Abstract
The claudin family of integral membrane proteins was identified as the major protein component of the tight junctions in all vertebrates. Since their identification, claudins, and their associated pfam00822 superfamily of proteins have been implicated in a wide variety of cellular processes. Claudin homologs have been identified in invertebrates as well, including Drosophila and C. elegans. Recent studies demonstrate that the C. elegans claudins, clc-1-clc- 5, and similar proteins in the greater PMP22/EMP/claudin/voltage-gated calcium channel γ subunit family, including nsy-4, and vab-9, while highly divergent at a sequence level from each other and from the vertebrate claudins, in many cases play roles similar to those traditionally assigned to their vertebrate homologs. These include regulating cell adhesion and passage of small molecules through the paracellular space, channel activity, protein aggregation, sensitivity to pore-forming toxins, intercellular signaling, cell fate specification and dynamic changes in cell morphology. Study of claudin superfamily proteins in C. elegans should continue to provide clues as to how claudin family protein function has been adapted to perform diverse functions at specialized cell-cell contacts in metazoans.
Keywords: claudin, junctions, C. elegans, epithelia, morphogenesis, actomyosin, neuronal symmetry, VAB-9, CLC-1, NSY-4
Introduction: Claudins Regulate Barrier Functions in Vertebrate Epithelia
Epithelia function as regulated barriers in tissue and organ architecture, defining compartment boundaries within organisms and forming boundaries apposed to the external environment. The proper physiological state of epithelia depend on cellular contacts, or junctions, that mediate cell-cell adhesion, function as selective gates for the ingress or egress of specific ions and small molecules, and maintain unique membrane identities on apical and basal sides of the cellular layer, thus contributing to epithelial polarity. The tight junction is the most apical of cellular junctions in vertebrate epithelia. Freeze fracture TEM reveals one of the most striking features of tight junctions, a network of intertwined strands that occlude the intercellular or paracellular space at points of contact that often circumscribe cells. In this way, tight junction strands function as gaskets that separate and isolate the two sides of an epithelium (Fig. 1). The major protein component of the tight junction was identified 15 y ago as the claudin family of proteins. Claudins were originally identified in a search for the protein component of tight junction strands in vertebrate epithelia following a painstaking purification scheme.1,2 Claudins 1 and 2 were shown to localize to tight junctions, to generate tight junction strands when expressed in fibroblasts, and to mediate homotypic and heterotypic cell-cell adhesion.1,2 Consistent with these findings, loss of claudin activity results in paracellular barrier dysfunction.3-8 Overall tissue polarity does not appear to be heavily dependent on claudin activity, since gross cell polarity is maintained following loss of claudin function.6 The vertebrate claudin family has expanded over the years to include about 27 members, all of which are predicted four-pass integral membrane proteins with cytoplasmic N- and C-termini, two extracellular loops, and one short intracellular loop.9-12

Figure 1. Schematic of various cell junction arrangements in vertebrates, Drosophila, and C. elegans epithelia. Corresponding cell junction regions either in relative location, function, and/or molecular make-up are indicated with similar colors. Green indicates the tight junction in vertebrates the SAR-like region in C. elegans, the sub-apical region (SAR) in Drosophila. Blue indicates the cadherin-based adherens junction. Red indicates the septate junction in Drosophila and the AJM-1/DLG-1 region in C. elegans. Vertebrates have no precisely analogous structure to the septate junction in epithelia, but share a molecularly similar barrier at the paranodal junction. Vertebrate desmosomes are shown in yellow (A). Lateral views (cutaways) show the nature of the junctional structures in the membranes. Adherens junctions appear as solid bands in the membrane, pleated septate junctions are characterized by regular wave-like strands, and tight junctions appear as irregular but connected anastomosing strands. For simplicity, only the composition of cell junction components in the paracellular space is shown. (B) The most common cellular junctions and representative protein components are listed. Transmembrane proteins are indicated in color (bold) and components unique to vertebrates (other than desmosomes) are underlined.
As integral membrane proteins, claudins may function as adhesion receptors, or to regulate paracellular traffic. Since claudins have been shown to form oligomers in vitro, with hexamers being favored, one possibility is that claudins, like gap junctions, might generate pores through the lipid bilayer; at present there is no clear evidence this is the case.13-15 Rather, claudins appear to establish charged pores within the paracellular space that regulate the traffic of charged ions across the epithelial barrier. Charged residues in the extracellular domain are responsible for regulating charge selectivity. As a result, claudins with different charged amino acids can, by virtue of their expression in overlapping and non-overlapping patterns, define the paracellular charge selectivity of a wide variety of tissues.16 For example, in the vertebrate kidney, different claudins are expressed along the length of the nephron, suggesting that the differential expression correlates and may help generate that different charge selectivity among the distinct regions of the nephron.17 Supporting this hypothesis, claudin 16 maintains a negative charge required for the re-uptake of Mg2+ and Ca2+ ions in the thick ascending part of Henle’s loop in the nephron and loss of claudin 16 activity results in magnesium and calcium wasting.18 Similarly, in the longitudinal axis of the intestine as well as the crypt to villus axis of the intestine, the claudins are expressed in complex patterns, and expression of claudin types can change temporally during epidermal development as well.19,20 Charge selectivity of claudins is most dramatically illustrated by experiments in which the charge of the extracellular loops is reversed by swapping amino acids in this loop; in this case the charge selectivity is reversed, as was demonstrated for claudins 10 and 15.21,22 In general, amino acid charge in extracellular loop 1 dramatically influences paracellular charge selectivity. The extracellular Claudin loops are also targets of viruses, and binding of viruses can disrupt the epithelial barrier, open the paracellular space, and allow viral entry.23-25
The claudin C-terminus functions in trafficking of the claudin protein, binding to junctional plaque proteins through PDZ domain binding motifs, or regulation of the paracellular space through phosphorylation.8,26-30 While the PDZ binding peptide is found in most claudins, this domain is absent from many of the C-termini in related protein families of both vertebrates and invertebrates, suggesting that there may be as many functions as there are unique C-terminal domains.
While much is now known about the function of vertebrate claudins, less is known about similar molecules in invertebrates. Since tight junctions structures in invertebrates are uncommon, claudin-like molecules in invertebrates, although likely having conserved essential functions with their vertebrate relatives, may be exploited in unique ways to achieve distinct outcomes in different tissue types.31 The roles of claudin family proteins in other invertebrates will be covered in detail elsewhere; this review will detail the known functions of this class of proteins in C. elegans.
Sequence Conservation among Claudin Family Proteins
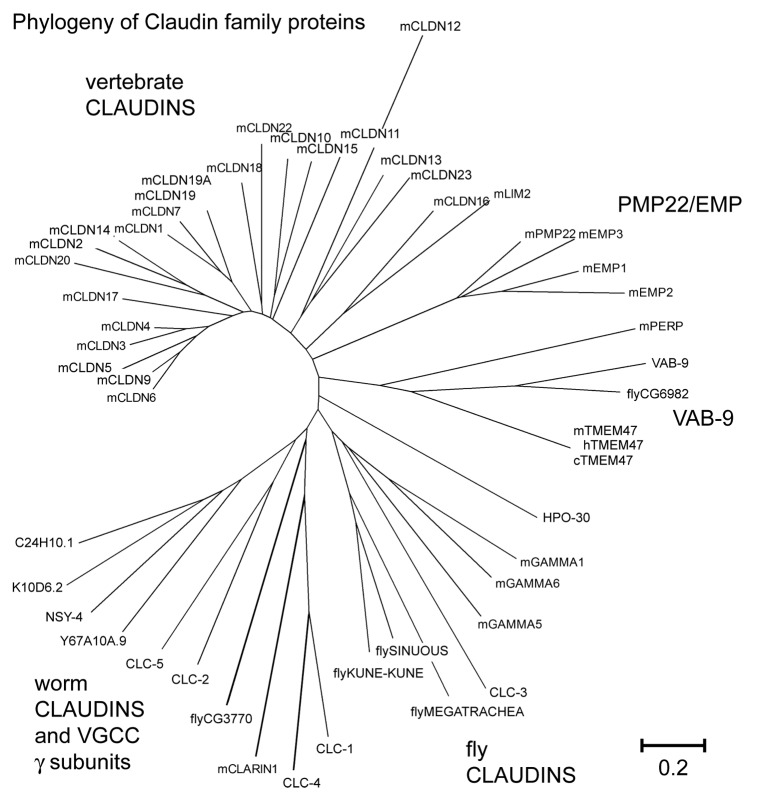
The sequences of C. elegans and vertebrate claudin family proteins are so divergent that it was proposed sequence similarity was insufficient to establish homology or that claudins only exist in chordates.9,32 Despite these initial assessments, claudin family proteins were ultimately identified in C. elegans by sequence homology searches and also through the identification of claudin-related proteins participating in diverse processes. Here will be reviewed what is known about the claudin/PMP22/EMP superfamily of proteins (NCBI pfam00822) encoded by vab-9, nsy-4, clc-1–5, and related genes. The comparison of C. elegans sequences with each other and with claudin family proteins from vertebrates indicates that the sequences are indeed highly divergent (Fig. 2). A similar observation was made when Drosophila claudins, including Sinuous and Megatrachea, were placed into a claudin family phylogenetic tree.33 A previous analysis of Stargazin indicated there were at least 3 distinct clades within this greater tetraspan family: PMP22/EMP, Claudin, and the gamma subunits of voltage-gated calcium channels.34 Analysis of these protein groupings, in conjunction with VAB-9 and VAB-9 orthologs from Drosophila and vertebrates, suggests that a separate VAB-9 clade exists, which is distinct from other subgroups (Fig. 2). Previously PERP, a tetraspan protein required for desmosomal structure and epithelial integrity, was recognized as the closest ortholog to TM4SF10 (or TMEM47) in the mouse35,36 (JSS and L. Attardi, unpublished observations). In our phylogenetic analysis, PERP is weakly associated with the VAB-9 clade, (Fig. 2). Despite being closer to VAB-9 in homology, expression of mouse PERP in C. elegans epithelia is incapable of rescuing vab-9 phenotypes, while similar expression of TM4SF10 rescues vab-9 37 (JSS and L. Attardi, unpublished observations). Bootstrap analysis shows that the frequency of particular association nodes for C. elegans claudins are often lower than 50%, highlighting the diversity of this protein family.
Figure 2. pfam00822 proteins from vertebrates and invertebrates are highly divergent. Bootstrap analysis of pfam00822 proteins using the MEGA (Molecular Evolutionary Genetic Analysis) software program available at http://www.megasoftware.net.108 The phylogenetic tree resulting from ClustalW2 analysis is shown. Several different subgroup clades are observed. Note that NSY-4 clusters with other putative gamma subunits and C. elegans claudins, while the vertebrate VGCC gamma subunits are loosely associated with fly claudins. Scale bar indicates amino acid substitutions per residue. Accession numbers for proteins included in the alignments are: NSY-4 (NP_500189.4), K10D6.2(NP_505843), R04F11.1 (NP_506087), C24H10.1(NP_508863), Y67A10A.9(NP_502746.1), mGamma5 (voltage-dependent calcium channel gamma subunit 5) (NP_542375), mGamma1 (NP_031608), mGamma6 (NP_573446.1), mGamma3 (NP_062303,)CLC-1(NP_509847), CLC-2(NP_509257), CLC-3 isoform a(NP_001024993), CLC-4 (NP_509800), CLC-5(NP_509258), VAB-9 (NP_495836), CG6982 (dVAB-9) (NP_001097876), mTMEM47(NP_620090), cTMEM47 (NP_001003045.1), hTm47 (NP_113630.1), xtmem47 (NP_001085134.1), mclaudin1 (NP_057883), mclaudin2 (NP_057884), mclaudin3 (NP_034032), mclaudin4 (NP_034033), mclaudin5 (NP_038833), mclaudin6 (NP_0247), mclaudin7 (NP_058583), mclaudin9 (NP_064689), mclaudin10 (a) (NP_076367), mclaudin11 (NP_032796), mclaudin12 (NP_075028), mclaudin13 (NP_065250), mclaudin14 (NP_001159398), mclaudin15 (NP_068365), mclaudin16 (NP_444471), claudin17 (NP_852467), mclaudin18(NP_062789), mclaudin19(1)(NP_001033679), Claudin19(2)(NP_694745), mclaudin20 (NP_001095030), mclaudin22 (NP_083659), mclaudin23 (NP_082274), fly_Sinuous (a) (NP_647971), fly_Megatrachea (NP_726742), fly_CG3770 (NP_611985.), fly_Kune-kune (NP_610179), mEMP2 (NP_031955), mPMP22 (NP_032911), mEMP3(NP_001139818), mEMP1(NP_034258), mPERP(NP_071315), LIM2 (NP_808361)
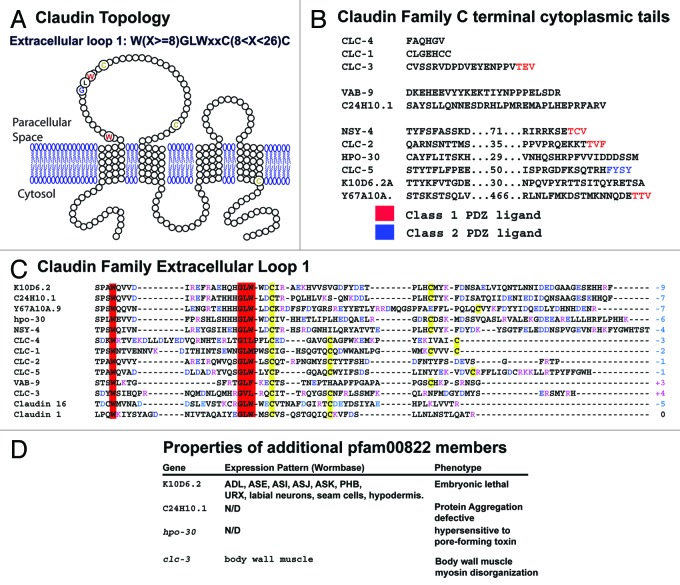
Previously, a consistent motif was found in the first extracellular loop of the Claudin superfamily of proteins, as noted by others (e.g., 38). In C. elegans, this consensus roughly corresponds to W(X > 8)GLWXXC(8–10X)C (Fig. 3A). Direct sequence comparison shows that VAB-9 is not entirely conserved in this region, since VAB-9 lacks the highly conserved tryptophan in the GLW tripeptide (Fig. 3C). Nevertheless, the predicted topology of VAB-9, as well as conservation of many of the residues in the motif, suggests it may share structural features with other members of the claudin superfamily. Other proteins from C. elegans tend to lack one or more key residues but retain a broad similarity. Whether this similarity translates to a conserved structure will require functional tests.
Figure 3. Sequence conservation and motifs in C. elegans pfam00822 proteins. (A) The predicted membrane spanning topology of VAB-9 is shown; pfam00822 proteins have the same membrane-spanning topology. Highly conserved residues among family members are indicated in enlarged, colored circles. (B) The C-terminal cytoplasmic tails of the C. elegans pfam00822 proteins are shown, from the end of the final transmembrane domain to the final residue. Sequences of longer proteins are abbreviated; numbers indicate intervening residues not shown. Terminal sequences conforming to class 1 and 2 PDZ binding domain ligand rules are indicated in red and blue, respectively. (C) The first extracellular loop domain of C. elegans proteins and murine claudin are shown. Postiviely charged residues are indicated in red, negative charged residues in blue. The sum of extracellular loop charge is indicated at the end of the sequence. Cysteines are indicated in yellow and the conserved tryptophan and GLW motif is indicated in red. (D) Additional pfam00822 proteins, for which some preliminary information is known, are shown. K10D6.2 expression, C24H10 function in protein aggregation, hpo-30 function in toxin resistance, and clc-3 expression and phenotype are referenced in 109, 110, 111, 112, and 113 respectively. Additional unpublished results, particularly regarding clc-3, can be found at http://wormbase.org.
The first extracellular loop has also been identified as the essential domain for regulating paracellular charge selectivity, as described above. The EL1 loops and their overall charge are indicated in Figure 3C. It will be interesting to see whether any of these proteins play a role in regulating charge selectivity; currently there is no evidence for regulation of paracellular ion transport by C. elegans claudins.
Another striking feature of most claudins is the presence of a C-terminal PDZ domain ligand. Figure 3B shows that several of the C. elegans claudin family proteins contain PDZ motifs, but the PDZ binding motif appears to be lacking in others, such as VAB-9, CLC-1, CLC-4, and HPO-30. Either these proteins lack a true PDZ ligand, or the ligand has diverged so as to not be easily recognized as such using the simple rules that identify class 1, 2 and 3 ligands. Interestingly, peptide scanning and network approaches to understanding the structural variation in PDZ ligands indicates that many PDZ binding domains diverge from the more static class identifications and reinforces the caution that characterization of a PDZ ligand optimally requires direct demonstration of binding and a functional in vivo requirement for that binding.39,40
Since the claudin family has undergone extensive divergence, the study of claudin-like proteins in C. elegans and other invertebrates is likely to reveal both essential functions of the claudin family of proteins as well as adaptations of members of this divergent family to unique functions.
Tight vs. Septate Junctions: Cell Junctions in Vertebrates, Drosophila and C. elegans
While cadherin-based junctions appear to be present in epithelia throughout the animal kingdom, the position and morphology of occluding junctions vary among metazoans. The most apparent distinction is the location and structure of cell junctions (Fig. 1). In vertebrates, the tight junction is apical to the adherens junction, while invertebrate junctions typically have no significant occluding junction apical to the adherens junction. Although some lower chordates and invertebrates have tight junction structures, most invertebrate epithelia have instead a different junctional structure, the septate junction, localized basal to the adherens junction.41-43 Septate junctions consist of at least two types: pleated septate junctions, which are characterized by ladder-like bridges, and smooth septate junctions, in which bridges are not detected using electron microscopy.31,44,45 The separation across the paracellular space at the septate junction is about 10–20nm and there are no “kissing points,” as in vertebrate tight junctions, where there is effectively no paracellular separation and opposing membranes appear to directly touch.46 Although pleats in septate junctions are organized into precise parallel rows, freeze fracture TEM of insect and Ascaris epithelia typically reveals the rows are composed of spaced, contacting puncta, rather than the continuous strands characteristic of many vertebrate tight junctions.47,48 Despite the freeze fracture appearance of a more porous and incomplete barrier, the septate junction structure still fulfills the function of a barrier, since the loss of proteins that localize to the septate junction in Drosophila, including claudin-like proteins Megatrachea, Sinuous, and Kune-kune result in the disruption of junctional structure, epithelial cell adhesion, and the paracellular barrier gate function.33,49-52
Structural and morphological differences between TJs and SJs, are reflected in differences at the molecular level. For example, the vertebrate tight junction protein complexes containing PAR-3/PAR-6/aPKC/CDC-42 or CRB3/Pals1/PATJ have homologs in Drosophila that localize to the sub-apical region (also known as the apical marginal zone), a region apical to the adherens junction.53 In contrast, a distinct complex, including the MAGUKs Varicose and Dlg, the Erm protein Coracle, Neurexin IV, and other proteins localizes basal to the adherens junction at the SJ.50,51,54-57 Thus, although tight junctions and septate junctions share some common functions, they are not analogous structures at the structural and molecular levels.
In contrast to vertebrates and other invertebrates, the C. elegans epidermis contains a single discernable electron dense junctional region.58 Although the spatial distribution of specific molecular complexes within this single electron density has not been performed, C. elegans epithelia contain many of the same proteins as in Drosophila, with similar spatial ordering. C. elegans possesses a single homolog of classical cadherins, HMR-1, and associated proteins HMP-2/β-catenin, HMP-1/α-catenin, and a divergent p120ctn, JAC-1.59,60 These proteins appear to localize to the nematode equivalent of adherens junctions. Similarly, in C. elegans, the discs large homolog DLG-1 and its binding partner AJM-1 localize basal to the adherens junction. Co-staining demonstrates that these cell junction proteins localize to different regions of the lateral cell membrane, with the adherens junction proteins being more apical.61-64 A third localization domain, apical to the adherens junction, which may extend from the junctions across the apical surface, contains of the familiar Par-3-Par-6-aPKC complex (Fig. 1).61,63-66 Other proteins, not part of the Par complex, such as CHE-14 and EAT-20 (a crumbs homolog) are also localized across the apical surface.67,68
As in Drosophila, overall polarity of epithelial junctions is established and maintained by LET-413, the LAP (LRR and PDZ) domain containing protein homolog of Drosophila Scribble.69,70 LET-413 localizes along the basolateral surface of epithelial cells, establishes global cell polarity and therefore is required for the localization of cellular junction components in epithelial cells and the structure of the intestinal terminal web.70,71 The apical junctional complex appears to be composed of two distinct and independent protein localization domains, since even though loss of dlg-1 gene function can completely eliminate the lone electron dense structure, and cause the mislocalization of the AJM-1 protein, classical adherens junction proteins such as HMR-1 (cadherin), HMP-1 (α-catenin), and VAB-9 (TM4SF10) remain essentially localized, if not completely aggregated, in a narrow band. Similarly, loss of adherens junction proteins does not disrupt the localization of DLG-1 or AJM-1, providing further evidence for the existence of at least two independent junctional regions within or near the apical electron dense junctional structure.61-65,70 Furthermore, cell polarity and adhesion are only mildly disrupted in ajm-1 and dlg-1 RNAi animals, and these adhesion defects are enhanced by vab-9 65. Notably, the localization of apical surface proteins is also not affected by the loss adherens junction or DLG-1/AJM-1 domain proteins.64,67 Loss of dlg-1 can affect the localization of the Drosophila Crumbs homolog (CRB-1) protein, which is a component of a separate apical protein complex, but this protein is not required for polarity in C. elegans as is the case for Drosophila Crumbs.61 These findings indicate that there are at least three distinct apical cell junctional complexes in C. elegans epithelia that together contribute to cell adhesion and morphogenesis, and demonstrate that even though C. elegans epidermal cells lack the ultrastructural signatures of vertebrate and Drosophila tight and septate junctions, respectively, at least some of the core properties of those junctions are conserved in the nematode.
Spermathecal Junctions: Specialized Cellular Junctions that Facilitate Extreme, Rapid, and Reversible Changes in Tissue Morphology
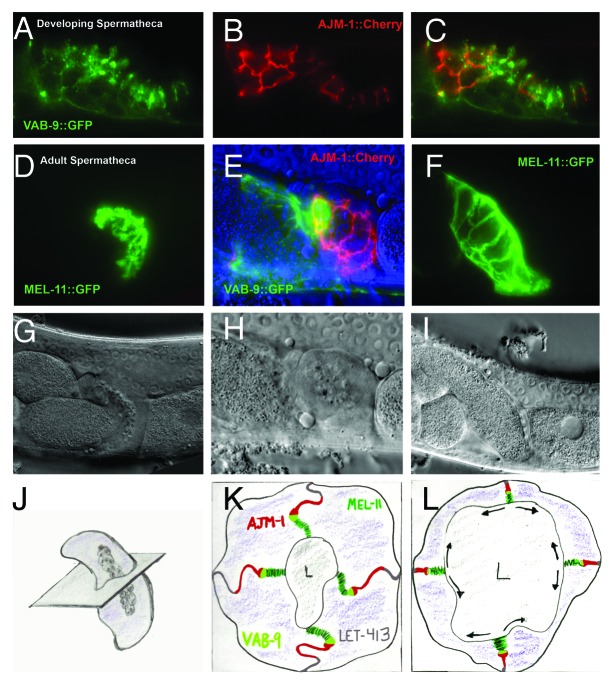
C. elegans cellular junctions arising from the mesoderm, such as the cell junctions in the spermatheca of the somatic gonad, are distinct from the epidermal or intestinal epithelia. The spermatheca stores sperm in the hermaphrodite. During fertilization, the spermatheca expands dramatically to allow the passage of an oocyte from the syncytial gonad into the uterus.72-74 During this expansion, cell junctions must “unzip” on both sides of a stable adherens junction to increase the amount of cell membranes contributing to the expanded luminal and basal membranes.75 Apical to the adherens junction is a junction with an appearance similar to a pleated septate junction, characterized by ladder-like crossbridges between cells, and basal is a junction with a smooth or continuous septate junction appearance. Based on AJM-1::Cherry localization (4E), AJM-1 expression appears to be strongest in the region of the adherens junction, and the smooth or continuous septate junction, while MEL-11::GFP localizes to the pleated septate junction (Fig. Four D and F). The finding that AJM-1 decorates the electron dense adherens junction structure in immuno EM preparations of the spermatheca (D. Hall, personal communication) may seem to be a paradox, but can be explained by the observation that the loss of dlg-1 results in a loss of this electron dense structure and AJM-1 localization in epidermal cells, without loss of adherens junction protein localization (e.g,64 suggesting that adherens junction proteins (such as VAB-9 and HMR-1) localize just apical to the electron dense structure.
MEL-11 regulates actomyosin-mediated contraction during cytokinesis and embryonic elongation, and also is required in the spermatheca for fertility.76-78 Since actomyosin at cellular junctions regulates both apical constriction and the organization of cellular junctions, it is exciting to speculate that mel-11 regulates actomyosin contraction required for unzipping and zipping of the pleated spermathecal junctions during fertilization. How similar are the spermathecal junctions to those of Drosophila pleated septate junctions based on expression of claudin family proteins? Of the claudin-like proteins in C. elegans, both VAB-9 and CLC-1 are expressed in the spermatheca, with VAB-9 being expressed during the larval stages of development and concentrated apically during tissue polarization (Fig. 4A). As in epidermal tissues, VAB-9 localization in the spermatheca is apical to AJM-1 at the subcellular level (Fig. 4B,C).65 In the fourth larval stage and in young adults, VAB-9 localizes just basal to MEL-11, which is concentrated in the pleated septate junctions. Thus, MEL-11, VAB-9 and AJM-1 roughly specify three regions of spermathecal cell junctions (Fig. 4K). In adults, during ovulation, expression levels and localization are altered: VAB-9 levels are reduced in spermathecal junctions and are prominent in the spermatheca-uterine valve (Fig. 4D-F), suggesting that changes in tissue and expression levels may be required for fertilization. One hypothesis is that VAB-9 has a unique role in regulating the function of MEL-11 and therefore actomyosin contraction during fertilization; specifically, VAB-9 may regulate the “unzippering” of the MEL-11 decorated pleated septate junction. Since CLC-1 is also expressed in the spermatheca, but its subcellular localization remains unknown, it remains a possibility that CLC-1, or one of the other C. elegans claudin-like proteins localize to the pleated septate junction and may regulate spermathecal development or function during fertilization79.
Figure 4. Cell junctions in the spermatheca. VAB-9::GFP, AJM-1::Cherry and MEL-11::GFP expression decorate distinct junctional regions of the spermathecal membrane. (A-C) During development, VAB-9::GFP (A) localizes to more a more apical (luminal) position relative to AJM-1::Cherry (B). The merged image is presented in (C). (D-E) In adult spermatheca, MEL-11::GFP (D) appears to localize to the most apical, folded regions of the lateral membranes (corresponding to the pleated septate junctions), while AJM-1::Cherry (E) decorates the more basal membranes (corresponding to the electron dense adherens junction and smooth or continuous cell junction). LET-413 (gray) is localized along the most basal portion of the lateral surface.66 VAB-9::GFP expression is reduced in the adult spermatheca, but is increased in the spermatheca-uterine valve (E). During ovulation (F), the MEL-11::GFP-decorated pleated septate junctions “unzip” to allow the passage of the oocyte. (G-H) show the matching DIC images for (D-F), and (J-L) are representative illustrations of (G-I). The plane passing through the center of the spermatheca in (J) is projected in (K) and (L). Arrows in (L) indicate the expanding apical (luminal) surface of spermathecal cells at the expense of the lateral (pleated junctional) domain. Similar expansion may take place reducing the smooth/continuous junction in favor of basolateral surface are (not shown).
The Function of VAB-9: A Claudin-Like Protein at Adherens Junctions
VAB-9 is an approximately 22kDa protein with similarity to the PMP22/EMP/claudin/gas3 family of 4 pass membrane spanning proteins. VAB-9 is expressed in all C. elegans epithelia and co-localizes with the adherens junction proteins HMR-1 (cadherin) and HMP-1 and 2 (α- and β-catenin, respectively). HMR-1 is required for VAB-9 membrane localization and HMP-1 is required to maintain uniform circumferential VAB-9 distribution about the adherens junction. vab-9 mutants have defects in body morphology, likely due to defective filamentous actin organization in epidermal cells. Mutations in vab-9 enhance the morphological defects of weak hmp-1 loss of function and enhance cell adhesion defects in ajm-1 and dlg-1 animals. Thus, vab-9 participates in the organization of F-actin at the adherens junction and, alongside with the AJM-1/DLG-1 complex, maintains proper epithelial adhesion.
Clues to the general functions of VAB-9 may come from the vertebrate ortholog, TM4SF10, previously known as BCMP1. TM4SF10 is expressed strongly in the canine brain, and based on SAGE analysis, is expressed in human brain astrocytoma, ependymoma, and normal spinal cord. This finding, along with the genetic position of TM4SF10 on the X chromosome at p21.1, suggests an association with hereditary X-linked mental retardation loci; however, no disease-associated changes were detected in the TM4SF10 locus in XLMR patients from 14 unrelated families.80,81 Thus, mutations in TM4SF10 are either rare among affected individuals or TM4SF10 is involved in separate processes in the brain. In the developing mouse kidney, TM4SF10 is expressed transiently in podocyte precursors, and expression diminishes as the cell junctions of these precursors transition from an occluding type junction typical of columnar epithelia to the specialized adherens junctions of slit diaphragms.37 Slit diaphragms are organized around the transmembrane proteins of the nephrin family.82,83 Nephrin and the related protein, Neph1, participate in homo- and heterotypic intercellular adhesion along interdigitated podocyte cell extensions.84,85 Slit diaphragm development results from signaling through Nephrin family protein cytoplasmic domains which in turn effect changes in the underlying actin cytoskeleton, cell process extension, and cell process interdigitation, ultimately generating the podocyte side of the filtration barrier in the glomerulus. Following phosphorylation of Nephrin and Neph1 by the Src family kinase Fyn, Nck, PI3K and Grb-2 bind specific phosphotyrosines and signal to reorganize the actin cytoskeleton.86-93 TM4SF10 expression regulates the activity of Fyn and Nephrin maturation in podocytes suggesting that TM4SF10 may regulate the ability of nephrin to control actin dynamics and cell process extension, possibly by influencing Fyn phosphorylation of Neprhin.94 Consistent with this hypothesis is the finding that overexpression of TM4SF10 phenocopies inhibition of neurite outgrowth by RhoA overexpression following NGF stimulation of PC12 cells.95 Similarly, overexpression of TM4SF10 in MDCK cells blocks Fyn-dependent cell process extension.94 These results indicate that TM4SF10 may have a general role in maintaining a columnar epithelia phenotype indirectly by preventing the formation of actin-dependent cell extensions. To date, interactions between VAB-9 and homologs of Nephrin and Fyn in C. elegans have not been investigated, although FRK-1, a non-receptor FER-like kinase, is required for embryonic morphogenesis.96-98 It will be of future interest to explore the interactions between TM4SF10, Fyn, and Nephrin and determine whether such interactions are unique to vertebrate podocytes or if they are conserved in all epithelial cell types that express these proteins.
Further insight into the function of vab-9 comes from an analysis of the MAGUK protein ZOO-1. ZOO-1 is the C. elegans ortholog of the tight junction membrane-associated guanylate kinase (MAGUK) protein ZO-1.99 ZOO-1 is a predicted 129kDa protein with three PDZ motifs in the N terminus, an SH3 domain, a guanylate kinase domain (predicted to be inactive), and a C-terminal ZU-5 domain, unique to this class of MAGUKS and netrins.51 Several data indicate that zoo-1 closely interacts with vab-9. First, ZOO-1 localizes to cell junctions in the epidermis and, like VAB-9 requires HMR-1 (cadherin), but not HMP-1 or HMP-2, for junctional localization. Second, zoo-1 localization requires VAB-9. Third, similar to vab-9 mutants, loss of zoo-1 function enhances the morphogenetic phenotypes of a weak hmp-1 loss of function allele. Fourth, loss of zoo-1 and vab-9 function affects the organization of filamentous actin in the epidermis of elongating embryos. Fifth, loss of zoo-1 function does not enhance the phenotypes of vab-9 mutants, suggesting they act in a common pathway. Sixth, both vab-9 and zoo-1 interact with mutations in genes of the actomyosin contractile machinery. Loss of zoo-1 activity enhances mel-11 (myosin light chain phosphatase) mutants and suppresses let-502 (ROCK) mutants, whereas vab-9 mutations suppress mel-11 alleles (JSS, unpublished). Surprisingly, zoo-1 mutants alone have virtually no effect on morphogenetic phenotypes, although very mild abnormalities of F-actin accumulation at junctions were noted (Lockwood). Together these results suggest a regulatory pathway in which HMR-1 regulates VAB-9 and vab-9 in turn regulates a subset of epidermal morphogenetic processes; a subset of these processes are in turn mediated by zoo-1 and mel-11 either in common or independent pathways. Since unpublished data from my laboratory suggests VAB-9 may be required for MEL-11 localization within the epidermis, it should be interesting to determine the nature of the interactions between these protein classes in regulating F-actin organization at epidermal cell junctions and within epidermal epithelia. Vertebrate ZO-1 and ZO-2 have been shown to influence the paracellular barrier and epithelial morphology through regulation of perijunctional actomyosin, suggesting TM4SF10 in vertebrates and VAB-9 in C. elegans may regulate cell morphology in concert with actomyosin and ZO-1-like proteins.100
Claudin Like Genes clc-1–clc-5
Claudin family proteins exist in all metazoans. In humans and mice there are as many as 24 different claudins. To date, five claudin-like proteins have been identified in C. elegans by virtue of sequence identity to mammalian claudins or to identified C. elegans claudins.79 Not surprisingly, sequence homology was relatively low, with CLC-1, CLC-2 and CLC-3 sharing 25%, 23%, and 26% identity with mouse claudin-6, 5 and 4, respectively. CLC-4 shares just 36% identity with CLC-1. CLC-5 is the paralogue of CLC-2, probably resulting from a gene duplication, being located just 3′ of clc-2 in the genome. Despite this location, CLC-2 and CLC-5 are only 31% identical. Of the five putative C. elegans claudins, two have been further analyzed to determine the extent of their function as classical claudins. GFP fusions proteins were constructed for both CLC-1 and CLC-2. CLC-1 is localized at the apical region of the lateral cell membranes in all four regions of the C. elegans pharynx, the procorpus, metacorpus, isthmus, and terminal bulb. In the isthmus region, six cellular junctions connect three pharyngeal myoepithelial and three marginal cells. Freeze substitution electron microscopy reveals a thick subapical junction—like a classic adherens junction—and a thinner apical region with a narrower paracellular space. It is possible that CLC-1, and the cell junction protein AJM-1, localize to this more closely opposed region and adherens junction proteins localize to the thicker basal junction. In the epidermis and intestine AJM-1 and DLG-1 localize basal to the adherens junction so that in the pharynx, the localization of the two junction types may be reversed; however that has not been conclusively demonstrated. It remains for immuno TEM with different diameter gold particles linked to secondary antibodies against HMR-1 and AJM-1/CLC-1 (for example) or super resolution microscopy of fluorescently labeled/tagged proteins to resolve this issue. An alternative possibility is that the narrow electron dense junction is the locale for one or more CLC proteins and this may define a separate lateral region more typical of tight junctions in mammals, rather than the septate junctions of invertebrates.
CLC-1 expression was also reported in the vulva, spermatheca and pore cell of the excretory system, but was not assigned to a specific junctional complex. CLC-2::GFP shows expression in the hypodermal seam cells of adults. Following RNAi of clc-1 and clc-2 a 10,000MW TRITC-dextran is able to infiltrate to the interior of the pharynx and body cavity (clc-1 RNAi) or the body cavity (clc-2 RNAi), suggesting that both of these barriers are maintained by claudin-like proteins. clc-1 and clc-2 expression only account for a subset of epithelial tissues with cell junctions in nematodes. Further studies should indicate whether clc-3–5 are expressed in epithelia lacking clc-1 and clc-2 and whether inactivation of these claudins in other tissues has similar effects on the epithelial barrier function of such tissues or whether there are alternative phenotypes. One expectation is that the various C. elegans claudins will be localized at distinct regions along the lateral cell membrance, since vertebrates claudins display this property. It will also be interesting to determine whether any of the CLC proteins can generate tight junction strands when expressed in L cells, like their mammalian counterparts. Expression of VAB-9 in L cells does not result in the formation of tight junction strands (JSS unpublished). Further data regarding the function of the other claudins and the voltage-gated calcium channel gamma subunits have been uncovered in genome-wide RNAi screens or expression studies and are tabulated in Figure 3D.
nsy-4 and Neuronal Cell Fate Specification
nsy-4 (Neuronal SYmmetry gene 4) was identified for its role in specifying the distinct fate of one AWC olfactory neuron; specifically, nsy-4 is required for one AWC neuron to express the G protein-coupled odorant receptor gene str-2 and detect the odor 2-butanone.101 Without nsy-4 activity, both AWC neurons fail to express str-2 and detect the odor 2,3-pentadione.101 These alternate fates, based on expression of str-2, are referred to as AWCon and AWCoff, respectively. In C. elegans, NSY-4 is most similar to uncharacterized genes K10D6.2 and C24H10.1 as well as claudin genes clc-1 – clc-5 and hpo-30.1. Outside C. elegans, NSY-4 is most similar to Drosophila Stargazin, TARPS, and gamma subunits of voltage-gated calcium channels. Surprisingly, nsy-4 rescue by human claudin-1 expression was weak, but was stronger than stargazin/γ2 expression while rescue by human γ7 channel was complicated by additional phenotypes. NSY-4 is thought to function as a gamma channel based on the genetic data, which indicate that nsy-4 represses the function of calcium channel genes unc-2 and unc-36. Together these findings suggest that NSY-4 may have additional functions in AWC specification beyond regulation of gamma channels. Additional functions may involve claudin-like adhesive functions. Genetic and expression studies demonstrate that NSY-4 levels correlate with the AWCon fate: nsy-4 expression in a single AWC is sufficient for the AWCon fate, while overexpression of nsy-4 in both AWC cells results in an increased frequency of animals with AWCon fates in both AWC cells. NSY-4 regulates the choice of cell fate by influencing the activity of a signaling pathway consisting of voltage-gated calcium channels encoded by unc-2 (α1 subunit) and unc-36(α2δ subunit), CAMKII encoded by unc-43, a Toll-interleukin 1 repeat protein encoded by tir-1, a MAPKKK encoded by nsy-1, a MAPKK encoded by sek-1, and a homeodomain protein encoded by nsy-7.102-105 The innexin protein, NSY-5, acts in parallel with NSY-4 to regulate the activity of this pathway, so that overexpression of nsy-4 can partially compensate for loss of nsy-5 and vice versa.106 While both proteins are localized at cell membranes (NSY-4 broadly and NSY-5 in puncta at junctions), neither protein affects the other’s localization. The nsy-5 gap junction network also regulates calcium in non-AWC cells to promote the AWCon fates, including left-right side bias as well as feedback inhibition of AWCon fates from surrounding cells.106,107 Similarly, nsy-4 is proposed to regulate communication between AWC cells in contacting axons across the midline.
Normally, the determination of which cell will express the AWCon fate is stochastic, suggesting that some small fluctuation in activity upstream or parallel to NSY-4 tips the choice of signaling resulting in str-2 expression in favor of one cell over another. Feedback signaling to reinforce this decision resulting in one, and only one, cell expressing the AWCon fate is supported by genetic mosaic experiments. Such experiments show that overexpression of NSY-4 in one AWC cell suppresses the expression of the AWCon fate in the neighboring wild type AWC cell. Since claudin family members are known to form homotypic and heterotypic pairs and since the nsy-4 homolog Stargazin has been shown to mediate both homotypic and heterotypic cell adhesion when expressed in L cells, it seems likely that NSY-4 is one half of a signaling pair and interacts with itself and/or a different claudin superfamily protein on neighboring AWC (and perhaps other) opposing cell membranes.34 Binding to NSY-4 then influences nsy-4 dependent signaling by refining forward and feedback signaling. Thus, NSY-4 related proteins, claudin family members, or VAB-9 may participate in heterotypic interactions with NSY-4 and mediate axon interaction and communication, participating in specification of AWC cell fate and possibly the fates of other neurons. It is interesting to note that VAB-9 is expressed in the nerve ring, although the specific cells have not been identified.65 While clc-1 and clc-2 are not expressed in the nerve ring, The expression patterns and phenotypes of the other claudin-related genes are not completely known (Fig. 3D). Given the combinatorial possibilities, multiple distinct axonal signaling events could be mediated by these proteins. Specific cell-cell interactions mediated by distinct claudin pairs may activate multiple signaling events within the same or among closely related cells. In a similar fashion, NSY-4 may participate in functions distinct from neuronal specification, since NSY-4 is expressed in epithelial cells and some alleles show morphological defects and embryonic lethality.101 It is tempting to speculate that NSY-4 could function redundantly for C. elegans pfam00822 proteins in various cellular processes.
When is a Claudin a Claudin?
The superfamily of verebrate proteins that fall into the general category of similarity to claudins has grown, begging the question of what exactly are the essential properties of a claudin, as opposed to other closely-related four-pass integral membrane proteins. Most claudins generate a charge-selective pore in the paracellular space that regulates electrical properties of a cell or groups of cells due to specific charge permeability. Most claudins mediate adhesion and this adhesion can be homotypic or heterotypic between various claudin types. The formation of paired strands within the membrane by claudins reduces the paracellular space to zero and the number and arrangement of the strands correlates with the tightness of the epithelium. Most claudins appear able to traffic to cell membranes and generate paired strands in the absence of other cell adhesion molecules, but require targeting proteins to designate the proper membrane location. Like other transmembrane proteins, claudins transduce extracellular information, such as the binding of ligands, small molecules, and viruses, to the nucleus in order to change gene expression or to transduce signals to junctional proteins (including themselves) to modulate the nature of the paracellular space. Claudin superfamily proteins are likely to have at least a majority of these properties, however due to the various organisms, organs, tissues, and developmental and physiological systems in which claudin-like proteins have been shown to function, along with the dramatic divergence of the family, it is highly likely that the functional repertoire of vertebrate claudin-like proteins will continue to expand. The claudin family proteins in C. elegans described in this review support this hypothesis, since they are involved in diverse roles such as maintaining tissue integrity, epithelial morphogenesis, regulating cell junction dynamics through filamentous actin, signal transduction, and cell fate specification. Our understanding of the roles for all the members of this family in C. elegans is incomplete, suggesting that future research will reveal novel functions.
Acknowledgments
This work was supported by NIH grant DK095832. I thank Jeff Hardin and David Hall for discussion and personal communications, and the anonymous reviewers for insightful critiques that together helped shape the ideas in this paper.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/tissuebarriers/article/25502
References
- 1.Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S. Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J Cell Biol. 1998;141:1539–50. doi: 10.1083/jcb.141.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furuse M, Sasaki H, Fujimoto K, Tsukita S. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J Cell Biol. 1998;143:391–401. doi: 10.1083/jcb.143.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morita K, Sasaki H, Furuse M, Tsukita S. Endothelial claudin: claudin-5/TMVCF constitutes tight junction strands in endothelial cells. J Cell Biol. 1999;147:185–94. doi: 10.1083/jcb.147.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nitta T, Hata M, Gotoh S, Seo Y, Sasaki H, Hashimoto N, et al. Size-selective loosening of the blood-brain barrier in claudin-5-deficient mice. J Cell Biol. 2003;161:653–60. doi: 10.1083/jcb.200302070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gow A, Davies C, Southwood CM, Frolenkov G, Chrustowski M, Ng L, et al. Deafness in Claudin 11-null mice reveals the critical contribution of basal cell tight junctions to stria vascularis function. J Neurosci. 2004;24:7051–62. doi: 10.1523/JNEUROSCI.1640-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furuse M, Hata M, Furuse K, Yoshida Y, Haratake A, Sugitani Y, et al. Claudin-based tight junctions are crucial for the mammalian epidermal barrier: a lesson from claudin-1-deficient mice. J Cell Biol. 2002;156:1099–111. doi: 10.1083/jcb.200110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilcox ER, Burton QL, Naz S, Riazuddin S, Smith TN, Ploplis B, et al. Mutations in the gene encoding tight junction claudin-14 cause autosomal recessive deafness DFNB29. Cell. 2001;104:165–72. doi: 10.1016/S0092-8674(01)00200-8. [DOI] [PubMed] [Google Scholar]
- 8.Tatum R, Zhang Y, Lu Q, Kim K, Jeansonne BG, Chen YH. WNK4 phosphorylates ser(206) of claudin-7 and promotes paracellular Cl(-) permeability. FEBS Lett. 2007;581:3887–91. doi: 10.1016/j.febslet.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 9.Kollmar R, Nakamura SK, Kappler JA, Hudspeth AJ. Expression and phylogeny of claudins in vertebrate primordia. Proc Natl Acad Sci U S A. 2001;98:10196–201. doi: 10.1073/pnas.171325898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krause G, Winkler L, Mueller SL, Haseloff RF, Piontek J, Blasig IE. Structure and function of claudins. Biochim Biophys Acta. 2008;1778:631–45. doi: 10.1016/j.bbamem.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 11.Morita K, Furuse M, Fujimoto K, Tsukita S. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc Natl Acad Sci U S A. 1999;96:511–6. doi: 10.1073/pnas.96.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turksen K, Troy TC. Junctions gone bad: claudins and loss of the barrier in cancer. Biochim Biophys Acta. 2011;1816:73–9. doi: 10.1016/j.bbcan.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Blasig IE, Winkler L, Lassowski B, Mueller SL, Zuleger N, Krause E, et al. On the self-association potential of transmembrane tight junction proteins. Cell Mol Life Sci. 2006;63:505–14. doi: 10.1007/s00018-005-5472-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coyne CB, Gambling TM, Boucher RC, Carson JL, Johnson LG. Role of claudin interactions in airway tight junctional permeability. Am J Physiol Lung Cell Mol Physiol. 2003;285:L1166–78. doi: 10.1152/ajplung.00182.2003. [DOI] [PubMed] [Google Scholar]
- 15.Mitic LL, Unger VM, Anderson JM. Expression, solubilization, and biochemical characterization of the tight junction transmembrane protein claudin-4. Protein Sci. 2003;12:218–27. doi: 10.1110/ps.0233903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Itallie CM, Anderson JM. Claudins and epithelial paracellular transport. Annu Rev Physiol. 2006;68:403–29. doi: 10.1146/annurev.physiol.68.040104.131404. [DOI] [PubMed] [Google Scholar]
- 17.Kiuchi-Saishin Y, Gotoh S, Furuse M, Takasuga A, Tano Y, Tsukita S. Differential expression patterns of claudins, tight junction membrane proteins, in mouse nephron segments. J Am Soc Nephrol. 2002;13:875–86. doi: 10.1681/ASN.V134875. [DOI] [PubMed] [Google Scholar]
- 18.Simon DB, Lu Y, Choate KA, Velazquez H, Al-Sabban E, Praga M, et al. Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science. 1999;285:103–6. doi: 10.1126/science.285.5424.103. [DOI] [PubMed] [Google Scholar]
- 19.Holmes JL, Van Itallie CM, Rasmussen JE, Anderson JM. Claudin profiling in the mouse during postnatal intestinal development and along the gastrointestinal tract reveals complex expression patterns. Gene Expr Patterns. 2006;6:581–8. doi: 10.1016/j.modgep.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 20.Troy TC, Li Y, O’Malley L, Turksen K. The temporal and spatial expression of Claudins in epidermal development and the accelerated program of epidermal differentiation in K14-CaSR transgenic mice. Gene Expr Patterns. 2007;7:423–30. doi: 10.1016/j.modgep.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 21.Colegio OR, Van Itallie CM, McCrea HJ, Rahner C, Anderson JM. Claudins create charge-selective channels in the paracellular pathway between epithelial cells. Am J Physiol Cell Physiol. 2002;283:C142–7. doi: 10.1152/ajpcell.00038.2002. [DOI] [PubMed] [Google Scholar]
- 22.Van Itallie CM, Rogan S, Yu A, Vidal LS, Holmes J, Anderson JM. Two splice variants of claudin-10 in the kidney create paracellular pores with different ion selectivities. Am J Physiol Renal Physiol. 2006;291:F1288–99. doi: 10.1152/ajprenal.00138.2006. [DOI] [PubMed] [Google Scholar]
- 23.Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wölk B, et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–5. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 24.Meertens L, Bertaux C, Cukierman L, Cormier E, Lavillette D, Cosset FL, et al. The tight junction proteins claudin-1, -6, and -9 are entry cofactors for hepatitis C virus. J Virol. 2008;82:3555–60. doi: 10.1128/JVI.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng A, Yuan F, Li Y, Zhu F, Hou P, Li J, et al. Claudin-6 and claudin-9 function as additional coreceptors for hepatitis C virus. J Virol. 2007;81:12465–71. doi: 10.1128/JVI.01457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamazaki Y, Itoh M, Sasaki H, Furuse M, Tsukita S. Multi-PDZ domain protein 1 (MUPP1) is concentrated at tight junctions through its possible interaction with claudin-1 and junctional adhesion molecule. J Biol Chem. 2002;277:455–61. doi: 10.1074/jbc.M109005200. [DOI] [PubMed] [Google Scholar]
- 27.Itoh M, Furuse M, Morita K, Kubota K, Saitou M, Tsukita S. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J Cell Biol. 1999;147:1351–63. doi: 10.1083/jcb.147.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Müller D, Kausalya PJ, Bockenhauer D, Thumfart J, Meij IC, Dillon MJ, et al. Unusual clinical presentation and possible rescue of a novel claudin-16 mutation. J Clin Endocrinol Metab. 2006;91:3076–9. doi: 10.1210/jc.2006-0200. [DOI] [PubMed] [Google Scholar]
- 29.D’Souza T, Agarwal R, Morin PJ. Phosphorylation of claudin-3 at threonine 192 by cAMP-dependent protein kinase regulates tight junction barrier function in ovarian cancer cells. J Biol Chem. 2005;280:26233–40. doi: 10.1074/jbc.M502003200. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka M, Kamata R, Sakai R. EphA2 phosphorylates the cytoplasmic tail of Claudin-4 and mediates paracellular permeability. J Biol Chem. 2005;280:42375–82. doi: 10.1074/jbc.M503786200. [DOI] [PubMed] [Google Scholar]
- 31.Lane NJ, Skaer HI. Intercellular junctions in insect tissues. Adv Insect Physiol. 1980;15:35–213. doi: 10.1016/S0065-2806(08)60141-1. [DOI] [Google Scholar]
- 32.Hua VB, Chang AB, Tchieu JH, Kumar NM, Nielsen PA, Saier MH., Jr. Sequence and phylogenetic analyses of 4 TMS junctional proteins of animals: connexins, innexins, claudins and occludins. J Membr Biol. 2003;194:59–76. doi: 10.1007/s00232-003-2026-8. [DOI] [PubMed] [Google Scholar]
- 33.Wu VM, Schulte J, Hirschi A, Tepass U, Beitel GJ. Sinuous is a Drosophila claudin required for septate junction organization and epithelial tube size control. J Cell Biol. 2004;164:313–23. doi: 10.1083/jcb.200309134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price MG, Davis CF, Deng F, Burgess DL. The alpha-amino-3-hydroxyl-5-methyl-4-isoxazolepropionate receptor trafficking regulator “stargazin” is related to the claudin family of proteins by Its ability to mediate cell-cell adhesion. J Biol Chem. 2005;280:19711–20. doi: 10.1074/jbc.M500623200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ihrie RA, Marques MR, Nguyen BT, Horner JS, Papazoglu C, Bronson RT, et al. Perp is a p63-regulated gene essential for epithelial integrity. Cell. 2005;120:843–56. doi: 10.1016/j.cell.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 36.Attardi LD, Reczek EE, Cosmas C, Demicco EG, McCurrach ME, Lowe SW, et al. PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev. 2000;14:704–18. [PMC free article] [PubMed] [Google Scholar]
- 37.Bruggeman LA, Martinka S, Simske JS. Expression of TM4SF10, a Claudin/EMP/PMP22 family cell junction protein, during mouse kidney development and podocyte differentiation. Dev Dyn. 2007;236:596–605. doi: 10.1002/dvdy.21052. [DOI] [PubMed] [Google Scholar]
- 38.Katoh M, Katoh M. CLDN23 gene, frequently down-regulated in intestinal-type gastric cancer, is a novel member of CLAUDIN gene family. Int J Mol Med. 2003;11:683–9. [PubMed] [Google Scholar]
- 39.Kim J, Kim I, Yang JS, Shin YE, Hwang J, Park S, et al. Rewiring of PDZ domain-ligand interaction network contributed to eukaryotic evolution. PLoS Genet. 2012;8:e1002510. doi: 10.1371/journal.pgen.1002510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tonikian R, Zhang Y, Sazinsky SL, Currell B, Yeh JH, Reva B, et al. A specificity map for the PDZ domain family. PLoS Biol. 2008;6:e239. doi: 10.1371/journal.pbio.0060239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lane NJ. Tight junctions in invertebrates. In: Cereijido MaA, J., ed. Tight Junctions. New York: CRC Press, 2001:39-59. [Google Scholar]
- 42.Spiegel E, Howard L. Development of cell junctions in sea-urchin embryos. J Cell Sci. 1983;62:27–48. doi: 10.1242/jcs.62.1.27. [DOI] [PubMed] [Google Scholar]
- 43.Tepass U, Hartenstein V. The development of cellular junctions in the Drosophila embryo. Dev Biol. 1994;161:563–96. doi: 10.1006/dbio.1994.1054. [DOI] [PubMed] [Google Scholar]
- 44.Noirot-Timothee C, Noirot C. Septate and scalariform junctions in arthropods. Int Rev Cytol. 1980;63:97–140. doi: 10.1016/S0074-7696(08)61758-1. [DOI] [PubMed] [Google Scholar]
- 45.Gilula NB, Branton D, Satir P. The septate junction: a structural basis for intercellular coupling. Proc Natl Acad Sci U S A. 1970;67:213–20. doi: 10.1073/pnas.67.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Satir P, Gilula NB. The fine structure of membranes and intercellular communication in insects. Annu Rev Entomol. 1973;18:143–66. doi: 10.1146/annurev.en.18.010173.001043. [DOI] [PubMed] [Google Scholar]
- 47.Davidson LA. A freeze fracture and thin section study of intestinal cell membranes and intercellular junctions of a nematode, Ascaris. Tissue Cell. 1983;15:27–37. doi: 10.1016/0040-8166(83)90031-9. [DOI] [PubMed] [Google Scholar]
- 48.Lane NJ, Swales LS. Stages in the assembly of pleated and smooth septate junctions in developing insect embryos. J Cell Sci. 1982;56:245–62. doi: 10.1242/jcs.56.1.245. [DOI] [PubMed] [Google Scholar]
- 49.Behr M, Riedel D, Schuh R. The claudin-like megatrachea is essential in septate junctions for the epithelial barrier function in Drosophila. Dev Cell. 2003;5:611–20. doi: 10.1016/S1534-5807(03)00275-2. [DOI] [PubMed] [Google Scholar]
- 50.Genova JL, Fehon RG. Neuroglian, Gliotactin, and the Na+/K+ ATPase are essential for septate junction function in Drosophila. J Cell Biol. 2003;161:979–89. doi: 10.1083/jcb.200212054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu VM, Yu MH, Paik R, Banerjee S, Liang Z, Paul SM, et al. Drosophila Varicose, a member of a new subgroup of basolateral MAGUKs, is required for septate junctions and tracheal morphogenesis. Development. 2007;134:999–1009. doi: 10.1242/dev.02785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nelson KS, Furuse M, Beitel GJ. The Drosophila Claudin Kune-kune is required for septate junction organization and tracheal tube size control. Genetics. 2010;185:831–9. doi: 10.1534/genetics.110.114959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suzuki A, Ohno S. The PAR-aPKC system: lessons in polarity. J Cell Sci. 2006;119:979–87. doi: 10.1242/jcs.02898. [DOI] [PubMed] [Google Scholar]
- 54.Fehon RG, Dawson IA, Artavanis-Tsakonas S. A Drosophila homologue of membrane-skeleton protein 4.1 is associated with septate junctions and is encoded by the coracle gene. Development. 1994;120:545–57. doi: 10.1242/dev.120.3.545. [DOI] [PubMed] [Google Scholar]
- 55.Baumgartner S, Littleton JT, Broadie K, Bhat MA, Harbecke R, Lengyel JA, et al. A Drosophila neurexin is required for septate junction and blood-nerve barrier formation and function. Cell. 1996;87:1059–68. doi: 10.1016/S0092-8674(00)81800-0. [DOI] [PubMed] [Google Scholar]
- 56.Woods DF, Wu JW, Bryant PJ. Localization of proteins to the apico-lateral junctions of Drosophila epithelia. Dev Genet. 1997;20:111–8. doi: 10.1002/(SICI)1520-6408(1997)20:2<111::AID-DVG4>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 57.Schulte J, Tepass U, Auld VJ. Gliotactin, a novel marker of tricellular junctions, is necessary for septate junction development in Drosophila. J Cell Biol. 2003;161:991–1000. doi: 10.1083/jcb.200303192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Knust E, Bossinger O. Composition and formation of intercellular junctions in epithelial cells. Science. 2002;298:1955–9. doi: 10.1126/science.1072161. [DOI] [PubMed] [Google Scholar]
- 59.Costa M, Raich W, Agbunag C, Leung B, Hardin J, Priess JR. A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J Cell Biol. 1998;141:297–308. doi: 10.1083/jcb.141.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pettitt J, Cox EA, Broadbent ID, Flett A, Hardin J. The Caenorhabditis elegans p120 catenin homologue, JAC-1, modulates cadherin-catenin function during epidermal morphogenesis. J Cell Biol. 2003;162:15–22. doi: 10.1083/jcb.200212136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bossinger O, Klebes A, Segbert C, Theres C, Knust E. Zonula adherens formation in Caenorhabditis elegans requires dlg-1, the homologue of the Drosophila gene discs large. Dev Biol. 2001;230:29–42. doi: 10.1006/dbio.2000.0113. [DOI] [PubMed] [Google Scholar]
- 62.Firestein BL, Rongo C. DLG-1 is a MAGUK similar to SAP97 and is required for adherens junction formation. Mol Biol Cell. 2001;12:3465–75. doi: 10.1091/mbc.12.11.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Köppen M, Simske JS, Sims PA, Firestein BL, Hall DH, Radice AD, et al. Cooperative regulation of AJM-1 controls junctional integrity in Caenorhabditis elegans epithelia. Nat Cell Biol. 2001;3:983–91. doi: 10.1038/ncb1101-983. [DOI] [PubMed] [Google Scholar]
- 64.McMahon L, Legouis R, Vonesch JL, Labouesse M. Assembly of C. elegans apical junctions involves positioning and compaction by LET-413 and protein aggregation by the MAGUK protein DLG-1. J Cell Sci. 2001;114:2265–77. doi: 10.1242/jcs.114.12.2265. [DOI] [PubMed] [Google Scholar]
- 65.Simske JS, Köppen M, Sims P, Hodgkin J, Yonkof A, Hardin J. The cell junction protein VAB-9 regulates adhesion and epidermal morphology in C. elegans. Nat Cell Biol. 2003;5:619–25. doi: 10.1038/ncb1002. [DOI] [PubMed] [Google Scholar]
- 66.Aono S, Legouis R, Hoose WA, Kemphues KJ. PAR-3 is required for epithelial cell polarity in the distal spermatheca of C. elegans. Development. 2004;131:2865–74. doi: 10.1242/dev.01146. [DOI] [PubMed] [Google Scholar]
- 67.Michaux G, Gansmuller A, Hindelang C, Labouesse M. CHE-14, a protein with a sterol-sensing domain, is required for apical sorting in C. elegans ectodermal epithelial cells. Curr Biol. 2000;10:1098–107. doi: 10.1016/S0960-9822(00)00695-3. [DOI] [PubMed] [Google Scholar]
- 68.Shibata Y, Fujii T, Dent JA, Fujisawa H, Takagi S. EAT-20, a novel transmembrane protein with EGF motifs, is required for efficient feeding in Caenorhabditis elegans. Genetics. 2000;154:635–46. doi: 10.1093/genetics/154.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bilder D, Perrimon N. Localization of apical epithelial determinants by the basolateral PDZ protein Scribble. Nature. 2000;403:676–80. doi: 10.1038/35001108. [DOI] [PubMed] [Google Scholar]
- 70.Legouis R, Gansmuller A, Sookhareea S, Bosher JM, Baillie DL, Labouesse M. LET-413 is a basolateral protein required for the assembly of adherens junctions in Caenorhabditis elegans. Nat Cell Biol. 2000;2:415–22. doi: 10.1038/35017046. [DOI] [PubMed] [Google Scholar]
- 71.Bossinger O, Fukushige T, Claeys M, Borgonie G, McGhee JD. The apical disposition of the Caenorhabditis elegans intestinal terminal web is maintained by LET-413. Dev Biol. 2004;268:448–56. doi: 10.1016/j.ydbio.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 72.McCarter J, Bartlett B, Dang T, Schedl T. Soma-germ cell interactions in Caenorhabditis elegans: multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev Biol. 1997;181:121–43. doi: 10.1006/dbio.1996.8429. [DOI] [PubMed] [Google Scholar]
- 73.Schedl T. Developmental genetics of the germ line In: D. Riddle TB, B. Meyer and J. Priess, ed. C elegans II. Cold Spring Harbor: Cold Spring Harbor Laboratory Press., 1997:241-70. [PubMed] [Google Scholar]
- 74.Hall DH, Winfrey VP, Blaeuer G, Hoffman LH, Furuta T, Rose KL, et al. Ultrastructural features of the adult hermaphrodite gonad of Caenorhabditis elegans: relations between the germ line and soma. Dev Biol. 1999;212:101–23. doi: 10.1006/dbio.1999.9356. [DOI] [PubMed] [Google Scholar]
- 75.Hall DH, Altun ZF. Reproductive System In: Press CSHL, ed. C elegans Atlas. Cold Spring Harbor, 2008:243-91. [Google Scholar]
- 76.Piekny AJ, Johnson JL, Cham GD, Mains PE. The Caenorhabditis elegans nonmuscle myosin genes nmy-1 and nmy-2 function as redundant components of the let-502/Rho-binding kinase and mel-11/myosin phosphatase pathway during embryonic morphogenesis. Development. 2003;130:5695–704. doi: 10.1242/dev.00807. [DOI] [PubMed] [Google Scholar]
- 77.Piekny AJ, Mains PE. Rho-binding kinase (LET-502) and myosin phosphatase (MEL-11) regulate cytokinesis in the early Caenorhabditis elegans embryo. J Cell Sci. 2002;115:2271–82. doi: 10.1242/jcs.115.11.2271. [DOI] [PubMed] [Google Scholar]
- 78.Wissmann A, Ingles J, Mains PE. The Caenorhabditis elegans mel-11 myosin phosphatase regulatory subunit affects tissue contraction in the somatic gonad and the embryonic epidermis and genetically interacts with the Rac signaling pathway. Dev Biol. 1999;209:111–27. doi: 10.1006/dbio.1999.9242. [DOI] [PubMed] [Google Scholar]
- 79.Asano A, Asano K, Sasaki H, Furuse M, Tsukita S. Claudins in Caenorhabditis elegans: their distribution and barrier function in the epithelium. Curr Biol. 2003;13:1042–6. doi: 10.1016/S0960-9822(03)00395-6. [DOI] [PubMed] [Google Scholar]
- 80.Christophe-Hobertus C, Kooy F, Gecz J, Abramowicz MJ, Holinski-Feder E, Schwartz C, et al. TM4SF10 gene sequencing in XLMR patients identifies common polymorphisms but no disease-associated mutation. BMC Med Genet. 2004;5:22. doi: 10.1186/1471-2350-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Christophe-Hobertus C, Szpirer C, Guyon R, Christophe D. Identification of the gene encoding Brain Cell Membrane Protein 1 (BCMP1), a putative four-transmembrane protein distantly related to the Peripheral Myelin Protein 22 / Epithelial Membrane Proteins and the Claudins. BMC Genomics. 2001;2:3. doi: 10.1186/1471-2164-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Holzman LB, St John PL, Kovari IA, Verma R, Holthofer H, Abrahamson DR. Nephrin localizes to the slit pore of the glomerular epithelial cell. Kidney Int. 1999;56:1481–91. doi: 10.1046/j.1523-1755.1999.00719.x. [DOI] [PubMed] [Google Scholar]
- 83.Huber TB, Benzing T. The slit diaphragm: a signaling platform to regulate podocyte function. Curr Opin Nephrol Hypertens. 2005;14:211–6. doi: 10.1097/01.mnh.0000165885.85803.a8. [DOI] [PubMed] [Google Scholar]
- 84.Barletta GM, Kovari IA, Verma RK, Kerjaschki D, Holzman LB. Nephrin and Neph1 co-localize at the podocyte foot process intercellular junction and form cis hetero-oligomers. J Biol Chem. 2003;278:19266–71. doi: 10.1074/jbc.M301279200. [DOI] [PubMed] [Google Scholar]
- 85.Khoshnoodi J, Sigmundsson K, Ofverstedt LG, Skoglund U, Obrink B, Wartiovaara J, et al. Nephrin promotes cell-cell adhesion through homophilic interactions. Am J Pathol. 2003;163:2337–46. doi: 10.1016/S0002-9440(10)63590-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Verma R, Wharram B, Kovari I, Kunkel R, Nihalani D, Wary KK, et al. Fyn binds to and phosphorylates the kidney slit diaphragm component Nephrin. J Biol Chem. 2003;278:20716–23. doi: 10.1074/jbc.M301689200. [DOI] [PubMed] [Google Scholar]
- 87.Verma R, Kovari I, Soofi A, Nihalani D, Patrie K, Holzman LB. Nephrin ectodomain engagement results in Src kinase activation, nephrin phosphorylation, Nck recruitment, and actin polymerization. J Clin Invest. 2006;116:1346–59. doi: 10.1172/JCI27414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Garg P, Verma R, Nihalani D, Johnstone DB, Holzman LB. Neph1 cooperates with nephrin to transduce a signal that induces actin polymerization. Mol Cell Biol. 2007;27:8698–712. doi: 10.1128/MCB.00948-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lahdenperä J, Kilpeläinen P, Liu XL, Pikkarainen T, Reponen P, Ruotsalainen V, et al. Clustering-induced tyrosine phosphorylation of nephrin by Src family kinases. Kidney Int. 2003;64:404–13. doi: 10.1046/j.1523-1755.2003.00097.x. [DOI] [PubMed] [Google Scholar]
- 90.Li H, Lemay S, Aoudjit L, Kawachi H, Takano T. SRC-family kinase Fyn phosphorylates the cytoplasmic domain of nephrin and modulates its interaction with podocin. J Am Soc Nephrol. 2004;15:3006–15. doi: 10.1097/01.ASN.0000146689.88078.80. [DOI] [PubMed] [Google Scholar]
- 91.Liu XL, Kilpeläinen P, Hellman U, Sun Y, Wartiovaara J, Morgunova E, et al. Characterization of the interactions of the nephrin intracellular domain. FEBS J. 2005;272:228–43. doi: 10.1111/j.1432-1033.2004.04408.x. [DOI] [PubMed] [Google Scholar]
- 92.Harita Y, Kurihara H, Kosako H, Tezuka T, Sekine T, Igarashi T, et al. Neph1, a component of the kidney slit diaphragm, is tyrosine-phosphorylated by the Src family tyrosine kinase and modulates intracellular signaling by binding to Grb2. J Biol Chem. 2008;283:9177–86. doi: 10.1074/jbc.M707247200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhu J, Sun N, Aoudjit L, Li H, Kawachi H, Lemay S, et al. Nephrin mediates actin reorganization via phosphoinositide 3-kinase in podocytes. Kidney Int. 2008;73:556–66. doi: 10.1038/sj.ki.5002691. [DOI] [PubMed] [Google Scholar]
- 94.Azhibekov TA, Wu Z, Padiyar A, Bruggeman LA, Simske JS. TM4SF10 and ADAP interaction in podocytes: role in Fyn activity and nephrin phosphorylation. Am J Physiol Cell Physiol. 2011;301:C1351–9. doi: 10.1152/ajpcell.00166.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Laketa V, Simpson JC, Bechtel S, Wiemann S, Pepperkok R. High-content microscopy identifies new neurite outgrowth regulators. Mol Biol Cell. 2007;18:242–52. doi: 10.1091/mbc.E06-08-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shen K, Fetter RD, Bargmann CI. Synaptic specificity is generated by the synaptic guidepost protein SYG-2 and its receptor, SYG-1. Cell. 2004;116:869–81. doi: 10.1016/S0092-8674(04)00251-X. [DOI] [PubMed] [Google Scholar]
- 97.Shen K, Bargmann CI. The immunoglobulin superfamily protein SYG-1 determines the location of specific synapses in C. elegans. Cell. 2003;112:619–30. doi: 10.1016/S0092-8674(03)00113-2. [DOI] [PubMed] [Google Scholar]
- 98.Putzke AP, Hikita ST, Clegg DO, Rothman JH. Essential kinase-independent role of a Fer-like non-receptor tyrosine kinase in Caenorhabditis elegans morphogenesis. Development. 2005;132:3185–95. doi: 10.1242/dev.01900. [DOI] [PubMed] [Google Scholar]
- 99.Lockwood C, Zaidel-Bar R, Hardin J. The C. elegans zonula occludens ortholog cooperates with the cadherin complex to recruit actin during morphogenesis. Curr Biol. 2008;18:1333–7. doi: 10.1016/j.cub.2008.07.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fanning AS, Van Itallie CM, Anderson JM. Zonula occludens-1 and -2 regulate apical cell structure and the zonula adherens cytoskeleton in polarized epithelia. Mol Biol Cell. 2012;23:577–90. doi: 10.1091/mbc.E11-09-0791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vanhoven MK, Bauer Huang SL, Albin SD, Bargmann CI. The claudin superfamily protein nsy-4 biases lateral signaling to generate left-right asymmetry in C. elegans olfactory neurons. Neuron. 2006;51:291–302. doi: 10.1016/j.neuron.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 102.Lesch BJ, Gehrke AR, Bulyk ML, Bargmann CI. Transcriptional regulation and stabilization of left-right neuronal identity in C. elegans. Genes Dev. 2009;23:345–58. doi: 10.1101/gad.1763509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tanaka-Hino M, Sagasti A, Hisamoto N, Kawasaki M, Nakano S, Ninomiya-Tsuji J, et al. SEK-1 MAPKK mediates Ca2+ signaling to determine neuronal asymmetric development in Caenorhabditis elegans. EMBO Rep. 2002;3:56–62. doi: 10.1093/embo-reports/kvf001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wes PD, Bargmann CI. C. elegans odour discrimination requires asymmetric diversity in olfactory neurons. Nature. 2001;410:698–701. doi: 10.1038/35070581. [DOI] [PubMed] [Google Scholar]
- 105.Chuang CF, Bargmann CIA. A Toll-interleukin 1 repeat protein at the synapse specifies asymmetric odorant receptor expression via ASK1 MAPKKK signaling. Genes Dev. 2005;19:270–81. doi: 10.1101/gad.1276505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chuang CF, Vanhoven MK, Fetter RD, Verselis VK, Bargmann CI. An innexin-dependent cell network establishes left-right neuronal asymmetry in C. elegans. Cell. 2007;129:787–99. doi: 10.1016/j.cell.2007.02.052. [DOI] [PubMed] [Google Scholar]
- 107.Schumacher JA, Hsieh YW, Chen S, Pirri JK, Alkema MJ, Li WH, et al. Intercellular calcium signaling in a gap junction-coupled cell network establishes asymmetric neuronal fates in C. elegans. Development. 2012;139:4191–201. doi: 10.1242/dev.083428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 109.Kunitomo H, Uesugi H, Kohara Y, Iino Y. Identification of ciliated sensory neuron-expressed genes in Caenorhabditis elegans using targeted pull-down of poly(A) tails. Genome Biol. 2005;6:R17. doi: 10.1186/gb-2005-6-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hamamichi S, Rivas RN, Knight AL, Cao S, Caldwell KA, Caldwell GA. Hypothesis-based RNAi screening identifies neuroprotective genes in a Parkinson’s disease model. Proc Natl Acad Sci U S A. 2008;105:728–33. doi: 10.1073/pnas.0711018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kao CY, Los FC, Huffman DL, Wachi S, Kloft N, Husmann M, et al. Global functional analyses of cellular responses to pore-forming toxins. PLoS Pathog. 2011;7:e1001314. doi: 10.1371/journal.ppat.1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Meissner B, Rogalski T, Viveiros R, Warner A, Plastino L, Lorch A, et al. Determining the sub-cellular localization of proteins within Caenorhabditis elegans body wall muscle. PLoS One. 2011;6:e19937. doi: 10.1371/journal.pone.0019937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Meissner B, Warner A, Wong K, Dube N, Lorch A, McKay SJ, et al. An integrated strategy to study muscle development and myofilament structure in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000537. doi: 10.1371/journal.pgen.1000537. [DOI] [PMC free article] [PubMed] [Google Scholar]