Abstract
Proton beam irradiation is a form of advanced radiotherapy providing superior distributions of a low LET radiation dose relative to that of photon therapy for the treatment of cancer. Even though this clinical treatment has been developing for several decades, the proton radiobiology critical to the optimization of proton radiotherapy is far from being understood. Proteomic changes were analyzed in human melanoma cells treated with a sublethal dose (3 Gy) of proton beam irradiation. The results were compared with untreated cells. Two-dimensional electrophoresis was performed with mass spectrometry to identify the proteins. At the dose of 3 Gy a minimal slowdown in proliferation rate was seen, as well as some DNA damage. After allowing time for damage repair, the proteomic analysis was performed. In total 17 protein levels were found to significantly (more than 1.5 times) change: 4 downregulated and 13 upregulated. Functionally, they represent four categories: (i) DNA repair and RNA regulation (VCP, MVP, STRAP, FAB-2, Lamine A/C, GAPDH), (ii) cell survival and stress response (STRAP, MCM7, Annexin 7, MVP, Caprin-1, PDCD6, VCP, HSP70), (iii) cell metabolism (TIM, GAPDH, VCP), and (iv) cytoskeleton and motility (Moesin, Actinin 4, FAB-2, Vimentin, Annexin 7, Lamine A/C, Lamine B). A substantial decrease (2.3 x) was seen in the level of vimentin, a marker of epithelial to mesenchymal transition and the metastatic properties of melanoma.
Introduction
Proton therapy is used worldwide to treat several types of cancer due to superior targeting and energy deposition [1], [2]. Uveal melanoma is especially well suited for this kind of radiotherapy, as precise dose delivery is crucial to maintain eye function. In spite of the fact that proton therapy is used clinically with great success, not much is known about the biological effects of proton radiation. A substantial body of data has been accumulated on the biological effectiveness of proton radiation [3]–[13], and on some mechanisms of proton-induced cell death [14]–[17], but many other biological effects of the proton beam are unclear [1]. As proton irradiation is considered to be low-LET radiation (<20 keV/µm), its biological effects are assumed to be similar to those induced by photon radiation. However, there is some experimental data demonstrating that this is not always the case [1], [18].
As a stressor and a factor in cell death, proton radiation, as with other types of radiation, induces DNA damage, followed by DNA repair and a cellular stress response cascade. However, in comparison with photon radiation, proton therapy has been observed to be more effective against photon-radioresistant cell lines [4], [7], [12], [19] and different DNA repair responses were triggered, e.g. no ATR (ataxia-telangiectasia and Rad3-related protein kinases) activation being observed [20]. In terms of DNA damage, there was a prevalence of oxidative base damage [14], more double strand breaks [21]–[23] and larger DNA repair foci [24], [25]. Apoptosis was induced at doses above 10 Gy [14], [17] and cell cycle arrest in the G2-M phase was observed [11]. Interestingly, a differential oxidative stress response gene expression was recorded [16], [18].
Several cellular signaling cascades were induced by proton irradiation. For example, in PC3 cells irradiated with 10 Gy, it induced the cell cycle checkpoint ataxia-telangiectasia, mutated gene (ATM) and its downstream targets, such as p53, p21, and bax-α (Bcl-2–associat GTPase KRas protein and cyclin F, in contrast to gamma radiation which upregulates cell cycle blockers, such as Cyclin-dependent kinase inhibitor 2A [26]. p38, c-Jun N-terminal kinases (JNK) and Mitogen-activated protein kinases (MAP) signaling turned out to be crucial for proton irradiation–induced apoptosis, whereas pro-survival ERK (Extracellular Signal-regulated Kinase) activation, although typical for gamma-radiation, was absent after proton irradiation [1]. Other differences were also found in the regulation of angiogenesis, metastasis and migration properties, and inflammation [1].
The goal of this study was to characterize the cellular response to a sublethal dose of proton beam irradiation in a comprehensive way at protein level. Proteomic analysis was performed using BLM cells irradiated with 3 Gy of a 60 keV proton beam, passaged for 28–35 days to allow cellular repair. Our hypothesis was that a low dose of proton beam irradiation affects the proteins implicated in DNA repair, cellular stress response and survival even as a delayed outcome of proton beam irradiation. A significant (more than 1.5 × change) upregulation of 13 proteins and a downregulation of 4 proteins was found.
Materials and Methods
Cells
Human melanoma BLM is a highly metastasizing cell line. It was derived from a lung metastasis of the human melanoma Bro subline implanted in a nude mouse as earlier described [27], [28] and was cultured in an RPMI medium with 10% fetal bovine serum and antibiotics. The BLM cells were a kind gift from Prof. Martine J. Jager from Leiden University.
Proton Beam Irradiation
Cells in suspension in phosphate buffer saline (PBS) at 1×106 cells/ml were transported on ice to the proton beam facility. 1.5 ml of the cell suspension was irradiated at RT in an Eppendorf vial placed in a positioning holder. The source of the 58 MeV proton beam was the AIC-144 cyclotron at The Institute of Nuclear Physics, Polish Academy of Sciences, Kraków. The cells were irradiated with the dose of 3.0 Gy (CGE). The dose rate was 0.15 Gy/s. Irradiated cells were transferred to the medium and plated at 1×105 cell/ml in 10 cm Petri dishes. After 7 passages of irradiated BLM cells (4–5 weeks in culture after irradiation), they were isolated and were used for proteomic analysis. Non-irradiated BLM cells served as the control.
Cell Growth and Comet Assay
For the proliferation assay, 1,000 or 5,000 cells were seeded in each well of a 96-well plate. At selected time points cells were removed by trypsin, stained with trypan blue and the number of living cells was counted using a Burker cytometer. The level of DNA damage was determined by the electrophoresis of single cells in agarose gel as earlier described [29], [30]. Briefly, the cell suspension was mixed with low melting point agarose, set on slides, lysed and neutralized in appropriate buffers. Electrophoresis was performed at 23 V (0.74 V/cm, 300 mA) for 30 min at 4°C. All stages of the experiment were carried out in the dark to eliminate any extra DNA damages. Prior to analysis the slides were stained with propidium iodide (2.5 µg/ml). The analysis of DNA damage was carried out with COMET PLUS 2.9 software (Comet Plus, Theta System Gmbh, Germany). The percentage content of DNA in the comet’s tail (TDC) was determined from 100 random images of comets per slide. The analysis was done in two replicates.
Proteomic Analysis
Sample preparation
The cells were washed two times in 1 ml of a wash solution (10 mM tris(hydroxymethyl)-aminomethane (Tris), 5 mM magnesium acetate, pH 8.0) and centrifuged at 400 g for 10 min at 20°C. The cell pellet was lysed in an ice-cold buffer that contained 7 M urea, 2 M thiourea, 4% 3-[(3- cholamidopropyl) dimethylammonio] -1-propanesulfonate (CHAPS), 40 mM Tris pH 8.5, 65 mM Dithiothreitol (DTT), 1 mM EDTA and 1 mM phenylmethylsulphonyl fluoride. Cell disruption was achieved by sonication at 320 W, 20 kHz, 30 s ON/30 s OFF, two times for 10 minutes with Bioruptor UCD-200 (Diogenode, Liege, Belgium). The cell lysate was centrifuged at 15 000 g for 30 min at 12°C to remove debris. Proteins were precipitated using methanol and chloroform according to Wessel and Flügge [31]. The protein pellet was solubilized in a rehydration buffer that was composed of 7 M urea, 2 M thiourea, 2% CHAPS, 0.002% bromophenol blue, 0.5% immobilized pH gradient (IPG) buffer and 20 mM DTT (the IPG buffer and DTT were added just before use). Protein concentration was measured by Bradford assay [32].
Two-dimensional electrophoresis
Isoelectric focusing (IEF) was carried out with an Ettan IPGphor 3 IEF System (GE Healthcare, Uppsala, Sweden). 24-cm strips with an immobilized pH 3–10 nonlinear gradient (GE Healthcare) were passively rehydrated for 16 h with 450 µl rehydration buffer containing 200 µg of total protein. The IEF was performed at 50 µA/strip for 80 000 Vh at 20°C. Prior to Sodium dodecyl sulfate - Polyacrylamide gel electrophoresis the IPG strips were equilibrated using a two-step procedure, each being gently shaken for 10 min in a 75 mM Tris–HCl pH 8.8 buffer that contained 30% glycerol, 6 M urea and 2% SDS. In the first step, the proteins were reduced by the addition of 1% DTT to the buffer. In the second, they were alkylated with 2.5% iodoacetamide. After IEF the proteins were separated on 12.5% polyacrylamide gels (255 · 296 · 1 · mm). The gels were cast using an Ettan DALTsix Gel Caster (GE Healthcare). Electrophoresis was performed on an Ettan DALTsix Large Vertical System (GE Healthcare) at 2 W/gel for 45 min and 16 W/gel for 3.5 h at 20°C. After electrophoresis, the gels were fixed overnight in 50% methanol, 3% phosphoric acid and stained with Colloidal Coomassie Blue according to the following protocol. The gels were washed 3×30 min with water. In the first step of staining, the gels were incubated in a 400 ml solution consisting of 34% methanol, 3% phosphoric acid and 17% ammonium sulfate. After 1 hour, 140 mg Coomassie Blue G-250 was added to each gel and incubated for the next 5 days. Before scanning on ImageScanner III (GE Healthcare, Uppsala, Sweden) the gels were washed 3×30 minutes with water. The gel images were analyzed by ImageMaster 2D Platinum v 7.0 (GE Healthcare).
Protein identification by mass spectrometry
To identify the content of the protein spots of interest, the gel pieces were manually excised, destained at 37°C by washing several times in 25% and 50% acetonitrile in a 25 mM ammonium bicarbonate buffer (NH4HCO3). The gel pieces were dehydrated in 100% acetonitrile (ACN) and dried in a Speedvac for 5 min, then 15 µL trypsin (Biocentrum) solution (10 ng/µL in 25 mM NH4HCO3, pH 8.0) was added and incubated for 15 min. After that an additional 20 µL of 25 mM NH4HCO3 was added. Digestion was carried out overnight at 37°C. Peptides were extracted by sonication and drying with 100% ACN. The extracts were evaporated to dryness and resuspended in 2% ACN with 0,05% TFA. Peptides were analyzed using the UltiMate 3000RS LCnanoSystem (Dionex) coupled with a MicrOTOF-QII mass spectrometer (Bruker) using nano online ESIsprayer.
To briefly describe the of peptide analysis, the peptides were injected on a C18 precolumn (Acclaim PepMap Nano trap Column) using 2% ACN with 0,05% TFA as the mobile phase. They were further separated on a 15 cm×75 µm RP column (Acclaim PepMap 75 µm 100 Å Nano Series TM Column) using gradient 2–40% ACN in 0.05% FA for 30 minutes. MS was operated in standard DDA (data dependent acqusition) MS/MS mode with fragmentation of the most intensive precursor ions.
The MS spectra measured were recalibrated using the fragment ions of trypsin derived peptides. The Mascot Generic format (.mgf) was generated by pre-processing the raw data with Data Analysis 4.0 software (Bruker, Germany). The resulting lists of peaks were used to search the non-redundant protein database Swissprot with the taxonomic restriction – Homo sapiens (20 321 sequences) using an in-house Mascot server (v.2.3.0, Matrix Science, London, UK). The following search parameters were applied: enzyme specificity – trypsin, permitted number of missed cleavages –1, fixed modification – carbamidomethylation (C), variable modifications – oxidation (M), phosphorylation (STY), methylation (DE), protein mass – unrestricted, peptide mass tolerance – ±20 ppm, fragment mass tolerance – ±0.05 Da.
Results
Cell Growth Inhibition and DNA Damage
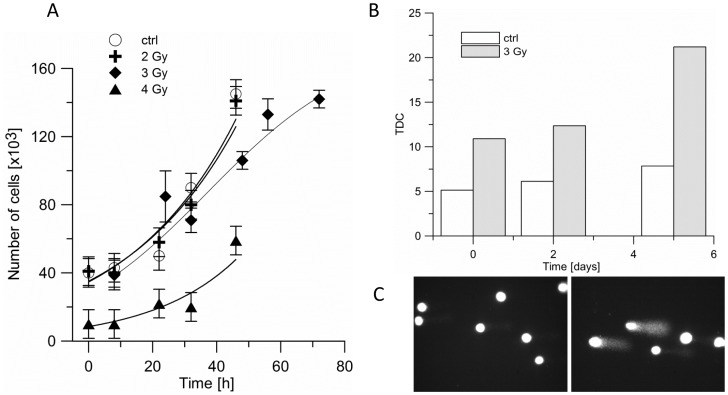
3 Gy of proton beam irradiation slowed down cell growth in culture by app. 15% (Fig. 1 A). Starting from day 4, the number of viable cells in culture decreased. This is in agreement with the increase in DNA damage shortly after the treatment (Fig. 1B,C). The cells were kept in culture for several passages (4–5 weeks) before the proteomic analysis was performed, in order to allow for cellular repair.
Figure 1. The effect of proton beam irradiation on BLM melanoma cells.
A. Proliferation of BLM cells after proton beam irradiation with doses 0 (open circle), 2 (cross), 3 (diamond) and 4 Gy (triangle). Cells were irradiated in suspension, and then plated in 96-well plates. B, C. DNA damage in untreated (white bar), and irradiated with 3 Gy of proton beam (stripped bar) BLM cells are presented in terms of the percentage of DNA that left the comet's head and was found in the comet’s tail after electrophoresis (TDC).
Proteomic Analysis
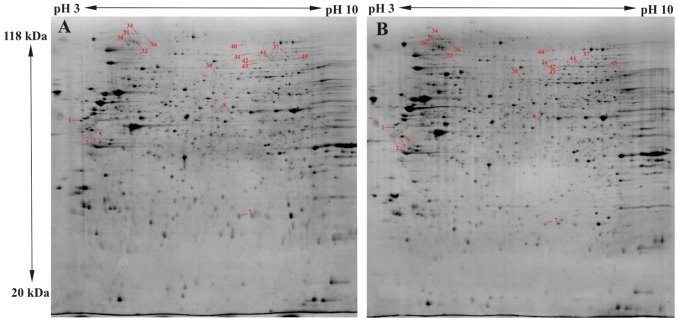
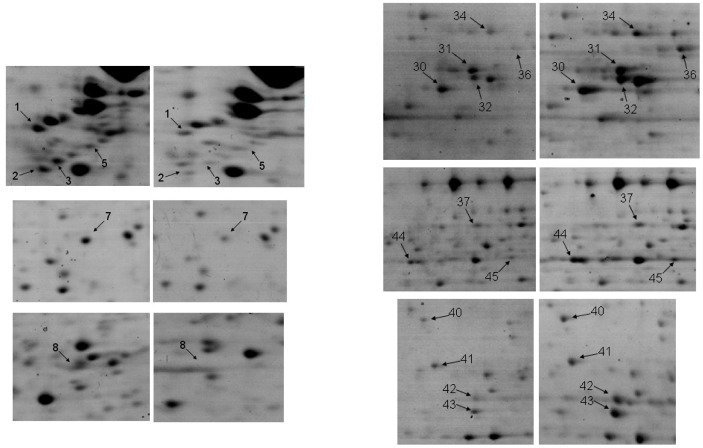
The mean number of identified spots per gel was 1200 and the average number of matches per gel was 860 (71%). 22 differential spots were found which fulfilled the Student t-test (Table 1). A representative spot pattern is depicted in Fig. 2. The protein contents of all differential spots were identified by LC-MS/MS on the basis of peptide mass matching following digestion with trypsin, and their peptide sequences which were obtained in a fragmentation process. The SwissProt accession numbers, the abbreviated and full names of the proteins, their theoretical pI and MW values are presented in Table 1. It also shows data from the mass spectrometry analysis, such as their score and the protein amino acid sequence coverage by matching peptides and the number of unique peptides. For the majority the identification was very reliable with a very high score. Only in the case of two spots were two proteins identified where it was not possible to specify the source of the difference. Finally, 17 regulated proteins were identified, 4 of them being downregulated, and 13 upregulated in the proton irradiated samples in comparison to the control group. The protein which showed the most pronounced downregulation after proton irradiation was vimentin, which was found in four spots. Detailed analysis of the MS/MS spectra for vimentin revealed differences in the methylation pattern of the protein isoforms. In spot number 1 methylation was recognized at 7 glutamic acid residues (109, 134, 136, 151, 151, 153, 225), in spots no. 2 and 3 only at glutamic acid in position 151, while in spot number 5 no modification was found. Although methylation typically occurs at lysine and arginine, there are also reports of the important biological role of the methylation of glutamic acid [33], [34]. In Fig. 3 the differential spots are shown.
Table 1. List of differentially regulated proteins from a comparison of proton beam irradiated and unirradiated BLM cells.
| Spot noa | Accession nrb | Protein | MW [kDa] calculated | pI | MW [kDa]in gel | Scores | #Peptides | SCc [%] | Ratiod ir/ctrl | fold change | p-value |
| 1 | VIME_HUMAN | Vimentin | 53.6 | 4.9 | 52.6 | 2736.8 | 41 | 60.3 | 0.49 | −2.0 | 0.018 |
| 2 | VIME_HUMAN | Vimentin | 53.6 | 4.9 | 48.7 | 1866.5 | 29 | 51.9 | 0.46 | −2.2 | 0.031 |
| 3 | VIME_HUMAN | Vimentin | 53.6 | 4.9 | 49.7 | 2230.9 | 36 | 65.5 | 0.29 | −3.4 | 0.001 |
| 5 | VIME_HUMAN | Vimentin | 53.6 | 4.9 | 51.1 | 781.7 | 15 | 36.9 | 0.64 | −1.6 | 0.015 |
| 7 | TPIS_HUMAN | Triosephosphate isomerase | 30.8 | 5.6 | 31.4 | 889.2 | 10 | 52.1 | 0.46 | −2.2 | 0.017 |
| 8 | ANXA7_HUMAN | Annexin A7 | 52.7 | 5.4 | 55.0 | 637.2 | 8 | 15.0 | 0.40 | −2.5 | 0.032 |
| 46 | G3P_HUMAN | Glyceraldehyde-3-phosphate dehydrogenase | 36.0 | 9.3 | 35.0 | 932.7 | 47 | 24.0 | 0.42 | −2.4 | 0.003 |
| 30 | TERA_HUMAN | Transitional endoplasmic reticulum ATPase | 89.3 | 5.0 | 104.3 | 3308.2 | 46 | 59.3 | 1.89 | 1.9 | 0.015 |
| 31 | LMNB2_HUMAN | Lamin-B2 | 67.6 | 5.2 | 108.8 | 2304.7 | 35 | 48.3 | 1.38 | 1.4 | 0.005 |
| 32 | ACTN4_HUMAN | Alpha-actinin-4 | 104.8 | 5.2 | 106.7 | 4204.7 | 61 | 69.9 | 1.88 | 1.9 | 0.043 |
| 34 | CAPR1_HUMAN | Caprin-1 | 78.3 | 5.0 | 117.1 | 192.5 | 3 | 4.8 | 1.78 | 1.8 | 0.038 |
| 36 | MVP_HUMAN | Major vault protein | 99.3 | 5.2 | 112.6 | 1937.7 | 29 | 43.9 | 1.95 | 1.9 | 0.012 |
| 37 | FUBP2_HUMAN | Far upstream element-binding protein 2 | 73.1 | 7.0 | 94.6 | 1464.6 | 26 | 32.1 | 1.72 | 1.7 | 0.042 |
| 38 | ACTN1_HUMAN | Alpha-actinin-1 | 103.0 | 5.1 | 75.5 | 2786.0 | 43 | 59.2 | 1.85 | 1.8 | 0.049 |
| LMNA_HUMAN | Prelamin-A/C | 74.1 | 6.6 | 2069.0 | 32 | 42.9 | |||||
| 40 | PDC6I_HUMAN | Programmed cell death 6-interacting protein | 96.0 | 6.1 | 104.1 | 1739.9 | 25 | 37.9 | 1.55 | 1.6 | 0.029 |
| 41 | MCM7_HUMAN | DNA replication licensing factor MCM7 | 81.3 | 6.1 | 94.1 | 1574.3 | 25 | 37.0 | 1.66 | 1.7 | 0.047 |
| 42 | LMNA_HUMAN | Prelamin-A/C | 74.1 | 6.6 | 87.1 | 2285.0 | 30 | 47.0 | 1.59 | 1.6 | 0.028 |
| 43 | MOES_HUMAN | Moesin | 67.8 | 6.0 | 84.5 | 1807.4 | 29 | 42.6 | 2.36 | 2.4 | 0.013 |
| 44 | LMNA_HUMAN | Prelamin-A/C | 74.1 | 6.6 | 86.6 | 1881.7 | 23 | 37.3 | 2.45 | 2.5 | 0.027 |
| 45 | LMNA_HUMAN | Prelamin-A/C | 74.1 | 6.6 | 87.0 | 2508.5 | 33 | 50.8 | 1.46 | 1.5 | 0.033 |
| 47 | HSP71_HUMAN | Heat shock 70 kDa protein 1 | 70.0 | 5.4 | 64.5 | 1611.1 | 61 | 35.0 | 1.81 | 1.8 | 0.082 |
| G3BP1_HUMAN | Ras GTPase-activating protein-bindingprotein 1 | 52.1 | 5.3 | 969.4 | 55 | 24.0 | |||||
| 48 | STRAP_HUMAN | Serine-threonine kinase receptor-associated protein | 38.4 | 4.8 | 42.0 | 866.1 | 58 | 15.0 | 4.16 | 4.2 | 0.006 |
Spots were compared by 2D stained with Colloidal Coomassie and proteins were identified using LC-MS/MS.
a The spot location is shown in Figure 2 ad 3.
b Protein accession number from the UniProtKB/Swiss-Prot nonredundant protein database.
c obtained sequence coverage.
d Ratio calculated in relation to unirradiated control group.
Figure 2. Representative 2D electrophoresis maps stained with Colloidal Commassie.
The results obtained for each experimental group are shown: control BLM cells (untreated) (A), 3 Gy proton beam irradiated BLM cells (B).
Figure 3. The zoomed changes in the level of spots identified by MS in the two groups.
Left panel – control group, Right panel –3 Gy irradiated group. Spot numbers as listed in Table 1.
Discussion
Our initial hypothesis was that a sublethal dose of proton beam irradiation would upregulate the proteins implicated in DNA repair, inflammation, stress response and the regulation of survival and, that it would possibly downregulate those connected to the metabolism, or to protein production. However, the data revealed a slightly different picture. First of all, relatively few protein levels were changed after irradiation in comparison with ionizing radiation [35], [36]. Moreover, relatively few proteins engaged in DNA repair were upregulated. These unexpected results may arise from the long period given for recuperation after the irradiation insult. Another unanticipated result is the lack of inflammation-related proteins, as well as the number of regulated proteins connected to the cytoskeleton and motility.
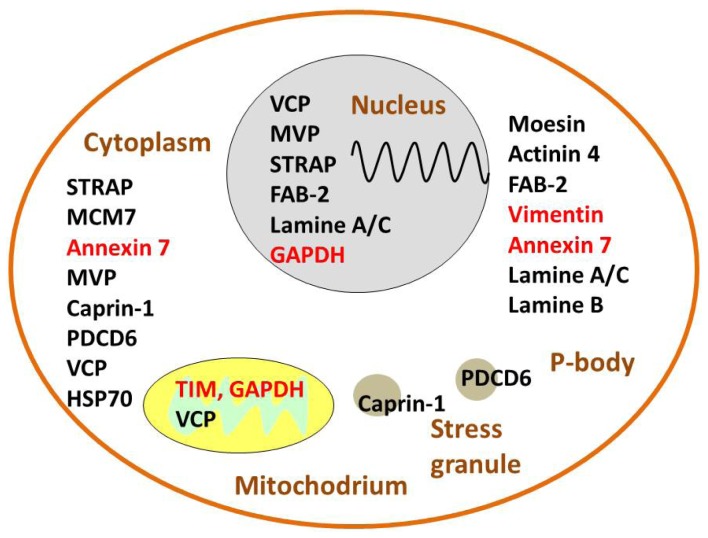
A closer look at the functions of proteins’ regulation after proton beam irradiation suggests that it is possible to classify them, simply for streamlining purposes, into four groups: i) DNA repair and stress, ii) proliferation and survival control, iii) metabolism and iv) connected to motility and the cytoskeleton (Fig. 4). This is an arbitrary classification, as some proteins have multiple functions and can play a role in more than one group.
Figure 4. A diagram showing all proteins regulated in proton-irradiated BLM cells.
The level of 17 proteins changed (>1.5×) in comparison with control. Thirteen proteins were upregulated (in black) and 4 were downregulated (in red). Here they are presented in four, not exclusive, groups: i) DNA repair and stress, ii) proliferation and survival control, iii) metabolic and iv) connected to motility and the cytoskeleton. ACTN 4 - α Actinin 4, Caprin-1 - Cytoplasmic activation/proliferation-associated protein-1, FAB-2 - Far upstream element binding protein 2, G3BP1 - RasGAP SH3-domain-binding protein 1, GADPH - Glyceraldehyde 3-phosphate dehydrogenase, MCM-7– Minichromosome Maintenance Protein 7, Moesin - Actin-regulatory protein, MVP - Major Vault Protein, PDCD6 - Programmed cell death 6, or apoptosis-linked gene-2, STRAP - Serine-threonine kinase receptor-associated protein, TIM - Triosephosphate isomerase, VCP – Transitional endoplasmic reticulum ATPase.
Allowing cells to undergo several passages in culture to repair proton irradiation damage before carrying out the proteomic analysis ensures that the effects seen are long-term. Even a low dose of 3 Gy results in proteomic changes in the next generations of cells.
Inducing DNA Repair and Stress Response
As expected after any kind of radiation treatment, proton beam irradiation upregulated protein levels engaged in DNA repair, such as MVP, Lamine A/C and Glyceraldehyde 3-phosphate dehydrogenase (GADPH). The last one participates in DNA repair via APE1 endonuclease [37]. What is more, the level of Transitional endoplasmic reticulum ATPase (VCP), known to be involved in chromatin associated degradation, was also increased. These changes are consistent with DNA repair after radiation.
The primary known function of MVP (Major Vault Protein) is multidrug resistance, but recent studies have demonstrated that it is also engaged in transport mechanisms, signal transmission, and the immune response. MVP plays a role in cytoplasmatic signal transduction cascades, and is involved in signal transduction through the nucleocytoplasmic transport of PTEN (nuclear phosphatase and tensin homologue deleted on chromosome 10). Nuclear PTEN plays a role in chromosome stability (reducing spontaneous double strand breaks), DNA repair, cell cycle arrest and cellular stability. MVP is engaged in the DNA repair mechanism (nonhomologous end joining) by interaction with Ku70/80. The level of MVP increased in response to various DNA damaging agents including irradiation [38]. MVP is recognized as a prognostic biomarker of radiotherapy resistance [39].
Lamine A/C A-type lamins control the transcription and degradation of proteins, play key roles in cell cycle regulation and DNA double-strand break repair. Importantly, the proteins regulated by A-type lamins– Rb family members, 53BP1 (Tumor suppressor p53-binding protein 1), BRCA1(breast cancer type 1 susceptibility protein) and RAD51– exert tumor suppressor functions, with their loss being associated with cancer susceptibility [40]. High levels of lamin A/C are followed by stimulation of cell growth, migration and invasion [41].
VCP (Transitional endoplasmic reticulum ATPase, or p97 or Cdc48) is highly abundant, accounting for about 1% of total cellular protein. Conserved hexameric p97 belongs to the type II AAA+ (ATPases associated with diverse cellular activities) family [42]. VCP is postulated to be an ubiquitin-selective chaperone, whose key function is to disassemble protein complexes. VCP is involved in a wide variety of cellular activities e.g., cell cycle, apoptosis, gene transcription, proteostasis, and DNA damage response autophagy and the regulation of chromatine. In response to UV, phosphorylated VCP modulates the function of RNA polymerase [43].
Several proteins related to mRNA regulation increased after proton beam irradiation. Caprin-1 (Cytoplasmic activation/proliferation-associated protein-1), involved in stress granules, and programmed cell death 6 (PDCD6) engaged in P-bodies were upregulated. Both constitutive P-bodies and stress-induced stress granules (SGs) are important for mRNA regulation upon stress, inhibiting protein synthesis to conserve energy for the repair of molecular damage. They contain mRNA and numerous proteins, and their content is highly dynamic. SGs also are known to modulate signaling balancing apoptosis and cell survival [44], [45]. Interestingly, our results show the upregulation of G3BP1 and/or HSP70 (Heat shock protein 70 kDa). Both these proteins are tightly connected to stress granules, as HSP70 is directly correlated with SG formation, and G3BP1 (RasGAP SH3-domain-binding protein 1) is a protein regulating mRNA stability and stress granule formation [44]. Furthermore, MVP and VCP are connected to transcription regulation, and Serine-threonine kinase receptor-associated protein (STRAP) and Far upstream element binding protein 2 (FAB-2) are associated with mRNA regulation.
Cytoplasmic activation/proliferation-associated protein-1 (Caprin-1) is a cytoplasmic phosphoprotein. Caprin-1 binds G3BP-1 (RasGAP SH3 domain binding protein-1), in cytoplasmic stress granules [46], formed in response to stress like heat, arsenite, unfolded protein response or toxins. G3BP-1 is marker for SGs, which stabilize specific mRNAs, control their translation and degradation and also control apoptosis by sequestering and nullifying apoptosis-promoting factors [47]. Overexpression of Caprin-1 induced the formation of cytoplasmic stress granules. Caprin-1 directly and selectively binds mRNA for c-Myc and cyclin D2 [46].
PDCD6 (programmed cell death protein 6, or apoptosis-linked gene-2, ALG-2)
PDCP6 interacts with PATL1, a component of the P-body, which is a cytoplasmic non-membranous granule composed of translation-inactive mRNAs and proteins involved in mRNA decay. PDCP6 co-localizes also with mRNA-decapping enzyme 1A, a marker of P-bodies [48]. PDCD6 in complex with Alix (ALG-2 (apoptosis-linked gene 2)-interacting protein X, known also as AIP1 or p95) induces calcium-dependent apoptosis and reduces tumorigenicity [49].
STRAP (Serine-threonine kinase receptor-associated protein) plays a role in mRNA splicing and cap-independent translation [50].
FAB-2/FBP-2 (Far upstream element binding protein 2 (FBP-2, KHSRP) belongs to the FBP family, engaged in transcription mechanisms i.e. splicing, mRNA stabilization, and degradation. FBPs are special transcription factors; they recognize the ssFUSE, single strand DNA cis element located upstream of the promoter of a target gene (e.g c-myc) and bind torsionally stressed pre-existing single strand regions and up-regulate gene expression. FBP2 can promote the decay of labile mRNA machinery or favor the maturation of a select group of microRNA precursors. FBP2 serves as a component of both Drosha and Dicer complexes and regulates the biogenesis of a subset of miRNAs [51], [52].
Proliferation and Survival Control
The largest group of proteins affected by proton beam irradiation in BLM cells is associated with the proliferation and pro-survival response, including STRAP, MCM-7, Annexin 7, MVP, Caprin-1, PDCD6, and VCP. However, it is not clear whether the overall effect is pro-survival or pro-apoptotic.
STRAP (Serine-threonine kinase receptor-associated protein), which regulates both transforming growth factor beta (TGF-β) and p53 signaling [53], is localized in both the cytoplasm and nucleus. On the one hand STRAP as regulator of Phosphoinositide-dependent kinase-1 and Apoptosis signal-regulating kinase 1, stimulates cell growth and can contribute to tumor progression by blocking TGF-β-mediated signaling, but on the other hand it potentiates p53-induced apoptosis through direct interaction with p53. These findings suggest that STRAP might have an ambivalent role in the regulation of cell growth [53]. However, the mechanism of STRAP regulation is not completely understood [50]. STRAP also physically interacts with B-MYB. B-myb is a member of the Myb family of transcription factors, which is ubiquitously expressed and involved in controlling cell proliferation and differentiation [53].
MCM-7 (Minichromosome Maintenance Protein 7) is a member of the group of protein complexes that are essential for DNA replication licensing and the control of cell cycle progression from the G1 to the S phase. MCM belongs to the family of AAA+ proteins. MCM7 overexpression and amplification occurs in several human malignancies [54], [55], although MCM7 takes part in both oncogenic and tumor suppressor signaling pathways. MCM7 binds Rb, p107, p130 proteins, which exert control over the entry into the S phase of DNA replication and cellular proliferation [56]. MCM7 also binds androgen receptor (AR) which regulates cell growth and proliferation. MCM7 serves as a co-transcriptional and co-translational enhancing factor of AR [57]. A unique feature of the MCM7 genome is that it contains an intronic cluster of miRNA in intron 13. These miRNAs shut down the expression of multiple tumor suppressor genes like p21, transcription factor E2F1, Bcl-2-like protein 11, PTEN [58]. PTEN deficiency causes Akt (Protein Kinase B, or PKB) hyperactivition and in consequence tumor initiation and progression. MCM7 overexpreession fosters tumorigenesis in combination with the action of miRNAs [59]. MCM7 is also involved in the activation of the ATR-dependent S-phase checkpoint by agents that induce DNA replication stress e.g. UV or X radiation [60].
Annexin A7 (ANXA7 or synexin) is a ubiquitously expressed member of the multifunctional Ca/phospholipid-binding annexin family. It shows Ca2+ dependent GTPase activity and is involved in membrane fusion and exocytosis. Annexin A7 is a tumor suppressor in human prostate and breast cancers. It overcomes pathologic androgen-receptor (AR) dependent proliferation via retinoblastoma protein (Rb1/p105) [61]. On the other hand Annexin A7 correlates with tumor malignancy and lymph node metastasis. High levels of Annexin A7 in a tumor correlate strongly with poor survival of patients. Repression of Annexin A7 results in increased levels of cell apoptosis, down-regulated cell proliferation, inhibited cell motility ability, and decreased cell invasive capacity [62].
Major vault protein, MVP
Recent studies have shown that MVP can cooperate with Insulin-like Growth Factor 1 (IGF-1R) in preventing apoptosis by upregulation of B-cell lymphoma 2 family of proteins (Bcl-2) and downregulation of Bax. Tumor progression and resistance to chemotherapy and radiotherapy may also be activated through the suppression of Bax and upregulation of IGF-1R, resulting in increased proliferation and reduced apoptosis caused by upregulation of Bcl-2 and overexpression of altered p53 [63]. It was also shown that MVP binds to COP1 (constitutively photomorphogenic 1 ubiquitin ligase). Mammalian COP1 causes prooncogene c-Jun proteasome-mediated degradation but COP1 can also act as a monomeric E3 ubiquitin ligase to directly ubiquitinate tumor suppressor p53. Under unstressed conditions MVP enters into direct physical contact with COP1 and suppress c-Jun-mediated transcription of activator protein 1 (AP-1) pathway. After UV irradiation, MVP becomes phosphorylated, releases COP1, and does not inhibit the AP-1 transcription anymore [64].
Cytoplasmic activation/proliferation-associated protein-1 (Caprin-1) level correlates with cellular proliferation [65], [66]. Suppression of the expression of human Caprin-1 resulted in a slowing of the proliferation rate, due to a prolongation of the G1 phase of the cell cycle [65]. The Caprin-1 gene is suppressed in response to the overexpression of the p53 gene, which suggests that this gene could be an important mediator of p53-dependent tumor growth suppression [66].
PDCD6 (programmed cell death 6, or apoptosis-linked gene-2, ALG-2) is a calcium-binding protein involved in cell proliferation and death. PDCD6 can induce calcium-dependent apoptosis and reduce tumorigenicity [67]. PDCD6 is ubiquitously localized in the cytoplasm; however, PDCD6 is translocated to the nucleus in response to DNA damage. PDCD6 is involved in the signaling cascade of the p53-responsive apoptotic machinery [68]. PDCD6 also plays a significant role in modulating cellular angiogenesis and it can inhibit tumor growth via the suppression of tumor angiogenesis [69]. There are conflicting reports concerning the level of PDCD6 in tumors. PDCD6 expression is up-regulated in lung cancer patients, while reduced PDCD6 expression is observed in gastric cancer, ovarian cancer tissues and cancer cell lines. Moreover, recent studies indicate that PDCD6 has a synergic pro-apoptotic effect with anti-cancer drugs through the activation of NF-κB pathway [70].
VCP (Transitional endoplasmic reticulum ATPase, or p97 or Cdc48) is a substrate for protein tyrosine phosphatase (PTPL1), which is important in cellular transformations [71]. PTPL1 can have either a pro-apoptotic effect (via the death receptor, Fas) or an anti-apoptotic impact (via Proto-oncogene tyrosine-protein kinase Src, Insulin-like growth factor 1 receptor, Human Epidermal Growth Factor Receptor 2 kinases) depending on the cellular context [43]. Elevated expression of VCP is observed in certain types of cancer tissues, e.g., colorectal carcinomas, pancreatic endocrine neoplasms, follicular thyroid cancer, hepatocellular and lung carcinoma [72], [73]. VCP increases the protection of cells against apoptotic stimuli through activation of the NFκB and Akt signaling pathways [74].
G3BP1 (RasGAP SH3-domain-binding protein 1) or HSP70 have been observed, although it is not clear which one gives the signal. G3BP1 is a protein regulating mRNA stability and stress granule formation and HSP70 is often induced in stress. Both proteins are connected to the cellular stress response. As mentioned above, Caprin-1 binds G3BP-1 stress granules [46].
Downregulation of Glycolysis
We observed the downregulation of two glycolytic enzymes as a result of proton beam irradiation, namely TIM and GADPH. TIM and GADPH are enzymes catalyzing two consecutive reactions of glycolysis, step 5 (conversion of dihydroxyacetone phosphate to glyceraldehyde 3-phosphate by TIM) and step 6 (simultaneous phosphorylation and oxidation of glyceraldehyde-3-phosphate to 1,3-biphosphoglycerate). GADPH is also involved in membrane transport, tubulin bundling and cytoskeletal dynamics, stem cell regulation and DNA repair. The downregulation of two consecutive steps in glycolysis might lead to a switch to mitochondrial oxidation for energy production and a less oncogenic fenotype [75].
TIM (Triosephosphate isomerase) is a glycolytic enzyme which catalyses the conversion of dihydroxyacetone phosphate to glyceraldehyde 3-phosphate. Different isoformes of the protein were found in melanoma [76]. In our study TIM was identified as a phosphorylated form (at serine 58). Earlier proteomic studies revealed TIM upregulation (1.6 fold change) in melanoma patients [77].
GADPH (Glyceraldehyde 3-phosphate dehydrogenase) is a glycolytic enzyme tightly regulated at both transcriptional and posttranslational levels. GAPDH specifically catalyzes the simultaneous phosphorylation and oxidation of glyceraldehyde-3-phosphate to 1,3-biphosphoglycerate. GAPDH is also involved in membrane transport, tubulin bundling and cytoskeletal dynamics. GAPDH interacts with tRNA and a transcription factor Oct-1 implicated in stress responses, metabolic control, and poised transcription states, regulates normal and pathological stem cell function. Many factors such as calcium, insulin or hypoxia up-regulate GAPDH expression. The level of GAPDH is elevated in many cancers [78].
Paradoxical Changes in Motility and Cytoskeleton
In metastatic melanoma cells, many proteins connected to cell motility, adhesion and migration are upregulated. For example, matrix metalloproteinase-2 and integrin αvβ3 are implicated in melanoma metastases and progression [79]. The BLM cell line, used in this study, is a metastatic melanoma, widely studied for its high invasive capabilities [28], [79], [80]. For example, the chemokine receptors CXCR3 and CXCR4 were shown to be expressed in the BLM cell line [80].
Proton ion irradiation decreased cell migration and invasion in a dose-dependent manner, strongly inhibited matrix metaloproteinase-2 activity in highly aggressive HT1080 human fibrosarcoma cells in vitro, and significantly decreased pulmonary metastasis in mouse osteosarcoma in vivo [81].
In the present work, several proteins known to influence cellular motility were affected after proton beam irradiation. Most distinctly, Vimentin was strongly downregulated, along with the downregulation of Annexin 7, whereas Moesin, Lamine A/C, Lamine B and α Actinin 4 were upregulated.
Moesin (Actin-regulatory protein moesin) belongs to the ezrin/radixin/moesin (ERM) family which provides a regulated linkage between the plasma membrane and the actin cytoskeleton. Moesin plays an important role in cell motility by connecting the actin cytoskeleton to a variety of membrane anchoring proteins. Moesin is an essential mediator for actin cytoskeleton remodeling [82]. Moesin controls the adhesion dependent activation of Rho and subsequent myosin II contractility during 3D collagen invasion by melanoma cells [83]. Increased moesin expression also promotes the epithelial–mesenchymal transition (EMT) by regulating adhesion and contractile elements for changes in actin filament organization [84].
ACTN4 (α Actinin 4) is an actin-binding protein associated with cell motility and metastasis in breast, colorectal, pancreatic, hepatocellular carcinoma, and ovarian cancers. ACTN4 crosslinks F-actin filaments into bundles to form filopodia. Overexpression of ACTN4 increases the migratory potential of cells [85].
FAB-2/FBP-2 (Far upstream element binding protein 2 (FBP-2, KHSRP)
FBPs are co-regulated with the microtubule destabilizer, cytosolic phosphoprotein stathmin (known as oncoprotein 18). FBP-2 primarily supports migration in different hepatocellular carcinoma cells [86].
Lamins A/C and B type are the main constituents of a polymeric network known as a nuclear lamina which plays a key role in the maintenance of genome localization, structure and function. Overexpression of Lamin A/C results in the stimulation of cell growth, migration and invasion. This oncogenic behaviour of lamin A/C is regulated through modulation of the PI3K/AKT/PTEN signaling pathway [41]. Lamin A regulates actin dynamics, and its overexpression leads to a loss of cell adhesion. This in turn increases cell motility and consequently increases the invasive potential of the tumor [87]. A-type lamins also influence the activity of oncogene β-catenin (via the interaction of the membrane protein emerin with the Wnt/β-catenin pathway) which also points to a possible role of A-type lamins in tumor progression [88]. In turn, B lamin deficiency is involved in the development of prostate cancer. B-deficient microdomains correlate with prostate cancer cell line aggressiveness and increased cell motility [89].
Annexin A7 (ANXA7 or synexin)
Repression of Annexin A7 results in increased levels of cell apoptosis, down-regulated cell proliferation, inhibited cell motility ability, and decreased cell invasive capacity [62].
Vimentin is one of the most widely expressed mammalian intermediate filament proteins. It is a multifunctional protein, regulating several different physiological functions, such as the structural integrity of cells and tissues, adhesion and migration, signal transduction, apoptotic and immune defense and the regulation of genomic DNA [90]. In the majority of cancers vimentin is overexpressed and associated with a metastatic phenotype and a poor prognosis for the disease outcome. Vimentin is a marker, as well as a key factor in regulating EMT, a critical event in the induction of cell motility [91]. Recent studies have revealed that vimentin is not only a diagnostic marker but also a hematogenous metastasis clinical predictor for melanoma [92].
A decrease in vimentin may lead to diminished metastatic potential. For example, in B16 melanoma cells treated with cyclohexamide a decrease in vimentin mRNa was seen, together with a decrease in the formation of lung metastastes in mice. What is more, this effect was reversible [93]. Similarly, withaferin A caused the dose-dependent inhibition of lung metastases in a breast cancer model [94].
Post-translation modifications of vimentin, specifically phosphorylation, are especially important for vimentin functioning. Vimentin contains a highly complex phosphorylation pattern involving a multitude of kinase specific sites which are recognized by many kinases, including Rho kinase, protein kinase C (PKC), cGMP kinase, Yes kinase, Raf-1 kinase, PAK kinase, and Aurora B kinase. Phosphorylation enhances the disassembly of vimentin into nonfilamentous (monomeric, dimeric, and tetrameric) particles, shifting the equilibrium between polymeric and depolymerized vimentin. Dephosphorylated vimentin exerts its influence on motility, adhesion, cell signaling, and cell survival. Similarly, phosphorylated, disassembled vimentin has been demonstrated to enhance the recycling of integrins subjected to endocytosis to the plasma membrane during cell migration [90]. The results of a proteomic 2DE experiment on melanoma cells confirm the multiisofomic pattern of vimentin [76]. It was attempted in the present study to confirm the changes in the level of vimetin by means of Western Blot analysis (data not shown), although the results were not statistically significant. The antibody applied (Vimentin (R28) Antibody #3932, Cell Signaling Technology) recognizes a synthetic peptide of vimetin which includes arginine at position 28. This residue is surrounded by serine and threonine amino acids, which are very often modified (phosphorylated). Because of the multiplicity of possible vimentin post-translational modifications, which may alter the affinity of the antibody for a protein, the Western Blot analysis is hampered especially in terms of quantitative analysis.
A decrease in vimentin level, as well as in Annexin 7, might suggest cell motility impairment, and decreased cell invasive capacity [62]. However, other proteins involved in motility and the cytoskeleton were upregulated. It is difficult, therefore, to assess the overall effect on cellular motility and migration. These proteins may change as the response of highly motile cells to stress, as part of their prosurvival response.
Signalling Pathways Affected by Proton Beam Irradiation
Disturbances in several signaling pathways, such as MAPK-ERK, RAS, AKT/PI3, G1/S Cyclin/Cyclin-dependent kinases, p53, B-RAF are characteristic of melanoma progression [95], [96]. These pathways are connected with the modulation of proliferation, survival, the cell cycle, and cell differentiation. It is not surprising that some of the proteins upregulated after proton beam irradiation are also involved in these signaling pathways, critical for survival and proliferation (Table 2).
Table 2. Signaling pathways involving proteins upregulated after sublethal proton beam irradiation.
| Pathway | Regulated proteins (reference) |
| MAPK | MVP (Valenciano et al., 2012) [63] |
| AKT/PI3 | MVP (Blanco-Aparicio et al., 2007) [103] |
| PDCD6 (Rho et al., 2012) [69] | |
| VCP (Braun and Zischka, 2008) [74] | |
| Lamin A/C (Kong et al., 2012) [41] | |
| PTEN | MVP (Lara et al., 2011) [104] |
| MCM-7 (Luo et al., 2011) [58], (Poliseno et al., 2010) [59] | |
| Lamin A/C (Kong et al., 2012) [41] | |
| p-53 | MVP (Lloret et al., 2009) [105] |
| STRAP (Seong et al., 2007) [106] | |
| PDCD6 (Suzuki et al., 2012) [68] | |
| Caprin-1 (Saffari et al., 2009) [66] | |
| Cell cycle | MCM-7, G1/S phase (Mukherjee et al., 2009) [56] |
| Caprin-1, G1/S phase (Wang et al., 2005) [65] | |
| Caprin-1, cyclin D2 (Solomon S. et al., 2007) [46] | |
| TGF-β | STRAP (Seong et al., 2007) [106] |
| nFκB | VCP (Braun and Zischka, 2008) [74] |
| PDCD6 (Park et al., 2012) [70] |
TGFβ acts as a tumor-derived immunosuppressor, an inducer of tumor mitogens, a promoter of carcinoma invasion, and a trigger of prometastatic cytokine secretion [97]. Photon radiation causes the upregulation of TGFβ secretion into the microenvironment and can induce fibro- sis by promoting the excessive synthesis of matrix components [98]. STRAP inhibits TGF-β signalling by stabilizing the association between TGF-β receptors and Smad7 or Smad3 (proteins that transduce extracellular signals from transforming growth factor beta ligands to the nucleus) [53], [99], [100]. The interaction of MCM7 with Rb is essential for transforming the growth factor (TGF)-β-induced blockade of entry into the S phase [56].
The relatively low dose of 3 Gy used in our studies, the time allowed for cell repair after irradiation might be the reasons we did not see the induction of signaling pathways described by others after proton irradiation, such as the direct apoptosis induction pathways p38/JNK [26].
Comparison with other Melanoma Studies
Proteomic analysis of uveal melanoma from patients with distant metastases has shown increased expression of vimentin and TIM, among other proteins [77], in comparison to patients without metastases. In metastases of uveal melanomas, several proteins were found to be upregulated: Colony stimulating factor 2, Ficolin precursor, Haptoglobin -2 precursor, Hemopexin precursor, α-1-antitrypsin precursor, α-1-antichymotrypsin precursor, Vitronectin precursor, and Down syndrome cell adhesion molecule [101]. However, none of them were found to be modulated in our experiments. Proteomic analysis of melanoma metastases in comparison to primary tumors has shown upregulation of several proteins, implicated as metastasis-related proteins, such as lactate dehydrogenase, heat shock protein 90 KDa, glucose transporter 1, Macrophage migration inhibitory factor, Protein DJ-1, pyruvate kinase isozyme 2 and prohibitin-2 [102]. Again, none of these proteins were modulated in our study. Only GADPH, elevated in metastasis, was downregulated after proton beam irradiation in our cell line. Another comparative proteome analysis of two genetically, very closely related, melanoma cell lines with low- and high-metastatic potentials identified 42 proteins with increased levels in highly metastatic melanoma cells, while 68 showed decreased levels [98]. Four of the proteins with increased levels in highly metastatic melanoma cells were also seen in our study: Vimentin and TIM levels decreased after proton beam treatment, and STRAP and Actinin 4 increased.
Future work should cover other cancer cell lines, such as uveal melanoma or prostate cancer, both of which respond well to proton beam therapy. Perhaps proteomic analysis should also be performed a shorter time after irradiation.
Conclusions
Proteomic analysis of the BLM melanoma cell line irradiated with a low dose of 3 Gy of proton beam shows a significant (more than 1.5×change) upregulation of 13 proteins and downregulation of 4 proteins. These proteins might be roughly grouped into four categories by function: (i) DNA repair and RNA regulation (VCP, MVP, STRAP, FAB-2, Lamine A/C, GAPDH), (ii) cell survival and stress response (STRAP, MCM7, Annexin 7, MVP, Caprin-1, PDCD6, VCP, HSP70), (iii) cell metabolism (TIM, GAPDH, VCP), and (iv) cytoskeleton and motility (Moesin, Actinin 4, FAB-2, Vimentin, Annexin 7, Lamine A/C, Lamine B). Of particular interest is the substantial decrease (2.3×) in vimentin, a marker of EMT and of the metastatic properties of melanoma [92].
Funding Statement
The MS measurements were performed using the spectrometer MicroTOF purchased by the European Regional Development Fund in the framework of the Polish Innovation Economy Operational Program (contract No. POIG.02.01.00-12-167/08, project Małopolska Center of Biotechnology, http://www.poig.gov.pl/english/Strony/Introduction.aspx), supported by grant Nr 2023/B/P01/2010/39 (K/PBW/000676) from the Polish Ministry of Science and Higher Education (http://www.nauka.gov.pl/en/), and by grants UMO-2012/05/B/NZ4/02428 (K/PBO/000134) and 012/07/B/NZ4/01657 from the National Science Center, Krakow, Poland (http://www.ncn.gov.pl/?language=en). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Girdhani S, Sachs R, Hlatky L (2013) Biological Effects of Proton Radiation: What We Know and Don’t Know. Radiat Res 179: 1–16. [DOI] [PubMed] [Google Scholar]
- 2. Fokas E, Kraft G, An H, Engenhart-Cabillic R (2009) Ion beam radiobiology and cancer: time to update ourselves. Biochim Biophys Acta 1796: 216–229. [DOI] [PubMed] [Google Scholar]
- 3. Ristić-Fira AM, Petrović IM, Korićanac LB, Valastro LM, Privitera G, et al. (2008) Assessment of the inhibitory effects of different radiation qualities or chemotherapeutic agents on a human melanoma cell line. Phys medica 24: 187–195. [DOI] [PubMed] [Google Scholar]
- 4. Petrović IM, Korićanac LB, Todorović DV, Ristić-Fira AM, Valastro LM, et al. (2007) Viability of a human melanoma cell after single and combined treatment with fotemustine, dacarbazine, and proton irradiation. Ann N Y Acad Sci 1095: 154–164. [DOI] [PubMed] [Google Scholar]
- 5. Ristić-fira AM, Korićanac LB, Žakula JJ, Valastro LM, Iannolo G, et al. (2009) Effects of fotemustine or dacarbasine on a melanoma cell line pretreated with therapeutic proton irradiation. J Exp Clin Cancer Res 28: 50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Korićanac LB, Zakula JJ, Petrović IM, Valastro LM, Cirrone G a P, et al. (2010) Anti-tumour activity of fotemustine and protons in combination with bevacizumab. Chemotherapy 56: 214–222. [DOI] [PubMed] [Google Scholar]
- 7. Petrović I, Ristić-Fira A, Todorović D, Korićanac L, Valastro L, et al. (2010) Response of a radioresistant human melanoma cell line along the proton spread-out Bragg peak. Int J Radiat Biol 86: 742–751. [DOI] [PubMed] [Google Scholar]
- 8. Ristic-Fira A, Nikolic D, Petrovic I (2001) The late effects of proton irradiation on cell growth, cell cycle arrest and apoptosis in a human melanoma cell line. J Exp Clin Cancer Res 20: 135–143. [PubMed] [Google Scholar]
- 9. Ristic-Fira A, Petrovic I, Todorovic D, Koricanac L (2004) Inactivation of HTB63 human melanoma cells by irradiation with protons and gamma rays. Oncol Rep 12: 1323–1328. [PubMed] [Google Scholar]
- 10. Todorović D, Petrović I, Todorović M, Cuttone G, Ristić-Fira a (2008) Early effects of gamma rays and protons on human melanoma cell viability and morphology. J Microsc 232: 517–521. [DOI] [PubMed] [Google Scholar]
- 11. Ristic-Fira AM, Todorovic D V, Koricanac LB, Petrovic IM, Valastro LM, et al. (2007) Response of a human melanoma cell line to low and high ionizing radiation. Ann N Y Acad Sci 1095: 165–174. [DOI] [PubMed] [Google Scholar]
- 12. Petrovic I, Ristic-Fira A, Todorovic D, Valastro L, Cirrone P, et al. (2006) Radiobiological analysis of human melanoma cells on the 62 MeV CATANA proton beam. Int J Radiat Biol 82: 251–265. [DOI] [PubMed] [Google Scholar]
- 13. Korićanac L, Petrović I, Privitera G, Cuttone G, Ristić-Fira a (2007) HTB140 melanoma cells under proton irradiation and/or alkylating agents. Russ J Phys Chem A 81: 1467–1470. [Google Scholar]
- 14. Di Pietro C, Piro S, Tabbì G, Ragusa M, Di Pietro V, et al. (2006) Cellular and molecular effects of protons: Apoptosis induction and potential implications for cancer therapy. Apoptosis 11: 57–66. [DOI] [PubMed] [Google Scholar]
- 15. Ianzini F, Cherubini R, Mackey MA (1999) Mitotic catastrophe induced by exposure of V79 Chinese hamster cells to low-energy protons. Int J Radiat Biol 75: 717–723. [DOI] [PubMed] [Google Scholar]
- 16. Lee KB, Lee JS, Park JW, Huh TL, Lee YM (2008) Low energy proton beam induces tumor cell apoptosis through reactive oxygen species and activation of caspases. Exp Mol Med 40: 118–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee KB, Kim K-R, Huh T, Lee YM (2008) Proton induces apoptosis of hypoxic tumor cells by the p53-dependent and p38/JNK MAPK signaling pathways. Int J Oncol 33: 1247–1256. [PubMed] [Google Scholar]
- 18. Gridley DS, Freeman TL, Makinde AY, Wroe AJ, Luo-Owen X, et al. (2011) Comparison of proton and electron radiation effects on biological responses in liver, spleen and blood. Int J Radiat Biol 87: 1173–1181. [DOI] [PubMed] [Google Scholar]
- 19. Sgura A, Antoccia A, Cherubini R, Dalla Vecchia M, Tiveron P, et al. (2000) Micronuclei, CREST-positive micronuclei and cell inactivation induced in Chinese hamster cells by radiation with different quality. Int J Radiat Biol 76: 367–374. [DOI] [PubMed] [Google Scholar]
- 20. Ghosh S, Bhat NN, Santra S, Thomas RG, Gupta SK, et al. (2010) Low energy proton beam induces efficient cell killing in A549 lung adenocarcinoma cells. Cancer Invest 28: 615–622. [DOI] [PubMed] [Google Scholar]
- 21. Calugaru V, Nauraye C, Noël G, Giocanti N, Favaudon V, et al. (2011) Radiobiological characterization of two therapeutic proton beams with different initial energy spectra used at the Institut Curie Proton Therapy Center in Orsay. Int J Radiat Oncol Biol Phys 81: 1136–1143. [DOI] [PubMed] [Google Scholar]
- 22. Leloup C, Garty G, Assaf G, Cristovão A, Breskin A, et al. (2005) Evaluation of lesion clustering in irradiated plasmid DNA. Int J Radiat Biol 81: 41–54. [DOI] [PubMed] [Google Scholar]
- 23. Hada M, Sutherland BMB (2006) Spectrum of complex DNA damages depends on the incident radiation. Radiat Res 165: 223–230. [DOI] [PubMed] [Google Scholar]
- 24. Ibañez IL, Bracalente C, Molinari BL, Palmieri MA, Policastro L, et al. (2009) Induction and rejoining of DNA double strand breaks assessed by H2AX phosphorylation in melanoma cells irradiated with proton and lithium beams. Int J Radiat Oncol Biol Phys 74: 1226–1235. [DOI] [PubMed] [Google Scholar]
- 25. Leatherbarrow EL, Harper J V, Cucinotta FA, O’Neill P (2006) Induction and quantification of gamma-H2AX foci following low and high LET-irradiation. Int J Radiat Biol 82: 111–118. [DOI] [PubMed] [Google Scholar]
- 26. Finnberg N, Wambi C (2008) Gamma-radiation (GR) triggers a unique gene expression profile associated with cell death compared to proton radiation (PR) in mice in vivo. Cancer Biol Ther 7: 2023–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lockshin A, Giovanella B, Ipolyi P De (1985) Exceptional lethality for nude mice of cells derived from a primary human melanoma. Cancer Res 45: 345–350. [PubMed] [Google Scholar]
- 28. Van Muijen GN, Cornelissen LM, Jansen CF, Figdor CG, Johnson JP, et al. (1991) Antigen expression of metastasizing and non-metastasizing human melanoma cells xenografted into nude mice. Clin Exp Metastasis 9: 259–272. [DOI] [PubMed] [Google Scholar]
- 29. Tice RR, Agurell E, Anderson D, Burlinson B, Hartmann A, et al. (2000) Single cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testing. Environ Mol Mutagen 35: 206–221. [DOI] [PubMed] [Google Scholar]
- 30. Singh NP, Stephens RE, Schneider EL (1994) Modifications of alkaline microgel electrophoresis for sensitive detection of DNA damage. Int J Radiat Biol 66: 23–28. [DOI] [PubMed] [Google Scholar]
- 31. Wessel D, Flügge UI (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 138: 141–143. [DOI] [PubMed] [Google Scholar]
- 32. Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254. [DOI] [PubMed] [Google Scholar]
- 33. Sprung R, Chen Y, Zhang K (2008) Identification and validation of eukaryotic aspartate and glutamate methylation in proteins. J Proteome Res 7: 1001–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wloga D, Gaertig J (2010) Post-translational modifications of microtubules. J Cell Sci 124: 154–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Azzam EI, Jay-Gerin J-P, Pain D (2012) Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer Lett 327: 48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marchetti F, Coleman MA, Jones IM, Wyrobek AJ (2006) Candidate protein biodosimeters of human exposure to ionizing radiation. Int J Radiat Biol 82: 605–639. [DOI] [PubMed] [Google Scholar]
- 37. Colell A, Green DR, Ricci J-E (2009) Novel roles for GAPDH in cell death and carcinogenesis. Cell Death Differ 16: 1573–1581. [DOI] [PubMed] [Google Scholar]
- 38. Shimamoto Y, Sumizawa T, Haraguchi M, Gotanda T, Jueng H-C, et al. (2006) Direct activation of the human major vault protein gene by DNA-damaging agents. Oncol Rep 15: 645–652. [PubMed] [Google Scholar]
- 39. Silva P, West CM, Slevin N, Valentine H, Ryder WDJ, et al. (2007) Tumor expression of major vault protein is an adverse prognostic factor for radiotherapy outcome in oropharyngeal carcinoma. Int J Radiat Oncol Biol Phys 69: 133–140. [DOI] [PubMed] [Google Scholar]
- 40. Redwood AB, Gonzalez-Suarez I, Gonzalo S (2011) Regulating the levels of key factors in cell cycle and DNA repair: new pathways revealed by lamins. Cell Cycle 10: 3652–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kong L, Schäfer G, Bu H, Zhang YY, Klocker H (2012) Lamin A/C protein is overexpressed in tissue-invading prostate cancer and promotes prostate cancer cell growth, migration and invasion through the PI3K/AKT/PTEN pathway. Carcinogenesis 33: 751–759. [DOI] [PubMed] [Google Scholar]
- 42. Yamanaka K, Sasagawa Y, Ogura T (2012) Recent advances in p97/VCP/Cdc48 cellular functions. Biochim Biophys Acta 1823: 130–137. [DOI] [PubMed] [Google Scholar]
- 43. Meyer H (2012) P97 Complexes As Signal Integration Hubs. BMC Biol 10: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thomas M, Loschi M, Desbats M, Boccaccio G (2011) RNA granules: the good, the bad and the ugly. Cell Signal 23: 324–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Anderson P, Kedersha N (2008) Stress granules: the Tao of RNA triage. Trends Biochem Sci 33: 141–150. [DOI] [PubMed] [Google Scholar]
- 46. Solomon S, Xu Y, Wang B, David MD, Schubert P, et al. (2007) Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2alpha, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol Cell Biol 27: 2324–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Buchan JR, Parker R (2009) Eukaryotic stress granules: the ins and outs of translation. Mol Cell 36: 932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Osugi K, Suzuki H, Nomura T, Ariumi Y, Shibata H, et al. (2012) Identification of the P-body component PATL1 as a novel ALG-2-interacting protein by in silico and far-Western screening of proline-rich proteins. J Biochem 151: 657–666. [DOI] [PubMed] [Google Scholar]
- 49. Wu Y, Pan S, Luo W, Lin S-H, Kuang J (2002) Hp95 promotes anoikis and inhibits tumorigenicity of HeLa cells. Oncogene 21: 6801–6808. [DOI] [PubMed] [Google Scholar]
- 50. Reiner JE, Datta PK (2011) TGF-beta-dependent and -independent roles of STRAP in cancer. Front Biosci (Landmark Ed 16: 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, et al. (2009) The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 459: 1010–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cukier CD, Hollingworth D, Martin SR, Kelly G, Díaz-Moreno I, et al. (2010) Molecular basis of FIR-mediated c-myc transcriptional control. Nat Struct Mol Biol 17: 1058–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Seong H-A, Manoharan R, Ha H (2011) B-MYB positively regulates serine-threonine kinase receptor-associated protein (STRAP) activity through direct interaction. J Biol Chem 286: 7439–7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Toyokawa G, Masuda K, Daigo Y, Cho H-S, Yoshimatsu M, et al. (2011) Minichromosome Maintenance Protein 7 is a potential therapeutic target in human cancer and a novel prognostic marker of non-small cell lung cancer. Mol Cancer 10: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhou Y-M, Zhang X-F, Cao L, Li B, Sui C-J, et al. (2012) MCM7 expression predicts post-operative prognosis for hepatocellular carcinoma. Liver Int 32: 1505–1509. [DOI] [PubMed] [Google Scholar]
- 56. Mukherjee P, Cao T V, Winter SL, Alexandrow MG (2009) Mammalian MCM loading in late-G(1) coincides with Rb hyperphosphorylation and the transition to post-transcriptional control of progression into S-phase. PLoS One 4: e5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shi Y-K, Yu YP, Zhu Z-H, Han Y-C, Ren B, et al. (2008) MCM7 interacts with androgen receptor. Am J Pathol 173: 1758–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Luo J-H (2011) Oncogenic activity of MCM7 transforming cluster. World J Clin Oncol 2: 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, et al. (2010) Identification of the miR-106b∼25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal 3: ra29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tsao C-C, Geisen C, Abraham RT (2004) Interaction between human MCM7 and Rad17 proteins is required for replication checkpoint signaling. EMBO J 23: 4660–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Torosyan Y, Simakova O, Naga S, Mezhevaya K, Leighton X, et al. (2009) Annexin-A7 protects normal prostate cells and induces distinct patterns of RB-associated cytotoxicity in androgen-sensitive and -resistant prostate cancer cells. Int J Cancer 125: 2528–2539. [DOI] [PubMed] [Google Scholar]
- 62. Sun M-Z, Liu S, Tang J, Wang Z, Gong X, et al. (2009) Proteomics analysis of two mice hepatocarcinoma ascites syngeneic cell lines with high and low lymph node metastasis rates provide potential protein markers for tumor malignancy attributes to lymphatic metastasis. Proteomics 9: 3285–3302. [DOI] [PubMed] [Google Scholar]
- 63. Valenciano A, Moreno M, Lloret M, Lara PC (2012) Role of IGF-1 receptor in radiation response. Transl Oncol 5: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yi C, Li S, Chen X, Wiemer EA, Wang J, et al. (2005) Major vault protein, in concert with constitutively photomorphogenic 1, negatively regulates c-Jun-mediated activator protein 1 transcription in mammalian cells. Cancer Res 65: 5835–5840. [DOI] [PubMed] [Google Scholar]
- 65. Wang B, David M, Schrader J (2005) Absence of caprin-1 results in defects in cellular proliferation. J Immunol 175: 4274–4282. [DOI] [PubMed] [Google Scholar]
- 66. Saffari M, Dinehkabodi OS, Ghaffari SH, Modarressi MH, Mansouri F, et al. (2009) Identification of novel p53 target genes by cDNA AFLP in glioblastoma cells. Cancer Lett 273: 316–322. [DOI] [PubMed] [Google Scholar]
- 67. Maki M, Suzuki H, Shibata H (2011) Structure and function of ALG-2, a penta-EF-hand calcium-dependent adaptor protein. Sci China Life Sci 54: 770–779. [DOI] [PubMed] [Google Scholar]
- 68. Suzuki K, Dashzeveg N, Lu Z-G, Taira N, Miki Y, et al. (2012) Programmed cell death 6, a novel p53-responsive gene, targets to the nucleus in the apoptotic response to DNA damage. Cancer Sci 103: 1788–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rho SB, Song YJ, Lim MC, Lee S-H, Kim B-R, et al. (2011) Programmed cell death 6 (PDCD6) inhibits angiogenesis through PI3K/mTOR/p70S6K pathway by interacting of VEGFR-2. Cell Signal 24: 131–139. [DOI] [PubMed] [Google Scholar]
- 70. Park SH, Lee JH, Lee G-B, Byun H-J, Kim B-R, et al. (2012) PDCD6 additively cooperates with anti-cancer drugs through activation of NF-κB pathways. Cell Signal 24: 726–733. [DOI] [PubMed] [Google Scholar]
- 71. Abaan OD, Hendriks W, Uren A, Toretsky JA, Erkizan H V (2013) Valosin containing protein (VCP/p97) is a novel substrate for the protein tyrosine phosphatase PTPL1. Exp Cell Res 319: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Liu Y, Hei Y, Shu Q, Dong J, Gao Y, et al. (2012) VCP/p97, down-regulated by microRNA-129–5p, could regulate the progression of hepatocellular carcinoma. PLoS One 7: e35800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Valle CW, Min T, Bodas M, Mazur S, Begum S, et al. (2011) Critical role of VCP/p97 in the pathogenesis and progression of non-small cell lung carcinoma. PLoS One 6: e29073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Braun RJ, Zischka H (2008) Mechanisms of Cdc48/VCP-mediated cell death: from yeast apoptosis to human disease. Biochim Biophys Acta 1783: 1418–1435. [DOI] [PubMed] [Google Scholar]
- 75. Berardi MJ, Fantin VR (2011) Survival of the fittest: metabolic adaptations in cancer. Curr Opin Genet Dev 21: 59–66. [DOI] [PubMed] [Google Scholar]
- 76. Pardo M, García A, Thomas B, Piñeiro A, Akoulitchev A, et al. (2005) Proteome analysis of a human uveal melanoma primary cell culture by 2-DE and MS. Proteomics 5: 4980–4993. [DOI] [PubMed] [Google Scholar]
- 77. Linge A, Kennedy S, O’Flynn D, Beatty S, Moriarty P, et al. (2012) Differential expression of fourteen proteins between uveal melanoma from patients who subsequently developed distant metastases versus those who did Not. Invest Ophthalmol Vis Sci 53: 4634–4643. [DOI] [PubMed] [Google Scholar]
- 78. Formentini L, Martínez-Reyes I, Cuezva J (2010) The mitochondrial bioenergetic capacity of carcinomas. IUBMB Life 62: 554–560. [DOI] [PubMed] [Google Scholar]
- 79. Hofmann UB, Westphal JR, Waas ET, Becker ÈC, Ruiter DJ, et al. (2000) Coexpression of Integrin alphabeta and matrix metalloproteinase-2 (MMP-2) coincides with mmp-2 activation: correlation with melanoma progression. J Invest Dermatol 2: 0–7. [DOI] [PubMed] [Google Scholar]
- 80. Robledo MM, Bartolome RA, Longo N, Rodríguez-Frade JM, Mellado M, et al. (2001) Expression of functional chemokine receptors CXCR3 and CXCR4 on human melanoma cells. J Biol Chem 276: 45098–45105. [DOI] [PubMed] [Google Scholar]
- 81. Ogata T, Teshima T, Kagawa K, Hishikawa Y, Takahashi Y, et al. (2005) Particle irradiation suppresses metastatic potential of cancer cells. Cancer Res 65: 113. [PubMed] [Google Scholar]
- 82. He M, Cheng Y, Li W, Liu Q, Liu J, et al. (2010) Vascular endothelial growth factor C promotes cervical cancer metastasis via up-regulation and activation of RhoA/ROCK-2/moesin cascade. BMC Cancer 10: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Estecha A, Sánchez-Martín L, Puig-Kröger A, Bartolomé RA, Teixidó J, et al. (2009) Moesin orchestrates cortical polarity of melanoma tumour cells to initiate 3D invasion. J Cell Sci 122: 3492–3501. [DOI] [PubMed] [Google Scholar]
- 84. Haynes J, Srivastava J, Madson N, Wittmann T, Barber DL (2011) Dynamic actin remodeling during epithelial-mesenchymal transition depends on increased moesin expression. Mol Biol Cell 22: 4750–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sen S, Dong M, Kumar S (2009) Isoform-specific contributions of alpha-actinin to glioma cell mechanobiology. PLoS One 4: e8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Malz M, Weber A, Singer S, Riehmer V, Bissinger M, et al. (2009) Overexpression of far upstream element binding proteins: a mechanism regulating proliferation and migration in liver cancer cells. Hepatology 50: 1130–1139. [DOI] [PubMed] [Google Scholar]
- 87. Willis ND, Cox TR, Rahman-Casañs SF, Smits K, Przyborski S a, et al. (2008) Lamin A/C is a risk biomarker in colorectal cancer. PLoS One 3: e2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Markiewicz E, Tilgner K, Barker N, van de Wetering M, Clevers H, et al. (2006) The inner nuclear membrane protein emerin regulates beta-catenin activity by restricting its accumulation in the nucleus. EMBO J 25: 3275–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Helfand BT, Wang Y, Pfleghaar K, Shimi T, Taimen P, et al. (2012) Chromosomal regions associated with prostate cancer risk localize to lamin B-deficient microdomains and exhibit reduced gene transcription. J Pathol 226: 735–745. [DOI] [PubMed] [Google Scholar]
- 90. Ivaska J, Pallari H-M, Nevo J, Eriksson JE (2007) Novel functions of vimentin in cell adhesion, migration, and signaling. Exp Cell Res 313: 2050–2062. [DOI] [PubMed] [Google Scholar]
- 91. Ivaska J (2011) Vimentin: Central hub in EMT induction? Small GTPases 2: 51–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Li M, Zhang B, Sun B, Wang X, Ban X, et al. (2010) A novel function for vimentin: the potential biomarker for predicting melanoma hematogenous metastasis. J Exp Clin Cancer Res 29: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ben-Ze’ev A, Raz A (1985) Relationship between the organization and synthesis of vimentin and the metastatic capability of B16 melanoma cells. Cancer Res 45: 2632–2641. [PubMed] [Google Scholar]
- 94. Thaiparambil JT, Bender L, Ganesh T, Kline E, Patel P, et al. (2011) Withaferin A inhibits breast cancer invasion and metastasis at sub-cytotoxic doses by inducing vimentin disassembly and serine 56 phosphorylation. Int J cancer 129: 2744–2755. [DOI] [PubMed] [Google Scholar]
- 95. Fernandez-Flores A (2012) Prognostic factors for melanoma progression and metastasis: from Hematoxylin-Eosin to genetics. Rom J Morphol Embryol 53: 449–459. [PubMed] [Google Scholar]
- 96. Rodríguez-Cerdeira C, Molares-Vila A (2011) New Perspectives of “omics” Applications in Melanoma Research. Open Biochem J 5: 60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Massagué J (2008) TGF-beta in cancer. Cell 134: 215–230.18662538 [Google Scholar]
- 98.Atkinson MJ (2013) Radiation Treatment Effects on the Proteome of the Tumour Microenvironment. In: Leszczynski D, editor. Radiation Proteomics. The effects of ionizing and non-ionizing radiation on cells and tissues. Dordrecht Heidelberg New York London. 49–60.
- 99. Anumanthan G, Halder SK, Friedman DB, Datta PK (2006) Oncogenic serine-threonine kinase receptor-associated protein modulates the function of Ewing sarcoma protein through a novel mechanism. Cancer Res 66: 10824–10832. [DOI] [PubMed] [Google Scholar]
- 100. Kim W, Seok Kang Y, Soo Kim J, Shin N-Y, Hanks SK, et al. (2008) The integrin-coupled signaling adaptor p130Cas suppresses Smad3 function in transforming growth factor-beta signaling. Mol Biol Cell 19: 2135–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Meir T, Dror R, Yu X, Qian J, Simon I, et al. (2007) Molecular characteristics of liver metastases from uveal melanoma. Invest Ophthalmol Vis Sci 48: 4890–4896. [DOI] [PubMed] [Google Scholar]
- 102. Huang SK, Darfler MM, Nicholl MB, You J, Bemis KG, et al. (2009) LC/MS-based quantitative proteomic analysis of paraffin-embedded archival melanomas reveals potential proteomic biomarkers associated with metastasis. PLoS One 4: e4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Blanco-Aparicio C, Renner O, Leal JFM, Carnero A (2007) PTEN, more than the AKT pathway. Carcinogenesis 28: 1379–1386. [DOI] [PubMed] [Google Scholar]
- 104. Lara PC, Pruschy M, Zimmermann M, Henríquez-Hernández LA (2011) MVP and vaults: a role in the radiation response. Radiat Oncol 6: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Lloret M, Lara PC, Bordón E, Fontes F, Rey A, et al. (2009) Major vault protein may affect nonhomologous end-joining repair and apoptosis through Ku70/80 and bax downregulation in cervical carcinoma tumors. Int J Radiat Oncol Biol Phys 73: 976–979. [DOI] [PubMed] [Google Scholar]
- 106. Seong H-A, Jung H, Ha H (2007) NM23-H1 tumor suppressor physically interacts with serine-threonine kinase receptor-associated protein, a transforming growth factor-beta (TGF-beta) receptor-interacting protein, and negatively regulates TGF-beta signaling. J Biol Chem 282: 12075–12096. [DOI] [PubMed] [Google Scholar]