Abstract
The mammalian diaphragm muscle is essential for respiration, and thus it is among the most critical of the skeletal muscles in the human body. Defects in diaphragm development, leading to congenital diaphragmatic hernias (CDH), are common birth defects and result in severe morbidity or mortality. Given its functional importance and the frequency of congenital defects, an understanding of diaphragm development normally and during herniation is important. We review the current knowledge of the embryological origins of the diaphragm, diaphragm development and morphogenesis, and the genetic and developmental etiology of diaphragm birth defects.
Keywords: diaphragm, Congenital Diaphragmatic Hernia, CDH, muscle, development, tendon
Introduction
Of all the skeletal muscles in the human body, the diaphragm is one of the most essential. It is a domed muscle separating the thoracic and abdominal cavities and is critical for respiration. Birth defects and diseases that affect diaphragm structure and function reveal its critical function. Congenital diaphragmatic hernias (CDH) result from defects in diaphragm development and are both common and frequently lethal [1]. In addition, the lethality associated with many myopathies, such as Duchenne muscular dystrophy, is due to failure of the diaphragm and other respiratory muscles [2]. However, despite its critical function, knowledge of the molecular and cellular mechanisms regulating diaphragm development normally and during herniation is limited. In this review, we will describe current knowledge about diaphragm development, review the genetic and developmental etiology of diaphragm birth defects, and finally compare the developmental processes regulating limb and diaphragm muscle morphogenesis.
Diaphragm Structure, Function, and Evolution
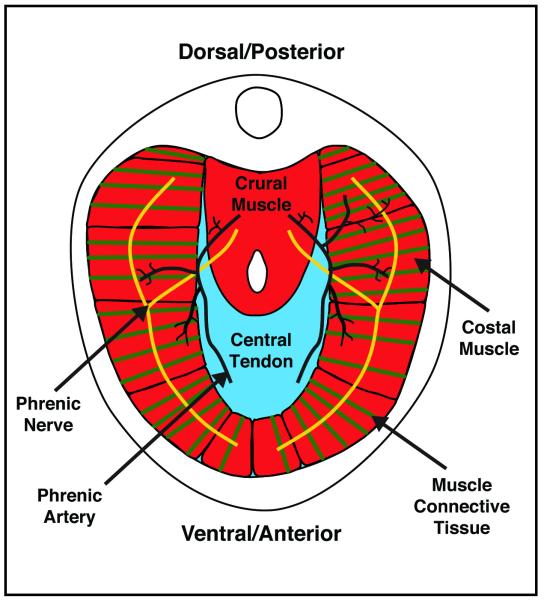
The diaphragm muscle is composed of two domains [3]. The costal diaphragm is a thin domed sheet of muscle composed of a radial array of myofibers extending laterally from the ribs and medially to a central tendon (Figure 1). The crural diaphragm is thicker and located more posteriorly (dorsally), where it attaches to the vertebrae and surrounds the esophagus and aorta (Figure 1). Medially, the myofibers of both the costal and crural muscles insert into the central tendon. The central tendon is located at the apex of the domed diaphragm, holding the diaphragm muscle domains together. Caudally, it attaches to the liver via the falciform and coronary ligaments. The tendon is a connective tissue sheet composed of extracellular matrix and the tendon cells that secrete it [3]. Although less visible in whole mount, each of the myofibers of the costal and crural muscles is surrounded by muscle connective tissue. The right and left halves of the diaphragm are innervated by the right and left phrenic motor nerves [4]. These nerves originate from the cervical nerves, C3-C5, descend along the interior of the vertebrae, pierce the left and right diaphragm, and spread posteriorly (dorsally) and anteriorly (ventrally) to innervate myofibers of the crural and costal muscles. The diaphragm is vascularized by the phrenic, internal thoracic, and intercostal arteries [5].
Figure 1.
Major components of the adult diaphragm.
The diaphragm has multiple functions, with its principle function being its critical role in respiration. During respiration, the actions of the diaphragm are central for inspiration. Contraction of the diaphragm muscle flattens the dome-shaped diaphragm and central tendon, which in turn expands the volume of the thoracic cavity, reduces thoracic pressure, and allows air to flow into the lungs [6]. Interestingly, the diaphragm is not strictly required for respiration when humans or animals are resting. However, analysis of rats, dogs, and humans with bilateral diaphragmatic paralysis (in which the phrenic nerves are severed), demonstrate that the diaphragm is required for respiration and lung ventilation during supine posture (particularly during rapid-eye-movement sleep) and vigorous activity [7-9]. In addition to respiration, the diaphragm (particularly the crural domain) has functional roles in swallowing and emesis [10]. Finally, the diaphragm also has a passive functional role. The diaphragm serves as a barrier between the thoracic and abdominal cavities. The importance of this barrier function is dramatically apparent in newborns with Congenital Diaphragmatic Hernias (CDH), whereby a weak or incompletely formed diaphragm allows abdominal contents to herniate into the thoracic cavity and impair lung development [1].
The presence of a muscularized diaphragm is unique to and, in fact, a defining characteristic of mammals [11, 12]. While a muscularized diaphragm is unique to mammals, the presence of a septum separating the lungs from the abdominal viscera is an ancient character, and some variant of this septum is present in reptiles and birds (but not in fish and amphibians) [12, 13]. In mammals, this septum becomes muscularized to form the diaphragm. It has been proposed that the diaphragm evolved in mammals as a stabilizer of the abdominal viscera and an inspiratory muscle [12]. Together these functions of the diaphragm allowed mammals to evolve as high-performance homeotherms, capable of concomitant respiration and locomotion. Thus the question of how the mammalian diaphragm evolved is an important question. Presumably, developmental innovations were critical for the development of a muscularized diaphragm, but the molecular and cellular nature of these innovations is currently unknown.
Embryological Sources of the Diaphragm
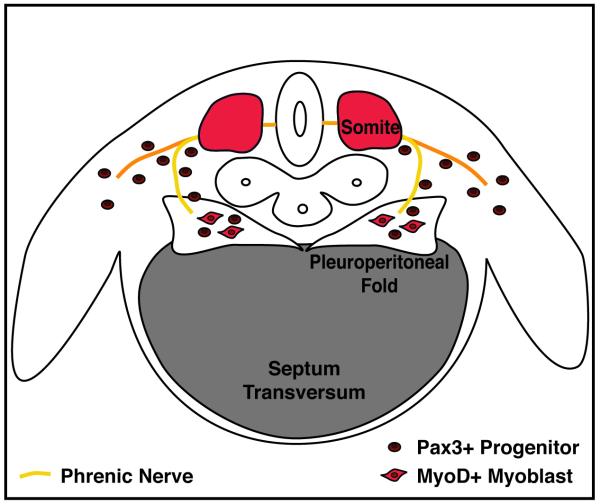
The diaphragm develops from multiple embryonic sources. The muscle and its associated connective tissue and central tendon develop from three sources: the septum transversum, the pleuroperitoneal folds, and the somites.
The septum transversum is the first structure present in the developing diaphragm and serves as the initial barrier between the thoracic and abdominal cavities (Figure 2). In all vertebrates, the septum transversum is a thin, mesodermal sheet of tissue that separates the heart from the liver [12]. In many reptiles and all birds and mammals, septa separate the heart and lungs from the liver and the rest of the abdominal contents [12]. Expression analysis of Cited2 (originally called Mrg1), which is expressed in the mouse septum transversum, suggests that the septum originates as the rostral-most mesoderm in the embryo [14]. During the process of foregut invagination, folding causes the Cited2+ tissue to lie in a position caudal to the heart and form the septum transversum, which can be detected by the 9-12 somite stage in mice. Other genes, such as α4-integrin [15] and Pb×3 [16] have also been shown to be expressed in the mouse septum transversum by embryonic day (E) 9. It is unclear what the septum gives rise to (if anything) in the adult, although some investigators have suggested that the septum gives rise to the non-muscle and central tendon components of the diaphragm [17]. However, without markers or genetic tools to follow the fate of the septum, it is still unclear whether or what the septum contributes to in the adult diaphragm. Although not explicitly tested, during development the septum is likely to provide a scaffold for diaphragm morphogenesis.
Figure 2.
Embryonic sources of the diaphragm. Mouse E12.5 developing diaphragm. Diaphragm develops from the septum transversum, pleuroperitoneal folds, and the somites and is innervated by the phrenic nerve.
The pleuroperitoneal folds (also referred to as the posthepatic mesenchymal plate) are the second important component of the developing diaphragm. The pleuroperitoneal folds are two transient, pyramidal-shaped structures lying on either side of the esophagus that protrude from the body wall between the pleural and peritoneal cavities (Figure 2). The pleuroperitoneal folds have been identified both histologically in sections [4, 17] and through scanning electron microscopy [18, 19]. Molecularly, the cells comprising the pleuroperitoneal folds express the nuclear receptor NR2F2 (also known as Coup-TFII) and the transcription factors Wt1 and Gata4 [20]. In mouse, the PPF are first present at E11 [17], proliferate (to be most prominent at E12.5), then appear to spread ventrally, and eventually are thought to fuse with the septum [4, 17]. It is currently unclear whether the pleuroperitoneal folds are simply transient embryonic structures with no adult derivatives or if they give rise to cells or tissues of the adult diaphragm.
Similar to trunk and limb muscles, the somites are the source of the diaphragm’s muscle cells [Figure 2; 4, 21, 22]. Evidence that the diaphragm’s muscle originates from the somites comes from analysis of the transcription factor Pa×3 and the receptor tyrosine kinase Met (also known as c-Met). Both of these genes are strongly expressed in the muscle progenitors within the somites [23-25]. Furthermore, these genes are required for the muscle progenitors to migrate from the somites to form muscles in the limb and trunk. Strikingly, mice with null mutations in either Pa×3 or Met have no limb or diaphragm muscles [including both the costal and crural muscle domains; 21, 26, 27, 28]. Thus the diaphragm derives from migratory muscle progenitors originating from the somites. Expression studies [4, 21, 22] show that the diaphragm’s muscle progenitors originate from the cervical somites, likely C3-C5.
Migration of Muscle and Nerve to the Developing Diaphragm
The muscle progenitors and the phrenic nerve axons migrate from the somites and neural tube, respectively, to the developing diaphragm (Figure 2). The target of their migration is the pleuroperitoneal folds [4, 22]. In mice, the phrenic nerve axons and muscle progenitors migrate toward the folds by E10.5 [4, 21, 22]. The phrenic nerve migrates after the muscle progenitors and initially migrates with nerves of the brachial plexus before separating and targeting the folds [29]. When the nerves and muscle progenitors reach the folds, the folds are located at the level of cervical somites. However, gradually the folds and the developing diaphragm with its nerves and muscle descend caudally, ultimately to lie at the thoracic/lumbar boundary [4].
The pleuroperitoneal folds are likely to be an important source of signals guiding the migration of nerves and muscle to the developing diaphragm. Our knowledge of the molecular nature of these signals is limited. Neural cell adhesion molecule (NCAM) and low-affinity nerve growth factor receptor (NGFR) are expressed along the path from the neural tube to the pleuroperitoneal folds and may guide outgrowth of the phrenic nerve [4]. In addition, Hepatocyte Growth Factor (HGF), the ligand for the Met receptor, is expressed along the migratory path for both the muscle progenitors and the nerves [21]. The finding that HGF null mice have no diaphragm muscle strongly suggests that HGF is important for guidance of the myogenic progenitors to the developing diaphragm [30]. HGF may also be critical for phrenic nerve development [31, 32].
Diaphragm Morphogenesis
To produce a functional diaphragm, the morphogenesis of the muscle, muscle connective tissue, and tendon derived from these different embryonic sources must not only be coordinated with each other, but with the nerves and vasculature.
After reaching the pleuroperitoneal folds, the muscle progenitors undergo the processes of myogenesis and morphogenesis. During myogenesis, the muscle progenitors differentiate into multinucleate myofibers. Similar to trunk and limb muscle, diaphragm progenitors initially express the transcription factors Pa×3 and Pa×7. These progenitors become committed myoblasts expressing the transcription factors Myf5 and MyoD, differentiate into Myogenin+ myocytes, and finally fuse to form multinucleate myofibers [22, 33]. Pa×3/7+ muscle progenitors and MyoD+ myoblasts are present in the pleuroperitoneal folds in mice by E12.5 [E13.5 in rats; 22]. Subsequently the progenitors differentiate into myoblasts and then fuse into myofibers, which are assembled into the costal and crural muscles. The morphogenesis of the costal diaphragm has been described in more detail [22]. Myofibers first begin differentiating in posterolateral regions of each left and right hemi-diaphragms. Between E12.5 and E15.5 a wave of differentiation expands both ventrally and dorsally as well as medially (towards the central tendon) and laterally (towards the ribs) of each hemi-diaphragm. Morphogenesis of the costal diaphragm is largely complete by E15.5. It is still unclear what drives this morphogenetic process, what orients the myofibers to form a radial array, and why the central tendon region is devoid of muscle.
The morphogenesis of the diaphragm’s muscle connective tissue and central tendon and their relationship to the transverse septum and the pleuroperitoneal folds remain poorly understood. Based on histological analysis of sections through developing mouse embryos, it has been proposed that the septum transversum, initially present on the cranial surface of the liver, remains in place and gives rise to the central tendon [17]. The pleuroperitoneal folds have a complicated morphogenesis. After E12.5 the folds, initially located adjacent to the cervical somites, descend caudally (along with the phrenic nerves and muscle progenitors) to reach the thoracic/lumbar boundary [4]. Using scanning electron micrographs of a series of developing embryos, the pleuroperitoneal folds then appear to spread to cover the cranial surface of the liver and fuse with the septum transversum [18, 19]. The molecular and cellular nature of the interactions between the folds and the septum is not known. Also, it is unclear what the ultimate fate of the folds is; the folds may simply be transient embryonic structures or they may give rise to the non-muscular parts of the diaphragm. Thus it is currently unknown whether the muscle connective tissue and central tendon arise from the septum transversum, the pleuroperitoneal folds, or some other embryonic source.
The close proximity of the muscle and non-muscle cells, which presumably give rise to the muscle connective tissue and central tendon, suggest that cell-cell interactions between these tissues may be critical for proper diaphragm development. Interestingly, analysis of Met null mice (in which muscle progenitors do not migrate into the developing diaphragm) demonstrate that the connective tissue is present even in the absence of muscle [34]. Thus the connective tissue develops, at least initially, independent of muscle, although it is unclear whether distinct tissues such as the central tendon still form. Whether the diaphragm’s muscle requires signals from the developing muscle connective tissue and/or central tendon has not yet been tested.
Innervation of the diaphragm is critical for the development of a fully functional diaphragm. After reaching the pleuroperitoneal folds, the phrenic nerves must spread, branch, and innervate the developing diaphragm by forming neuromuscular junctions with differentiated myofibers. After reaching the pleuroperitoneal folds, by E13.5 the phrenic nerve splits into three branches: sternocostal, dorsocostal, and crural branches [35]. The sternal branches extend and cross past the midline of the diaphragm before retracting to reach their final positions [22]. Subsequently, the three major branches send out short secondary branches, arborize, and then form neuromuscular junctions with the costal and crural muscle. The molecular signals and cellular interactions controlling axon outgrowth, branching, and formation of neuromuscular junctions are beginning to be elucidated. The receptor protein tyrosine phosphatases σ and δ are required for the phrenic nerve branching, as the phrenic nerves reach the pleuroperitoneal folds but fail to extend and branch appropriately in mice mutant for these phosphatases [35]. Additionally, Hoxa5 and Hoxc5 are required for proper secondary branching of the phrenic nerve, as deletion of these genes in the motor neurons results in a severe reduction of branching as well as limited synapse formation with the muscle [36]. Interactions between muscle and nerve are also critical for secondary branching and neuromuscular junction formation [37]. Conditional mutagenesis experiments in mice found that secondary branching and arborization of the phrenic nerve is regulated via β-catenin within the muscle, demonstrating that muscle-derived signals regulate phrenic nerve development [38, 39]. In addition, a multitude of studies have shown that formation of phrenic nerve neuromuscular junctions involves a complex interplay of muscle-nerve retrograde and anterograde signaling [40, 41].
Finally, vascularization of the diaphragm is also critical for diaphragm development. This is an area of limited research. However, recent research using XLacZ4 transgenic mice, which label the nuclei of vascular smooth muscle cells, shows the vascularization of the diaphragm by the phrenic, intercostal, and internal thoracic arteries. Use of the XLacZ4 transgenic mice permits visualization of the complex branching structures of both the arteries and veins. This work provides the first detailed description of diaphragm vascularization and holds promise for future research [5].
Congenital Diaphragmatic Hernias
Congenital diaphragmatic hernias (CDH) are common birth defects (1:3000) that often have severe medical consequences [1]. CDH occurs from a failure of the diaphragm to form properly, resulting in weak or incomplete regions of muscle. Through these weakened or incomplete regions the abdominal contents herniate into the thoracic cavity. In turn, the herniated abdominal contents impede lung development, leading to hypoplastic lungs. Although it is generally thought that lung hypoplasia simply results from physical impedance of lung growth by the herniated tissue, some genetic defects associated with CDH directly affect both diaphragm and lung development [42, 43]. The lung hypoplasia accompanying CDH is the main cause of the high morbidity and mortality associated with CDH. In spite of medical intervention, the mortality rate for CDH is 50% and results from respiratory failure [1, 44-46]. For patients that survive, chronic respiratory and neurodevelopmental problems are common [42].
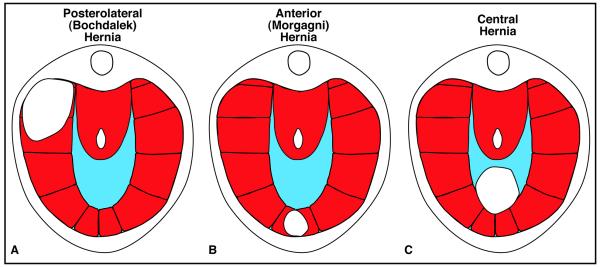
Diaphragmatic hernias vary both in the region of the diaphragm in which they form and in size. In the majority of cases (90%), hernias form in the posterior lateral diaphragm, and these posterolateral hernias (termed Bochdalek hernias) develop predominantly on the left side of the diaphragm [Figure 3A; 1, 46, 47]. Posterolateral hernias are the form of CDH most commonly associated with lung hypoplasia because the abdominal contents herniate into the posterior pleural cavity where the developing lungs are forming. Hernias also form in anterior regions (termed Morgagni hernias) or in the central tendon (termed central hernias) [Figure 3B and C;1]. However, hernias in these regions generally have less severe consequences [48]. Diaphragmatic hernias also vary in size [49], with larger hernias having more critical impacts on health, while smaller hernias may be asymptomatic.
Figure 3.
Major types of congenital diaphragmatic hernias.
Diaphragmatic hernias can develop in isolation or in association with other developmental abnormalities [1]. CDH that arises in association with other abnormalities are often associated with recognized syndromes. Syndromes that commonly include CDH are Fryns Syndrome, Denys-Drash syndrome, Cornelia de Lange syndrome, Donnai-Barrow syndrome, Wolf-Hirschhorn syndrome, Ehlers-Danlos Syndrome, and focal dermal hypoplasia [50-56].
Diaphragm morphogenesis requires muscle progenitors to migrate to the developing diaphragm and differentiate into muscle. Deletion of genes required for delamination and migration of muscle precursors from the somite, such as HGF and Met, result in a failure of muscle to migrate into the diaphragm from the somite and leads to an amuscularized diaphragm [21, 26]. Similarly, deletion of genes required for myogenesis, such as MyoD or Myogenin, result in thinner or absent diaphragm muscle [57, 58]. Interestingly, the complete loss of muscle in mouse Met mutants does not result in herniation of abdominal contents through the diaphragm [34]. Thus the lack of diaphragm muscle is not sufficient to allow herniation of abdominal contents through the diaphragm.
Formation of the diaphragm also requires that the connective tissue forms with proper structural integrity, and mutations that inhibit development of the extracellular matrix result in CDH. Mice with null mutations for lysyl oxidase, an enzyme responsible for cross-linking collagen, develop central tendon hernias [59]. The central tendon of these mice is unable to withstand pressure from the growing liver and allows herniation of abdominal contents through the weakened connective tissue. Similarly, patients with mutations in Collagen 3a1 develop CDH [54]. In these patients, reoccurrence of herniation can occur because of the weakened state of the diaphragm. Together, the lysyl oxidase and Collagen 3a1 mutations indicate that loss of connective tissue integrity can be a cause of CDH.
The molecular pathway most frequently associated with CDH is the retinoic acid (RA) signaling pathway. Much of the initial work investigating the development of CDH in rodent models used the teratogen nitrofen, which inhibits RA signaling [17, 60, 61]. Administration of nitrofen, as well as other teratogens that inhibit RA signaling, to pregnant mice or rats induces the development of CDH in their fetuses [62]. Furthermore, simultaneous administration of RA with these teratogens is sufficient to rescue CDH [62]. This verifies that the production of RA is critical for the developing diaphragm. Further evidence for the importance of RA signaling comes from the finding that mutations in Stra6, which is involved in retinol uptake and the production of retinoic acid, are present in some humans with CDH [63, 64]. Also, mutations in the retinoic acid receptors RARa and RARb cause CDH in a small percentage of mice [65]. Production of RA requires processing by Retinal Dehydrogenase (RALDH). RALDH2, the only RALDH enzyme present in the diaphragm, has been localized by immunofluorescence to the non-muscle cells of the pleuroperitoneal folds [62]. This suggests that the non-muscle cells are the source of RA in the developing diaphragm. An intriguing hypothesis is that RA signals from the non-muscle cells to the muscle cells are important for proper diaphragm morphogenesis.
In addition to mutations associated with RA signaling, other genetic causes underlying CDH have been identified by examining chromosomal abnormalities in CDH patients. Three of the frequently identified chromosomal regions are 8p23.1, 8q22-23, and 15q26.1-26.2, with the most commonly suggested candidate genes in these regions being Gata4, Zfpm2 (also known as Fog2), and Nr2f2 (also known as Coup-TFII), respectively. Significantly, these three genes can interact with one another and have also been proposed to interact with the RA signaling pathway [66, 67]. Although Gata4 has long been implicated in the formation of CDH, until recently no coding variants had been identified within this gene in CDH patients. Whole exome sequencing of a familial case of CDH has identified a novel variant in a highly conserved arginine residue in the zinc finger domain of Gata4 that is predicted to be pathogenic [68]. Additional enhancer and intronic variants in Gata4 in CDH have also been recently identified [69, 70]. Functional analysis of the role of Gata4 in the formation of CDH in mice is limited, as mice with homozygous null mutations of Gata4 die prior to the formation of the diaphragm due to heart defects. However, in a study of mice heterozygous for a deletion of Gata4, 14% of the mice developed anterior congenital diaphragmatic hernias, and 29% of mice had some type of diaphragm defect [43], providing further evidence for Gata4 as a candidate gene in the 8p23.1 region. Zfpm2 has also been implicated as important for CDH and is a binding partner of Gata4 [Fog2 is an abbreviation for Friend of Gata 2; 71]. Mice with null mutations of Zfpm2 develop CDH [42]. Further evidence for a role of Zfpm2 in CDH comes from the finding of a de novo mutation resulting in a premature stop in Zfpm2 in a child with CDH [42]. Finally, Nr2f2 is located in the 15q26.1-26.2 chromosomal region that is frequently deleted in CDH patients [72]. Although no causal variants within Nr2f2 have been reported, mice with a conditional deletion of Nr2f2 in the foregut mesentery develop CDH [73]. In addition to these genes, a multitude of other genes have been implicated by human genetic studies [1]. Also, a recent screen of genes expressed in the developing diaphragm in mouse has identified a list of 27 candidate CDH-causing genes [74].
Despite the frequency with which CDH occurs and extensive research identifying genes implicated in the formation of CDH, little is known about the molecular and cellular mechanisms by which these genetic mutations cause the diaphragm to form aberrantly. First, it is unclear in which tissues of the developing diaphragm mutations in the CDH genes are causative. Immunofluorescence of candidate CDH genes shows that these genes are predominantly expressed in non-muscle cells of the pleuroperitoneal folds (Clugston 2007). This suggests that expression of CDH genes in these cells is critical. However, conditional deletion of these genes in particular cells (e.g. the non-muscle cells of the folds) will be necessary to determine in which cells CDH-genes are important. Second, while CDH is undoubtedly caused by defects in diaphragm morphogenesis, what particular cells or tissues behave aberrantly is not known. The incomplete development of the diaphragm could be due to defects in proliferation and/or survival of myogenic progenitors, aberrant migration of myogenic cells, or defects in muscle differentiation or morphogenesis. Alternatively, the incomplete development of the diaphragm could result from aberrant development of the septum transversum or defects in the formation or spread of the pleuroperitoneal folds. Finally, another unknown aspect of diaphragmatic herniation is the question of why hernias only form in local regions of the diaphragm, as opposed to throughout the entire diaphragm muscle. The local and variable nature of the defects suggests that local changes in levels, timing, or site of gene expression during diaphragm development may regulate the size and location of hernias.
Comparison of Diaphragm and Limb Muscle Development
Comparison of diaphragm with limb muscle development reveals many similarities in their regulation, a few differences, and suggests avenues of future research. Both diaphragm and limb muscles derive from populations of migratory muscle progenitors. These progenitors delaminate from the somites and migrate into either the pleuroperitoneal folds or the limb buds. Migration of muscle precursors to both sites relies on the signaling of HGF to Met+ muscle precursors to induce delamination and migration [26]. However, while Lb×1 marks progenitors migrating either into the developing diaphragm or limb [21], surprisingly, null mutations in Lb×1 affect only the migration of limb progenitors and not diaphragm progenitors [75, 76]. Once in the developing diaphragm or limb, myogenic progenitors go through a similar process of myogenesis whereby progenitors become committed myoblasts, differentiate into myocytes, and fuse into myofibers. In the limb, the processes regulating muscle morphogenesis and patterning are beginning to be elucidated. Lateral plate-derived muscle connective tissue is critical for determining the pattern of individual limb muscles [77, 78]. How the pattern of costal and crural diaphragm muscles is established is currently unknown. If diaphragm muscle morphogenesis is similar to that in the limb, the diaphragm’s muscle connective tissue may be an important determinant of the diaphragm’s muscle pattern.
Conclusion
The diaphragm is a unique mammalian muscle, essential for respiration. Unfortunately, defects in diaphragm development, leading to congenital diaphragmatic hernias, are common birth defects and result in severe morbidity and mortality. Given its functional importance and the frequency of congenital defects, an understanding of the genetic, cellular, and morphogenetic mechanisms regulating diaphragm development normally and during herniation is critical. However, many fundamental questions about diaphragm development remain unanswered. Embryonically, the diaphragm derives from three sources: the septum transversum, the pleuroperitoneal folds, and the somites. Currently, it is unclear whether the septum and folds are only transient embryonic structures or whether they contribute to tissues of the mature diaphragm. Also unknown is whether the central tendon or muscle connective tissue arise from these structures. Muscularization of the diaphragm requires muscle progenitors to migrate from the somites to the developing diaphragm, proliferate and differentiate, and form costal and crural muscles. While a few molecular signals (e.g. HGF/Met) have been identified which regulate progenitor migration, the signals and cell-cell interactions regulating muscle differentiation and morphogenesis have not been established. Finally, although a multitude of genes have been identified as likely important in CDH, how mutations in these genes mechanistically lead to hernias has not been determined. Elucidation of the molecular, cellular, and morphogenetic processes that CDH genes regulate will reveal both how the diaphragm develops normally and how defects in diaphragm development lead to these devastating birth defects.
Acknowledgements
Diaphragm research in the Kardon lab is supported by March of Dimes and NICHD/NIH grants to GK. AJM is supported by a University of Utah Graduate Research Fellowship.
References
- 1.Pober BR. Overview of epidemiology, genetics, birth defects, and chromosome abnormalities associated with CDH. Am J Med Genet C Semin Med Genet. 2007;145C:158–71. doi: 10.1002/ajmg.c.30126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finsterer J. Cardiopulmonary support in duchenne muscular dystrophy. Lung. 2006;184:205–15. doi: 10.1007/s00408-005-2584-x. [DOI] [PubMed] [Google Scholar]
- 3.Pearce JM. Henry Gray’s Anatomy. Clin Anat. 2009;22:291–5. doi: 10.1002/ca.20775. [DOI] [PubMed] [Google Scholar]
- 4.Allan DW, Greer JJ. Embryogenesis of the phrenic nerve and diaphragm in the fetal rat. J Comp Neurol. 1997;382:459–68. doi: 10.1002/(sici)1096-9861(19970616)382:4<459::aid-cne3>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 5.Stuelsatz P, Keire P, Almuly R, Yablonka-Reuveni Z. A contemporary atlas of the mouse diaphragm: myogenicity, vascularity, and the Pax3 connection. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 2012;60:638–57. doi: 10.1369/0022155412452417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell EJM, Agostoni E, Newsom Davis J. The Respiratory Muscles: Mechanics and Neural Control. Lloyd-Luke; London: 1970. [Google Scholar]
- 7.Davis J, Goldman M, Loh L, Casson M. Diaphragm function and alveolar hypoventilation. The Quarterly journal of medicine. 1976;45:87–100. [PubMed] [Google Scholar]
- 8.Ruben JA, Bennett AF, Hisaw FL. Selective Factors in the Origin of the Mammalian Diaphragm. Paleobiology. 1987;13:54–59. [Google Scholar]
- 9.Stradling JR, Kozar LF, Dark J, Kirby T, Andrey SM, Phillipson EA. Effect of acute diaphragm paralysis on ventilation in awake and sleeping dogs. Am Rev Respir Dis. 1987;136:633–7. doi: 10.1164/ajrccm/136.3.633. [DOI] [PubMed] [Google Scholar]
- 10.Pickering M, Jones JF. The diaphragm: two physiological muscles in one. Journal of anatomy. 2002;201:305–12. doi: 10.1046/j.1469-7580.2002.00095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchholtz EA, Bailin HG, Laves SA, Yang JT, Chan MY, Drozd LE. Fixed cervical count and the origin of the mammalian diaphragm. Evolution & development. 2012;14:399–411. doi: 10.1111/j.1525-142X.2012.00560.x. [DOI] [PubMed] [Google Scholar]
- 12.Perry SF, Similowski T, Klein W, Codd JR. The evolutionary origin of the mammalian diaphragm. Respir Physiol Neurobiol. 2010;171:1–16. doi: 10.1016/j.resp.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Goodrich ES. Studies on the structure & development of vertebrates. Macmillan and co., limited; 1930. [Google Scholar]
- 14.Dunwoodie SL, Rodriguez TA, Beddington RS. Msg1 and Mrg1, founding members of a gene family, show distinct patterns of gene expression during mouse embryogenesis. Mech Dev. 1998;72:27–40. doi: 10.1016/s0925-4773(98)00011-2. [DOI] [PubMed] [Google Scholar]
- 15.Pinco KA, Liu S, Yang JT. alpha4 integrin is expressed in a subset of cranial neural crest cells and in epicardial progenitor cells during early mouse development. Mech Dev. 2001;100:99–103. doi: 10.1016/s0925-4773(00)00503-7. [DOI] [PubMed] [Google Scholar]
- 16.Di Giacomo G, Koss M, Capellini TD, Brendolan A, Popperl H, Selleri L. Spatio-temporal expression of Pbx3 during mouse organogenesis. Gene Expr Patterns. 2006;6:747–57. doi: 10.1016/j.modgep.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Iritani I. Experimental study on embryogenesis of congenital diaphragmatic hernia. Anat Embryol (Berl) 1984;169:133–9. doi: 10.1007/BF00303142. [DOI] [PubMed] [Google Scholar]
- 18.Kluth D, Keijzer R, Hertl M, Tibboel D. Embryology of congenital diaphragmatic hernia. Semin Pediatr Surg. 1996;5:224–33. [PubMed] [Google Scholar]
- 19.Kluth D, Tenbrinck R, von Ekesparre M, Kangah R, Reich P, Brandsma A, Tibboel D, Lambrecht W. The natural history of congenital diaphragmatic hernia and pulmonary hypoplasia in the embryo. J Pediatr Surg. 1993;28:456–62. doi: 10.1016/0022-3468(93)90248-j. discussion 462-3. [DOI] [PubMed] [Google Scholar]
- 20.Clugston RD, Zhang W, Greer JJ. Gene expression in the developing diaphragm: significance for congenital diaphragmatic hernia. Am J Physiol Lung Cell Mol Physiol. 2008;294:L665–75. doi: 10.1152/ajplung.00027.2008. [DOI] [PubMed] [Google Scholar]
- 21.Dietrich S, Abou-Rebyeh F, Brohmann H, Bladt F, Sonnenberg-Riethmacher E, Yamaai T, Lumsden A, Brand-Saberi B, Birchmeier C. The role of SF/HGF and c-Met in the development of skeletal muscle. Development. 1999;126:1621–9. doi: 10.1242/dev.126.8.1621. [DOI] [PubMed] [Google Scholar]
- 22.Babiuk RP, Zhang W, Clugston R, Allan DW, Greer JJ. Embryological origins and development of the rat diaphragm. J Comp Neurol. 2003;455:477–87. doi: 10.1002/cne.10503. [DOI] [PubMed] [Google Scholar]
- 23.Buckingham M. Skeletal muscle progenitor cells and the role of Pax genes. C R Biol. 2007;330:530–3. doi: 10.1016/j.crvi.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 24.Relaix F, Rocancourt D, Mansouri A, Buckingham M. Divergent functions of murine Pax3 and Pax7 in limb muscle development. Genes Dev. 2004;18:1088–105. doi: 10.1101/gad.301004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang XM, Vogan K, Gros P, Park M. Expression of the met receptor tyrosine kinase in muscle progenitor cells in somites and limbs is absent in Splotch mice. Development. 1996;122:2163–71. doi: 10.1242/dev.122.7.2163. [DOI] [PubMed] [Google Scholar]
- 26.Bladt F, Riethmacher D, Isenmann S, Aguzzi A, Birchmeier C. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature. 1995;376:768–71. doi: 10.1038/376768a0. [DOI] [PubMed] [Google Scholar]
- 27.Tajbakhsh S, Rocancourt D, Cossu G, Buckingham M. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell. 1997;89:127–38. doi: 10.1016/s0092-8674(00)80189-0. [DOI] [PubMed] [Google Scholar]
- 28.Tremblay P, Dietrich S, Mericskay M, Schubert FR, Li Z, Paulin D. A crucial role for Pax3 in the development of the hypaxial musculature and the long-range migration of muscle precursors. Dev Biol. 1998;203:49–61. doi: 10.1006/dbio.1998.9041. [DOI] [PubMed] [Google Scholar]
- 29.Allan DW, Greer JJ. Polysialylated NCAM expression during motor axon outgrowth and myogenesis in the fetal rat. J Comp Neurol. 1998;391:275–92. doi: 10.1002/(sici)1096-9861(19980216)391:3<275::aid-cne1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 30.Maina F, Casagranda F, Audero E, Simeone A, Comoglio PM, Klein R, Ponzetto C. Uncoupling of Grb2 from the Met receptor in vivo reveals complex roles in muscle development. Cell. 1996;87:531–42. doi: 10.1016/s0092-8674(00)81372-0. [DOI] [PubMed] [Google Scholar]
- 31.Ebens A, Brose K, Leonardo ED, Hanson MG, Bladt F, Birchmeier C, Barres BA, Tessier-Lavigne M. Hepatocyte growth factor/scatter factor is an axonal chemoattractant and a neurotrophic factor for spinal motor neurons. Neuron. 1996;17:1157–72. doi: 10.1016/s0896-6273(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto Y, Livet J, Pollock RA, Garces A, Arce V, deLapeyriere O, Henderson CE. Hepatocyte growth factor (HGF/SF) is a muscle-derived survival factor for a subpopulation of embryonic motoneurons. Development. 1997;124:2903–13. doi: 10.1242/dev.124.15.2903. [DOI] [PubMed] [Google Scholar]
- 33.Murphy M, Kardon G. Origin of vertebrate limb muscle: the role of progenitor and myoblast populations. Curr Top Dev Biol. 2011;96:1–32. doi: 10.1016/B978-0-12-385940-2.00001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Babiuk RP, Greer JJ. Diaphragm defects occur in a CDH hernia model independently of myogenesis and lung formation. Am J Physiol Lung Cell Mol Physiol. 2002;283:L1310–4. doi: 10.1152/ajplung.00257.2002. [DOI] [PubMed] [Google Scholar]
- 35.Uetani N, Chagnon MJ, Kennedy TE, Iwakura Y, Tremblay ML. Mammalian motoneuron axon targeting requires receptor protein tyrosine phosphatases sigma and delta. J Neurosci. 2006;26:5872–80. doi: 10.1523/JNEUROSCI.0386-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Philippidou P, Walsh CM, Aubin J, Jeannotte L, Dasen JS. Sustained Hox5 gene activity is required for respiratory motor neuron development. Nature neuroscience. 2012;15:1636–44. doi: 10.1038/nn.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis J, Chevallier A, Kieny M, Wolpert L. Muscle nerve branches do not develop in chick wings devoid of muscle. J Embryol Exp Morphol. 1981;64:211–32. [PubMed] [Google Scholar]
- 38.Li XM, Dong XP, Luo SW, Zhang B, Lee DH, Ting AK, Neiswender H, Kim CH, Carpenter-Hyland E, Gao TM, Xiong WC, Mei L. Retrograde regulation of motoneuron differentiation by muscle beta-catenin. Nature neuroscience. 2008;11:262–8. doi: 10.1038/nn2053. [DOI] [PubMed] [Google Scholar]
- 39.Wu H, Lu Y, Barik A, Joseph A, Taketo MM, Xiong WC, Mei L. beta-Catenin gain of function in muscles impairs neuromuscular junction formation. Development. 2012;139:2392–404. doi: 10.1242/dev.080705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burden SJ. SnapShot: Neuromuscular Junction. Cell. 2011;144:826–826 e1. doi: 10.1016/j.cell.2011.02.037. [DOI] [PubMed] [Google Scholar]
- 41.Wu H, Xiong WC, Mei L. To build a synapse: signaling pathways in neuromuscular junction assembly. Development. 2010;137:1017–33. doi: 10.1242/dev.038711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ackerman KG, Herron BJ, Vargas SO, Huang H, Tevosian SG, Kochilas L, Rao C, Pober BR, Babiuk RP, Epstein JA, Greer JJ, Beier DR. Fog2 is required for normal diaphragm and lung development in mice and humans. PLoS Genet. 2005;1:58–65. doi: 10.1371/journal.pgen.0010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jay PY, Bielinska M, Erlich JM, Mannisto S, Pu WT, Heikinheimo M, Wilson DB. Impaired mesenchymal cell function in Gata4 mutant mice leads to diaphragmatic hernias and primary lung defects. Dev Biol. 2007;301:602–14. doi: 10.1016/j.ydbio.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ackerman KG, Pober BR. Congenital diaphragmatic hernia and pulmonary hypoplasia: new insights from developmental biology and genetics. Am J Med Genet C Semin Med Genet. 2007;145C:105–8. doi: 10.1002/ajmg.c.30133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keller RL. Antenatal and postnatal lung and vascular anatomic and functional studies in congenital diaphragmatic hernia: implications for clinical management. Am J Med Genet C Semin Med Genet. 2007;145C:184–200. doi: 10.1002/ajmg.c.30130. [DOI] [PubMed] [Google Scholar]
- 46.Harrison MR, Adzick NS, Estes JM, Howell LJ. A prospective study of the outcome for fetuses with diaphragmatic hernia. JAMA. 1994;271:382–4. [PubMed] [Google Scholar]
- 47.Torfs CP, Curry CJ, Bateson TF, Honore LH. A population-based study of congenital diaphragmatic hernia. Teratology. 1992;46:555–65. doi: 10.1002/tera.1420460605. [DOI] [PubMed] [Google Scholar]
- 48.Stokes KB. Unusual varieties of diaphragmatic herniae. Prog Pediatr Surg. 1991;27:127–47. doi: 10.1007/978-3-642-87767-4_8. [DOI] [PubMed] [Google Scholar]
- 49.Ackerman KG, Greer JJ. Development of the diaphragm and genetic mouse models of diaphragmatic defects. Am J Med Genet C Semin Med Genet. 2007;145C:109–16. doi: 10.1002/ajmg.c.30128. [DOI] [PubMed] [Google Scholar]
- 50.Antonius T, van Bon B, Eggink A, van der Burgt I, Noordam K, van Heijst A. Denys-Drash syndrome and congenital diaphragmatic hernia: another case with the 1097G > A(Arg366His) mutation. Am J Med Genet A. 2008;146A:496–9. doi: 10.1002/ajmg.a.32168. [DOI] [PubMed] [Google Scholar]
- 51.Schrier SA, Sherer I, Deardorff MA, Clark D, Audette L, Gillis L, Kline AD, Ernst L, Loomes K, Krantz ID, Jackson LG. Causes of death and autopsy findings in a large study cohort of individuals with Cornelia de Lange syndrome and review of the literature. Am J Med Genet A. 2011;155A:3007–24. doi: 10.1002/ajmg.a.34329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Donnai D, Barrow M. Diaphragmatic hernia, exomphalos, absent corpus callosum, hypertelorism, myopia, and sensorineural deafness: a newly recognized autosomal recessive disorder? Am J Med Genet. 1993;47:679–82. doi: 10.1002/ajmg.1320470518. [DOI] [PubMed] [Google Scholar]
- 53.Tautz J, Veenma D, Eussen B, Joosen L, Poddighe P, Tibboel D, de Klein A, Schaible T. Congenital diaphragmatic hernia and a complex heart defect in association with Wolf-Hirschhorn syndrome. Am J Med Genet A. 2010;152A:2891–4. doi: 10.1002/ajmg.a.33660. [DOI] [PubMed] [Google Scholar]
- 54.Lin IC, Ko SF, Shieh CS, Huang CF, Chien SJ, Liang CD. Recurrent congenital diaphragmatic hernia in Ehlers-Danlos syndrome. Cardiovasc Intervent Radiol. 2006;29:920–3. doi: 10.1007/s00270-005-0154-5. [DOI] [PubMed] [Google Scholar]
- 55.Smigiel R, Jakubiak A, Lombardi MP, Jaworski W, Slezak R, Patkowski D, Hennekam RC. Co-occurrence of severe Goltz-Gorlin syndrome and pentalogy of Cantrell - Case report and review of the literature. Am J Med Genet A. 2011;155A:1102–5. doi: 10.1002/ajmg.a.33895. [DOI] [PubMed] [Google Scholar]
- 56.Slavotinek AM. Fryns syndrome: a review of the phenotype and diagnostic guidelines. Am J Med Genet A. 2004;124A:427–33. doi: 10.1002/ajmg.a.20381. [DOI] [PubMed] [Google Scholar]
- 57.Inanlou MR, Dhillon GS, Belliveau AC, Reid GA, Ying C, Rudnicki MA, Kablar B. A significant reduction of the diaphragm in mdx:MyoD-/-(9th) embryos suggests a role for MyoD in the diaphragm development. Dev Biol. 2003;261:324–36. doi: 10.1016/s0012-1606(03)00319-1. [DOI] [PubMed] [Google Scholar]
- 58.Tseng BS, Cavin ST, Booth FW, Olson EN, Marin MC, McDonnell TJ, Butler IJ. Pulmonary hypoplasia in the myogenin null mouse embryo. Am J Respir Cell Mol Biol. 2000;22:304–15. doi: 10.1165/ajrcmb.22.3.3708. [DOI] [PubMed] [Google Scholar]
- 59.Hornstra IK, Birge S, Starcher B, Bailey AJ, Mecham RP, Shapiro SD. Lysyl oxidase is required for vascular and diaphragmatic development in mice. J Biol Chem. 2003;278:14387–93. doi: 10.1074/jbc.M210144200. [DOI] [PubMed] [Google Scholar]
- 60.Kluth D, Kangah R, Reich P, Tenbrinck R, Tibboel D, Lambrecht W. Nitrofen-induced diaphragmatic hernias in rats: an animal model. J Pediatr Surg. 1990;25:850–4. doi: 10.1016/0022-3468(90)90190-k. [DOI] [PubMed] [Google Scholar]
- 61.Allan DW, Greer JJ. Pathogenesis of nitrofen-induced congenital diaphragmatic hernia in fetal rats. J Appl Physiol. 1997;83:338–47. doi: 10.1152/jappl.1997.83.2.338. [DOI] [PubMed] [Google Scholar]
- 62.Clugston RD, Zhang W, Alvarez S, de Lera AR, Greer JJ. Understanding abnormal retinoid signaling as a causative mechanism in congenital diaphragmatic hernia. Am J Respir Cell Mol Biol. 2010;42:276–85. doi: 10.1165/rcmb.2009-0076OC. [DOI] [PubMed] [Google Scholar]
- 63.Chassaing N, Golzio C, Odent S, Lequeux L, Vigouroux A, Martinovic-Bouriel J, Tiziano FD, Masini L, Piro F, Maragliano G, Delezoide AL, Attie-Bitach T, Manouvrier-Hanu S, Etchevers HC, Calvas P. Phenotypic spectrum of STRA6 mutations: from Matthew-Wood syndrome to non-lethal anophthalmia. Hum Mutat. 2009;30:E673–81. doi: 10.1002/humu.21023. [DOI] [PubMed] [Google Scholar]
- 64.Chitayat D, Sroka H, Keating S, Colby RS, Ryan G, Toi A, Blaser S, Viero S, Devisme L, Boute-Benejean O, Manouvrier-Hanu S, Mortier G, Loeys B, Rauch A, Bitoun P. The PDAC syndrome (pulmonary hypoplasia/agenesis, diaphragmatic hernia/eventration, anophthalmia/microphthalmia, and cardiac defect) (Spear syndrome, Matthew-Wood syndrome): report of eight cases including a living child and further evidence for autosomal recessive inheritance. Am J Med Genet A. 2007;143A:1268–81. doi: 10.1002/ajmg.a.31788. [DOI] [PubMed] [Google Scholar]
- 65.Mendelsohn C, Lohnes D, Decimo D, Lufkin T, LeMeur M, Chambon P, Mark M. Function of the retinoic acid receptors (RARs) during development (II). Multiple abnormalities at various stages of organogenesis in RAR double mutants. Development. 1994;120:2749–71. doi: 10.1242/dev.120.10.2749. [DOI] [PubMed] [Google Scholar]
- 66.Brady PD, Srisupundit K, Devriendt K, Fryns JP, Deprest JA, Vermeesch JR. Recent developments in the genetic factors underlying congenital diaphragmatic hernia. Fetal Diagn Ther. 2011;29:25–39. doi: 10.1159/000322422. [DOI] [PubMed] [Google Scholar]
- 67.Goumy C, Gouas L, Marceau G, Coste K, Veronese L, Gallot D, Sapin V, Vago P, Tchirkov A. Retinoid pathway and congenital diaphragmatic hernia: hypothesis from the analysis of chromosomal abnormalities. Fetal Diagn Ther. 2010;28:129–39. doi: 10.1159/000313331. [DOI] [PubMed] [Google Scholar]
- 68.Yu L, Wynn J, Cheung YH, Shen Y, Mychaliska GB, Crombleholme TM, Azarow KS, Lim FY, Chung DH, Potoka D, Warner BW, Bucher B, Stolar C, Aspelund G, Arkovitz MS, Chung WK. Variants in GATA4 are a rare cause of familial and sporadic congenital diaphragmatic hernia. Hum Genet. 2012 doi: 10.1007/s00439-012-1249-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arrington CB, Bleyl SB, Matsunami N, Bowles NE, Leppert TI, Demarest BL, Osborne K, Yoder BA, Byrne JL, Schiffman JD, Null DM, Digeronimo R, Rollins M, Faix R, Comstock J, Camp NJ, Leppert MF, Yost HJ, Brunelli L. A family-based paradigm to identify candidate chromosomal regions for isolated congenital diaphragmatic hernia. Am J Med Genet A. 2012;158A:3137–47. doi: 10.1002/ajmg.a.35664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Longoni M, Lage K, Russell MK, Loscertales M, Abdul-Rahman OA, Baynam G, Bleyl SB, Brady PD, Breckpot J, Chen CP, Devriendt K, Gillessen-Kaesbach G, Grix AW, Rope AF, Shimokawa O, Strauss B, Wieczorek D, Zackai EH, Coletti CM, Maalouf FI, Noonan KM, Park JH, Tracy AA, Lee C, Donahoe PK, Pober BR. Congenital diaphragmatic hernia interval on chromosome 8p23.1 characterized by genetics and protein interaction networks. Am J Med Genet A. 2012;158A:3148–58. doi: 10.1002/ajmg.a.35665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cantor AB, Orkin SH. Coregulation of GATA factors by the Friend of GATA (FOG) family of multitype zinc finger proteins. Seminars in cell & developmental biology. 2005;16:117–28. doi: 10.1016/j.semcdb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 72.Klaassens M, van Dooren M, Eussen HJ, Douben H, den Dekker AT, Lee C, Donahoe PK, Galjaard RJ, Goemaere N, de Krijger RR, Wouters C, Wauters J, Oostra BA, Tibboel D, de Klein A. Congenital diaphragmatic hernia and chromosome 15q26: determination of a candidate region by use of fluorescent in situ hybridization and array-based comparative genomic hybridization. Am J Hum Genet. 2005;76:877–82. doi: 10.1086/429842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.You LR, Takamoto N, Yu CT, Tanaka T, Kodama T, Demayo FJ, Tsai SY, Tsai MJ. Mouse lacking COUP-TFII as an animal model of Bochdalek-type congenital diaphragmatic hernia. Proc Natl Acad Sci U S A. 2005;102:16351–6. doi: 10.1073/pnas.0507832102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Russell MK, Longoni M, Wells J, Maalouf FI, Tracy AA, Loscertales M, Ackerman KG, Pober BR, Lage K, Bult CJ, Donahoe PK. Congenital diaphragmatic hernia candidate genes derived from embryonic transcriptomes. Proc Natl Acad Sci U S A. 2012;109:2978–83. doi: 10.1073/pnas.1121621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brohmann H, Jagla K, Birchmeier C. The role of Lbx1 in migration of muscle precursor cells. Development. 2000;127:437–45. doi: 10.1242/dev.127.2.437. [DOI] [PubMed] [Google Scholar]
- 76.Gross MK, Moran-Rivard L, Velasquez T, Nakatsu MN, Jagla K, Goulding M. Lbx1 is required for muscle precursor migration along a lateral pathway into the limb. Development. 2000;127:413–24. doi: 10.1242/dev.127.2.413. [DOI] [PubMed] [Google Scholar]
- 77.Hasson P, DeLaurier A, Bennett M, Grigorieva E, Naiche LA, Papaioannou VE, Mohun TJ, Logan MP. Tbx4 and tbx5 acting in connective tissue are required for limb muscle and tendon patterning. Developmental cell. 2010;18:148–56. doi: 10.1016/j.devcel.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kardon G, Harfe BD, Tabin CJ. A Tcf4-positive mesodermal population provides a prepattern for vertebrate limb muscle patterning. Developmental cell. 2003;5:937–44. doi: 10.1016/s1534-5807(03)00360-5. [DOI] [PubMed] [Google Scholar]