Abstract
The phase II detoxification system glutathione transferase (GST) is associated with the establishment of parasitic nematode infections within the gastrointestinal environment of the mammalian host. We report the functional analysis of a GST from an important worldwide parasitic nematode of small ruminants, Haemonchus contortus. This GST shows limited activity with a range of classical GST substrates but effectively binds hematin. The high-affinity binding site for hematin was not present in the GST showing the most identity, CE07055 from the free-living nematode Caenorhabditis elegans. This finding suggests that the high-affinity binding of hematin may represent a parasite adaptation to blood or tissue feeding from the host.
The gastrointestinal blood-feeding nematode Haemonchus contortus represents a major economic burden to agricultural communities worldwide, causing infections resulting in anemia, weight loss, and ultimately death in small ruminants. There are presently no commercial vaccines available for H. contortus infections, and the most effective method of control is a combination of pasture management and the use of chemical agents (anthelmintics). Increasing reports of drug-resistant H. contortus indicate that this current control strategy is not sustainable (26), with chemicals no longer effectively controlling H. contortus infections in several parts of the world (33). Understanding the host-parasite relationship at the mucosal feeding surface is important in identifying new therapeutic approaches. The levels of the phase II detoxification system glutathione transferase (GST) have been shown to increase in parasitic helminths during chronic infection (3). Previous research has attempted to correlate this overexpression with the ability of GST to detoxify immune-initiated cytotoxic products of lipid peroxidation (8, 10) or has associated the overexpression of GST, including H. contortus GST, with drug resistance (18, 19). In this paper, we analyze a new GST from the sheep strongylid H. contortus and show that this GST does not appear to have a broad immune defense or drug metabolism role but possibly has a more focused detoxification function within the nematode.
MATERIALS AND METHODS
Isolation, recombinant expression, and purification of H. contortus GST.
mRNA was isolated from adult H. contortus nematodes (CAVR, a drug-resistant strain) with a Quickprep Micro mRNA purification kit (Pharmacia). H. contortus cDNA was obtained with a First Strand cDNA synthesis kit (Pharmacia). The H. contortus GST-encoding DNA was isolated by an established strategy with an upstream primer derived from the N-terminal sequence of the native H. contortus GST protein (27) and a downstream oligo(dT)-based anchor primer (1, 5). The PCR product (approximately 650 bp) was cloned into pUC18 (SureClone ligation kit; Pharmacia), and the insert was sequenced. The H. contortus GST was directionally cloned into pET23d and sequenced in both directions (Long-read LI-COR, GenBank accession number AF281663). The recombinant H. contortus GST protein (HcGST-1) was expressed in Escherichia coli BL21(DE3) by induction with isopropylthio-β-d-galactoside (IPTG). The protein was purified by glutathione (GSH)-Sepharose affinity chromatography (6). Purity was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electrospray mass spectrometry. Prior to electrospray mass spectrometry, salts were removed with a ZipTipC18 (Millipore). The protein was eluted in 50 μl of 50% (vol/vol) acetonitrile-0.1% (vol/vol) formic acid and injected into a LCT-TOF mass spectrometer (Micromass, Manchester, United Kingdom) at 5 μl/min with a 100-μl syringe pump (40 V).
Modeling of HcGST-1.
Homology models of HcGST-1 were initially obtained from the SWISS-MODEL protein-modeling server (14, 15, 24), and the model was based on three templates (PDB accession numbers 1PD2, 2GSQ, and 1GUK). This preliminary model was refined by minimization to 0.1 kJ/mol/A by using a Polak-Ribiere conjugate gradients algorithm and the OPLS-AA force field (17) with MaestroT (version 3.0027; Schrodinger Inc.).
Southern blot analysis.
Genomic DNA was extracted from H. contortus adults (CAVR strain) or mixed-stage wild-type Caenorhabditis elegans nematodes (N2-Bristol) by using a DNeasy tissue kit (QIAGEN). After single or double digestion with BamHI, EcoRI, and/or HindIII, the DNA fragments were separated in 0.8% Tris-acetate-EDTA agarose and transferred in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate [pH 8]) to a Zeta-Probe GT membrane (Bio-Rad). H. contortus GST-encoding cDNA was used as a probe following isolation with the Qiaex II gel extraction kit (QIAGEN). The probe was radiolabeled with [α-32P]dCTP by using the Random Primers DNA labeling kit (Gibco). Southern blots were blocked with stopping solution (5× SSC, 5× Denhardt's solution, 0.5% SDS, 0.1 mg/ml preboiled herring sperm DNA) (catalog number D3159; Sigma), incubated overnight with the radiolabeled probe (55°C), and washed with wash A (0.1% SDS/2× SSC [wt/vol]), wash B (0.1% SDS/1× SSC [wt/vol]), and wash C (0.1% SDS/0.1× SSC [wt/vol]) twice for 10 min each time. After each wash, the filter was exposed to a phosphor storage screen (GP SO230, Kodak) for up to 24 h and images were acquired by using a Typhoon 8600 variable-mode imager (Molecular Dynamics).
Enzymatic and ligand binding assays.
GST enzymatic activity in 100 mM potassium phosphate buffer (pH 6.5) containing reduced GSH at a concentration of 1 mM and the model GST substrate 1-chloro-2,4-dinitrobenzene (CDNB) at a concentration of 1 mM (11) was measured spectrophotometrically at 340 nm (25°C). Assays with natural and other model substrates were completed as previously described (11). Both CDNB and GSH were utilized as competing substrates for a Dixon plot analysis as outlined by Cortes et al. (12). Protein concentrations were determined by the Bradford method (Bio-Rad, commercial kit). Ligand binding to rHcGST-1 was estimated by measuring changes in intrinsic or competitive protein fluorescence. In all ligand binding assays, a final concentration of 1 μM HcGST-1 was used at 25°C in 20 mM potassium phosphate buffer (pH 6.5) containing 100 mM sodium chloride (31). Changes in fluorescence were recorded with a Shimadzu spectrofluorometer (RF-5301 PC) with excitation and emission wavelengths for intrinsic protein fluorescence (tryptophan) of 280 and 320 nm, respectively. Increasing concentrations of hematin or anthelmintic were added, followed by 3 min of incubation prior to measurement. For competitive fluorescent assays, the competitive GST ligand 8-anilino-1-naphthalene-sulfonic acid (ANS) was used at a final concentration of 10 μM as previously described (11). Excitation and emission wavelengths for ANS were established at 365 and 480 nm, respectively.
Two-dimensional gel electrophoresis and Western blotting.
Two-dimensional electrophoresis (13) was undertaken with the Multiphor II system (Pharmacia). Somatic protein extracts or GSH-Sepharose affinity-purified proteins from C. elegans or H. contortus were separated by isoelectric focusing overnight on commercially available immobilized pH gradient (IPG) strips (11 cm, pH 3 to 10; Pharmacia) followed by size separation with precast SDS-8 to 18% PAGE gels (ExcelGel XL; Pharmacia). The gels were silver-stained with an automated gel-staining system (Hoefer). Images were scanned into Adobe Photoshop and analyzed by using Phoretix 5.01 software.
Polyclonal antibodies to HcGST-1 were raised in rabbits by standard procedures (4, 7) and used for Western blot analysis. After SDS-12% PAGE, proteins were electrotransferred to a polyvinylidene difluoride membrane. The membrane was blocked with blocking buffer (5% [wt/vol] Marvel milk powder and 0.1% [vol/vol] Tween-20 in phosphate buffered saline [pH 7.3]) and subsequently incubated in blocking buffer containing GST antisera for 1 h (1:250 dilution), followed by four 15-min washes in 0.1% (vol/vol) Tween 20 in phosphate-buffered saline. After 1 h of incubation with the secondary antibody (alkaline phosphatase conjugated anti-rabbit immunoglobulin G; 1:10,000 dilution), blots were washed again four times for 15 min each time. The blots were developed with BCIP/NBT (5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium; Sigma).
Nucleotide sequence accession number.
The nucleotide sequence for the H. contortus GST has been deposited in the GenBank database, under GenBank accession no. AF281663.
RESULTS AND DISCUSSION
GST informatics.
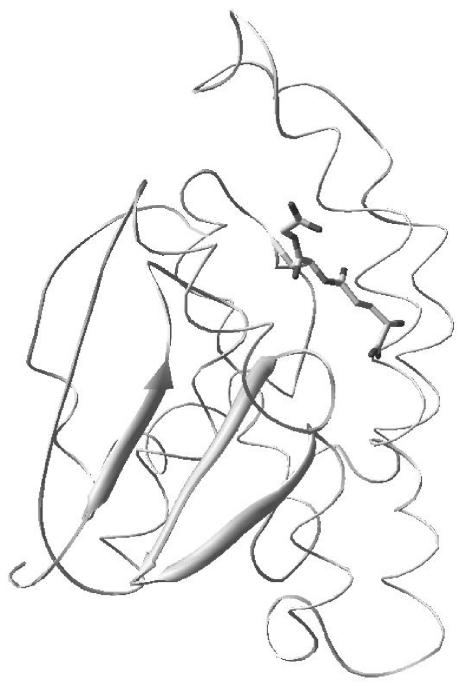
Protein database analysis at EMBL-EBI (http://www.ebi.ac.uk; July 2003) indicated that the submitted protein sequence (GenBank accession number AF281663) is a member of the GST superfamily. InterPro (EMBL-EBI) database searching demonstrated that the putative H. contortus GST contains the typical GST N-terminal (IPR004045) and C-terminal (IPR004046) domain signatures. Homology modeling with known GST crystal structures as templates confirmed that the protein is a GST, with the N-terminal domain possessing the classical GST βαβαββα fold (Fig. 1). An α2β3 fold was indicative of the cofactor GSH-binding (G) site, and the presence of a GST hydrophobic (H) site along the interdomain cleft was highlighted by two parallel β strands and the N-terminal βαβ motif. The C terminus of the H. contortus GST also possesses an α helix that is generally presumed to contribute to the binding of hydrophobic ligands to GSTs.
FIG. 1.
Structure homology model of H. contortus GST (HcGST-1) with GSH constructed with SWISS-MODEL and viewed with Rasmol. In the model, the GSH thiol is within hydrogen-bonding distance of Tyr 8, the GSH Gly carboyl is within hydrogen-bonding distance of Trp 39, the γ-Glu carboxyl is within hydrogen bonding distance of Gln 63, and the GSH backbone hydrogen bonds to Leu 51.
The HcGST-1 displays 70% similarity to a predicted GST (HcGST-E) recently demonstrated in the excretory-secretory products of H. contortus by Yatsuda et al. (35) by use of global proteomic screening approaches. Phylogenetic analysis suggests that the H. contortus GST-1 in this study and the HcGST-E demonstrated by Yatsuda et al. (35) form part of a new nematode GST class together with GSTs predicted from the C. elegans proteome (Fig. 2). At 60% identity, the closest C. elegans sequence is CE07055 (GST 7; P91253), expressed in all life cycle stages. The nearest nonnematode sequences are from German cockroach GST (O18598; major antigen Bla g5) and GSH-dependent prostaglandin D synthase (PGDS) from chicken (O73888; sigma class GST), both at 38% identity. However, the predicted H. contortus GST contains only 4 of the 15 amino acids in the proposed active site of PGDS. Furthermore, PGDS displays broad substrate specificity (29), unlike the predicted H. contortus GST.
FIG.2.
Phylogenetic tree showing the relationship of HcGST-1 to all C. elegans proteins containing an N-terminal and/or C-terminal GST domain, as indicated by InterPro (domains IPR004045 and IPR004046, respectively). The initial alignment of protein sequences was achieved with BioEdit (16) and CLUSTAL W (28). The final tree was constructed by cluster analysis with TreeCon (30). The numbers indicate the bootstrap values for 100 replicates. All C. elegans protein sequences were obtained from the Sanger Institute (http://www.sanger.ac.uk/Projects/C_elegans/wormpep/), and accession numbers are indicated by the prefix “CE.” The nucleotide sequence of the H. contortus-excreted GST (HcGST-E) was obtained from GenBank (National Center for Biology Information; http://www.ncbi.nlm.nih.gov/) (accession no. BM138779).
Recombinant expression.
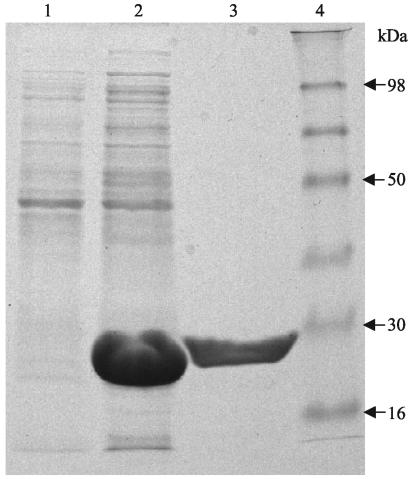
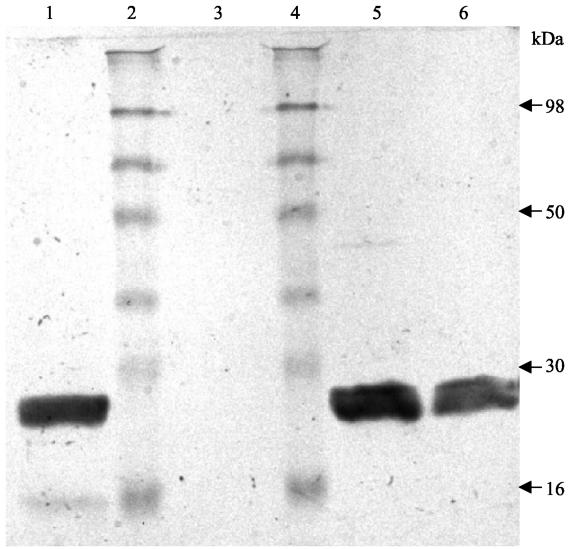
HcGST-1 was overexpressed as an active soluble protein (50 to 100 mg/liter of culture) in E. coli. After GSH affinity purification, the protein was confirmed as a single band at approximately 26 kDa by SDS-PAGE (Fig. 3). Mass spectrometry established the mass of the recombinant GST subunit as 23,603 Da (Fig. 4), identical to the theoretical mass predicted from the coding cDNA. Western blot analysis illustrated that the polyclonal antibodies raised against HcGST-1 recognized this GST as well as a specific protein band of approximately 26 kDa in native preparations of adult H. contortus worms (Fig. 5).
FIG. 3.
Production and purification of rHcGST-1: Coomassie-stained SDS-12% PAGE. Lane 1, somatic extract of E. coli BL21(DE3) plus pET23d (control); lane 2, somatic extract of E. coli BL21(DE3) plus pAR10 (containing HcGST-1); lane 3, GSH-agarose affinity-purified rHcGST-1; lane 4, molecular mass protein standard.
FIG. 4.
Electrospray mass spectrometer analysis of purified recombinant H. contortus GST, establishing the mass at 23,603 Da.
FIG. 5.
Western blot probed with polyclonal antibodies to H. contortus GST-1, showing the expression of GST in native H. contortus tissue. Lane 1, somatic extract of H. contortus; lanes 2 and 4, molecular mass protein standard; lane 3, somatic extract of E. coli BL21(DE3) plus pET23d (control); lane 5, somatic extract of E. coli BL21(DE3) plus pAR10 (containing HcGST-1); lane 6, GSH-agarose affinity-purified rHcGST-1.
Gene and protein structure.
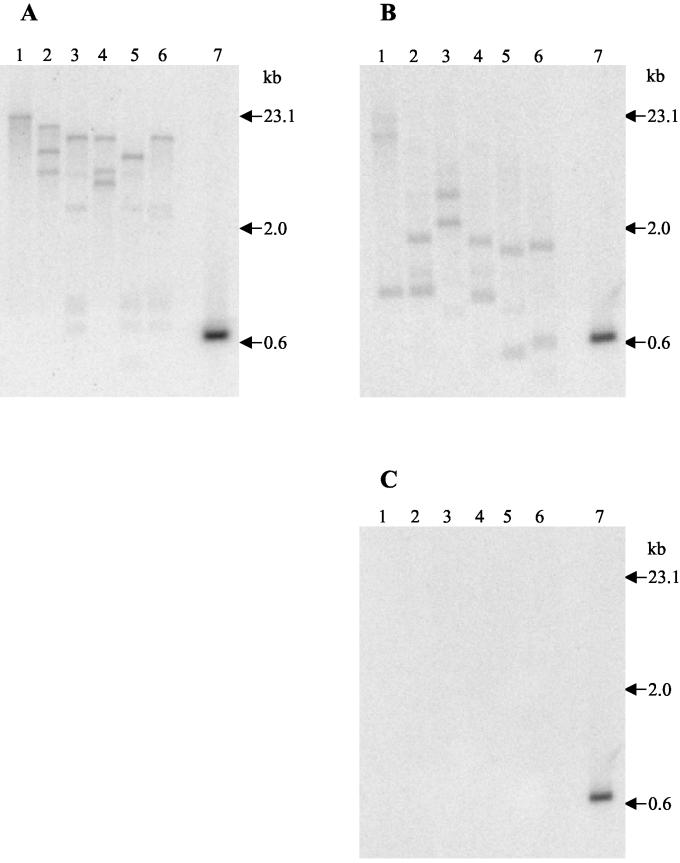
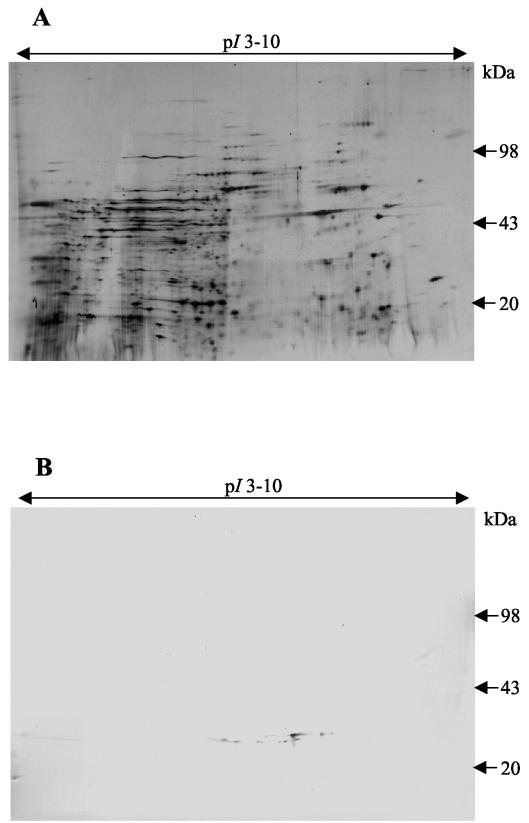
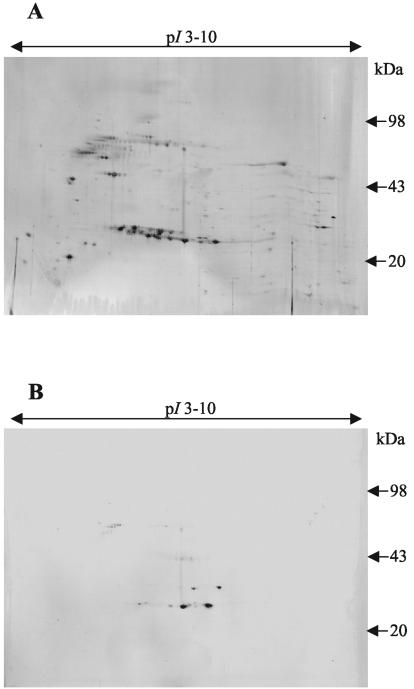
Southern blot analysis supported the idea that the GST sequence is part of the H. contortus genome (Fig. 6). Since the intron sequences and similarities between the recombinant and native H. contortus GSTs have not yet been determined, the copy number of this GST on the H. contortus genome could not be conclusively established, but it appears to be between 1 and 3. Southern blotting also indicated that the H. contortus GST gene has a close sequence relationship with the verified GST genes in the C. elegans genome, since only stringent washes can deter the probe from binding to the C. elegans genomic DNA (Fig. 6C). Two-dimensional Western blotting of somatic H. contortus native protein extract demonstrated that eight similar GSTs were expressed in adult worms (Fig. 7). In C. elegans, 26 individual protein spots, represented by 12 separate GSTs, were identified by peptide mass fingerprinting following GSH affinity chromatography, suggesting posttranslational modification (32). In H. contortus, some of the eight GST subunits identified following GSH affinity chromatography may also be posttranslationally modified versions of identical proteins. Cross-reactivity between the recombinant GST derived from H. contortus and four C. elegans GSH affinity-purified proteins supports the presence of nematode generic classes of GSTs previously indicated only by bioinformatics (Fig. 8). The close sequence relationship suggests that C. elegans can be used as a transgenic expression tool to further investigate the function of this H. contortus GST, as previously described for parasite β-tubulin (20).
FIG. 6.
Southern blots on H. contortus (A) and C. elegans (B and C) genomic DNA digests, confirming a close relationship between H. contortus and C. elegans at the genomic level. Lanes 1, BamHI; lanes 2, EcoRI; lanes 3, HindIII; lanes 4, BamHI and EcoRI; lanes 5, BamHI and HindIII; lanes 6, EcoRI and HindIII; lanes 7, positive control (HcGST-1 gene). (A) H. contortus genomic DNA digests probed with radiolabeled HcGST-1 gene (60°C; high-stringency washes: twice with wash A, twice with wash B, and twice with wash C). (B) C. elegans genomic DNA digests probed with radiolabeled HcGST-1 gene (60°C; low-stringency washes: twice with wash A). (C) C. elegans genomic DNA digests probed with radiolabeled HcGST-1 gene (60°C; high-stringency washes: twice with wash A, twice with wash B, and twice with wash C).
FIG. 7.
Two-dimensional gel and Western blot on somatic H. contortus protein, showing the expression of up to eight similar GSTs, or modified forms of GST(s), in adult worms. (A) Two-dimensional gel of somatic H. contortus protein. The first dimension was run at 11 cm with an IPG of pH 3 to 10, and the second dimension was run on SDS-8 to 18% PAGE. (B) Western blot on two-dimensional gel of somatic H. contortus protein probed with polyclonal antibodies to rHcGST-1.
FIG. 8.
Two-dimensional gel and Western blot on somatic C. elegans protein, supporting the presence of nematode generic classes of GSTs. (A) Two-dimensional gel of glutathione-agarose affinity-purified C. elegans protein extract. The first dimension was run at 11 cm with an IPG of pH 3 to 10, and the second dimension was run on SDS-8 to 18% PAGE. (B) Western blot on two-dimensional gel of affinity-purified C. elegans protein extract probed with polyclonal antibodies to HcGST-1.
Functional analysis.
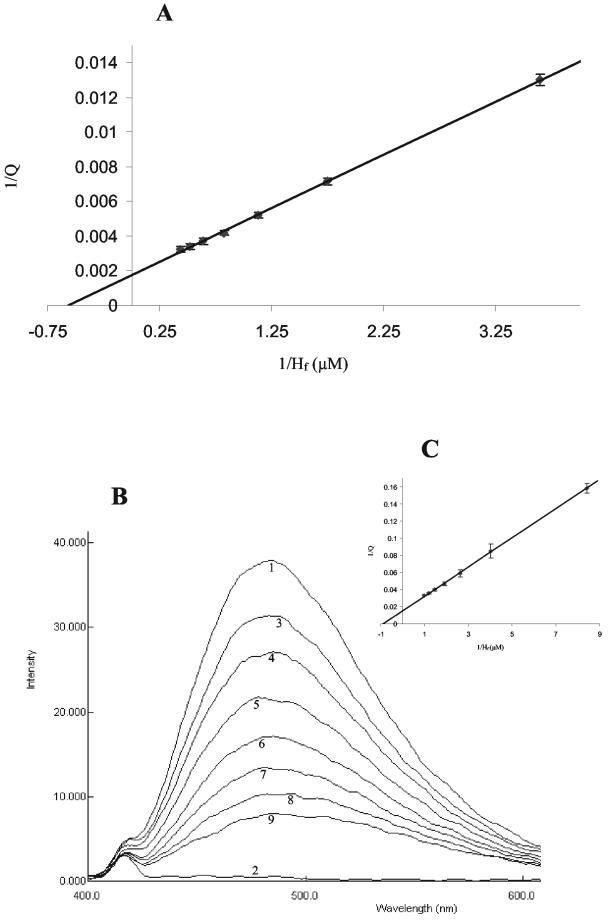
The function of the HcGST-1 was investigated by using a panel of model and potential natural GST substrates and inhibitors (Tables 1 and 2). HcGST-1 shows limited activity with model substrates and parasite GST natural substrates (the reactive carbonyls), indicating a minor role in host immune-initiated lipid peroxidation defense. In addition, somatic extracts of cestode and digenean parasites have higher activity towards lipid peroxidation products than nematodes, including H. contortus, do (2, 9). Parasite GSTs have also been associated with drug resistance (18, 19). HcGST-1, isolated from a drug-resistant strain of H. contortus, only weakly bound the majority of commercial anthelmintics, with no evidence of enzymatic conjugation (results not shown). The binding of hematin to HcGST-1 was demonstrated by both intrinsic fluorescence and competitive fluorescence assays with the hydrophobic binding site probe ANS. The dissociation constant (KD) values for hematin, determined by double-reciprocal plots, was in good agreement for both methods, with an intrinsic fluorescence KD of 1.72 ± 0.10 μM and a competitive fluorescence KD of 1.13 ± 0.07 μM (Fig. 9). Analysis of the data by Dixon plot suggested that the binding of hematin is noncompetitive (results not shown). Fitting the inhibition data directly by nonlinear regression to the rate equation for noncompetitive inhibition gives the following estimates: Ki apparent (hematin/CDNB) = 262 ± 39 nM, Ki apparent (hematin/GSH) = 208 ± 30 nM, Km apparent (CDNB) = 211 ± 68 μM, and Km apparent (GSH) = 367 ± 94 μM, where Ki apparent is the inhibition constant at a fixed substrate level and Km apparent is the Km determined at a fixed concentration of the second substrate.
TABLE 1.
Substrate specificities of rHcGST-1, showing limited activity with model and natural GST substrates under standard assay conditions
| Substrate | Substrate concn (mM) | GSH concn (mM) | pH | λmax (nm) | Δɛa | Sp act (μmol/min/mg of protein)b |
|---|---|---|---|---|---|---|
| 1-Chloro-2,4-dinitrobenene | 1 | 1 | 6.5 | 340 | 9.6 × 106 cm2 · mol−1 | 1,517 ± 42 |
| Cumene hydroperoxide | 0.064 | 1 | 7.0 | 340 | 6.22 × 106 cm2 · mol−1 | 536 ± 24 |
| trans-2-Nonenal | 0.023 | 1 | 6.5 | 225 | −19.2 mM−1 · cm−1 | 41 ± 4.5 |
| Ethacrynic acid | 0.08 | 0.25 | 6.5 | 270 | 5.0 mM−1 · cm−1 | 14 ± 1.2 |
| 0.08 | 1 | 6.5 | 270 | 15 ± 1.6 | ||
| Bromosulfophthalein | 0.03 | 5 | 7.5 | 330 | 4.5 mM−1 · cm−1 | ND (<0.5) |
| 1,2-Dichloro-4-nitrobenzene | 1 | 5 | 7.5 | 345 | 9.6 × 106 cm2 · mol−1 | ND (<0.5) |
| 1,2-Epoxy-3-(p-nitrophenoxy)propane | 1 | 5 | 6.5 | 360 | 4.5 mM−1 · cm−1 | ND (<0.5) |
| trans-4-Phenyl-3-buten-2-one | 0.05 | 0.25 | 6.5 | 290 | −24.8 mM−1 · cm−1 | ND (<0.5) |
| trans,trans-2, 4-Decadienal | 0.023 | 1 | 6.5 | 280 | −29.7 mM−1 · cm−1 | ND (<0.5) |
Δɛ, extinction coefficient.
Mean ± standard deviation of at least six determinations. ND, not detected.
TABLE 2.
IC50s of commercial nematocides and hematin for the conjugation of CDNB by rHcGST-1
| Compound | IC50 at 340 nma |
|---|---|
| Hematin | 180 ± 13 nM |
| Albendazole | 662 ± 51 μM |
| Diethylcarbamazine | ND (>5 mM) |
| Fenbendazole | 84.4 ± 6.0 μM |
| Imidazole | ND (>5 mM) |
| Ivermectin | ND (>5 mM) |
| Levamisole | ND (>5 mM) |
| Mebendazole | 851 ± 55 μM |
| Methimazole | ND (>5 mM) |
| Morantel tartrate salt | 215 ± 16 μM |
| Oxibendazole | 3.19 ± 0.3 mM |
| Piperazine | ND (>5 mM) |
| Pyrantel tartrate salt | 358 ± 24 μM |
| Thiabendazole | ND (>5 mM) |
Mean ± standard deviation of at least four determinations. ND, not detected. All values were determined from direct plots of activity against the inhibitor concentration fitted by nonlinear regression.
FIG. 9.
Binding of hematin to HcGST-1 as demonstrated by intrinsic fluorescence and competitive fluorescence spectrometry. (A) Double-reciprocal plot of the quenching of intrinsic fluorescence in rHcGST-1 (Q) against the concentration of free hematin (Hf) (calculated as previously described [31]), with an intrinsic fluorescence Kd value (rHcGST-1) of 1.72 ± 0.10 μM for hematin. Shown are averages of at least three determinations, with error bars representing standard deviations. (B) Competitive fluorescence of rHcGST-1 with the fluorescent ligand ANS and hematin. Lines 1 and 2 indicate the fluorescence of the rHcGST-1 protein with and without ANS, respectively. Lines 3 to 9 show the quenching of fluorescence due to the addition of hematin (with increments of 0.2 μM). (C) Competitive fluorescence of rHcGST-1 with ANS presented as a double-reciprocal plot, with a competitive fluorescence Kd value (rHcGST-1) of 1.13 ± 0.07 μM for hematin. Shown are averages of at least three determinations, with error bars representing standard deviations.
The 50% inhibitory concentration (IC50) of hematin for the GSH-CDNB conjugation rate was estimated to be 180 ± 13 nM (average of four replicates ± standard deviation) (Table 2) and was determined from direct plots of activity against inhibitor concentration, fitted by nonlinear regression. One anthelmintic, fenbendazole, inhibited GST at physiological concentrations (Table 2). This drug also competed with hematin for the same binding site on the GST in intrinsic fluorescence analysis (results not shown). The binding of fenbendazole to nematode GSTs with key feeding roles may be another mode of action for this benzimidazole drug in addition to presumed parasite tubulin inhibition (26).
The recombinant form of the closest relative in the free-living nematode C. elegans, CE07055 (Fig. 2), was expressed in E. coli following cloning from a cDNA library and successfully purified from the soluble native protein fraction by GSH affinity chromatography (results not shown). The specific activity of CE07055 with the model substrate CDNB was found to be 5,034.72 ± 223 nmol · min−1 · mg of protein−1 (average of four replicates ± standard deviation). Thus, the purification of recombinant CE07055 by GSH affinity chromatography and the activity with CDNB confirmed the functional integrity of the protein. The activity obtained is in the range of those of other nematode GSTs (22, 21). CE07055 did not appear to have a site with high affinity for hematin (IC50 = 32.7 ± 4.3 μM; average of four replicates ± standard deviation).
The HcGST hematin binding range is similar for hematin interaction with a number of mammalian liver tissue alpha class GSTs, which have proposed roles in heme detoxification and/or transport (23). Thirteen investigated human liver GSTs exhibit variation as hematin-binding proteins, with inhibition levels ranging from noninteraction to near 20 nM for GST III, with some of the GSTs showing a simple inhibition pattern and others showing a more complex, two-phase inhibition pattern (31). In contrast, mammalian extrahepatic Pi GSTs bind hematin in the range of 4 to 5 μM (23). The values for intrinsic and competitive fluorescent binding (Kd values of 1.72 ± 0.10 and 1.13 ± 0.07 μM, respectively) compared to the IC50 of hematin (180 ± 13 nM) suggest that fluorescence binding detects a different hematin site on the HcGST, distinct from the enzymatic active site. A number of mammalian liver GSTs also have proposed secondary binding sites, distinct from the substrate site, for hydrophobic ligands (25).
A universal heme signature sequence for heme binding proteins could not be determined by using the MEME motif discovery and search program (version 3.0; http://meme.sdsc.edu/meme/website/meme.html). Crystals of HcGST-1 bound to hematin have recently been produced (unpublished data) and this development will ultimately lead to the location of the hematin binding site and the investigation of potential heme binding signatures.
A high affinity for hematin and limited other activities imply a focused role for this GST from H. contortus in the detoxification or transport of heme or related compounds. In contrast to cestode and digenean parasites, nematodes may express GSTs with specific physiological roles rather than general detoxification activities. Compared to rHcGST-1, GSTs from the blood-dwelling digenean parasites Schistosoma mansoni (Sm28GST) and Schistosoma japonicum (Sj26GST) displayed a lower affinity for hematin (up to 23-fold lower) and a broader substrate specificity (34). This finding indicates that the digenean GSTs have more generalized detoxification roles than the more specialized GST from the voracious blood feeder H. contortus.
In conclusion, a GST from the drug-resistant parasitic nematode H. contortus may have a focused, nonenzymatic role involving binding and/or transporting heme-related compounds and is not apparently associated with immune defense or with drug metabolism or resistance. The function appears to be adapted to parasitism or, specifically, blood or tissue feeding, as this biochemical feature is not evident in the closest GST relative from the free-living nematode C. elegans. The expression of a parasitic nematode protein with a focused role may have implications for future drug and vaccine discovery. The C. elegans nematode model will now be utilized to assess the physiological significance of this GST class under external hematin stress via reverse and transgenic expression strategies.
Acknowledgments
We thank James K. Heald and Robert M. Darby for technical assistance.
We thank the EU and BBSRC for financial support.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Barrett, J., N. Saghir, A. Timanova, K. Clarke, and P. M. Brophy. 1997. Characterisation and properties of an intracellular lipid-binding protein from the tapeworm Moniezia expansa. Eur. J. Biochem. 250:269-275. [DOI] [PubMed] [Google Scholar]
- 2.Brophy, P. M., and J. Barrett. 1990. Strategies for detoxification of aldehydic products of lipid-peroxidation in helminths. Mol. Biochem. Parasitol. 42:205-211. [DOI] [PubMed] [Google Scholar]
- 3.Brophy, P. M., A. Bensmith, A. Brown, J. M. Behnke, and D. I. Pritchard. 1995. Differential expression of glutathione-S-transferase (GST) by adult Heligmosomoides polygyrus during primary infection in fast and slow responding hosts. Int. J. Parasitol. 25:641-645. [DOI] [PubMed] [Google Scholar]
- 4.Brophy, P. M., A. Bensmith, A. Brown, J. M. Behnke, and D. I. Pritchard. 1994. Glutathione S-transferases from the gastrointestinal nematode Heligmosomoides polygyrus and mammalian liver compared. Comp. Biochem. Physiol. Part B 109:585-592. [DOI] [PubMed] [Google Scholar]
- 5.Brophy, P. M., A. Brown, and D. I. Pritchard. 1994. A PCR strategy for the isolation of glutathione S-transferases (GSTs) from nematodes. Int. J. Parasitol. 24:1059-1061. [DOI] [PubMed] [Google Scholar]
- 6.Brophy, P. M., A. Papadopoulos, M. Touraki, B. Coles, W. Korting, and J. Barrett. 1989. Purification of cytosolic glutathione transferases from Schistocephalus solidus (plerocercoid)—interaction with anthelmintics and products of lipid-peroxidation. Mol. Biochem. Parasitol. 36:187-196. [DOI] [PubMed] [Google Scholar]
- 7.Brophy, P. M., L. H. Patterson, A. Brown, and D. I. Pritchard. 1995. Glutathione-S-transferase (GST) expression in the human hookworm Necator americanus—potential roles for excretory-secretory forms of GST. Acta Trop. 59:259-263. [DOI] [PubMed] [Google Scholar]
- 8.Brophy, P. M., and D. I. Pritchard. 1992. Immunity to helminths—ready to tip the biochemical balance? Parasitol. Today 8:419-422. [DOI] [PubMed] [Google Scholar]
- 9.Brophy, P. M., and D. I. Pritchard. 1992. Metabolism of lipid-peroxidation products by the gastrointestinal nematodes Necator americanus, Ancylostoma ceylanicum and Heligmosomoides polygyrus. Int. J. Parasitol. 22:1009-1012. [DOI] [PubMed] [Google Scholar]
- 10.Brophy, P. M., and D. I. Pritchard. 1994. Parasitic helminth glutathione S-transferases—an update on their potential as targets for immunotherapy and chemotherapy. Exp. Parasitol. 79:89-96. [DOI] [PubMed] [Google Scholar]
- 11.Brophy, P. M., C. Southan, and J. Barrett. 1989. Glutathione transferases in the tapeworm Moniezia expansa. Biochem. J. 262:939-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cortes, A., M. Cascante, M. L. Cardenas, and A. Cornish-Bowden. 2001. Relationships between inhibition constants, inhibitor concentrations for 50% inhibition and types of inhibition: new ways of analysing data. Biochem. J. 357:263-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorg, A., C. Obermaier, G. Boguth, A. Harder, B. Scheibe, R. Wildgruber, and W. Weiss. 2000. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 21:1037-1053. [DOI] [PubMed] [Google Scholar]
- 14.Guex, N., A. Diemand, and M. C. Peitsch. 1999. Protein modelling for all. Trends Biochem. Sci. 24:364-367. [DOI] [PubMed] [Google Scholar]
- 15.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 16.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 17.Jorgensen, W. L., and J. Tiradorives. 1988. The OPLS potential functions for proteins—energy minimizations for crystals of cyclic-peptides and crambin. J. Am. Chem. Soc. 110:1666-1671. [DOI] [PubMed] [Google Scholar]
- 18.Kawalek, J. C., R. S. Rew, and J. Heavner. 1984. Glutathione S-transferase, a possible drug-metabolizing enzyme, in Haemonchus contortus—comparative activity of a cambendazole-resistant and a susceptible strain. Int. J. Parasitol. 14:173-175. [DOI] [PubMed] [Google Scholar]
- 19.Kerboeuf, D., and J. Aycardi. 1999. Unexpected increased thiabendazole tolerance in Haemonchus contortus resistant to anthelmintics by modulation of glutathione activity. Parasitol. Res. 85:713-718. [DOI] [PubMed] [Google Scholar]
- 20.Kwa, M. S. G., J. G. Veenstra, M. Vandijk, and M. H. Roos. 1995. Beta-tubulin genes from the parasitic nematode Haemonchus contortus modulate drug-resistance in Caenorhabditis elegans. J. Mol. Biol. 246:500-510. [DOI] [PubMed] [Google Scholar]
- 21.Liebau, E., V. H. O. Eckelt, G. Wildenburg, P. Teesdale-Spittle, P. M. Brophy, R. D. Walter, and K. Henkle-Duhrsen. 1997. Structural and functional analysis of a glutathione S-transferase from Ascaris suum. Biochem. J. 324:659-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liebau, E., G. Wildenburg, P. M. Brophy, R. D. Walter, and K. Henkle-Duhrsen. 1996. Biochemical analysis, gene structure and localization of the 24 kDa glutathione S-transferase from Onchocerca volvulus. Mol. Biochem. Parasitol. 80:27-39. [DOI] [PubMed] [Google Scholar]
- 23.Mannervik, B., P. Alin, C. Guthenberg, H. Jensson, M. K. Tahir, M. Warholm, and H. Jornvall. 1985. Identification of three classes of cytosolic glutathione transferase common to several mammalian species: correlation between structural data and enzymatic properties. Proc. Natl. Acad. Sci. USA 82:7202-7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peitsch, M. C. 1995. Protein modelling by e-mail. Bio/Technology 13:658-660. [Google Scholar]
- 25.Salinas, A. E., and M. G. Wong. 1999. Glutathione S-transferases—a review. Curr. Med. Chem. 6:279-309. [PubMed] [Google Scholar]
- 26.Sangster, N. C. 1999. Anthelmintic resistance: past, present and future. Int. J. Parasitol. 29:115-124. [DOI] [PubMed] [Google Scholar]
- 27.Sharp, P. J., D. R. J. Smith, W. Bach, B. M. Wagland, and G. S. Cobon. 1991. Purified glutathione S-transferases from parasites as candidate protective antigens. Int. J. Parasitol. 21:839-846. [DOI] [PubMed] [Google Scholar]
- 28.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomson, A. M., D. J. Meyer, and J. D. Hayes. 1998. Sequence, catalytic properties and expression of chicken glutathione-dependent prostaglandin D-2 synthase, a novel class Sigma glutathione S-transferase. Biochem. J. 333:317-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows—a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 31.Vander Jagt, D. L., L. A. Hunsaker, K. B. Garcia, and R. E. Royer. 1985. Isolation and characterization of the multiple glutathione S-transferases from human liver: evidence for unique heme-binding sites. J. Biol. Chem. 260:11603-11610. [PubMed] [Google Scholar]
- 32.Van Rossum, A. J., P. M. Brophy, A. Tait, J. Barrett, and J. R. Jefferies. 2001. Proteomic identification of glutathione S-transferases from the model nematode Caenorhabditis elegans. Proteomics 1:1463-1468. [DOI] [PubMed] [Google Scholar]
- 33.Van Wyk, J. A., M. O. Stenson, J. S. Van der Merwe, R. J. Vorster, and P. G. Viljoen. 1999. Anthelmintic resistance in South Africa: surveys indicate an extremely serious situation in sheep and goat farming. Onderstepoort J. Vet. Res. 66:273-284. [PubMed] [Google Scholar]
- 34.Walker, J., P. Crowley, A. D. Moreman, and J. Barrett. 1993. Biochemical properties of cloned glutathione S-transferases from Schistosoma mansoni and Schistosoma japonicum. Mol. Biochem. Parasitol. 61:255-264. [DOI] [PubMed] [Google Scholar]
- 35.Yatsuda, A. P., J. Krijgsveld, A. W. C. A. Cornelissen, A. J. R. Heck, and E. De Vries. 2003. Comprehensive analysis of the secreted proteins of the parasite Haemonchus contortus reveals extensive sequence variation and differential immune recognition. J. Biol. Chem. 278:16941-16951. [DOI] [PubMed] [Google Scholar]