Abstract
Translation of mRNA into proteins by the ribosome is universally conserved in all cellular life. The composition and complexity of the translation machinery differ markedly between the three domains of life. Organisms from the domain Archaea show an intermediate level of complexity, sharing several additional components of the translation machinery with eukaryotes that are absent in bacteria. One of these translation factors is initiation factor 6 (IF6), which associates with the large ribosomal subunit. We have reconstructed the 50S ribosomal subunit from the archaeon Methanothermobacter thermautotrophicus in complex with archaeal IF6 at 6.6 Å resolution using cryo-electron microscopy (EM). The structure provides detailed architectural insights into the 50S ribosomal subunit from a methanogenic archaeon through identification of the rRNA expansion segments and ribosomal proteins that are shared between this archaeal ribosome and eukaryotic ribosomes but are mostly absent in bacteria and in some archaeal lineages. Furthermore, the structure reveals that, in spite of highly divergent evolutionary trajectories of the ribosomal particle and the acquisition of novel functions of IF6 in eukaryotes, the molecular binding of IF6 on the ribosome is conserved between eukaryotes and archaea. The structure also provides a snapshot of the reductive evolution of the archaeal ribosome and offers new insights into the evolution of the translation system in archaea.
Keywords: ribosome, IF6, cryo-EM, CCD detector, evolution
Introduction
The ribosome is the macromolecular machine responsible for protein synthesis in all living cells.1,2 Eukaryotes feature the most complex translation systems. Their ribosomes are constituted of 4 rRNAs and approximately 80 proteins, forming a particle of about 3.5 MDa sedimenting at 80S (Svedberg units). In contrast, the prokaryotic ribosome is composed of only 3 rRNAs and approximately 50 proteins. Even though both archaeal and bacterial ribosomes sediment at 70S, the archaeal translation system is more closely related to its eukaryotic counterpart than to the bacterial one.3 The comparative similarity of the archaeal and eukaryotic translation systems is reflected in phylogenetic trees based on rRNA sequence comparison.4,5
The ribosomal proteome exhibits considerable diversity across the archaeal domain of life. Ribosomes of species belonging to the phylum Crenarchaeota contain the highest number of ribosomal proteins that are shared between Archaea and Eukaryota.6,7 Within the phylum Euryarchaeota, there seems to be a gradual reduction of the number of shared ribosomal proteins, with early-branching lineages, such as Pyrococcus, retaining more ribosomal proteins than late-branching ones, such as Halobacterium. This is an example of domain-scale reductive evolution.6,7 Methanothermobacter thermautotrophicus, which belongs to the Methanobacteria, is located at an intermediate stage, both phylogenetically8 and in terms of its ribosomal protein content.6 The sole high-resolution three-dimensional (3D) structure of an archaeal ribosomal subunit available so far is that of the late-branching halophilic euryarchaeon Haloarcula marismortui,9 which is among the most strongly reduced archaeal ribosomes in terms of its protein and rRNA content. Many archaeal organisms are extremophiles, such as (hyper)thermophiles, acidophiles, or halophiles. Therefore, some of the proteomic diversity of archaeal ribosomes might reflect adaptations of the biological macromolecules to these extreme lifestyles, highlighting the need for experimental study of archaeal ribosomes to understand how the ribosome may adapt to such conditions.7
The archaeal and eukaryotic translation systems share several translation initiation factors that are absent in bacteria, such as initiation factor 6 (IF6). IF6 was initially characterized as a molecular anti-association factor inhibiting ribosomal subunit joining.10,11 The gene coding for eIF6 is essential in yeast,12,13 and a homozygous deletion of eIF6 in mice is embryonic lethal,14 highlighting the importance of eIF6 for the eukaryotic protein synthesis machinery. Current data indicate that eIF6 has more than one major cellular function in eukaryotes: (1) In mammalian cells, eIF6 is required in vivo for the stimulation of translation initiation in response to growth factor signaling, which affects global translation regulation and cell proliferation.14 In this role, the anti-association activity of eIF6 may be required to avoid the sequestration of large ribosomal subunits into unproductive 80S complexes, thereby keeping them available for translation.14,15 (2) In both yeast and higher eukaryotes, eIF6 is involved in ribosome biogenesis,12,13,15,16 more specifically in pre-rRNA processing17 and ribosome export.18,19 (3) More recently, eIF6 has been shown to aid in vitro ribosome recycling by the ABC family protein ABCE1.20 The role of eIF6 in this process in vivo remains to be determined. The cellular function of the archaeal homolog aIF6 is considerably less well understood. The expression of aIF6 has been reported to be upregulated under stress conditions, such as cold and heat shock.21 Therefore, it has been suggested that IF6 originally served to regulate translation under unfavorable conditions in organisms ancestral to both eukaryotes and archaea and acquired additional functions after the split of the eukaryotic and archaeal lineages.21 As in eukaryotes, an ABCE1-dependent ribosome recycling pathway has also been described in archaea,22 and an involvement of aIF6 in this process has been proposed.23 Interestingly, the activities of both aIF6 and eIF6 seem to be modulated by posttranslational modifications: Phosphorylation of eIF6 is required for the regulation of the nucleocytoplasmic shuttling of the factor,17,24 while an uncharacterized chemical modification seems to accompany release of aIF6 from the 50S subunit.21
The high-resolution X-ray structures of aIF6 and eIF6 in isolation revealed a pentein fold, the arrangement of five quasi-identical αββαβ-subdomains around a 5-fold axis of pseudo-symmetry.25 More recently, the structure of eIF6 bound to the eukaryotic 60S ribosomal subunit has been solved by both cryo-EM26 and X-ray crystallography.27 These structures show yeast and Tetrahymena thermophila eIF6 bound to rpL23e in the sarcin–ricin loop region at the edge of the interface side of the 60S subunit.26,27 This localization on the 60S ribosomal subunit is inhibitory to intersubunit bridge formation and explains the mechanism of the eIF6-mediated inhibition of subunit joining by steric exclusion of the small ribosomal subunit.26 No 3D structural information on the archaeal 50S–aIF6 complex is available so far. The binding site of aIF6 has been localized to a similar region of the large ribosomal subunit by protein–protein interaction data and RNA footprinting.21 However, RNA footprinting data indicating that aIF6 is located in proximity to rRNA helix 69 suggested that aIF6 utilizes a different interaction interface with the 50S ribosomal subunit compared to eIF6.21,23
We have reconstructed the 3D structure of the natively purified 50S–aIF6 complex from the methanogenic euryarchaeon M. thermautotrophicus28,29 using single-particle cryo-EM, revealing that the IF6 binding site on the large ribosomal subunit is highly conserved between the archaeal and eukaryotic domains of life. Furthermore, with the availability of high-resolution crystallographic data for the eukaryotic 60S ribosomal subunit,27 the molecular interpretation of the M. thermautotrophicus 50S ribosomal subunit has now become possible also for protein and rRNA components not present in H. marismortui, enabling us to gain insight into the structural changes that accompany the evolution of the archaeal ribosome.
Results
The purification and biochemical analysis of M. thermautotrophicus ribosomes reveal the isolation of a native 50S–aIF6 complex
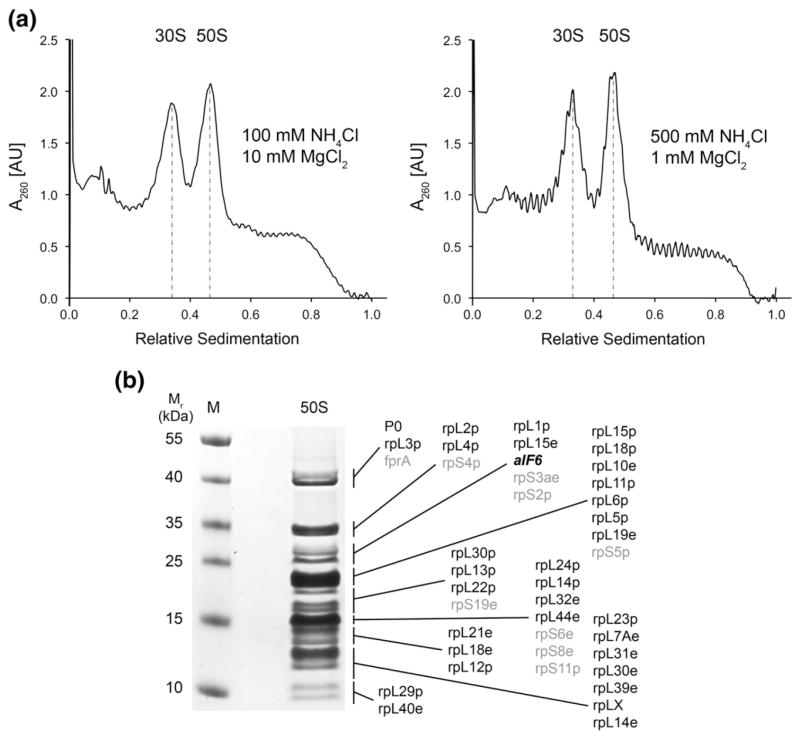
To obtain new insights into the conservation of archaeal ribosomes and their interaction with translation factors, we purified ribosomal subunits from M. thermautotrophicus for structural analysis. M. thermautotrophicus is a halotolerant, thermophilic methanogen, growing ideally at temperatures ranging from 65 °C to 72 °C under anoxic conditions.29,30 The preparation of ribosomal particles from M. thermautotrophicus resulted in the purification of separated 30S and 50S ribosomal subunits, without formation of a significant fraction of 70S ribosomes even at low monovalent salt and high magnesium concentrations, which strongly favor 70S formation in bacterial systems (Fig. 1a). We therefore concluded that ribosomes were present predominantly as dissociated or only weakly associated subunits in the cell lysate, as observed previously for ribosomal samples from Crenarchaeota.7,31,32 The protein content of the ribosomal preparations obtained under standard conditions was then analyzed by LC-MS/MS (liquid chromatography–tandem mass spectrometry) (Fig. 1b). Consistent with the prevalence of dissociated ribosomal subunits in our preparations of M. thermautotrophicus ribosomes, the anti-association factor aIF6 was found with several unique peptides (not shown). Therefore, this ribosome preparation provided an opportunity to study a natively purified archaeal 50S–aIF6 complex. Along with aIF6, our mass spectrometry analysis identified approximately 90% of the known M. thermautotrophicus 50S ribosomal subunit proteins (Table S1). Co-purification of aIF6 with archaeal ribosomes has also been reported previously for other species.7,33
Fig. 1.
Biochemical characterization of the M. thermautotrophicus 50S ribosomal subunit. (a) Ribosome profiles from M. thermautotrophicus crude ribosomes applied onto high and low magnesium and monovalent salt 10–40% (w/v) sucrose gradients. Under both associating (left) and dissociating (right) conditions, only 30S and 50S peaks, but no 70S peaks, were observed. (b) Mass spectrometric analysis of M. thermautotrophicus ribosomes. Large ribosomal subunit proteins are printed in black, aIF6 in bold italics, and proteins most likely co-sedimenting with the 50S subunit unspecifically or due to 30S contamination in gray.
3D reconstruction of the M. thermautotrophicus 50S ribosomal subunit by single-particle cryo-EM
To determine the structure of the M. thermautotrophicus 50S ribosomal subunit using single-particle cryo-EM, we imaged the specimen in frozen-hydrated state using a FEI Tecnai F20 electron microscope. From 553 exposures on a CCD camera, we extracted a data set of 101,438 particle images. The data set was subjected to alignment onto reference projections, multivariate statistical analysis, and 3D reconstruction using the angular reconstitution approach, followed by higher-resolution refinement by projection matching (see Methods). The resulting cryo-EM map shows clear density for bound aIF6 (Fig. 2a), with density values comparable to ribosomal proteins, suggesting a high occupancy of the 50S subunit with the bound factor. This indicates that the preparation of ribosomal subunits from M. thermautotrophicus indeed yielded a native 50S–aIF6 complex. The initial reconstruction indicated that the 50S subunits exhibit preferential orientation on the cryo-EM grid. Therefore, we limited the number of particle images entering the 3D reconstruction step for each reference projection during the refinement of the reconstruction to higher resolution to avoid reconstruction artifacts.34 The resulting cryo-EM map of the 50S–aIF6 complex is at 6.6 Å resolution according to the FSC=0.5 criterion (Fig. 2b; Fig. S1) and shows density features typical for subnanometer cryo-EM reconstructions, such as rRNA major and minor grooves, as well as tubular densities corresponding to protein α-helices (Fig. 2c and d).
Fig. 2.
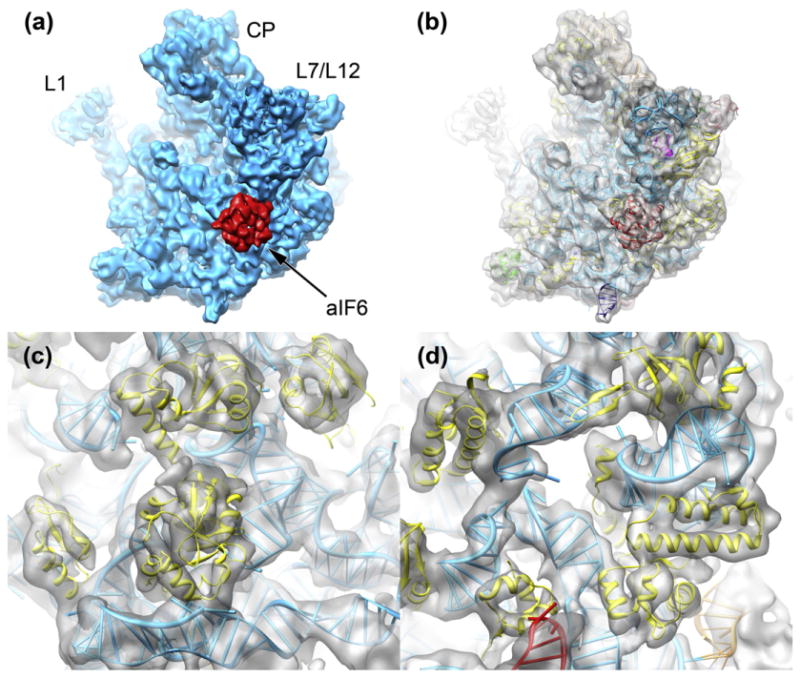
The 3D reconstruction of the M. thermautotrophicus 50S ribosomal subunit. (a) The cryo-EM structure of the M. thermautotrophicus 50S subunit (cyan) reveals density for aIF6 (red) bound in the sarcin–ricin loop region (L1: L1 stalk; L7/L12: L7/L12 stalk; CP: central protuberance). (b) The reconstruction of the M. thermautotrophicus 50S subunit (transparent gray) fitted with molecular structures and models for aIF6 and the ribosomal subunit. See the text for details. (c and d) The 6.6-Å cryo-EM density shows features typical for subnanometer-resolution cryo-EM reconstructions, such as rRNA major and minor grooves and tubular density for protein α-helices.
The ribosomal binding site of IF6 is conserved across the archaeal and eukaryotic domains of life
The cryo-EM density fitted with the high-resolution structure of the H. marismortui 50S subunit35 [Protein Data Bank (PDB) ID: 3CC2] and a homology model of the M. thermautotrophicus aIF6 generated from the Methanococcus jannaschii aIF6 X-ray crystal structure25 (PDB ID:1G61) allowed the unambiguous assignment of the rotational position of aIF6 on the archaeal 50S subunit by both visual inspection (Fig. 3a and b) and cross-correlation (Table S2). The rotational orientation of the pseudo-5-fold symmetrical factor on the archaeal large ribosomal subunit is identical with the orientation of eukaryotic eIF6 in 60S–eIF6 complexes.26,27 In agreement with protein–protein interaction data,21 aIF6 forms a large contact surface with its main binding partner rpL14p (rpL23e in eukaryotes) (Fig. 3c and d). The aIF6 cryo-EM density also shows smaller and weaker connections to ribosomal proteins rpL3p and rpL24e and to the base of the highly conserved sarcin–ricin loop of the 23S rRNA (Fig. 3c and d; Fig. S2). The comparison of the binding surface of IF6 on archaeal rpL14p and eukaryotic rpL23e reveals a conserved patch of interacting amino acids on both interaction partners (Fig. 4a and b). The remainder of the binding surface shows considerable sequence variability, mostly due to variability of the archaeal sequences (Fig. 4c and d).
Fig. 3.
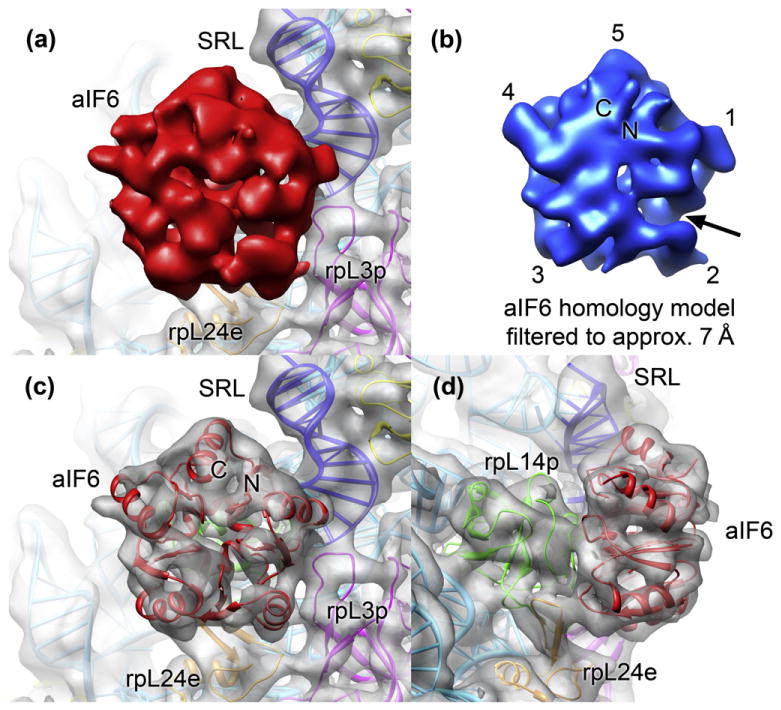
The binding site of aIF6 on the archaeal 50S ribosomal subunit. (a) View onto aIF6 (solid red) in the M. thermautotrophicus cryo-EM map. The binding site of aIF6 (rpL14p is invisible because it is hidden behind aIF6) is in the vicinity of the sarcin–ricin loop of the 23S rRNA (blue) and ribosomal proteins rpL24e (orange) and rpL3p (pink). (b) View of the M. thermautotrophicus aIF6 homology model filtered to the resolution of the cryo-EM map. The rotational positioning of aIF6 can be assigned both by correlation (Table S2) and visually, based on the asymmetry within the pseudo-5-fold symmetric aIF6. The most pronounced asymmetric feature is an indentation near α-helix 2 (arrow). (c) The fitting of the M. thermautotrophicus aIF6 model into the cryo-EM map revealed an excellent fit of the model to the map. The positions of the N- and C-termini of aIF6 are indicated. (d) Side view of the aIF6 binding site. The main interaction surface connects aIF6 to ribosomal protein rpL14p (green).
Fig. 4.
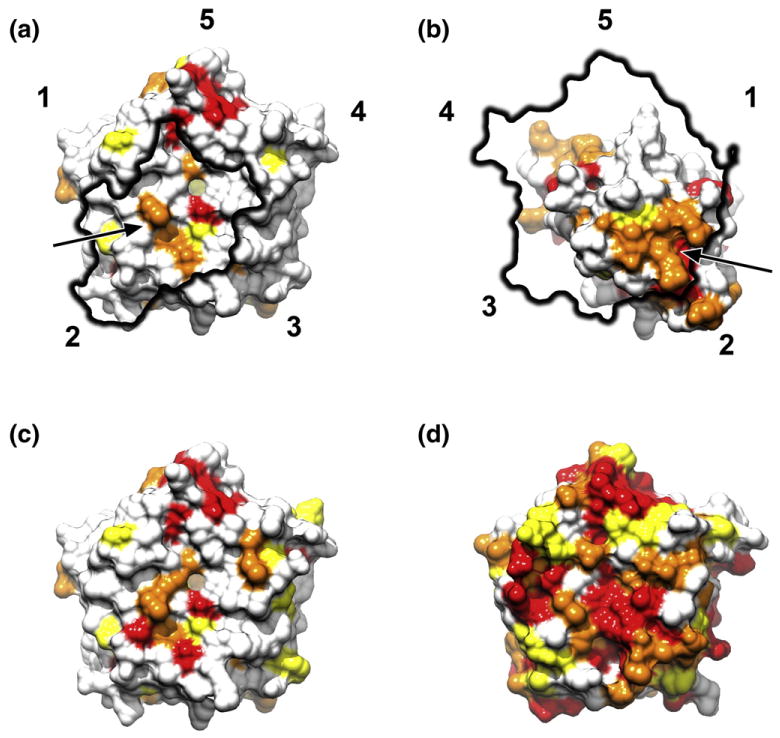
Sequence conservation of the interaction surfaces of IF6 and rpL14p/rpL23e in eukaryotes and archaea. Surface views of the M. jannaschii aIF6 (a) and H. marismortui rpL14p (b) structures were colored according to sequence conservation based on the multiple sequence alignment in Fig. S3a (all organisms). Conservation coloring was applied according to Clustal characters. Patches of more highly conserved residues (arrow) in both IF6 and rpL14p/rpL23e form a part of the interaction surface, along with less conserved surface areas. The highly conserved patch (red) near the C-terminal helix (5) on the interaction surface of IF6 does not participate in ribosome binding. Coloration of the M. jannaschii and S. cerevisiae IF6 structures based on the aligned archaeal (c) or eukaryotic (d) sequences reveals that the archaeal factor shows more sequence variability than the eukaryotic one.
Analysis of the protein and rRNA content of the M. thermautotrophicus 50S subunit
Apart from the density for the bound aIF6, the cryo-EM reconstruction of the M. thermautotrophicus 50S subunit shows several other areas of additional density compared to the H. marismortui X-ray crystal structure,9 reflecting the presence of extensions of the 23S rRNA and additional proteins. A comparison of the proteins assigned to the M. thermautotrophicus and H. marismortui 50S ribosomal subunits in the Swis-Prot database36 revealed that rpL14e, rpL30e, and rpL34e are present in M. thermautotrophicus but not in H. marismortui (Table S3). Further, rpL1p, rpLX, and rpL40e are also present in the H. marismortui genome but were not included in the X-ray crystal structure of the 50S ribosomal subunit from that organism.9 Therefore, in addition to the L1 stalk density encompassing rpL1p, unassigned density for five proteins was expected in the cryo-EM map of the M. thermautotrophicus 50S subunit. A comparison of the rRNA sequences of M. thermautotrophicus and H. marismortui by multiple sequence alignment37 indicates that the extensions in the M. thermautotrophicus 23S rRNA coincide with the location of known rRNA expansion segments (Fig. S4).
A molecular model of the M. thermautotrophicus 50S subunit reveals that eukaryotic-like rRNA structures and ribosomal proteins were lost during archaeal evolution
The recently solved crystal structure of the eukaryotic 60S subunit27 facilitated the assignment of eukaryotic-like rRNA structures and proteins in the reconstruction of the M. thermautotrophicus 50S subunit. Superposition of the 5.8S and 26S rRNA coordinates of the T. thermophila X-ray crystal structure onto the H. marismortui structure allowed the assignment of the rRNA expansion segments in M. thermautotrophicus. Inspection of the cryo-EM density revealed that a major fraction of the previously unassigned density in the M. thermautotrophicus 50S subunit cryo-EM map is located in the region below the L1 stalk (Figs. 5a and 6a), forming a density feature formerly termed the eocytic lobe.38 Expansion segments ES3, ES20, and ES26 as well as an extension of ES5 are located in this region (Fig. 5a). These expansion segments are found in all domains of life, as some bacteria also show corresponding sequence extensions. However, M. thermautotrophicus ES20 and ES26 are structurally more closely related to the eukaryotic extensions than to the known bacterial structures.39–41 In the immediate vicinity of these rRNA segments, ribosomal proteins rpL34e27 and rpL30e27,42 could be localized in the cryo-EM density (Fig. 6a).
Fig. 5.
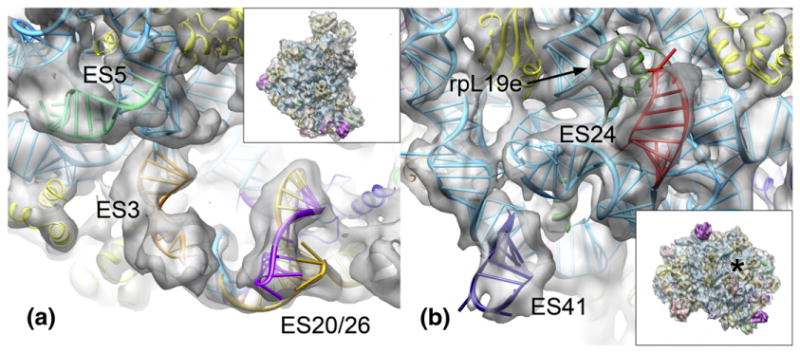
Expansion segments shared between eukaryotes and early-branching archaea, as seen in the M. thermautotrophicus 50S ribosomal subunit. Insets show the 50S subunit in the same orientation as the panels. An asterisk denotes the polypeptide tunnel exit. (a) View of the region below the L1 stalk. ES3 is colored orange, the ES5 extension in pale green, ES20 in gold, and ES26 in purple. (b) View of the bottom of the 50S subunit near the polypeptide exit tunnel. ES24 is colored red, ES40 in deep navy blue, and rpL19e in forest green.
Fig. 6.
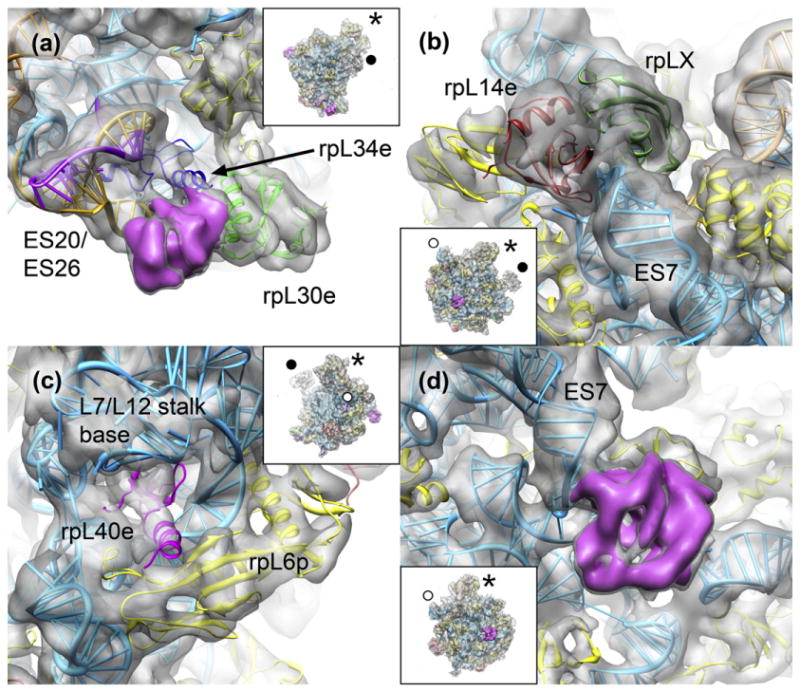
Newly assigned and unassigned protein densities in the M. thermautotrophicus 50S subunit. Insets show the 50S subunit in the same orientation as the panels (asterisk, central protuberance; filled circle, L1 stalk; open circle, L7/L12 stalk). (a) In the region below the L1 stalk, ribosomal proteins rpL30e (green) and rpL34e (blue) could be placed in the 50S cryo-EM map. An unassigned density is shown as a pink density segment. (b) On the solvent side of the large ribosomal subunit, rpL14e (dark red) and rpLX (forest green) could be placed in the M. thermautotrophicus 50S density. (c) The small ribosomal protein rpL40e (pink) is bound in the L7/L12 stalk region. (d) The unassigned density at the base of ES7 on the solvent side of the 50S subunit is shown as a pink density segment.
Further additional density features are scattered across the solvent side of the 50S subunit. An unoccupied density area near the polypeptide tunnel exit could be assigned to rRNA expansion segment ES24 (Fig. 5b). ES24 is present in both eukaryotes and bacteria but not in H. marismortui. In M. thermautotrophicus, ES24 adopts the eukaryotic-like configuration27,43 required to accommodate rpL19e, which is not present in bacteria. Enlarged density for ES41 is present at the edge of the interface side of the subunit (Fig. 5b). Compared to H. marismortui, ES7 and ES9 are elongated by a few nucleotides in M. thermautotrophicus, which is reflected in the cryo-EM density (not shown).
Two density features between the tip of expansion segment ES7 and the L7/L12 stalk base could be assigned to ribosomal proteins rpL14e7,27 and rpLX27 (Fig. 6b), respectively. The gene coding for rpL14e is missing in the H. marismortui genome but present in M. thermautotrophicus, while both genomes encode rpLX. However, rpLX was not observed in the H. marismortui X-ray crystal structure.9 The assignment of rpLX is based on the homology of M. thermautotrophicus rpLX (PDB ID: 2JXT) to the C-terminal SH3-like domain of the eukaryotic rpL18Ae (Fig. S5), which is a tandem repeat of two SH3-like folds.27 Density is present also for the very small protein rpL40e27 sitting in a cavity formed by rRNA, located between the L7/L12 stalk base and rpL6p (Fig. 6c). This localization of rpL40e was predicted previously by comparative structural analysis of the binding mode of Escherichia coli L36 to 23S rRNA.44 The solution structure of S. solfataricus rpL40e45 (PDB ID: 2AYJ) shows a disordered N-terminus, while, in agreement with the features observed in the M. thermautotrophicus cryo-EM map (Fig. S6), this part of the protein adopts an α-helical conformation when bound to the ribosome.27 Therefore, based on our cryo-EM map, we conclude that the disordered N-terminus observed in the solution structure of rpL40e does not reflect an archaea-specific conformation of this protein but that the N-terminus of rpL40e may fold only upon contact with the rRNA, as observed previously for other ribosomal proteins.46–48 We did not interpret the highly mobile L1 and L7/L12 stalks, as they showed only very weak or very fragmented density due to their high level of flexibility.
Evidence for the presence of two additional ribosomal proteins in the M. thermautotrophicus 50S subunit
After the completion of the analysis based on homology to known eukaryotic structures, a prominent extra density on the back of the ribosomal subunit in proximity to rpL32e and the base of ES7, corresponding in volume to approximately 15 kDa of protein density, remained unassigned (Fig. 6d). This finding was unexpected, as all proteins annotated to the M. thermautotrophicus 50S subunit in the SwissProt database36 had been placed in the density at this point (with the exception of proteins localized to the highly mobile elements of the subunit, the L1 and L7/L12 stalks). The presence of this density has also been reported for the cryo-EM reconstruction of the 50S subunit of Sulfolobus acidocaldarius,7 while there was no such density visible in the cryo-EM map of the Pyrobaculum aerophilum 50S subunit7 and the X-ray crystal structure of the H. marismortui 50S subunit.9 This suggests a heterogeneous distribution of this structural feature throughout both Crenarchaeota and Euryarchaeota. As there is no uninterpreted 23S rRNA sequence of sufficient size in this area (Fig. S7), this additional density most likely corresponds to an undetermined ribosome-associated protein.
A second unassigned density corresponding to a protein of approximately 8 kDa is located near ribosomal proteins rpL30e and rpL34e (Fig. 6a). This site was previously assigned to rpL34e in lower-resolution reconstructions of P. aerophilum and S. acidocaldarius 50S ribosomal subunits.7 However, in the X-ray structure of the eukaryotic 60S subunit,27 this site is occupied by rpL27e, and rpL34e occupies a binding site in its immediate vicinity but buried more deeply within rRNA. There is no homologous gene for rpL27e annotated in the M. thermautotrophicus genome49 or in the SwissProt database.36
Discussion
Purification of the native 50S–aIF6 complex
Most 50S subunits in the M. thermautotrophicus 50S ribosomal subunit sample are associated with aIF6, and only a minor fraction of 50S subunits is present in 70S ribosomes. The presence of large quantities of aIF6 in our ribosomal preparations might originate from cellular stress during growth or harvest of the M. thermautotrophicus cells, as aIF6 expression has been shown to be upregulated by heat and cold stress in archaea.21 In this case, the isolation of the 50S–aIF6 complex and the lack of 70S ribosomes would reflect the physiological state of the M. thermautotrophicus cells at the time of ribosome purification. The lack of 70S particles has been observed previously in preparations of archaeal ribosomes.7,31,32 Given that unbound proteins are separated very early from ribosomal complexes in our purification procedure (see Methods), it is likely that the large ribosomal subunits were already present in complex with aIF6 in the M. thermautotrophicus cells. Consistent with a physiological stress reaction, aIF6 was present in our M. thermautotrophicus cellular samples in significantly higher abundance than the 1:10 stoichiometry relative to 50S subunits observed in S. solfataricus during normal growth conditions.21 This can be inferred from two observations: (i) The analyzed 50S ribosomal subunits are occupied with aIF6 in nearly stoichiometric amounts, and (ii) there is very likely no other pool of 50S subunits, as only very low amounts of 70S ribosomes were present.
Refinement of the cryo-EM reconstruction of the M. thermautotrophicus 50S–aIF6 complex to subnanometer resolution
Preferential orientation of the ribosomal particles on the cryo-EM grid was suggested to limit the resolution of other recent reconstructions of large ribosomal subunits from both eukaryotic26 and archaeal7 organisms. If excessive numbers of particles from preferred views, such as the crown view of the large ribosomal subunit, are included in the 3D reconstruction, the resulting maps may be distorted or show artifacts.34 Therefore, we limited the number of particle images entering the 3D reconstruction step for each reference projection during the high-resolution refinement of the 50S–aIF6 reconstruction to ensure a more even coverage of all projection directions in the reconstruction process. Employing this correction method, the M. thermautotrophicus 50S–aIF6 complex could be refined to 6.6 Å resolution. The presence of bound ribosome-associated factor may also have contributed to a less pronounced preferential orientation of the particles. To our knowledge, the structure of the M. thermautotrophicus 50S–aIF6 complex is the highest-resolution EM reconstruction achieved for an asymmetric particle imaged on a CCD detector and the most highly resolved cryo-EM reconstruction of an isolated ribosomal subunit. The high resolution that was achieved is probably in a large part due to the high structural stability and rigidity of the archaeal 50S subunit isolated from a thermophilic species. Furthermore, we could use fewer particles for the reconstruction because the large ribosomal subunit does not exhibit large-scale conformational flexibility, as is the case for the head–body movements of the small ribosomal subunit or the ratchet movement of the ribosomal subunits relative to each other in the translating ribosome.50
The location of the binding site of IF6 on the large ribosomal subunit is highly conserved
The structure of the M. thermautotrophicus 50S–aIF6 complex reveals that the location of the binding site and the rotational orientation of IF6 on the large ribosomal subunit are highly conserved between the eukaryotic and archaeal domains of life, which is not fully consistent with a recently proposed model based on biochemical data, localizing aIF6 on the face, rather than at the edge, of the interface side of the large ribosomal subunit.21,23 Indeed, when we superimposed as a reference rpL23e of the T. thermophila X-ray crystal structure with its homolog rpL14p in the M. thermautotrophicus cryo-EM model, we obtained a Cα backbone RMSD of 2.2 Å for the ribosome-bound IF6 molecules. In comparison, the Cα backbone RMSD of the directly superposed M. thermautotrophicus and T. thermophila IF6 molecules is 1.6 Å. This indicates that the binding of aIF6 and eIF6 to the large ribosomal subunit is identical within the experimental error of our cryo-EM map. Given that the physiological mechanism of IF6 loading onto the large ribosomal subunit is not well understood, the high level of similarity of the native archaeal 50S–aIF6 complex to the in vitro reconstituted eukaryotic 60S–eIF6 complexes confirms the functional relevance of the data obtained from these in vitro reconstituted complexes. The structural elements of the ribosome in the vicinity of the IF6 binding site—rpL14p, rpL3p, rpL24e, and the sarcin–ricin loop—are conserved between archaea and eukaryotes. Although the location and the mode of binding of IF6 to the large ribosomal subunit are conserved, the interaction interface is less conserved on the sequence level and shows significant amino acid sequence variability in the Archaea (Fig. 4c). Possibly, some sequence variation was required to fine-tune the affinity of the aIF6–50S interaction to the unusual growth conditions of some archaeal species, such as high temperature or high salinity.
The conservation of the IF6 binding site on the ribosome may be a requirement for its function in conserved molecular pathways
In both eukaryotes and archaea, IF6 blocks the formation of 70S or 80S ribosomes by steric clashes with the small ribosomal subunit. Our results show that archaeal aIF6 directly blocks the formation of the intersubunit bridge between rpL14p of the 50S subunit and 16S rRNA helix 14 of the 30S subunit.41 Thus, the molecular mechanism of the anti-association activity of IF6 is highly conserved across archaea and eukaryotes, indicating that the exact positioning of IF6 on the large ribosomal subunit is critical for its cellular function. Whereas the inhibition of subunit joining could be achieved by IF6 bound in any conformation on the interface side of the large ribosomal subunit, the interaction with other translation factors, or factors regulating the association of IF6 with the large ribosomal subunit, might impose more stringent constraints on IF6 positioning on the ribosomal subunit.
The interaction of IF6 with factors involved in its release very likely requires a highly specific orientation of IF6 on the large ribosomal subunit. In eukaryotes, eIF6 is removed from the large ribosomal subunit by the concerted action of the translation elongation factor GTPase EF-2/EF-G homolog Efl118,51,52 and the Shwachman–Bodian–Diamond syndrome (SBDS) protein (Sdo1 in yeast).16,51 Archaeal genomes encode an SBDS protein53,54 but do not seem to code for an Efl1 homolog distinct from EF-2. Therefore, it has been proposed that the archaeal EF-2 may fulfill dual roles in archaea, both in translation elongation and in the release of aIF6 from the 50S subunit.55 The conservation of the binding site of IF6 in eukaryotes and archaea is consistent with such a mechanistically conserved pathway for the release of IF6 from the large ribosomal subunit. Furthermore, the conservation of the binding site also indicates that the evolution of a specialized release factor in eukaryotes may not be due to a difference in the binding of eIF6 to the large ribosomal subunit, but possibly to enable the regulation of IF6 release by modification of Efl1 activity without affecting EF-2 and basal translation.
A nuclear pool of eIF6 functions in ribosome biogenesis in eukaryotes, where eIF6 is required for normal pre-rRNA processing56 and remains bound to the ribosome until the passage through the nuclear pore has been completed.18,19 Whether aIF6 is involved in archaeal ribosome biogenesis remains to be determined. In eukaryotic ribosome biogenesis, there seems to be a functional linkage between the eukaryotic 60S nuclear export receptor Nmd357 and eIF6. Nmd3 and eIF6 bind to the 60S ribosomal subunit in close proximity to each other,26,27,58 and dissociation of eIF6 is a requirement for the release of Nmd3 from the pre-60S particle after nuclear export.19 The archaeal Nmd3 homolog59 lacks the C-terminal nuclear transport signals of eukaryotic Nmd3.57 It may function in an ancient pathway conserved between archaea and eukaryotes, possibly in a step of large ribosomal subunit biogenesis distinct from nuclear export, or in ribosome recycling.57,60 The involvement of aIF6 in such a pathway in archaea has not been demonstrated experimentally but might be conceivable given the conservation of the involved protein factors.
The conserved molecular mechanism of the IF6 anti-association activity can give rise to a variety of cellular functions
In archaea, aIF6 represses translation under stress conditions.21 In contrast, it has been shown that eIF6 stimulates translation initiation in mammalian cells in vivo, possibly by mediating growth factor signaling.14 It is likely that these seemingly contradictory roles are both based on the conserved molecular anti-association activity of IF6, which might be regulated in response to different stimuli to mediate either repression or upregulation of translation in archaea and eukaryotes, respectively.
Additional proteins in the M. thermautotrophicus 50S ribosomal subunit
Two density features in the cryo-EM map of the M. thermautotrophicus 50S–aIF6 complex could not be assigned to corresponding proteins. The first density is located near the base of ES7 (Fig. 6d), where density was also observed in the reconstruction of the S. acidocaldarius 50S ribosomal subunit.7 Considering that all predicted proteins of the 50S ribosomal subunit in M. thermautotrophicus have been accounted for in our structure, this density may indicate the presence of a loosely attached and therefore unassigned additional protein in the archaeal large ribosomal subunit. However, our mass spectrometry analysis (Fig. 1; Table S1) detected all known M. thermautotrophicus ribosomal proteins above a molecular mass of approximately 10.5 kDa, and a novel 15-kDa protein would likely have been detected. No hit for an additional ribosomal protein that could convincingly explain the additional density was found in the mass spectrometric analysis of the S. acidocaldarius 50S ribosomal subunit, either.7 Therefore, one of the known ribosomal proteins might be present in two copies in the M. thermautotrophicus large ribosomal subunit. The 13.1-kDa protein rpL7Ae has been reported to be chemically modified,7 which might enable its binding to two different binding sites on the ribosomal subunit, but to interpret this density feature with confidence, further experimental evidence or more highly resolved structural data are required.
The second unassigned density in our M. thermautotrophicus 50S cryo-EM map is located in the location occupied by rpL27e in the eukaryotic 60S ribosomal subunit, adjacent to rpL34e and rpL30e27 (Fig. 6a). Based on the observation that rpL6e, rpL27e, and rpL14e share an SH3-like fold, it has been suggested61 that gene duplication, creating separate gene copies that could subsequently evolve to bind to the ribosome in distinct locations, was involved at the origin of these ribosomal proteins. In M. thermautotrophicus, the site occupied by rpL27e in eukaryotes might, therefore, be occupied by an as-yet unidentified protein, possibly as a result of gene duplication, or alternatively by a second molecule of the archaeal SH3-like fold containing proteinrpL14e.
Implications for ribosome evolution
Comparison of the H. marismortui and M. thermautotrophicus large ribosomal subunit structures reveals the reduction of mostly eukaryotic-like protein and rRNA structures in the ribosomes of the phylogenetically late-branching H. marismortui. The structural similarity of these features in M. thermautotrophicus to eukaryotic ribosomes suggests that they represent an ancestral state in the common ancestor of archaea and eukaryotes. Indeed, although archaeal ribosomes contain the prokaryotic-type 16S, 23S, and 5S rRNAs, the M. thermautotrophicus large subunit rRNAs show clear structural similarities to eukaryotic rRNAs. It is intriguing that after the split of the eukaryotic and archaeal lineages, the eukaryotic ribosome acquired both novel ribosomal proteins and rRNA expansion segments, while the evolutionary trend for the archaeal ribosome was the opposite, with progressively increasing and apparently uncompensated losses of both protein and rRNA content from the ribosome (Fig. 7). Extensive loss of rRNA has also occurred during the evolution of the mitochondrial ribosome, with particularly large reduction occurring in mammalian mitochondria.63,64 In the mitochondrial ribosome, however, the loss of rRNA is structurally compensated by enlargement of already-present ribosomal proteins and recruitment of novel mitochondria-specific ribosomal proteins.65–69 In contrast, the loss of protein components in the archaeal ribosome is not compensated by enlargement of rRNA. Instead, rRNA segments were also shortened, sometimes in the immediate vicinity of the proteins that were lost or reduced in size. It will be interesting to investigate the causes and selection pressures—or lack thereof—underlying these highly divergent evolutionary trajectories (see Supplementary Text).
Fig. 7.
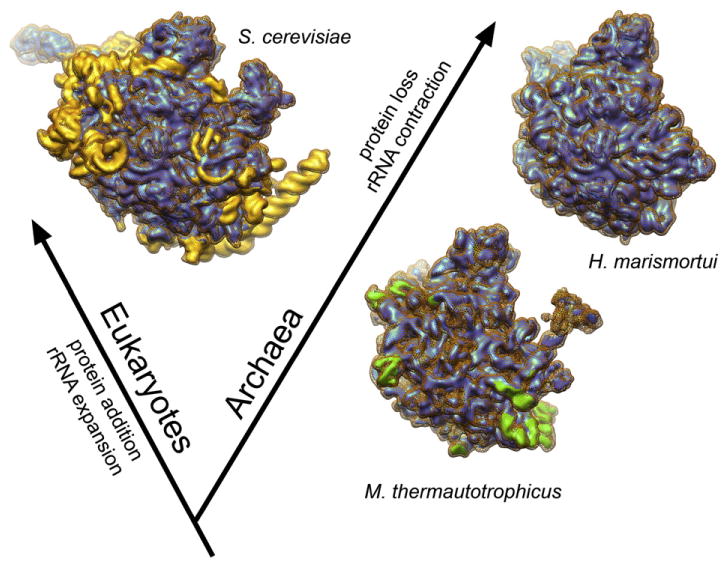
Snapshots of the structural evolution of the large ribosomal subunit. Structural elements missing in the M. thermautotrophicus 50S ribosomal subunit are shown in yellow in the S. cerevisiae 60S62 depiction. Structural elements missing in the H. marismortui 50S subunit9 are shown in green in the M. thermautotrophicus 50S subunit reconstruction. The densities of the H. marismortui and S. cerevisiae large ribosomal subunits were simulated in Chimera from atomic coordinates [PDB IDs: 3CC2 (Hm); 3IZS and 3IZF (Sc)].
Methods
Cell culture
M. thermoautotrophicus ΔH cultures were grown at 65 °C in a mineral salts medium supplied with an 80% H2:20% CO2 gas mixture, as previously described (Zeikus et al. 1982).
Biochemistry
For ribosome purification, frozen M. thermautotrophicus cells were thawed and resuspended in lysis buffer [20 mM Hepes–NaOH, pH 7.6, 200 mM NH4Cl, 20.5 mM MgCl2, 0.5 mM ethylenediaminetetraacetic acid (EDTA), and 5 mM β-mercaptoethanol] and lysed by five passages through a French press at 1.1 kpsi. The lysate was cleared using a Beckman Type 70Ti rotor (2×25 min, 25 krpm). The resulting cleared lysate was centrifuged trough a 40% (w/v) sucrose cushion (in 20 mM Hepes–NaOH, pH 7.6, 500 mM NH4Cl, 10.5 mM MgCl2, 0.5 mM EDTA, and 5 mM β-mercaptoethanol) in a Beckman Type 70Ti rotor (17 h, 34 krpm). The ribosomal pellets were resuspended (20 mM Hepes–NaOH, pH 7.6, 6.5 mM MgCl2, 0.5 mM EDTA, 60 mM NH4Cl, and 2 mM DTT), layered onto 10–40% sucrose gradients (in 20 mM Hepes–NaOH, pH 7.6, 60 mM NH4Cl, 6.5 mM MgCl2, 0.5 mM EDTA, and 2 mM DTT) and spun in an SW32Ti rotor (17 h, 19 krpm). The ribosomal bands were visualized using light scattering and collected with a syringe. The sample was concentrated to approximately 10 mg/ml, flash frozen in liquid nitrogen, and stored at −80 °C. For mass spectrometry analysis of the 50S ribosomal subunit preparation, a sample of M. thermautotrophicus 50S subunits was separated on a 4–12% Bis-Tris NuPage SDS-PAGE gel. Gel slices were sent for mass spectrometry (FGCZ protein service, commercial service†) and analyzed by LC-MS/MS.
Ribosome profiles of M. thermautotrophicus were obtained from samples of 100 μl of approximately 200 nM crude ribosomes obtained after purification on a sucrose cushion as described above, with minor modifications [the cells were lysed using a cell cracker (Constant Cell Disruption Systems, UK) at 30 kpsi and the buffer molecule was Hepes–KOH, pH 7.6, instead of Tris–HCl, pH 8.0]. Crude ribosome samples were loaded onto 4.8-ml sucrose gradients [10–30% (w/v) sucrose] in a selection of †http://www.fgcz.ch buffers (low Mg2+: 20 mM Hepes–NaOH, pH 7.6, 1 mM MgCl2, and 2 mM DTT combined with 100, 500, and 700 mM NH4Cl; high Mg2+: 20 mM Hepes–NaOH, pH 7.6, 10.5 mM MgCl2, 0.5 mM EDTA, and 2 mM DTT combined with 100, 500, and 700 mM NH4Cl). The gradients were spun (SW55Ti, 45 krpm, 90 min), and the ribosome profiles were analyzed by UV absorbance.
Cryo-EM
Samples containing 150 nM 50S ribosomal subunit complex in 20 mM Tris–HCl, pH 8.0, 50 mM NH4Cl, and 20 mM MgCl2 were applied to Quantifoil R2/1 Holey Carbon EM grids. The grids were flash frozen in liquid ethane by manual plunging. Image acquisition was performed at a FEI Tecnai F20 (FEI, Hillsboro, OR, USA) transmission electron microscope at 200 kV acceleration voltage at 83,000× magnification (1.81 Å per pixel on the object scale) and defocus values ranging from 1.2 μm to 3.5 μm. A Gatan US 4000 Special 4 k ×4 k CCD detector (Gatan, Pleasanton, CA, USA) was used for image acquisition. A total of 101,438 particles were windowed semiautomatically using batchboxer (EMAN 1.6)70 and inspected manually in boxer (EMAN 1.6). After defocus correction,71 the particle images were coarsened and high-and low-pass filtered (Imagic-5 or SPIDER).72,73 The angular reconstitution approach in Imagic-5 was applied during the first refinement rounds using an initial data set of approximately 45,000 particle images. The refinement in Imagic-5 started from a cryo-EM map of the E. coli 50S ribosomal subunit74 filtered to approximately 20 Å resolution as an initial reference. Further refinement to higher resolution was performed in SPIDER using the final data set. The final refinement rounds were carried out using full-sized particle images, employing correction for projection angle distribution, and using Fourier amplitude enhancement based on SAXS data75 after each refinement cycle. The resolution of the final map computed from 70,364 particles was estimated according to the FSC=0.5 criterion based on subvolumes obtained from the BP 32F command in SPIDER.73
Modeling and interpretation of maps
M. thermautotrophicus aIF6 was modeled in MODELLER76 using the M. jannaschii aIF6 crystal structure25 (PDB ID: 1G61) as a template. The last two residues of the M. thermautotrophicus sequence were truncated, as there is no structural information available that would allow the modeling of these residues. The M. thermautotrophicus ribosomal proteins were modeled using the H. marismortui crystal structure35 (PDB ID: 3CC2) as a template for the proteins shared between the two organisms. The X-ray crystal structure of the T. thermophila 60S subunit27 was used as a template for the modeling of rpL34e, rpL40e, rpL30e, and rpL14e. As the resolution of the data did not allow for detailed model building and refinement, the modeling was kept to a minimum and only included the truncation of the template structures to the length of the corresponding M. thermautotrophicus sequences followed by the removal of the side chains. The solution structure of the M. thermautotrophicus rpLX is known (PDB ID: 2JXT), and generation of the model only required the removal of the residues corresponding to the His6-tag. The 23S rRNA core and the 5S rRNA are based on the H. marismortui X-ray crystal structure35 (PDB ID: 3CC2). The rRNA expansion segments not present in H. marismortui are based on the T. thermophila 60S–eIF6 X-ray crystal structure27 (ES3, ES5, ES20, ES26, and ES41) and the Saccharomyces cerevisiae 80S ribosome crystal structure (PDB ID: 3O5843) (ES24). The nucleotide bases were removed from the rRNAs. To generate the final ribosome model, we aligned the crystal structures of the H. marismortui 50S subunit and the T. thermophila 60S–eIF6 complex in PyMOL (DeLano, 2002‡) and used them as templates for positioning of all M. thermautotrophicus rRNAs and ribosomal proteins. The position of aIF6 was refined according to the cryo-EM density in Chimera.77,78 Detailed information on the model components is given in Tables S4 and S5.
Multiple sequence alignment
The protein sequences were retrieved from UniProt§,36 aligned using ClustalW,79 and visualized in Jalview||.80
Preparation of figures
Depictions of biomolecules were generated in Chimera78 using POV-Ray (Persistence of Vision Pty. Ltd., 2004¶) for rendering of atomic surfaces.
Supplementary Material
Acknowledgments
We thank Sebastian Klinge, Felix Voigts-Hoffmann, and Marc Leibundgut for sharing of data on the T. thermophila 60S–eIF6 X-ray crystal structure and Marc Leibundgut and Felix Voigts-Hoffmann for the critical reading of the manuscript. We thank Electron Microscopy ETH Zurich for data collection and Peter Tittmann for support. This work was supported by the Swiss National Science Foundation, the National Center of Excellence in Research Structural Biology Program of the Swiss National Science Foundation, and European Research Council grant 250071 under the European Community’s Seventh Framework Programme (to N.B.). The visit of V.G.M. to ETH and A.C. to Ohio State University was supported by the Unity through Knowledge Fund, project 10/07.
Abbreviations used
- IF6
initiation factor 6
- EM
electron microscopy
- 3D
three-dimensional
- PDB
Protein Data Bank
- EDTA
ethylenediaminetetraacetic acid
Footnotes
Accession numbers
The M. thermautotrophicus 50S–aIF6 cryo-EM map has been deposited at the EM Data Bank with accession number EMD-2012. The coordinates of the Cα and phosphate backbones of the M. thermautotrophicus 50S–aIF6 model have been deposited at the PDB with accession number 4ADX.
Supplementary data to this article can be found online at doi:10.1016/j.jmb.2012.01.018
References
- 1.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 2.Steitz TA. A structural understanding of the dynamic ribosome machine. Nat Rev, Mol Cell Biol. 2008;9:242–253. doi: 10.1038/nrm2352. [DOI] [PubMed] [Google Scholar]
- 3.Dennis PP. Ancient ciphers: translation in Archaea. Cell. 1997;89:1007–1010. doi: 10.1016/s0092-8674(00)80288-3. [DOI] [PubMed] [Google Scholar]
- 4.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lecompte O, Ripp R, Thierry JC, Moras D, Poch O. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res. 2002;30:5382–5390. doi: 10.1093/nar/gkf693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Márquez V, Fröhlich T, Armache JP, Sohmen D, Dönhöfer A, Mikolajka A, et al. Proteomic characterization of archaeal ribosomes reveals the presence of novel archaeal-specific ribosomal proteins. J Mol Biol. 2011;405:1215–1232. doi: 10.1016/j.jmb.2010.11.055. [DOI] [PubMed] [Google Scholar]
- 8.Brochier-Armanet C, Forterre P, Gribaldo S. Phylogeny and evolution of the Archaea: one hundred genomes later. Curr Opin Microbiol. 2011;14:274–281. doi: 10.1016/j.mib.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 9.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 10.Russell DW, Spremulli LL. Purification and characterization of a ribosome dissociation factor (eukaryotic initiation factor 6) from wheat germ. J Biol Chem. 1979;254:8796–8800. [PubMed] [Google Scholar]
- 11.Valenzuela DM, Chaudhuri A, Maitra U. Eukaryotic ribosomal subunit anti-association activity of calf liver is contained in a single polypeptide chain protein of Mr = 25,500 (eukaryotic initiation factor 6) J Biol Chem. 1982;257:7712–7719. [PubMed] [Google Scholar]
- 12.Sanvito F, Piatti S, Villa A, Bossi M, Lucchini G, Marchisio PC, Biffo S. The beta4 integrin interactor p27(BBP/eIF6) is an essential nuclear matrix protein involved in 60S ribosomal subunit assembly. J Cell Biol. 1999;144:823–837. doi: 10.1083/jcb.144.5.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Si K, Maitra U. The Saccharomyces cerevisiae homologue of mammalian translation initiation factor 6 does not function as a translation initiation factor. Mol Cell Biol. 1999;19:1416–1426. doi: 10.1128/mcb.19.2.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gandin V, Miluzio A, Barbieri AM, Beugnet A, Kiyokawa H, Marchisio PC, Biffo S. Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation. Nature. 2008;455:684–688. doi: 10.1038/nature07267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miluzio A, Beugnet A, Volta V, Biffo S. Eukaryotic initiation factor 6 mediates a continuum between 60S ribosome biogenesis and translation. EMBO Rep. 2009;10:459–465. doi: 10.1038/embor.2009.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finch AJ, Hilcenko C, Basse N, Drynan LF, Goyenechea B, Menne TF, et al. Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman–Diamond syndrome. Genes Dev. 2011;25:917–929. doi: 10.1101/gad.623011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basu U, Si K, Deng H, Maitra U. Phosphorylation of mammalian eukaryotic translation initiation factor 6 and its Saccharomyces cerevisiae homologue Tif6p: evidence that phosphorylation of Tif6p regulates its nucleocytoplasmic distribution and is required for yeast cell growth. Mol Cell Biol. 2003;23:6187–6199. doi: 10.1128/MCB.23.17.6187-6199.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Senger B, Lafontaine DL, Graindorge JS, Gadal O, Camasses A, Sanni A, et al. The nucle(ol) ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol Cell. 2001;8:1363–1373. doi: 10.1016/s1097-2765(01)00403-8. [DOI] [PubMed] [Google Scholar]
- 19.Lo KY, Li Z, Bussiere C, Bresson S, Marcotte EM, Johnson AW. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol Cell. 2010;39:196–208. doi: 10.1016/j.molcel.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, et al. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benelli D, Marzi S, Mancone C, Alonzi T, La Teana A, Londei P. Function and ribosomal localization of aIF6, a translational regulator shared by archaea and eukarya. Nucleic Acids Res. 2009;37:256–267. doi: 10.1093/nar/gkn959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barthelme D, Dinkelaker S, Albers SV, Londei P, Ermler U, Tampé R. Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. Proc Natl Acad Sci USA. 2011;108:3228–3233. doi: 10.1073/pnas.1015953108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benelli D, Londei P. Translation initiation in Archaea: conserved and domain-specific features. Biochem Soc Trans. 2011;39:89–93. doi: 10.1042/BST0390089. [DOI] [PubMed] [Google Scholar]
- 24.Biswas A, Mukherjee S, Das S, Shields D, Chow CW, Maitra U. Opposing action of casein kinase 1 and calcineurin in nucleo-cytoplasmic shuttling of mammalian translation initiation factor eIF6. J Biol Chem. 2011;286:3129–3138. doi: 10.1074/jbc.M110.188565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groft CM, Beckmann R, Sali A, Burley SK. Crystal structures of ribosome anti-association factor IF6. Nat Struct Biol. 2000;7:1156–1164. doi: 10.1038/82017. [DOI] [PubMed] [Google Scholar]
- 26.Gartmann M, Blau M, Armache JP, Mielke T, Topf M, Beckmann R. Mechanism of eIF6-mediated inhibition of ribosomal subunit joining. J Biol Chem. 2010;285:14848–14851. doi: 10.1074/jbc.C109.096057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klinge S, Voigts-Hoffmann F, Leibundgut MA, Arpagaus S, Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334:941–948. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- 28.Wasserfallen A, Nölling J, Pfister P, Reeve J, Conway de Macario E. Phylogenetic analysis of 18 thermophilic Methanobacterium isolates supports the proposals to create a new genus, Methanothermobacter gen. nov., and to reclassify several isolates in three species, Methanothermobacter thermautotrophicus comb nov., Methanothermobacter wolfeii comb nov., and Methanothermobacter marburgensis sp nov. Int J Syst Evol Microbiol. 2000;50:43–53. doi: 10.1099/00207713-50-1-43. [DOI] [PubMed] [Google Scholar]
- 29.Zeikus JG, Wolfe RS. Methanobacterium thermoautotrophicus sp n., an anaerobic, autotrophic, extreme thermophile. J Bacteriol. 1972;109:707–715. doi: 10.1128/jb.109.2.707-713.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ciulla R, Clougherty C, Belay N, Krishnan S, Zhou C, Byrd D, Roberts MF. Halotolerance of Methanobacterium thermoautotrophicum delta H and Marburg. J Bacteriol. 1994;176:3177–3187. doi: 10.1128/jb.176.11.3177-3187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Londei P, Altamura S, Cammarano P, Petrucci L. Differential features of ribosomes and of poly(U)-programmed cell-free systems derived from sulphur-dependent archaebacterial species. Eur J Biochem. 1986;157:455–462. doi: 10.1111/j.1432-1033.1986.tb09689.x. [DOI] [PubMed] [Google Scholar]
- 32.Londei P, Altamura S, Caprini E, Martayan A. Translation and ribosome assembly in extremely thermophilic archaebacteria. Biochimie. 1991;73:1465–1472. doi: 10.1016/0300-9084(91)90179-5. [DOI] [PubMed] [Google Scholar]
- 33.Benelli D, Londei P. In vitro studies of archaeal translational initiation. Methods Enzymol. 2007;430:79–109. doi: 10.1016/S0076-6879(07)30005-0. [DOI] [PubMed] [Google Scholar]
- 34.Boisset N, Penczek P, Taveau J, You V, de Haas F, Lamy J. Overabundant single-particle electron microscope views induce a three-dimensional reconstruction artifact. Ultramicroscopy. 1998;74:201–207. [Google Scholar]
- 35.Blaha G, Gürel G, Schroeder SJ, Moore PB, Steitz TA. Mutations outside the anisomycin-binding site can make ribosomes drug-resistant. J Mol Biol. 2008;379:505–519. doi: 10.1016/j.jmb.2008.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.UniProt Consortium. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D’Souza LM, Du Y, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henderson E, Oakes M, Clark MW, Lake JA, Matheson AT, Zillig W. A new ribosome structure. Science. 1984;225:510–512. doi: 10.1126/science.6429855. [DOI] [PubMed] [Google Scholar]
- 39.Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, et al. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 40.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JHD. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 41.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 42.Chandramouli P, Topf M, Ménétret JF, Eswar N, Cannone JJ, Gutell RR, et al. Structure of the mammalian 80S ribosome at 8.7 Å resolution. Structure. 2008;16:535–548. doi: 10.1016/j.str.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 44.Roberts E, Sethi A, Montoya J, Woese CR, Luthey-Schulten Z. Molecular signatures of ribosomal evolution. Proc Natl Acad Sci USA. 2008;105:13953–13958. doi: 10.1073/pnas.0804861105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu B, Lukin J, Yee A, Lemak A, Semesi A, Ramelot TA, et al. Solution structure of ribosomal protein L40E, a unique C4 zinc finger protein encoded by archaeon Sulfolobus solfataricus. Protein Sci. 2008;17:589–596. doi: 10.1110/ps.073273008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williamson JR. Induced fit in RNA–protein recognition. Nat Struct Biol. 2000;7:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 47.Markus MA, Hinck AP, Huang S, Draper DE, Torchia DA. High resolution solution structure of ribosomal protein L11-C76, a helical protein with a flexible loop that becomes structured upon binding to RNA. Nat Struct Biol. 1997;4:70–77. doi: 10.1038/nsb0197-70. [DOI] [PubMed] [Google Scholar]
- 48.Timsit Y, Allemand F, Chiaruttini C, Springer M. Coexistence of two protein folding states in the crystal structure of ribosomal protein L20. EMBO Rep. 2006;7:1013–1018. doi: 10.1038/sj.embor.7400803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith DR, Doucette-Stamm LA, Deloughery C, Lee H, Dubois J, Aldredge T, et al. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 51.Menne TF, Goyenechea B, Sánchez-Puig N, Wong CC, Tonkin LM, Ancliff PJ, et al. The Shwachman–Bodian–Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat Genet. 2007;39:486–495. doi: 10.1038/ng1994. [DOI] [PubMed] [Google Scholar]
- 52.Bécam AM, Nasr F, Racki WJ, Zagulski M, Herbert CJ. Ria1p (Ynl163c), a protein similar to elongation factors 2, is involved in the biogenesis of the 60S subunit of the ribosome in Saccharomyces cerevisiae. Mol Genet Genomics. 2001;266:454–462. doi: 10.1007/s004380100548. [DOI] [PubMed] [Google Scholar]
- 53.Boocock GRB, Marit MR, Rommens JM. Phylogeny, sequence conservation, and functional complementation of the SBDS protein family. Genomics. 2006;87:758–771. doi: 10.1016/j.ygeno.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 54.Boocock GRB, Morrison JA, Popovic M, Richards N, Ellis L, Durie PR, Rommens JM. Mutations in SBDS are associated with Shwachman–Diamond syndrome. Nat Genet. 2003;33:97–101. doi: 10.1038/ng1062. [DOI] [PubMed] [Google Scholar]
- 55.Johnson AW, Ellis SR. Of blood, bones, and ribosomes: is Swachman–Diamond syndrome a ribosomopathy? Genes Dev. 2011;25:898–900. doi: 10.1101/gad.2053011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Basu U, Si K, Warner JR, Maitra U. The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol Cell Biol. 2001;21:1453–1462. doi: 10.1128/MCB.21.5.1453-1462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ho JH, Kallstrom G, Johnson AW. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J Cell Biol. 2000;151:1057–1066. doi: 10.1083/jcb.151.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sengupta J, Bussiere C, Pallesen J, West M, Johnson AW, Frank J. Characterization of the nuclear export adaptor protein Nmd3 in association with the 60S ribosomal subunit. J Cell Biol. 2010;189:1079–1086. doi: 10.1083/jcb.201001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ho JH, Johnson AW. NMD3 encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2389–2399. doi: 10.1128/mcb.19.3.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ho JH, Kallstrom G, Johnson AW. Nascent 60S ribosomal subunits enter the free pool bound by Nmd3p. RNA. 2000;6:1625–1634. doi: 10.1017/s1355838200001291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, et al. Localization of eukaryote-specific ribosomal proteins in a 5.5-Å cryo-EM map of the 80S eukaryotic ribosome. Proc Natl Acad Sci USA. 2010;107:19754–19759. doi: 10.1073/pnas.1010005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, et al. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-Å resolution. Proc Natl Acad Sci USA. 2010;107:19748–19753. doi: 10.1073/pnas.1009999107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smits P, Smeitink JAM, van den Heuvel LP, Huynen MA, Ettema TJG. Reconstructing the evolution of the mitochondrial ribosomal proteome. Nucleic Acids Res. 2007;35:4686–4703. doi: 10.1093/nar/gkm441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O’Brien TW. Properties of human mitochondrial ribosomes. IUBMB Life. 2003;55:505–513. doi: 10.1080/15216540310001626610. [DOI] [PubMed] [Google Scholar]
- 65.Suzuki T, Terasaki M, Takemoto-Hori C, Hanada T, Ueda T, Wada A, Watanabe K. Proteomic analysis of the mammalian mitochondrial ribosome. Identification of protein components in the 28 S small subunit. J Biol Chem. 2001;276:33181–33195. doi: 10.1074/jbc.M103236200. [DOI] [PubMed] [Google Scholar]
- 66.Suzuki T, Terasaki M, Takemoto-Hori C, Hanada T, Ueda T, Wada A, Watanabe K. Structural compensation for the deficit of rRNA with proteins in the mammalian mitochondrial ribosome. Systematic analysis of protein components of the large ribosomal subunit from mammalian mitochondria. J Biol Chem. 2001;276:21724–21736. doi: 10.1074/jbc.M100432200. [DOI] [PubMed] [Google Scholar]
- 67.Koc EC, Burkhart W, Blackburn K, Moseley A, Spremulli LL. The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. J Biol Chem. 2001;276:19363–19374. doi: 10.1074/jbc.M100727200. [DOI] [PubMed] [Google Scholar]
- 68.Koc EC, Burkhart W, Blackburn K, Moyer MB, Schlatzer DM, Moseley A, Spremulli LL. The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J Biol Chem. 2001;276:43958–43969. doi: 10.1074/jbc.M106510200. [DOI] [PubMed] [Google Scholar]
- 69.Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 70.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 71.Sander B, Golas MM, Stark H. Automatic CTF correction for single particles based upon multivariate statistical analysis of individual power spectra. J Struct Biol. 2003;142:392–401. doi: 10.1016/s1047-8477(03)00072-8. [DOI] [PubMed] [Google Scholar]
- 72.Van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 73.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 74.Matadeen R, Patwardhan A, Gowen B, Orlova EV, Pape T, Cuff M, et al. The Escherichia coli large ribosomal subunit at 7.5 Å resolution. Structure. 1999;7:1575–1583. doi: 10.1016/s0969-2126(00)88348-3. [DOI] [PubMed] [Google Scholar]
- 75.Gabashvili IS, Agrawal RK, Spahn CM, Grassucci RA, Svergun DI, Frank J, Penczek P. Solution structure of the E. coli 70S ribosome at 11.5 Å resolution. Cell. 2000;100:537–549. doi: 10.1016/s0092-8674(00)80690-x. [DOI] [PubMed] [Google Scholar]
- 76.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 77.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 78.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 79.Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2002;Chapter 2(Unit 2.3) doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- 80.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.