Summary
Host-associated microbial communities are unique to individuals, they affect host health, and they correlate with disease states. Although advanced technologies capture detailed snapshots of microbial communities, high within- and between-subject variation hampers discovery of microbial signatures in diagnostic or forensic settings. We suggest turning to machine learning and discuss key directions toward harnessing human associated microbial signatures.
Keywords: microbial communities, machine learning, source tracking, indicator species
Introduction
Different people harbor radically different microbial communities, which likely play key roles in a wide range of chronic diseases. If we can identify groups of bacterial taxa present in a human body habitat that are consistently predictive of host phenotype for different illnesses or treatments, then these biological signatures can be used to build models that predict therapeutic outcomes based on an individual’s specific microbiota. This approach, based on predictive models, has implications for diverse diseases that may benefit by modulation of the microbiota (e.g. through pre-biotics, pro-biotics, or targeted antibiotics), such as Inflammatory Bowel Diseases (IBD), obesity, diabetes, or diseases that are associated with malnutrition. Furthermore, given the recent finding that humans leave a signature of a distinctive skin microbiota on their keyboards (Fierer et al., 2010), this work also has implications for forensic identification. The crux of the problem is coping with the complexity and high-dimensionality of human-associated microbiota. Some progress has been made towards establishing the feasibility of supervised classification of these communities (Knights et al., 2011a), but there has been limited development of novel approaches, and many challenges remain. We discuss several of these challenges and important areas for future research into predictive modeling of human-associated microbial communities, as well as the potential applications that motivate this research.
Discovery of microbial signatures
Many human diseases are caused by single species or strains of bacteria, such as tuberculosis (Mycobacterium tuberculosis), tetanus (Clostridium tetani), and diphtheria (Corynebacterium diphtheriae); these specific taxa, along with their associations to host phenotypes, are sometimes referred to as biomarkers. Diagnosis and prevention of these types of diseases is relatively simple: if you have the biomarker, you have the disease. Similarly, tracking pathogens and contaminants in environmental samples has traditionally focused on counts of a single species, such as E. coli, or group of species, such as coliforms (Simpson et al., 2002). In the age of high-throughput DNA sequencing, discovery and verification of individual biomarkers for various host phenotypes is straightforward: collect and sequence enough data from hosts with and without the phenotype, and a classical hypothesis test (e.g. t-test or Mann-Whitney U test) will detect differential abundance of the biomarker. But there may be other cases when there is no single biomarker for a phenotype. We know now that host-associated bacterial communities are composed of hundreds or thousands of unique species, and many host phenotypes are associated with shifts in bacterial communities, but not with specific causative agents. For example, let us consider a hypothetical enteric disease state that is associated with concurrent overrepresentation of the phylum Bacteroidetes, the genus Shigella and the species Helicobacter pylori. We now have a three-way interaction between three different lineages of varying phylogenetic depth. We could refer to this set of interacting biomarkers, and the relationship that they have with the host phenotype, as a microbial signature. Such a signature need not be limited to taxonomic characterizations of communities (e.g. surveys of marker genes such as 16S rRNA) but may also include genes or functional categories.
As illustrated in the example above, a microbial signature may be arbitrarily complex, involving simultaneous over- and under-representations of multiple taxa at multiple taxonomic levels. In some cases, the traits that lead to disease may be limited to a single bacterial strain (perhaps one that has acquired virulent factors on a plasmid), while in others these traits may be more phylogenetically conserved, such that treating a whole genus or family as a feature would be optimal for dimensionality reduction. Given a hypothetical data set containing 1,000 unique species (pragmatically defined as 97% OTUs, or organisms with at least 97% identity in their 16S rRNA sequences), we would have to perform approximately one billion classical hypothesis tests to explore all such interactions at all taxonomic ranks, and controlling the rate of false positives would be next to impossible. Within these complex communities, how can we determine which lineages or genes matter, and at what taxonomic level, for a given host phenotype?
The discovery of such relationships is the goal of supervised learning: we use a set of communities with known phenotype to train a machine learning algorithm; the algorithm identifies discriminative independent variables and produces a predictive model which can then be used to predict the phenotype associated with other microbial communities. The machine learning community refers to this approach as “supervised learning”, or “supervised classification” (this use of the term “classification” is not to be confused with taxonomic classification of individual sequences or OTUs). Supervised learning is essentially a formalization of the implicit goal of most exploratory scientific research: based on the results of an experiment, we propose a descriptive model (e.g. a linear regression) that we believe will hold true for similar experiments in the future. What distinguishes supervised learning from classical hypothesis testing is that supervised learning deals explicitly with estimating and improving the expected future accuracy of a predictive model at the same time that it is discovering predictive signatures—they are two parts of the same process. There are extensive and varied approaches within machine learning devoted to building predictive models and maximizing their expected accuracy (reviewed in the context of microbial community classification in (Knights et al., 2011a)).
For simplicity we have focused so far on scenarios involving diagnosis of disease states, but we also envision potential applications in prognosis of treatment response, forensic identification of the host, and detection and sourcing of environmental sample contamination. In the context of these potential applications, we now discuss several remaining challenges in the discovery of predictive microbial signatures.
Improving discovery with existing biological knowledge
In many ways, studies of the microbiome can be informed by the extensive work that has been done in the closely related area of microarray classification (Lee et al., 2005), although there are some important distinctions (Knights et al., 2011a). Both microarrays and high-throughput characterizations of microbial communities such as marker-gene surveys or shotgun metagenomics produce high-dimensional data. However, unlike gene expression data, the low degree of overlap in species among subjects, for example, in the human gut, also leads to very sparse data matrices (i.e. matrices that contain many zeros) in marker gene surveys. The dual challenges of high dimensionality and high sparsity make it hard to identify individual biomarkers. Much of the work on predictive modeling of microarray data has focused on removing noisy or irrelevant independent variables (genes) from the data (Lee et al., 2005). In the field of machine learning this process of identifying and discarding noisy independent variables (e.g., taxa or genes) is often referred to as “feature selection”. Feature selection is similar to controlling the Type I error rate for multiple individual hypothesis tests, but the underlying motivation is to reduce the expected error of the model when it classifies novel communities.
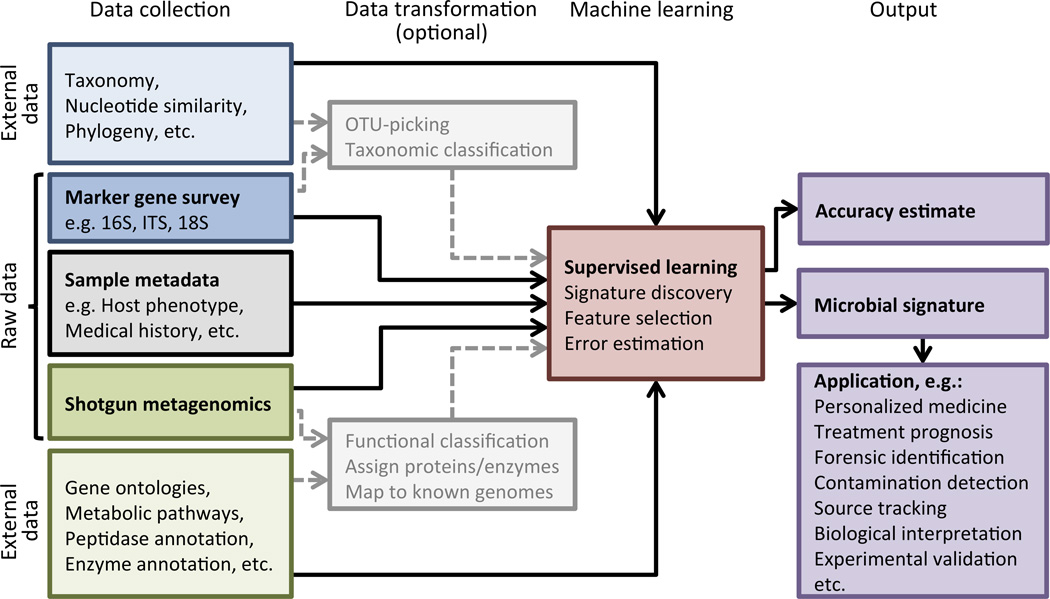
Several existing feature selection techniques are helpful for classifying microbial communities (Knights et al., 2011a). However, it is likely that we can also take advantage of relational or hierarchical structures in the data such as taxonomies, gene ontologies, metabolic pathways, etc. (Fig. 1) to share statistical strength between weakly predictive independent variables. One important consideration is that the abundance of taxa or genes is usually measured in relative terms. In this case the data are compositional, that is, when the relative abundance of one taxon increases, the relative abundance of the rest of the community must necessarily decrease. Consequently, explicit modeling of compositional distributions may be appropriate. One such probability distribution, the Dirichlet, has already been effective for community-wide microbial source tracking (Knights et al., 2011b).
Figure 1. Processes for microbial signature discovery.
The process begins with the collection of a large set of sequencing data from various bacterial communities associated with different environments or different host phenotypes. These sequences can serve directly as input to a machine learning algorithm, or they can be transformed through a preprocessing step (data transformation). Although for microbial community analysis data transformation and supervised learning are typically performed as separate steps, we suggest that predictive models will be improved by the development of novel machine learning techniques that are informed by the potential data transformations. For example, constructing a good predictive model using metabolic characterizations of metagenomics sequences might be easier if the algorithm has knowledge of the hierarchical relationships between metabolic functions. In the case of marker-gene surveys, a machine learning algorithm may benefit from knowledge of the phylogenetic relationships of the observed lineages, or the network of average nucleotide similarities between the input sequences. These structures may allow models to share statistical strength across related independent variables in cases where there is high variability within a given environment or host phenotype (i.e. lack of a “core microbiome”).
The hardest part of detecting microbial signatures is overcoming the high variability in microbial community composition both between and within hosts (or environmental habitats). Thus, transforming the raw data by collapsing or clustering the observed taxa or genes according to similarity is key. In the case of shotgun metagenomic sequences, we might first filter the sequences for known genes, and then assign them to functional or metabolic groups according to established databases prior to downstream analysis (Fig. 1). For surveys of marker genes (such as 16S rRNA), we commonly cluster sequences into operational taxonomic units (OTUs) based on a pre-determined threshold of nucleotide similarity (e.g. 97%). However, when we perform data transformation as a fixed pre-processing step, we may be making incorrect assumptions about the best way to collapse input data for a given predictive task. Alternatively, we propose that the next generation of predictive models must be able to integrate external information sources into the process of feature selection to determine the appropriate levels of collapsing, filtering, or clustering.
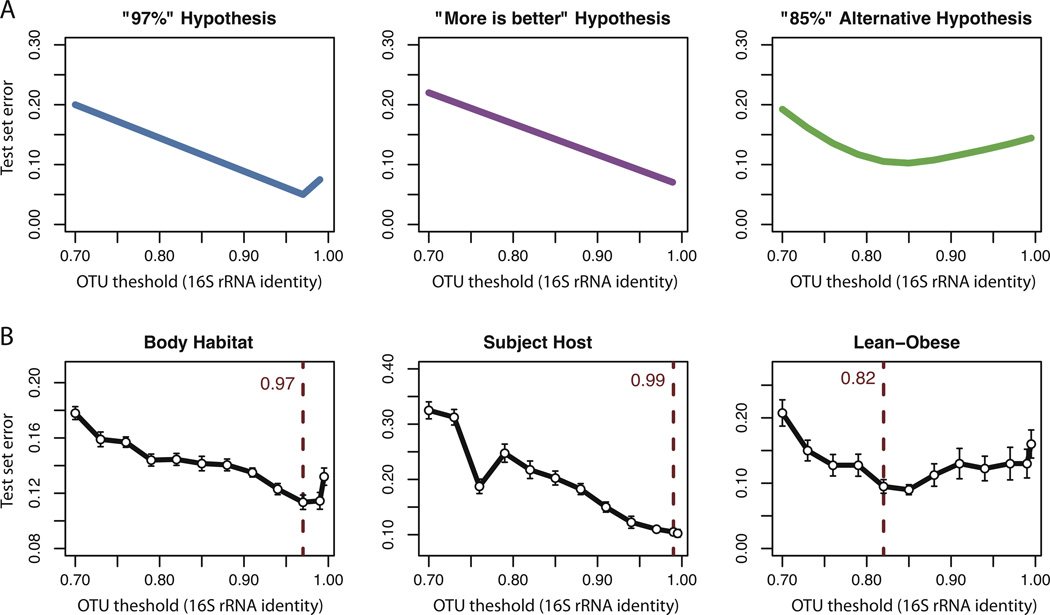
For example, when we pick OTU clusters for marker-gene sequences at a fixed threshold, potentially discriminative taxa may lose their signal if we make the clusters either too specific (e.g. 99% similarity), or too broad (e.g. 80% similarity). In the case where the clusters are too specific, any conclusions made about those clusters may not generalize well to future data sets due to high variability between communities. This potential pitfall is referred to as “overfitting”. Many published studies use a within-cluster similarity threshold of 97%, but we have found that this is not necessarily the best level for predictive modeling. In the context of predictive modeling, it is possible to estimate the best OTU threshold empirically as the one that minimizes the expected future error of a classifier. We studied six human-associated microbial communities with well-understood clustering patterns to determine their optimal OTU thresholds for predictive modeling. Three examples are shown in Figure 2. For a given benchmark, we estimated the generalization error of the Random Forests classifier (Breiman, 2001) using as input features OTUs picked at thresholds ranging from 60% to 99.5% nucleotide similarity. We then chose the optimal threshold for a given benchmark as the one giving the most parsimonious model (fewest OTUs) within one standard error of the best model (Figure 2). Optimal thresholds for the six tasks were surprisingly variable, ranging from 76% to 99%). This implies that predictive models are likely to benefit from a flexible approach to picking predictive OTU clusters, instead of the current practice of clustering at a fixed, pre-defined threshold of 97%.
Figure 2. Are we overfitting with 97% OTUs?
Many microbial ecology studies use operational taxonomic units (OTUs) defined at 97% 16S SSU rRNA sequence identity, consistent with the conventional bacterial species threshold. However, it is possible that either more specific, or more general OTU definitions may be useful for machine learning studies. Panel A shows hypothetical error curves for the case that the commonly used 97% 16S SSU rRNA identity threshold represents an optimal OTU definition for a given classification task, the case that more specific OTUs are always better, and the case that the optimal identity threshold is lower, for example 85%. The hypothetical error curves illustrate the concepts of “overfitting” and “underfitting”: if the clusters are too specific, then a predictive model cannot observe general trends in the data (overfitting); if they are too general, then the predictive features are getting buried during the clustering (underfitting). Panel B relates the choice of OTU threshold to empirical error in correctly classifying samples using a random forest classifier (Breiman, 2001) trained on two-thirds of the data and tested on the remaining third, for 10 randomly chosen train/test splits of the data. Three classification benchmarks are shown: the Body Habitat benchmark categorizes host-associated microbial communities by general body habitat; the Host Subject benchmark categorizes communities from the forearm, palm, and index finger by host subject; the Lean-Obese benchmark categorizes gut communities by host phenotype. Vertical dashed lines indicate the most parsimonious model (i.e. fewest OTUs) whose mean generalization error is within one standard error of the best model. The empirical error curves (B) suggest that different classification tasks may be best accomplished with different OTU definitions. This is a demonstration of our more general suggestion that existing knowledge about raw input data, whether marker genes or shotgun metagenomic sequences, must be incorporated into the next generation of predictive algorithms.
Furthermore, a recent exploratory study found that several host quantitative trait loci influenced the relative abundance of taxonomic groups of variable breadth (Benson et al., 2010), indicating that even within a given classification task, a single threshold for taxonomic clustering may be insufficient to capture the relevant habitat-related adaptations of microbial communities. For this reason, we believe that information about the nucleotide similarity or phylogenetic relationships of the input 16S rRNA sequences should be supplied directly to the machine learning algorithm, as shown in Figure 1. This will require the development of novel algorithms, but it has the benefit that the algorithm may select the appropriate levels of specificity for clustering input sequences given a particular predictive task. In the case of shotgun metagenomic sequences, we may cluster according to existing ontologies (Fig. 1).
Biological considerations and validation
Assuming that we are able to identify microbial signatures that are predictive of, for example, a diseased host phenotype, it may still be difficult to determine whether differences in "discriminating" taxa are a cause or a consequence of disease without large prospective longitudinal studies. As an example, although the composition of the vaginal microbiota may impact the rate at which HIV is transmitted, subsequent changes to the vaginal microbiota due to immune-dysfunction would make it impossible to characterize a community signature that may pre-dispose an individual to HIV infection by comparing the vaginal microbiota of HIV positive women to healthy controls. Similarly, individuals with IBD and celiac disease are believed to have increased intestinal permeability prior to the onset of disease (Groschwitz and Hogan, 2009), and it is reasonable to expect that corresponding changes, such as alterations in the phospholipid composition in the intestinal mucous barrier (Braun et al., 2009), may be associated with characteristic changes in particular bacterial species (e.g. promoting particular mucolytic species). Studies of how the microbiota differ with IBD, however, have generally compared people who have already developed the disease to those who have not (Frank et al., 2007). Consequently, taxa that differ may be those that can tolerate inflammation in the gut, and not those that are causing it, or those whose presence could predict disease onset.
Assuming that microbial signatures can be successfully associated with host traits, there are still many issues of interpretation that complicate attempts to make biological or mechanistic conclusions from those associations. The most reliable microbial markers for hard-to-observe host conditions will be backed both by extensive correlation data across studies and well-understood mechanisms that relate phenotype to particular genes, organisms, or community features. Two particularly noteworthy approaches to supplementing correlation data with mechanism include experimental confirmation, and genomic studies of microbial lineages. As an example of the first approach, Sharon et al. (Sharon et al., 2010) applied a combination of correlation studies and experimental confirmation to uncover a bacterium involved in Drosophila melanogaster mate preference. It had previously been observed that Drosophila raised on different media interbred less than those raised on the same medium. Investigation of the fly microbiota revealed that some lineages, in particular the Lactobacilli, differed in flies raised on different media, indicating that this could be either a cause or secondary marker of the observed difference in mate preferences. To distinguish between these possibilities, Sharon et al., demonstrated that broad-spectrum antibiotics could abolish the observed mate preference. Adding Lactobacillus plantarum could rescue the mate preference effect in antibiotic-treated flies. Such experimental confirmation greatly strengthens the case for approaches that would seek to use L. plantarum levels as a marker for mate preference in wild Drosophila populations beyond what could be said from correlation data alone. Further characterization of the mechanism involved in L. plantarum modification of mate preference (e.g. does it affect Drosophila pheromones?) would make this an even stronger candidate as a marker.
In cases where experimental manipulation is difficult, additional mechanistic information into the role of a putative marker microbe can be gained by examination of genome sequences. For example, Turnbaugh et al. (Turnbaugh et al., 2009) used a combination of genomic and transcriptomic approaches to study members of class Erysipelotrichi that increased when gnotobiotic mice, transplanted with a human microbial community, were switched from a low-fat diet rich in vegetables to a high-fat, high-sugar diet. These analyses found the genome of the cultured isolate to be enriched in phosphotransferase system (PTS) transporters, and identified PTS genes involved in the import of simple sugars as upregulated following the switch to a sucrose- and fat- rich western diet. Such genomic and transcriptomic findings supported the hypothesis that the observed increase in Erysipelotrichi was caused by changes in diet.
Discussion
In some cases, models of human-associated microbial communities can already give reasonably accurate predictions of important traits such as host phenotype, forensic identification of the host (Fierer et al., 2010), and environmental sources of sample contamination (Knights et al., 2011b). There is likely an enormous potential for improvement, however, with the increased availability of training data from a broad variety of prospective studies and the development of novel theoretical approaches that account for latent structures such as the phylogeny and behavioral characteristics of a microbiome. Experimental validation and biological interpretation of predictive models is also essential as the field moves toward high-stakes applications including personalized medicine and the early diagnosis of disease.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci U S A. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun A, Treede I, Gotthardt D, Tietje A, Zahn A, Ruhwald R, Schoenfeld U, Welsch T, Kienle P, Erben G, et al. Alterations of phospholipid concentration and species composition of the intestinal mucus barrier in ulcerative colitis: a clue to pathogenesis. Inflamm Bowel Dis. 2009;15:1705–1720. doi: 10.1002/ibd.20993. [DOI] [PubMed] [Google Scholar]
- Breiman L. Random Forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- Fierer N, Lauber CL, Zhou N, McDonald D, Costello EK, Knight R. Forensic identification using skin bacterial communities. Proc Natl Acad Sci U S A. 2010;107:6477–6481. doi: 10.1073/pnas.1000162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groschwitz KR, Hogan SP. Intestinal barrier function: molecular regulation and disease pathogenesis. The Journal of allergy and clinical immunology. 2009;124:3–20. doi: 10.1016/j.jaci.2009.05.038. quiz 21-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knights D, Costello EK, Knight R. Supervised classification of human microbiota. FEMS Microbiol Rev. 2011a;35:343–359. doi: 10.1111/j.1574-6976.2010.00251.x. [DOI] [PubMed] [Google Scholar]
- Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, Bushman FD, Knight R, Kelley ST. Bayesian community-wide culture-independent microbial source tracking. Nat Methods. 2011b;8:761–763. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Lee JB, Park M, Song SH. An extensive comparison of recent classification tools applied to microarray data. Computational Statistics & Data Analysis. 2005;48:869–885. [Google Scholar]
- Sharon G, Segal D, Ringo JM, Hefetz A, Zilber-Rosenberg I, Rosenberg E. Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc Natl Acad Sci U S A. 2010;107:20051–20056. doi: 10.1073/pnas.1009906107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JM, Santo Domingo JW, Reasoner DJ. Microbial source tracking: state of the science. Environ Sci Technol. 2002;36:5279–5288. doi: 10.1021/es026000b. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Science translational medicine. 2009;1:6ra14–16ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]