Abstract
Streptococcus pneumoniae causes several diseases, including otitis media, pneumonia, and meningitis. Although little is known about the regulation of or how individual pneumococcal factors contribute to these disease states, there is evidence suggesting that some factors are regulated by a cell-density-dependent mechanism (quorum sensing). Quorum sensing allows bacteria to couple transcription with changes in cell density; bacteria achieve this by sensing and responding to small diffusible signaling molecules. We investigated how the LuxS signaling system impacts the biology of S. pneumoniae. An analysis of the transcriptional profiles of a serotype 2 strain and an isogenic luxS deletion strain utilizing an S. pneumoniae-specific microarray indicated that LuxS regulates gene expression involved in discrete cellular processes, including pneumolysin expression. Contrary to the paradigm for quorum sensing, we observed pronounced effects on transcription in early log phase, where gene expression was repressed in the mutant. Assessing the mutant for its ability to infect and cause disease in animals revealed a profound defect in ability to persist in the nasopharyngeal tissues. Our analysis of an S. pneumoniae transcriptome revealed a function for LuxS in gene regulation that is not dependent upon high cell density and is likely involved in the maintenance of pneumococcal load in susceptible hosts.
Less than 40 years ago, the first study supporting the existence of bacterial cell-to-cell communication systems was published (63). This work put forth evidence that the gram-positive diplococcus Streptococcus pneumoniae produced a hormone-like substance at a particular cell density that, when detected, affected the precise temporal regulation of genetic competence in S. pneumoniae. Since that time, bacterial cell-density-dependent signaling, termed quorum sensing (23), has been described and characterized to varying extents in many diverse bacterial species (reviewed in references 4, 21, 40, and 58). It is widely believed that these regulatory systems evolved to coordinate the expression of target genes whose products would be most advantageous when a critical mass of bacteria were expressing them but would provide little benefit when expressed by an individual bacterium.
Quorum-sensing-mediated signaling can be achieved through a variety of regulatory mechanisms, and all of the systems described to date involve the production and secretion or diffusion of low-molecular-weight signaling molecules (autoinducers) into the extracellular milieu. Quorum sensing in gram-negative bacteria is most commonly mediated by derivatives of N-acyl homoserine lactones (recently reviewed in reference 21), while gram-positive organisms typically utilize unmodified or posttranslationally modified oligopeptide pheromones (15, 35, 37). When these molecules accumulate to a critical threshold concentration, they interact with their cognate receptors. Once activated, these proteins either directly or indirectly control transcription of target genes. Moreover, there is a growing body of evidence suggesting that many bacteria elaborate multiple signal sensing systems, offering an elegant and economical means of orchestrating the coordinated expression of diverse sets of genes in response to specific environmental cues.
While the signaling molecules, the proteins that mediate transcriptional regulation, and the target genes controlled by these sensing pathways are not universally conserved among bacteria, it is now apparent that the ability to detect changes in concentration of these extracellular cues and transduce this information into the cell is responsible for regulating a wide variety of biological processes (for reviews, see references 10, 14, 22, 26, 50, 66, and 69). A connection between quorum sensing and virulence has been made for a few pathogens, in particular Pseudomonas aeruginosa (45), Staphylococcus aureus (31, 43), Erwinia carotovora (32), Vibrio cholerae (41, 72) and, recently, S. pneumoniae (36).
For most of the described quorum-sensing systems, recognition of any particular autoinducer is restricted to the species that produced it, conferring a high level of specificity for these types of cell-to-cell communication systems that ensures an accurate flow of information from initial detection (sensing) to molecular response (modulation of gene expression). Recently, however, a novel quorum-sensing system in the bioluminescent marine bacterium Vibrio harveyi was described (60). In V. harveyi, bioluminescence is controlled by two quorum-sensing systems, each of which responds to a different autoinducer. The HAI-1 autoinducer is species specific, being synthesized and recognized solely by V. harveyi. However, the second autoinducer, AI-2, is synthesized by the product of the luxS gene, which is present in the genomes of over 40 bacterial species (2, 60), including S. pneumoniae. Since AI-2 isolated from several diverse species has been shown to complement a luxS defect in V. harveyi, this molecule is proposed to act as an interspecies cellular communication molecule. In addition to its role in controlling bioluminescence, there is evidence that the AI-2 signaling system may play a role in regulating expression of several genes that affect the pathogenic capabilities of several organisms, including modulating motility (18, 24), proteolytic activity (7, 38), hemolytic activity (38), expression of a type III secretion system, Tir, and intimin in Escherichia coli (54), antibiotic production (12), and iron acquisition (19).
Since peptide-mediated cell-to-cell signaling is known to be an important regulatory mechanism for gene expression in S. pneumoniae, we were interested in determining if the LuxS signaling system contributed to the regulation of pneumococcal genes. To accomplish this, we compared the in vitro transcriptional profiles over time of S. pneumoniae D39 with that of isogenic deletion mutant a ΔluxS using a spotted DNA microarray. Microarray analyses reveal at least five pneumococcal operons that are aberrantly regulated in the ΔluxS strain. These operons encode proteins for fatty acid biosynthesis, a putative hemolysin, and pneumolysin, a major virulence determinant of S. pneumoniae. Unexpectedly, this regulation did not occur at high cell density, the paradigm for quorum sensing. Results of infection studies in animals revealed a significant defect for the ΔluxS strain in its ability to persist in a murine model of nasopharyngeal carriage. Taken together, these data suggest that LuxS activity modulates the fitness of the organism in a discrete host niche and that mechanistic explanations other than quorum sensing for how LuxS functions need to be considered.
MATERIALS AND METHODS
Bacterial strains and culture.
E. coli DH5α (Bio-Rad Laboratories) and RR1 were used for cloning experiments. S. pneumoniae strain D39 and its derivatives (Table 1) were grown in brain heart infusion (BHI) broth (Difco), Todd-Hewitt broth (Difco) supplemented with 5% horse serum (Gibco), and on tryptic soy agar (TSA; Difco) supplemented with 5% defibrinated sheep blood (Hemostat Laboratories, Dixon, Calif.). Both broth and agar media were supplemented with antibiotics as indicated.
TABLE 1.
Strains, plasmids, and primer sequences used in this study
| Strain, plasmid, or primer | Relevant genotype or primer sequence (5′ to 3′) | Reference, source, or primer purpose |
|---|---|---|
| E. coli strains | ||
| DH5α | F− Φ80dlacZM15 endA1 recA1 hsdR17 (rK− mK+) supE44 thi-1 gyrA96 relA1 (lacZYA-argF)U169 deoR λ− | |
| RR1 | HB101 recA+ | |
| S. pneumoniae strains | ||
| D39 | Serotype 2 | 62 |
| EJ1 | D39 streptomycin-resistant derivative | This study |
| EJ2 | D39 ΔluxS::spect | This study |
| EJ3 | EJ1ΔluxS::spect | This study |
| EJ5 | EJ1ΔSP1923::spect | This study |
| S. enterica serovar Typhimurium LT2 | 51 | |
| V. harveyi BB170 | luxN::Tn5 | 59 |
| Plasmids | ||
| pCZA342 | 29 | |
| pCR4-TOPO | Cloning vector; ampicillin and kanamycin resistance | Invitrogen |
| pEJ1 | Suicide vector to delete luxS | This study |
| pEJ2 | Suicide vector to delete pneumolysin | This study |
| pBADluxSSP | This study | |
| Primers | ||
| LuxS1 | GGTATCACACCTGCAGACCGT | Deletion construction |
| LuxS2 | CTCGAGGTTGCTCCTGAGACAGAGAG | Deletion construction |
| LuxS3 | GAGCTCTCTAGCCCCTCTCACACC | Deletion construction |
| LuxS4 | GGATCCCTTCTTCCGTCCAGAATTC | Deletion construction |
| SP1923A | CTGCAGCTAGTGCAGATGCTCCAG | Deletion construction |
| SP1923B | CTCGAGCTACCTCCTAATAAGTTCCTGG | Deletion construction |
| SP1923C | GAGCTCGACTAGGAGAGGAGAAT | Deletion construction |
| SP1923D | GGATCCATTTACGTCCCATTAGGAATC | Deletion construction |
| LuxSOX1 | ATGTCAAAAGAAGTTATTGTCGAAAGTTTTG | luxS overexpression construction |
| LuxSOX2 | AATCACATGACGTTCAAAGGC | luxS overexpression construction |
| rpoB RT2 forward | AGATGGAGGTTTGGGCTCTT | Real-time RT-PCR analysis |
| rpoB RT2 reverse | CCGTTGATATCGTCCGACTT | Real-time RT-PCR analysis |
| 1923 RT4 forward | CAGTCGCCTCTATCCTGGAG | Real-time RT-PCR analysis |
| 1923 RT4 reverse | CCGCAAGAAGAGTGGGATTA | Real-time RT-PCR analysis |
| 1466 RT6 forward | GCTGGTTGGTTCTGGCTATC | Real-time RT-PCR analysis |
| 1466 RT6 reverse | AGTCCGCCAGTTACCATGAG | Real-time RT-PCR analysis |
| AccD RT3 forward | CAATCGTTCGGTTAGGGAAA | Real-time RT-PCR analysis |
| AccD RT3 reverse | TGCTTACAGCCTGGACACTG | Real-time RT-PCR analysis |
For all experiments, cultures were prepared by serially diluting glycerol stocks of pneumococcal strains into BHI broth and incubating for 12 to 14 h at 37°C in a 5% CO2 atmosphere. For in vitro time course experiments, tubes with optical densities between ∼0.01 and 0.02 were selected, and the cells were collected by centrifugation, washed once with sterile phosphate-buffered saline (PBS), back diluted to ∼3 × 105 CFU/ml into fresh BHI broth, and immediately incubated at 37°C in a 5% CO2 chamber for 45 min. At this point, the first sample was collected. Subsequently, growth was monitored for approximately eight generations both by measuring the optical density at 600 nm (OD600) and by determining viable counts every 30 to 45 min. At these points samples were also harvested by centrifugation for RNA isolation and stored at −80°C until processing. In addition, cell-free supernatants were obtained by filtering culture supernatants through a 0.22-μm-pore-size filter (Fisher) and stored at −20°C until the density-dependent bioluminescence assays were performed. For animal infections, mid-exponential bacterial cells (OD600, ∼0.4) were centrifuged, washed once with PBS, and resuspended in PBS to the appropriate cell density for animal infection (∼108 CFU/ml for murine respiratory tract infections and ∼109 CFU/ml for the murine carriage model).
DNA manipulations and strain constructions.
To generate gene replacement mutant strains, approximately 1 kb of DNA fragments flanking the coding sequences was PCR amplified from D39 genomic DNA using primers pairs that had unique restriction sites designed into their 5′ ends. The amplified products were cloned into pCR4-TOPO (Invitrogen, Carlsbad, Calif.). Subsequently, the resulting plasmids were digested with the appropriate restriction enzymes to release the products, which were gel purified. A four-part ligation comprised of these two DNA fragments, a DNA cassette encoding resistance to spectinomycin, and the linearized S. pneumoniae suicide vector pCZA342 (29) was prepared. After a 2-h room temperature incubation, one-fifth of the ligation mixture was electroporated into E. coli DH5α, and transformants containing the resulting plasmid (pEJ1) were selected for on Luria-Bertani agarose plates containing 100 mg of apramycin/ml and 200 mg of spectinomycin/ml. Expression of the spectinomycin gene is driven by its own promoter. Correct orientation of the fragments of the resulting plasmid (pEJ2) was confirmed by restriction digests. To express LuxS, the S. pneumoniae luxS gene was amplified and the resulting 480-bp DNA fragment was cloned into the pBAD Topo TA expression vector (Invitrogen), resulting in pBADluxSSP, which was expressed in the luxS mutant E. coli strain DH5α.
Transformation protocol for S. pneumoniae.
Competence induction medium was prepared as follows: 9 ml of BHI broth, 1 ml of horse serum, 100 μl of 1 M glucose, and 1 μl of competence-stimulating peptide 1 (1 μg/μl) were combined. A 900-μl aliquot of competence induction medium was added to glass test tubes, and 100 μl of pneumococcal early-log (OD, 0.05 to 0.1) BHI broth-grown culture was added to this. After a 30-min, 37°C, 5% CO2 incubation, approximately 1 μg of transforming DNA was added. The tubes were reincubated under the above conditions for 1 h, after which cells were plated on selective medium. All resulting mutant strains were confirmed by both PCR and Southern blot analysis. In addition, Western analysis using a monoclonal antibody directed against pneumolysin (generously provided by J. Paton) was performed on cell lysates isolated from the pneumolysin deletion strain to confirm the absence of the protein.
RNA isolation and real-time PCR assay.
Bacterial pellets were thawed on ice and treated with 400 mg of lysozyme ml−1 in 100 μl of PBS for 5 min at room temperature. RNA was purified according to the QIAGEN RNeasy kit instructions with the following modifications. After the buffer RLT was added, cell suspensions were mixed by pipetting and transferred to a 2-ml Eppendorf tube containing approximately 300 μl of 0.1-mm zirconia-silica beads (Biospec Products, Inc., Bartlesville, Okla.) and vortexed for 45 s. The tubes were spun briefly, and the lysed cell extract was loaded onto an RNeasy column. Samples were treated on the column with DNase I (QIAGEN), as recommended by the manufacturer. RNA quality was determined by agarose gel electrophoresis and by OD260/OD280 ratios. All RNA samples were subjected to PCR amplification to ensure the absence of contaminating chromosomal DNA. Real-time PCR was carried out with a Bio-Rad iCycler by using gene-specific primers and rTth polymerase (Perkin-Elmer) as directed by the manufacturer. Cyber Green was used to detect specific signal. A standard curve was plotted for each primer set with Ct values obtained from amplification of a dilution series of samples known to contain the specific messages of interest. The standard curves were used to determine relative quantities of cDNA for each experimental gene. To compare between samples and strains, these values were normalized to the quantity of rpoB-specific cDNA in each sample.
Microarray design.
We constructed a 4,420-element S. pneumoniae-specific spotted DNA microarray based on the preannotated TIGR4 genome sequence (61). In order to minimize the potential for cross-hybridization on the array, primer pairs (Illumina, Inc., San Diego, Calif.) were designed using the Primer3 program (http://www-genome.wi.mit.edu/genome_software/other/primer3.html) (49) based on a sequence that was determined to be unique to each open reading frame (ORF). This was accomplished using an algorithm that identifies the most-3′ region of an ORF that contains no significant homology to any other ORF in the genome as identified by NCBI BLAST (49). In addition, primer pairs were also designed for 975 noncoding putative regulatory sequences. DNA fragments were PCR amplified using predominantly D39 genomic DNA as a template; however, for 65 ORFs that are not present in this strain (61), TIGR4 genomic DNA was used as a template for amplification. Amplified products were analyzed on agarose gels and ranged from 70 to 300 bp, with 210 bp as the median length and 198 bp as the average length. PCR amplification methods, polylysine glass slide preparation, printing, and array postprocessing were performed as previously reported (16). The final array was composed of 3,620 elements corresponding to 98% of all TIRG4 ORFs and 975 elements corresponding to intergenic sequences. Many elements were represented by multiple spots.
Probe synthesis.
RNA was converted to cDNA in 20-μl reaction mixtures by combining 0.5 μg of RNA and 0.5 μg of random hexamers (Amersham), heating to 65°C for 10 min, and then snap-cooling the reactions on ice. The following were then added: 2 μl of 0.1 M dithiothreitol, 0.5 μl of 10 mM deoxynucleoside triphosphates, 4 μl of 5× RT buffer (Invitrogen), and 1 μl (200 U) of Superscript II (Gibco BRL). This mixture was incubated at 42°C for 150 min. RNA was hydrolyzed with 1 μl of 1 M NaOH at 65°C for 10 min and neutralized with 1 μl of 1 M HCl. Samples were purified over a Qia-Quick PCR column (QIAGEN) according to the manufacturer's instructions and eluted with 40 μl of elution buffer. Amino-allyl dUTP was incorporated into the cDNA samples as follows. For each sample, 40 μl of the eluted DNA was incubated for 5 min at 99°C and then for 5 min on ice. A 5-μl aliquot of 10× random octamer buffer (1550-2; NEB), 3 μl of deoxynucleoside triphosphate-dUTP mix (3 mM dGTP, dATP, and dCTP; 1.8 mM amino-allyl-dUTP [A-0410; Sigma-Aldrich], 1.2 mM dTTP), and 2 μl of Exo− Klenow (NEB) were added, and the mixture was incubated for 150 min at 37°C and then stored at 4°C overnight. Free amines were removed with a Qia-Quick PCR purification kit (QIAGEN), and the eluted sample was dried in a speed-vac. Samples were resuspended in 4.5 μl of distilled water and incubated with a 1 μM concentration of either Cy3 or Cy5 monofunctional reactive dye (Amersham) for 1 h at room temperature in the dark. The time point samples were incubated with Cy5, and the reference samples were incubated with Cy3. The reference sample for each time course was generated from the D39 time zero RNA. The reactions were quenched with 4.5 μl of 4 M hydroxylamine for 15 min at room temperature, and then each Cy5-labeled sample was mixed with a Cy3-labeled reference. Unincorporated dye was removed with a Qia-Quick PCR purification kit, and probes were eluted with 40 μl of elution buffer and dried in a speed-vac. To hybridize, the samples were resuspended in 11.3 μl of Tris-EDTA (pH 7.5), 1 μl of a 10-mg/ml solution of yeast tRNA, 2.25 μl of 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and 0.45 μl of 10% sodium dodecyl sulfate. The mixture was heated to 99°C for 2 min and immediately centrifuged for 2 min at maximum speed. The probe was applied to a microarray and incubated for at least 24 h at 60°C.
Data analysis.
Arrays were washed and then scanned using a GenePix 4000A scanner (Axon Instruments, Foster City, Calif.), and images were analyzed with GenePix Pro software. The raw data were loaded into the Stanford Microarray database (25), where they were normalized according to the default computed normalization calculation. All data are publicly available (http://genome-www.stanford.edu/microarray). Data were filtered to remove poor-quality measurements, and the red/green ratios were log2 transformed. The data were subsequently zero transformed for each time course by subtracting the average time zero value (n = 3) for each strain from all subsequent time points measured. This allowed us to identify genes whose patterns of expression differed between the two strains through the time courses relative to that at time zero.
Density-dependent bioluminescence assays.
The V. harveyi reporter strain, BB170 (Sensor 1− and Sensor 2+), was grown for 13 h at 30°C with aeration in AB broth, a minimal medium (3), at which point it was diluted 1:5,000 into fresh AB medium and aliquoted in 90-μl amounts into 96-well microtiter plates (Dynex Microlite; Fisher Scientific) containing 10 μl of the various substances to be tested for AI-2 activity. Microtiter plates were incubated with aeration at 30°C, and light production was measured every 30 min using a Wallac model 1450 Microbeta Plus liquid scintillation counter.
Virulence assays.
Animals were housed according to federal, state, and local guidelines for laboratory animal care in the Research Animal Facility in the Department of Comparative Medicine at Stanford University School of Medicine. Male CD-1 mice (35 g; Charles River Laboratories, Wilmington, Mass.) were used in these assays and were provided with sterile water and food ad libitum. Prior to infection, mutant (EJ3) and parent (EJ1) strains were grown separately as described above and were subsequently mixed in a 1:1 ratio. The mice were anaesthetized with isoflurane (4% in O2). For lung infection, mice were individually anesthetized for 2.5 to 3 min each. Because isoflurane depresses respiration, this amount of anesthetic results in a compensatory gasping reaction, facilitating aspiration of the inocula. At this point, mice were removed from the anesthetic apparatus, and 50 μl of a 108-CFU/ml bacterial suspension (∼5 × 106 cells total) was introduced by intranasal instillation. For nasopharyngeal inoculation, mice were lightly anesthetized for 45 to 60 s only, followed by the slow intranasal instillation of 10 μl of a ∼109-CFU/ml bacterial suspension (∼107 cells total). Animals were allowed to recover, given food and water ad libitum, and housed according to the Stanford University Department of Comparative Medicine guidelines. Lung infections were carried out over 3 days, while nasopharyngeal colonization studies were carried out over 15 days. At designated time points, animals were sacrificed by CO2 overdose. Blood was collected by cardiac puncture; the lungs were aseptically removed to 2 ml of PBS and homogenized using a Tissue Tearor (Biospect Inc). The nasopharyngeal cavities were washed by transecting the trachea, inserting a 1-in. 24-gauge feeding needle (Popper and Sons, New Hyde Park, N.Y.) into the larynx, and irrigating the nasal passage with 500 μl of sterile PBS. The wash was collected through the nares. All samples were assayed for viable bacteria by serial dilution onto both TSA plus blood agar plates containing streptomycin-oxacillin-colistin (selects for both pneumococcal strains) and TSA plus blood agar plates containing streptomycin-spectinomycin-oxacillin-colistin (selects for EJ3). Each experiment was performed a minimum of two times, and the data shown are composites of all experiments. In conjunction with each in vivo competition, an in vitro competition was carried out as follows: 50 μl of each mixture was inoculated into 10 ml of BHI broth and grown to mid-log phase for ∼5 h, at which point the cell suspension was serially diluted and plated on the above media. Following each experiment, the ratio of mutant to wild-type bacteria, for both in vitro and in vivo competitions, was determined. Competitive indices were calculated as the ratio of mutant to wild-type bacteria recovered under each in vivo or in vitro condition adjusted for the input ratio. Competition experiments carried out with EJ1 and EJ5 were done similarly.
Statistical analysis.
Student's unpaired, two-tailed t test was used to assess the statistical significance of any apparent differences observed in the animal challenge experiments. To determine statistically significant changes, for all experiments, data collected subsequent to day 1 were compared with the data sets collected on day 1. For competitions in which no mutant bacteria were recovered from a particular animal, 1 was substituted as the numerator when determining the in vivo ratio for that animal. P values of <0.05 were considered significant.
RESULTS
Presence of the V. harveyi luxS homolog in S. pneumoniae.
An examination of the published S. pneumoniae genome databases (28, 61) and a prior study (60) revealed an ORF in each of the S. pneumoniae strains sequenced to date whose predicted protein product is 57% homologous and 36% identical to LuxS from V. harveyi. While the luxS gene has been shown to be highly conserved among both gram-positive and gram-negative bacteria (3, 60), its chromosomal location is not conserved between different bacterial species. In both the TIGR4 and the R6 genomes, luxS is situated 98 bp downstream of and in the same transcriptional orientation as a gene of unknown function and 302 bp upstream of an ORF with homology to a putative Clp protease ATP-binding subunit, which is convergently transcribed.
AI-2 activity in S. pneumoniae.
Production of AI-2 requires a functional LuxS enzyme (9, 52, 60). It has been previously shown that AI-2-containing cell-free supernatants harvested from cultures expressing luxS can activate V. harveyi AI-2-dependent luciferase expression when supplemented into the V. harveyi growth medium. To assess whether the S. pneumoniae luxS gene encodes an AI-2-producing enzyme, we constructed a ΔluxS mutant strain by replacing the luxS coding sequence with a spectinomycin cassette (EJ2). This mutation was also moved into EJ1, the streptomycin-resistant laboratory strain used to infect animals, to create EJ3. Since the closest downstream gene to luxS is over 300 bp away and in the opposite transcriptional orientation and there is a predicted rho-independent terminator downstream of the luxS coding sequence, mutant construction is unlikely to have polar effects on transcription. The parents and ΔluxS derivatives showed virtually identical generation times throughout the growth curve in BHI broth (data not shown). We performed two sets of experiments isolating cell-free supernatants over the course of growth from cultures of the parent S. pneumoniae and the ΔluxS strains grown in BHI broth and tested for the ability of these supernatants to stimulate light production in an AI-2-dependent manner in a V. harveyi indicator strain, BB170 (3). The supernatants from both strains failed to induce luminescence above background levels in the reporter strain, raising the possibility that S. pneumoniae lacked the ability to produce functional AI-2. However, we also noticed that the levels of luminescence in a set of experimental control reactions, BB170 supplemented with sterile BHI broth, consistently showed severalfold less luminescence than that from BB170 supplemented with sterile AB (data not shown). BB170 actually exhibited a faster growth rate in BHI broth than in AB medium, suggesting that this phenotype was not due to a growth deficiency (data not shown). Therefore, we were concerned that the lack of observable induction from the parent supernatants might be due to an inhibitory effect from the BHI medium rather than a lack of functional AI-2. Recently, Merritt et al. (39) described a similar phenotype in Streptococcus mutans; although supernatants derived from BHI broth-grown S. mutans failed to complement bioluminescence in BB170, supernatants collected from E. coli DH5α expressing S. mutans luxS could. Because S. pneumoniae does not grow in AB medium, to further investigate whether S. pneumoniae produces a molecule capable of inducing luminescence in V. harveyi we collected supernatants from wild-type S. pneumoniae grown in a variety of media, including one (THBS) that has been reported to allow the detection of AI-2 in another streptococcal species (5). In addition, we cloned the pneumococcal luxS gene into a pBAD expression vector (Invitrogen). The resulting plasmid construct, pBADluxSSP, was transformed into an E. coli background (DH5α) known to be deficient for AI-2 production (59). pBADluxSSP/DH5α was grown in Luria-Bertani, BHI, and THBS and induced with 2% arabinose to express luxSSP. The supernatants were collected after 3 h of induction and tested in BB170. Despite several attempts, we were unable to detect the AI-2 molecule from actively growing wild-type S. pneumoniae in any of the media (data not shown). Importantly, however, the supernatants harvested from induced strains of pBADluxSSP grown in BHI resulted in a 67-fold induction of luminescence over background. While this level of stimulation was not as great as that seen in the positive control supernatant (>1,000-fold) harvested from LT2, these data indicated that the S. pneumoniae luxS gene product is capable of synthesizing the autoinducer, AI-2, and that AI-2 activity is not inhibited by BHI broth. It appears that S. pneumoniae either (i) does not produce active AI-2, (ii) can efficiently degrade any AI-2 that is made, (iii) does not release AI-2 into the supernatant, or (iv) if it does release AI-2, has an efficient system in place to transport AI-2 back into the cell. Regardless, these data suggest that the mechanism of LuxS-dependent signaling in S. pneumoniae is not dependent upon accumulation of AI-2 in the extracellular medium.
The ΔluxS deletion strain shows aberrant transcriptional profiles for several groups of genes.
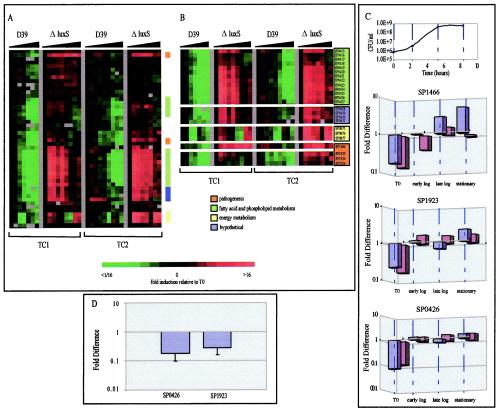
To obtain a comprehensive view of the extent to which LuxS activity impacts pneumococcal gene expression, we performed two individual time course experiments (TC1 and TC2) and compared the transcriptional profiles of the S. pneumoniae D39 parent strain and the ΔluxS deletion strain over the course of growth using a S. pneumoniae-specific spotted DNA microarray. We first examined the time zero transformed data to identify genes whose patterns of expression differed between the two strains over the duration of the time course. We then assembled these genes according to similarities in their expression patterns using a hierarchical clustering program (17). Figure 1A depicts one of the nodes resulting from this analysis. Within this node, one can clearly see distinct clusters of genes whose transcript abundance increases relative to time zero over time in ΔluxS. Arranging the genes in chromosomal order revealed that several of the most highly dysregulated genes appear to be organized into distinct operons (Fig. 1B) that could be grouped into several functional categories, including fatty acid and phospholipid biosynthesis, energy metabolism, and pathogenesis. The annotations for all of these genes are listed in Table 2.
FIG. 1.
The expression profiles of the luxS mutant and the isogenic parent strain over two time courses (TC1 and TC2). (A) Cluster diagram showing the expression profiles of the zero-transformed data for 46 genes from both time courses which met the filter criteria as explained in Materials and Methods. Experiments are organized by increasing time of culture as noted by the black triangles. Cultures were sampled at an initial cell density of 5 × 105 CFU/ml for each strain in TC1 and then at regular periodic intervals thereafter until a density of 4 × 108 CFU/ml was reached. Cultures were similarly sampled for TC2. Initial and final cell densities were 2 × 106 and 6.5 × 108 CFU/ml, respectively. Functional categories are noted and are based on the TIGR4 annotation. (B) A subset of the regulated genes arranged in chromosomal order. Putative operons composed of groups of consecutive genes are shown. (C) The difference in gene expression between mutant and parent strains at various time points for the non-zero-transformed data from SP1466 (putative hemolysin), SP1923 (pneumolysin), and SP0426 (accD), expressed on a log scale. Time zero, early-log, late-log, and stationary-phase time points were chosen. The blue and purple bars represent data from TC1 and TC2, respectively. (D) Real-time RT-PCR results. Differences in gene expression between mutant and parent strains at time zero were confirmed by real-time RT-PCR. RNA was prepared from at least two independent experiments, and the real-time assays were carried out in triplicate. The relative abundance of each message of interest in each sample was normalized to the level of rpoB-specific transcript. The data are depicted as the fold difference of the mutant versus that of the parent for each gene.
TABLE 2.
Putative annotations and cellular functions of genes affected in the luxS mutant strain
| TIGR annotationa | Gene symbol or putative identification | Cellular role |
|---|---|---|
| SP0024 | Conserved hypothetical protein | Unknown function |
| SP0025 | Hypothetical protein | Unknown function |
| SP0026 | Hypothetical protein | Unknown function |
| SP0173 | hexB (mutL) | DNA mismatch repair protein HexB |
| SP0415 | Enoyl-CoAb hydratase/isomerase family protein | Fatty acid and phospholipid metabolism, degradation |
| SP0416 | Transcriptional regulator, MarR family | Regulatory functions |
| SP0417 | 3-Oxoacyl-(acyl-carrier-protein) synthase III | Fatty acid and phospholipid metabolism |
| SP0418 | Acyl carrier protein | Fatty acid and phospholipid metabolism |
| SP0419 | Enoyl-(acyl-carrier-protein) reductase | Fatty acid and phospholipid metabolism |
| SP0420 | Malonyl CoA-acyl carrier protein transacylase | Fatty acid and phospholipid metabolism |
| SP0421 | 3-Oxoacyl-(acyl-carrier protein) reductase | Fatty acid and phospholipid metabolism |
| SP0422 | 3-Oxoacyl-(acyl-carrier-protein) synthase II | Fatty acid and phospholipid metabolism |
| SP0423 | Acetyl-CoA carboxylase, bitoin carboxyl carrier protein | Fatty acid and phospholipid metabolism |
| SP0424 | (3R)-Hydroxymyristoyl-(acyl-carrier-protein) dehydratase | Fatty acid and phospholipid metabolism |
| SP0425 | Acetyl-CoA carboxylase, biotin carboxylase | Fatty acid and phospholipid metabolism |
| SP0426 | Acetyl-CoA carboxylase, carboxyl transferase, beta subunit | Fatty acid and phospholipid metabolism |
| SP0427 | Acetyl-CoA carboxylase, carboxyl transferase, alpha subunit | Fatty acid and phospholipid metabolism |
| SP0429 | Hypothetical protein | Unknown function |
| SP0430 | Hypothetical protein | Unknown function |
| SP0431 | Conserved domain protein | Unknown function |
| SP0520 | Hypothetical protein | Unknown function |
| SP0521 | HIT family protein | Unknown function |
| SP0742 | Conserved domain protein | Unknown function |
| SP0744 | Conserved domain protein | Cytidine and deoxycytidylate deaminase family protein |
| SP0804 | 4-Methyl-5(β-hydroxyethyl)-thiazole monophosphate biosynthesis protein | Biosynthesis of cofactors, prosthetic groups, and carriers, thiamine |
| SP0875 | lacR | Regulatory functions, DNA interactions |
| SP0876 | 1-Phosphofructokinase, putative | Energy metabolism |
| SP0877 | PTS system, fructose-specific IIABC components | Signal transduction |
| SP0892 | Type I restriction-modification system, R subunit, putative | DNA metabolism, restriction/modification |
| SP1045 | Conserved hypothetical protein | Unknown function |
| SP1232 | Membrane protein | Cell envelope |
| SP1390 | murB | Cell envelope, biosynthesis of murein sacculus and peptidoglycan |
| SP1447 | Membrane protein | Cell envelope |
| SP1466 | Hemolysin, putative | Pathogenesis |
| SP1588 | Oxidoreductase, pyridine nucleotide-disulfide | Unknown function |
| SP1599 | truA | Protein synthesis, tRNA and rRNA base modification |
| SP1632 | Sensor histidine kinase | Regulatory functions, protein interactions |
| SP1644 | Conserved hypothetical protein | Unknown function |
| SP1725 | scrR | Regulatory functions, sucrose operon repressor |
| SP1922 | Conserved hypothetical protein | Unknown function |
| SP1923 | Pneumolysin | Toxin production, pathogenesis |
| SP1924 | Hypothetical protein | Unknown function |
| SP1926 | Hypothetical protein | Unknown function |
| SP2011 | Ribosomal large-subunit pseudouridine synthase, RluD subfamily | Protein synthesis, tRNA and rRNA base modification |
| SP2045 | Conserved hypothetical protein | Unknown function |
| SP2106 | Glycogen phosphorylase family protein | Energy metabolism, biosynthesis and degradation of polysaccharides |
| SP2173 | dltD | Cell envelope, biosynthesis and degradation of surface polysaccharides and lipopolysaccharides |
Putative operons are indicated in bold.
CoA, coenzyme A.
While zero transformation facilitates the comparison and identification of differential expression patterns between strains over time, it does not reveal baseline time zero differences in gene expression that may exist between strains. To address this, we examined the non-zero-transformed data for this cluster of genes and found that the greatest difference between the two strains existed at the very earliest points in the time course. The luxS mutant strain showed 5- to 15-fold decreases in transcript abundance for all genes in this cluster compared to that in the parent strain at time zero. The magnitude of these differences diminished as the time course progressed, but by the end of the time course, the expression levels of almost every gene in the luxS mutant had actually exceeded those in the parent strain. To illustrate this, the fold change in expression between mutant and parent strains for data from three genes, representing three of the putative operons identified in Fig. 1B, was graphed. Four points in the time courses were chosen where both cultures were at equivalent growth phases and cell densities, which is schematically represented by the top graph in Fig. 1C. The three remaining graphs depict the gene expression data for both time courses for SP1466, a putative hemolysin, SP1923, pneumolysin, and SP0426, accD, a gene involved in fatty acid biosynthesis. To confirm the differences in gene expression between the two strains revealed by microarray analysis, we performed real-time reverse transcription-PCR (RT-PCR) with gene-specific primers for two of the differentially regulated genes using the time zero RNA samples from the time courses. The results of this analysis demonstrated that the fold change in gene expression between mutant and parent was approximately the same as or greater than that seen by microarray measurement (Fig. 1D), validating our microarray as a reliable tool for accurately detecting relative changes in gene expression between two samples.
Quorum sensing is not the mechanism mediating the differential gene expression seen in the ΔluxS strain.
Quorum sensing refers to the ability of a bacterium to sense and respond to signaling molecules generated by other cells in a population when they reach a critical concentration. Typically, this occurs at relatively high cell density. However, the largest-magnitude differences in transcription that we observed occurred at relatively low cell density, suggesting that LuxS is affecting gene expression independent of high cell density and presumably a diffusible signaling molecule. To test this, we repeated the in vitro time course experiments in triplicate, harvesting cell-free supernatants from both the parent and the ΔluxS mutant strains at time zero and late log phase. We then tested these supernatants for the presence of a signaling molecule that could complement the ΔluxS transcriptional defect. Time zero was chosen because this is the time point at which we observed the greatest magnitude difference in differential gene expression between the parent and the mutant strain as determined by microarray and real-time RT-PCR analyses. If a small diffusible molecule, such as AI-2, were responsible for the observed transcriptional dysregulation, then we would expect that supernatant harvested from the parent strain would complement the ΔluxS transcriptional defect. Supernatants from late log phase were also tested, since this is typically the growth phase and time point when the effects of diffusible quorum-sensing molecules are observed.
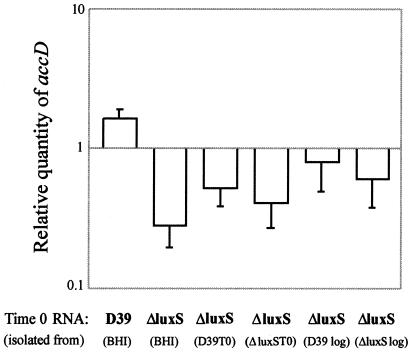
The parent strain was resuspended in BHI broth, and aliquots containing the same number of the ΔluxS mutant were resuspended in BHI, as well as the cell-free supernatants described above, and the strains were subsequently grown exactly as described for the microarray experiments. At time zero, RNA was isolated and was used to quantify one of the transcripts (accD) identified to be differentially regulated. As evidenced in Fig. 2, we did not observe complementation of the transcriptional defect, supporting our hypothesis that LuxS is affecting gene regulation in a cell-density-independent manner.
FIG. 2.
Cell-free supernatants harvested from the luxS mutant or D39 are unable to complement the transcriptional defect in the ΔluxS mutant. RNA was isolated at time zero from D39 or the ΔluxS mutant after incubation with either BHI medium or various cell-free supernatants (in parentheses) isolated from mutant and parent cultures at two different growth phases. RNA was prepared from three independently collected sets of supernatants, and the real-time assays were carried out in triplicate. The relative quantity of accD transcript from each strain under each condition was determined, and the values were normalized to the quantity of rpoB transcript. The average relative value under each condition is shown.
The ΔluxS mutant shows a persistence defect in the murine model of nasopharyngeal carriage.
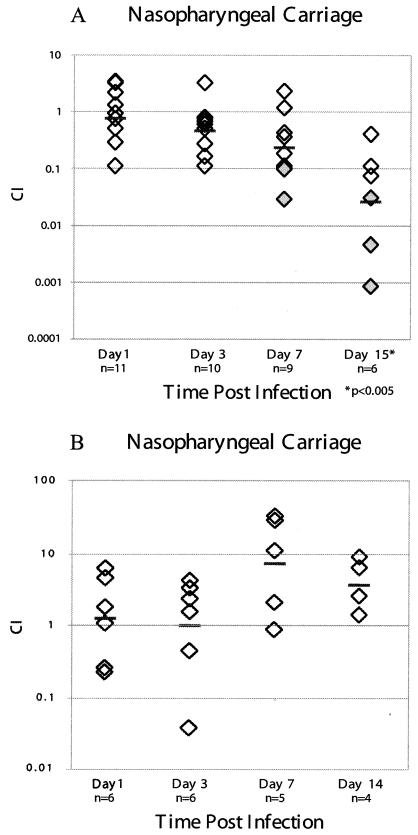
Because microarray analysis suggested that pneumolysin, a confirmed virulence factor of S. pneumoniae, and an additional putative hemolysin were dysregulated in the luxS mutant, we tested the ΔluxS mutant for defects in two animal models of pneumococcal infection. Both in vitro and in vivo mixed-infection experiments were carried out. Equivalent numbers of EJ1 and EJ3 cells grown to low cell density were mixed and grown together for >8 generations. The culture was sampled at several points during growth and plated for viable EJ1 and EJ3 colonies. At each time point sampled, equivalent numbers of both strains were recovered, suggesting that there is no in vitro competition phenotype. Animal infections were performed with CD-1 mice to determine the ability of the EJ3 mutant strain to persist asymptomatically in the nasopharynx of infected animals in competition with the EJ1 parent strain. Mice were challenged by nasal instillation with equivalent numbers of EJ1 and EJ3 bacteria (∼5 × 106 CFU of each strain; n = 35 mice). At four time points over a period of 2 weeks after infection, mice were sacrificed and nasal washes, lung tissue, and blood were assayed for colonization. No organisms were recovered from the lungs or blood at any of the time points, highlighting the utility of this infection to specifically examine asymptomatic carriage. The luxS mutant strain showed no statistically significant colonization differences after days 1 and 3, suggesting that the luxS mutant showed no defect in the ability to initially interact and colonize this host niche (Fig. 3A). However, indications that the mutant was less fit were found on day 7, when the parent was clearly beginning to outcompete the luxS mutant strain. By day 15, mutant bacteria were outcompeted by a factor of 53 (P < 0.005), with three of the six mice completely clearing the EJ3 bacteria while remaining colonized with EJ1 (day 15, mean log10 EJ1 = 2.98 CFU/ml of nasal wash [range, 1.19 to 3.78]). These data demonstrate a role for luxS in long-term, persistent carriage of pneumococcus in the nasopharynx.
FIG. 3.
Analysis of the ΔluxS mutant (A) and the pneumolysin mutant (B) in an animal model of nasopharyngeal carriage. Mutant and/or parent bacteria were recovered from the nasopharynx, lung, and blood after challenge of CD-1 mice with the parent (EJ1) and EJ3 or EJ5 mutants. The in vivo competitive index (CI) was calculated as described in the text; each diamond represents the CI for a single mouse in each set of competitions. A CI of less than 1 suggests that the mutant is less fit than the parent. Shaded diamonds indicate that no mutant bacteria were recovered from that animal and, therefore, 1 was substituted in the numerator when calculating the CI. The geometric mean of the CIs for all mice in a set of competitions is shown, and statistically significant data are indicated (*, P < 0.05). n is the number of animals examined per day.
Competition experiments were also performed with CD-1 mice to determine if a luxS deletion was impaired for causing invasive pneumococcal disease. Mice were challenged with a large volume (50 μl) by nasal instillation with equivalent numbers of EJ1 and EJ3 bacteria (∼2.5 × 105 CFU of each strain; n = 29 mice). Mice were sacrificed at three time points over a period of 3 days postinfection, after which the animals became moribund. Lung tissue and blood were assayed for colonization, and similar numbers of both mutant and parents strains were recovered from each location (data not shown), suggesting that luxS does not play a significant role in the disease process once the organism becomes invasive.
There has been a report that pneumolysin is required for nasopharyngeal colonization (33), and since the array data suggested that LuxS affects the expression of the pneumolysin locus, we constructed a strain bearing a deletion of this gene (EJ5) and performed competition experiments to determine whether this mutant strain would show a persistence defect similar to that for the luxS mutation during carriage. Figure 3B clearly shows that EJ5 was able to persist in the nasopharynx at least as well as the parent strain, suggesting that the persistence defect we observed for the luxS mutant is not due to the lack of pneumolysin expression.
DISCUSSION
Quorum sensing has been studied in several microbes, and a variety of cleverly employed bacterial genetic approaches has identified a multitude of genes and phenotypes that respond to this mode of regulation. However, timing is everything, and the success of these approaches in identifying target genes is dependent upon examining the relevant conditions at the appropriate time. The importance of this notion is evidenced from studies on luxS; although mutations in this gene have been constructed in at least 20 organisms, only a few of these mutants have shown significant phenotypic changes under the conditions examined. An alternative method to identify genes regulated by a particular process employs microarray technology, which profiles transcription on a genome-wide level. Because this technology allows the simultaneous examination of the transcription profiles for every gene in a genome, it is particularly well suited for identifying genes for which the impact of a specific condition is unknown. This technology has been used to experimentally scrutinize quorum-sensing responses in a few bacterial species (11, 12, 46, 48, 53, 55, 64). Importantly, the results of these analyses have in large part both confirmed data generated by previous genetically based studies and extended their findings, providing new insights into this complex biological process.
To assess the impact of LuxS on pneumococcal biology, we initiated our studies by constructing a strain bearing a deletion of luxS and used an S. pneumoniae-specific microarray to compare the expression profiles of this strain with that of its parent. Previously reported studies employing microarray technology to identify quorum-sensing-responsive genes have utilized similar types of strains for comparison. However, the experimental designs of these studies varied in several parameters, including whether RNA was isolated from a time course experiment or from a single time point and whether or not exogenous autoinducer was added to the medium. We performed two independent time course experiments without adding exogenous AI-2 to the medium. This was done for two reasons. First, the concentration of AI made by an organism in the wild that is necessary to induce quorum-sensing-regulated gene expression is not known for any quorum-sensing system, and thus the decision of how much inducer to add is arbitrary and not necessarily biologically relevant. Second, isolating AI-2 is problematic; it is currently not feasible to synthesize pure preparations of this molecule (56), and the specific activity can be highly variable among different preparations. Thus, making definitive statements about the direct effects of AI-2 on alterations in gene expression that occur after addition of in vitro-synthesized preparations of AI-2 or spent medium containing AI-2 is difficult. We anticipated that if LuxS were playing a role in quorum sensing in S. pneumoniae, comparison of two strains, one expressing luxS and the other not over a time course of growth, would distinguish genes displaying a LuxS-dependent pattern of expression. Our analysis of the data allowed us to readily identify a distinct cluster of 46 genes (∼2% of all S. pneumoniae genes) that reproducibly showed a differential pattern of expression over time between the two strains (Fig. 1A). Surprisingly, our data suggest that this regulation is not initiated at high cell density; to the contrary, the actual transcript abundance for all of these genes was 5- to 15-fold lower in the mutant than in the parent strain at early time points, when the cell density is low (Fig. 1C). Arranging the genes in chromosomal order and examining their annotations (http://www.tigr.org/tigr-scripts/CMR2/GenomePage3.spl?database=bsp) revealed that several of these genes are organized into at least five putative operon structures (Fig. 1B and Table 2) whose genes can be categorized into discrete functional classes dedicated to fatty acid and phospholipid metabolism, energy metabolism, and pathogenesis.
The biosynthesis of fatty acids is essential for all living organisms and is required for membrane lipid biogenesis. In bacteria, it is catalyzed by the type II fatty acid synthase system, which is composed of several small soluble proteins encoded by a discrete set of genes. Eleven out of the 19 genes assigned to the fatty acid and lipid biosynthesis category on the TIGR4 database exhibited markedly lower transcript abundance in the ΔluxS strain at early points during the time courses (Fig. 1B). Bacteria must carry out hundreds of biosynthetic reactions to produce basic building blocks—including amino acids, sugars, coenzymes, and nucleic acids—to sustain and promote growth. With the exception of the fructose-specific sugar transport operon, the only other biosynthetic process that is substantially and uniquely modulated in the ΔluxS strain is fatty acid biosynthesis. It is important to emphasize that great care was taken to ensure that the inocula for these time courses were taken from low-density (<5 × 106 CFU/ml) bacterial cultures that were approximating steady-state growth. Thus, one would not expect to observe acute changes in the biosynthetic capacity of these cells early during the time course; rather, biosynthesis of basic cellular components should remain relatively constant.
Of interest to us was the effect of the LuxS status in the cell on the transcription of both the pneumolysin operon and a putative hemolysin operon. Nothing has been reported on the putative hemolysin operon in the literature, and we are currently investigating the genes in this operon, both to characterize their functions and to define any potential contributions they may make to infection. Pneumolysin is a 53-kDa cholesterol-dependent cytolysin that is universally produced by pneumococcal clinical isolates and is required for virulence in animals (reviewed in reference 42). In contrast to other characterized cytolysins, it is localized to the cytoplasm and is not actively secreted by the cell. For most pneumococcal strains, including D39, it is released from the cell in stationary phase when S. pneumoniae undergoes cell lysis. While the protein and the multiple functions it carries out during invasive disease have been studied extensively, very little is known about the temporal and spatial regulation of the pneumolysin operon or how this impacts pneumococcal biology (33, 44).
What can be gleaned from these experimental results about how LuxS is impacting gene expression, particularly with regard to quorum sensing? While there is a preponderance of data supporting a role for AI-2 as a quorum-sensing signaling molecule in V. harveyi (4, 20, 52, 60), until recently this was the only organism for which luxS had been functionally linked to a particular and specific pathway. Importantly, despite the presence of luxS in over 30 different bacterial species to date (2, 71), homologs of the other genes involved in the V. harveyi AI-2 signaling pathway (luxP, -Q, -U, and -O) have not been found in other species, including S. pneumoniae. This has understandably led some investigators to question whether AI-2 signaling should be generally regarded as a quorum-sensing system in organisms other than V. harveyi (67, 68). Recently Schauder et al. (52) and Winzer et al. (67) have presented evidence that, in addition to the quorum-sensing role that LuxS plays in V. harveyi, LuxS also functions in the S-adenosylmethionine utilization pathway (the AdoMet pathway), where it catalyzes the conversion of S-ribosylhomocysteine to homocysteine and 4,5-dihydroxy-2,3-pentanedione, which spontaneously forms AI-2. This pathway plays an essential and dynamic role in modulating DNA, RNA, and protein synthesis in all organisms; the enzymes in the AdoMet pathway are highly active when cells are actively growing and, in general, their activity diminishes as cells enter stationary phase. Thus, as the end product of this pathway, AI-2 may serve as a barometer of cellular health. While the results of both our microarray expression analyses and the experiments assessing the ability of cell-free supernatants isolated from D39 to complement the ΔluxS phenotype imply that the differential regulation we observed between the two strains is not mediated by a quorum-sensing mechanism, they do suggest a previously unrecognized link between cellular well-being or growth phase and regulation of the above-mentioned operons. The transient nature of these expression differences could indicate that LuxS-dependent signaling acts as a first-line defense that can immediately provide the cell with vital information regarding environmental changes that could impact growth. Since this response is transient, subsequent additional signals are also integrated to catalog metabolic status. Indeed, there is a great deal of evidence for cells coordinating gene expression with nutritional status using a variety of small molecules (1, 6, 8, 30, 65, 70). Further studies examining the role of AI-2 in gauging metabolic status will help to validate its addition to this list.
Precise temporal gene expression is crucial for the survival of any organism, particularly in organisms that encounter and respond to several environments. While S. pneumoniae can asymptomatically colonize the nasopharynx of healthy adults, it also causes a range of diseases at a variety of host sites. Given the gene expression changes that we observed, particularly those seen in virulence factor expression, we challenged mice with a 1:1 ratio of parent to ΔluxS mutant to determine if and where in the host LuxS might play a role during infection. In an independent study, Stroeher et al. (57) have examined the effects of a defined luxS mutation on the virulence of S. pneumoniae and determined that their mutant (also a D39 derivative) was less able to access the lungs and the blood from the nasopharynx than the parent strain and was reduced in virulence when delivered intraperitoneally. Our data corroborate these observations to some extent and also extend these findings considerably. There are, however, differences between the studies. Unlike Stroeher et al., we did not see any statistically significant differences in the lung or the blood when mice were challenged with both parent and mutant strains in the pneumonia model of infection. This discrepancy may be due to differences in mouse strains used between the two studies, differences in luxS mutant construction, variations in the method used to infect animals, and/or the amount of time postinfection that animals were monitored. Based on the results of single strain infections by intraperitoneal inoculation, Stroeher et al. claim a role for luxS in survival and proliferation in the blood. While it is true that we did not observe a similar phenotype in CD-1 mice, their most compelling data were obtained from experiments, which were also performed as coinfections, done in a different mouse strain (BALB/c). More interesting to us, however, was the inoculum used: for the intraperitoneal infections (but not for the intranasal infections), bacteria were grown to a low cell density prior to infection, which is exactly when we observed transcriptional dysregulation as measured by microarray analysis.
Our data support Stroeher et al.'s (59) conclusion that luxS is not required during the initial stages of nasopharyngeal colonization. However, whereas Stroeher et al. examined carriage for 2 days postinfection, we followed asymptomatic infection for a full 15 days and found that the luxS mutant showed a statistically significant defect in its ability to persist in this niche.
Since our array data suggest that the luxS mutation has pleiotropic effects on gene transcription, we constructed a deletion in one of the affected genes in the luxS mutant, the pneumolysin gene, and used it to challenge mice to determine if the differential regulation of this gene was responsible for the carriage defect that we observed in the luxS mutant. Our results clearly showed that a pneumolysin-deficient strain could colonize and persist in the nasopharynx at least as well as the parent strain, indicating that the persistence defect we observed in the ΔluxS mutant must be due to something other than pneumolysin expression.
One question that remains unanswered is why some organisms have evolved the ability to use AI-2 as a means for taking census of the population, thereby coupling gene expression with cell density, while other AI-2-producing organisms seemingly have not. Of particular interest to us are the compelling data in support of a signaling role for LuxS activity in Vibrio vulnificus (34) and a close relative of pneumococcus, group A Streptococcus (GAS). In both of these organisms, a luxS mutation resulted in alterations in hemolytic activity, which occurs as the cells enter stationary phase, and also altered protease activity. In GAS, the expression of neither the hemolysin gene, streptolysin S (SLS), nor the gene encoding the protease (speB) was affected in a luxS mutant. Rather, the aberrant expression of SLS activity appeared to be due in part to the observation that in a luxS mutant background sagA, which encodes a transcriptional regulator required for SLS hemolytic activity, is overexpressed (38). In addition, the proteolytic SpeB defect was at the level of secretion and processing of the SpeB precursor. In V. vulnificus, the effects of a luxS mutation are more direct: Kim et al. observed an increase in hemolytic activity and a decrease in protease activity in a luxS mutant of V. vulnificus, which corresponded to overexpression of the hemolysin transcript and repression of the protease transcript (34). Both sets of authors hypothesized that the mechanism of this regulation is cell density dependent. However, their data do not preclude the possibility that this regulation could be due to the impacts of a luxS mutation on the abilities to sense and assess the growth potential of the environment. In our study, the effects of LuxS activity on pneumococcal transcription were seen very early on in the growth phase; perhaps in GAS and V. vulnificus, the activity of LuxS is likewise critical for monitoring cellular health at the exponential growth-stationary phase transition. Future studies aimed at resolving exactly how LuxS activity is integrated into specific gene regulatory networks will elucidate this.
While the mechanism of exactly how LuxS activity influences pneumococcal gene expression remains elusive, it is clear from the present studies that it does impact it. It also affects the ability of the organism to be carried in the nasopharynx, which is clearly important from both a medical and a biological standpoint. S. pneumoniae is transmitted person to person by asymptomatically infected individuals, not from those with invasive disease. A critical first step in understanding the pneumococcal disease process, therefore, is identifying those features that lead to in vivo survival, growth, and transmission. Because S. pneumoniae is a strict human pathogen, discriminating between tissue-specific factors required for establishing and maintaining the carriage state from those solely involved in the manifestation of invasive disease will facilitate this. The latter category has historically been more straightforward to study. Within the past several years, three large-scale screens aimed at identifying tissue-specific factors have been carried out (27, 36, 47). All three studies identified subsets of genes required for lung infection and bacteremia. However, Hava and Camilli also found that several, though not all, of the genes required for invasive disease were required for full levels of carriage in the nasopharynx (27), suggesting that these niche-specific distinctions are potentially important and can be distinguished. From the perspective of the host, nasopharyngeal carriage is considered to be a relatively quiescent state; however, from the perspective of the pneumococcus, this host niche is far from benign. It is precisely within this niche that S. pneumoniae must compete with other colonizing organisms and the host immune system. The ability to orchestrate the expression of a precise set of genes in this environment is crucial for the survival of this species, and identifying the class of genes, which includes luxS, whose products are important for survival on mucosal surfaces will significantly contribute to our understanding of the pneumococcal disease process.
Acknowledgments
E.A.J. is supported by NRSA grant 1F32 A151859-01. C.C.K. is supported by a Howard Hughes Predoctoral Fellowship and a Stanford Graduate Fellowship. This work was supported by a generous grant from Chiron Vaccines.
We thank Denise Monack, Erin Gaynor, Manuel Amieva, and Scotty Merrell for critical review of the manuscript. We also acknowledge David Hava for thoughtful scientific discussions and the Schneider and Myers laboratories for reagents and technical advice.
Editor: A. D. O'Brien
REFERENCES
- 1.Barker, M. M., T. Gaal, C. A. Josaitis, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305:673-688. [DOI] [PubMed] [Google Scholar]
- 2.Bassler, B. L. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2:582-587. [DOI] [PubMed] [Google Scholar]
- 3.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179:4043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. [DOI] [PubMed] [Google Scholar]
- 5.Blehert, D. S., R. J. Palmer, Jr., J. B. Xavier, J. S. Almeida, and P. E. Kolenbrander. 2003. Autoinducer 2 production by Streptococcus gordonii DL1 and the biofilm phenotype of a luxS mutant are influenced by nutritional conditions. J. Bacteriol. 185:4851-4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruckner, R., and F. Titgemeyer. 2002. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209:141-148. [DOI] [PubMed] [Google Scholar]
- 7.Burgess, N. A., D. F. Kirke, P. Williams, K. Winzer, K. R. Hardie, N. L. Meyers, J. Aduse-Opoku, M. A. Curtis, and M. Camara. 2002. LuxS-dependent quorum sensing in Porphyromonas gingivalis modulates protease and haemagglutinin activities but is not essential for virulence. Microbiology 148:763-772. [DOI] [PubMed] [Google Scholar]
- 8.Chang, D. E., D. J. Smalley, and T. Conway. 2002. Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Mol. Microbiol. 45:289-306. [DOI] [PubMed] [Google Scholar]
- 9.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 10.de Kievit, T. R., and B. H. Iglewski. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLisa, M. P., C. F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183:5239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derzelle, S., E. Duchaud, F. Kunst, A. Danchin, and P. Bertin. 2002. Identification, characterization, and regulation of a cluster of genes involved in carbapenem biosynthesis in Photorhabdus luminescens. Appl. Environ. Microbiol. 68:3780-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Saizieu, A., C. Gardes, N. Flint, C. Wagner, M. Kamber, T. J. Mitchell, W. Keck, K. E. Amrein, and R. Lange. 2000. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J. Bacteriol. 182:4696-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunny, G. M., and S. C. Winans. 1999. Cell-cell signaling in bacteria. ASM Press, Washington, D.C.
- 15.Dunny, G. M., and B. A. Leonard. 1997. Cell-cell communication in gram-positive bacteria. Annu. Rev. Microbiol. 51:527-564. [DOI] [PubMed] [Google Scholar]
- 16.Eisen, M. B., and P. O. Brown. 1999. DNA arrays for analysis of gene expression. Methods Enzymol. 303:179-205. [DOI] [PubMed] [Google Scholar]
- 17.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elvers, K. T., and S. F. Park. 2002. Quorum sensing in Campylobacter jejuni: detection of a luxS encoded signalling molecule. Microbiology 148:1475-1481. [DOI] [PubMed] [Google Scholar]
- 19.Fong, K. P., W. O. Chung, R. J. Lamont, and D. R. Demuth. 2001. Intra- and interspecies regulation of gene expression by Actinobacillus actinomycetemcomitans LuxS. Infect. Immun. 69:7625-7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freeman, J. A., and B. L. Bassler. 1999. Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J. Bacteriol. 181:899-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuqua, C., and E. P. Greenberg. 2002. Listening in on bacteria: acyl-homoserine lactone signalling. Nat. Rev. Mol. Cell Biol. 3:685-695. [DOI] [PubMed] [Google Scholar]
- 22.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 23.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giron, J. A., A. G. Torres, E. Freer, and J. B. Kaper. 2002. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol. Microbiol. 44:361-379. [DOI] [PubMed] [Google Scholar]
- 25.Gollub, J., C. A. Ball, G. Binkley, J. Demeter, D. B. Finkelstein, J. M. Hebert, T. Hernandez-Boussard, H. Jin, M. Kaloper, J. C. Matese, M. Schroeder, P. O. Brown, D. Botstein, and G. Sherlock. 2003. The Stanford Microarray Database: data access and quality assessment tools. Nucleic Acids Res. 31:94-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardman, A. M., G. S. Stewart, and P. Williams. 1998. Quorum sensing and the cell-cell communication dependent regulation of gene expression in pathogenic and non-pathogenic bacteria. Antonie Leeuwenhoek 74:199-210. [DOI] [PubMed] [Google Scholar]
- 27.Hava, D. L., and A. Camilli. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45:1389-1406. [PMC free article] [PubMed] [Google Scholar]
- 28.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoskins, J., P. Matsushima, D. L. Mullen, J. Tang, G. Zhao, T. I. Meier, T. I. Nicas, and S. R. Jaskunas. 1999. Gene disruption studies of penicillin-binding proteins 1a, 1b, and 2a in Streptococcus pneumoniae. J. Bacteriol. 181:6552-6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hung, S. P., P. Baldi, and G. W. Hatfield. 2002. Global gene expression profiling in Escherichia coli K12. The effects of leucine-responsive regulatory protein. J. Biol. Chem. 277:40309-40323. [DOI] [PubMed] [Google Scholar]
- 31.Ji, G., R. C. Beavis, and R. P. Novick. 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA 92:12055-12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones, S., B. Yu, N. J. Bainton, M. Birdsall, B. W. Bycroft, S. R. Chhabra, A. J. Cox, P. Golby, P. J. Reeves, S. Stephens, et al. 1993. The lux autoinducer regulates the production of exoenzyme virulence determinants in Erwinia carotovora and Pseudomonas aeruginosa. EMBO J. 12:2477-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kadioglu, A., S. Taylor, F. Iannelli, G. Pozzi, T. J. Mitchell, and P. W. Andrew. 2002. Upper and lower respiratory tract infection by Streptococcus pneumoniae is affected by pneumolysin deficiency and differences in capsule type. Infect. Immun. 70:2886-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, S. Y., S. E. Lee, Y. R. Kim, C. M. Kim, P. Y. Ryu, H. E. Choy, S. S. Chung, and J. H. Rhee. 2003. Regulation of Vibrio vulnificus virulence by the LuxS quorum-sensing system. Mol. Microbiol. 48:1647-1664. [DOI] [PubMed] [Google Scholar]
- 35.Kleerebezem, M., and L. E. Quadri. 2001. Peptide pheromone-dependent regulation of antimicrobial peptide production in gram-positive bacteria: a case of multicellular behavior. Peptides 22:1579-1596. [DOI] [PubMed] [Google Scholar]
- 36.Lau, G. W., S. Haataja, M. Lonetto, S. E. Kensit, A. Marra, A. P. Bryant, D. McDevitt, D. A. Morrison, and D. W. Holden. 2001. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40:555-571. [DOI] [PubMed] [Google Scholar]
- 37.Lazazzera, B. A., and A. D. Grossman. 1998. The ins and outs of peptide signaling. Trends Microbiol. 6:288-294. [DOI] [PubMed] [Google Scholar]
- 38.Lyon, W. R., J. C. Madden, J. C. Levin, J. L. Stein, and M. G. Caparon. 2001. Mutation of luxS affects growth and virulence factor expression in Streptococcus pyogenes. Mol. Microbiol. 42:145-157. [DOI] [PubMed] [Google Scholar]
- 39.Merritt, J., F. Qi, S. D. Goodman, M. H. Anderson, and W. Shi. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 71:1972-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 41.Miller, M. B., K. Skorupski, D. H. Lenz, R. K. Taylor, and B. L. Bassler. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303-314. [DOI] [PubMed] [Google Scholar]
- 42.Mitchell, T. J., and P. W. Andrew. 2000. Biological properties of pneumolysin, p. 279-286. In A. Tomasz (ed.), Streptococcus pneumoniae. Mary Ann Liebert, Inc., Larchmont, N.Y.
- 43.Novick, R. P. 1999. Regulation of pathogenicity in Staphylococcus aureus by a peptide-based density-sensing system, p. 129-146. In G. M. Dunny and S. C. Winans (ed.), Cell-to-cell signaling in bacteria. ASM Press, Washington, D.C.
- 44.Ogunniyi, A. D., P. Giammarinaro, and J. C. Paton. 2002. The genes encoding virulence-associated proteins and the capsule of Streptococcus pneumoniae are upregulated and differentially expressed in vivo. Microbiology 148:2045-2053. [DOI] [PubMed] [Google Scholar]
- 45.Pesci, E. C., and B. H. Iglewski. 1999. Quorum sensing in Pseudomonas aeruginosa, p. 147-158. In G. M. Dunny and S. C. Winans (ed.), Cell-to-cell signaling in bacteria. ASM Press, Washington, D.C.
- 46.Peterson, S., R. T. Cline, H. Tettelin, V. Sharov, and D. A. Morrison. 2000. Gene expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays. J. Bacteriol. 182:6192-6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polissi, A., A. Pontiggia, G. Feger, M. Altieri, H. Mottl, L. Ferrari, and D. Simon. 1998. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66:5620-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rimini, R., B. Jansson, G. Feger, T. C. Roberts, M. de Francesco, A. Gozzi, F. Faggioni, E. Domenici, D. M. Wallace, N. Frandsen, and A. Polissi. 2000. Global analysis of transcription kinetics during competence development in Streptococcus pneumoniae using high density DNA arrays. Mol. Microbiol. 36:1279-1292. [DOI] [PubMed] [Google Scholar]
- 49.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 50.Salmond, G. P., B. W. Bycroft, G. S. Stewart, and P. Williams. 1995. The bacterial “enigma”: cracking the code of cell-cell communication. Mol. Microbiol. 16:615-624. [DOI] [PubMed] [Google Scholar]
- 51.Sanderson, K. E., A. Hessel, and B. A. D. Stocker. 1996. Strains of Salmonella typhimurium and other Salmonella species used in genetic analysis, p. 2496-2503. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 52.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 53.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sperandio, V., A. G. Torres, J. A. Giron, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:5187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sperandio, V., A. G. Torres, B. Jarvis, J. P. Nataro, and J. B. Kaper. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. USA 100:8951-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stroeher, U. H., A. W. Paton, A. D. Ogunniyi, and J. C. Paton. 2003. Mutation of luxS of Streptococcus pneumoniae affects virulence in a mouse model. Infect. Immun. 71:3206-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sturme, M. H., M. Kleerebezem, J. Nakayama, A. D. Akkermans, E. E. Vaugha, and W. M. de Vos. 2002. Cell to cell communication by autoinducing peptides in gram-positive bacteria. Antonie Leeuwenhoek 81:233-243. [DOI] [PubMed] [Google Scholar]
- 59.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 62.Tiraby, G., M. S. Fox, and H. Bernheimer. 1975. Marker discrimination in deoxyribonucleic acid-mediated transformation of various Pneumococcus strains. J. Bacteriol. 121:608-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tomasz, A. 1965. Control of the competent state in Pneumococcus by a hormone-like cell product: an example for a new type of regulatory mechanism in bacteria. Nature 208:155-159. [DOI] [PubMed] [Google Scholar]
- 64.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wanner, B. L., and M. R. Wilmes-Riesenberg. 1992. Involvement of phosphotransacetylase, acetate kinase, and acetyl phosphate synthesis in control of the phosphate regulon in Escherichia coli. J. Bacteriol. 174:2124-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams, P., M. Camara, A. Hardman, S. Swift, D. Milton, V. J. Hope, K. Winzer, B. Middleton, D. I. Pritchard, and B. W. Bycroft. 2000. Quorum sensing and the population-dependent control of virulence. Philos. Trans. R. Soc. Lond. Ser. B 355:667-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Winzer, K., K. R. Hardie, N. Burgess, N. Doherty, D. Kirke, M. T. Holden, R. Linforth, K. A. Cornell, A. J. Taylor, P. J. Hill, and P. Williams. 2002. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 148:909-922. [DOI] [PubMed] [Google Scholar]
- 68.Winzer, K., K. R. Hardie, and P. Williams. 2002. Bacterial cell-to-cell communication: sorry, can't talk now—gone to lunch! Curr. Opin. Microbiol. 5:216-222. [DOI] [PubMed] [Google Scholar]
- 69.Winzer, K., and P. Williams. 2001. Quorum sensing and the regulation of virulence gene expression in pathogenic bacteria. Int. J. Med. Microbiol. 291:131-143. [DOI] [PubMed] [Google Scholar]
- 70.Wolfe, A. J., D. E. Chang, J. D. Walker, J. E. Seitz-Partridge, M. D. Vidaurri, C. F. Lange, B. M. Pruss, M. C. Henk, J. C. Larkin, and T. Conway. 2003. Evidence that acetyl phosphate functions as a global signal during biofilm development. Mol. Microbiol. 48:977-988. [DOI] [PubMed] [Google Scholar]
- 71.Xavier, K. B., and B. L. Bassler. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6:191-197. [DOI] [PubMed] [Google Scholar]
- 72.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]