents the last point at which translational control mechanisms can act to prevent aberrant protein synthesis (Jackson et al., 2010; Kong and Lasko, 2012). The mTOR pathway derepresses eIF4E activity by phosphorylation of inhibitory 4E-binding proteins (Gingras et al., 1999), coupling important components of cellular homeostasis with modulation of translation efficiency through direct effects on eIF4E dynamics in the eIF4F complex. In humans, this regulatory mechanism malfunctions in disease states such as cancer (Zoncu et al., 2011), autism (Gkogkas et al., 2013), and viral infection (Kobayashi et al., 2012). Yeast remains an attractive model organism for elucidating general mechanistic principles that can be applied to the study of translational regulation in humans, as it allows hypotheses derived from in vitro experiments to be tested in vivo with genetic methods (Altmann and Trachsel, 1994).
eIF4E has been extensively studied by genetic, biochemical, biophysical, and structural techniques. The structural basis for cap binding and interaction with eIF4G through the eIF4G 4E-binding domain (eIF4G-4EBD) is known (Gross et al., 2003, Yanagiya et al., 2009); NMR data have also highlighted the importance of dynamics in this interaction (Volpon et al., 2006). Yeast 4EBD allosterically increases the affinity of eIF4E for the cap structure (von der Haar et al., 2006, von der Haar et al., 2000) through a coupled conformational change in the two proteins and wrapping of the 4EBD polypeptide around the eIF4E N-terminus in what has been termed a “molecular bracelet” (Gross et al., 2003). Yet no information on structure or dynamics is available for complexes containing either full-length eIF4G, or a capped oligoribonucleotide or mRNA. Pre-steady-state rapid-mixing methods have explored the thermodynamics and kinetics of eIF4E-cap binding and their modulation by 4EBD, particularly for mammalian eIF4E (Slepenkov et al., 2008, Slepenkov et al., 2006); detailed kinetic information on binding of eIF4E to capped oligoribonucleotides or full-length RNAs is limited for human eIF4E and non-existent for yeast eIF4E. Moreover, no data are available for formation of an eIF4E•eIF4G•mRNA complex with full-length eIF4G and mRNA, or for the eIF4F complex including eIF4A. The absence of structural and kinetic data hinders interpretation of in vivo genetic and in vitro biochemical experiments to investigate the mechanism of initiation.
In addition to protein factors, RNA secondary structures in the 5′ untranslated region modulate the efficiency of translation of mRNAs (Kozak, 1992). In general, hairpin structures in the 5′-UTR inhibit translation. However, there are differences between yeast and higher eukaryotes in terms of the dependence of inhibition on the position, thermal stability, and GC content of inhibitory structures. While inhibition of translation by 5′-UTR hairpins in yeast mRNAs shows little or no dependence on the distance from the cap, higher eukaryotes exhibit a greater sensitivity to structures near the mRNA 5′ end (McCarthy, 1998). Hairpin structures located ~12 nucleotides away from the cap increased translation efficiency in vivo in mammalian cells, while the same hairpin positioned immediately adjacent to the cap had an inhibitory effect (Babendure et al., 2006). Moreover, in vitro equilibrium binding studies with mammalian eIF4E and such hairpin-containing RNAs showed that secondary structures near the cap can increase RNA affinity for eIF4E, even when steric considerations suggest that they should do the opposite (Carberry et al., 1992). Integrated structural and kinetic data are needed to understand the molecular mechanism by which these interactions influence translation, and to understand the mechanistic basis for the observed differences between yeast and higher eukaryotes.
The polyadenylated 3′ end of eukaryotic mRNAs may also play a role in modulating translation efficiency, and the eukaryotic poly(A)-binding protein (Pab1p in S. cerevisiae) has been shown to act as a true initiation factor (Otero et al., 1999, Kahvejian et al., 2005). The activity of Pab1p in initiation is described in terms of its ability to bind both eIF4G and the poly(A) tail. Poly(A)-bound Pab1p is thought to allosterically increase the affinity of eIF4E for the cap through its interaction with eIF4G (Safaee et al., 2012). Simultaneous binding of Pab1p to the 3′ poly(A) tail and eIF4G1 has been shown statically but directly by atomic force microscopy (Wells et al., 1998), and indirectly by genetic and biochemical experiments, to produce a circularized, “closed loop” mRNA (Tarun et al., 1997). Commonly invoked hypotheses implicate Pab1p-induced conformational changes in eIF4G or an increased effective concentration of eIF4F near the cap caused by Pab1p-poly(A) binding as the basis for Pab1p-mediated stimulation of translation. However, these conformational changes or the altered kinetics of binding of eIF4E to capped RNAs in the presence of Pab1p have not been directly confirmed and observed. On the other hand, Pab1p has been shown to stimulate translation in vitro in a manner that is independent of the presence of the poly(A) tail or binding to eIF4G1 (Otero et al., 1999, Borman et al., 2002). No experiments have directly observed the dynamics of eIF4F-mRNA-Pab1p interaction to understand their impact on translation.
Single-molecule fluorescence studies have provided substantial insights into the mechanism of translation, predominantly in prokaryotes (Petrov et al., 2012) A recent study also examined the activity of mammalian eIF4AI at the single-molecule level (Sun et al., 2012). Biomolecular conformational and compositional trajectories are followed free from obfuscation by bulk averaging, such that complex, multi-step processes may be observed in real time (Tsai et al., 2012, Uemura et al., 2010). Conformational trajectories may be deduced from the time- and composition-dependence of FRET (Förster Resonance Energy Transfer), combined with structural information that allows precise knowledge of the relative locations of fluorophores (Aitken and Puglisi, 2010). FRET signals between biomolecules labeled with Cy3 and Cy5 report on complexes in which the dyes are presented to each other at distances between 20 and 80 Å, a length scale appropriate for the study of eIF4F•mRNA complexes.
We report here single-molecule fluorescence analyses of the dynamics of Saccharomyces cerevisiae eIF4E-mRNA interaction, studied using total internal reflection fluorescence (TIRF) microscopy. By observing FRET between eIF4E and mRNA, we determined kinetics and structural dynamics of eIF4E-mRNA interaction. We observed the effects of eIF4G1 and its 4E-binding domain, RNA secondary structure, and Pab1p on eIF4E-cap interaction. To examine the 5′-end-specific dynamics, we made measurements with capped RNA oligonucleotides lacking a poly(A) tail. We measured the kinetics of binding of yeast eIF4E to capped RNAs in the presence of the eIF4F factors and Pab1p at equilibrium at the single-molecule level. Our results show how factor composition and mRNA secondary structure near the 5′ cap influence eIF4E-mRNA interaction, and how the poly(A) binding protein Pab1p can act to stimulate cap-dependent translation initiation, even in the absence of a poly(A) tail.
Results
Cy5-eIF4E functions in cap and eIF4G binding
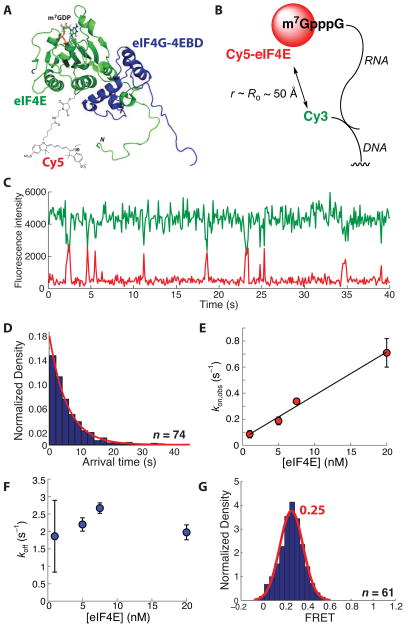
To label eIF4E specifically with a fluorescent dye, we exploited the absence of cysteine in wild-type S. cerevisiae eIF4E to generate a eIF4E(A124C) variant for thiol-maleimide labeling. We reasoned that introduction of a label at this position should not impair eIF4E activity, based on the solution structure of eIF4E bound to m7GDP and the eIF4E-binding domain of eIF4G1 (Gross et al., 2003); when conjugated to C124 the fluorophore would be located distal to the cap-binding site, away from the site of interaction with eIF4G (Figure 1A). While we did not test the function of eIF4E(A124C) in mutant yeast strains, Ala124 is not conserved and is located in a loop region based on the structure; there is thus no evidence to suggest that the A124C variant will impair translation. After overexpression in E. coli and purification, N-terminally polyhistidine-tagged eIF4E(A124C) was labeled with Cy5 maleimide. Under the labeling conditions (typically ~5 μM protein), eIF4E multimerizes (readily observed in size exclusion chromatograms and by gel shift assays with capped mRNAs, Figure S1A), resulting consistently in a labeling efficiency of between 50 and 53%. Monomeric Cy5-eIF4E was readily isolated by size exclusion chromatography. Observed association rates measured in kinetics experiments were adjusted to account for the labeling efficiency, to compute true rate constants. We also expressed and purified recombinant yeast eIF4G1 eIF4E-binding domain (4EBD) flanked by additional eIF4G1 sequence, (amino acids 348–513) with an N-terminal polyhistidine tag (von der Haar et al., 2000). A fluorescence-based native gel shift assay to determine the apparent dissociation constant for Cy5-eIF4E in the Cy5-eIF4E(A124C)•eIF4G-4EBD•cap-RNA complex, containing the model unstructured poly(CU) RNA sequence 5′-m7GpppGG(CU)10GCG(CGA)3-3′, indicated an apparent KD of < 100 nM for Cy5-eIF4E, which is indistinguishable from the unlabeled protein (Figure S1B–D). This value is considerably lower than the ~1 μM measured previously by SPR for binding of the yeast protein to a capped RNA (von der Haar et al., 2000), and closer to the value of 110 ± 10 nM measured by a stopped-flow fluorescence technique for the human protein binding to a capped RNA oligonucleotide at 300 mM KCl, which reduces the affinity (Slepenkov et al., 2006). This stopped-flow approach has not been applied to yeast eIF4E. RNA-protein complex formation was abrogated by lack of an RNA cap, or by addition of 1 mM m7GpppG cap structure analogue, confirming that complex formation requires the presence of the RNA cap. Cy5-eIF4E is therefore active in both cap- and eIF4G-4EBD binding.
Figure 1.
A single-molecule FRET assay for eIF4E activity. (A) Schematic of the Cy5-eIF4E•eIF4G-4EBD complex based on the solution structure in the presence of m7GDP (Gross et al., 2003; PDB 1RF8). The schematic indicates the location of the Cy5 label attached to residue 124. (B) Experimental design for single-molecule FRET, showing immobilization of the capped RNA by annealing to a 5′-Cy3-labeled, 3′-biotinylated DNA oligonucleotide. (C) Representative single-molecule FRET trace showing FRET events due to binding of Cy5-eIF4E to the immobilized, Cy3-labeled RNA. (D) Representative arrival time distribution for binding of Cy5-eIF4E to the Cy3-RNA, with single-exponential fit. (E) Dependence of the rate of formation of the Cy5-eIF4E•cap-RNA complex on the eIF4E concentration, with linear fit. (F) Dependence of dissociation rate of the Cy5-eIF4E•cap-RNA complex on the Cy5-eIF4E concentration. (G) FRET distribution for the Cy5-eIF4E•cap-RNA complex, fit to a single Gaussian function. The number of single-molecule traces (n) used to construct distributions is given where appropriate. Error bars in kinetic plots represent standard errors for three separate measurements. See also Figure S1.
Cy5-eIF4E cap binding assayed by single-molecule FRET
We next measured Cy5-eIF4E activity in a single-molecule FRET experiment. To allow immobilization and single-molecule fluorescence detection of the model mRNA, a DNA oligonucleotide bearing a 5′-biotin and a 3′-Cy3 was annealed to the RNA 3′ end. This dye-labeled, capped RNA was immobilized on a biotin-PEG quartz surface through neutravidin (Figure 1B).
The system was designed to allow RNA-eIF4E FRET upon delivery of Cy5-eIF4E and excitation of Cy3 fluorescence with a 532 nm laser. Analysis of hundreds of fluorescence traces from this experiment revealed pulses of red (Cy5) fluorescence, concomitant with anticorrelated drops in the Cy3 fluorescence intensity, characteristic of FRET (Figure 1C). In contrast, no FRET events were observed with similar numbers of molecules in control experiments where the RNA was uncapped, or with capped RNA in the presence of m7GpppG, confirming that the FRET signal reports specifically on the interaction of Cy5-eIF4E with RNA cap. The arrival time distributions for formation of the Cy5-eIF4E•mRNA complex – that is, the distribution of times between the end of one FRET event and the beginning of the next, obtained from observation of many molecules and events – were fit well by single exponential functions at concentrations of Cy5-eIF4E between 1 and 20 nM (Figure 1D). The rate of appearance of FRET, determined from the fits as the reciprocal of the exponential mean of the arrival time distribution, was linearly dependent on the Cy5-eIF4E concentration (kon = 33 μM−1 s−1 from the slope of the line; Figure 1E), while the FRET lifetime of the bound state was independent of the Cy5-eIF4E concentration (Figure 1F). Taken together, these data are consistent with a simple on-off equilibrium-binding model for assembly of the eIF4E•cap-RNA complex. In this model, the ratio koff/kon defines the equilibrium dissociation constant (KD) for the complex (~90 nM). The association rate is slower than the diffusion-controlled limit of ~109 M−1 s−1. The smFRET assay not only gives us kinetics, but also structural information from the FRET values. The FRET distribution (Figure 1G) of the bound state could be fit by a single Gaussian function centered at 0.25-FRET, suggesting that the eIF4E•cap-RNA complex adopts a uniform conformation with a distance of 65 – 85 Å between RNA label and the protein Cy5 label, assuming a Förster distance of ~53 Å. In this model, we subsume intermolecular orientation under conformation.
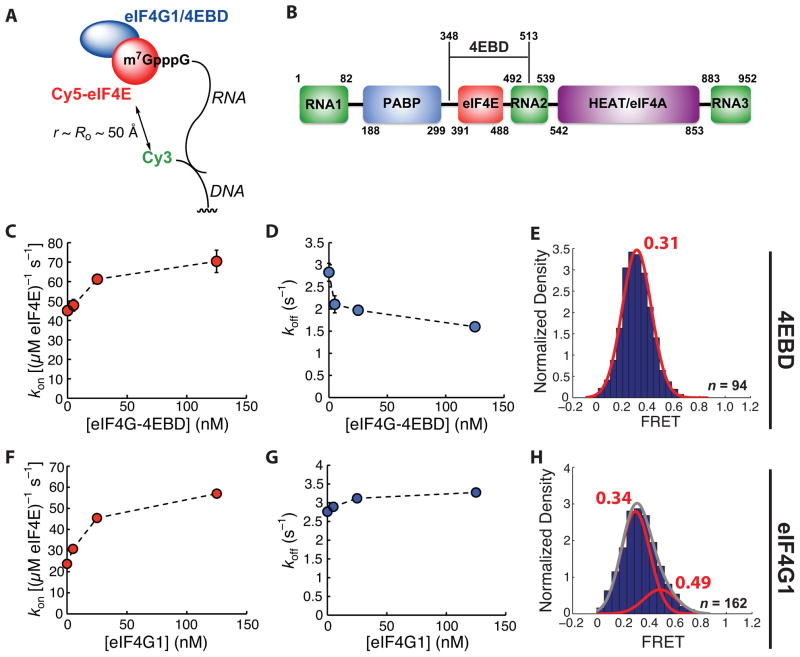
Modulation of Cy5-eIF4E activity by eIF4G-4EBD and full-length eIF4G1
We next used the smFRET assay to compare the dynamics of Cy5-eIF4E interaction with a capped oligoribonucleotide in the presence of eIF4G-4EBD (Figure 2A) or full-length eIF4G1, which has additional RNA- and protein-binding domains (Figure 2B). In the presence of increasing concentrations of eIF4G-4EBD, there was a saturable increase in the rate of association of Cy5-eIF4E with immobilized, capped RNA (Figure 2C). The dependence of the observed association rate on eIF4G-4EBD concentration was fit by a hyperbolic function, which we interpret as reporting on the equilibrium abundance of the free Cy5-eIF4E•eIF4G-4EBD complex in solution. We estimated a value of 20 ± 13 nM for the KD of the Cy5-eIF4E•eIF4G-4EBD protein-protein interaction based on the half-height of the hyperbola, at least 1.3 times larger than the value of <15 nM measured for the equilibrium binding of full-length eIF4G1 to eIF4E by equilibrium fluorescence titration (Mitchell et al., 2010). Extrapolation of the observed association rate to saturating eIF4G-4EBD concentrations provided an estimate for the rate of binding of the Cy5-eIF4E•eIF4G-4EBD complex of ~75 μM−1 s−1, approximately double the rate observed for formation of the binary eIF4E•mRNA complex (33 μM−1 s−1).
Figure 2.
Modulation of eIF4E•cap-poly(CU) binding kinetics by the eIF4G 4E-binding domain and by eIF4G1. (A) Schematic of experimental design. (B) Domain architecture of S. cerevisiae eIF4G1, showing relative positions of RNA-, eIF4E-, Pab1p-, and eIF4A-binding domains, and the region of the protein constituting eIF4E-4EBD. (C) Dependence of Cy5-eIF4E-RNA association rate on eIF4G-4EBD concentration, measured at 7.5 nM Cy5-eIF4E. (D) Dependence of observed rate of dissociation of the Cy5-eIF4E•RNA complex on the concentration of eIF4G-4EBD. (E) Representative FRET distribution for the Cy5-eIF4E•RNA complex (in the presence of 125 nM eIF4G-4EBD), fit to a single Gaussian function. (F) Dependence of Cy5-eIF4E-RNA association rate on full-length eIF4G1 concentration, measured at 7.5 nM Cy5-eIF4E. (G) Dependence of observed rate of dissociation of the Cy5-eIF4E•RNA complex on the concentration of eIF4G1. (H) Representative FRET distribution for the Cy5-eIF4E•RNA complex (in the presence of 125 nM eIF4G1), fit to the sum (grey line) of two Gaussian functions (red lines). The number of single-molecule traces (n) used to construct distributions is given where appropriate. Error bars in kinetic plots represent standard errors for single-exponential fits. See also Figure S2.
The rate of dissociation of Cy5-eIF4E from the capped RNA decreased with increasing eIF4G-4EBD concentration and was approximately halved at saturating concentrations (Figure 2D). The ratio koff/kon, defining the apparent dissociation constant (KD) for the protein-RNA complex, was thus decreased from ~90 nM for the Cy5-eIF4E•RNA binary complex to ~15 nM for the Cy5-eIF4E•eIF4G-4EBD•RNA ternary complex. This decrease in KD is consistent with the results of the gel shift assay and previous measurements made on the eIF4E•eIF4G-4EBD•cap ternary complex (von der Haar et al., 2000).
The FRET distribution for the Cy5-eIF4E•eIF4G-4EBD•RNA ternary complex was also best fit by a single Gaussian function (Figure 2E). The mean FRET was increased slightly (0.31) relative to the eIF4E•RNA binary complex (0.25). Binding of eIF4G1-4EBD therefore at most induces subtle conformational changes impacting the distance between the RNA-bound FRET donor and the FRET acceptor on the protein. This is consistent with existing data for the eIF4E•eIF4G-4EBD complex bound to cap structure analogues, which show that 4EBD binding stabilizes the structure of the cap-binding site and adds additional structure to the eIF4E N-terminus, rather than altering the protein conformation in the region of our label (Gross et al., 2003).
We next examined dynamics in the presence of eIF4G1. In contrast to the situation for eIF4G-4EBD, detailed kinetic measurements of the binding of eIF4E to capped RNAs in the presence of full-length eIF4G1 from any organism have not been reported. eIF4G1 represents a significant challenge for in vitro assays, as it undergoes extensive, spontaneous proteolysis during expression and upon storage after purification (Berset et al., 2003). Solutions of eIF4G1 thus contain appreciable quantities of degradation products that potentially may (in the best case) convolute kinetic binding data, or (in the worst case) competitively inhibit the activity of the full-length protein. To circumvent these issues, we cloned, expressed, and purified recombinant N-terminally His6-tagged S. cerevisiae eIF4G1 and immediately used the most homogeneous fractions of full-length protein in single-molecule experiments (Supplementary Experimental procedures, and Figure S2A). The full-length protein was judged to be >90% pure on the basis of Coomassie-stained SDS-PAGE. We also expressed and purified N-terminally His6-tagged S. cerevisiae eIF4A (Figure S2B), in order to reconstitute the full eIF4F complex.
Binding of eIF4E to capped RNAs in the presence of eIF4G1 showed similar kinetic characteristics to those observed with eIF4G1-4EBD. Increasing concentrations of eIF4G1 saturably increased the association rate (Figure 2F), however the magnitude of the increase at saturation was ~3.2-fold, larger than the doubling observed with eIF4G1-4EBD. Fits of the rate-concentration dependence to a hyperbolic function indicated an apparent dissociation constant for the Cy5-eIF4E•eIF4G1 complex of 20 ± 10 nM, similar to the value of ~25 nM we measured for the isolated eIF4G-4EBD and the value of <15 nM measured by equilibrium fluorescence titration. In contrast with eIF4G-4EBD, there was essentially no measurable dependence of the dissociation rate of the Cy5-eIF4E•eIF4G1•RNA complex on the concentration of eIF4G1 (Figure 2G). The net effect on koff/kon = KD,app observed with full-length eIF4G is still an increase in apparent eIF4E affinity for the RNA, but the kinetics underlying this effect differ between the isolated 4EBD and full-length protein.
The FRET distribution for the Cy5-eIF4E•RNA complex in the presence of saturating concentrations of eIF4G1 differed from that seen with eIF4E alone or with the eIF4E•eIF4G-4EBD complex; an additional FRET state with a mean value of 0.49 was observed (Figure 2H). This provides further evidence to suggest that domains of eIF4G1 other than the eIF4G-4EBD engage the RNA in a manner that leads to conformational rearrangement of the RNA and altered binding kinetics. We did not observe transitions between the two FRET states in fluorescence traces.
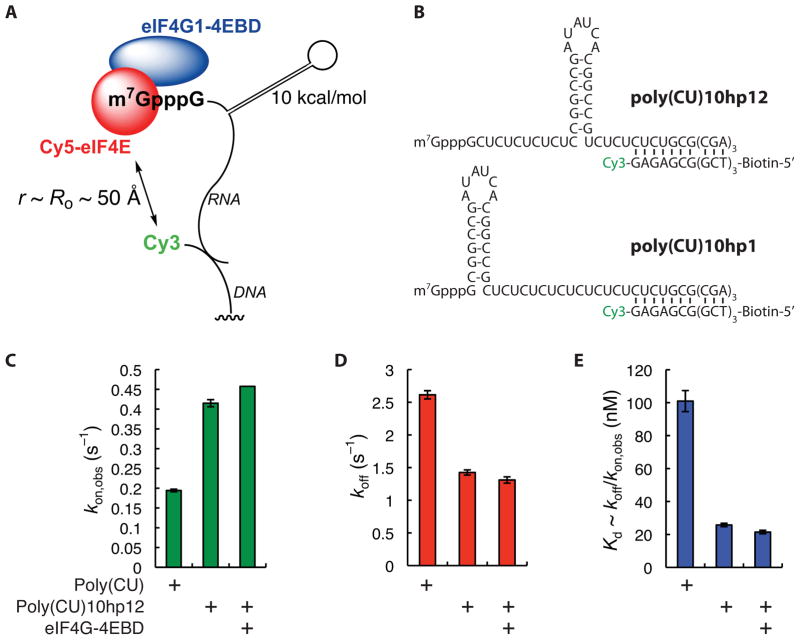
mRNA secondary structure modulates Cy5-eIF4E cap binding dynamics
To investigate how cap-proximal RNA secondary structure modulates the eIF4E-cap interaction, we used our FRET assay to observe the dynamics of binding of eIF4E to our unstructured model RNA modified by inclusion of a 10 kcal/mol hairpin shown previously to modulate translation efficiency in vivo in mammalian cells (Figure 3A). The hairpin was inserted at the +12 position (stimulatory, poly(CU)10hp12) or +1 position (inhibitory, poly(CU)10hp1) (Figure 3B).
Figure 3.
Effects of RNA secondary structure on kinetics of binding of Cy5-eIF4E to immobilized RNA. (A) Schematic of experimental design. (B) Sequences of structured RNAs and hybrid duplex with labeled DNA oligonucleotides, indicating positions of hairpins, Cy3 label, and biotin. (C) Rates of association of Cy5-eIF4E (7.5 nM) with immobilized poly(CU) and poly(CU)10hp12 RNAs, and with poly(CU)10hp12 RNA in the presence of eIF4G-4EBD (150 nM). (D) Rates of dissociation of the Cy5-eIF4E•poly(CU), Cy5-eIF4E•poly(CU)10hp12, and Cy5-eIF4E•4EBD•poly(CU)10hp12 complexes. (E) Apparent dissociation constants for the Cy5-eIF4E•poly(CU), Cy5-eIF4E•poly(CU)10hp12, and Cy5-eIF4E•4EBD•poly(CU)10hp12 complexes, computed from the quotient koff/kon. Error bars in kinetic plots represent standard errors for single-exponential fits. See also Figure S3.
Where a hairpin was present at the +12 position, both the association rate and lifetime of the eIF4E•RNA complex increased by approximately two-fold relative to the unstructured RNA, resulting in a four-fold increase in apparent affinity (Figures 3C–3E). A further small increase in both parameters was observed in the presence of eIF4G-4EBD. The FRET distribution for the Cy5-eIF4E•poly(CU)10hp12 complex was best fit by a single Gaussian function centered at ~0.35-FRET, slightly larger than the value for the unstructured RNA (0.25) (Figure S3A). In the presence of full-length eIF4G1, a second, higher-FRET state was observed, similar to the situation with the unstructured mRNA (Figure S3B).
In contrast, no eIF4E-RNA FRET was observed in the case of hundreds of RNA molecules where the hairpin was inserted at the +1 position. Gel shift analysis confirmed that eIF4E bound poly(CU)10hp1 in bulk (Figure S3C). Moreover, TIRF experiments with direct Cy5 excitation also indicated that Cy5-eIF4E bound to the immobilized poly(CU)10hp1 RNA. Even though events due to non-specific Cy5-eIF4E interaction with the surface are convoluted into the measurement of association and dissociation rates in a direct Cy5 excitation experiment, both the frequency and duration of directly-observed Cy5 fluorescence bursts increased when poly(CU)10hp1 RNA was present, compared with when no RNA was immobilized (Figure S3D, E). The lack of FRET seems therefore to result from a significantly different trajectory of the oligoribonucleotide as it exits the cap binding site, such that the Cy3 donor on the RNA is no longer within FRET distance of the Cy5 acceptor on the eIF4E protein.
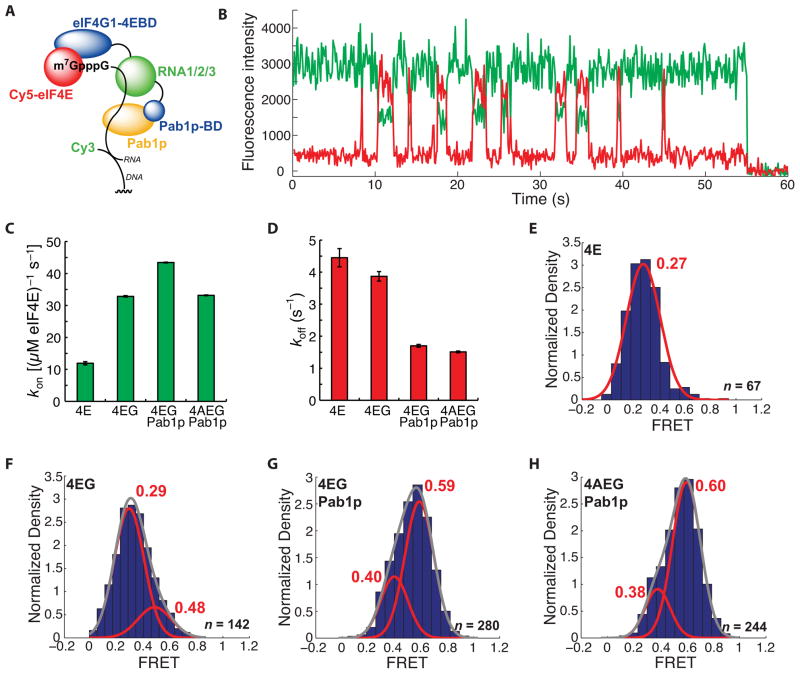
Pab1p alters RNA conformation and eIF4E binding dynamics in the absence of a poly(A) tail
We next used our single-molecule FRET assay to examine the eIF4E-cap interaction in the presence of full-length eIF4G1 and Pab1p (Figure 4A). We chose to study RNAs that lacked a poly(A) tail in order to measure directly the effects of Pab1p on the eIF4F complex related only to the 5′ mRNA end. Addition of 2 μM Pab1p caused a striking change in the FRET behavior of the Cy5-eIF4E•eIF4G1•RNA complex with our model unstructured RNA (Figure 4B). This concentration was chosen as it approximates the best available estimate for the concentration of Pab1p not associated with poly(A) mRNA tails in vivo (Görlach et al., 1994). While the rate of binding of Cy5-eIF4E•eIF4G1 to the mRNA showed a small but reproducible increase in the presence of Pab1p (Figure 4C), the dissociation rate of Cy5-eIF4E from its cap-bound complex was more than halved (Figure 4D). Compared with the FRET distributions for complexes formed in the presence of Cy5-eIF4E (Figure 4E) or Cy5-eIF4E•eIF4G1 (Figure 4F), a higher-, ~0.6-FRET state was formed for the Pab1p-bound Cy5-eIF4E•eIF4G1•RNA complex (Figure 4G). In all cases where eIF4G1 was present, FRET distributions were fit best by double Gaussian functions. No transitions between the higher- and lower-FRET states were observed, indicating that the altered conformation induced by Pab1p is established prior to, or within ~100 ms of, Cy5-eIF4E binding and persists until dissociation of eIF4E from the cap. Addition of eIF4A (400 nM) resulted in a FRET distribution similar to that observed for the Cy5-eIF4E•eIF4G1•Pab1p•RNA complex (Figure 4H).
Figure 4.
A long-lived, high-FRET state for the Cy5-eIF4E•RNA complex observed in the presence of Pab1p. (A) Schematic of experimental design. The domain architecture of eIF4G1 is abridged from the full disposition of domains (Figure 2B) by combining the three separate RNA binding domains and omitting the eIF4A-binding domain. Pab1p-bd refers to the eIF4G1 Pab1p-binding domain. (B) Representative single-molecule FRET trace showing binding of Cy5-eIF4E to immobilized, cap-poly(CU) RNA in the presence of 2 μM Pab1p. (C) Dependence of Cy5-eIF4E-mRNA association rate on factor composition. (D) Dependence of dissociation rate of the Cy5-eIF4E•mRNA complex in the presence of eIF4G1 (250 nM), Pab1p (2 μM), and eIF4A (400 nM). (E) – (H) FRET intensity distributions for the Cy5-eIF4E complexes in (D). The number of single-molecule traces (n) used to construct distributions is given where appropriate. Error bars in kinetic plots represent standard errors for single-exponential fits. See also Figure S4.
The Pab1p-induced shift to higher FRET was observed for the unstructured poly(CU) RNA, and the hairpin-containing poly(CU)10hp12 RNA, suggesting that it may be a general phenomenon (Figures S4A and B). We also observed a similar effect of Pab1p with RPL41A mRNA – chosen as it is a true cellular RNA used previously in experiments on translation initiation – though this highly structured RNA exhibited a more complex FRET distribution for the Pab1p-bound state than the model RNAs (Figures S4C and D).
Importantly, the effects of Pab1p on FRET value and lifetime were also observed for the eIF4E•RNA or eIF4G-4EBD•RNA complexes, suggesting that their origin is not due to interaction with eIF4G1 and its Pab1p-binding domain (Figures S4E and F).
Discussion
A single-molecule FRET assay for eIF4E activity
Despite great strides in understanding eukaryotic translation initiation, a dynamic mechanism integrating detailed kinetics with structural data remains elusive. The utility of single-molecule methods to address this deficit has been demonstrated for prokaryotic translation. eIF4E represents an excellent starting point to apply single-molecule methods to eukaryotic translation. Its cap-binding activity is the first step in canonical initiation, this activity is readily measurable in bulk assays, and a significant body of structural, biochemical, and biophysical data exists, allowing for validation of single-molecule reagents.
Our single-molecule FRET assay allowed us to measure the kinetics of binding of Cy5-labeled yeast eIF4E to single capped RNA oligonucleotides at equilibrium in real time. Complex formation involves relatively fast cycling of the apo- and cap-bound states of eIF4E, as seen previously in bulk kinetic studies with cap structure analogues and for the human protein with short oligoribonucleotides. Our FRET data for yeast eIF4E agree with results from rapid-mixing kinetic studies on human eIF4E that support a simple on-off equilibrium model for the eIF4E•cap-RNA complex. Cy5-eIF4E binds capped RNAs at a rate that depends on additional initiation factors, and on RNA secondary structure near the cap, in the 20 – 100 μM−1 s−1 range, close to the diffusion limit. This is much faster than rates of binding to an immobilized, capped oligoribonucleotide measured by surface plasmon resonance experiments (0.5 – 1 μM−1 s−1; von der Haar et al., 2000), but slower than the rates reported for binding of human eIF4E to a capped oligoribonucleotide in stopped-flow pre-steady-state experiments (>300 μM−1 s−1). The increased association rates observed on addition of eIF4G1 and other factors may thus reflect the effects of pre-forming the eIF4F complex, or of altered electrostatics in the binding of the larger complexes to the RNA. Sequence differences between yeast and human eIF4E may also alter the rate of the folding transition associated with cap binding (Volpon et al., 2006).
Our experiments add an additional structural dimension to existing kinetic models of capped-RNA recognition by eIF4E. FRET is a function of the inverse sixth power of the distance between donor and acceptor, and is most sensitive to changes in this distance around the Förster radius for a given donor-acceptor pair (~53 Å for Cy3 and Cy5). FRET values for eIF4E-RNA complexes probe the range of conformers within the RNA-bound states. For the binary Cy5-eIF4E•cap-RNA(Cy3) complex with a model unstructured RNA, the FRET distribution was consistent with a relatively narrow range of inter-dye distances, encompassing 65–85 Å between the fixed label on the protein and the Cy3 label on the RNA, which may fluctuate with the motion of the RNA polymer chain on a timescale faster than the resolution of our single-molecule experiments (<100 ms). Thus, while the eIF4E•mRNA binary complex is in dynamic equilibrium with the free components, the bound state is conformationally homogeneous with respect to the RNA, as reported by smFRET.
Dynamics of the eIF4E•eIF4G1•mRNA complex
Our assay also allowed us to measure the kinetics underlying the increased affinity of eIF4E for capped RNA due to eIF4G-4EBD and full-length eIF4G1. Our data suggest that the two proteins stabilize the eIF4E•RNA complex by different mechanisms; 4EBD increases the rate of formation and lifetime of the eIF4E•RNA complex, while eIF4G1 increases the association rate but not the lifetime. Engagement of the additional domains of eIF4G1 with the RNA likely accelerates association of the protein-RNA complex, but must also alter the dissociation kinetics. Consistent with this hypothesis, FRET distributions show a difference between 4EBD and eIF4G1: formation of a new conformational state of the eIF4E•RNA complex with the full-length protein, represented by a more condensed RNA-protein complex showing higher FRET. For our model unstructured RNA, the FRET states are closely spaced at 0.3- and 0.5-FRET. The low-FRET state is similar to that of the eIF4E•4EBD•RNA complex. The lack of observable interconversion of the two states suggests a model where the RNA-binding domains of eIF4G1 must engage the RNA prior to, or within 100 ms of, eIF4E-cap association to produce the condensed RNA conformation. If this does not occur, eIF4E may still bind to the RNA cap and the lifetime of the protein-RNA complex is prolonged through the eIF4E-eIF4G interaction, but the conformational rearrangement of the RNA is not observed. This conclusion is not without precedent – an altered RNA conformation for eIF4F-bound mRNA was proposed previously to account for stimulation by eIF4F of binding of capped RNAs to the 43S pre-initiation complex, resulting in enhanced initiation complex formation (Mitchell et al., 2010). Our results lend further weight to this hypothesis. Additionally, other components of the intracellular environment may modulate the dynamics of eIF4F-RNA recognition in ways that impact on measured translation efficiency. This might include proteins such as Ded1p, which can remodel the mRNA structure or otherwise potentially alter the eIF4G1-mRNA interaction dynamics (Firczuk et al., 2013).
RNA structure modulates eIF4E-RNA binding kinetics
Our data emphasize the impact of RNA structure on the efficiency of binding of eIF4E or eIF4F to capped RNAs. We show directly that a hairpin positioned 12 nucleotides from the cap results in faster binding of RNA to eIF4E, and slower dissociation of the eIF4E•RNA complex. The increase in both association rate and lifetime suggests that dynamic interactions between RNA elements other than those immediately proximal to the cap are important determinants of recognition by eIF4E. The same hairpin located immediately adjacent to the cap, still permits RNA binding to the eIF4E•eIF4G-4EBD complex, but results in altered RNA-eIF4E FRET: the hairpin structure likely changes the path of the ssRNA chain moving away from the cap-binding site. Such RNAs containing cap-proximal secondary structures are translated inefficiently in vivo. For higher eukaryotes, inhibition is reduced as the structure is located increasingly further from the cap; however, this has not been observed in yeast. Secondary interactions between eIF4E and RNA remote from the RNA cap have been suggested previously, but were thought to be transient (Volpon et al., 2006; Slepenkov et al., 2006). Our results suggest that hairpin structures alter the conformation of the RNA remote from the cap in a way that could change the efficiency of formation of protein-RNA interactions important in later stages of initiation. More specifically, the geometry of the eIF4E•mRNA complex with such RNAs could orient the RNA in space such that it cannot be efficiently bound by eIF4G and eIF4A. However, any attempt to relate these dynamics directly to observed physiological effects of mRNA structure is challenging with our current knowledge and understanding of the mechanism of translation. This highlights the need for further experiments to understand the dynamic molecular underpinnings of translational control.
Pab1p significantly alters eIF4F•cap-RNA binding dynamics in the absence of a poly(A) tail
Our results show that Pab1p increases the apparent affinity of eIF4E for the cap, consistent with previous studies, and also show that it is kinetically competent to do so in the absence of a poly(A) tail. Pab1p increases eIF4E•mRNA affinity primarily by increasing the lifetime of the eIF4E•mRNA complex, which is at least doubled across the range of RNAs used in our present study. Moreover, the enhanced lifetime is associated with significant changes in the conformation of the RNA-protein complex, and in particular due to a more compact structure with higher FRET values than the eIF4E•eIF4G1•RNA complex. Our data also suggest that the effects of Pab1p are independent of the Pab1p-eIF4G1 interaction, as they are observed in the absence of eIF4G.
What might be the nature of this structural rearrangement? Pab1p can bind to RNA through its RNA recognition motif domains. While one of these (RRM2) has been shown to convey specificity for poly(A) sequences (Deardorff and Sachs, 1997), the protein shows appreciable non-specific binding to single-stranded RNA, with dissociation constants in the 0.5 μM range (Görlach et al., 1994). Proteins that possess multiple RNA binding domains have been shown to play a role in conformational remodeling of RNA. For example, the human pyrimidine tract binding protein (PTB) contains two adjacent RRMs that loop RNA around themselves, resulting in a more compact RNA structure (Oberstrass et al., 2005). RNA binding activity similar to this could account for the increased mean FRET value observed for our RNAs in the presence of Pab1p. In turn, conformational changes induced by RNA binding to Pab1p could mediate an interaction with eIF4G that leads to stabilization of eIF4E-cap interaction. We did not observe transitions between FRET states in this complex, suggesting that Pab1p may bind to the RNA prior to the arrival of the eIF4E•eIF4G complex and remain bound until it departs. We saw a small but reproducible antagonistic effect of eIF4A on the rate of binding of eIF4E•eIF4G to the RNA in the presence Pab1p, suggesting that either eIF4A and Pab1p compete for eIF4G binding, or that eIF4A competes with Pab1p for binding to the RNA. In humans, the cellular concentration of poly(A)-binding protein is thought to be ~4 μM, of which only ~30% is thought to be associated with mRNAs (Görlach et al., 1994). Our experimental conditions, where Pab1p is present at 2 μM, are thus a reasonable recapitulation of the cellular environment. Our results with Pab1p reinforce the idea that compositional dynamics play a central role in early initiation.
A dynamic model for early initiation
Our data show how kinetics and conformation are intimately linked in cap-dependent translation initiation. Structural studies have revealed how eIF4G-4EBD alters eIF4E conformation to modulate eIF4E affinity for the cap. We show here that the relationship between conformation and kinetics is a theme that extends further through early initiation. RNA structure and conformation influence the kinetics of formation and dissociation of the eIF4F•mRNA complex. At the same time, factor binding to the mRNA can change RNA conformation to further alter eIF4F•mRNA dynamics, such as in the cases of eIF4G1 and Pab1p. This all occurs along with protein-protein interactions that allosterically impact the RNA- and cap-binding activities of the factors themselves. Isolated effects are relatively small, changing individual rates by factors of two or three. However the product of all of the effects for a given mRNA can alter the probability of the eIF4F complex being bound to the RNA by an order of magnitude or more. Cumulative effects of factor-RNA and factor-factor dynamics may thus modulate the efficiency of initiation to produce significant biological outcomes.
The RNA binding activity of initiation factors is central to these allosteric effects in early initiation. In the presence of the initiation factors that possess multiple RNA-binding domains (eIF4G, Pab1p), both the FRET value and lifetime of the eIF4E•RNA complex are increased. Presumably the increase in FRET value is due to formation of a more compact ribonucleoprotein complex. Considering that our model hairpin-containing poly(CU)10hp12 RNA doubles the association rate and lifetime of the eIF4E•RNA complex compared with the equivalent unstructured RNA, and given that electrostatics play such an important role in eIF4E-RNA binding (Niedzwiecka et al., 2004), RNA structures or conformations that present a greater density of negative charge near the cap may interact more favorably with eIF4E. In the case of the eIF4F•Pab1p•mRNA complex, the looping of the RNA chain around successive RNA-binding domains of the initiation factors could also create this situation, resulting in the observed increased lifetime of the bound state. An electrostatic scaffolding function for proteins involved in translation initiation was recently proposed on the basis of bioinformatics and gene ontology studies in yeast (Cawley and Warwicker, 2012).
The eIF4F•mRNA complex is characterized by rapid-equilibrium binding on the timescale of initiation (8 – 10 s, Ciandrini et al., 2013). Our data suggest that elements of RNA structure or sequence that act to prolong the eIF4F•mRNA complex lifetime will be key determinants of initiation efficiency, because this efficiency is determined by kinetic forward partitioning of eIF4F•mRNA along the initiation pathway. Stated otherwise, the eIF4F•mRNA complex must live sufficiently long in the correct conformation to allow the subsequent step in initiation to occur. Further single-molecule studies with labeled components that can track initiation dynamics from cap recognition through to subunit joining will provide valuable mechanistic information on this intricate process.
Experimental Procedures
Protein expression and purification
Proteins were expressed in a variety of E. coli strains (see Supplementary Experimental Procedures). A saturated 10 mL overnight starter culture of the relevant strain was used to inoculate a 1.5 L expression culture. This was grown at 37 °C until the O.D.600 was ~0.6 (typically 3.5–4.5 h), then the temperature was lowered to 30 °C and IPTG was added to a final concentration of 1 mM. Expression was then allowed to proceed for 5 h, after which the cells were harvested by centrifugation (4,800 ×g, 25 minutes) and stored at −80 °C until purification. A two-step purification strategy was employed for all proteins, consisting of Ni2+-NTA chromatography followed by size-exclusion chromatography. Ni2+–NTA chromatography was carried out at room temperature with chilled buffers. FPLC size exclusion chromatography was carried out at 6 °C. Cells were lysed by sonication in the presence of lysozyme (~0.3 mg/mL). For protein-specific details, see the Supplementary Experimental Procedures.
Labeling of eIF4E with Cy5-maleimide
Labeling was carried out between the Ni2+-NTA and size exclusion purification steps. After removal of imidazole, 1.5 mL of protein solution (typically ~5 μM) was added to 1 mg of Cy5-maleimide (GE Healthcare Life Sciences). The reaction mixture was incubated in darkness overnight at 6 °C, then separated from unreacted dye by two rounds of gel filtration (Bio-Rad 10 DG columns) in the size exclusion column buffer.
RNA preparation
RNAs were prepared using the AmpliCap-Max T7 transcription kit (CellScript, Inc.), using the manufacturer’s instructions. DNA templates were designed using a previously-described approach (Puglisi and Wyatt, 1995). RNA was purified from the transcription mixture by anion exchange chromatography (Q HP, 5 mL, GE Healthcare Life Sciences), eluting with a linear gradient from 50 mM to 1 M KCl in 50 mM Tris-HCl, pH 8. TBE-Urea PAGE was used to confirm purity (>90% single RNA band in each case). RNAs eluted between 500 and 750 mM KCl, in proportion to their length. The model cap-unstructured poly(CU) RNA was also purchased from TriLink Biotechnologies. RNAs were annealed to Cy3-labeled, biotinylated DNAs by heating to 90 °C for 3 minutes, then slow-cooling to room temperature.
Single-molecule experiments
The home-built TIRF apparatus has been described previously (Aitken and Puglisi, 2010). Samples were prepared at 20× the intended assay concentration and diluted into assay buffer (50 mM HEPES-KOH, pH 7.4, 100 mM KCl) containing the PCD/PCA oxygen-scavenging system (2.5 mM 3,4-dihydroxybenzoic acid, 250 nM protocatechuate dioxygenase; Aitken et al., 2008), and Trolox (Dave et al., 2009). For Pab1p, where the protein was present at 2 μM final concentration, it was included at 2 μM in the 20× sample, and then supplemented into the final sample at 2 μM. Data were acquired at 10 frames per second for 60 s, with three separate movies per sample, processed independently. Data were analyzed by in-house spot-picking and colocalization scripts implemented in MATLAB (Mathworks). The locations of FRET events in fluorescence traces were manually assigned after initial estimates obtained from a hidden-Markov-based algorithm. Arrival time, lifetime, and FRET distributions were normalized such that the area under the fitted curve was equal to one. The area of each bin is thus numerically equal to the probability of an event occurring within the bin width.
Supplementary Material
Highlights.
A single-molecule assay to observe directly S. cerevisiae eIF4E-mRNA interaction.
Measured the kinetics of assembly of the eIF4E•eIF4G1•eIF4A•mRNA complex.
Measured the modulation of eIF4E-RNA binding kinetics by RNA secondary structures.
Observed Pab1p-mediated, poly(A)-independent stimulation of eIF4E-RNA binding.
Acknowledgments
This work was supported by NIH grants GM073999 and GM099687 (to JDP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol. 2012;19:568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- Aitken CE, Puglisi JD. Following the intersubunit conformation of the ribosome during translation in real time. Nat Struct Mol Biol. 2010;17:793–800. doi: 10.1038/nsmb.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann M, Trachsel H. The yeast Saccharomyces cerevisiae system: a powerful tool to study the mechanism of protein synthesis initiation in eukaryotes. Biochimie. 1994;76:853–861. doi: 10.1016/0300-9084(94)90187-2. [DOI] [PubMed] [Google Scholar]
- Babendure JR, Babendure JL, Ding JH, Tsien RY. Control of mammalian translation by mRNA structure near caps. RNA. 2006;12:851–861. doi: 10.1261/rna.2309906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berset C, Zurbriggen A, Djafarzadeh S, Altmann M, Trachsel H. RNA-binding activity of translation factor eIF4G1 from Saccharomyces cerevisiae. RNA. 2003;9:871–880. doi: 10.1261/rna.5380903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borman AM, Michel YM, Malnou CE, Kean KM. Free poly(A) stimulates capped mRNA translation in vitro through the eIF4G-poly(A)-binding protein interaction. J Biol Chem. 2002;277:36818–36824. doi: 10.1074/jbc.M205065200. [DOI] [PubMed] [Google Scholar]
- Carberry SE, Friedland DE, Rhoads RE, Goss DJ. Binding of protein synthesis initiation factor 4E to oligoribonucleotides: effects of cap accessibility and secondary structure. Biochemistry. 1992;31:1427–1432. doi: 10.1021/bi00120a020. [DOI] [PubMed] [Google Scholar]
- Cawley A, Warwicker J. eIF4E-binding protein regulation of mRNAs with differential 5′-UTR secondary structure: a polyelectrostatic model for a component of protein mRNA-interactions. Nucl Acids Res. 2012;40:7666–7675. doi: 10.1093/nar/gks511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson BK, Gilbert WV, Doudna JA. Functional overlap between eIF4G isoforms in Saccharomyces cerevisiae. PLoS One. 2010;5:e9114. doi: 10.1371/journal.pone.0009114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciandrini L, Stansfeld I, Romano MC. Ribosome traffic on mRNAs maps to gene ontology: genome-wide quantification of translation initiation rates and polysome size regulation. PLoS Comput Biol. 2013;9:e1002866. doi: 10.1371/journal.pcbi.1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave R, Terry DS, Munro JB, Blanchard SC. Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys J. 2009;96:2371–2381. doi: 10.1016/j.bpj.2008.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff JA, Sachs AB. Differential effects of aromatic and charged residue substitutions in the RNA binding domains of the yeast poly(A)-binding protein. J Mol Biol. 1997;269:67–81. doi: 10.1006/jmbi.1997.1013. [DOI] [PubMed] [Google Scholar]
- Firczuk H, Kannambath S, Pahle J, Claydon A, Beynon R, Duncan J, Westerhoff H, Mendes P, McCarthy JE. An in vivo control map for the eukaryotic mRNA translational machinery. Mol Sys Biol. 2013;9:635. doi: 10.1038/msb.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A-C, Gygi SP, Raught BP, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkogkas CG, Khoutorsky A, Ran I, Rampakakis E, Nevarko T, Weatherill DB, Vasuta C, Yee S, Truitt M, Dallaire P, et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature. 2013;493:371–377. doi: 10.1038/nature11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlach M, Burd CG, Dreyfuss G. The mRNA poly(A)-binding protein: localization, abundance, and RNA-binding specificity. Exp Cell Res. 1994;211:400–407. doi: 10.1006/excr.1994.1104. [DOI] [PubMed] [Google Scholar]
- Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JEG, Wagner G. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell. 2003;115:739–750. doi: 10.1016/s0092-8674(03)00975-9. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CUT, Pestova T. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;10:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahvejian A, Svitkin YV, Sukarieh R, M’Boutchou MN, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Wilson AC, Chao MV, Mohr I. Control of viral latency in neurons by axonal mTOR signaling and the 4E-BP translational repressor. Genes Dev. 2012;26:1527–1532. doi: 10.1101/gad.190157.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J, Lasko P. Translational control in cellular and developmental processes. Nat Rev Genet. 2012;13:383–394. doi: 10.1038/nrg3184. [DOI] [PubMed] [Google Scholar]
- Kozak M. Regulation of translation in eukaryotic systems. Annu Rev Cell Biol. 1992;8:197–225. doi: 10.1146/annurev.cb.08.110192.001213. [DOI] [PubMed] [Google Scholar]
- McCarthy JEG. Posttranscriptional control of gene expression in yeast. Microbiol Mol Biol Rev. 1998;62:1492–1553. doi: 10.1128/mmbr.62.4.1492-1553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SF, Walker SE, Algire MA, Park E-H, Hinnebusch AG, Lorsch JR. The 5′-7-methylguanosine cap on eukaryotic mRNAs serves both to stimulate canonical translation initiation and to block an alternative pathway. Mol Cell. 2010;39:950–962. doi: 10.1016/j.molcel.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiecka A, Darzynkiewicz E, Stolarski R. Thermodynamics of mRNA 5′ cap binding by eukaryotic initiation factor eIF4E. Biochemistry. 2004;43:13305–13317. doi: 10.1021/bi0491651. [DOI] [PubMed] [Google Scholar]
- Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, Allain FH. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- Otero LJ, Ashe MP, Sachs AB. The yeast poly(A)-binding protein Pab1p stimulates in vitro poly(A)-dependent and cap-dependent translation by distinct mechanisms. EMBO J. 1999;18:3153–3163. doi: 10.1093/emboj/18.11.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov A, Chen J, O’Leary S, Tsai A, Puglisi JD. Single-molecule analysis of translational dynamics. Cold Spring Harb Perspect Biol. 2012 doi: 10.1101/cshperspect.a011551. Ptushkina. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi JD, Wyatt JR. Biochemical and NMR studies of RNA conformation with an emphasis on RNA pseudoknots. Methods Enzymol. 1995;261:323–350. doi: 10.1016/s0076-6879(95)61016-2. [DOI] [PubMed] [Google Scholar]
- Rajagopal V, Park EH, Hinnebusch AG, Lorsch JR. Specific domains in yeast translation factor eIF4G strongly bias RNA unwinding activity of the eIF4F complex toward duplexes with 5′ overhangs. J Biol Chem. 2012;267:20301–20312. doi: 10.1074/jbc.M112.347278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safaee N, Kozlov G, Noronha AM, Xie J, Wilds CJ, Gehring K. Interdomain allostery promotes assembly of the poly(A) mRNA complex with PABP and eIF4G. Mol Cell. 2012;48:375–386. doi: 10.1016/j.molcel.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Slepenkov SV, Korneeva NL, Rhoads RE. Kinetic mechanism for assembly of the m7GpppG•eIF4E•eIF4G complex. J Biol Chem. 2008;12:25227–25237. doi: 10.1074/jbc.M801786200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slepenkov SV, Darzynkiewicz E, Rhoads RE. Stopped-flow kinetic analysis of eIF4E and phosphorylated eIF4E binding to cap analogs and capped oligoribonucleotides. J Biol Chem. 2006;281:14927–14938. doi: 10.1074/jbc.M601653200. [DOI] [PubMed] [Google Scholar]
- Sun Y, Atas E, Lindqvist L, Sonenberg N, Pelletier J, Meller A. The eukaryotic initiation factor eIF4H facilitates loop-binding, repetitive RNA unwinding by the eIF4A DEAD-box helicase. Nucleic Acids Res. 2012;40:6199–6207. doi: 10.1093/nar/gks278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun SZ, Jr, Wells SE, Deardorff JA, Sachs AB. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci USA. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai A, Petrov A, Marshall RA, Korlach J, Uemura S, Puglisi JD. Heterogeneous pathways and timing of factor departure during translation initiation. Nature. 2012;487:390–393. doi: 10.1038/nature11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura S, Aitken CE, Korlach J, Flusberg BA, Turner SW, Puglisi JD. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010;464:1012–1017. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SE, Zhou F, Mitchell SF, Larson VS, Valasek L, Hinnebusch AG, Lorsch JR. Yeast eIF4B binds to the head of the 40S ribosomal subunit and promotes mRNA recruitment through its N-terminal and internal repeat domains. RNA. 2012;19:191–207. doi: 10.1261/rna.035881.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpon L, Osborne M, Toposirovic I, Siddiqui N, Borden KLB. Cap-free structure of eIF4E suggests a basis for conformational regulation by ligands. EMBO J. 2006;25:5138–5149. doi: 10.1038/sj.emboj.7601380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von der Haar T, Oku Y, Ptushkina M, Moerke N, Wagner G, Gross JD, McCarthy JE. Folding transitions during assembly of the eukaryotic mRNA cap-binding complex. J Mol Biol. 2006;356:982–992. doi: 10.1016/j.jmb.2005.12.034. [DOI] [PubMed] [Google Scholar]
- Von der Haar T, Gross JD, Wagner G, McCarthy JEG. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat Struct Mol Biol. 2004;11:503–511. doi: 10.1038/nsmb779. [DOI] [PubMed] [Google Scholar]
- Von der Haar T, Ball PD, McCarthy JEG. Stabilization of eukaryotic initiation factor 4E binding to the mRNA 5′-cap by domains of eIF4G. J Biol Chem. 2000;275:30551–30555. doi: 10.1074/jbc.M004565200. [DOI] [PubMed] [Google Scholar]
- Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- Yanagiya A, Svitkin YV, Shibata S, Mikami S, Imataka H, Sonenberg N. Requirement of RNA binding of mammalian eukaryotic translation initiation factor 4GI (eIF4GI) for efficient interaction of eIF4E with the mRNA cap. Mol Cell Biol. 2009;29:1661–1669. doi: 10.1128/MCB.01187-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth and signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.