Abstract
Cancer stem cells (CSCs) have been reported in many human tumors and are proposed to drive tumor initiation and progression. CSCs share a variety of biological properties with normal somatic stem cells such as the capacity for self-renewal, the propagation of differentiated progeny, and the expression of specific cell surface markers and stem cell genes. However, CSCs differ from normal stem cells in their chemoresistance and tumorigenic and metastatic activities. Despite their potential clinical importance, the regulation of CSCs at the molecular level is not well-understood. MicroRNAs (miRNAs) are a class of endogenous non-coding RNAs that play an important role in the regulation of several cellular, physiological, and developmental processes. Aberrant miRNA expression is associated with many human diseases including cancer. miRNAs have been implicated in the regulation of CSC properties; therefore, a better understanding of the modulation of CSC gene expression by miRNAs could aid the identification of promising biomarkers and therapeutic targets. In the present review, we summarize the major findings on the regulation of CSCs by miRNAs and discuss recent advances that have improved our understanding of the regulation of CSCs by miRNA networks and may lead to the development of miRNA therapeutics specifically targeting CSCs.
Keywords: microRNA, cancer stem cells (CSCs), tumor initiation, therapy resistance, metastasis
Background
The CSC theory, which is based on the concept that cancer might arise from a rare population of cells with stem cell properties, was proposed approximately 150 years ago (Cohnheim, 1875; Wicha et al., 2006). Recent technological developments (flow cytometry analysis and cell sorting) and the establishment of new animal models have provided evidence supporting the CSC theory. Moreover, CSCs are resistant to conventional treatments and are therefore not only of academic interest, but may also be an important consideration in clinical practice. Therefore, a better understanding of the characteristics of CSCs and the identification of therapeutic agents capable of targeting the CSC population are critical issues. Cancer researchers have investigated protein-coding genes and products, including surface markers that are involved in the self-renewal and asymmetric cell division of CSCs. Recently, in addition to alterations in protein-coding genes, abnormalities in non-coding RNAs [miRNAs and long intergenic non-coding RNAs] have been observed in various types of cancers and have been shown to play important roles in the regulation of CSC properties such as asymmetric cell division, tumorigenicity, and drug resistance. In the present review, we discuss the general features of CSCs and the role of miRNAs in the regulation of CSC properties, and summarize the current therapeutic strategies targeting miRNAs for CSC therapy.
Biogenesis and functions of miRNAs
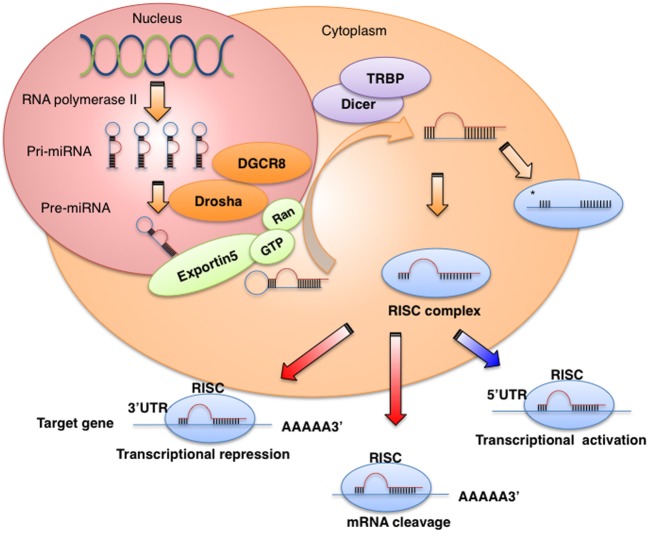
miRNAs are 21–25 nucleotides long, non-coding RNAs that regulate gene expression at the post-transcriptional level by binding to the 3′-untranslated regions (3′UTRs) or the open reading frames of target genes, leading to the degradation of target mRNAs or repression of mRNA translation (Iorio and Croce, 2012). miRNAs are transcribed for the most part by RNA polymerase II as long primary transcripts characterized by hairpin structures (pri-miRNA), and are processed in the nucleus by RNase III Drosha into 70–100 nucleotide long precursor miRNAs (pre-miRNAs) in combination with cofactors such as DGCR8, an evolutionarily conserved protein that interacts with proline-rich peptides through its WW domain (Gregory et al., 2004; Lee et al., 2004) (Figure 1). DGCR8 is located on chromosome region 22q11.2, whose heterozygous deletion results in the most common human genetic deletion syndrome, known as DiGeorge syndrome. The clinical symptoms of the disease are highly variable and in approximately 75% of patients, congenital heart defects are observed (Shiohama et al., 2003; Yamagishi and Srivastava, 2003). The product of pri-miRNA cleavage, the pre-miRNA, is exported to the cytoplasm by exportin-5, a member of the Ran-dependent nuclear transport receptor family (Lee et al., 2004) and further cleaved in a complex composed of RNase III Dicer and the transactivating response RNA- binding protein (TRBP) into a miRNA:miRNA* complex. While one of the two strands is selected as a guide strand, the complementary strand (miRNA*) is usually degraded (Iorio and Croce, 2012). miRNA* was originally considered to have no function and to be degraded; however, recent evidence suggests that it can be used as a functional strand and may play significant biological roles (Uchino et al., 2013; Yang et al., 2013).
Figure 1.
miRNA biogenesis and function. miRNAs are transcribed by RNA polymerase II or III as pri-miRNA, and are processed in the nucleus by Drosha-DGCR8 into pre-miRNAs. The product of pri-miRNA cleavage, the pre-miRNA, is exported to the cytoplasm by exportin-5 and further cleaved in a complex composed of Dicer and TRBP. The functional strand of mature miRNA is incorporated into the RNA-induced silencing complex (RISC), which contains GW182 and Argonaute protein. As a part of this complex, the mature miRNA regulates gene expression by binding to partially complementary sequences in the 3′UTRs of target mRNAs, leading to mRNA degradation or translation inhibition.
The mature miRNA is incorporated into a complex known as the RNA-induced silencing complex (RISC), which contains the GW182 and Argonaute proteins. As a part of this complex, the mature miRNA regulates gene expression by binding to partially complementary sequences in the 3′UTRs of target mRNAs, leading to mRNA degradation or translation inhibition (Iorio and Croce, 2012). Several studies have reported that miRNAs also bind to the 5′UTR or the open reading frame (Orom et al., 2008; Mandke et al., 2012) and can promote the translation of their target genes under growth arrest conditions (Vasudevan et al., 2007). Recently, Nishi et al. showed that TNRC6A, a human GW182 paralog, shuttles Ago2 into the nucleus and the colocalization of Ago2-TNRC6A with miRNAs mediates gene silencing (Nishi et al., 2013).
MicroRNAs regulate pluripotency and differentiation
The discovery of two miRNAs, lin-4 and let-7, in Caenorhabditis elegans suggested that miRNAs are important regulators of embryonic development and stem cell functions in mammals (Lee et al., 1993; Pasquinelli et al., 2000; Reinhart et al., 2000). The function of miRNAs in mouse and human embryonic stem cells (ESCs) has been investigated using cells lacking Dicer1 and DGCR8, which are critical for miRNA biogenesis. Deletion of Dicer1 leads to embryonic lethality in mice (Bernstein et al., 2003) and DGCR8-deficient mouse ESCs show alterations in the regulation of the cell cycle and differentiation that are associated with failure to silence stemness markers, such as Oct4, Rex1, Sox2, and Nanog, as well as delayed expression of differentiation markers (Wang et al., 2007).
In a comparative transcriptome analysis, Dicer1-deficient mouse ESCs lacking miRNAs showed a significant increase in transcripts containing a GCACUU motif in the 3′UTR (Sinkkonen et al., 2008). This sequence is complementary to the AAGUGC seed sequence of the miR-290-295 cluster (miR-290, miR-291a, miR-292, miR-291b, miR-294, and miR-295) and the miR-302/367 cluster (miR-302a, miR-302b, miR-302c, miR-302d, and miR-367) in mouse ESCs. Using a similar approach, novel stem cell-specific miRNAs were initially identified in human ESCs. These miRNAs include two clusters: miR-302/367 and the miR-371 cluster (miR-372 and miR-373). The expression of the miR-371 cluster is downregulated before that of the miR-302/367 cluster, suggesting a temporal hierarchy in the duration of specific miRNA activity (Stadler et al., 2010; Kim et al., 2011). Members of the miR-302 family rescue the proliferation defects of DGCR8-mutant mouse ESCs (Wang et al., 2008) and reprogram human skin cancer cells into a pluripotent ESC-like state (Lin et al., 2008).
The Let-7 family is another critical regulator of ESC differentiation. Mature let-7 family members are essentially absent in ESCs and accumulate only upon ESC differentiation (Viswanathan et al., 2008). Melton et al. reported that whereas transfection of let-7c into wild-type cells had no effect on the expression of pluripotency genes, let-7c rescued the differentiation defect in DGCR8−/− cells by downregulating Oct4, Sox2, and Nanog (Melton et al., 2010). Lin-28, a marker of undifferentiated ESCs, is also used to induce pluripotent stem cells (Yu et al., 2007b). A negative feedback loop between Lin-28 and let-7 family members precisely controls the levels of these miRNAs. Although Lin-28 regulates the expression of let-7 miRNAs by binding to the precursors and blocking their maturation, the let-7 family is highly expressed and targets Lin-28 mRNA in mouse differentiated cells and embryonic carcinoma cells (Yu et al., 2007b) (Figure 2). Members of the miR-34 family of miRNAs are direct targets of p53 and function as tumor suppressors, inhibiting reprogramming through the repression of pluripotency genes such as Nanog, Sox2, and N-myc (Choi et al., 2011) (Figure 2). Since the cell cycle regulator p21 also represses reprogramming efficiency, these findings suggest that p53 represses pluripotency via two distinct mechanisms. Evidence that let-7 and miR-34 family members are tumor suppressor miRNAs (Takamizawa et al., 2004; Johnson et al., 2005; Tazawa et al., 2007) suggests that stem cell-specific miRNAs play important roles in tumor initiation and development.
Figure 2.
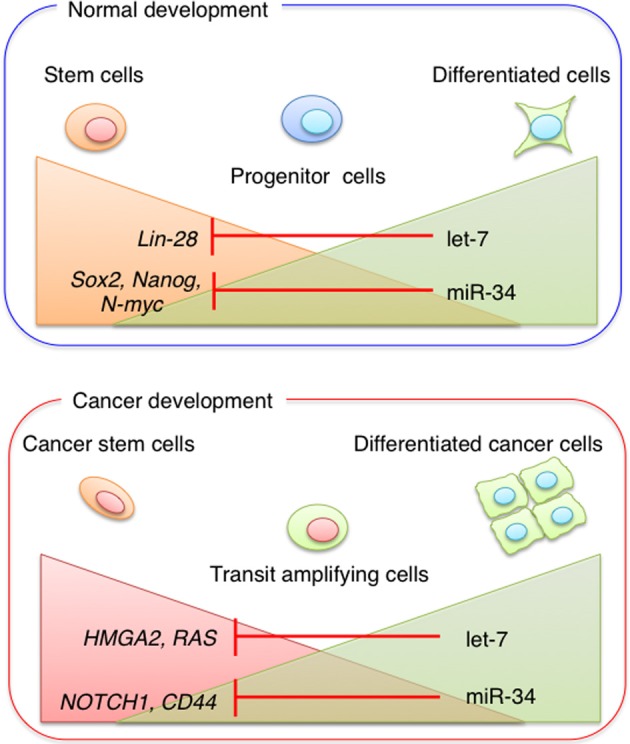
miRNA in stem cells and cancer stem cells. Stem cell-specific miRNAs play important roles in tumor initiation and development. During normal development, pluripotent stem cells become more restricted to specific cell lineages. Progenitor cells are committed to generating different cell types, whereas fully differentiated cells have a low potential for self-renewal. The expression levels of miR-34 and let-7 family members increase during differentiation. During cancer development, CSC properties are regulated by the balance between miRNA expression and the expression of miRNA target genes.
miRNA regulation in cancer
miRNAs play a crucial role in the progression of human cancer, and expression profiling in human malignancies has identified signatures associated with cancer development, progression, and prognosis (Liu et al., 2012; Volinia and Croce, 2013). Chromosomal regions coding for oncogenic miRNAs that are involved in the negative regulation of a tumor suppressor gene can be amplified in association with cancer development. This amplification would result in the upregulation of oncogenic miRNAs and silencing of tumor suppressor genes (He et al., 2005). On the other hand, miRNAs targeting oncogenes are often located in fragile site, where deletions or mutations can occur, leading to the reduction or loss of miRNAs and the overexpression of their target oncogenes. Dysregulation of miRNA expression affects processes associated with cancer progression such as the induction of anti-apoptotic activity, drug resistance, tissue invasion, and metastasis (Cimmino et al., 2005; Tavazoie et al., 2008; To et al., 2008). Recent evidence suggests that miRNAs are involved in tumor initiation through the regulation of CSC properties such as self-renewal ability, tumorigenicity and drug-resistance (Yu et al., 2007a; Shimono et al., 2009; Song et al., 2013a,b).
CSCs
Accumulating lines of evidence suggest that CSCs share a variety of biological properties with normal somatic stem cells such as the capacity for self-renewal, the propagation of differentiated progenitors, and the expression of specific stem cell genes (Colmont et al., 2012). However, CSCs differ from normal stem cells in their chemoresistance and tumorigenic and metastatic activities (Colmont et al., 2012 and Table 1). In addition, recently glycosylation patterns are found to be different between normal stem cells and CSCs (Karsten and Goletz, 2013). The CSC theory is generally accepted in the field of cancer research, not only in basic research but also with regard to cancer drug discovery.
Table 1.
Representative cell surface markers for human CSCs.
| Cancer type | CSC marker | References |
|---|---|---|
| AML | CD34+/CD38− | Bonnet and Dick, 1997 |
| Breast | CD44+/CD24−/low | Al-Hajj et al., 2003 |
| ALDH1 | Ginestier et al., 2007 | |
| Glioma | CD133 | Singh et al., 2003, 2004 |
| Colon | CD133 | O'brien et al., 2007; Ricci-Vitiani et al., 2007 |
| CD44/EpCAM/CD166 | Dalerba et al., 2007 | |
| Metastatic Colon | CD133+/CD26+ | Pang et al., 2010 |
| Melanoma | CD20 | Fang et al., 2005 |
| CD271 | Boiko et al., 2010 | |
| Pancreatic | ESA/CD44/CD24 | Hermann et al., 2007 |
| Metastatic Pancreatic | CD133/CXCR4 | Li et al., 2007a |
| Prostate | CD44/a2β1/CD133 | Collins et al., 2005 |
| Lung | CD133 | Eramo et al., 2008 |
| Hepatic | EpCAM/AFP | Yamashita et al., 2010 |
| Gastric | CD44 | Takaishi et al., 2009 |
AML, acute myelogenous leukemia; ALDH, aldehyde dehydrogenase; EpCAM, epithelial cell adhesion molecule; CXCR4, CXC chemokine receptor 4; AFP, alpha-fetoprotein.
Normal stem cells and CSCs act via common signaling pathways that regulate self-renewal activity, including Wnt, Notch, and Sonic Hedgehog, and dysregulation of these pathways plays a role in tumor initiation and development (Reya et al., 2001). Jamieson et al. showed that aberrations in the Wnt/β-catenin pathway enhance self-renewal activity during leukemia stem cell propagation (Jamieson et al., 2004). Korkaya et al. reported that the Wnt/β-catenin pathway is involved in the regulation of normal and malignant mammary stem/progenitor cell populations (Korkaya et al., 2009). Several studies have shown that the Notch pathway is activated in breast, glioblastoma, and colon CSCs (Hoey et al., 2009; Taketo, 2011). Alterations in Hedgehog signaling have been reported in colon, breast, and glioblastoma CSCs (Liu et al., 2006; Varnat et al., 2009; Takezaki et al., 2011).
The development of fluorescent antibodies, flow cytometry, and cell sorting techniques enabled the identification of cell populations possessing CSC properties. Furthermore, the development of severely immunodeficient mouse strains facilitated the evaluation of tumor formation ability. These methods have enabled the identification and isolation of CSCs from various cancers (Bonnet and Dick, 1997; Al-Hajj et al., 2003; Collins et al., 2005; Fang et al., 2005; Ginestier et al., 2007; Hermann et al., 2007; Li et al., 2007a; Eramo et al., 2008; Takaishi et al., 2009; Boiko et al., 2010; Pang et al., 2010; Yamashita et al., 2010) (Table 1). In this review, we discuss the major findings of recent studies highlighting the roles of certain “CSC-specific” miRNAs in representative cancer types (Table 2). From these discussions, we present an emerging theme that several miRNAs may exert a functional role in the regulation of the key biological properties of CSCs.
Table 2.
The regulatory roles of miRNAs in CSCs.
| Cancer Type | miRNA | Target gene | Role of miRNA in CSC properties | References |
|---|---|---|---|---|
| Leukemia (AML and MDS) | miR-22 | TET2 | Promotion of self-renewal | Song et al., 2013a |
| Breast | Let-7 | RAS and HMGA2 | Inhibition of self-renewal and de-differentiation | Yu et al., 2007a |
| miR-200 family | ZEB1/ZEB2 | Inhibition of EMT | Gregory et al., 2008 | |
| BMI-1 | Inhibition of self-renewal | Shimono et al., 2009 | ||
| SUZ12 | Inhibition of mammosphere formation | Iliopoulos et al., 2010 | ||
| miR-22 | TET family (TET1 -3) | Suppression of miR-200 family expression | Song et al., 2013b | |
| Brain | miR-9/9*, miR-17 | CAMTA1 | Promotion of CD133+ cell proliferation | Schraivogel et al., 2011 |
| miR-128 | BMI-1 | Inhibition of self-renewal | Godlewski et al., 2008 | |
| miR-199b-5p | HES1 | Reduction of the CD133+ cell fraction | Garzia et al., 2009 | |
| Colon | miR-193 | PLAU and K-RAS | Inhibition of tumorigenicity and invasiveness | Iliopoulos et al., 2011 |
| miR-451 | MIF and COX-2 | Inhibition of self-renewal and tumorigenicity | Bitarte et al., 2011 | |
| miR-34a | NOTCH 1 | Suppression of asymmetric cell division | Bu et al., 2013 | |
| Prostate | miR-34a | CD44 | Inhibition of self-renewal and metastasis | Liu et al., 2011 |
| miR-320 | β-catenin | Inhibition of Wnt/β-catenin pathway | Hsieh et al., 2013 |
AML, acute myelogenous leukemia; MDS, myelodysplastic syndrome.
Leukemia stem cells
Through an integrated approach that combined miRNA expression analysis and bioinformatic prediction of mRNA targets, distinct miRNA signatures were shown to fine-tune each step of hematopoiesis, including the reconstitution potential of hematopoietic stem cells (Arnold et al., 2011). The miR-17-92 cluster functions as an oncogenic miRNA by enhancing the formation of Myc-driven B-cell lymphomas in a mouse model (He et al., 2005). Single miRNAs function as oncogenes. The overexpression of miR-155 in early B-cells leads to polyclonal expansion of the pro-B-cell compartment (Costinean et al., 2006), and retroviral expression of miR-155 in immature mouse hematopoietic cells resulted in the expansion of granulocyte/monocyte populations displaying pathological features characteristic of myeloid neoplasia without progression to acute myeloid leukemia (AML) (O'connell et al., 2008). Recently, dysregulation of single miRNAs was shown to contribute to hematological malignancies, including AML and myelodysplastic syndrome (Han et al., 2010; Song et al., 2013a). Han et al. reported that miR-29a regulates early hematopoiesis and induces AML by converting myeloid progenitors into self-renewing leukemia stem cells via targeting several tumor suppressors and cell cycle regulators (Han et al., 2010). miR-22-induced inhibition of the ten-eleven-translocation gene 2 (TET2) tumor suppressor increased the methylation of TET2 target genes, such as Aim2, Hal, Igbt2, and Sp140, and resulted in positive effects on hematopoietic stem cell self-renewal and transformation. This has led to the suggestion that mir-22 is associated with myelodysplastic syndrome and hematological malignancies (Song et al., 2013a).
Breast CSCs
The first solid tumor CSCs were identified in and isolated from breast tumors in 2003 (Al-Hajj et al., 2003). Al-Hajj et al. described a CD44+/CD24−/low cell population that had a markedly high tumor-initiating capacity. In 2007, Yu et al. identified let-7 as a master regulator of breast CSC properties (Yu et al., 2007a). In breast CSCs, reduced let-7 expression controls self-renewal and differentiation through RAS and HMGA2, respectively (Figure 2). Since HMGA2 plays a role in the control of differentiation and proliferation of both human and mouse ESCs (Li et al., 2007b), these findings also suggest that let-7 is involved in the growth and differentiation of ESCs beyond tumorigenesis.
Epithelial-to-mesenchymal transition (EMT) is an evolutionarily conserved process that occurs during embryonic development in many species of mammals (Liu et al., 2006). Since the EMT program is often activated during tumor invasion and metastasis, the genetic controls and biochemical mechanisms underlying the acquisition of invasiveness and the subsequent systemic spread of cancer cells have been areas of intensive research. The EMT phenotype is characterized by the downregulation of epithelial markers such as E-cadherin, the expression of mesenchymal markers such as N-cadherin and vimentin, the loss of cell-cell contact and cell polarity, and the acquisition of cell invasive capabilities. Mani et al. reported that EMT is also associated with the acquisition of CSC properties (Mani et al., 2008). A CD44+/CD24−/low cell population purified from cancer tissues shows the features of an EMT phenotype, and human cancer cells induced to undergo EMT exhibit a CD44+/CD24−/low antigen phenotype and high tumorigenicity.
Recently, two studies reported the clinical relevance of CSCs in breast cancer specimens (Giordano et al., 2013; Yu et al., 2013). In early breast cancer patients, the presence of CD44+/CD24−/low cells in bone marrow was indicative of a poor prognosis (Giordano et al., 2013). Circulating tumor cells (CTCs) in breast cancer patients also showed the EMT phenotype (Yu et al., 2013). Progressive disease patients undergoing therapy had a higher number of mesenchymal marker positive CTCs than epithelial marker positive CTCs. These results suggest that the CSC phenotype is clinically important not only as a therapeutic target but also as a potential biomarker for the prognostic evaluation of patients undergoing cancer treatment.
A molecular link between EMT and the miR-200 family is provided by the zinc-finger E-box-binding homeobox protein encoding genes (ZEB1/ZEB2) (Gregory et al., 2008; Park et al., 2008). The miR-200 family consists of five members that are classified into two clusters: miR−200a, miR−200b, and miR−429 on human chromosome 1; and miR−200c and miR−141 on human chromosome 12 (Gregory et al., 2008). Expression of the miR-200 family strongly inhibits the EMT phenotype induced by TGF-β, and a reciprocal feedback loop between the miR-200 family and the ZEB family of transcription factors tightly regulates both EMT and mesenchymal-to-epithelial transition (Burk et al., 2008). MiR-200 family members are downregulated in normal human and mouse mammary stem cells and breast CSCs, and miR-200c inhibits the formation of mammary ducts from mammary stem cells and tumor formation from breast CSCs (Shimono et al., 2009). Members of the miR-200 family also modulate the self-renewal ability of CSCs by targeting B-lymphoma Mo-MLV insertion region 1 homolog (BMI-1) and SUZ12, a subunit of a polycomb repressor complex (Iliopoulos et al., 2010). BMI-1 regulates the self-renewal and differentiation of several types of stem cells, including hematopoietic, brain, and mammary stem cells (Molofsky et al., 2003; Park et al., 2003; Pietersen et al., 2008). Therefore, modulation of the activity of the miR-200 family using conventional therapy could be a promising approach to improve the effectiveness of breast cancer treatments.
Normal human and mouse mammary stem cells can be isolated and characterized on the basis of their aldehyde dehydrogenase (ALDH) activities (Ginestier et al., 2007). Using ALDH activity, Ibara et al. determined that miR-205 and miR-22 were highly expressed in mouse mammary progenitor cells (Ibarra et al., 2007). MiR-22 was recently shown to be an epigenetic modifier that promotes stemness and metastasis in breast cancer by directly targeting enzymes in the TET family, which regulate DNA demethylation (Song et al., 2013b). The TET family is involved in the demethylation of the miR-200 promoter, and miR-22 promotes CSC properties such as EMT and a metastatic phenotype through the suppression of the miR-200 family. This provides the first evidence that chromatin-remodeling systems with opposing effects on cell fate (self-renewal vs. differentiation) are regulated by opposing sets of miRNAs.
Brain CSCs
The pentaspan membrane glycoprotein CD133, also known as Prominin-1, was first identified as a marker of hematopoietic stem cells and progenitor cells, and was subsequently used to detect malignancies (Miraglia et al., 1997; Yin et al., 1997). In solid cancers, CD133 was first used to identify CSCs in different types of human brain tumors including glioblastoma, medulloblastoma, and ependymomas (Singh et al., 2003, 2004; Yu et al., 2010). In these studies, patient tumor cells were separated based on the expression of CD133. The CD133+ cell population is highly tumorigenic in vivo, whereas CD133− cells do not form tumors even at high numbers (Singh et al., 2003, 2004; Yu et al., 2010). CD133+ cells are also resistant to radiation and chemotherapy. These findings led to the hypothesis that glioblastomas are maintained by CSCs, and that this treatment-resistant subpopulation is a promising target for effective therapies. CD133 has been instrumental for the identification of CSCs in colorectal (Ricci-Vitiani et al., 2007) and pancreatic (Hermann et al., 2007) carcinomas. CD133 itself is a marker of normal neural stem cells in both humans (Uchida et al., 2000) and mice (Lee et al., 2005).
In cancer cells, the deacetylase HDAC6 directly interacts with and regulates the intracellular localization of CD133 (Mak et al., 2012). CD133 forms a stable protein complex with HDAC6 and β-catenin, which leads to the activation of β-catenin signaling targets in different types of cancer. CD133 is also associated with phosphoinositide 3-kinase (PI3K) 85 kDa regulatory subunit (p85) in glioma stem cells (GSCs) (Wei et al., 2013). The PI3K pathway is a key regulator of tumorigenesis in glioblastoma and other cancers (Godlewski et al., 2010). Therefore, activation of the PI3K/Akt pathway by the physical interaction between CD133 and p85 promotes tumorigenicity in GSCs. The function of CD133 in brain tumors should be fully characterized in the near future, which may shed light on the role of CD133 as a functional marker of GSCs.
Schraivogel et al. reported that miR-9, miR-9* (miR-9/9*), miR-17, and miR-106b are highly abundant in the CD133+ cell population in glioblastoma cell lines. Among the upregulated miRNAs in the CD133+ cell population, inhibition of miR-9/9* or miR-17 leads to reduced neurosphere formation and stimulates cell differentiation. Functional analysis of these miRNAs showed that miR-9/9* and miR-17 target calmodulin-binding transcription activator 1 (CAMTA1), a putative transcription factor of the anti-proliferative cardiac hormone natriuretic peptide A (NPPA). Clinical studies also demonstrated that CAMTA1 and NPPA expression is correlated with patient survival. These findings could provide a basis for the design of novel treatment strategies for glioblastoma (Schraivogel et al., 2011).
MiR-124 and miR-128 are the most highly expressed miRNAs in the adult brain and are preferentially expressed in neurons (Smirnova et al., 2005). Patients with high-grade glioma show significant downregulation of miR-128 expression. Functional analyses showed that miR-128 expression inhibits glioma cell proliferation in vitro and glioma xenograft growth in vivo (Godlewski et al., 2008). In addition, miR-128 specifically inhibits the self-renewal capacity of GSCs by directly targeting BMI-1, a polycomb family transcriptional repressor required for postnatal maintenance of neural stem cells in the peripheral and central nervous system (Molofsky et al., 2003). Since BMI-1 maintains neural stem cells in an undifferentiated self-renewing state, the regulation of BMI-1 by miR-128 may contribute to normal stem cell regulation.
Another study showed that miR-199b-5p downregulation was associated with metastatic spread in medulloblastoma. In medulloblastoma cells, miR-199b-5p directly targets HES1, a transcription factor of the Notch signaling pathway (Garzia et al., 2009). During brain development, Notch functions as a critical regulator of cell fate, by which gliogenesis can only occur when Notch signaling specifically represses the neuronal pathway in progenitor cells (Karamboulas and Ailles, 2013). MiR-199b-5p blocks Notch signaling, inhibiting the self-renewal capacity of medulloblastoma cells by reducing the CD133+ subpopulation (Garzia et al., 2009). Recently, miR-34a was shown to regulate Notch signaling by targeting Notch-1 and Notch-2 in medulloblastoma cells (Li et al., 2009). Therefore, miR-199b-5p and miR-34a are important for the self-renewal potential of GSCs via the Notch signaling pathway.
Colon CSCs
CD133 was initially used to identify and isolate colon CSCs (O'brien et al., 2007; Ricci-Vitiani et al., 2007), which was followed by the identification of CD44, epithelial surface antigen (EpCAM), and CD166 as alternative colon CSC markers (Dalerba et al., 2007). CD166 is a mesenchymal stem cell marker whose expression is correlated with poor prognosis in colon cancer patients (Weichert et al., 2004). Compared to CD44−/EpCAMlow cells, CD44+/EpCAMhigh cells from primary tumors show high tumorigenic activity in NOD/SCID mice. Moreover, CD166+ cells in the CD44+/EpCAMhigh cell fraction contribute to the tumorigenic activity of colon CSCs. In addition to CD133, CD44, EpCAM, and CD166, the expression of leucine-rich repeat-containing G-protein-coupled receptor 5 (Lgr5) varies among colorectal cancer (CRC) cases and is significantly correlated with lymphatic and vascular invasion, lymph node metastasis, and drug resistance (Vermeulen et al., 2008; Merlos-Suarez et al., 2011; Kobayashi et al., 2012).
Iliopoulos et al. reported that the expression of miR-193a is inversely correlated with K-RAS and plasminogen activator urokinase (PLAU) expression in human colon adenocarcinomas, and that miR-193 expression inhibits tumorigenicity and invasiveness by directly targeting K-RAS and PLAU, respectively (Iliopoulos et al., 2011). MiR-451 is another regulator of CSC properties such as self-renewal, tumorigenicity, and drug resistance. In spheroid cell culture, downregulation of miR-451 induces the upregulation of macrophage migration inhibitory factor (MIF) and COX-2, resulting in the acquisition of self-renewal and tumorigenic properties (Bitarte et al., 2011). MIF and Cox-2 are involved in the activation of the Wnt pathway, which is functionally essential for the maintenance of colon CSCs (Vermeulen et al., 2010), suggesting that miR-451 could regulate the properties of colon CSCs by suppressing the Wnt pathway.
Notch signaling is frequently activated in CRCs, and is dysregulated directly by epigenetic and genetic changes and indirectly by synergistic interactions with the Wnt pathway, which is also activated in CRC (Taketo, 2011). Notch signaling promotes the self-renewal activity of intestine and colon stem cells (Taketo, 2011). Therefore, colon CSCs in CRC are thought to arise from, or at least share common properties with, normal colon stem cells (Clevers, 2011; O'brien et al., 2012). Bu et al. reported that miR-34a determines whether colon CSCs undergo symmetric or asymmetric division, and that inhibition of asymmetric cell division suppresses tumorigenicity (Bu et al., 2013). MiR-34a inhibits Notch signaling by directly targeting Notch receptors (Li et al., 2009), suggesting that the upregulation of miR-34a weakens Notch signaling and promotes the generation of daughter cells (non-CSCs), whereas low miR-34a levels promote Notch signaling and lead to the maintenance of CSCs. This study also demonstrated that the expression level of miR-34a correlates more closely with the differentiation of daughter cells than the presence of Numb, which also suppresses Notch signaling by promoting the degradation of membrane-bound Notch and its intracellular domain (Bu et al., 2013).
Prostate CSCs
In prostate cancer (PCa), α2 β1 integrin, CD133, and CD44 were initially used to identify and isolate CSCs (Collins et al., 2005; Patrawala et al., 2006, 2007). Patrawala et al. reported that CD44+ PCa cells have higher proliferative, tumorigenic, and metastatic potentials than CD44− PCa cells (Patrawala et al., 2006), and showed that androgen receptor (AR)-negative CD44+ PCa cells differentiate into AR-positive CD44− PCa cells. Consistent with this report, prostate-specific antigen (PSA)-negative or -low PCa cells that are resistant to androgen ablation have a highly tumorigenic phenotype (Qin et al., 2012). In addition, PSA−/low PCa cells generate PSA+ PCa cells through asymmetric cell division, and highly tumorigenic PSA−/low PCa cells are characterized by an ALDH+/CD44+/α2β1 integrin+ phenotype (Qin et al., 2012).
Liu et al. reported that miR-34a is downregulated in CD44+ PCa cells purified from xenografts and primary tumors, and that miR-34a directly regulates the expression of CD44 at the post-transcriptional level by binding to its 3′UTR (Liu et al., 2011). Expression of miR-34a in CD44+ PCa cells inhibits tumor migration and metastasis in a xenograft model (Liu et al., 2011), and miR-34a inhibits Notch and AR signaling in PCa cells (Li et al., 2009; Kashat et al., 2012), suggesting that miR-34a suppresses the self-renewal activity of CSCs in PCa cells.
Another miRNA that regulates CSC properties is miR-320, which acts by directly targeting β-catenin in PCa cells (Hsieh et al., 2013). miR-320 and β-catenin expression is inversely correlated in CD44+ PCa cells. Furthermore, gene expression profiling of miR-320-overexpressing PCa cells showed a significant decrease in downstream target genes of the Wnt/β-catenin pathway and CSC markers (Hsieh et al., 2013).
Therapeutic approaches to target CSCs
The development of therapies against CSCs has resulted in the establishment of a new generation of cancer therapeutics, which is particularly important in the treatment of intractable cancers. Since CSCs are molecularly distinct from non-CSCs and bulk tumor cells, a high-throughput screening approach was used to identify small compounds that eliminate or reduce levels of CSCs (Gupta et al., 2009; Sachlos et al., 2012). Gupta et al. identified salinomycin as a selective inhibitor of breast CSCs (Gupta et al., 2009) by screening a library of 16,000 natural and commercial chemical compounds in a search for small compounds capable of killing breast CSCs. Although the precise molecular mechanisms underlying the elimination of CSCs by salinomycin are not fully understood, several studies have improved our understanding of the mechanisms and pharmacological action of salinomycin in human CSCs (Fuchs et al., 2010; Lu et al., 2011; Tang et al., 2011). Systemic salinomycin therapy induces a marked regression of subcutaneous thoracal metastases of breast cancer, and combination therapy of salinomycin with erlotinib resulted in significant tumor regression in metastatic squamous cell carcinoma (Naujokat and Steinhart, 2012).
High-throughput screening using neoplastic and normal human pluripotent stem cells (hPSC) showed that among 590 compounds, only thioridazine significantly promoted differentiation of neoplastic hPSCs but not of normal hPSCs (Sachlos et al., 2012). Thioridazine acts through dopamine receptors (dopamine receptor1-5) (Seeman and Lee, 1975), indicating that its selective interference with human CSCs is mediated by dopamine receptor antagonism.
The development of therapies against CSCs is challenging because both bulk cancer cells and CSCs must be eliminated. As CSCs are molecularly distinct from bulk tumor cells, they can be targeted by exploiting their molecular differences as described above (Tables 1, 2). One of the most promising approaches is the cell based delivery of miRNAs or miRNA inhibitors. Several studies demonstrated that miRNAs are secreted through “exosomes,” which are small endosome-derived vesicles (30–100 nm) secreted from different cell types, such as dendritic cells, hepatocyte, and tumor cells (Mittelbrunn et al., 2011; Luga et al., 2012; Ramakrishnaiah et al., 2013). The exosome secreted from mesenchymal stem cells (MSC) is selectively transferred to the glioblastoma multiforme (GBM) (Munoz et al., 2013). Since miR-9 is involved in the upregulation of p-glycoprotein, Munoz et al. developed an MSC derived exosome containing anti-miR-9 that efficiently suppressed p-glycoprotein expression in the temozolomide-resistant GBM.
The glycosylation pattern of CSC markers on CSCs is different from normal stem cells (Karsten and Goletz, 2013). Some CSC markers such as CD44 and CD133 are also expressed in normal stem and progenitor cells (Karsten and Goletz, 2013), which might have negative implications for the development of CSC-targeted delivery. This problem could be addressed by the development of liposomes or nanoparticles conjugated to antibodies against CSC specific glycans that permit the selective delivery of CSC suppressive miRNAs or small molecules.
Recent studies have shown that several dietary compounds can directly or indirectly affect the properties of CSCs (Li et al., 2011). Therefore, natural dietary compounds have received increasing attention in cancer chemoprevention, and several natural compounds that induce the elimination or differentiation of breast CSCs have been identified (Kakarala et al., 2010; Li et al., 2010; Hagiwara et al., 2012). Resveratrol is a non-toxic natural product that is found in grapes, berries, peanuts and red wine (Aziz et al., 2003). Nowadays, resveratrol is widely consumed as a nutritional supplement (Prasad, 2012), and its multifaceted biological effects include anti-mutagenic and anti-cancer properties (Prasad, 2012; Patel et al., 2013). Hagiwara et al. found that resveratrol enhances miRNA functions through the upregulation of Ago2 expression, which leads to the suppression of CSC properties (Hagiwara et al., 2012). These results suggest that the identification of non-toxic natural compounds capable of suppressing the properties of CSCs through the regulation of miRNA expression is a promising approach to support conventional chemotherapy.
Conclusions
Accumulating lines of evidence have shown that the heterogeneity and plasticity of cancer cells is reflected in the transition from a non-CSC to a CSC phenotype. Therefore, clinical oncologists and cancer researchers need to determine which cancer cells have the potential to contribute to tumor initiation and progression, including therapeutic resistance and metastasis. Several studies reviewed here have shown that miRNAs can function as tumor suppressors or oncogenes and play important roles in various aspects of CSC properties. In this regard, miRNAs are considered to be functional markers of CSCs. Therefore, a more detailed understanding of the function of miRNAs in CSC biology may improve cancer treatments and possibly lead to the clinical application of miRNAs in cancer diagnosis, treatment, and prognosis.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported in part by a grant-in-aid for the Third-Term Comprehensive 10-Year Strategy for Cancer Control of Japan; a grant-in-aid for Scientific Research on Priority Areas Cancer from the Japanese Ministry of Education, Culture, Sports, Science, and Technology; and the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation of Japan.
References
- Al-Hajj M., Wicha M. S., Benito-Hernandez A., Morrison S. J., Clarke M. F. (2003). Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 100, 3983–3988 10.1073/pnas.0530291100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold C. P., Tan R., Zhou B., Yue S. B., Schaffert S., Biggs J. R., et al. (2011). MicroRNA programs in normal and aberrant stem and progenitor cells. Genome Res. 21, 798–810 10.1101/gr.111385.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz M. H., Kumar R., Ahmad N. (2003). Cancer chemoprevention by resveratrol: in vitro and in vivo studies and the underlying mechanisms (review). Int. J. Oncol. 23, 17–28 [PubMed] [Google Scholar]
- Bernstein E., Kim S. Y., Carmell M. A., Murchison E. P., Alcorn H., Li M. Z., et al. (2003). Dicer is essential for mouse development. Nat. Genet. 35, 215–217 10.1038/ng1253 [DOI] [PubMed] [Google Scholar]
- Bitarte N., Bandres E., Boni V., Zarate R., Rodriguez J., Gonzalez-Huarriz M., et al. (2011). MicroRNA-451 is involved in the self-renewal, tumorigenicity, and chemoresistance of colorectal cancer stem cells. Stem Cells 29, 1661–1671 10.1002/stem.741 [DOI] [PubMed] [Google Scholar]
- Boiko A. D., Razorenova O. V., Van De Rijn M., Swetter S. M., Johnson D. L., Ly D. P., et al. (2010). Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature 466, 133–137 10.1038/nature09161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D., Dick J. E. (1997). Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 3, 730–737 10.1038/nm0797-730 [DOI] [PubMed] [Google Scholar]
- Bu P., Chen K. Y., Chen J. H., Wang L., Walters J., Shin Y. J., et al. (2013). A microRNA miR-34a-regulated bimodal switch targets notch in colon cancer stem cells. Cell Stem Cell 12, 602–615 10.1016/j.stem.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk U., Schubert J., Wellner U., Schmalhofer O., Vincan E., Spaderna S., et al. (2008). A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 9, 582–589 10.1038/embor.2008.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. J., Lin C. P., Ho J. J., He X., Okada N., Bu P., et al. (2011). miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat. Cell Biol. 13, 1353–1360 10.1038/ncb2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A., Calin G. A., Fabbri M., Iorio M. V., Ferracin M., Shimizu M., et al. (2005). miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. U.S.A. 102, 13944–13949 10.1073/pnas.0506654102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. (2011). The cancer stem cell: premises, promises and challenges. Nat. Med. 17, 313–319 10.1038/nm.2304 [DOI] [PubMed] [Google Scholar]
- Cohnheim J. (1875). Congenitales, quergestreiftes Muskelsarkon der Nireren. Virchows Arch. 65:64 10.1007/BF01978936 [DOI] [Google Scholar]
- Collins A. T., Berry P. A., Hyde C., Stower M. J., Maitland N. J. (2005). Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 65, 10946–10951 10.1158/0008-5472.CAN-05-2018 [DOI] [PubMed] [Google Scholar]
- Colmont C. S., Harding K. G., Piguet V., Patel G. K. (2012). Human skin cancer stem cells: a tale of mice and men. Exp. Dermatol. 21, 576–580 10.1111/j.1600-0625.2012.01533.x [DOI] [PubMed] [Google Scholar]
- Costinean S., Zanesi N., Pekarsky Y., Tili E., Volinia S., Heerema N., et al. (2006). Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 103, 7024–7029 10.1073/pnas.0602266103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalerba P., Dylla S. J., Park I. K., Liu R., Wang X., Cho R. W., et al. (2007). Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. U.S.A. 104, 10158–10163 10.1073/pnas.0703478104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eramo A., Lotti F., Sette G., Pilozzi E., Biffoni M., Di Virgilio A., et al. (2008). Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 15, 504–514 10.1038/sj.cdd.4402283 [DOI] [PubMed] [Google Scholar]
- Fang D., Nguyen T. K., Leishear K., Finko R., Kulp A. N., Hotz S., et al. (2005). A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 65, 9328–9337 10.1158/0008-5472.CAN-05-1343 [DOI] [PubMed] [Google Scholar]
- Fuchs D., Daniel V., Sadeghi M., Opelz G., Naujokat C. (2010). Salinomycin overcomes ABC transporter-mediated multidrug and apoptosis resistance in human leukemia stem cell-like KG-1a cells. Biochem. Biophys. Res. Commun. 394, 1098–1104 10.1016/j.bbrc.2010.03.138 [DOI] [PubMed] [Google Scholar]
- Garzia L., Andolfo I., Cusanelli E., Marino N., Petrosino G., De Martino D., et al. (2009). MicroRNA-199b-5p impairs cancer stem cells through negative regulation of HES1 in medulloblastoma. PLoS ONE 4:e4998 10.1371/journal.pone.0004998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestier C., Hur M. H., Charafe-Jauffret E., Monville F., Dutcher J., Brown M., et al. (2007). ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 1, 555–567 10.1016/j.stem.2007.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano A., Gao H., Cohen E. N., Anfossi S., Khoury J., Hess K., et al. (2013). Clinical relevance of cancer stem cells in bone marrow of early breast cancer patients. Ann. Oncol. 24, 2515–2521 10.1093/annonc/mdt223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godlewski J., Newton H. B., Chiocca E. A., Lawler S. E. (2010). MicroRNAs and glioblastoma; the stem cell connection. Cell Death Differ. 17, 221–228 10.1038/cdd.2009.71 [DOI] [PubMed] [Google Scholar]
- Godlewski J., Nowicki M. O., Bronisz A., Williams S., Otsuki A., Nuovo G., et al. (2008). Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 68, 9125–9130 10.1158/0008-5472.CAN-08-2629 [DOI] [PubMed] [Google Scholar]
- Gregory R. I., Yan K. P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., et al. (2004). The Microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240 10.1038/nature03120 [DOI] [PubMed] [Google Scholar]
- Gregory P. A., Bert A. G., Paterson E. L., Barry S. C., Tsykin A., Farshid G., et al. (2008). The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593–601 10.1038/ncb1722 [DOI] [PubMed] [Google Scholar]
- Gupta P. B., Onder T. T., Jiang G., Tao K., Kuperwasser C., Weinberg R. A., et al. (2009). Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 138, 645–659 10.1016/j.cell.2009.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara K., Kosaka N., Yoshioka Y., Takahashi R. U., Takeshita F., Ochiya T. (2012). Stilbene derivatives promote Ago2-dependent tumour-suppressive microRNA activity. Sci. Rep. 2, 314 10.1038/srep00314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y. C., Park C. Y., Bhagat G., Zhang J., Wang Y., Fan J. B., et al. (2010). microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J. Exp. Med. 207, 475–489 10.1084/jem.20090831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Thomson J. M., Hemann M. T., Hernando-Monge E., Mu D., Goodson S., et al. (2005). A microRNA polycistron as a potential human oncogene. Nature 435, 828–833 10.1038/nature03552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann P. C., Huber S. L., Herrler T., Aicher A., Ellwart J. W., Guba M., et al. (2007). Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell 1, 313–323 10.1016/j.stem.2007.06.002 [DOI] [PubMed] [Google Scholar]
- Hoey T., Yen W. C., Axelrod F., Basi J., Donigian L., Dylla S., et al. (2009). DLL4 blockade inhibits tumor growth and reduces tumor-initiating cell frequency. Cell Stem Cell 5, 168–177 10.1016/j.stem.2009.05.019 [DOI] [PubMed] [Google Scholar]
- Hsieh I. S., Chang K. C., Tsai Y. T., Ke J. Y., Lu P. J., Lee K. H., et al. (2013). MicroRNA-320 suppresses the stem cell-like characteristics of prostate cancer cells by downregulating the Wnt/beta-catenin signaling pathway. Carcinogenesis 34, 530–538 10.1093/carcin/bgs371 [DOI] [PubMed] [Google Scholar]
- Ibarra I., Erlich Y., Muthuswamy S. K., Sachidanandam R., Hannon G. J. (2007). A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev. 21, 3238–3243 10.1101/gad.1616307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D., Lindahl-Allen M., Polytarchou C., Hirsch H. A., Tsichlis P. N., Struhl K. (2010). Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol. Cell 39, 761–772 10.1016/j.molcel.2010.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D., Rotem A., Struhl K. (2011). Inhibition of miR-193a expression by Max and RXRalpha activates K-Ras and PLAU to mediate distinct aspects of cellular transformation. Cancer Res. 71, 5144–5153 10.1158/0008-5472.CAN-11-0425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio M. V., Croce C. M. (2012). MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 4, 143–159 10.1002/emmm.201100209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson C. H., Ailles L. E., Dylla S. J., Muijtjens M., Jones C., Zehnder J. L., et al. (2004). Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N. Engl. J. Med. 351, 657–667 10.1056/NEJMoa040258 [DOI] [PubMed] [Google Scholar]
- Johnson S. M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., et al. (2005). RAS is regulated by the let-7 microRNA family. Cell 120, 635–647 10.1016/j.cell.2005.01.014 [DOI] [PubMed] [Google Scholar]
- Kakarala M., Brenner D. E., Korkaya H., Cheng C., Tazi K., Ginestier C., et al. (2010). Targeting breast stem cells with the cancer preventive compounds curcumin and piperine. Breast Cancer Res. Treat. 122, 777–785 10.1007/s10549-009-0612-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamboulas C., Ailles L. (2013). Developmental signaling pathways in cancer stem cells of solid tumors. Biochim. Biophys. Acta 1830, 2481–2495 10.1016/j.bbagen.2012.11.008 [DOI] [PubMed] [Google Scholar]
- Karsten U., Goletz S. (2013). What makes cancer stem cell markers different? Springerplus 2, 301 10.1186/2193-1801-2-301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashat M., Azzouz L., Sarkar S. H., Kong D., Li Y., Sarkar F. H. (2012). Inactivation of AR and Notch-1 signaling by miR-34a attenuates prostate cancer aggressiveness. Am. J. Transl. Res. 4, 432–442 [PMC free article] [PubMed] [Google Scholar]
- Kim H., Lee G., Ganat Y., Papapetrou E. P., Lipchina I., Socci N. D., et al. (2011). miR-371-3 expression predicts neural differentiation propensity in human pluripotent stem cells. Cell Stem Cell 8, 695–706 10.1016/j.stem.2011.04.002 [DOI] [PubMed] [Google Scholar]
- Kobayashi S., Yamada-Okabe H., Suzuki M., Natori O., Kato A., Matsubara K., et al. (2012). LGR5-positive colon cancer stem cells interconvert with drug-resistant LGR5-negative cells and are capable of tumor reconstitution. Stem Cells 30, 2631–2644 10.1002/stem.1257 [DOI] [PubMed] [Google Scholar]
- Korkaya H., Paulson A., Charafe-Jauffret E., Ginestier C., Brown M., Dutcher J., et al. (2009). Regulation of mammary stem/progenitor cells by PTEN/Akt/beta-catenin signaling. PLoS Biol. 7:e1000121 10.1371/journal.pbio.1000121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A., Kessler J. D., Read T. A., Kaiser C., Corbeil D., Huttner W. B., et al. (2005). Isolation of neural stem cells from the postnatal cerebellum. Nat. Neurosci. 8, 723–729 10.1038/nn1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L., Ambros V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- Lee Y., Kim M., Han J., Yeom K. H., Lee S., Baek S. H., et al. (2004). MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23, 4051–4060 10.1038/sj.emboj.7600385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Heidt D. G., Dalerba P., Burant C. F., Zhang L., Adsay V., et al. (2007a). Identification of pancreatic cancer stem cells. Cancer Res. 67, 1030–1037 10.1158/0008-5472.CAN-06-2030 [DOI] [PubMed] [Google Scholar]
- Li O., Li J., Droge P. (2007b). DNA architectural factor and proto-oncogene HMGA2 regulates key developmental genes in pluripotent human embryonic stem cells. FEBS Lett. 581, 3533–3537 10.1016/j.febslet.2007.06.072 [DOI] [PubMed] [Google Scholar]
- Li Y., Guessous F., Zhang Y., Dipierro C., Kefas B., Johnson E., et al. (2009). MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 69, 7569–7576 10.1158/0008-5472.CAN-09-0529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wicha M. S., Schwartz S. J., Sun D. (2011). Implications of cancer stem cell theory for cancer chemoprevention by natural dietary compounds. J. Nutr. Biochem. 22, 799–806 10.1016/j.jnutbio.2010.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang T., Korkaya H., Liu S., Lee H. F., Newman B., et al. (2010). Sulforaphane, a dietary component of broccoli/broccoli sprouts, inhibits breast cancer stem cells. Clin. Cancer Res. 16, 2580–2590 10.1158/1078-0432.CCR-09-2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. L., Chang D. C., Chang-Lin S., Lin C. H., Wu D. T., Chen D. T., et al. (2008). Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA 14, 2115–2124 10.1261/rna.1162708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Kelnar K., Liu B., Chen X., Calhoun-Davis T., Li H., et al. (2011). The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat. Med. 17, 211–215 10.1038/nm.2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Chen N. Y., Cui R. X., Li W. F., Li Y., Wei R. R., et al. (2012). Prognostic value of a microRNA signature in nasopharyngeal carcinoma: a microRNA expression analysis. Lancet Oncol. 13, 633–641 10.1016/S1470-2045(12)70102-X [DOI] [PubMed] [Google Scholar]
- Liu S., Dontu G., Mantle I. D., Patel S., Ahn N. S., Jackson K. W., et al. (2006). Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 66, 6063–6071 10.1158/0008-5472.CAN-06-0054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Choi M. Y., Yu J., Castro J. E., Kipps T. J., Carson D. A. (2011). Salinomycin inhibits Wnt signaling and selectively induces apoptosis in chronic lymphocytic leukemia cells. Proc. Natl. Acad. Sci. U.S.A. 108, 13253–13257 10.1073/pnas.1110431108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luga V., Zhang L., Viloria-Petit A. M., Ogunjimi A. A., Inanlou M. R., Chiu E., et al. (2012). Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 151, 1542–1556 10.1016/j.cell.2012.11.024 [DOI] [PubMed] [Google Scholar]
- Mak A. B., Nixon A. M., Kittanakom S., Stewart J. M., Chen G. I., Curak J., et al. (2012). Regulation of CD133 by HDAC6 promotes beta-catenin signaling to suppress cancer cell differentiation. Cell Rep. 2, 951–963 10.1016/j.celrep.2012.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandke P., Wyatt N., Fraser J., Bates B., Berberich S. J., Markey M. P. (2012). MicroRNA-34a modulates MDM4 expression via a target site in the open reading frame. PLoS ONE 7:e42034 10.1371/journal.pone.0042034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani S. A., Guo W., Liao M. J., Eaton E. N., Ayyanan A., Zhou A. Y., et al. (2008). The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C., Judson R. L., Blelloch R. (2010). Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 463, 621–626 10.1038/nature08725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlos-Suarez A., Barriga F. M., Jung P., Iglesias M., Cespedes M. V., Rossell D., et al. (2011). The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 8, 511–524 10.1016/j.stem.2011.02.020 [DOI] [PubMed] [Google Scholar]
- Miraglia S., Godfrey W., Yin A. H., Atkins K., Warnke R., Holden J. T., et al. (1997). A novel five-transmembrane hematopoietic stem cell antigen: isolation, characterization, and molecular cloning. Blood 90, 5013–5021 [PubMed] [Google Scholar]
- Mittelbrunn M., Gutierrez-Vazquez C., Villarroya-Beltri C., Gonzalez S., Sanchez-Cabo F., Gonzalez M. A., et al. (2011). Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2, 282 10.1038/ncomms1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky A. V., Pardal R., Iwashita T., Park I. K., Clarke M. F., Morrison S. J. (2003). Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 425, 962–967 10.1038/nature02060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz J. L., Bliss S. A., Greco S. J., Ramkissoon S. H., Ligon K. L., Rameshwar P. (2013). Delivery of functional anti-miR-9 by mesenchymal stem cell-derived exosomes to glioblastoma multiforme cells conferred chemosensitivity. Mol. Ther. Nucleic Acids 2, e126 10.1038/mtna.2013.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naujokat C., Steinhart R. (2012). Salinomycin as a drug for targeting human cancer stem cells. J. Biomed. Biotechnol. 2012, 950658 10.1155/2012/950658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi K., Nishi A., Nagasawa T., Ui-Tei K. (2013). Human TNRC6A is an Argonaute-navigator protein for microRNA-mediated gene silencing in the nucleus. RNA 19, 17–35 10.1261/rna.034769.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'brien C. A., Kreso A., Ryan P., Hermans K. G., Gibson L., Wang Y., et al. (2012). ID1 and ID3 regulate the self-renewal capacity of human colon cancer-initiating cells through p21. Cancer Cell 21, 777–792 10.1016/j.ccr.2012.04.036 [DOI] [PubMed] [Google Scholar]
- O'brien C. A., Pollett A., Gallinger S., Dick J. E. (2007). A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 445, 106–110 10.1038/nature05372 [DOI] [PubMed] [Google Scholar]
- O'connell R. M., Rao D. S., Chaudhuri A. A., Boldin M. P., Taganov K. D., Nicoll J., et al. (2008). Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J. Exp. Med. 205, 585–594 10.1084/jem.20072108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom U. A., Nielsen F. C., Lund A. H. (2008). MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 30, 460–471 10.1016/j.molcel.2008.05.001 [DOI] [PubMed] [Google Scholar]
- Pang R., Law W. L., Chu A. C., Poon J. T., Lam C. S., Chow A. K., et al. (2010). A subpopulation of CD26+ cancer stem cells with metastatic capacity in human colorectal cancer. Cell Stem Cell 6, 603–615 10.1016/j.stem.2010.04.001 [DOI] [PubMed] [Google Scholar]
- Park I. K., Qian D., Kiel M., Becker M. W., Pihalja M., Weissman I. L., et al. (2003). Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423, 302–305 10.1038/nature01587 [DOI] [PubMed] [Google Scholar]
- Park S. M., Gaur A. B., Lengyel E., Peter M. E. (2008). The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 22, 894–907 10.1101/gad.1640608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli A. E., Reinhart B. J., Slack F., Martindale M. Q., Kuroda M. I., Maller B., et al. (2000). Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408, 86–89 10.1038/35040556 [DOI] [PubMed] [Google Scholar]
- Patel K. R., Andreadi C., Britton R. G., Horner-Glister E., Karmokar A., Sale S., et al. (2013). Sulfate metabolites provide an intracellular pool for resveratrol generation and induce autophagy with senescence. Sci. Transl. Med. 5, 205ra133 10.1126/scitranslmed.3005870 [DOI] [PubMed] [Google Scholar]
- Patrawala L., Calhoun-Davis T., Schneider-Broussard R., Tang D. G. (2007). Hierarchical organization of prostate cancer cells in xenograft tumors: the CD44+alpha2beta1+ cell population is enriched in tumor-initiating cells. Cancer Res. 67, 6796–6805 10.1158/0008-5472.CAN-07-0490 [DOI] [PubMed] [Google Scholar]
- Patrawala L., Calhoun T., Schneider-Broussard R., Li H., Bhatia B., Tang S., et al. (2006). Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene 25, 1696–1708 10.1038/sj.onc.1209327 [DOI] [PubMed] [Google Scholar]
- Pietersen A. M., Evers B., Prasad A. A., Tanger E., Cornelissen-Steijger P., Jonkers J., et al. (2008). Bmi1 regulates stem cells and proliferation and differentiation of committed cells in mammary epithelium. Curr. Biol. 18, 1094–1099 10.1016/j.cub.2008.06.070 [DOI] [PubMed] [Google Scholar]
- Prasad K. (2012). Resveratrol, wine, and atherosclerosis. Int. J. Angiol. 21, 7–18 10.1055/s-0032-1306417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Liu X., Laffin B., Chen X., Choy G., Jeter C. R., et al. (2012). The PSA(-/lo) prostate cancer cell population harbors self-renewing long-term tumor-propagating cells that resist castration. Cell Stem Cell 10, 556–569 10.1016/j.stem.2012.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnaiah V., Thumann C., Fofana I., Habersetzer F., Pan Q., De Ruiter P. E., et al. (2013). Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc. Natl. Acad. Sci. U.S.A. 110, 13109–13113 10.1073/pnas.1221899110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B. J., Slack F. J., Basson M., Pasquinelli A. E., Bettinger J. C., Rougvie A. E., et al. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906 10.1038/35002607 [DOI] [PubMed] [Google Scholar]
- Reya T., Morrison S. J., Clarke M. F., Weissman I. L. (2001). Stem cells, cancer, and cancer stem cells. Nature 414, 105–111 10.1038/35102167 [DOI] [PubMed] [Google Scholar]
- Ricci-Vitiani L., Lombardi D. G., Pilozzi E., Biffoni M., Todaro M., Peschle C., et al. (2007). Identification and expansion of human colon-cancer-initiating cells. Nature 445, 111–115 10.1038/nature05384 [DOI] [PubMed] [Google Scholar]
- Sachlos E., Risueno R. M., Laronde S., Shapovalova Z., Lee J. H., Russell J., et al. (2012). Identification of drugs including a dopamine receptor antagonist that selectively target cancer stem cells. Cell 149, 1284–1297 10.1016/j.cell.2012.03.049 [DOI] [PubMed] [Google Scholar]
- Schraivogel D., Weinmann L., Beier D., Tabatabai G., Eichner A., Zhu J. Y., et al. (2011). CAMTA1 is a novel tumour suppressor regulated by miR-9/9* in glioblastoma stem cells. EMBO J. 30, 4309–4322 10.1038/emboj.2011.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman P., Lee T. (1975). Antipsychotic drugs: direct correlation between clinical potency and presynaptic action on dopamine neurons. Science 188, 1217–1219 10.1126/science.1145194 [DOI] [PubMed] [Google Scholar]
- Shiohama A., Sasaki T., Noda S., Minoshima S., Shimizu N. (2003). Molecular cloning and expression analysis of a novel gene DGCR8 located in the DiGeorge syndrome chromosomal region. Biochem. Biophys. Res. Commun. 304, 184–190 10.1016/S0006-291X(03)00554-0 [DOI] [PubMed] [Google Scholar]
- Shimono Y., Zabala M., Cho R. W., Lobo N., Dalerba P., Qian D., et al. (2009). Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 138, 592–603 10.1016/j.cell.2009.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. K., Clarke I. D., Terasaki M., Bonn V. E., Hawkins C., Squire J., et al. (2003). Identification of a cancer stem cell in human brain tumors. Cancer Res. 63, 5821–5828 [PubMed] [Google Scholar]
- Singh S. K., Hawkins C., Clarke I. D., Squire J. A., Bayani J., Hide T., et al. (2004). Identification of human brain tumour initiating cells. Nature 432, 396–401 10.1038/nature03128 [DOI] [PubMed] [Google Scholar]
- Sinkkonen L., Hugenschmidt T., Berninger P., Gaidatzis D., Mohn F., Artus-Revel C. G., et al. (2008). MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 15, 259–267 10.1038/nsmb.1391 [DOI] [PubMed] [Google Scholar]
- Smirnova L., Grafe A., Seiler A., Schumacher S., Nitsch R., Wulczyn F. G. (2005). Regulation of miRNA expression during neural cell specification. Eur. J. Neurosci. 21, 1469–1477 10.1111/j.1460-9568.2005.03978.x [DOI] [PubMed] [Google Scholar]
- Song S. J., Ito K., Ala U., Kats L., Webster K., Sun S. M., et al. (2013a). The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell 13, 87–101 10.1016/j.stem.2013.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S. J., Poliseno L., Song M. S., Ala U., Webster K., Ng C., et al. (2013b). MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell 154, 311–324 10.1016/j.cell.2013.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler B., Ivanovska I., Mehta K., Song S., Nelson A., Tan Y., et al. (2010). Characterization of microRNAs involved in embryonic stem cell states. Stem Cells Dev. 19, 935–950 10.1089/scd.2009.0426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaishi S., Okumura T., Tu S., Wang S. S., Shibata W., Vigneshwaran R., et al. (2009). Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells 27, 1006–1020 10.1002/stem.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., et al. (2004). Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 64, 3753–3756 10.1158/0008-5472.CAN-04-0637 [DOI] [PubMed] [Google Scholar]
- Taketo M. M. (2011). Reflections on the spread of metastasis to cancer prevention. Cancer Prev. Res. (Phila.) 4, 324–328 10.1158/1940-6207.CAPR-11-0046 [DOI] [PubMed] [Google Scholar]
- Takezaki T., Hide T., Takanaga H., Nakamura H., Kuratsu J., Kondo T. (2011). Essential role of the Hedgehog signaling pathway in human glioma-initiating cells. Cancer Sci. 102, 1306–1312 10.1111/j.1349-7006.2011.01943.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q. L., Zhao Z. Q., Li J. C., Liang Y., Yin J. Q., Zou C. Y., et al. (2011). Salinomycin inhibits osteosarcoma by targeting its tumor stem cells. Cancer Lett. 311, 113–121 10.1016/j.canlet.2011.07.016 [DOI] [PubMed] [Google Scholar]
- Tavazoie S. F., Alarcon C., Oskarsson T., Padua D., Wang Q., Bos P. D., et al. (2008). Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451, 147–152 10.1038/nature06487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazawa H., Tsuchiya N., Izumiya M., Nakagama H. (2007). Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc. Natl. Acad. Sci. U.S.A. 104, 15472–15477 10.1073/pnas.0707351104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K. K., Zhan Z., Litman T., Bates S. E. (2008). Regulation of ABCG2 expression at the 3′ untranslated region of its mRNA through modulation of transcript stability and protein translation by a putative microRNA in the S1 colon cancer cell line. Mol. Cell. Biol. 28, 5147–5161 10.1128/MCB.00331-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N., Buck D. W., He D., Reitsma M. J., Masek M., Phan T. V., et al. (2000). Direct isolation of human central nervous system stem cells. Proc. Natl. Acad. Sci. U.S.A. 97, 14720–14725 10.1073/pnas.97.26.14720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchino K., Takeshita F., Takahashi R. U., Kosaka N., Fujiwara K., Naruoka H., et al. (2013). Therapeutic effects of microRNA-582-5p and -3p on the inhibition of bladder cancer progression. Mol. Ther. 21, 610–619 10.1038/mt.2012.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnat F., Duquet A., Malerba M., Zbinden M., Mas C., Gervaz P., et al. (2009). Human colon cancer epithelial cells harbour active HEDGEHOG-GLI signalling that is essential for tumour growth, recurrence, metastasis and stem cell survival and expansion. EMBO Mol. Med. 1, 338–351 10.1002/emmm.200900039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S., Tong Y., Steitz J. A. (2007). Switching from repression to activation: microRNAs can up-regulate translation. Science 318, 1931–1934 10.1126/science.1149460 [DOI] [PubMed] [Google Scholar]
- Vermeulen L., De Sousa E. M. F., Van Der Heijden M., Cameron K., De Jong J. H., Borovski T., et al. (2010). Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat. Cell Biol. 12, 468–476 10.1038/ncb2048 [DOI] [PubMed] [Google Scholar]
- Vermeulen L., Todaro M., De Sousa Mello F., Sprick M. R., Kemper K., Perez Alea M., et al. (2008). Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc. Natl. Acad. Sci. U.S.A. 105, 13427–13432 10.1073/pnas.0805706105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan S. R., Daley G. Q., Gregory R. I. (2008). Selective blockade of microRNA processing by Lin28. Science 320, 97–100 10.1126/science.1154040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S., Croce C. M. (2013). Prognostic microRNA/mRNA signature from the integrated analysis of patients with invasive breast cancer. Proc. Natl. Acad. Sci. U.S.A. 110, 7413–7417 10.1073/pnas.1304977110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Baskerville S., Shenoy A., Babiarz J. E., Baehner L., Blelloch R. (2008). Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet. 40, 1478–1483 10.1038/ng.250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Medvid R., Melton C., Jaenisch R., Blelloch R. (2007). DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 39, 380–385 10.1038/ng1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Jiang Y., Zou F., Liu Y., Wang S., Xu N., et al. (2013). Activation of PI3K/Akt pathway by CD133-p85 interaction promotes tumorigenic capacity of glioma stem cells. Proc. Natl. Acad. Sci. U.S.A. 110, 6829–6834 10.1073/pnas.1217002110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichert W., Knosel T., Bellach J., Dietel M., Kristiansen G. (2004). ALCAM/CD166 is overexpressed in colorectal carcinoma and correlates with shortened patient survival. J. Clin. Pathol. 57, 1160–1164 10.1136/jcp.2004.016238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicha M. S., Liu S., Dontu G. (2006). Cancer stem cells: an old idea–a paradigm shift. Cancer Res. 66, 1883–1890 discussion: 1895–1886. 10.1158/0008-5472.CAN-05-3153 [DOI] [PubMed] [Google Scholar]
- Yamagishi H., Srivastava D. (2003). Unraveling the genetic and developmental mysteries of 22q11 deletion syndrome. Trends Mol. Med. 9, 383–389 10.1016/S1471-4914(03)00141-2 [DOI] [PubMed] [Google Scholar]
- Yamashita T., Honda M., Nio K., Nakamoto Y., Yamashita T., Takamura H., et al. (2010). Oncostatin m renders epithelial cell adhesion molecule-positive liver cancer stem cells sensitive to 5-Fluorouracil by inducing hepatocytic differentiation. Cancer Res. 70, 4687–4697 10.1158/0008-5472.CAN-09-4210 [DOI] [PubMed] [Google Scholar]
- Yang X., Du W. W., Li H., Liu F., Khorshidi A., Rutnam Z. J., et al. (2013). Both mature miR-17-5p and passenger strand miR-17-3p target TIMP3 and induce prostate tumor growth and invasion. Nucleic Acids Res. 41, 9688–9704 10.1093/nar/gkt680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin A. H., Miraglia S., Zanjani E. D., Almeida-Porada G., Ogawa M., Leary A. G., et al. (1997). AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood 90, 5002–5012 [PubMed] [Google Scholar]
- Yu F., Yao H., Zhu P., Zhang X., Pan Q., Gong C., et al. (2007a). let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131, 1109–1123 10.1016/j.cell.2007.10.054 [DOI] [PubMed] [Google Scholar]
- Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., et al. (2007b). Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 10.1126/science.1151526 [DOI] [PubMed] [Google Scholar]
- Yu L., Baxter P. A., Voicu H., Gurusiddappa S., Zhao Y., Adesina A., et al. (2010). A clinically relevant orthotopic xenograft model of ependymoma that maintains the genomic signature of the primary tumor and preserves cancer stem cells in vivo. Neuro Oncol. 12, 580–594 10.1093/neuonc/nop056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Bardia A., Wittner B. S., Stott S. L., Smas M. E., Ting D. T., et al. (2013). Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339, 580–584 10.1126/science.1228522 [DOI] [PMC free article] [PubMed] [Google Scholar]