Background: Cpa, the pilus adhesin from Streptococcus pyogenes, has an active thioester domain.
Results: Cpa has a second thioester domain reactive toward biological amines.
Conclusion: The crystal structure of spermidine-bound Cpa provides a model for covalent adhesion.
Significance: Thioester domains and covalent adhesion may be common in Gram-positive pathogens.
Keywords: Bacterial Adhesion, Bacterial Pathogenesis, Crystal Structure, Mass Spectrometry (MS), Post-translational Modification, Protein Cross-linking, Protein Domains, Streptococcus pyogenes, Bacterial Pilus, Thioester Domain
Abstract
The human pathogen Streptococcus pyogenes produces pili that are essential for adhesion to host surface receptors. Cpa, the adhesin at the pilus tip, was recently shown to have a thioester-containing domain. The thioester bond is believed to be important in adhesion, implying a mechanism of covalent attachment analogous to that used by human complement factors. Here, we have characterized a second active thioester-containing domain on Cpa, the N-terminal domain of Cpa (CpaN). Expression of CpaN in Escherichia coli gave covalently linked dimers. These were shown by x-ray crystallography and mass spectrometry to comprise two CpaN molecules cross-linked by the polyamine spermidine following reaction with the thioester bonds. This cross-linked CpaN dimer provides a model for the covalent attachment of Cpa to target receptors and thus the streptococcal pilus to host cells. Similar thioester domains were identified in cell wall proteins of other Gram-positive pathogens, suggesting that thioester domains are more widely used and provide a mechanism of adhesion by covalent bonding to target molecules on host cells that mimics that used by the human complement system to eliminate pathogens.
Introduction
The human pathogen Streptococcus pyogenes elaborates long, hairlike structures called pili (1). In many pathogenic bacteria, pili mediate adherence to the host organism and are thus adhesins (2). They can also play a role in biofilm formation, mediating interactions with other bacteria (2).
S. pyogenes commonly infects the human skin, throat, and tonsils and is the major cause of tonsillitis (3). It has been shown that pili are essential for the binding of S. pyogenes to human tonsils, primary and immortalized keratinocytes, and epithelial cells of throat and lung (4–7). The specificity of pili as adhesins to epithelial cells is underlined by findings that they are dispensable for streptococcal adhesion to HEp-2 cells (6–8), which are often referred to as human epithelial cells but are actually derived from the HeLa carcinoma cell line (7). Thus, the primary role of S. pyogenes pili appears to be in the colonization of epithelia of the human skin, throat, and tonsils.
Structurally, Gram-positive pili are covalent polymers that are built from many repeats of a backbone protein (BP)5 with ancillary protein 1 (AP1) at the pilus tip and ancillary protein 2 (AP2) at the base (9, 10). One or more pilus-specific sortases catalyze the covalent linkage of the BP units and the ancillary proteins. A so-called housekeeping sortase anchors the pilus to the bacterial cell wall (11).
In S. pyogenes, the pilus proteins are encoded in the fibronectin- and collagen-binding protein and T-antigen (FCT) island (1). Nine types of FCT islands have been described so far (9, 12, 13). The T-antigen is the BP of the pilus in all FCT types, the collagen-binding protein is the AP1 in most FCT types, and the fibronectin-binding protein is a protein outside of the pilus operon proper, designated PrtF1 or PrtF2 depending on the FCT type (12). The majority of S. pyogenes strains belong to FCT types 2, 3, and 4 (9, 12, 14) with orthologous pilins in each. FCT type 2 is represented by the strain typed as T-antigen serotype 1/M1-serotype 1 (T1/M1), whereas FCT type 3 and 4 islands can be found in various T- and M-serotypes (9, 12). The pilin proteins are generally called FctA (BP), Cpa (AP1), and FctB (AP2).
The BP, AP1, and AP2 components of the S. pyogenes pilus have all been structurally characterized (15–18). Their structures have led to the discovery of intramolecular, stabilizing isopeptide bonds in the BP and AP1 proteins (16, 18, 19) and of conserved lysine residues that are used in linking the BP, AP1, and AP2 units together (4, 15, 16, 20).
The crystal structure of a three-domain C-terminal fragment of the AP1 protein Cpa from the T1/M1 strain SF370 unexpectedly revealed a thioester bond joining the side chains of a cysteine residue (Cys426) and a glutamine residue (Gln575) in its “top” domain (18). In this Cpa molecule, also referred to as Spy0125, the thioester bond is found in a groove on the protein surface and is thus solvent-accessible. It does not contribute to protein stability (18, 21). However, because Cpa is located at the pilus tip where it acts as the pilus adhesin (4, 20, 22), the occurrence of a thioester bond is highly suggestive.
Cys-Gln thioester bonds have been discovered previously in the human complement proteins C3 and C4 (23). Upon proteolytic activation, C3 and C4 go through a conformational rearrangement that exposes their thioester bond, enabling nucleophilic attack on it by bacterial cell wall components and protein amino and hydroxyl groups. This results in the formation of a covalent bond between the Gln residue and the attacking amine or hydroxyl group (23). By analogy with the C3/C4 thioester reaction mechanism, it was suggested that the Cpa thioester moiety may also enable covalent bonding with a target human receptor (18).
The thioester sequence motif is conserved in an equivalent position in all AP1 proteins of S. pyogenes FCT types 2, 3, and 4. Interestingly, a similar motif is also found in the N-terminal domain of Cpa (CpaN), which was missing from the construct used for the crystal structure determined by Pointon et al. (18). Here, we have characterized the complete Cpa structure by small angle x-ray scattering (SAXS) analysis and modeling and carried out detailed structural and functional studies of CpaN. We show that CpaN does indeed contain a thioester bond and demonstrate its reactivity to biological amines by crystallographic and mass spectral analysis of a spermidine-linked covalent dimer. The Cpa present in most S. pyogenes strains thus has two reactive thioester bonds, i.e. one in its CpaN domain and the other in its top domain (CpaT), and thereby presents an intriguing example of a two-headed adhesin. Finally, we show that similar domains appear to be present in other bacterial adhesins, implying that these bind to host cells in mimicry of the mechanism used by the complement system to eliminate pathogens.
EXPERIMENTAL PROCEDURES
Cloning of Cpa
The cpa gene was cloned from S. pyogenes strain 90/306S. The emm genotype of this strain was determined as ST6030.1 using established protocols (24, 25). Using the primers Cpa_forw2 and Cpa_rev3, which encode conserved regions of cpa, the gene was PCR-amplified from S. pyogenes genomic DNA. A resulting 2-kb product was used as a template for a second PCR round using primers Cpa_forw and Cpa_rev1 and cloning into the vector pProEx using KasI and EcoRI restriction sites (construct pProEx_Cpa1). The cpa sequence was determined by the Centre for Genomics and Proteomics of the University of Auckland. In the following, residue referencing refers to the deposited Cpa sequence AGQ45688. Mutations were introduced into CpaN using site-directed mutagenesis by inverse PCR. All primers are listed in Table 1.
TABLE 1.
Primers used
Restriction sites and Gateway adapter sequences are underlined, stop codons are in italics, and mutations are in bold.
| Cloning primers | |
| Cpa_forw2 | 5′-GTA GGG TTT TCT ATC AGA GCG TTC GG-3′ |
| Cpa_rev3 | 5′-GTC AAA CCA GTT GGT GGG ACA AGA TC-3′ |
| Cpa_forw | 5′-GAA GGC GCC GAA GAA AAA TCT ACT GAA ACT AAA-3′ |
| Cpa_rev1 | 5′-GAA GAA TTC TTA AGT TGG TGG GAC AAG ATC TTT-3′ |
| Cpa_forw4 | 5′-GGC AGC GGC GCG GAA GAA CAA TCA GTG CCA AAT A-3′ |
| Cpa_rev7 | 5′-GAA AGC TGG GTGTTA ATC TTT TCG GTT TTC AAA AGC TA-3′ |
| attB1 | 5′-GGGG ACA AGT TTG TAC AAA AAA GCA GGC TTC GAA AAC CTGTAT TTT CAG GGC AGC GGC GCG-3′ |
| attB2 | 5′-GGGG AC CAC TTT GTA CAA GAA AGC TGG GTG-3′ |
| Cpa_forw9 | 5′-CGG CCG G GGCGCCGAA GAA CAA TCA GTG CCA AAT A-3′ |
| Cpa_rev12 | 5′-CGG CCG G CTCGAGTTA ATC TTT TCG GTT TTC AAA AGC TA-3′ |
| Cpa_forw7 | 5′-CGG CCG G GGCGCCGAA GAA CAA TCA GTG CCA AAT A-3′ |
| Cpa_rev13 | 5′-CGG CCG G CTCGAGTTA AGT ATC CGG AAC ATA CTC AGC-3′ |
| Mutagenesis primers | |
| cpaN_C62/AFwd | 5′-GCG TTT AAT TTA ACA AAA CAC TTT CCA TC-3′ |
| cpaN_C62/A Rev | 5′-GTATGCTTGATACTCCTTACTTC-3′ |
| cpaN_Q138/AFwd | 5′-GCG AAT GCT ATT TGG TAC TAT ACT GAT A-3′ |
| cpaN_Q138/A Rev | 5′-AGT CAC TAA AAT AGC GTT TAG AGG-3′ |
| cpaN_W142/LFwd | 5′-CTG TAC TAT ACT GAT AGT TCC TAT ATT TC-3′ |
| cpaN_W142/L Rev | 5′-AATAGCATTTTGAGTCACTAAAATAG-3′ |
Two full-length expression constructs were subcloned from pProEx_Cpa1. A construct, pProEx_Cpa (residues 8–670), was generated using the primers Cpa_forw9 and Cpa_rev12 and restriction enzymes KasI and XhoI. A second construct, pDEST17_Cpa (residues 8–670), was generated in two rounds of PCR using first primers Cpa_forw4 and Cpa_rev7 and then primers attB1 and attB2 and Gateway cloning. A construct for CpaN (residues 8–222) was generated using primers Cpa_forw7 and Cpa_rev13 and introduced into the vector pProEx using KasI/XhoI. All constructs contain an N-terminal His6 tag and a tobacco etch virus protease recognition site. After removal of the His6 tag, Cpa expressed in pDEST17_Cpa has an N-terminal Gly-Ser extension.
Protein Expression
For native proteins, Escherichia coli BL21 DE3 cells transformed with pProEx_Cpa, pDEST17_Cpa, pProEx_CpaN, or pProEx_CpaN mutants were grown to an optical density of 0.6–0.8 in LB medium when expression was induced using 1 mm isopropyl 1-thio-β-d-galactopyranoside. Cells were further incubated at 18 °C for 16 h and harvested. Selenomethionine-substituted CpaN (SeMet-CpaN) was produced using a metabolic repression protocol. E. coli BL21 DE3 pProEx_CpaN cells were grown in M9 medium at 37 °C to an optical density of 0.4 when the following amino acids were added: 25 mg/liter l-lysine, 25 mg/liter l-phenylalanine, 25 mg/liter l-threonine, 20 mg/liter l-isoleucine, 20 mg/liter l-leucine, 20 mg/liter l-valine, and 15 mg/liter l-selenomethionine. After 15 min, expression was induced using isopropyl 1-thio-β-d-galactopyranoside (final concentration, 1 mm), and the cultures were incubated at 28 °C for 16 h.
Protein Purification
For both Cpa and CpaN, cells were resuspended using buffer A (50 mm Tris-HCl, pH 8.0, 500 mm NaCl, 20 mm imidazole, 5% (v/v) glycerol, 0.002% (w/v) NaN3) including Complete protease inhibitors (Roche Applied Science) and lysed using a cell disruptor. After clearing by centrifugation, the lysate was applied to a 5-ml HiTrap Chelating column (GE Healthcare) charged with Ni2+. After washing, protein was eluted in a gradient from 0 to 100% buffer B (buffer A + 500 mm imidazole). In the case of CpaN, the monomer form eluted slightly earlier than the dimer. Cpa and CpaN were treated using tobacco etch virus protease and a second immobilized metal affinity chromatography step to remove the His6 tag. Cpa was further purified by anion exchange chromatography using a Q Sepharose FF column (GE Healthcare) and a gradient from 50 to 500 mm NaCl (in 25 mm Tris-HCl, pH 8.0). Both Cpa and CpaN were then subjected to size exclusion chromatography using an S200 (Cpa) or S75 (CpaN) 16/60 column (GE Healthcare) and buffer C (10 mm Tris-HCl, pH 7.4, 137 mm NaCl, 2.7 mm KCl, 0.002% (w/v) NaN3) + 1 mm β-mercaptoethanol (Cpa) or 1 mm tris(2-carboxyethyl)phosphine (CpaN). Finally, both proteins were shown to be monodisperse by dynamic light scattering.
Crystallization and X-ray Data Collection
CpaN and SeMet-CpaN crystals were grown by hanging drop vapor diffusion, mixing equal volumes of CpaN (27–36 mg/ml) and 16–21% (w/v) PEG3350, 0.2–0.3 m KH2PO4 and incubating at 18 °C overnight. For cryoprotection, crystals were briefly soaked in 18–20% (v/v) glycerol, 22% (w/v) PEG3350, 0.2 m KH2PO4, 10 mm Tris-HCl, pH 7.4, 137 mm NaCl, 2.7 mm KCl and flash cooled in liquid nitrogen. Diffraction data were collected at 100 K on beamline PX-2 of the Australian Synchrotron, Clayton, Victoria, Australia. All data sets were indexed and integrated using XDS (26), reindexed using POINTLESS (27), and scaled using SCALA (27).
Phase Determination and Model Building
Many crystals displayed signs of twinning. For phasing, an SeMet-CpaN crystal was identified that was untwinned, and a four-wavelength multiwavelength anomalous dispersion data set was collected. The data set was combined using POINTLESS. Using AutoSHARP (28), selenium sites (two per CpaN monomer; correlation coefficient, 0.74) were identified, refined, and used to derive initial phases, which were improved by density modification. The resulting electron density map was suitable for model building using Buccaneer (29). This initial model was used for molecular replacement to solve the crystal structure of native CpaN (untwinned) using Phaser (30). Initially, the model of native CpaN was refined in a unit cell with dimensions a = b = 132.2 Å and c = 68.4 Å, but high R/Rfree values suggested underlying issues with the choice of cell or space group. Closer inspection of the frames revealed weak reflections along the c axis, a strong indicator of pseudotranslational symmetry, that were unrecorded. Systematic analysis of the c axis length was performed with Zanuda (31), and the true length of the c axis was determined to be 136.6 Å with a 0.5 × c pseudotranslation vector, giving a unit cell with dimensions a = b = 132.2 Å and c = 136.6 Å. The final model with six CpaN dimers in the asymmetric unit was refined in iterative cycles of manual building using Coot (32) and maximum likelihood refinement with non-crystallographic symmetry (NCS) and translation/libration/screw (TLS) using BUSTER (33). Data collection and refinement statistics are in Tables 2 and 3, respectively. Model geometry was assessed using MolProbity (34). Figures were created with PyMOL (35).
TABLE 2.
Diffraction data collection and phasing statistics
Values in parentheses are for the outermost resolution shell. Molecules per asymmetric unit (a.u.) refers to CpaN dimers.
| Data set |
|||||
|---|---|---|---|---|---|
| Selenium peak | Selenium inflection | Selenium low energy remote | Selenium high energy remote | Native | |
| Crystal data | |||||
| Space group | P31 | P31 | P31 | P31 | P31 |
| Unit cell parameters (Å; °) | a = b = 133.1, c = 137.0; α = β = 90, γ = 120 | a = b = 132.5, c = 136.5; α = β = 90, γ = 120 | a = b = 133.0, c = 136.9; α = β = 90, γ = 120 | a = b = 132.7, c = 136.5; α = β = 90, γ = 120 | a = b = 133.3, c = 137.8; α = β = 90, γ = 120 |
| Molecules per a.u. | 6 | 6 | 6 | 6 | 6 |
| Data collection | |||||
| Wavelength (Å) | 0.97930 | 0.97940 | 0.99190 | 0.95370 | 0.95370 |
| Resolution range (Å) | 137.0–2.16 (2.20–2.16) | 136.5–2.16 (2.19–2.16) | 136.9–2.19 (2.22–2.19) | 136.5–2.10 (2.14–2.10) | 47.9–2.00 (2.03–2.00) |
| Total observations | 4,444,356 (67,414) | 4,420,571 (72,390) | 2,300,192 (33,516) | 4,797,172 (87,116) | 1,069,788 (52,132) |
| Unique reflections | 144,449 (6,115) | 143,506 (6,064) | 138,294 (5,454) | 155,569 (6,861) | 185,886 (9,148) |
| Redundancy | 30.8 (11.0) | 30.8 (11.9) | 16.6 (6.1) | 30.8 (12.7) | 5.8 (5.7) |
| Completeness (%) | 98.7 (84.6) | 98.7 (74.9) | 98.3 (69.2) | 99.3 (87.3) | 100.0 (100.0) |
| Mean I/σ(I) | 8.0 (0.2) | 8.8 (0.3) | 6.9 (0.2) | 10.8 (0.7) | 7.6 (0.6) |
| Rmerge (%) | 80.1 (2112.0) | 44.0 (808.8) | 39.8 (677.7) | 34.3 (487.2) | 16.2 (331.2) |
| Rp.i.m. (%) | 21.5 (920.4) | 11.6 (327.7) | 14.6 (394.9) | 9.1 (197.8) | 8.3 (174.4) |
| CC1/2 | 0.98 (0.10) | 1.0 (0.11) | 0.99 (0.08) | 1.0 (0.17) | 1.0 (0.16) |
| Resolution/Rmerge (Å/%) when I/σI drops below 1 | 3.22/56.6 | 3.00/67.8 | 3.04/66.2 | 2.83/61.0 | 2.54/61.0 |
| Phasing | |||||
| Selenium sites | 14 | 14 | 14 | 14 | |
| Phasing power overall | 0.391 | 0.261 | 0.034 | 0.369 | |
| Phasing power >1.0 (Å) | 3.85 | 5.43 | 4.21 | ||
TABLE 3.
Refinement statistics for the native CpaN structure
r.m.s., root mean square.
| Resolution range (Å) | 47.50–2.00 |
| Number of reflections | 180,308 |
| R/Rfree (%) | 20.1/23.2 |
| Cutoff criterion (σF) | 0 |
| Number of atoms | |
| Protein | 20,388 |
| Ligand | 107 |
| Water | 985 |
| r.m.s. deviations from standard values | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.05 |
| Wilson B (Å2) | 36.1 |
| Mean B, protein (Å2) | 49.6 |
| Mean B, ligand (Å2) | 49.8 |
| Mean B, solvent (Å2) | 45.2 |
| Ramachandran statistics | |
| Favored (%) | 96.8 |
| Allowed (%) | 99.6 |
| Outliers (%) | 0.4 |
Alignments
Sequences were aligned with Clustal Ω (36). Protein structures were aligned using secondary structure matching (37). The NCBI protein sequence database was searched using Delta-BLAST (38). Alignment figures were created using Jalview (39) and colored according to a BLOSUM62 score.
Mass Spectrometry
Protein molecular weights were determined by electrospray ionization-time of flight (ESI-TOF) mass spectrometry. Proteins were diluted in 50% (v/v) acetonitrile, 0.1% (v/v) formic acid and analyzed using a Q-STAR XL Hybrid system (Applied Biosystems, Foster City, CA). Raw data were deconvoluted using the Bayesian Protein Reconstruct Tool within Analyst QS 1.1 (Applied Biosystems).
For peptide mass fingerprinting and identification of cross-linked peptides, protein samples were separated by SDS-PAGE. Protein bands were diced; destained in 25 mm NH4HCO3, 50% (v/v) acetonitrile; dried using acetonitrile; optionally reduced and alkylated with DTT and iodoacetamide; and then subjected to either trypsin, chymotrypsin (all sequencing grade), or both dissolved in 25 mm NH4HCO3. Peptide fragments were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) on a Q-STAR XL Hybrid MS/MS system. Linear peptides were identified using the Mascot search engine v2.0.05 (Matrix Science, London, UK); cross-linked peptides were identified by manual inspection of MS/MS spectra.
Samples of the tryptic CpaN dimer digest were subjected to high resolution accurate mass analysis using a ThermoFinnigan LTQ-FT hybrid ion trap ion cyclotron resonance mass spectrometer. A coated glass PicoTip emitter (New Objective Inc., Woburn, MA) was filled with the peptide solution in 25% (v/v) acetonitrile and mounted on the nanospray source set at a voltage of 1.4 kV and passing through the heated capillary (200 °C). Full scan spectra were obtained at both 100,000 and 500,000 resolution at m/z 400.
SAXS Data Collection
SAXS data were collected at the SAXS beamline of the Australian Synchrotron, Clayton, Victoria, Australia. A monodisperse solution of Cpa (verified by dynamic light scattering) was analyzed over a concentration range of 0.8–3.3 mg/ml in buffer C at 4 °C. Diffraction data were recorded using a Pilatus 1M detector with a camera length of 3400 mm and a slit size of 0.220 mm. Pure water was used to normalize intensities for absolute scaling. After averaging and buffer subtraction, scattering curves were analyzed using PRIMUS (40). The distance distribution function was determined using GNOM (41). Using GasborP (42), an independent set of 16 ab initio dummy atom models were calculated of which 15 were averaged using DAMAVER (43). For building a model of Cpa, CORAL (44) was used to fit CpaN and a model of the C-terminal part of Cpa (calculated using Phyre (45) and the Spy0125 structure, Protein Data Bank code 2XIC) to the Cpa scattering data. Two flexible linkers were assumed, one between the middle domain and CpaN and one between the middle and top domains. Linker length was varied stepwise to account for the unknown degree of flexibility and to obtain the best fit (final χ2 of 0.37). The theoretical diffraction of the final model was calculated and fitted to the scattering data using CRYSOL (46). An electron density-like envelope of the final filtered bead model was calculated using the Situs package (47). Docking of the model Cpa model was performed using Chimera (48). Further experimental details are given in Table 4.
TABLE 4.
SAXS data collection and scattering-derived parameters for Cpa
| Data collection parameters | ||
| X-ray source | Australian Synchrotron | |
| Wavelength (Å) | 1.127 | |
| Beam geometry | 0.22 mm slit | |
| q range (Å−1) | 0.004–0.20 | |
| Sample flow rate (μl s−1) | 4 | |
| Exposure time (s) | 2 | |
| Concentration range (mg ml−1) | 0.8–3.3 | |
| Temperature (K) | 277 | |
| Structural parameters | ||
| I(0) (cm−1) | (from Guinier plot) | (from P(r)) |
| 3.3 mg ml−1 | 0.178 ± 0.0001 | 0.179 ± 0.0006 |
| 1.6 mg ml−1 | 0.091 ± 0.0001 | 0.092 ± 0.0005 |
| 1.2 mg ml−1 | 0.067 ± 0.0001 | 0.068 ± 0.0005 |
| 0.8 mg ml−1 | 0.047 ± 0.0001 | 0.048 ± 0.0004 |
| Rg (Å) | (from Guinier plot) | (from P(r)) |
| 3.3 mg ml−1 | 38.4 ± 0.7 | 39.4 ± 0.2 |
| 1.6 mg ml−1 | 38.8 ± 0.8 | 40.0 ± 0.3 |
| 1.2 mg ml−1 | 38.9 ± 2.3 | 39.9 ± 0.4 |
| 0.8 mg ml−1 | 39.3 ± 1.2 | 40.2 ± 0.4 |
| Dmax (Å)a | 134.5 | |
| Molecular mass (from I(0))a (Da) | 74,758 | |
| Calculated monomeric mass from sequence (Da) | 74,608 | |
| Normalized spatial discrepancy of averaged dummy atom modelsa | 1.306 ± 0.077 | |
a Reported for 3.3 mg ml−1 measurement, which was used for model building.
Ligand Binding Assays for Cpa and CpaN
The binding of Cpa and CpaN to spermidine and related ligands was analyzed using ESI-TOF mass spectrometry (see above). CpaN (10 μm) was incubated with 10 mm spermidine, spermine, or lysine at 18 °C for 20 h. Cpa (0.5 mm) was incubated with 50 mm spermidine at 18 °C for 20 h. After incubation, protein samples were stored frozen until ESI-TOF analysis.
RESULTS
Cloning and Characterization of Cpa from S. pyogenes 90/306S
Cpa was cloned from a local S. pyogenes strain, 90/306S. The T-type of this strain was identified as T9, and the emm genotype was identified as ST6030.1 (data not shown). This Cpa is almost identical to Cpa from the S. pyogenes M3/T3 strain 315 with only two amino acid changes and is homologous with Spy0125, the Cpa protein from S. pyogenes serotype M1/T1 with ∼50% sequence identity. In contrast to Spy0125 (4), however, the full-length Cpa in our study was stable throughout purification and for many weeks at 4 °C.
The molecular mass of our full-length recombinant Cpa deviated from the expected mass. ESI-TOF mass spectrometry determined its molecular mass to be 74,683.8 Da, which is 68.4 Da less than expected from its sequence (74,752.2 Da). As the C-terminal fragment of Spy0125 contains two intramolecular isopeptide bonds and one thioester bond (18) and the residues involved in these bonds are conserved, we tested the hypothesis that our full-length Cpa might contain similar bonds. This was confirmed by LC-MS/MS analysis, which identified two cross-linked peptides that contain isopeptide bonds between Lys243 and Asp546 and between Lys562 and Asn667 (data not shown). The formation of these bonds accounts for the loss of one water molecule and one ammonia molecule (18 and 17 Da, respectively). As was the case for the C-terminal fragment of Spy0125 (18), we did not find direct evidence for a thioester bond between Cys374 and Gln526, but instead full conversion of Gln526 to Glu was seen in our LC-MS/MS analysis (data not shown). This deamidation is consistent with the presence of a thioester bond, which eliminates the Gln amino group and is subject to subsequent hydrolysis during reduction and alkylation prior to LC-MS/MS analysis. Two isopeptide bonds and a thioester bond thus account for a loss of 52 Da as was the case for Spy0125 (18), leaving an additional loss of 16.4 Da unaccounted for.
CpaN Contains a Thioester Bond and Forms a Covalent Dimer
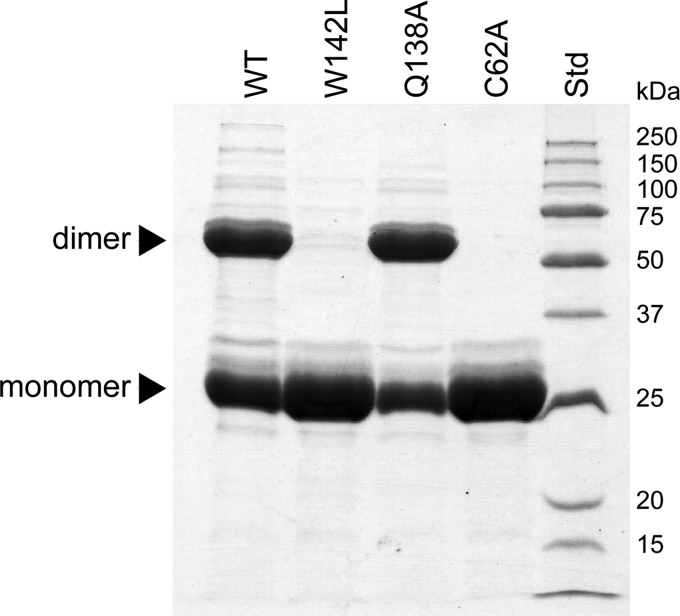
When CpaN was purified by immobilized metal affinity chromatography and analyzed by SDS-PAGE under reducing conditions, two bands were observed (Fig. 1). One band corresponds to the 25-kDa CpaN monomer, whereas the upper, stronger band corresponds to a protein of 50 kDa. Using LC-MS/MS, both bands were confirmed as belonging to CpaN, indicating that CpaN forms a very stable dimer. The monomer and dimer could be separated by size exclusion chromatography.
FIGURE 1.
Dimerization of CpaN. Shown is reducing SDS-PAGE of CpaN (WT) and its mutants W142L, Q138A, and C62A. Bands for the covalent dimer and the monomer are indicated. Std indicates standard proteins.
The putative CpaN dimer was found by ESI-TOF to have a molecular mass of 49,817.6 Da, whereas the mass of the monomer was determined to be 24,836.2 Da. As the expected molecular mass of CpaN is 24,853.6 Da, however, the dimer shows a gain of 110.4 Da, and the monomer shows a loss of 17.4 Da.
The reduced mass of the monomer can be explained by the presence of a thioester bond between residues Cys62 and Gln211. These residues are equivalent by sequence alignment (Fig. 2A) to Cys426 and Gln575, which form the thioester bond in the top domain of Spy0125 (18). In the CpaN monomer, a tryptic fragment with an m/z of 987.23+ was identified by LC-MS/MS. This fragment comprises the peptides EYQAYC62FNLTK and TFQ211NLLSAEYVPDT cross-linked through Cys62 and Gln211 with loss of one ammonia molecule (Fig. 3A), confirming the existence of a thioester bond between Cys62 and Gln211.
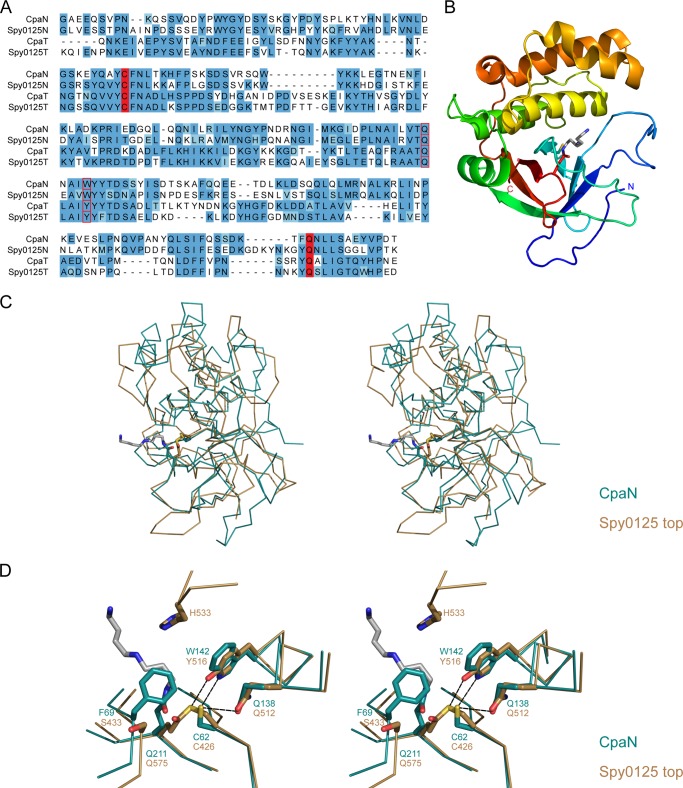
FIGURE 2.
CpaN comparisons with Spy0125. A, sequence alignment of the Cpa and Spy0125 N-terminal and top domains. Highlighted are Cys62 and Gln211, which form a thioester bond, and Gln138 and Trp142, which are close to the thioester bond. B, fold of the CpaN monomer. Cys62, Gln211, and the spermidine (gray) are indicated as stick models. C, superposition of the crystal structures of CpaN and the Spy0125 top domain (Protein Data Bank code 2XIC). D, superposition of the CpaN spermidine-binding site and the Spy0125 thioester site. Dashed lines indicate contacts ≤3.6 Å. Spermidine is shown in gray.
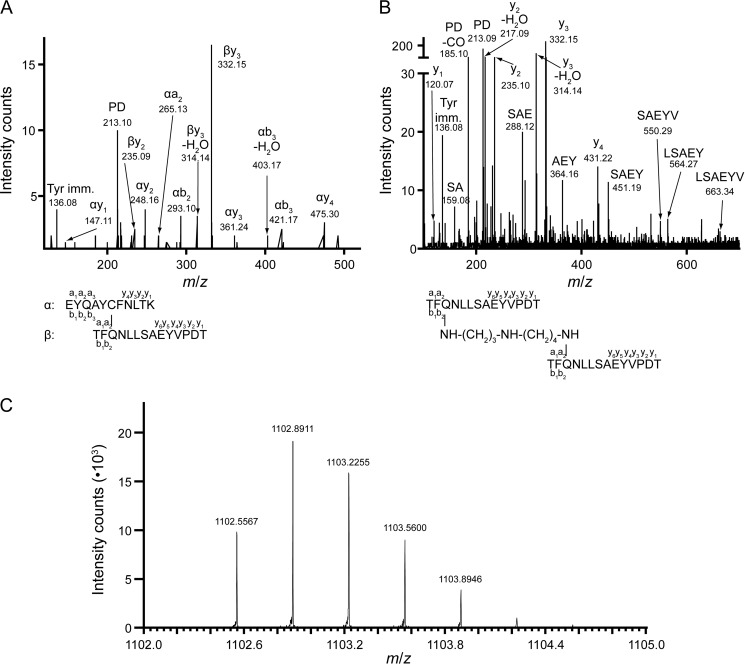
FIGURE 3.
The CpaN thioester bond and spermidine cross-link. Shown are MS/MS spectra of cross-linked peptide fragments involving the thioester linkage. A, thioester bond between Cys62 and Gln211. Shown are the fragments of a parent ion with an m/z of 987.23+. B, spermidine cross-link between Gln211 residues from the two monomers of a CpaN dimer. Shown are the fragments of a parent ion with an m/z of 1102.63+. In A and B, the annotation of the fragment peaks refers to the structure of the parent ion given at the bottom. Terminal fragments are indicated by a, b, and y with a number referring to the respective amino acid (given in the structure). Internal fragments are indicated by amino acid sequence. Tyr imm., tyrosine immonium ion. C, high accuracy FT-MS spectrum of the isotopic cluster of the parent ion shown in B. The first peak (m/z 1102.5567) corresponds to the monoisotopic parent ion; the other peaks represent isotope derivatives because of the natural incorporation of 13C and 15N.
Crystal Structure of the CpaN Dimer
To further characterize the CpaN dimer, we crystallized it and solved its structure using a multiwavelength anomalous dispersion protocol with a selenomethionine derivative (Tables 2 and 3). The nominal resolution is 2.5 Å, although reflections up to 2.0 Å were included for refinement of the CpaN dimer structure as this gave improved electron density maps. There are six dimers per asymmetric unit with good electron density for residues 13–219 in all but two chains.
The overall fold of the CpaN monomer comprises a β-sandwich packed against a bundle of α-helices decorated with extensive loops (Fig. 2B). The CpaN monomer fold is thus similar to that of the CpaT domain of Spy0125 (18) with a root mean square difference of 2.1 Å over 150 aligned Cα positions (Fig. 2C). Two CpaN monomers then form a globular dimer with P2 symmetry and an interface of 1119 Å2 per monomer (Fig. 4B).
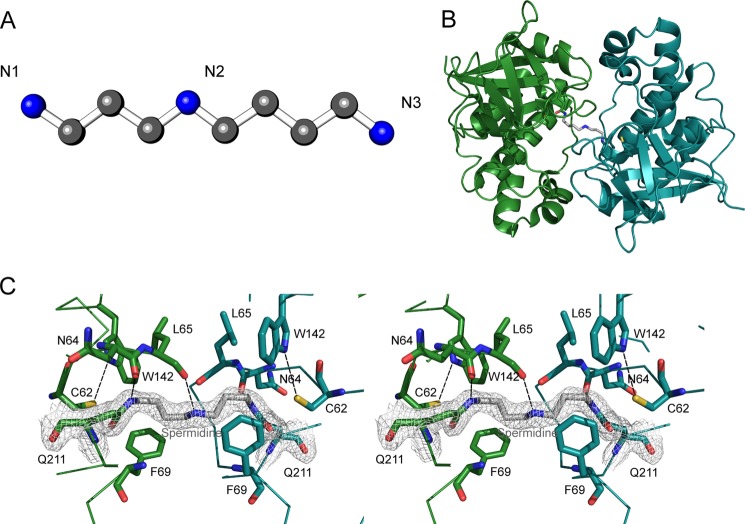
FIGURE 4.
Spermidine coordination in the CpaN dimer. A, structure of spermidine. B, the CpaN dimer. The spermidine cross-link, Cys62, and Gln211 are represented in stick mode; the cross-link is highlighted in gray. C, the final model of the spermidine-binding site and the 2Fo − Fc electron density at 1.5 σ around spermidine and the two covalently bound Gln211 residues from each CpaN monomer. Hydrogen bonds are indicated using dashes. Residues that belong to the same chain are drawn in the same color. For clarity, some of the surrounding loops and residues have been removed.
The positions of Cys62 and Gln211 in each molecule of the CpaN dimer match those of Cys426 and Gln575 in the top domain of Spy0125 when the domains are superimposed (Fig. 2D). The local environments are fairly weakly conserved, however, with Trp142 replacing Tyr516, Phe69 replacing Ser433, Leu48 replacing Tyr412, and His46 replacing Lys410. There is also a major difference in the absence in CpaN of a loop equivalent to residues 531–534, which in Spy0125 limits access to the thioester bond between Cys426 and Gln575. The most striking feature, however, is that although the positions of the Cys and Gln residues are conserved no thioester bond is present in the CpaN dimer for reasons explained below.
A Covalent Spermidine Cross-link in the CpaN Dimer
To our surprise, a stretch of continuous electron density was found connecting two Gln211 residues, which are 8.8 Å apart in the CpaN dimer (Fig. 4C). We modeled this density as a covalently bound spermidine molecule. Spermidine is a polyamine of molecular mass 145.25 Da that occurs naturally in E. coli (49) (Fig. 4A). Assuming a covalent bond with Gln211 at either end, we would expect the loss of two amino groups, i.e. 34 Da. Thus, the covalent binding of a spermidine cross-link would lead to a net addition of 111 Da to the CpaN dimer, exactly as was observed by the ESI-TOF analysis described above. Using LC-MS/MS, a peptide with an m/z of 1102.63+ was identified that contains two copies of the fragment TFQ211NLLSAEYVPDT and carries an additional 111 Da (Fig. 3B). Using Fourier transform mass spectrometry (FT-MS), we determined the exact m/z of this fragment as 1102.55673+, which corresponds very well with the theoretical m/z of 1102.55363+ (mass error, 2.7 ppm; Fig. 3C), thus supporting our interpretation of the density as spermidine.
The modeled spermidine molecule fits well within the environment of the CpaN dimer interface (Fig. 4C). The central spermidine nitrogen N2 is hydrogen-bonded to the main chain carbonyl oxygens of Leu65 from one protomer and Lys67 from the other. The terminal spermidine nitrogens, which form amide bonds with the Gln211 residues, interact with the main chain carbonyl oxygens of Asn64 of each protomer. Phe69 of each protomer makes van der Waals contacts with the methylene groups on either side of N2. These contacts are reminiscent of those in the spermidine-binding protein PotD from E. coli (50) in which spermidine is captured in a groove lined with both aromatic residues and hydrogen bond acceptors.
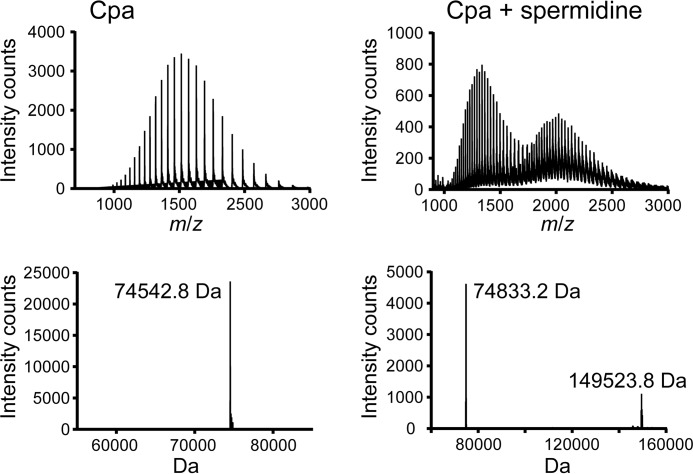
We then tested whether the binding of Cpa and CpaN to spermidine could be recapitulated in vitro. Full-length Cpa, which has two thioester bonds, was incubated with excess spermidine and analyzed by ESI-TOF mass spectrometry (Fig. 5). The molecular mass of Cpa was increased by 290 Da compared with the control (74,542.8 Da), consistent with the binding of spermidine to both thioesters of Cpa (145.25 Da per molecule). A small proportion formed a higher molecular mass complex of 149,523.8 Da (Fig. 5), which corresponds to two Cpa molecules plus three spermidines, thus indicating a spermidine-cross-linked Cpa dimer. When CpaN was incubated with excess spermidine, a mass shift of 145.2 Da was observed, consistent with the binding of one spermidine but no dimer formation under these conditions. The binding of two other naturally occurring amines, spermine and lysine, was also tested. Both compounds showed signs of partial binding (about 70% for spermine and 30% for lysine) when tested at the same concentration as spermidine. Thus, spermidine bound to the thioester moieties of both Cpa and CpaN in vitro and induced covalent dimerization of Cpa, indicating that spermidine binding is not restricted to the isolated CpaN domain.
FIGURE 5.
Binding of Cpa to spermidine. Shown is ESI-TOF mass spectrometry analysis of Cpa (left) and Cpa incubated with excess spermidine (right). The molecular mass is consistent with a bound spermidine (145.25 Da). A peak at 149,523.8 Da indicates dimer formation and binding of three spermidine molecules. Top, ESI-TOF spectrum; bottom, deconvoluted spectrum.
Conservation of CpaN-like Domains and Their Occurrence in Other Proteins
The CpaN domain from our T9 Cpa is the direct orthologue of the N-terminal domain of Spy0125 (47% sequence identity) and is homologous with the top domain of Spy0125 (Fig. 2C), sharing 24% sequence identity (Fig. 2A). It is clear that each of these two Cpa proteins has two homologous thioester-forming domains, which we will call CpaN and CpaT in further discussion. Most orthologues of the AP1 proteins Cpa and Spy0125 possess both a CpaN domain and a CpaT-like domain. However, Cpa homologs from certain S. pyogenes serotypes such as M5 and M18 lack a CpaN domain and are restricted to CpaT.
To explore the occurrence of CpaN-like domains in proteins other than Cpa, we performed Delta-BLAST searches (38). We identified a range of proteins from both streptococci and other Gram-positive bacteria such as Corynebacterium diphtheriae and Clostridium perfringens that share a CpaN-like domain including conserved thioester motifs (Fig. 6A). These proteins have in common a C-terminal LPXTG sortase and cell wall sorting motif, implying localization to the cell wall. In each case, the CpaN-like domain is at the N terminus of the protein, and in one instance, there is an additional CpaT domain (Fig. 6B). The C-terminal portions of the proteins are highly variable, however, and are built of various kinds of repetitive elements such as CnaB-type domains, fibronectin-binding repeats, or collagen-like repeats (Fig. 6B). Thus, CpaN-like domains are widespread building blocks in cell wall-attached proteins from Gram-positive bacteria.
FIGURE 6.

CpaN-like domains in non-pilin proteins. A, sequence alignment of the N-terminal domain of Cpa with N-terminal domains of selected cell wall-anchored proteins from Gram-positive bacteria. Cys62 and Gln211, which form a thioester bond in Cpa, are highlighted in red; Gln138 and Trp142 are boxed in red. B, the domain structure of the proteins aligned in A. Protein designations are as follows: Cpa, as sequenced in this study; PrtF1, fibronectin-binding protein I from S. pyogenes (NCBI accession number YP_005388204.1); Sequ, Sez_1457 from Streptococcus equi (YP_002123804.1); Cdip, putative collagen-binding protein from C. diphtheriae (YP_005134618.1); Bcer, TQXA domain-containing protein from Bacillus cereus (EOO76633.1); and Cper, putative surface-anchored protein from C. perfringens (WP_003474233.1). Fn, fibronectin-like domain; CnaB, Cna type B-like domain; FnB, fibronectin-binding domain; Col, collagen-like; Ig-like, immunoglobulin-like domain.
Notable among the proteins with CpaN-like domains is the fibronectin-binding protein PrtF1. PrtF1, like Cpa and Spy0125, is a component of certain FCT-type islands that encode distinct pilus types in the various S. pyogenes serotypes (12). PrtF1 has 52% sequence identity with CpaN (Fig. 6A) and is thus the closest homologue among non-pilin proteins.
The alignment of the CpaN-like domains identified in our BLAST search allows the identification of conserved elements of CpaN domains (Fig. 6A). The thioester bond-forming residues Cys62 and Gln211 are invariant, and the Cys residue is found in a conserved (A/V)YC(F/V/L) motif. There is also a highly conserved TQXA(I/V)W motif around residues Gln138 and Trp142. In the structure of CpaN, Gln138 and Trp142 are very close to Cys62 (3.5 and 3.6 Å distant, respectively). The TQXA(I/V)W motif is the signature of domains known as TQXA domains (PFAM identifier CHP03934_TQXA), and most but not all of these domains also seem to carry an (A/V)YC(F/V/L) thioester motif.
Interestingly, the Spy0125 top domain and other CpaT-like domains depart from this TQXA(I/V)W pattern with the Trp residue replaced by Tyr (Fig. 2D). Nevertheless, residues Gln138/Gln512, Trp142/Tyr516, and other residues surrounding Cys62/Cys426 are found in the same position in both CpaN and the Spy0215 top domain. Whereas most residues keep their relative positions, however, the Cys side chain moves further away from Gln138 when comparing CpaN (broken thioester) and Gln512 from the Spy0125 top domain (formed thioester). It can be speculated that the strictly conserved Gln138/Gln512 is close to the cysteine side chain before thioester bond formation with Gln211/Gln575 and may have a role in catalyzing the reaction.
Mutations of the Thioester Site
Conserved residues of the CpaN thioester site were mutated to test their role in thioester and dimer formation. All CpaN mutant proteins behaved like the wild type during expression and purification except for their ability to form dimers. The CpaN-C62A mutant was monomeric (Fig. 1) with its observed molecular mass as predicted from its sequence. This is further evidence that the 17-Da loss seen for both full-length Cpa and CpaN results from formation of a thioester bond. The thioester bond is also prerequisite to the formation of a spermidine-cross-linked dimer.
We also analyzed two residues of the TQ138XA(I/V)W142 motif. The CpaN-Q138A mutant was expressed as a mixture of monomers and covalent dimers (Fig. 1). The monomer mass (observed, 27,618.6 Da; predicted, 27,633.5 Da) is consistent with the formation of a thioester bond, and the dimer mass (observed, 55,378.8 Da) indicates a spermidine cross-link. Thus, Gln138 does not appear to influence thioester formation in vitro. However, the CpaN-W142L mutant was expressed purely as a monomer (Fig. 1). The mass of this monomer indicated the formation of a thioester bond (observed, 27,602.6 Da; predicted, 27,617.5 Da). Thus, Trp142 appears dispensable for the formation of a thioester bond but essential for reactivity with spermidine.
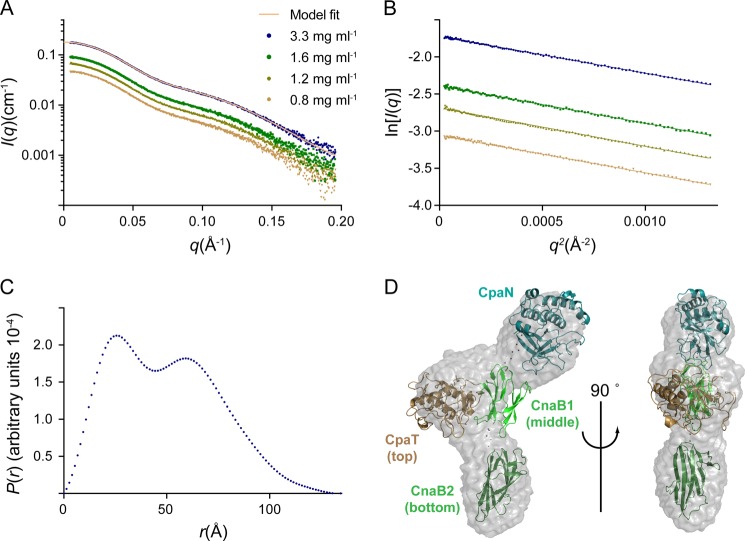
Structure of Full-length Cpa
The structure of full-length Cpa was analyzed by SAXS (Fig. 7 and Table 4) as crystals could not be grown. The radius of gyration (Rg) of 38.4 Å and the maximal extension (Dmax) of 135 Å are consistent with previous SAXS analysis of the orthologous intact Spy0125 (Rg of 40.5 Å and Dmax of 148 Å (17)). Rg and the molecular mass calculated from I0 indicate that Cpa (like Spy0125) is monomeric in solution (Table 4). The distance distribution function (Fig. 7C) suggests that Cpa forms a multidomain structure.
FIGURE 7.
SAXS analysis of Cpa. A, scattering curves at four Cpa concentrations. The model fit is the theoretical scattering from the final Cpa model shown in D (χ2 of 0.38). B, Guinier plot of the scattering data shown in A. C, the distance distribution function for Cpa at 3.3 mg/ml. D, envelope of the averaged, filtered dummy atom model reconstructed for Cpa. Fitted is a model of Cpa constructed using CORAL (43). The color coding is as follows: CpaN, turquoise; CpaT, brown; middle domain, light green; bottom domain, dark green.
To obtain a low resolution structure of Cpa, 16 low resolution dummy atom models of Cpa were reconstructed. Of these models, 15 were of very similar shape and were averaged (normalized spatial discrepancy, 1.3; Fig. 7D). The dimensions of the averaged model are 130 × 40 × 70 Å. Overall, three distinct domains are recognizable, and the Cpa structure appears similar to a model reconstructed for Spy0125 (17).
The crystal structure of CpaN and a model for the C-terminal fragment of Cpa were fitted to the Cpa SAXS data (Fig. 7D). The three C-terminal domains were modeled from the Spy0125 structure using Phyre (45). In the fitting, the proline-rich linker between CpaN and the middle domain was assumed to display some flexibility. Flexibility had been shown for the linker between the Spy0125 middle and bottom domains (18) and was also modeled during the fitting. The final model of Cpa fits well with the observed scattering for Cpa (χ2 of 0.38; Fig. 7A). In this model, the CpaN domain sits at the top of Cpa and thus the top of the pilus, whereas the CpaT domain is located slightly lower and facing the side of Cpa. Both domains are connected to the middle domain.
DISCUSSION
Biochemical and structural characterization of the pili from S. pyogenes and other Gram-positive bacteria has unearthed some unusual chemical features (11). These pili are covalent polymers in which the pilus subunits are covalently linked by sortases and are internally stabilized by isopeptide bond cross-links (11). The latest novel feature to be discovered was the thioester bond between Cys426 and Gln575 in the crystal structure of a C-terminal fragment of the adhesin subunit Cpa of S. pyogenes pili (18). Although thioester bonds are also formed during sortase-mediated assembly of Gram-positive pili (10, 11), these are transient reaction intermediates. The thioester bond in Cpa, like that in complement, is very different: it is relatively stable but reactive and thus able to bind to target cells.
In the M1/T1 strain of S. pyogenes studied by Pointon et al. (18), the N-terminal domain of the adhesin subunit Cpa (Spy0125) is labile and was not included in the construct used for structural analysis. Noting that this N-terminal domain is homologous with the thioester domain of the C-terminal fragment (referred to as the CpaT domain), we set out to characterize this missing domain to obtain a more complete picture of the pilus adhesin. We show here that CpaN is indeed homologous with the previously characterized Spy0125 thioester domain CpaT at the levels of both sequence and structure. The thioester-forming residues, Cys62 and Gln211, in CpaN are conserved and do form a thioester bond as shown by mass spectrometry. Thus, the S. pyogenes AP1 has two thioester domains. We propose that both domains can participate in cell adhesion through their thioester moieties and that the predicted thioester bonds in similar domains of other cell surface proteins imply a similar mechanism.
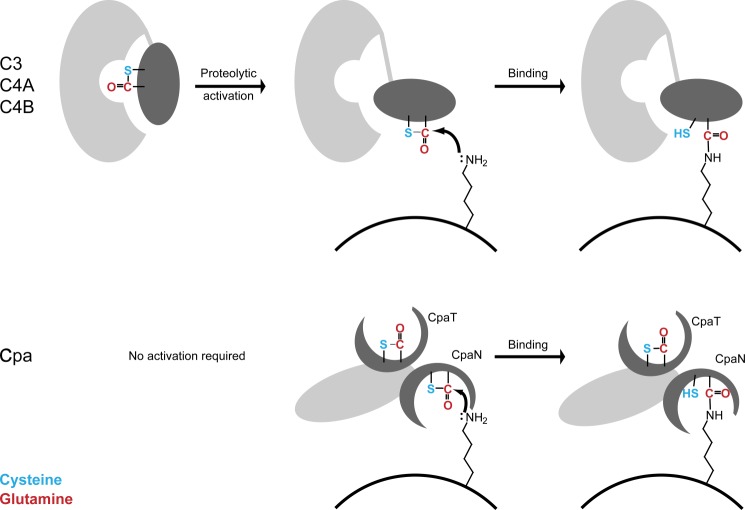
Thioester bonds are highly reactive. They are commonly observed as intermediates in the reactions of cysteine-dependent enzymes such as sortases (51) and ubiquitin ligases (52). The reactivity of thioester bonds is also exploited in protein synthesis for the chemical ligation of peptides (53). In the complement system, a part of the innate immune system, reactive thioester bonds are used for covalent attachment of the complement proteins C3 and C4 to target molecules or bacteria (23). C3 and C4 both have a thioester bond between Cys991 and Gln994 (human C4) that is buried within the inactive molecule. On activation, a major conformational rearrangement renders the thioester accessible to nucleophilic attack by functional groups on bacterial surfaces (Fig. 8) (23, 54, 55).
FIGURE 8.
Thioester reactions in Cpa and complement proteins. Shown is a simplified scheme for the reactions of the thioester bonds of complement proteins (upper panel) and Cpa (lower panel) with amino groups presented by target cells. The complement proteins C3, C4A, and C4B require activation by proteolysis and domain movement, which exposes the thioester bond (54, 55), whereas Cpa appears not to require activation. The complement proteins are here represented by C4A, which like Cpa has preference for amino groups (56, 57). In C4B and C3, a His/Asp substitution near the thioester bond changes the substrate specificity and allows reaction with hydroxyl groups.
In principle, the thioester bonds of C3 and the two isotypes of C4, C4A and C4B, can react with both amine and hydroxyl groups. They differ, however, in their reactivity to these two types of nucleophile (56). C3 and C4B have a conserved histidine (His1106 in C4) close to the thioester bond. His1106 executes a nucleophilic attack on the thioester bond, forming an acyl intermediate with Gln994 that is relieved by a hydroxyl group in a second step. In C4A in which His1106 is substituted by Asp, the thioester bond is preferentially attacked by amines, which react directly to form an amide with Gln994 (Fig. 8) (56, 57).
We have shown here that both thioester domains of Cpa react directly with the polyamine spermidine. This suggests a one-step reaction as in C4A (Fig. 8). In experiments with C4B, tyrosine can replace His1106 as a nucleophile in the two-step reaction with hydroxyl groups albeit at lower rates (56). Although Tyr516 in the Spy0125 top domain (Trp142 in CpaN) could be invoked as a nucleophile in a similar two-step reaction, it does not appear to be correctly positioned. We conclude that the Cpa thioester domains primarily bind amine groups in a one-step mechanism (Fig. 8).
A feature of the complement system is that large conformational changes, activated by proteolysis, are required to expose the thioester bond (Fig. 8) (23, 54); the system is highly regulated. Once exposed, the thioester bond is either bound by functional groups on the target receptor or hydrolyzed and inactivated within a very short time (56). In contrast, this does not seem to be the case for Cpa. The structures of CpaN and Spy0125 show that the thioester bonds in both the thioester domains are accessible, albeit not fully exposed, at the base of a groove (Fig. 8). Thus, residues surrounding the thioester bond may control access to it, similar to accepted models for the interplay between an enzyme and its substrate.
Our findings support this idea of controlled access to the thioester site. In monomeric CpaN and in the top domain of Spy0125, the thioester bonds are still intact after expression, purification and, in the case of Spy0125, crystallographic analysis. Thus, the thioester bonds of Cpa are not as readily hydrolyzed as those of activated C3 and C4 and seem relatively protected from water. In addition, cross-linking of the CpaN dimer is characterized by a surprising specificity for spermidine, which, as far as we can judge from our mass spectrometry data, seems to be the only compound co-purified from E. coli, although we also observed some degree of binding by other amines in vitro. Also, our CpaN W142L mutant forms a thioester bond, but not a spermidine-crosslinked dimer, underscoring the relevance of the environment of the thioester for its reactivity.
Our structure of the CpaN dimer cross-linked with spermidine highlights further residues that may be relevant for target recognition. Phe69 provides hydrophobic contacts with the hydrocarbon portions of the spermidine molecule, whereas the backbone carbonyl oxygens of Asn64, Leu65, and Lys67 act as hydrogen bond acceptors. Lysine side chains or a carbohydrate with a free amine might thus be candidates for Cpa binding. It is noteworthy that in the Spy0125 top domain, Phe69 is replaced by a serine and access to the thioester site is more restricted by a loop between Asp531 and Gly534. These structural differences may indicate distinct target patterns that remain to be analyzed.
We deem it unlikely that spermidine is the natural target of Cpa. Spermidine is an abundant polyamine, however, that is both synthesized and imported from the medium by E. coli (49). Depending on the growth conditions, intracellular concentrations between 100 μm and 6 mm have been reported for E. coli (49, 58). S. pyogenes may circumvent binding of Cpa by spermidine through export-coupled folding. Further work is required to identify target receptors of Cpa, but is beyond the scope of this work. Nevertheless, our structure of the CpaN dimer may at least narrow this search down to molecules with free amino groups and hydrophobic elements. The identification of a receptor for Cpa may also lead to development of an inhibitor of Cpa-mediated adhesion, which might be able to attack and thus inactivate the thioester bond.
The structures of CpaN and the SAXS model of Cpa also conclude the structural characterization of the S. pyogenes pilins of FCT types 2, 3, and 4. The architecture and assembly of the pilus stalk has been discussed elsewhere (11). We can now add that most Cpa orthologues have two homologous thioester-containing adhesin domains (Fig. 8), reminiscent of the two antigen-binding arms of an human antibody. In contrast to an antibody, however, the two thioester domains may have distinct specificities, thus enlarging the number of potential targets. These adhesin subunits are different from the AP1 pilins of some other streptococcal species, and certain rarer S. pyogenes FCT types. Both S. pneumoniae and S. agalactiae (and FCT type 6 S. pyogenes strains, whose pili are derived from S. agalactiae by horizontal gene transfer (9)) build their AP1 from another common adhesin domain, a von Willebrand factor-like domain (59, 60). However, even many S. pyogenes strains that do not have a Cpa orthologue apparently still encode another CpaN-like thioester domain in the form of PrtF1. We have also learnt that these thioester domains appear to be a common feature of adhesins from many Gram-positive pathogens, suggesting that adhesion by covalent attachment may be a widespread mechanism in the complex field of host-pathogen interactions.
In summary, we have shown that the N-terminal domain of the AP1 pilin Cpa (CpaN) contains a reactive thioester bond, which can form a covalent bond with free amines. Our crystal structure of the spermidine-cross-linked CpaN dimer may serve as a model for such a thioester-mediated interaction. Our results and those obtained previously for Spy0125 (18) lead to the conclusion that intact Cpa has two reactive thioester domains that can bind to host cells possibly with somewhat different specificities. Similar domains appear to be quite common in extracellular proteins in Gram-positive bacteria, implying that covalent binding to host cells via active thioester bonds may be a more widespread mechanism of attachment than recognized previously.
Acknowledgments
We thank Mike Herbert, Christopher Squire, and Yuliana Yosaatmadja for SAXS/x-ray data collection. The use of the Australian Synchrotron was supported by the New Zealand Synchrotron User Group.
This work was supported in part by the Marsden Fund of New Zealand, the Health Research Council of New Zealand, and the Tertiary Education Commission of New Zealand through funding of the Maurice Wilkins Centre for Molecular Biodiscovery.
The atomic coordinates and structure factors (code 4C0Z) have been deposited in the Protein Data Bank (http://wwpdb.org/).
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) KC622314.
The amino acid sequence of this protein can be accessed through NCBI Protein Database under NCBI accession number AGQ45688.
- BP
- backbone protein
- AP
- ancillary protein
- FCT
- fibronectin- and collagen-binding protein and T-antigen
- CpaN
- N-terminal domain of Cpa
- SAXS
- small angle x-ray scattering
- CpaT
- top domain of Cpa
- ESI-TOF
- electrospray ionization-time of flight
- FT
- Fourier transform.
REFERENCES
- 1. Mora M., Bensi G., Capo S., Falugi F., Zingaretti C., Manetti A. G., Maggi T., Taddei A. R., Grandi G., Telford J. L. (2005) Group A streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc. Natl. Acad. Sci. U.S.A. 102, 15641–15646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Proft T., Baker E. N. (2009) Pili in Gram-negative and Gram-positive bacteria—structure, assembly and their role in disease. Cell. Mol. Life Sci. 66, 613–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cunningham M. W. (2000) Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13, 470–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Smith W. D., Pointon J. A., Abbot E., Kang H. J., Baker E. N., Hirst B. H., Wilson J. A., Banfield M. J., Kehoe M. A. (2010) Roles of minor pilin subunits Spy0125 and Spy0130 in the serotype M1 Streptococcus pyogenes strain SF370. J. Bacteriol. 192, 4651–4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Manetti A. G., Zingaretti C., Falugi F., Capo S., Bombaci M., Bagnoli F., Gambellini G., Bensi G., Mora M., Edwards A. M., Musser J. M., Graviss E. A., Telford J. L., Grandi G., Margarit I. (2007) Streptococcus pyogenes pili promote pharyngeal cell adhesion and biofilm formation. Mol. Microbiol. 64, 968–983 [DOI] [PubMed] [Google Scholar]
- 6. Crotty Alexander L. E., Maisey H. C., Timmer A. M., Rooijakkers S. H., Gallo R. L., von Köckritz-Blickwede M., Nizet V. (2010) M1T1 group A streptococcal pili promote epithelial colonization but diminish systemic virulence through neutrophil extracellular entrapment. J. Mol. Med. 88, 371–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Abbot E. L., Smith W. D., Siou G. P., Chiriboga C., Smith R. J., Wilson J. A., Hirst B. H., Kehoe M. A. (2007) Pili mediate specific adhesion of Streptococcus pyogenes to human tonsil and skin. Cell. Microbiol. 9, 1822–1833 [DOI] [PubMed] [Google Scholar]
- 8. Nakata M., Köller T., Moritz K., Ribardo D., Jonas L., McIver K. S., Sumitomo T., Terao Y., Kawabata S., Podbielski A., Kreikemeyer B. (2009) Mode of expression and functional characterization of FCT-3 pilus region-encoded proteins in Streptococcus pyogenes serotype M49. Infect. Immun. 77, 32–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Falugi F., Zingaretti C., Pinto V., Mariani M., Amodeo L., Manetti A. G., Capo S., Musser J. M., Orefici G., Margarit I., Telford J. L., Grandi G., Mora M. (2008) Sequence variation in group A streptococcus pili and association of pilus backbone types with Lancefield T serotypes. J. Infect. Dis. 198, 1834–1841 [DOI] [PubMed] [Google Scholar]
- 10. Telford J. L., Barocchi M. A., Margarit I., Rappuoli R., Grandi G. (2006) Pili in Gram-positive pathogens. Nat. Rev. Microbiol. 4, 509–519 [DOI] [PubMed] [Google Scholar]
- 11. Kang H. J., Baker E. N. (2012) Structure and assembly of Gram-positive bacterial pili: unique covalent polymers. Curr. Opin. Struct. Biol. 22, 200–207 [DOI] [PubMed] [Google Scholar]
- 12. Kratovac Z., Manoharan A., Luo F., Lizano S., Bessen D. E. (2007) Population genetics and linkage analysis of loci within the FCT region of Streptococcus pyogenes. J. Bacteriol. 189, 1299–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bessen D. E., Kalia A. (2002) Genomic localization of a T serotype locus to a recombinatorial zone encoding extracellular matrix-binding proteins in Streptococcus pyogenes. Infect. Immun. 70, 1159–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Köller T., Manetti A. G., Kreikemeyer B., Lembke C., Margarit I., Grandi G., Podbielski A. (2010) Typing of the pilus-protein-encoding FCT region and biofilm formation as novel parameters in epidemiological investigations of Streptococcus pyogenes isolates from various infection sites. J. Med. Microbiol. 59, 442–452 [DOI] [PubMed] [Google Scholar]
- 15. Linke C., Young P. G., Kang H. J., Bunker R. D., Middleditch M. J., Caradoc-Davies T. T., Proft T., Baker E. N. (2010) Crystal structure of the minor pilin FctB reveals determinants of group A streptococcal pilus anchoring. J. Biol. Chem. 285, 20381–20389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kang H. J., Coulibaly F., Clow F., Proft T., Baker E. N. (2007) Stabilizing isopeptide bonds revealed in Gram-positive bacterial pilus structure. Science 318, 1625–1628 [DOI] [PubMed] [Google Scholar]
- 17. Solovyova A. S., Pointon J. A., Race P. R., Smith W. D., Kehoe M. A., Banfield M. J. (2010) Solution structure of the major (Spy0128) and minor (Spy0125 and Spy0130) pili subunits from Streptococcus pyogenes. Eur. Biophys. J. 39, 469–480 [DOI] [PubMed] [Google Scholar]
- 18. Pointon J. A., Smith W. D., Saalbach G., Crow A., Kehoe M. A., Banfield M. J. (2010) A highly unusual thioester bond in a pilus adhesin is required for efficient host cell interaction. J. Biol. Chem. 285, 33858–33866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kang H. J., Baker E. N. (2009) Intramolecular isopeptide bonds give thermodynamic and proteolytic stability to the major pilin protein of Streptococcus pyogenes. J. Biol. Chem. 284, 20729–20737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Quigley B. R., Zähner D., Hatkoff M., Thanassi D. G., Scott J. R. (2009) Linkage of T3 and Cpa pilins in the Streptococcus pyogenes M3 pilus. Mol. Microbiol. 72, 1379–1394 [DOI] [PubMed] [Google Scholar]
- 21. Walden M., Crow A., Nelson M., Banfield M. J. (2013) Intramolecular isopeptide but not internal thioester bonds confer proteolytic and significant thermal stability to the S. pyogenes pilus adhesin Spy0125. Proteins 10.1002/prot.24420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kreikemeyer B., Nakata M., Oehmcke S., Gschwendtner C., Normann J., Podbielski A. (2005) Streptococcus pyogenes collagen type I-binding Cpa surface protein: expression profile, binding characteristics, biological functions, and potential clinical impact. J. Biol. Chem. 280, 33228–33239 [DOI] [PubMed] [Google Scholar]
- 23. Law S. K., Dodds A. W. (1997) The internal thioester and the covalent binding properties of the complement proteins C3 and C4. Protein Sci. 6, 263–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Beall B., Facklam R., Thompson T. (1996) Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J. Clin. Microbiol. 34, 953–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Whatmore A. M., Kehoe M. A. (1994) Horizontal gene transfer in the evolution of group A streptococcal emm-like genes: gene mosaics and variation in vir regulons. Mol. Microbiol. 11, 363–374 [DOI] [PubMed] [Google Scholar]
- 26. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Evans P. R. (2011) An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D Biol. Crystallogr. 67, 282–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vonrhein C., Blanc E., Roversi P., Bricogne G. (2007) Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 [DOI] [PubMed] [Google Scholar]
- 29. Cowtan K. (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 30. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lebedev A. A., Isupov M. N. (2012) Space group validation with Zanuda. CCP4 Newsletter on Protein Crystallography, Number 48, Collaborative Computational Project No. 4, UK [Google Scholar]
- 32. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek C., Roversi P., Sharff A., Smart O. S., Vonrhein C., Womack T. O. (2011) BUSTER, version 2.11.5, Global Phasing Ltd., Cambridge, UK [Google Scholar]
- 34. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. DeLano W. L. (2002) The PyMOL Molecular Graphics System, Schrödinger, LLC, New York [Google Scholar]
- 36. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Ω. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krissinel E., Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 38. Boratyn G. M., Schäffer A. A., Agarwala R., Altschul S. F., Lipman D. J., Madden T. L. (2012) Domain enhanced lookup time accelerated BLAST. Biol. Direct 7, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J. (2009) Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H., Svergun D. I. (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 41. Svergun D. I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 42. Svergun D. I., Petoukhov M. V., Koch M. H. (2001) Determination of domain structure of proteins from x-ray solution scattering. Biophys. J. 80, 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Volkov V. V., Svergun D. I. (2003) Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G., Gajda M., Gorba C., Mertens H. D., Konarev P. V., Svergun D. I. (2012) New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 46. Svergun D., Barberato C., Koch M. H. (1995) CRYSOL—a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 47. Wriggers W. (2012) Conventions and workflows for using Situs. Acta Crystallogr. D Biol. Crystallogr. 68, 344–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 49. Tabor C. W., Tabor H. (1985) Polyamines in microorganisms. Microbiol. Rev. 49, 81–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sugiyama S., Matsuo Y., Maenaka K., Vassylyev D. G., Matsushima M., Kashiwagi K., Igarashi K., Morikawa K. (1996) The 1.8-Å x-ray structure of the Escherichia coli PotD protein complexed with spermidine and the mechanism of polyamine binding. Protein Sci. 5, 1984–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Frankel B. A., Kruger R. G., Robinson D. E., Kelleher N. L., McCafferty D. G. (2005) Staphylococcus aureus sortase transpeptidase SrtA: insight into the kinetic mechanism and evidence for a reverse protonation catalytic mechanism. Biochemistry 44, 11188–11200 [DOI] [PubMed] [Google Scholar]
- 52. Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 53. Dawson P. E., Muir T. W., Clark-Lewis I., Kent S. B. (1994) Synthesis of proteins by native chemical ligation. Science 266, 776–779 [DOI] [PubMed] [Google Scholar]
- 54. Janssen B. J., Christodoulidou A., McCarthy A., Lambris J. D., Gros P. (2006) Structure of C3b reveals conformational changes that underlie complement activity. Nature 444, 213–216 [DOI] [PubMed] [Google Scholar]
- 55. Janssen B. J., Huizinga E. G., Raaijmakers H. C., Roos A., Daha M. R., Nilsson-Ekdahl K., Nilsson B., Gros P. (2005) Structures of complement component C3 provide insights into the function and evolution of immunity. Nature 437, 505–511 [DOI] [PubMed] [Google Scholar]
- 56. Dodds A. W., Ren X. D., Willis A. C., Law S. K. (1996) The reaction mechanism of the internal thioester in the human complement component C4. Nature 379, 177–179 [DOI] [PubMed] [Google Scholar]
- 57. Gadjeva M., Dodds A. W., Taniguchi-Sidle A., Willis A. C., Isenman D. E., Law S. K. (1998) The covalent binding reaction of complement component C3. J. Immunol. 161, 985–990 [PubMed] [Google Scholar]
- 58. Tholl D., Harms R., Ludwig A., Kaiser A. (1998) Retarded growth of an Escherichia coli mutant deficient in spermidine synthase can be unspecifically repaired by addition of various polyamines. World J. Microbiol. Biotechnol. 14, 857–863 [Google Scholar]
- 59. Izoré T., Contreras-Martel C., El Mortaji L., Manzano C., Terrasse R., Vernet T., Di Guilmi A. M., Dessen A. (2010) Structural basis of host cell recognition by the pilus adhesin from Streptococcus pneumoniae. Structure 18, 106–115 [DOI] [PubMed] [Google Scholar]
- 60. Krishnan V., Dwivedi P., Kim B. J., Samal A., Macon K., Ma X., Mishra A., Doran K. S., Ton-That H., Narayana S. V. (2013) Structure of Streptococcus agalactiae tip pilin GBS104: a model for GBS pili assembly and host interactions. Acta Crystallogr. D Biol. Crystallogr. 69, 1073–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]