Abstract
The “Nucleotide-binding domain and Leucine-rich Repeat” (NLR) genes are a family of intracellular pattern recognition receptors (PRR) that are a critical component of the metazoan innate immune system, involved in both defense against pathogenic microorganisms and in beneficial interactions with symbionts. To investigate the origin and evolution of the NLR gene family, we characterized the full NACHT domain-containing gene complement in the genome of the sponge, Amphimedon queenslandica. As sister group to all animals, sponges are ideally placed to inform our understanding of the early evolution of this ancient PRR family. Amphimedon queenslandica has a large NACHT domain-containing gene complement that is dominated by bona fide NLRs (n = 135) with varied phylogenetic histories. Approximately half of these have a tripartite architecture that includes an N-terminal CARD or DEATH domain. The multiplicity of the A. queenslandica NLR genes and the high variability across the N- and C-terminal domains are consistent with involvement in immunity. We also provide new insight into the evolution of NLRs in invertebrates through comparative genomic analysis of multiple metazoan and nonmetazoan taxa. Specifically, we demonstrate that the NLR gene family appears to be a metazoan innovation, characterized by two major gene lineages that may have originated with the last common eumetazoan ancestor. Subsequent lineage-specific gene duplication, gene loss and domain shuffling all have played an important role in the highly dynamic evolutionary history of invertebrate NLRs.
Keywords: innate immunity, invertebrate genomics, NACHT domain, Porifera
Introduction
All animals have an innate immune system that differentiates self from nonself by using diverse, genome-encoded pattern recognition receptors (PRRs) (Hoffmann et al. 1999; Kurtz and Armitage 2006; Rosenstiel et al. 2009). PRRs recognize and bind to characteristic molecules that identify whole classes of microorganisms (Janeway and Medzhitov 2002); these are termed interchangeably as microbial- or pathogen-associated molecular patterns (MAMPs/PAMPs) (Boller and Felix 2009). A PRR binding event typically triggers a signaling cascade that results in the transcription of immune response effector genes encoding products such as antibacterial, antifungal, and antiviral proteins (Janeway and Medzhitov 2002). Several classes of PRRs are conserved among divergent animal lineages, both vertebrates and invertebrates (Sarrias et al. 2004; Yoneyama and Fujita 2009; Messier-Solek et al. 2010; Hansen et al. 2011; Buckley and Rast 2012). Notable among these are the Nucleotide-binding domain and Leucine-rich Repeat-containing genes (NLRs, known also as Nod-like receptors for nucleotide-binding oligomerization domain receptors).
The NLRs are a family of intracellular sentinels, capable of detecting a wide range of MAMPs that includes bacterial and viral RNA, bacterial flagellin, and peptidoglycan components (Kaparakis et al. 2007; Boller and Felix 2009; von Moltke et al. 2013). The cytosolic localization of NLRs suggests that they could respond to bacteria that escape extracellular detection and manage to invade the cell and also to bacterial products that are present in the cell following phagocytosis (Martinon et al. 2009; von Moltke et al. 2013). In addition to the detection of intracellular MAMPs, NLRs sense endogenous danger-associated molecular patterns (Sansonetti 2006). These are signals produced by the host following injury or cellular stress and include uric acid crystals, reactive oxygen species, and changes in ATP levels or intracellular potassium concentration (Boller and Felix 2009; Stuart et al. 2013). Most bacteria, however, are not pathogenic and many are in fact beneficial to the host (McFall-Ngai et al. 2013). How multicellular hosts differentiate between microbial friend and foe remains enigmatic, and it has been suggested that NLRs likely play an important role in mediating these animal–bacterial interactions (Robertson et al. 2012; Robertson and Girardin 2013).
Metazoan NLRs are defined by the presence of both a central NACHT (NAIP, CIITA, HET-E, and TP1) domain and a series of C-terminal leucine-rich repeats (LRRs) (Ting, Harton et al. 2008). The highly conserved central NACHT domain is a STAND P-loop NTPase that mediates self-oligomerization in the presence of ATP, hence it is also known interchangeably as a nucleotide oligomerization domain (NOD) or nucleotide-binding domain (NBD) (Koonin and Aravind 2002; Wilmanski et al. 2008). The C-terminal LRRs form the ligand sensing region, although it is presently unclear whether the LRRs interact with MAMPs/PAMPs directly or via an intermediate (Proell et al. 2008; Istomin and Godzik 2009). The LRRs also appear to play an autoregulatory role in maintaining the NLR in an inactive formation until a specific signal is detected (Martinon et al. 2002; Ting, Willingham et al. 2008).
In addition to these two highly conserved domains, the characteristic metazoan tripartite NLR architecture is completed by the presence of an N-terminal effector domain. The vast majority of N-terminal domains identified to date belong to the death-fold superfamily, which includes the caspase recruitment domain (CARD), pyrin domain (PYD), and DEATH domain (Laing et al. 2008; Proell et al. 2008; Messier-Solek et al. 2010). The N-terminal domain is responsible for homotypic protein–protein interactions that initiate immune signaling pathways (Kufer 2008; Shaw et al. 2010). The activation of at least some NLRs results in the formation of a multiprotein complex called an inflammasome and the activation of caspase-1, which ultimately leads to the production of inflammatory cytokines or the induction of apoptosis or pyroptosis (Schroder and Tschopp 2010; Aachoui et al. 2013). In addition, NLRs are also capable of activating NF-κB and p38 MAPK-dependent signaling via interactions with receptor interacting protein-2 (RIP2) kinase (Ting et al. 2010).
Vertebrate NLRs have been the focus of intense research, not least because of the link between dysfunctional NLRs and several human diseases (Franchi et al. 2009; Davis et al. 2011; Dunne 2011). Invertebrate NLRs, by contrast, are poorly characterized, probably in part due to the absence of NLRs in the classical invertebrate model organisms, Drosophila melanogaster and Caenorhabditis elegans (Zhang et al. 2010). Some existing ambiguity in the invertebrate NLR literature arises from the fact that plants have a similar family of PRRs with a tripartite architecture comprising a central NBD, C-terminal LRRs responsible for MAMP binding, and diverse N-terminal domains involved in signaling (Maekawa et al. 2011; Yue et al. 2012). However, the central NBD of plant NLRs is an NB-ARC domain—also a STAND P-loop NTPase—in place of the NACHT domain (Leipe et al. 2004). Despite the remarkable structural and functional similarities shared by plant and animal NLR families, these properties are thought to represent convergent evolution rather than shared ancestry (Yue et al. 2012). In addition, many of the C-terminal repeats associated with genes of the NACHT family are common to the NB-ARC family (Leipe et al. 2004).
Previous reports of metazoan NLRs have not necessarily been restricted to bona fide NLRs composed of NACHT and LRR domains but have more broadly discussed all genes containing a NACHT or an NB-ARC domain (Lange et al. 2011; Hamada et al. 2012). However, the shared characteristics of the NACHT and NB-ARC families is a potential source of confusion that has resulted in conflicting numbers of NACHT- and NB-ARC-containing genes being recorded within the same study, thus confounding interpretations of the reported NLR genome complements in the cnidarian Acropora digitifera and in the demosponge Amphimedon queenslandica (Hamada et al. 2012). Considering both NACHT and NB-ARC families together makes it difficult to ascertain the phylogenetic distribution of NLRs specifically and thus to discuss their origin and evolution. In this study, we thus focus only on bona fide invertebrate NLRs.
Indeed, a better understanding of the origin and evolution of this pivotal PRR gene family in animals awaits data from a greater number of animal lineages. Here, we increase the breadth of data by providing comprehensive annotation of the NLR genes in the sponge (phylum Porifera) A. queenslandica—a basal metazoan with a fully sequenced genome (Srivastava et al. 2010). The phylogenetic status of poriferans as sister group to the Eumetazoa makes them ideal for elucidating the origin and evolution of animal innate immunity, because traits shared between sponges and other animals likely reflect shared inheritance from the last common animal ancestor (Philippe et al. 2009; Srivastava et al. 2010). To reflect more broadly on the origin and evolution of this gene family, we also report the presence or absence of NLRs in several other organisms including nonmetazoan eukaryotes and eumetazoans. To avoid confusion with other NBD-containing genes that do not have LRRs, we at all times adhere strictly to the universal nomenclature proposed by Ting, Harton et al. (2008) and accepted by the HUGO Gene Nomenclature Committee. Specifically, we restrict ourselves to the definition of the acronym NLR as denoting a “Nucleotide-binding domain and Leucine-rich Repeat”-containing gene, as this highlights the two defining evolutionarily conserved domains while reflecting the (non-homologous) similarity of animal NLRs to the plant NLRs (Ting, Harton et al. 2008).
Results and Discussion
NLRs Are Abundant in the Amphimedon Genome and Likely Already Existed in the Last Common Ancestor to the Metazoa
Searches based upon the Pfam hidden Markov model (HMM) of the NACHT domain (PF05729) identified a total of 244 NACHT domain-containing gene models in the genome of A. queenslandica. A complementary Amphimedon-based HMM generated by us did not reveal any additional NACHT domains, and it is unlikely that any were missed given that the relaxed specificity that we used for the searches retrieved many non-NACHT P-loop NTPases. Of the 244 gene models that contain specifically a NACHT domain, 93 represent single genes on small contigs with high nucleotide sequence identity (>95%) to other NACHT domain-containing genes models, incomplete NACHT domains, or NACHT-only gene models (supplementary file S1, Supplementary Material online). This leads us to believe that these 93 models are more likely to represent erroneously assembled allelic variants rather than independent loci, and thus we excluded them from our final predictive count of 151 NACHT domain-containing genes in the A. queenslandica genome (table 1). Not surprisingly then, this number differs from that previously reported for A. queenslandica (Lange et al. 2011; Hamada et al. 2012); these differences are further confounded by the lack of clear discrimination between NACHT and other STAND P-loop NTPase domains in one of the prior analyses (Hamada et al. 2012).
Table 1.
The Full Complement of 151 Predicted NACHT Domain-Containing Genes Encoded by the Genome of Amphimedon queenslandica.
| Clade ID | Contig | Gene Model Codea | Assigned Nameb |
|---|---|---|---|
| AqDEATH-NACHT | Contig5503 | Aqu1.202261 | AqDN1 |
| AqNLR clade A | Contig8999 | Aqu1.204459 | AqNLRX5 |
| AqDEATH-NACHT | Contig9585 | Aqu1.204933 | AqDN2 |
| AqNLR clade A | Contig9959 | Aqu1.205319 | AqNLRX6 |
| AqDEATH-NACHT | Contig10075 | Aqu1.205456 | AqDN3 |
| AqNLR clade A | Contig10163 | Aqu1.205553-snap.11240 | AqNLRC1 |
| AqDEATH-NACHT | Contig10378 | Aqu1.206247 | AqDN5 |
| AqDEATH-NACHT | Contig10379 | Aqu1.205832 | AqDN4 |
| AqNLR clade A | Contig10757 | Aqu1.206272 | AqNLRX7 |
| AqNLR clade C | Contig10761 | Aqu1.206280 | AqNLRD1 |
| AqNLR clade A | Contig10879 | Aqu1.206401 | AqNLRX8 |
| AqNLR clade A | Contig10879 | hom.g7908.t1 | AqNLRC2c |
| AqNLR clade A | Contig11119 | Aqu1.206703 | AqNLRX9 |
| AqNLR clade A | Contig11252 | Aqu1.206887-hom.g8479.t1 | AqNLRD10c |
| AqNLR clade A | Contig11309 | Aqu1.206954 | AqNLRX10 |
| AqNLR clade A | Contig11352 | Aqu1.207007 | AqNLRX11 |
| AqNLR clade A | Contig11422 | Aqu1.207127 | AqNLRX12 |
| AqNLR clade A | Contig11546 | Aqu1.207322 | AqNLRX13 |
| AqNLR clade A | Contig11679 | Aqu1.207562 | AqNLRX14 |
| AqNLR clade A | Contig11719 | Aqu1.207649 | AqNLRX15 |
| AqNLR clade A | Contig11725 | Aqu1.207664 | AqNLRX16 |
| AqNLR clade A | Contig11740 | Aqu1.207697 | AqNLRX17 |
| AqDEATH-NACHT | Contig11763 | Aqu1.207748 | AqDN6 |
| AqNLR clade A | Contig11787 | Aqu1.207797 | AqNLRX18 |
| AqNLR clade A | Contig11837 | Aqu1.207890-hom.g9670.t1 | AqNLRC3c |
| AqNLR clade A | Contig11954 | Aqu1.208150 | AqNLRD11 |
| AqNLR clade A | Contig11961 | Aqu1.208167 | AqNLRX19 |
| AqNLR clade A | Contig11972 | Aqu1.208189 | AqNLRX20 |
| AqNLR clade A | Contig12017 | Aqu1.208293 | AqNLRD12c |
| AqNLR clade A | Contig12054 | Aqu1.208371 | AqNLRX21 |
| AqNLR clade A | Contig12055 | Aqu1.208372 | AqNLRX22 |
| AqNLR clade A | Contig12282 | Aqu1.208919 | AqNLRX23 |
| AqNLR clade A | Contig12315 | Aqu1.209017 | AqNLRC4 |
| AqNLR clade A | Contig12346 | Aqu1.209113 | AqNLRC5c |
| AqNLR clade A | Contig12356 | hom.g11149.t1-Aqu0.1446184 | AqNLRX24c |
| AqNLR clade A | Contig12364 | Aqu1.209168 | AqNLRX25 |
| AqNLR clade A | Contig12383 | hom.g11247.t1 | AqNLRD13 |
| AqNLR clade A | Contig12389 | Aqu1.209241 | AqNLRX26 |
| AqDEATH-NACHT | Contig12407 | Aqu1.209295 | AqDN7 |
| AqNLR clade A | Contig12431 | Aqu1.209372-hom.g11418.t1 | AqNLRD14 |
| AqNLR clade A | Contig12433 | Aqu1.209379 | AqNLRX27 |
| AqNLR clade A | Contig12433 | hom.g11429.t1 | AqNLRD15 |
| AqNLR clade A | Contig12489 | Aqu1.209557 | AqNLRX28 |
| AqNLR clade A | Contig12517 | Aqu1.209696 | AqNLRX29 |
| AqNLR clade A | Contig12522 | Aqu1.209720 | AqNLRX30 |
| AqNLR clade A | Contig12541 | Aqu1.209792 | AqNLRX31 |
| AqNLR clade A | Contig12563 | Aqu1.209880 | AqNLRX32 |
| AqNLR clade A | Contig12563 | Aqu1.209876 | AqNLRX33 |
| AqNLR clade A | Contig12595 | hom.g12176.t1 | AqNLRX34 |
| AqNLR clade A | Contig12595 | Aqu1.210033 | AqNLRD16 |
| AqNLR clade A | Contig12612 | Aqu1.210112 | AqNLRX35 |
| AqNLR clade A | Contig12676 | Aqu1.210376 | AqNLRX36 |
| AqNLR clade A | Contig12677 | Aqu1.210377 | AqNLRX37 |
| AqNLR clade C | Contig12691 | aq_ka12691x00220-12691x00230 | AqNLRD2 |
| AqNLR clade A | Contig12692 | Aqu1.210459 | AqNLRX38 |
| AqNLR clade A | Contig12704 | Aqu1.210507 | AqNLRX39 |
| AqNLR clade A | Contig12733 | Aqu1.210691 | AqNLRX40 |
| AqNLR clade A | Contig12734 | Aqu1.210692 | AqNLRX41 |
| AqNLR clade A | Contig12746 | Aqu1.210730 | AqNLRX42 |
| AqNLR clade A | Contig12746 | Aqu1.210737 | AqNLRD17 |
| AqNLR clade A | Contig12749 | Aqu1.210748-hom.g12993.t1 | AqNLRX43 |
| AqNLR clade A | Contig12812 | snap.24323-1447583 | AqNLRX44 |
| AqNLR clade A | Contig12829 | Aqu1.211218 | AqNLRX45 |
| AqNLR clade A | Contig12852 | Aqu1.211347 | AqNLRX46 |
| AqNLR clade A | Contig12852 | Aqu1.211348 | AqNLRD18 |
| AqNLR clade A | Contig12853 | Aqu1.211358 | AqNLRX47 |
| AqNLR clade A | Contig12862 | Aqu0.1447787 | AqNLRC6 |
| AqNLR clade A | Contig12862 | Aqu1.211413 | AqNLRX48 |
| AqNLR clade A | Contig12883 | Aqu1.211532 | AqNLRX49 |
| AqNLR clade A | Contig12887 | Aqu0.1447879-Aqu1.211549 | AqNLRD19c |
| AqNLR clade A | Contig12894 | Aqu1.211616-snap.25520 | AqNLRX50 |
| AqNLR clade A | Contig12897 | Aqu1.211634 | AqNLRX51 |
| AqNLR clade A | Contig12934 | Aqu1.211923 | AqNLRX52 |
| AqNLR clade A | Contig12934 | Aqu0.1448139 | AqNLRD20 |
| AqNLR clade A | Contig12950 | Aqu1.212035 | AqNLRX53 |
| AqNLR clade A | Contig12951 | Aqu0.1448222-snap.26675 | AqNLRX54 |
| AqNLR clade A | Contig12955 | Aqu1.212081-snap.26736 | AqNLRC7 |
| AqNLR clade A | Contig12956 | Aqu1.212082 | AqNLRC8 |
| AqNLR clade A | Contig12968 | Aqu1.212194-hom.g14686.t1 | AqNLRX55 |
| AqNLR clade A | Contig12968 | Aqu1.212193 | AqNLRD21 |
| AqNLR clade A | Contig12974 | Aqu1.212264 | AqNLRX56 |
| AqNLR clade A | Contig12983 | Aqu1.212346 | AqNLRD22 |
| AqNLR clade A | Contig12996 | Aqu1.212453 | AqNLRX57 |
| NACHT-WD40 | Contig13075 | hom.g15978.t1 | AqNWD40ii |
| AqNLR clade A | Contig13099 | Aqu1.213597 | AqNLRX58 |
| AqNLR clade A | Contig13103 | Aqu1.213662 | AqNLRX59 |
| AqNLR clade C | Contig13105 | aq_ka13105x00240 | AqNLRD9 |
| AqNLR clade C | Contig13105 | Contig13105:47,116-50,520 | AqNLRX2 |
| AqNLR clade C | Contig13105 | Aqu1.213698 | AqNLRX3 |
| AqNLR clade C | Contig13105 | Aqu1.213699-Aqu1.213700 | AqNLRX4 |
| AqNLR clade A | Contig13113 | Aqu1.213834 | AqNLRX60 |
| AqNLR clade A | Contig13117 | hom.g16620.t1 | AqNLRD23 |
| AqNLR clade A | Contig13133 | Aqu1.214051 | AqNLRD24 |
| AqNLR clade A | Contig13133 | hom.g16827.t1 | AqNLRD25 |
| AqNLR clade A | Contig13134 | Aqu1.214053-snap.31711 | AqNLRD26 |
| AqNLR clade A | Contig13140 | Aqu1.214168-snap.31947 | AqNLRX61 |
| AqNLR clade A | Contig13141 | Aqu1.214170 | AqNLRX62 |
| AqNLR clade A | Contig13142 | hom.g16965.t1 | AqNLRX63 |
| AqNLR clade A | Contig13142 | Aqu1.214186 | AqNLRX64 |
| AqNLR clade C | Contig13153 | aq_ka13153x00250 | AqNLRD6 |
| AqNLR clade C | Contig13153 | Aqu0.1449710 | AqNLRD7 |
| AqNLR clade C | Contig13153 | Aqu0.1449711 | AqNLRD8 |
| NACHT-WD40 | Contig13169 | hom.g17436.t1 | AqNWD40iii |
| AqNLR clade A | Contig13182 | hom.g17680.t1 | AqNLRC9c |
| AqNLR clade A | Contig13206 | Aqu1.215215 | AqNLRX65 |
| AqNLR clade A | Contig13206 | Aqu1.215210 | AqNLRD27 |
| AqNLR clade A | Contig13206 | hom.g18193.t1 | AqNLRD28 |
| AqNLR clade A | Contig13234 | hom.g18804.t1 | AqNLRX66 |
| AqNLR clade A | Contig13234 | Aqu1.215789 | AqNLRX67 |
| AqNLR clade A | Contig13234 | Aqu1.215792 | AqNLRX68 |
| AqNLR clade A | Contig13234 | Aqu1.215790 | AqNLRX69 |
| AqNLR clade A | Contig13234 | Aqu1.215785-snap.35932 | AqNLRC10 |
| AqNLR clade A | Contig13234 | Aqu1.215794 | AqNLRC11 |
| AqNLR clade A | Contig13245 | hom.g19060.t1 | AqNLRX70 |
| AqDEATH-NACHT | Contig13309 | Aqu1.217513 | AqDN8 |
| AqNLR clade A | Contig13332 | Aqu1.218194 | AqNLRX71 |
| AqNLR clade A | Contig13332 | Aqu1.218191 | AqNLRX72 |
| AqNLR clade A | Contig13332 | Aqu1.218192 | AqNLRX73 |
| AqNLR clade C | Contig13337 | Aqu1.218328 | AqNLRD3 |
| AqNLR clade A | Contig13346 | hom.g21949.t1 | AqNLRX74c |
| AqNLR clade A | Contig13346 | Aqu0.1452529-snap.42481 | AqNLRC12 |
| AqNLR clade A | Contig13346 | hom.g21925.t1-snap.42422 | AqNLRD29 |
| AqNLR clade A | Contig13354 | Aqu1.218980 | AqNLRX75 |
| AqNLR clade A | Contig13354 | Aqu1.218975 | AqNLRX76 |
| AqNLR clade A | Contig13354 | ab.g20734.t1 | AqNLRD30 |
| AqNLR clade A | Contig13358 | Aqu1.219134 | AqNLRX77 |
| AqNLR clade A | Contig13377 | Aqu1.219760 | AqNLRX78 |
| AqNLR clade A | Contig13377 | Aqu1.219777 | AqNLRX79 |
| AqNLR clade A | Contig13377 | Aqu1.219767-snap.44836 | AqNLRD31 |
| AqNLR clade A | Contig13379 | Aqu1.219814 | AqNLRX80 |
| AqNLR clade A | Contig13382 | Aqu0.1453371-hom.g23298.t1 | AqNLRC13c |
| AqNLR clade C | Contig13382 | aq_ka13382x00520 | AqNLRD4 |
| AqNLR clade C | Contig13382 | Aqu0.1453359 | AqNLRD5 |
| NACHT-WD40 | Contig13402 | hom.g24038.t1 | AqNWD40i |
| AqDEATH-NACHT | Contig13409 | Aqu1.220886 | AqDN10 |
| AqDEATH-NACHT | Contig13409 | Aqu1.220887 | AqDN11 |
| AqDEATH-NACHT | Contig13409 | Aqu1.220885 | AqDN9 |
| AqNLR clade A | Contig13412 | Aqu1.220996 | AqNLRX81 |
| AqNLR clade A | Contig13430 | hom.g25320.t1 | AqNLRX82c |
| AqNLR clade A | Contig13430 | Aqu1.221813 | AqNLRX83 |
| AqNLR clade A | Contig13430 | Aqu1.221810 | AqNLRC14c |
| AqNLR clade A | Contig13430 | hom.g25317.t1 | AqNLRC15 |
| AqNLR clade A | Contig13430 | snap.49468-Aqu1.221803 | AqNLRC16c |
| AqNLR clade A | Contig13430 | snap.49466-Aqu0.1454627 | AqNLRD32 |
| AqNLR clade B | Contig13467 | Aqu1.223871-Aqu0.1455876 | AqNLRX1 |
| AqNLR clade A | Contig13472 | Aqu1.224254 | AqNLRX84 |
| AqNLR clade A | Contig13473 | Aqu1.224303 | AqNLRX85 |
| AqNLR clade A | Contig13512 | Aqu1.228172 | AqNLRX86 |
| AqDEATH-NACHT | Contig13514 | Aqu1.228453 | AqDN12 |
| AqDEATH-NACHT | Contig13514 | Aqu1.228454 | AqDN13 |
| AqNLR clade A | Contig13518 | Aqu1.229088 | AqNLRX87 |
Note.—The list comprises 3 genes characterized by a WD40-NACHT domain combination, 13 genes characterized by a DEATH-NACHT domain combination, and 135 genes characterized by a NACHT-LRR domain combination. These latter 135 genes represent bona fide NLRs and include 48 characteristic tripartite NLR genes that also contain an N-terminal CARD or DEATH effector domain.
aGene model codes were obtained from the Joint Genome Institute Amphimedon queenslandica genome browser accessible at www.metazome.net/amphimedon (last accessed April 20, 2013) and include gene models derived from multiple different gene prediction algorithms and indicated as ab.g (Augustus ab initio); aq_ka (PASA and Augustus); snap (SNAP ab initio); hom (Augustus homology); Aqu0; Aqu1. Gene models that have been concatenated are separated by a hyphen.
bAqNLR nomenclature is based on the convention proposed by Ting, Harton et al. (2008) and accepted by the HUGO Gene Nomenclature Committee. Therefore, the name AqNLRD, for example, indicates a tripartite architecture of DEATH-NACHT-LRRs. Domain architecture: D, Death domain; C, Card domain; X, No N-terminal domain; N, NACHT domain.
cA transmembrane domain signal was detected at the N-terminus of this gene by TMHMM Server v.2.0 – CBS (available from: www.cbs.dtu.dk/services/TMHMM/, last accessed April 25, 2013).
Among the 151 NACHT domain-containing gene models that can confidently be assigned to discrete loci (table 1), we identified 135 bona fide NLR genes as defined by the presence of both a NACHT and an LRR domain (following Ting, Harton et al. 2008). We designate these as AqNLR genes. Of these 135 AqNLRs, 48 have the characteristic tripartite architecture that also includes an N-terminal CARD or DEATH domain. Although the presence of NACHT domain-containing genes in the A. queenslandica genome has previously been recognized, the gene numbers and domain architectures were either not supplied and thus not available for comparison (Lange et al. 2011) or were confounded by a lack of discrimination between the NACHT and other STAND P-loop NTPase domains (Hamada et al. 2012). We provide here, therefore, the first complete list of bona fide NLR genes in the basal metazoan phylum Porifera and also the first confirmed report of NLRs outside the eumetazoan lineage. This identification of NLRs in the genomes of both sponges and eumetazoans suggests that at least one ancestral NLR gene was already present in the last common ancestor of all animals.
NLR Genes Encoded by the Amphimedon Genome Have Diverse Phylogenetic Histories and Diverse LRRs
Three of the 151 A. queenslandica NACHT domain-containing genes comprise a NACHT domain coupled with C-terminal WD40 repeats. The NACHT domains of the NACHT-WD40 genes did not align well with the remaining 148 NACHT domains and thus were excluded from further analysis. Phylogenetic analyses of the remaining 148 A. queenslandica NACHT domain-containing genes reveal that they group into four discrete clades with high statistical support; the 135 AqNLRs are split among three of these clades (fig. 1). We designate these four clades as the DEATH-NACHT clade and AqNLR clades A, B, and C.
Fig. 1.
Phylogenetic analysis of the Amphimedon queenslandica NACHT domain-containing genes. The tree presented is a midpoint-rooted Bayesian tree, with branch lengths representing the number of substitutions per site. Posterior probabilities and ML bootstrap support values greater than 50% are indicated. AqNLRs form three discrete clusters: AqNLR clade A, AqNLR clade B, and AqNLR clade C. A small subset of the 122 AqNLR clade A genes was used in the final analysis. Summary domain architectures characteristic of each major clade are shown below the clade name. The intron/exon organizations of individual AqDEATH-NACHT clade, AqNLR clade B, and AqNLR clade C genes are depicted to the right of the tree. Exons are represented by boxes and are drawn to scale. Introns are represented as lines between exons; they range from 45 bp to 7 kbp in size but are all depicted as the same size (i.e., not to scale). Assembly gaps, represented by the line breaks, range from 386 bp to 1.975 kbp. Refer to supplementary file S3, Supplementary Material online, for the alignment used in the analyses.
AqNLR clade A is a large group of 122 genes that make up the vast majority of the A. queenslandica NACHT domain complement (figs. 1 and 2). The AqNLRs in this clade have LRRs that are recognized only by HMM profiles in the Superfamily (SSF52047) and Gene3D (G3DSA:3.80.10.10) protein structure libraries. Interestingly, these HMMs are based on the crystal structure of the LRR domain of the ribonuclease inhibitor-like (RNI-like) superfamily, unlike the sequence-based Pfam HMM models for LRRs. An N-terminal CARD (AqNLRC) or DEATH (AqNLRD) domain is encoded by almost one-third of the clade A genes (38 out of 122), but the remaining two-thirds lack the tripartite domain architecture typical of human NLRs and instead comprise just the NACHT-LRR domain combination (AqNLRX). In those genes where an N-terminal domain exists, its precise identity does not predict phylogenetic position of the gene; that is, AqNLRCs and AqNLRDs do not form discrete lineages within the clade (fig. 2). Although this pattern points to the role of N-terminal domain shuffling, gain, and loss in the evolution of the clade A AqNLRs, it comes with the caveat that the models for the clade A genes were frequently situated next to assembly gaps or at the edge of assembled scaffolds. Where possible, we interrogated adjacent genomic sequence N-terminal to an AqNLRX; however, the current poor quality of the assembly at many of these loci reduces our confidence in the reliability of models in this clade and suggests that the actual number of tripartite NLRs in the genome may be greater.
Fig. 2.
Phylogenetic analysis of the Amphimedon queenslandica NLR clade A gene expansion of 122 genes that make up the majority of the A. queenslandica NACHT domain complement. This unrooted Bayesian tree was constructed from an alignment of the A. queenslandica NACHT domains. Posterior probabilities for the major clades are indicated. Neither the presence nor the precise identity of an N-terminal domain—CARD (AqNLRC, shown in blue), DEATH (AqNLRD, shown in red), or absent (AqNLRX, shown in black)—appears to predict phylogenetic position of the gene. The alignment used to generate this phylogenetic tree is available upon request.
Clade A also contains three gene models that are predicted in the current genome version (gene models version Aqu1; Srivistava et al. 2010) to contain ankyrin (ANK) repeats N-terminal to the NACHT domain. Our closer inspection of these gene models indicates that the ANK repeats are more likely to be part of an adjacent gene upstream of the AqNLR, thus we suggest their inclusion in these gene model is erroneous and we exclude the ANK repeats from our characterization of these three AqNLR clade A genes (fig. 2 and table 1). Further, the combination of a NACHT domain coupled with C-terminal ANK repeats, which was previously reported as characteristic of the A. queenslandica NBD gene expansion (Hamada et al. 2012), cannot be confirmed at all in the genome. Instead, it is the bona fide NLRs (NACHT-LRR) that have undergone a major expansion, rather than the NACHT-ANK domain combination as previously concluded by Hamada et al. (2012).
The expansive AqNLR clade A is the sister group to AqNLR clade B (fig. 1). AqNLR clade B consists of a single NLR that lacks a known N-terminal domain but has LRRs that are recognized readily by the sequenced-based Pfam LRR clan HMMs (CL0022). Quite divergent from AqNLR clades A and B, AqNLR clade C contains 12 genes, 9 of which are tripartite NLRs characterized by an N-terminal DEATH domain, and with C-terminal LRRs also readily recognized by the Pfam LRR clan HMMs (CL0022). The NACHT domain and LRRs of the clade C NLRs are all encoded on one exon, whereas the LRRs of the single clade B NLR span multiple exons (fig. 1). It is worth highlighting that the exon/intron organization of the clade B gene AqNLRX1 reflects that of the human NLRC1 and NLRC2 genes and of Capitella NLRC - Capca1|214069. Similarly, the exon/intron organization of the clade C NLRs reflects that of Lottia NLRD - Lotgi1|152683, Capitella NLRC - Capca1|207210, and Nematostella NLRX - Nemve1|203213. The AqDEATH-NACHT clade—sister to AqNLR clades A and B—contains 13 genes that all share a common DEATH-NACHT domain structure defined by the absence of any detectable LRRs. Despite falling within the AqDEATH-NACHT clade, no DEATH domain was detected in the genomic sequence N-terminal to the NACHT domains of AqDN2 and AqDN6 (fig. 1). Notably, AqDN2 is a gene located on a small contig, and AqDN6 contains a small assembly gap in the exon where the DEATH domain might occur. In contrast with the AqNLR clade A gene models, those of the AqDEATH-NACHT clade, AqNLR clade B and C were mostly complete, with the exception of a few that contained only small assembly gaps (fig. 1). Importantly, there were no issues relating to poor assembly for the AqNLRX genes in AqNLR clades B and C, suggesting that our inability to identify a N-terminal DEATH domain in these particular genes reflects a genuine absence of this domain, rather than limitations of gene models (fig. 1).
The expansion of the AqNLR gene family (relative, for example, to mammalian NLRs) reflects similar reports of large numbers of NLRs—and indeed other PRRs—in other marine organisms including the scleractinian coral Acr. digitifera, the sea urchin Strongylocentrotus purpuratus, and the cephalochordate Branchiostoma floridae (supplementary file S2, Supplementary Material online; Messier-Solek et al. 2010; Lange et al. 2011; Hamada et al. 2012). The relatively short branch lengths of the clade A AqNLRs in particular (figs. 1 and 2) suggest a recent history of rapid expansion and diversification. Further, the facts that AqNLR gene models have proven difficult to predict (table 1 indicates multiple gene model versions that we have interrogated to identify the full complement of AqNLRs) and that AqNLR RNASeq data are equally difficult to assemble (personal observation) together suggest that there may be extensive intraspecific polymorphism in these genes. This would be consistent with reports of high intraspecific polymorphism in other PRR gene families, such as Toll-like receptors (TLRs) and Scavenger Receptor Cysteine-Rich genes, in the sea urchin S. purpuratus (Pancer 2001; Messier-Solek et al. 2010). Equally interesting is the observation that the AqNLR genes of clade A have LRRs that cannot be retrieved through searches based only on genomic sequence similarity, but that can only be recognized through conserved structural features, suggesting a high level of divergence from the Pfam LRR clan (CL0022). Furthermore, these LRRs display great within-clade diversity, ranging from close sequence identity to being unalignable with each other. The variation in the clade A AqNLRs, and their abundance, leads us to hypothesize that their evolution is being driven by a large and dynamic suite of ligand-binding conditions. Similar observations have been made for evolution of the large family of innate immunity TLR genes that display highly variable LRRs in echinoderms (Buckley and Rast 2012).
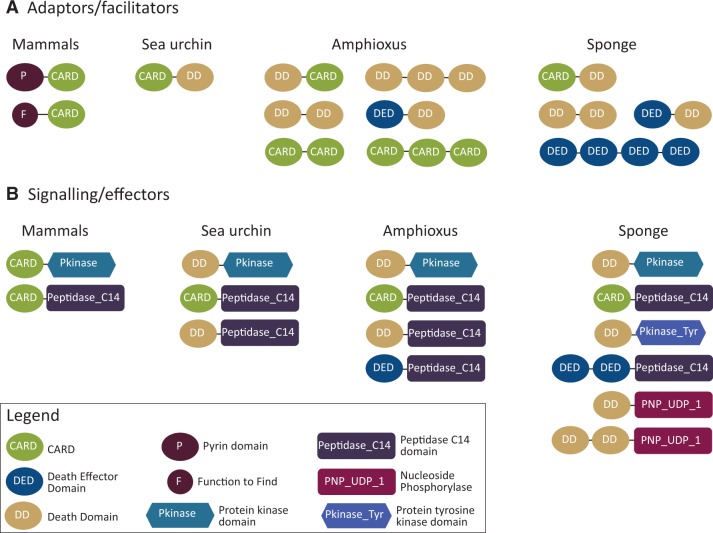
NLRs exert their functions through interactions of the N-terminal effector domain with downstream adaptor proteins, effector kinases, and caspases, often leading to inflammatory or apoptotic responses (Kaparakis et al. 2007; Schroder and Tschopp 2010; Damm et al. 2013). The N-terminal effector domain variation in AqNLR clade A, which includes both DEATH and CARD domains, provides an added level of complexity to the signaling potential of this large subfamily. It is noteworthy that the A. queenslandica genome contains a corresponding expansion in the variety of death-fold domain combinations that potentially could interact with the AqNLRs as downstream adaptor and effector proteins (fig. 3). Although this intriguing link requires empirical verification, there are certainly a great many death-fold domain-containing genes (∼460, excluding those associated with NLRs) in the A. queenslandica genome. This reflects a similar expansion of both NLR genes (n = 118) and death-fold domain-containing genes (n = 541) in the Branchiostoma genome (fig. 3) (Huang et al. 2008; Messier-Solek et al. 2010), where it has been proposed that the co-expansion and diversification of NLRs and death-fold domains are suggestive of enhanced signaling potential (Messier-Solek et al. 2010).
Fig. 3.
Architectures of potential NLR adaptor and signaling/effector proteins encoded by the Amphimedon queenslandica (sponge) genome, in comparison with those identified in other animal genomes. Data for mammals, sea urchin, and amphioxus have been copied directly from figure 3 in Messier-Solek et al. (2010). These proteins could be involved in NLR signaling pathways via homotypic interactions of the death-fold domains. The A. queenslandica (sponge) list is a conservative selection based on structural similarity to the mammalian ASC adaptor protein (Pyrin-CARD) and RIP2 kinase (Protein kinase-CARD). Not all A. queenslandica death-fold domain-containing gene models are depicted. The DEATH-UDP/PNP domain combination is a novel architecture identified in the A. queenslandica genome. Although proteins of this structure are not known to be involved in NLR signaling, they were included because the UDP/PNP domain has also been identified at the N-terminus of NLRs in Acropora digitifera (fig. 4) (Hamada et al. 2012).
The much smaller sizes of the other AqNLR clades (fig. 1) suggest that genes in these clades might have evolved divergent functional specializations relative to the genes in AqNLR clade A. Though we cannot predict the precise functions of the AqNLRs based on phylogenetics alone, it has become increasingly evident in other animals that some NLRs have evolved roles that go beyond pattern recognition (see reviews by Kufer and Sansonetti 2011 and Bonardi et al. 2012). For example, some human NLRs, such as CIITA, have no known role as PRRs but act as signaling platforms that activate other facets of the vertebrate immune system (Kufer and Sansonetti 2011). Thus, the presence of LRRs does not necessarily denote a role in MAMP binding, and this should be taken into consideration in future studies of NLR subfamilies in invertebrates. The absence of LRRs in genes of the AqDEATH-NACHT clade means that these genes are not strictly bona fide NLRs (fig. 1). However, their domain architecture and phylogenetic relationship suggest that their functions may be closely linked to those of the true AqNLRs, perhaps through their capacity for interactions involving oligomerization via the NACHT domain. Indeed, human NLRP10 is the only human NLR protein that similarly lacks LRRs, and it has been proposed to have a role as a regulatory or adaptor protein (see review by Damm et al. 2013).
The multiplicity and high level of overall variation of the A. queenslandica NLRs are consistent with an involvement in immunity (Hibino et al. 2006; Messier-Solek et al. 2010; Lange et al. 2011; Buckley and Rast 2012). Furthermore, the possibility of roles as regulatory proteins, the effector domain diversity, and the expansion of potential downstream components all lead us to hypothesize that sponges have an immune system with the capacity to recognize a vast array of ligands, coupled with complex regulatory potential (Messier-Solek et al. 2010).
NLRs Appear to be a Metazoan-Specific Invention Characterized by Two Major Gene Lineages That Each Contains Multiple Lineage-Specific Expansions
In addition to the AqNLRs, we report here for the first time bona fide NLRs in the genomes of other metazoan taxa: the polychaete Capitella teleta (n = 55); the molluscs Lottia gigantea (n = 1), Crassostrea gigas (n = 1), and Pinctada fucata (n = 45); and arthropods Strigamia maritima (n = 2) and Nasonia vitripennis (n = 1) (supplementary file S2, Supplementary Material online). In contrast, although we identified NACHT domains in the genomes of the placozoan Trichoplax adhaerens, the ctenophore Mnemiopsis leidyi, various arthropods (see supplementary file S2, Supplementary Material online), and the urochordate Oikopleura dioica, these are always in association with ANK, tetratricopeptide (TPR), or WD-40 repeats, and never with LRR domains. Thus, we find no evidence for the existence of bona fide NLRs in the genomes of those animals (supplementary file S2, Supplementary Material online).
A substantial gap in understanding the evolutionary origin of NLRs was not addressed in previously published studies because no nonmetazoan eukaryote genomes were included for comparison (Lange et al. 2011; Hamada et al. 2012). We therefore interrogated the genomes of multiple nonmetazoan eukaryotes (supplementary file S2, Supplementary Material online) in search of conserved NLR domain architectures. We identified multiple NACHT domains in the genomes of the holozoans Capsaspora owczarzaki, Salpingoeca rosetta, and Monosiga brevicolis and the non-holozoan eukaryotes Entamoeba histolytica, Thalassiosira pseudonana, Phytophthora ramorum, Toxoplasma gondii, Podospora aserina, and Dictyostelium pupureum, but again only in association with ANK, TPR, or WD-40 repeats. Thus, we conclude that bona fide NLRs appear not to exist outside of the Metazoa, including in the sister group to the metazoans, the choanoflagellates (represented here by Sal. rosetta and M. brevicolis). As such, we propose that NLRs are likely a metazoan-specific invention. The conservation of this ancient innate immune gene family in multiple animal lineages since the last common ancestor of all animals, combined with the absence of the gene family in choanoflagellates, suggests an important role for NLRs in the origin and evolution of metazoan multicellularity.
Previous studies have proposed that the evolutionary history of this ancient animal immune gene family has been characterized by lineage-specific expansions through multiple rounds of tandem gene duplication as well as by gene losses occurring independently in multiple taxa (Zhang et al. 2010; Lange et al. 2011; Hamada et al. 2012). The more recent phylogenetic analyses (Lange et al. 2011; Hamada et al. 2012) were not focused exclusively on bona fide NLRs but instead discussed the broader NBD gene complex; Hamada et al. (2012), in particular, did not discriminate between NACHT domain- and NB-ARC domain-containing genes. This lack of discrimination complicates discussion on the origin and evolution of the NLR family in Metazoa, because an NB-ARC-LRR gene architecture has thus far only been recorded in plants and, despite their superficially similarities, the NACHT and NB-ARC domains belong to distinct NTPase families (Leipe et al. 2004; Yue et al. 2012). By focusing specifically on NACHT-LRR gene architectures (the bona fide NLRs as defined by Ting, Harton et al. 2008), our results reveal novel, interesting patterns that were obscured by the inclusion of other genes of the NBD complex in previous analyses.
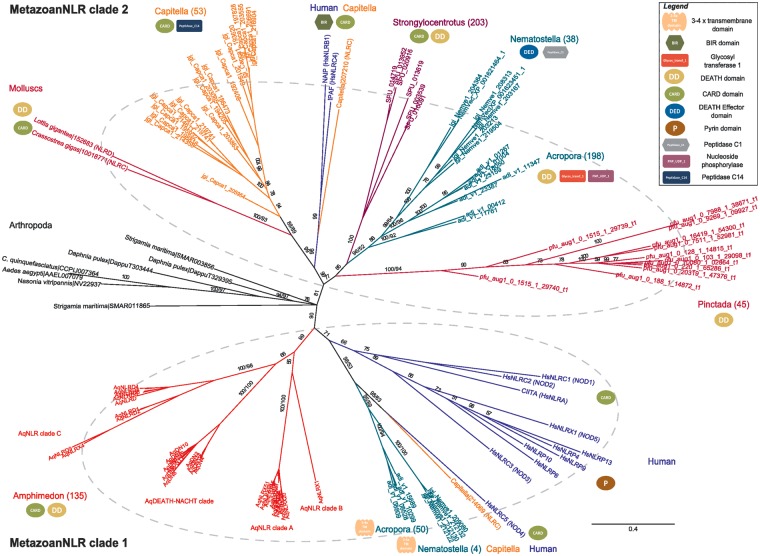
First, our phylogenetic analyses consistently identify two discrete groups of metazoan NLRs (fig. 4 and supplementary file S5, Supplementary Material online). We designate these two groups as MetazoanNLR clades 1 and 2. All of the AqNLRs fall as a single monophyletic group within MetazoanNLR clade 1. This major clade also contains some of the cnidarian (represented by Nematostella vectensis and Acr. digitifera) genes, one of the polychaete annelid Capitella telata genes, all of the human NLRP genes, and most of the human NLRC genes (those known as NODs). The other major grouping, MetazoanNLR clade 2, comprises all of the echinoderm S. purpuratus genes, the majority of C. telata genes, all genes from three molluscan taxa (Cra. gigas, L. gigantea, and P. fucata), the majority of the cnidarian (Acr. digitifera and N. vectensis) genes, and the two well-characterized human NLRs, NAIP and IPAF. The phylogenetic positions of the Pinctada and arthropod NLRs are difficult to resolve. The Pinctada NLR cluster is nested within MetazoanNLR clade 2 in the Bayesian tree (fig. 4) but is positioned as sister group to clade 2 in the maximum likelihood (ML) tree (supplementary file S5, Supplementary Material online). The arthropod cluster is not clearly associated with clade 1 or clade 2 in either the Bayesian (fig. 4) or the ML tree (supplementary file S5, Supplementary Material online). It has previously been reported that the human IPAF and NAIP genes cluster with S. purpuratus NLRs, indicating that the origin of at least these two genes likely predates the evolution of vertebrates (Laing et al. 2008; Zhang et al. 2010). The presence of two divergent NLR clades in the genomes of very divergent metazoan phyla (cnidarian, annelid, and human) strongly suggests that in fact all eumetazoan NLRs originated from at least two genes already present in the last common eumetazoan ancestor, as opposed to the single ancestral gene proposed previously (Zhang et al. 2010; Hamada et al. 2012). Indeed, previous metazoan NLR analyses also provide evidence for divergent NLR clades in N. vectensis and Acr. digitifera, although this was not explicitly discussed as evidence for more than one ancestral gene (Lange et al. 2011; Hamada et al. 2012). Interestingly, the genome of the cnidarian Hydra magnipapillata does not include any genuine NLR genes, but does include multiple DEATH-NACHT genes that cluster phylogenetically with vertebrate NLRs as represented by human genes in MetazoanNLR clade 1 (fig. 4; see also Lange et al. 2011; Hamada et al. 2012).
Fig. 4.
Phylogenetic analysis of the metazoan NLR genes constructed from an alignment of the NACHT domains (provided in supplementary file S4, Supplementary Material online). The tree presented is an unrooted Bayesian tree, with branch lengths representing the number of substitutions per site. Posterior probabilities and ML bootstrap support values greater than 50% are indicated for the clades of interest. The two major metazoan NLR clades are circled by dashed lines and are consistent in both Bayesian and ML trees (supplementary file S5, Supplementary Material online). N-terminal effector domain types are shown adjacent to the lineage in which they are observed. Amphimedon queenslandica lineage is in red; cnidarian lineages (Acropora digitifera and Nematostella vectensis) are in green; human NLRs are in blue; Capitella teleta NLRs are in orange; Strongylocentrotus purpuratus NLRs are in purple; mollusc NLRs are in dark pink; arthropod NLRs are in black. For clarity, only a subset of divergent representatives from each taxon was selected for inclusion in the alignment. The numbers to the right of the name of each taxon indicate the size of the NLR complement in that clade. Refer to supplementary file S5, Supplementary Material online, for the corresponding ML tree.
Second, reflecting outcomes of studies of NLR diversification in other animals (Lange et al. 2011; Hamada et al. 2012), our results strongly suggest that the large number of NLRs present in the Amphimedon genome have originated via a single lineage-specific expansion, all of which fall as a monophyletic group within MetazoanNLR clade 1 in our metazoan-wide phylogenetic analyses (fig. 4 and supplementary file S5, Supplementary Material online). Based on the current data, we cannot determine whether both ancestral genes were present in the common ancestor of all animals and one was lost after the divergence of A. queenslandica from the eumetazoan lineage, or whether the two ancestral genes arose only in the eumetazoan ancestor. This may be clarified as genomic data from other sponges becomes available.
Third, it is clear that the two major metazoan NLR clades have undergone differential expansion across the animal kingdom. Invertebrate NLR expansions predominantly occur in MetazoanNLR clade 2, whereas the vertebrate expansion occurred in MetazoanNLR clade 1 (fig. 4 and supplementary file S5, Supplementary Material online). The subsets of cnidarian and Capitella NLRs in clade 2 contain more genes than the corresponding taxonomic subsets in clade 1 (fig. 4 and supplementary file S5, Supplementary Material online). It is also interesting to note that the genome of the teleost fish, Danio rerio, contains an expanded subfamily of >70 NLRs orthologous to human NLRC3 (which phylogenetically falls in MetazoanNLR clade 1) but does not contain orthologs to human IPAF and NAIP (Laing et al. 2008). This is consistent with a more taxonomically limited study by Zhang et al. (2010), which found two discrete clades corresponding to invertebrate and vertebrate NLRs (with the exception of IPAF and NAIP, which nested in the invertebrate clade). The functional significance of this dichotomy is hard to infer given the lineage-specific nature of many NLR expansions. This dynamic evolutionary history is well captured by the substantial difference in numbers of NLRs between the pearl oyster P. fucata (45 genes) and the edible oyster Cra. gigas (1 gene), two members of the same class (Bivalvia) of molluscs (fig. 4 and supplementary files S2 and S5, Supplementary Material online). This disparity suggests that the Pinctada NLR expansion may be a unique response to specific selection pressures of currently unknown origin. This could provide a useful experimental system for future work aimed at investigating the selective pressures that drive NLR evolution. Similarly, it seems likely that at least one NLR gene was present in the ecdysozoan ancestor but has apparently been lost, perhaps multiple times independently, in many ecdysozoan lineages. In the handful of arthropods in which NLRs have been identified (supplementary file S2, Supplementary Material online), both the small numbers of NLRs and their apparent lack of effector domains together suggest that these genes may not be involved in arthropod immunity.
N-Terminal Domain of Metazoan NLRs is Highly Variable and Characterized by Convergent Evolution
There is substantial variation in the N-terminal domains of NLRs across the different animal lineages (fig. 4). At one end of the spectrum are the arthropod NLR genes, which occur only in very small numbers relative to other animals, and all of which comprise only the defining NACHT and LRR domains but no N-terminal domain. Our metazoan-wide phylogenetic analysis (fig. 4) reveals a lack of correlation between N-terminal effector domain type and phylogenetic position, which supports the suggestion of Zhang et al. (2010) that domain shuffling has been an important feature of the evolutionary history of this gene family. On multiple occasions, domain shuffling appears to have resulted in the independent evolution of identical domain combinations, even though it has been suggested that convergent evolution of domain architectures is probably a rare occurrence (Gough 2005). A plausible alternative is that the same domains have been lost multiple times from a common ancestral pool of N-terminal domains. Regardless, it is difficult to avoid the same conclusion of convergence on common gene structures (in this latter case, convergence on the loss of particular domains, rather than on gain). In particular, the presence of death-fold domains (DEATH, CARD, and DED) as N-terminal effector domains is prevalent across the different lineages (fig. 4). In contrast, multiple different domain combinations are apparent even within a single monophyletic anthozoan cnidarian clade in MetazoanNLR clade 2 (fig. 4; note Nematostella cf. Acropora N-terminal domains). This apparent plasticity in the combination of NACHT domains with various N-terminal effector domains makes it difficult to hypothesize on ancestral tripartite NLR domain architecture and equally difficult to infer function based on domain architecture (Istomin and Godzik 2009; Zhang et al. 2010).
Intriguingly, despite the large sizes of the NLR gene families in Amphimedon, Strongylocentrotus, and Branchiostoma, N-terminal effector domain types in these three organisms are limited exclusively to the death-fold domains (DEATH, CARD, and DED) (fig 4; Hibino et al. 2006; Huang et al. 2008; Messier-Solek et al. 2010) that are the most widespread effector domains across multiple independently derived expansions. Zhang et al. (2010) proposed that the apparent convergent evolution on certain domain combinations suggests constraints enforced by structure requirements for proper NACHT domain function. An alternative explanation for the repeated reinvention of the (death-fold)-NLR associations could be the importance of these effector domains to downstream signaling networks. The ability of death-fold domains to recruit other proteins via homotypic interactions facilitates the formation and regulation of multiprotein complexes that are central to cell death and inflammatory signaling pathways (Kersse et al. 2011). It is noteworthy that divergent human NLRs (particularly IPAF and NLRPs) form inflammasome protein complexes via homotypic interactions of their death-fold domains (Schroder and Tschopp 2010).
Little is known about invertebrate NLR function, including whether or not they form inflammasome-like complexes as their vertebrate counterparts do. However, members of the STAND class of P-loop NTPases, which includes NACHT domain-containing genes, are known to act as scaffolds for the assembly of protein complexes involved in regulatory networks (Leipe et al. 2004). It is possible that invertebrate NLRs may form multiprotein complexes via death-fold effector domain interactions, either through direct interactions to recruit effector proteins such as caspases or indirectly through an adaptor protein analogous in function to the vertebrate ASC adaptor (von Moltke et al. 2013). The co-immunoprecipitation of HyNLR (a Hydra DEATH-NACHT gene but not a bona fide NLR) with HyDD-caspase is consistent with the formation of such protein complexes. Furthermore, death-fold domains are important components of the apoptosis network (Kersse et al. 2011). The initiation of pyroptotic and apoptotic pathways of cell death is a vital component of immune defense (Aachoui et al. 2013). As awareness of the close integration between the innate immunity and apoptosis increases (Zmasek and Godzik 2013), the early branching position of A. queenslandica and its strikingly complex NLR repertoire make it an important system for providing new insights into the mechanics of cell death in basal metazoans and the evolution of the role of cell death in defense against pathogens.
The acquisition of novel NLR domain architectures in the anthozoan cnidarians Nematostella and Acropora suggests that functional convergence is not the whole story. These cnidarian NLRs display an unusual propensity for acquiring novel effector domains, as seen in both of the major metazoan NLR clades (fig. 4). The cnidarian genes in MetazoanNLR clade 1 are uniquely characterized by an N-terminal region containing three to four transmembrane domains; HMMER HMMscans identify a Gene3D profile match for the “gap junction channel protein cysteine-rich domain” (1.20.1440.80). Its presence in both Nematostella and Acropora suggests that this NLR combination may have been already present in the anthozoan ancestor. To our knowledge, this is the first report of a putative membrane-bound NLR, and its absence in the other eumetazoan taxa investigated herein suggests that this may be an anthozoan-specific innovation. Interestingly in this context, a small number of AqNLRs are also predicted to have one or two N-terminal transmembrane domains (table 1), but further investigation is necessary to confirm their presence because the signals are weak and inconclusive.
In the absence of a classical adaptive immunity, it has been proposed that highly specific immune responses could be generated in invertebrate animals through synergistic interactions among components of the innate immune system (Schulenburg et al. 2007). The multiplicity of the invertebrate NLRs and of their putative downstream signaling components, coupled with the potential for complex protein–protein interactions via the NACHT and death-fold domains, creates the potential for complex synergistic interactions to occur at the receptor, signaling, and effector levels of the NLR immune response (Schulenburg et al. 2007). This potential raises the possibility that invertebrate NLRs, although superficially similar at a structural level to vertebrate NLRs, might have the capacity for generating an innate immune response of greater specialization and diversity than vertebrate NLRs. As we learn more about the functions of the invertebrate NLRs, it is possible that the line that has conventionally separated our views of the metazoan innate and adaptive immune systems will become increasingly blurred.
Materials and Methods
A local version of HMMER 3.0 (Finn et al. 2011), available from http://hmmer.janelia.org/software (last accessed February 17, 2013), was used to interrogate the Joint Genome Institute (JGI) A. queenslandica genome database (www.metazome.net/amphimedon, last accessed April 20, 2013) for DEATH (PF00531), NACHT (PF05729), and LRR (PF12799) domains using Pfam HMMs available from http://pfam.sanger.ac.uk/ (last accessed February 17, 2013). The same genome is also available for interrogation at EnsemblMetazoa (http://metazoa.ensembl.org/Amphimedon_queenslandica/Info/Annotation/#about, last accessed October 24, 2013). As the seed sequences used to create the Pfam HMMs are vertebrate biased (particularly for the DEATH- and NACHT-domain), we also broadened our search space by constructing our own HMM profiles for each of the three domains of interest that incorporated sequences from A. queenslandica NLRs. We subsequently interrogated the sponge genome for potentially more divergent NLRs using these in-house HMMs. To investigate the origin of the bona fide NLRs (as defined by the NACHT-LRR domain combination; Ting, Harton et al. 2008), a number of fungal, plant, protozoan, and metazoan genomes were also interrogated (supplementary file S2, Supplementary Material online). All protein sequences identified by the HMM searches were further verified for the specific domains by scanning the PFAM, Gene3D (CATH), Superfamily (SCOP), SMART, and PROSITE databases using the following search tools: Pfam (http://pfam.sanger.ac.uk/search, last accessed March 4, 2013), Hmmer (http://hmmer.janelia.org/search, last accessed February 17, 2013), and InterProScan v1.05 plug-in for GeneiousPro v6.1.5 created by Biomatters (available from: http://www.geneious.com/, last accessed March 4, 2013).
Our search for NLRs in the genome of A. queenslandica was focused on the most current gene models (Aqu1). The list of NACHT domain-containing Aqu1 gene models were annotated as described above to identify other conserved domains. The complexity of NLR loci appears to pose problems for gene prediction algorithms, as has been reported for other PRR gene families in eumetazoans such as Hydra and Strongylocentrotus (Hibino et al. 2006; Lange et al. 2011). For Aqu1 gene models in which only a NACHT domain was detected, we expanded our search for N- and C-terminal domains by interrogating several different versions of Amphimedon gene models in the same location, as well as directly searching upstream and downstream genomic sequences. The alternate JGI gene models that we searched include Aqu0, Augustus, Augustus-PASA, SNAP, and GenomeScan (all available on the JGI browser www.metazome.net/amphimedon, last accessed April 20, 2013). Reciprocal Blast searches using tripartite AqNLRs were also incorporated to help identify NLRs. To retrieve the most accurate complement of NACHT domain-containing genes, we occasionally determined that concatenation of two gene models was warranted. As independent confirmation of these determinations, the genomic sequences spanning these concatenated models were submitted to the Augustus web server (http://bioinf.uni-greifswald.de/augustus, last accessed June 8, 2013) to predict the gene structure and coding sequence.
We conducted phylogenetic analyses of NLRs using only the highly conserved NACHT domains as identified by the PFAM HMM. All multiple alignments were performed through the Geneious Pro 6.1.5 MUSCLE plug-in and manually refined in Geneious Pro 6.1.5. The final alignments that we used for phylogenetic analysis are included as supplementary files S3 and S4, Supplementary Material online. For clarity, due to the large number of NACHT domain-containing genes and NLRs present in some genomes, only selected divergent representatives were included in the final trees presented here; full sets of identifiers included in the alignments are presented in supplementary files S3 and S4, Supplementary Material online. ML and Bayesian trees were estimated using PhyML3.1 and MrBayes3.2, respectively (Guindon et al. 2010; Ronquist et al. 2012). The appropriate models of evolution for each alignment were determined using the Bayesian Information Criterion implemented in ProtTest3.2 (Darriba et al. 2011). The best-fit model of evolution was determined to be CPREV + I + G for the Amphimedon NACHT alignment containing the reduced subset of clade A AqNLRs (Adachi et al. 2000), JTT + G for the Amphimedon NACHT alignment containing all the clade A AqNLRs (Jones et al. 1992), and WAG + I + G + F for the metazoan NLR alignment (Whelan and Goldman 2001). Statistical support for bipartitions in the ML analyses was estimated by 250 bootstrap replicates. Bayesian analyses were performed on two parallel runs, with distribution posterior probability of the generated trees estimated using Metropolis-Coupled Markov Chain Monte Carlo (MCMCMC) algorithm with four chains (1 cold, 3 heated) each and a subsampling frequency of 100. Runs were terminated when the average standard deviation of split frequencies of the two parallel runs was <0.01 (about 5,500,000 generations). ln L plots were assessed to determine the appropriate burn-in length (25%). A 50% majority rule tree was constructed from the remaining trees. The results presented are consistent with tree topologies generated by both phylogenetic reconstruction methods (ML and Bayesian inference). Phylogenetic trees were drawn using FigTree v1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/, last accessed May 16, 2013).
Supplementary Material
Supplementary files S1–S5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors are grateful to members of the Degnan lab for helpful advice and thoughtful input through the course of this study and to four anonymous reviewers for taking the time to help us greatly improve the original manuscript. This work was supported by a University of Queensland International Research Scholarship (to B.Y.), a UQ International Tuition Fee Scholarship (to J.B.), and Australian Research Council grants DP0985995 and DP110104601 (to S.M.D).
References
- Aachoui Y, Sagulenko V, Miao EA, Stacey KJ. Inflammasome-mediated pyroptotic and apoptotic cell death, and defense against infection. Curr Opin Microbiol. 2013;16:319–326. doi: 10.1016/j.mib.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi J, Waddell PJ, Martin W, Hasegawa M. Plastid genome phylogeny and a model of amino acid substitution for proteins encoded by chloroplast DNA. J Mol Evol. 2000;50:348–358. doi: 10.1007/s002399910038. [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- Bonardi V, Cherkis K, Nishimura MT, Dangl JL. A new eye on NLR proteins: focused on clarity or diffused by complexity? Curr Opin Immunol. 2012;24:41–50. doi: 10.1016/j.coi.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley KM, Rast JP. Dynamic evolution of toll-like receptor multigene families in echinoderms. Front Immunol. 2012;3:136. doi: 10.3389/fimmu.2012.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm A, Lautz K, Kufer TA. Roles of NLRP10 in innate and adaptive immunity. Microb Infect. 2013;15:516–523. doi: 10.1016/j.micinf.2013.03.008. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BK, Wen H, Ting JPY. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol. 2011;29:707–735. doi: 10.1146/annurev-immunol-031210-101405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunne A. Inflammasome activation: from inflammatory disease to infection. Biochem Soc Trans. 2011;39:669–673. doi: 10.1042/BST0390669. [DOI] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L, Eigenbrod T, Muñoz-Planillo R, Nuñez G. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol. 2009;10:241–247. doi: 10.1038/ni.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough J. Convergent evolution of domain architectures (is rare) Bioinformatics. 2005;21:1464–1471. doi: 10.1093/bioinformatics/bti204. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hamada M, Shoguchi E, Shinzato C, Kawashima T, Miller DJ, Satoh N. The complex NOD-like receptor repertoire of the coral Acropora digitifera includes novel domain combinations. Mol Biol Evol. 2012;30:167–176. doi: 10.1093/molbev/mss213. [DOI] [PubMed] [Google Scholar]
- Hansen JD, Vojtech LN, Laing KJ. Sensing disease and danger: a survey of vertebrate PRRs and their origins. Dev Comp Immunol. 2011;35:886–897. doi: 10.1016/j.dci.2011.01.008. [DOI] [PubMed] [Google Scholar]
- Hibino T, Loza-Coll M, Messier C, et al. (16 co-authors) The immune gene repertoire encoded in the purple sea urchin genome. Dev Biol. 2006;300:349–365. doi: 10.1016/j.ydbio.2006.08.065. [DOI] [PubMed] [Google Scholar]
- Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz RA. Phylogenetic perspectives in innate immunity. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- Huang S, Liu H, Ge J, et al. (17 co-authors) Genomic analysis of the immune gene repertoire of Amphioxus reveals extraordinary innate complexity and diversity. Genome Res. 2008;18:1112–1126. doi: 10.1101/gr.069674.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Istomin AY, Godzik A. Understanding diversity of human innate immunity receptors: analysis of surface features of leucine-rich repeat domains in NLRs and TLRs. BMC Immunol. 2009;10:48. doi: 10.1186/1471-2172-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway JCA, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kaparakis M, Philpott DJ, Ferrero RL. Mammalian NLR proteins; discriminating foe from friend. Immunol Cell Biol. 2007;85:495–502. doi: 10.1038/sj.icb.7100105. [DOI] [PubMed] [Google Scholar]
- Kersse K, Verspurten J, Vanden Berghe T, Vandenabeele P. The death-fold superfamily of homotypic interaction motifs. Trends Biochem Sci. 2011;36:541–552. doi: 10.1016/j.tibs.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Aravind L. Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Differ. 2002;9:394–404. doi: 10.1038/sj.cdd.4400991. [DOI] [PubMed] [Google Scholar]
- Kufer TA. Signal transduction pathways used by NLR-type innate immune receptors. Mol Biosyst. 2008;4:380–386. doi: 10.1039/b718948f. [DOI] [PubMed] [Google Scholar]
- Kufer TA, Sansonetti PJ. NLR functions beyond pathogen recognition. Nat Immunol. 2011;12:121–128. doi: 10.1038/ni.1985. [DOI] [PubMed] [Google Scholar]
- Kurtz J, Armitage SAO. Alternative adaptive immunity in invertebrates. Trends Immunol. 2006;27:493–496. doi: 10.1016/j.it.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Laing KJ, Purcell MK, Winton JR, Hansen JD. A genomic view of the NOD-like receptor family in teleost fish: identification of a novel NLR subfamily in zebrafish. BMC Evol Biol. 2008;8:42. doi: 10.1186/1471-2148-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C, Hemmrich G, Klostermeier UC, Lopez-Quintero JA, Miller DJ, Rahn T, Weiss Y, Bosch TCG, Rosenstiel P. Defining the origins of the NOD-like receptor system at the base of animal evolution. Mol Biol Evol. 2011;28:1687–1702. doi: 10.1093/molbev/msq349. [DOI] [PubMed] [Google Scholar]
- Leipe DD, Koonin EV, Aravind L. STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J Mol Biol. 2004;343:1–28. doi: 10.1016/j.jmb.2004.08.023. [DOI] [PubMed] [Google Scholar]
- Maekawa T, Kufer TA, Schulze-Lefert P. NLR functions in plant and animal immune systems: so far and yet so close. Nat Immunol. 2011;12:817–826. doi: 10.1038/ni.2083. [DOI] [PubMed] [Google Scholar]
- Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M, Gilbert SF, Hentschel U, et al. (26 co-authors) Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A. 2013;110:3229–3226. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messier-Solek C, Buckley KM, Rast JP. Highly diversified innate receptor systems and new forms of animal immunity. Semin Immunol. 2010;22:39–47. doi: 10.1016/j.smim.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Pancer Z. Individual-specific-repertoires of immune cells SRCR receptors in the Purple Sea Urchin (S. purpuratus) New York: Kluwer Academic/Plenum Publishing Co; 2001. pp. 31–40. [DOI] [PubMed] [Google Scholar]
- Philippe H, Derelle R, Lopez P, et al. (20 co-authors) Phylogenomics revives traditional views on deep animal relationships. Curr Biol. 2009;19:706–712. doi: 10.1016/j.cub.2009.02.052. [DOI] [PubMed] [Google Scholar]
- Proell M, Riedl SJ, Fritz JH, Rojas AM, Schwarzenbacher R. The Nod-like receptor (NLR) family: a tale of similarities and differences. PLoS One. 2008;3:e2119. doi: 10.1371/journal.pone.0002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson SJ, Girardin SE. Nod-like receptors in intestinal host defense: controlling pathogens, the microbiota, or both? Curr Opin Gastroenterol. 2013;29:15–22. doi: 10.1097/MOG.0b013e32835a68ea. [DOI] [PubMed] [Google Scholar]
- Robertson SJ, Rubino SJ, Geddes K, Philpott DJ. Examining host-microbial interactions through the lens of NOD: from plants to mammals. Semin Immunol. 2012;24:9–16. doi: 10.1016/j.smim.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP, Teslenko M, et al. (10 co-authors) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenstiel P, Philipp EER, Schreiber S, Bosch TCG. Evolution and function of innate immune receptors—insights from marine invertebrates. J Innate Immun. 2009;1:291–300. doi: 10.1159/000211193. [DOI] [PubMed] [Google Scholar]
- Sansonetti PJ. The innate signaling of dangers and the dangers of innate signaling. Nat Immunol. 2006;7:1237–1242. doi: 10.1038/ni1420. [DOI] [PubMed] [Google Scholar]
- Sarrias MR, Gronlund J, Padilla O, Madsen J, Holmskov U, Lozano F. The scavenger receptor cysteine-rich (SRCR) domain: an ancient and highly conserved protein module of the innate immune system. Crit Rev Immunol. 2004;24:1–37. doi: 10.1615/critrevimmunol.v24.i1.10. [DOI] [PubMed] [Google Scholar]
- Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Schulenburg H, Boehnisch C, Michiels NK. How do invertebrates generate a highly specific innate immune response? Mol Immunol. 2007;44:3338–3344. doi: 10.1016/j.molimm.2007.02.019. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Lamkanfi M, Kanneganti T-D. NOD-like receptor (NLR) signaling beyond the inflammasome. Eur J Immunol. 2010;40:624–627. doi: 10.1002/eji.200940211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Simakov O, Chapman J, et al. (33 co-authors) The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart LM, Paquette N, Boyer L. Effector-triggered versus pattern-triggered immunity: how animals sense pathogens. Nat Rev Immunol. 2013;13:199–206. doi: 10.1038/nri3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JPY, Duncan JA, Lei Y. How the noninflammasome NLRs function in the innate immune system. Science. 2010;327:286–290. doi: 10.1126/science.1184004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JPY, Harton JA, Hoffman HM, et al. (24 co-authors) The NLR gene family: a standard nomenclature. Immunity. 2008;28:285–287. doi: 10.1016/j.immuni.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JPY, Willingham SB, Bergstralh DT. NLRs at the intersection of cell death and immunity. Nat Rev Immunol. 2008;8:372–379. doi: 10.1038/nri2296. [DOI] [PubMed] [Google Scholar]
- von Moltke J, Ayres JS, Kofoed EM, Chavarría-Smith J, Vance RE. Recognition of bacteria by inflammasomes. Annu Rev Immunol. 2013;31:73–106. doi: 10.1146/annurev-immunol-032712-095944. [DOI] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Wilmanski JM, Petnicki-Ocwieja T, Kobayashi KS. NLR proteins: integral members of innate immunity and mediators of inflammatory diseases. J Leukoc Biol. 2008;83:13–30. doi: 10.1189/jlb.0607402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- Yue J-X, Meyers BC, Chen J-Q, Tian D, Yang S. Tracing the origin and evolutionary history of plant nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes. New phytol. 2012;193:1049–1063. doi: 10.1111/j.1469-8137.2011.04006.x. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Zmasek CM, Godzik A. Domain architecture evolution of pattern-recognition receptors. Immunogenetics. 2010;62:263–272. doi: 10.1007/s00251-010-0428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmasek CM, Godzik A. Evolution of the animal apoptosis network. Cold Spring Harb Perspect Biol. 2013;5:a008649. doi: 10.1101/cshperspect.a008649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.