Background: Biosynthesis and functions of Mycobacterium marinum lipooligosaccharides (LOSs) remain elusive.
Results: M. marinum mutants expressing various LOS profiles were generated and used to infect macrophages and amoebae.
Conclusion: Deep LOS mutants are more efficiently phagocytosed than those lacking only LOS-IV.
Significance: Three novel biosynthetic genes and the effect of the LOS content in modulating uptake by phagocytes are reported.
Keywords: Carbohydrate Biosynthesis, Glycolipid Structure, Macrophages, Mass Spectrometry (MS), Mycobacterium, Phagocytosis, Acanthamoeba, Lipooligosaccharide, Thin Layer Chromatography
Abstract
Mycobacterium marinum is a waterborne pathogen responsible for tuberculosis-like infections in ectotherms and is an occasional opportunistic human pathogen. In the environment, M. marinum also interacts with amoebae, which may serve as a natural reservoir for this microorganism. However, the description of mycobacterial determinants in the early interaction with macrophages or amoebae remains elusive. Lipooligosaccharides (LOSs) are cell surface-exposed glycolipids capable of modulating the host immune system, suggesting that they may be involved in the early interactions of M. marinum with macrophages. Herein, we addressed whether LOS composition affects the uptake of M. marinum by professional phagocytes. Mutants with various truncated LOS variants were generated, leading to the identification of several previously uncharacterized biosynthetic genes (wbbL2, MMAR_2321, and MMAR_2331). Biochemical and structural approaches allowed resolving the structures of LOS precursors accumulating in this set of mutants. These strains with structurally defined LOS profiles were then used to infect both macrophages and Acanthamoebae. An inverse correlation between LOS completeness and uptake of mycobacteria by phagocytes was found, allowing the proposal of three mutant classes: class I (papA4), devoid of LOS and highly efficiently phagocytosed; class II, accumulating only early LOS intermediates (wbbL2 and MMAR_2331) and efficiently phagocytosed but less than class I mutants; class III, lacking LOS-IV (losA, MMAR_2319, and MMAR_2321) and phagocytosed similarly to the control strain. These results indicate that phagocytosis is conditioned by the LOS pattern and that the LOS pathway used by M. marinum in macrophages is conserved during infection of amoebae.
Introduction
The hallmark of all members of the Mycobacterium genus is the presence of a unique and waxy cell envelope, which plays a key role in resistance to antimicrobial agents and in the physiopathology of mycobacterial infections by modulating the host immune response as well as phagocytic cell functions (1). This cell wall comprises mainly two types of lipids: (i) the very long-chain fatty acids, mycolic acids, covalently attached to the arabinogalactan/peptidoglycan backbone (2) and (ii) a vast panoply of structurally diverse and extractable compounds (3), including lipoarabinomannan, lipomannan, and related phosphatidylinositol mannosides; glycopeptidolipids; phthiocerol dimycocerosates and related phenolic glycolipids; triacylglycerols as well as different families of trehalose-based glycolipids, such as trehalose dimycolate, sulfolipids, di-, tri-, and pentaacyltrehalose; and lipooligosaccharides (LOSs).2 Although the structure and the distribution of these glycolipids have been well studied in various mycobacterial species, evidence for their participation in pathogenesis and virulence is now emerging (4).
LOSs have been found in many mycobacterial species, such as Mycobacterium kansasii, Mycobacterium gastri, Mycobacterium szulgai, the Mycobacterium canettii variant of Mycobacterium tuberculosis, and Mycobacterium marinum (5–10). However, among the different species, these glycolipids exhibit considerable structural variations in the glycan core as well as in the lipid moiety. Initial work identified the presence of four major LOSs in M. marinum that are designated LOS-I to LOS-IV (10). Partial characterization established the structure of LOS-I as a triacylated 3-O-Me-Rhap-(1–3)-Glcp-(1–3)-Glcp-(1–4)-Glcp-(1–1)-Glcp. This common oligosaccharide moiety is sequentially modified by additional monosaccharides, thus giving rise to more polar LOS species. In particular, an α-d-Xylp further substitutes the glycan core in LOS-II, LOS-III, and LOS-IV. In addition, LOS-II possesses a terminal α-caryophyllose residue (α-3,6-dideoxy-4-C-(d-altro-1,3,4,5-tetrahydroxyhexyl)-d-xylo-hexopyranose), whereas two α-caryophyllose units are present in LOS-III and LOS-IV (11). Caryophyllose may be substituted by the related monosaccharide hydroxylated in position C3 (α-6-deoxy-4-C-(d-altro-1,3,4,5-tetrahydroxyhexyl)-d-galactopyranose). The terminal monosaccharide of LOS-IV decorating the second caryophyllose residue has recently been identified as an unusual α-4-amino-4,6-dideoxy-Galp unit N-acylated by a 3-hydroxy-3-methylated pyrrolidone cycle (12). Subsequent studies established that all LOS subspecies are substituted by one 2-unsaturated 2,4-dimethyl fatty acid and two 2,4-dimethyl and/or 2,3,4-trimethyl fatty acids and demonstrated the critical role of the polyketide synthase-associated protein PapA4 in the assembly of LOS acylation (13). In this regard, the polyketide synthase Pks5, the fatty acid AMP ligase FadD25, and another polyketide synthase-associated protein, PapA3, have also been found to be involved in the biosynthesis of LOS fatty acids (14).
Albeit the structures of all LOSs from M. marinum now appear to be well described, the knowledge about the genes involved in LOS biogenesis is far from being complete, and until recently, only a few biosynthetic genes had been identified (10, 11, 13, 15, 16). However, in a recent search of additional components of the ESX-5 secretion system, a transposon screen in M. marinum for impaired PE_PGRS secretion (known as substrates of ESX-5) led to the identification of several mutants with transposon insertions in the LOS biosynthetic gene cluster (14). Intriguingly, in these LOS mutants, the PE_PGRS proteins appeared to be more firmly attached to the cell surface, thus putting forward an unexpected link between LOS and PE_PGRS release (14). This study not only brought to light a large variety of LOS biosynthetic genes whose disruption led to defects ranging from LOS-IV to complete LOS deficiency but also pointed out the transcriptional regulator WhiB4, which is located outside of the LOS region, as capable of regulating LOS production by controlling expression of several LOS biosynthetic genes (14).
LOSs are highly antigenic cell surface-exposed glycoconjugates and represent useful molecules to be targeted for serotyping in a given mycobacterial species. Early studies demonstrated that rough variants of M. kansasii lacking all LOSs are able to induce chronic systemic infections in mice, whereas smooth variants containing LOS are rapidly cleared from the organs of infected animals (17, 18), leading to the speculation that LOSs may be regarded as avirulence factors by masking other cell wall-associated virulence factors. Studies performed with M. marinum mutants suggested that LOSs may play a role in sliding motility and biofilm formation (15). In vitro studies indicated that purified LOSs inhibit the secretion of TNF-α in LPS-stimulated human macrophages, supporting the view that these glycolipids are key effectors susceptible to interfer with the induction of a proinflammatory response (11). In addition, it was demonstrated that the terminal monosaccharide conferred to LOS-IV important biological functions, such as stimulation of ICAM-1 and CD40 expression at the macrophage cell surface (12). That LOS-IV plays an important role in pathogenesis and/or modulation of M. marinum virulence was corroborated by a recent study demonstrating that a LOS-IV-deficient mutant (inactivated in wecE) showed increased virulence in infected zebrafish embryos compared with the wild-type strain (14). However, despite one study based on a restricted panel of mutants and suggesting that LOSs participate in phagocytosis of M. marinum by macrophages (15), our understanding and the precise contribution of LOS subspecies with respect to the early events between M. marinum and macrophages remain unknown. Such structure-function relationship studies are particularly difficult to assess mainly because of the limited number of isogenic LOS mutant strains and the lack of precise structural data of LOS variants.
To extend our understanding of LOS biosynthesis and to examine the potential role of LOSs in the early interactions of M. marinum with professional phagocytes, we selected/generated a panel of M. marinum mutants expressing a wide diversity of truncated LOS variants. This set of structurally defined mutants was used to decipher the role of LOSs in phagocytosis of M. marinum by macrophages and free-living amoebae.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Culture Conditions
Escherichia coli Top-10 (Invitrogen) was used as the host for cloning and was grown in LB medium. Plasmid selection and maintenance were performed using ampicillin (100 μg·ml−1), hygromycin (200 μg·ml−1), and kanamycin (50 μg·ml−1). The plasmids used in this study are listed and described in Table 1. M. marinum strain M isolated from a human patient as described previously (19) and the MmaM::Tn5370 transposon library (20) were grown/maintained in Sauton's broth medium at 30 °C or on Middlebrook 7H10 agar plates containing 10% oleic acid, albumin, dextrose, catalase (OADC) enrichment and supplemented with appropriate antibiotics. For selection, kanamycin (25 μg·ml−1), hygromycin (80 μg·ml−1), 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-Gal; 50 μg·ml−1), or 2% sucrose were used. For polar lipid extractions and infection experiments, M. marinum strains were grown at 30 °C on Sauton's agar plates containing 10% OADC enrichment and recovered by scraping off the bacterial lawn.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Source/Ref. |
|---|---|---|
| Cloning vectors | ||
| pJET1.2/blunt | PCR product cloning vector, AmpR | Thermo Scientific |
| p2NIL | Recombination vector, nonreplicating in mycobacteria, KanR | 22 |
| pGOAL17 | Source of PacI marker cassette, (sucB, lacZ), AmpR | 22 |
| pMV306Hyg | Mycobacterial integrating vector, HygR | MedImmune |
| pMV261 | Source of hsp60 promoter, KanR | 44 |
| Vectors used for gene replacement | ||
| pGR2349 | p2NIL-based recombination vector carrying the 5′-end of M. marinum MMAR_2349 and its upstream flanking sequence (1434 bp) cloned next to the 3′-end of the gene and its downstream flanking sequence (907 bp), enriched with the PacI cassette from pGOAL17, KanR | This study |
| pGR2331 | p2NIL-based recombination vector carrying the 5′-end of M. marinum MMAR_2331 and its upstream flanking sequence (1034 bp) cloned next to the 3′-end of the gene and its downstream flanking sequence (1277 bp), enriched with the PacI cassette from pGOAL17, KanR | This study |
| pGR2321 | p2NIL-based recombination vector carrying the 5′-end of M. marinum MMAR_2321 and its upstream flanking sequence (996 bp) cloned next to the 3′-end of the gene and its downstream flanking sequence (1666 bp), enriched with the PacI cassette from pGOAL17, KanR | This study |
| Vectors used for complementation | ||
| pPown2349-1kb | PCR fragment harboring whole MMAR_2349 gene (768 bp) together with 1014-bp sequence upstream from the gene, carrying the gene promoter, cloned into the KpnI/XbaI site of the pMV306Hyg integrating vector, HygR | This study |
| pPown2331 | PCR fragment harboring whole MMAR_2331 gene (801 bp) together with 603-bp sequence upstream from the gene, carrying the gene promoter, cloned into the XbaI/HindIII site of the pMV306Hyg integrating vector, HygR | This study |
| pPhsp2321 | PCR fragment harboring whole MMAR_2321 gene (666 bp) together with 435-bp sequence of the hsp60 promoter, cloned into the XbaI/HindIII site of the pMV306Hyg integrating vector, HygR | This study |
| Fluorescence-expressing vector for infection studies | ||
| pMV261_mCherry | pMV261 carrying mCherry under the control of the hsp60 promoter, KanR | 20 |
Screening of M. marinum Tn Library and Determination of the Transposon Insertion Site
We have previously screened a M. marinum Tn library in Dictyostelium discoideum aiming to identify new cell wall-defective mutants (20). During the course of this screening, a M. marinum mutant carrying a Tn5370 insertion in MMAR_2343/papA4 was identified and shown to be defective in LOS production (13). With the aim to identify new LOS biosynthetic genes and based on the observation that the MMAR_2343/papA4 exhibited a rough morphotype, additional mutants harboring a rough morphology were screened on agar plates. The Tn insertion site was determined as described previously (13, 21).
Gene Cloning Strategies
Standard molecular biology protocols were used for all cloning procedures. All PCR products were obtained using thermostable AccuPrimeTM Pfx DNA polymerase (Invitrogen). They were initially cloned into a pJET1.2/blunt vector (Thermo Scientific) followed by sequencing and digestion with the appropriate restriction enzymes. They were then cloned into the final vectors. To facilitate subcloning, most restriction enzyme recognition sites were incorporated into the primer sequences (Table 2), although in one case (2349GR1PstI-nat primer), a natural restriction site was used.
TABLE 2.
Primers used for PCR amplification
Underlined regions indicate the restriction sites used during the cloning steps.
| Primer | Sequence (5′ → 3′) | Description |
|---|---|---|
| Construction of the gene replacement vectors | ||
| 2349GR1PstI-nat | CCCCGAACCCCTTGGTATGCC | GR1-GR2 PCR fragment of MMAR_2349 |
| 2349GR2BamHI-rev | GCGGATCCCGCTCTCGCGCTCCAAGTCAC | |
| 2349GR3BamHI-sen | GCGGATCCCAGTAAGTGCGGTCGAGGGAACC | GR3-GR4 PCR fragment of MMAR_2349 |
| 2349GR4KpnI-rev | GGGGTACCACGCACAAACGGCCCAACAG | |
| 2331GR1HindIII-sen | CAAGCTTGATCCCGACCGGTGGCAG | GR1-GR2 PCR fragment of MMAR_2331 |
| 2331GR2BamHI-rev | CGGATCCGCCTTGACGCTCACTACGC | |
| 2331GR3BamHI-sen | CGGATCCACCCGCGAGACCATCAACTCACC | GR3-GR4 PCR fragment of MMAR_2331 |
| 2331GR4KpnI-rev | CGGTACCGGGCCGACATCGATGAACTC | |
| 2321GR1KpnI-sen | CGGTACCACGCCAGCAACCAGCACACCTG | GR1-GR2 PCR fragment of MMAR_2321 |
| 2321GR2HindIII-rev | GAAGCTTCGAGCTGGGCGATATCACCACTCC | |
| 2321GR3HindIII-sen | GAAGCTTTGTTGAACGACGGGCGGATC | GR3-GR4 PCR fragment of MMAR_2321 |
| 2321GR4ScaI-rev | GAGTACTGTCACTTCACCGCTCATTCACGC | |
| SCO/DCO mutant screening, Southern blot probe synthesis | ||
| 2349-dco-con-sen | CGGATCCATGTCCGTACTGATTATTGTCCCGG | ΔMMAR_2349 SCO/DCO mutant screening, Southern blot probe synthesis |
| 2349-dco-con-rev | CTCTAGAGTCACTGCTCGAGCTGGTGGG | |
| 2331-dco-con-sen | ACGGCGTAGTGAGCGTCAAGGC | ΔMMAR_2331 SCO/DCO mutant screening, Southern blot probe synthesis |
| 2331-dco-con-rev | GGCGTCCTCATCGGTGATGTGAC | |
| Construction of the complementation vectors | ||
| 2349PownXbaI-rev | GTCTAGAGTTGTCGTCAGATTGAGGAAGCGG | pPown2349-1kb vector construction |
| 2349P1kbKpnI-sen | CGGTACCGTAAAGAATCGGACACGGGCACC | |
| 2331PnatXbaI-sen | GTCTAGACAAGAACTGGGCACGCTTGCG | pPown2331 vector construction |
| 2331HindIII-rev | GAAGCTTCCGTTGCCTGCCAATCGTGATC | |
| 2321PhspBamHI-sen | CGGATCCAATGAAGAAACCATTGGTGATTTTCGGG | pPhsp2321 vector construction, ΔMMAR_2321 SCO/DCO mutant screening, Southern blot probe synthesis |
| 2321PhspHindIII-rev | CAAGCTTCCGTATTGATGCATCTCCACCGG | |
Construction of MMAR_2349, MMAR_2331, and MMAR_2321 Gene Replacement Vectors
All PCR primers utilized in this study are listed in Table 2. To create an unmarked deletion of the wbbL2, MMAR_2331, and MMAR_2321 genes, three suicidal recombination delivery vectors based on p2NIL were used (22). In each case, the recombination vector carried the region upstream of the gene together with its 5′-end (the GR1-GR2 PCR fragment; 1434 bp for MMAR_2349; 1034 bp for MMAR_2331, and 996 bp for MMAR_2321) cloned next to the 3′-end of the gene and its downstream region (the GR3-GR4 PCR fragment; 907 bp for MMAR_2349, 1277 bp for MMAR_2331, and 1666 bp for MMAR_2321). The GR1-GR2 and GR3-GR4 PCR fragments of each gene were ligated into p2NIL vector so that the resulting plasmid copies of each gene were devoid of an internal sequence (362 bp for MMAR_2349, 415 bp for MMAR_2331, and 302 bp for MMAR_2321). Finally, the PacI screening cassette from pGOAL17 (22) was inserted into the prepared constructs, yielding the suicide delivery vectors pGR2349, pGR2331, and pGR2321 (Table 1).
Disruption of MMAR_2349, MMAR_2331, and MMAR_2321 by Homologous Recombination
The two-step recombination protocol was used to disrupt the genes of interest at their native loci (22). The plasmid DNA of suicide delivery vectors pGR2349, pGR2331, and pGR2321 was UV-treated (100 mJ), and each of them was electroporated into competent M. marinum strain M where it integrated into the chromosome by homologous recombination. Resulting single crossover (SCO) recombinant mutant colonies were blue, KanR, and sensitive to sucrose (2%). A single MMAR_2349, MMAR_2331, and MMAR_2321 SCO colony was then picked, resuspended in fresh 7H9 medium with OADC, poured onto solid 7H10 medium with OADC without any selective markers, and incubated at 30 °C for 5 days to allow the second crossover to occur. Subsequently, serial dilutions were plated onto medium containing sucrose and X-Gal to select for double crossovers (DCOs). Potential double crossover colonies (white and sucrose-resistant) carrying either wild-type (WT-DCO) or the mutated (ΔMMAR_2349, ΔMMAR_2331, or ΔMMAR_2321) gene (mut-DCO) were screened for kanamycin sensitivity and confirmed by PCR and Southern blotting. The PCR analysis used to distinguish among SCO, WT-DCO, and mut-DCO strains was performed on chromosomal template DNA using primers 2349-dco-con-sen and 2349-dco-con-rev for MMAR_2349 mutants, 2331-dco-con-sen and 2331-dco-con-rev for MMAR_2331 mutants, or 2321PhspBamHI-sen and 2321PhspHindIII-rev for MMAR_2321 mutants. The probe for Southern blot hybridization was generated by PCR using the same primers with pGR2349 (ΔMMAR_2349), pGR2331 (ΔMMAR_2331), or pGR2321 (ΔMMAR_2321) vector as the template. Hybridization was performed on SalI-digested (MMAR_2349), PstI-digested (MMAR_2331), or BamHI/BglII-digested (MMAR_2321) chromosomal DNA. Probe labeling, hybridization, and signal detection were performed using the AlkPhos Direct labeling and detection system (GE Healthcare) according to the manufacturer's instructions.
Construction of Complementation Plasmids
For complementation of the ΔMMAR_2349 DCO mutant, we constructed the pPown2349-1kb (Table 1). A PCR fragment (1782 bp) carrying whole MMAR_2349 gene and a 1014-bp upstream sequence were amplified using primers 2349P1kbKpnI-sen and 2349PownXbaI-rev and cloned into the KpnI/XbaI site of the pMV306Hyg integrating vector. For complementation of the ΔMMAR_2331 DCO mutant, we constructed the pPown2331 carrying the MMAR_2331 gene together with an additional 603 bp of upstream sequence and cloned it into pMV306Hyg integrating vector (Table 1). For complementation of the ΔMMAR_2321 DCO mutant, the pPhsp2321 vector was constructed. The 690-bp fragment containing whole MMAR_2321 gene was PCR-amplified and cloned into the BamHI/HindIII site of the pMV261. The fragment carrying MMAR_2321 was then excised from pMV261 together with hsp60 promoter (435 bp) using XbaI/HindIII digestion and ligated into pMV306Hyg integrating vector (Table 1).
Drug Susceptibility Testing
Minimal inhibitory concentrations of several antimycobacterial agents were determined against M. marinum strains as described previously (20). Briefly, 10-fold serial dilutions of actively growing cultures were spotted on Middlebrook 7H10 agar plates supplemented with OADC and increasing drug concentrations and incubated around 12 days at 30 °C. The minimal inhibitory concentration was defined as the minimum concentration required to inhibiting 99% of mycobacterial growth.
Polar Lipid Extraction and Analysis
Bacteria were grown at 30 °C on Sauton's agar plates containing 10% OADC enrichment and recovered by scraping off the bacterial lawn. Polar lipids were extracted from the bacterial pellets according to established procedures (10) and separated by two-dimensional thin layer chromatography (TLC) on 10 × 10-cm2 plates of aluminum-backed silica Gel 60 (Merck) using chloroform/methanol/water (60:30:6, v/v/v) in the first direction and chloroform/acetic acid/methanol/water (40:25:3:6, v/v/v/v) in the second direction. Glycolipids were visualized by spraying plates with orcinol/sulfuric acid reagent (0.2% (w/v) orcinol in H2SO4/water (1:4, v/v)) followed by charring. For subsequent structural analyses, individual LOS species were purified by preparative two-dimensional TLC on 20 × 20-cm2 plates of glass-backed silica Gel 60 (Merck). The lipids were visualized with iodine labeling and recovered by scraping off the spots of interest prior to resuspension in chloroform/methanol/water (1:2:0.8, v/v/v). Solutions were filtered and dried under vacuum prior to structural determination.
Structural Analyses
Permethylation of LOSs was performed according to the procedure described by Ciucanu and Kerek (23). Briefly, glycolipids were incubated for 2 h in the presence of 200 mg·ml−1 NaOH in dry DMSO (300 μl) and iodomethane (200 μl). The methylated LOSs were extracted in chloroform and washed seven times with water. After evaporation of the reagents, the samples were dissolved in methanol prior to mass spectrometry analysis. The molecular masses of native and permethylated compounds were measured by MALDI-TOF on a Voyager Elite reflectron mass spectrometer (PerSeptive Biosystems, Framingham, MA) equipped with a 337-nm UV laser. Samples were prepared by mixing in a tube 5 μl of diluted native or permethylated LOS solutions in methanol and 5 μl of 2,5-dihydroxybenzoic acid matrix solution (10 mg·ml−1) dissolved in methanol/water (1:1, v/v).
Macrophage Culture and Phagocytosis Assays
The macrophage-like cell line J774 was maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) at 37 °C under atmosphere containing 5% CO2. Cells were allowed to adhere in a 24-well plate for 24 h at 37 °C at a density of 5 × 104 cells/well in 0.5 ml. For infections, mycobacteria were grown at 30 °C on Sauton's agar plates containing 10% OADC enrichment for 5–7 days. Bacterial lawns were scraped off the plates, and bacteria were resuspended in 1 ml of PBS. Bacterial clumps were disrupted by 10 successive passages through a 26-gauge needle. The remaining aggregates were then eliminated with a short spin for 1 min at 1200 rpm in a microcentrifuge. Homogeneity of the bacterial preparations and lack of eventual remaining bacterial clumps were assessed under the microscope following acid-fast staining. Bacterial suspensions were diluted in DMEM supplemented with 10% FCS at a density of 2 × 105 cells·ml−1 and plated onto Middlebrook 7H10 agar supplemented with 10% OADC to determine the exact number of bacteria in the inoculum. The macrophage medium was then replaced by 0.5 ml of the bacterial inoculum to reach a multiplicity of infection (m.o.i.) of 2. Cells were incubated for 3 h at 30 °C under atmosphere containing 5% CO2 to enable phagocytosis, and free extracellular bacteria were killed by the addition of 200 μg·ml−1 gentamycin for an additional 1 h. The residual extracellular bacteria were eliminated by washing the cells three times in PBS. To evaluate the number of phagocytized bacteria, cells were lysed in 0.1% Triton X-100 in PBS, and serial dilutions of the lysates were plated onto Middlebrook 7H10 medium containing 10% OADC. Colony forming units (cfu) were counted after 10 days of incubation at 30 °C. Each strain was tested in triplicate. The phagocytosis rate was expressed as the ratio of phagocytosed bacteria to the number of bacteria in the inoculum. Ratios were arbitrarily set to 1 for the wild-type M strain. When analyzing the intracellular growth rate, fresh DMEM supplemented with 10% FCS and 20 μg·ml−1 gentamycin was added to infected cells, which were further incubated for 4 days at 30 °C under atmosphere containing 5% of CO2. For cfu scoring, cells were washed three times in PBS and lysed in 0.1% Triton X-100 in PBS. Serial dilutions were plated onto Middlebrook 7H10 medium supplemented with 10% OADC.
To determine the number of bacterium-containing macrophages, the same procedure was applied except that macrophages were allowed to adhere on coverslips prior to infection with bacteria carrying the pMV261_mCherry (Table 1). Infected cells were washed three times in PBS to remove remaining extracellular bacteria and labeled with fluorescein-conjugated phalloidin (Sigma) according to the manufacturer's instructions to visualize the shape of the cells. Stained cells were then observed using a Zeiss Axioimager, and quantification of mycobacterium-residing cells was determined by counting 20 fields per coverslip. Each strain was analyzed in triplicates.
Amoeba Phagocytosis Assays
Acanthamoeba polyphaga and Acanthamoeba castellanii were grown and maintained axenically in peptone-yeast-glucose (PYG) broth (Biotechnologie Appliquée) at 30 °C for A. polyphaga and 28 °C for A. castellanii for 2 days in 150-cm3 culture flasks containing 30 ml of medium. Acanthamoeba were harvested when their average concentration reached 5 × 105 cells·ml−1. The harvested cells were then centrifuged at 2000 rpm for 10 min, and the pellet was suspended twice in 30 ml of Page's modified Neff's amoeba saline. One milliliter of this suspension was dropped into each well of a 12-well microplate. M. marinum strains were grown on Sauton's agar medium at 30 °C to late log phase, resuspended in PBS, syringe-filtered to remove clumps, and used to infect Acanthamoeba in 12-well culture plates at an m.o.i. of 1. Following incubation for 2 h at 30 °C, the medium containing extracellular bacteria was aspirated. The amoeba monolayer was washed three times with fresh PYG medium and treated with 150 μg·ml−1 gentamicin for 1 h to kill the remaining extracellular mycobacteria, allowing study of only internalized microorganisms. The medium was then removed and replaced with fresh PYG.
To assess the cfu, infected A. castellanii or A. polyphaga were treated with Mycoprep lysis buffer (BD Biosciences), and mycobacteria were removed by centrifugation at 13,000 rpm for 10 min at room temperature. The resulting bacterial pellet was washed three times with sterile PBS, diluted, and plated on Middlebrook 7H10 agar. cfu were counted after 8–10 days at 30 °C. The percentage of infected Acanthamoeba was determined based on counting >50 amoebae in 10 different microscopic fields. Infected amoebae were stained with Ziehl-Neelsen stain according to the manufacturer's instructions (BD TB stain kit).
RESULTS
Isolation of M. marinum LOS Mutants with Altered Colony Morphology
Initial studies dedicated to the genetics of LOS biosynthesis in mycobacteria were based on the isolation of a transposon-insertion mutant of M. marinum 1218R that exhibited altered colony morphology (24). This mutant, carrying a transposon in MMAR_2313/losA and originally thought to be defective in phosphatidylinositol mannoside synthesis, was later proven to be deficient in LOS biosynthesis (10). Similarly, inactivation of MMAR_2333 involved in addition of a caryophyllose moiety in LOSs was also reported to exhibit a rough texture (16). In addition, a recent study aimed to identify novel components involved in the secretion of PE_PGRS proteins led to the identification of several M. marinum mutants with transposon insertions in the LOS biosynthetic gene cluster (14). Most of these mutants also exhibited an altered morphotype (14). Based on these observations and with the aim to provide a large panel of LOS biosynthetic mutants to initiate structure-function relationship activities, we first screened an MmaM::Tn5370 transposon library (20) on Middlebrook 7H10 agar plates to select mutants with impaired colony morphology. This led to the recovery of six mutants exhibiting a “rough” morphotype compared with the parental strain (Fig. 1A). Determination of the Tn5370 insertion sites identified two independent losA mutants and single mutants in MMAR_2315, MMAR_2319, MMAR_2353, and MMAR_5103. In addition, we also included in this study an MMAR_2343/papA4 mutant previously identified as a rough mutant defective in LOS production (13).
FIGURE 1.
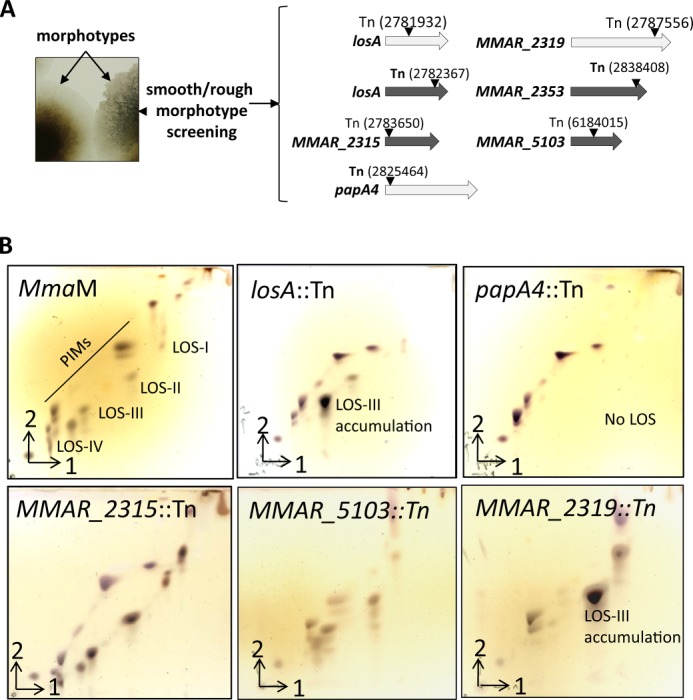
Screening for LOS biosynthetic mutants in M. marinum. A, selection of rough morphotypes. Screening the Tn library on Middlebrook 7H10 led to six rough variants carrying Tn insertions in losA (MMAR_2313) (two independent mutants), MMAR_2315, MMAR_2319, MMAR_2353, and MMAR_5103. Positions of the insertions are indicated in parentheses. White arrows indicate the Tn mutants that were further analyzed in this study. An additional papA4/MMAR_2343::Tn mutant picked up as a rough mutant and characterized previously (13) is also shown. B, LOS profiles of the M. marinum mutants exhibiting a rough morphotype on 7H10 agar. Following extraction, polar glycolipids from the parental strain (M. marinum strain M) and the losA, papA4, MMAR_2315, MMAR_2319, and MMAR_5103 Tn insertion mutants were separated by two-dimensional TLC using the solvent system chloroform/methanol/water (60:30:6, v/v/v) in the first direction and chloroform/acetic acid/methanol/water (40:25:3:6, v/v/v/v) in the second direction. Glycolipids were detected with orcinol/sulfuric acid staining and charring. PIMs, phosphatidylinositol mannosides.
Polar lipids were extracted from the parental M. marinum strain M as well as the various LOS mutants identified in this screen. Glycolipid profiles were recorded on two-dimensional TLC and revealed with orcinol/sulfuric acid. The glycolipid pattern of the wild-type strain was typically characterized by the presence of phosphatidylinositol mannosides and the four major LOS species designated LOS-I to LOS-IV (Fig. 1B) as reported earlier (10, 11). In agreement with previous work, the losA mutant was characterized by the absence of LOS-IV and the accumulation of LOS-III (10, 11), whereas the papA4 mutant completely lacked LOS production (13). The MMAR_2319 mutant, also identified in a previous screen (14), exhibited a profile similar to that of the losA mutant characterized by the accumulation of LOS-III (Fig. 1B). The MMAR_2353 mutant was characterized by a strongly reduced LOS profile as reported earlier (14) (data not shown). In addition, two new mutants with insertions in MMAR_2315, encoding a hypothetical methyltransferase, and in MMAR_5103, encoding a protein of unknown function, exhibited a LOS profile comparable with that of the parental strain (Fig. 1B) and therefore were not further investigated.
Targeted Inactivation of LOS Biosynthetic Genes
The above mentioned set of mutants comprises strains that are deficient either in the very early step of LOS biosynthesis (papA4 mutant) or in the final steps involved in the synthesis of LOS-IV (losA or MMAR_2319). However, to provide a wider panel of mutants expressing intermediate structural LOS variants, we opted for targeted gene disruption of three previously uncharacterized genes: MMAR_2349 (wbbL2), MMAR_2321, and MMAR_2331. wbbL2 was selected because it encodes a putative rhamnosyltransferase sharing homology with WbbL1, which uses dTDP-rhamnose to provide the rhamnosyl-containing linker unit responsible for the attachment of the cell wall polymer mycolyl arabinogalactan to the peptidoglycan (25). Methylrhamnose is a monosaccharide found in LOS-I to LOS-IV. MMAR_2331, encoding a hypothetical protein, was chosen because it is located in the close vicinity of MMAR_2333, reported to encode a glycosyltransferase involved in the generation of a lipid-linked caryophyllose donor (16), the caryophyllose consisting of a family of unusual 4-C-branched monosaccharides shared by LOS-II to LOS-IV (11). With respect to the biosynthesis/transfer of the very unusual aglycone moiety of LOS-IV, MMAR_2321 was proposed as a putative candidate for the transfer of the pyrrolidone cycle onto the dideoxy-Gal through an amide bond (12). Unmarked deletions of wbbL2, MMAR_2331, and MMAR_2321 were performed by generating the corresponding suicidal recombination delivery vectors, and disruption was achieved by homologous recombination using a two-step recombination protocol as described under “Experimental Procedures.” PCR analyses on chromosomal template DNA were done to distinguish single crossover, wild-type double crossover, and mutant double crossover strains (Fig. 2A). In addition, Southern blot hybridization was performed to confirm the deletion in mutated M. marinum strains (Fig. 2B).
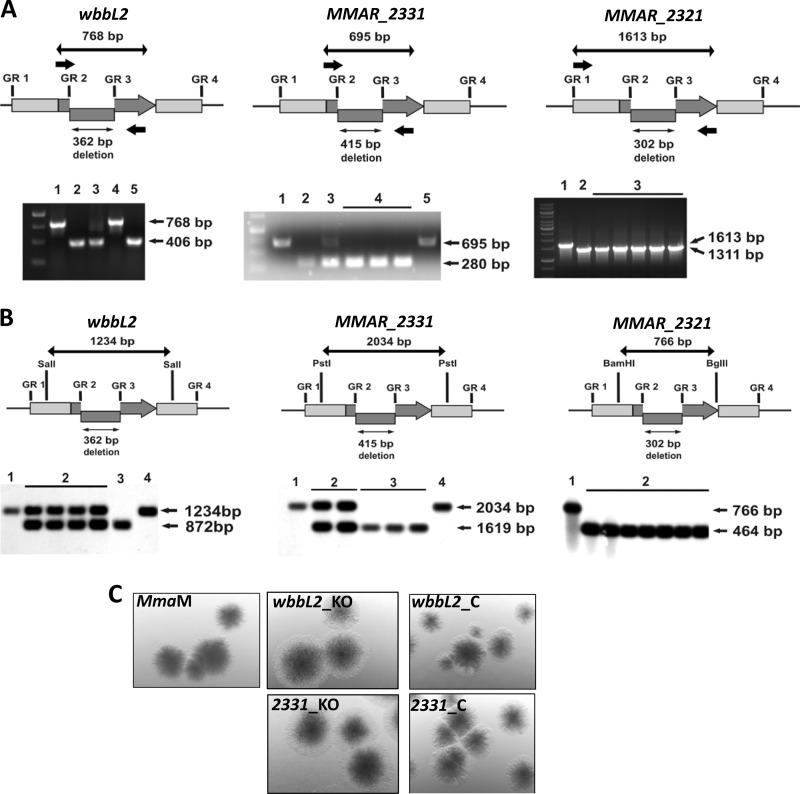
FIGURE 2.
Gene inactivation of wbbL2, MMAR_2331, and MMAR_2321 in M. marinum. A, PCR-based genotyping of the mutants. Top, schematic showing the genomic orientation of the wbbL2, MMAR_2331, and MMAR_2321 genes (thick gray arrows). The internal deletion introduced into a gene using a gene replacement vector is marked with a shifted rectangle. GR1-GR2/GR3-GR4 denote the gene-flanking fragments that were amplified and cloned together in the pGR2349, pGR2331, or pGR2321 suicide vector to obtain the mutated ΔwbbL2, ΔMMAR_2331, and ΔMMAR_2321 allele for gene replacement. The small thick black arrows indicate the binding sites for the primers that were used for PCR-based genotyping. The thick black arrows indicate the length of the product amplified from wild-type wbbL2 (768 bp), MMAR_2331 (695 bp), and MMAR_2321 (1613 bp). The thin arrows represent the length of the deleted fragment (362 bp, yielding a 406-bp ΔwbbL2 PCR product; 415 bp, yielding a 280-bp ΔMMAR_2331 PCR product; 302 bp, yielding a 1311-bp PCR product). Bottom, the genotypes of selected M. marinum wbbL2, MMAR_2331, and MMAR_2321 mutants were confirmed by PCR. For wbbL2, lanes are as follows: lane 1, control PCR conducted on wild-type M. marinum DNA; lane 2, control PCR conducted on pGR2349; lane 3, SCO strain carrying both wild-type and mutated genes; lane 4, the WT-DCO strain; lane 5, DCO strain ΔwbbL2. For MMAR_2331, lanes are as follows: lane 1, control PCR conducted on wild-type M. marinum DNA; lane 2, control PCR conducted on pGR2331; lane 3, SCO strain; lane 4, DCO strain ΔMMAR_2331; lane 5, WT-DCO strain. For MMAR_2321, lanes are as follows: lane 1, control PCR conducted on wild-type M. marinum DNA; lane 2, control PCR conducted on pGR2321; lane 3, DCO knock-out strain ΔMMAR_2321. B, Southern blot analysis of the mutants. In the schematics (upper panels), the long black arrow represents the restriction-digested DNA fragment (1234 bp for wbbL2, 2034 bp for MMAR_2331, and 766 bp for MMAR_2321), whereas the short black arrow indicates the size of the internal deletion in the mutated gene (362 bp for ΔwbbL2, 415 bp for ΔMMAR_2331, and 302 bp for ΔMMAR_2321). The wbbL2, MMAR_2331, and MMAR_2321 genes are represented by the gray arrows, and the internal deletion is represented by the shifted rectangles. Shown is a Southern blot (lower panels) confirming the deletion in mutated M. marinum strains. Lanes are as follows: for ΔwbbL2 and ΔMMAR_2331: lane 1, wild-type M. marinum; lane 2, SCO strains; lane 3, DCO knock-out ΔwbbL2 or ΔMMAR_2331 mutant; lane 4, wild-type DCO; for ΔMMAR_2321: lane 1, wild-type M. marinum; lane 2, DCO knock-out ΔMMAR_2321. C, effect of deletion of wbbL2 or MMAR_2331 on colony morphology. Shown are colonies of M. marinum strain M (wild type), wbbL2, or MMAR_2331 deletion mutants and their corresponding complemented strains (wbbL2_C and 2331_C) on 7H10 agar plates. Compared with wild-type M. marinum strain M, both mutants presented a rough and dried texture, a phenotype that was restored following complementation.
Both the wbbL2 and MMAR_2331 mutants exhibited altered colony morphology with a rough appearance, suggesting potential alterations in the cell wall and presumably in the LOS profile (Fig. 2C). For complementation of the wbbL2 mutant, pPown2349-1kb was constructed (Table 1) in which a DNA segment comprising the whole wbbL2 gene along with an additional 1014-bp fragment sequence upstream of wbbl2 was cloned in the pMV306Hyg integrating vector. For complementation of the MMAR_2331 mutant, pPown2331 was achieved by cloning MMAR_2331 together with its 603-bp upstream sequence cloned into pMV306Hyg (Table 1). In both cases, the colony morphology of the mutants was restored to that of the parental type upon introduction of the corresponding complementing plasmid, indicating that the phenotype observed was due to the loss of WbbL2 and MMAR_2331 activity (Fig. 2C). In contrast, the MMAR_2321 exhibited a colony morphology similar to that of the wild-type strain (data not shown).
LOS Content in the wbbL2, MMAR_2331, and MMAR_2321 Mutants
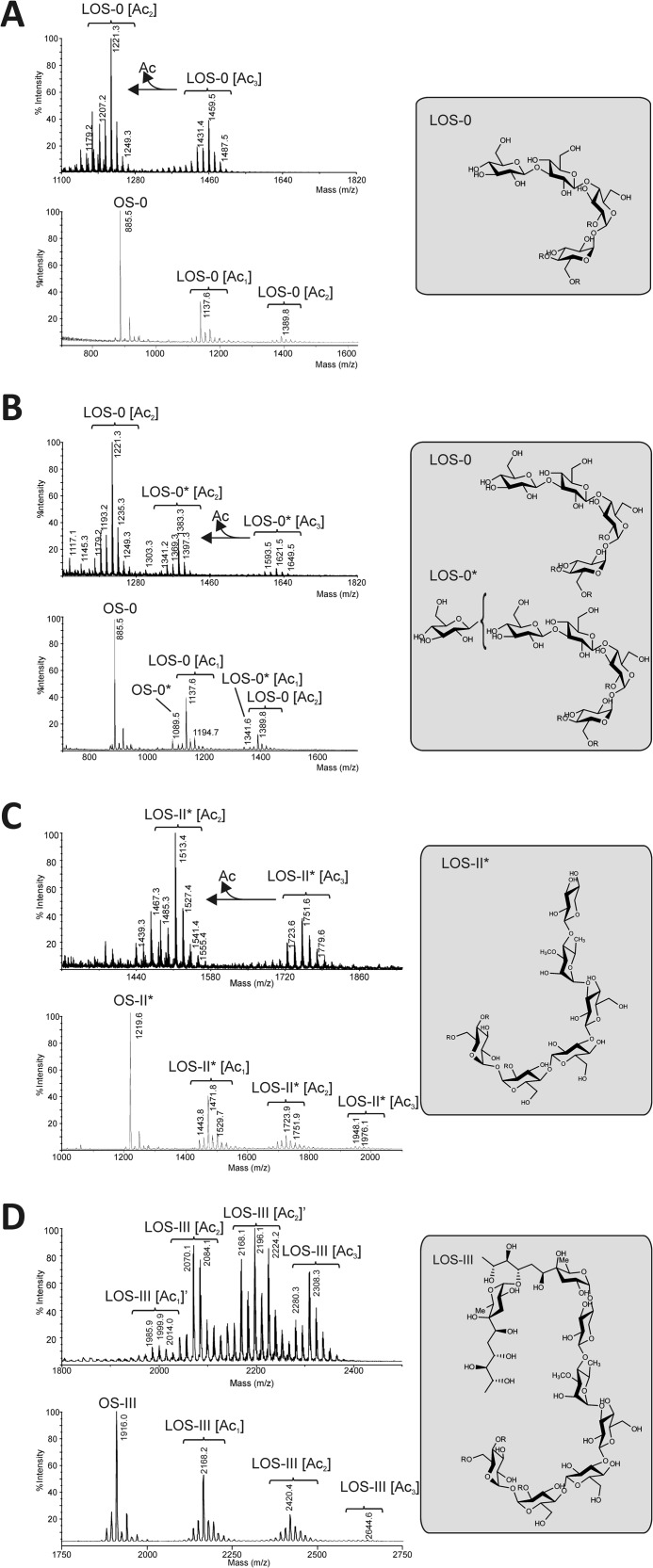
To determine whether disruptions of wbbL2, MMAR_2331, and MMAR_2321 play a role in LOS biosynthesis, the polar lipid profiles of the corresponding knock-out strains were examined by two-dimensional TLC (Fig. 3). The wbbL2 mutant clearly showed a distinctive LOS pattern with the absence of most major LOS species and a concomitant accumulation of two intermediates, designated spot 1 and spot 2 (Fig. 3). Both spots were purified by preparative TLC and analyzed by MALDI-MS in native and permethylated forms. The MALDI-MS spectrum of the native spot 1 exhibited two distinct 14-mass unit-incremented peak clusters with maximum intensities at m/z 1221 and 1459, establishing the presence of a di- and tri-acetylated Hex4, respectively, substituted by complex mixtures of lipids (Fig. 4A). The carbohydrate moiety attribution was confirmed by MS analysis of permethylated glycolipids that showed a sharp signal at m/z 885 corresponding to a Hex4 oligosaccharide. In addition, following the per-O-methylation procedure (13), the alkali-labile fatty acyl substituents are incompletely replaced by methyl groups, a fraction of which is retained on the trehalose moiety as demonstrated by clusters around signal at m/z 1137 and 1389 corresponding to partially deacetylated Hex4 (Fig. 4A). The MS profile is identical to that of LOS-I as reported previously (13) except for a loss of 174 mass units corresponding to a deHex residue. Altogether, these data strongly suggest that spot 1 contains a new biosynthetic intermediate Glc4 (Ac3) designated LOS-0 (Ac3) that corresponds to the immediate precursor of LOS-I. The observation of native LOS-0 (Ac2) along with LOS-0 (Ac3) was tentatively attributed to a partial deacylation of native LOS-0 (Ac3) that typically occurs during MALDI analysis. In contrast to spot 1, analysis of spot 2 showed a slightly more complicated pattern. In native form, the main signal at m/z 1221 was assigned to LOS-0 (Ac2), but no LOS-0 (Ac3) could be observed. Accordingly, the MALDI spectrum of the permethylated sample showed signals consistent with the presence of fully deacylated oligosaccharide OS-0 as well as partially deacylated LOS-0 (Ac1) and LOS-0 (Ac2) (Fig. 4B). Along with these signals attributed to LOS-0 (Ac2), another set of signals was found in native forms around m/z 1383 and 1621 that were attributed to Hex5 (Ac2) and Hex5 (Ac3). As described previously, observation of native Hex5 (Ac2) can be interpreted as a partial deacylation of native Hex5 (Ac3) during MALDI-MS analysis. This assignment was confirmed following permethylation, which led to signals at m/z 1089 and 1341 corresponding to Hex5 oligosaccharide and Hex5 (Ac1) glycolipid, respectively. This established spot 2 as being composed of LOS-0 (Ac2) and of a so far undescribed Hex5 (Ac3) glycolipid designated LOS-0*.
FIGURE 3.
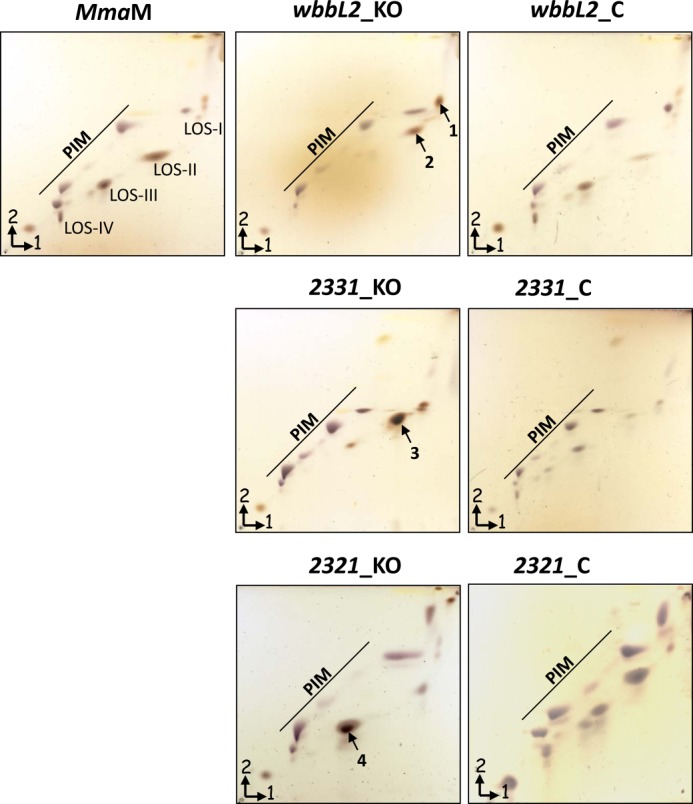
LOS content of the various M. marinum mutants. Following extraction, polar glycolipids from the wild type and the wbbL2, MMAR_2331, and MMAR_2321 deletion mutants as well as their corresponding complemented strains (wbbL2_C, 2331_C, and 2321_C) were separated by two-dimensional TLC using the solvent system chloroform/methanol/water (60:30:6, v/v/v) in the first direction and chloroform/acetic acid/methanol/water (40:25:3:6, v/v/v/v) in the second direction. Glycolipids were detected with orcinol/sulfuric acid staining and charring. In parallel, TLC plates were stained with iodine vapor, and the glycolipid intermediates accumulating in the different mutants (labeled 1 and 2 for wbbL2, 3 for MMAR_2331, and 4 for MMAR_2321) were individually extracted for further structural identification. PIM, phosphatidylinositol mannosides.
FIGURE 4.
Structural characterization of the LOS precursors accumulating in the various M. marinum mutant strains. Each purified component was analyzed by MALDI-TOF MS in their native (top panels) or their permethylated forms (bottom panels). A, mass spectrometry analysis of spot 1 from the wbbL2 mutant revealed the presence of a very early triacetylated LOS precursor, designated LOS-0 (Ac3), whose carbohydrate moiety consists of four Glc residues. B, spot 2 from the wbbL2 mutant consists of a mixture of LOS-0 (Ac2) and a new pentaglucosylated glycolipid designated LOS-0*. C, analysis of spot 3 from the MMAR_2331 mutant revealed the presence of a glycolipid whose m/z corresponds to LOS-II*, a LOS intermediate that has been reported to accumulate also in MMAR_2332 and MMAR_2333 mutants (15, 16). D, spot 4 from the MMAR_2321 mutant was identified as LOS-III (Ac3) based on the observation of fully acetylated and partially acetylated signal clusters in permethylated and native forms. The corresponding structures identified are shown in insets. R corresponds either to acylation position or hydrogen.
Overall, these results indicate that this strain corresponds to a high order LOS mutant and that functional inactivation of wbbL2 occurred in the very early biosynthetic steps. The LOS-0 precursor lacks the terminal methyl-Rha residue of LOS-I, confirming the involvement of WbbL2 with Rha metabolism. Accumulation of LOS-0 appears to trigger the synthesis of a so far undescribed LOS-0* most likely through the addition of a further glucose residue onto the tetraglucosyl backbone of LOS-0. However, the position of the extra glucose residue could not be fully determined due to low quantities.
The two-dimensional TLC profile of the MMAR_2331 mutant revealed the absence of LOS-II, LOS-III, and LOS-IV paralleled by the accumulation of a glycolipid absent in the parental strain and migrating to an intermediate position between LOS-I and LOS-II (Fig. 3, designated spot 3). However, this TLC profile was highly reminiscent of that reported in an MMAR_2332 mutant (also known as ilvB1_3) (14, 15) or in an MMAR_2333 mutant (16) characterized by the accumulation of a LOS-II* intermediate. To further demonstrate the nature of spot 3, the purified product was subjected to MALDI-MS (Fig. 4C). Analysis of the permethylated spot 3 revealed a major oligosaccharide signal at m/z 1219 attributed to Hex4deHex1Pent1 in agreement with the LOS-II* monosaccharide composition Glc4Me-Rha1Xyl1. Mono-, di-, and triacylated forms of this oligosaccharide substituted by various combinations of lipids were further observed as clusters around signals at m/z 1471, 1423, and 1976. Similarly, LOS-II* (Ac3) and its partially deacylated LOS-II* (Ac2) were observed in their native forms as clusters around major signals at m/z 1513 and 1751 (Fig. 4C). Overall, these results clearly establish the identity of spot 3 as LOS-II*, the immediate biosynthetic precursor of LOS-II, which contains d-Xylp attached to the Glc4Me-Rha1 core but lacks the caryophyllose found in LOS-II. These data indicate that the intermediate we isolated from the MMAR_2331 knock-out strain was identical to that produced in the MMAR_2332 (15) and MMAR_2333 (16) mutants, thus connecting MMAR_2331 to either the synthesis of caryophyllose or to its transfer on LOS-II* to generate LOS-II.
The MMAR_2321 mutant was characterized by the absence of LOS-IV and the concomitant accumulation of an intermediate glycolipid, designated spot 4 (Fig. 3). MALDI MS analysis (Fig. 4D) of permethylated spot 4 revealed a major signal at m/z 1916.0 tentatively attributed to Car2Hex4deHex1Pent1 in agreement with the oligosaccharide moiety OS-III identified in LOS-III (Fig. 4D) (11). The presence of two caryophyllose residues at the terminal non-reducing end was confirmed in MS/MS analysis of OS-III by observing two consecutive losses of 278 mass units at m/z 1791.7 and 1513.5 from signal at m/z 2070 (data not shown). Along with OS-III, three clusters of signals around m/z 2168, 2420, and 2644 were identified as partially deacylated LOS-III (Ac1) and LOS-III (Ac2) and as fully acetylated LOS-III (Ac3), respectively. The presence of LOS-III (Ac3) and deacetylated LOS-III (Ac2) was confirmed in the analysis of native spot 4 by the observation of two signals clusters around m/z 2308 (LOS-III (Ac3)) and 2070 (LOS-III (Ac2)). The acylation status of LOS-III (Ac3) and LOS-III (Ac2) are similar to those of LOS-II* and LOS-0. In addition, two additional clusters of signals were observed around m/z 2196 and 2000. The MS/MS fragmentation pattern of signal at m/z 2196 indicated that this compound is a diacylated LOS-III substituted by a set of fatty acids longer by an average of nine carbons than those of LOS-III (Ac2). These compounds are referred to as LOS-III (Ac2)′. Importantly, complementation of all the mutants restored a wild-type LOS profile (Fig. 3), indicating that alteration of the LOS pattern could be attributed to the inactivation of the targeted genes and not to an eventual polar effect on the downstream genes.
Altered LOS Production Facilitates Phagocytosis by Macrophages
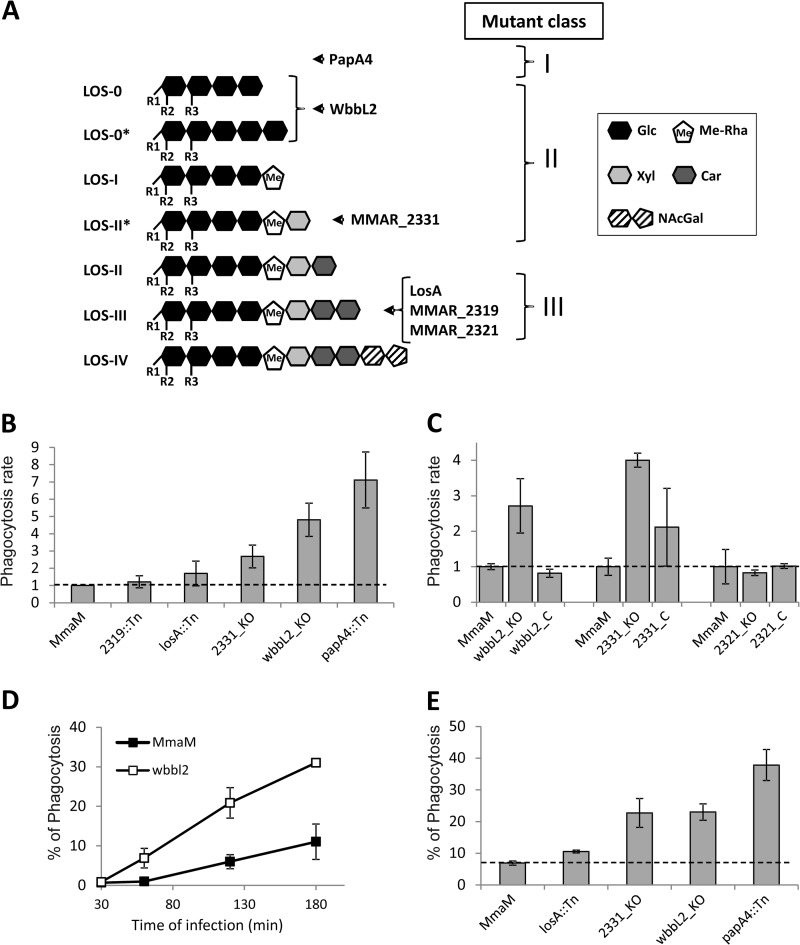
Fig. 5A relates the structures of the major LOS subspecies identified and described in the present study. The availability of a representative set of M. marinum mutants with various incomplete LOS variants allowed us to examine their potential biological role in the early interaction with professional phagocytes as would be expected for surface-attached molecules. To this aim, wild-type M. marinum strain M (expressing the whole range of LOSs), papA4 mutant (lacking the entire LOS profile), and other mutants expressing either only highly truncated LOS subspecies (wbbL2) or intermediate variants (MMAR_2331) or lacking only LOS-IV (losA, MMAR_2319, and MMAR_2321) were used to infect J774 macrophages to examine and compare their ability to enter into the cells. In Sauton's broth medium, all mutants exhibited growth rates similar to that of the parental strain (data not shown). To examine the entry of bacteria into macrophages, infection experiments were performed at an m.o.i. of 2. Immediately following infection (3 h at 30 °C), cells were treated with gentamycin to kill extracellular mycobacteria and washed three times with PBS. The cfu recovered from macrophages were enumerated by plating on 7H10 agar. Unexpectedly, our results showed an inverse correlation between the size of the LOS and an increased phagocytosis rate, culminating with a reproducibly 7-fold increased rate with the LOS-deficient papA4 mutant (Fig. 5B). Intermediate phagocytosis rates were observed with the wbbL2 mutant lacking most LOS subspecies and the MMAR_2331 mutant characterized by the accumulation of LOS-II* and the defect in more polar LOS subspecies. Conversely, the two LOS-IV-deficient strains (MMAR_2319 and losA) failed to demonstrate increased phagocytosis rates. Importantly, the increased phagocytosis rates of the wbbL2 and MMAR_2331 mutants were restored in the complemented mutant strains (Fig. 5C). Analyzing the kinetic of wbbL2 phagocytosis during the first 3 h of contact with macrophages showed that the mutant entered macrophages very efficiently and rapidly compared with the wild-type strain, resulting in around 30% of wbbL2-phagocytized mycobacteria versus 10% of wild type-phagocytized mycobacteria after 3 h of infection (Fig. 5D). That the absence of LOS-IV and/or accumulation of LOS-III failed to affect the uptake of M. marinum by macrophages was further supported by infection experiments with the MMAR_2321 mutant, which exhibited a phagocytosis rate similar to that of the wild-type strain (Fig. 5C). Based on these structure-function relationships, we propose that the different strains should be classified into three groups (Fig. 5A): (i) class I mutants (papA4) characterized by the absence of LOS are very efficiently phagocytosed, (ii) class II mutants (wbbL2 and MMAR2331) producing only the early and less polar LOS subspecies with less pronounced uptakes by macrophages than class I mutants, and (iii) class III mutants (losA, MMAR_2319, and MMAR_2321) synthesizing all LOS subspecies except the most polar LOS-IV and phagocytosed similarly to the wild-type strain.
FIGURE 5.
Phagocytosis of LOS mutants by murine macrophages. A, schematic representation of the different subspecies of LOSs and the intermediates identified in this study. NAc-Gal, N-acylated dideoxygalactose. R1, R2, and R3 represent the acyl chains. B, phagocytosis rate of the various LOS mutants by J774 macrophages normalized to 1 for the wild-type strain. Results are expressed as means ± S.E. (error bars) from three to five independent experiments. C, phagocytosis rates of the wbbL2, MMAR_2331, and MMAR_2321 mutants and complemented (_C) strains. D, kinetics of phagocytosis of the wbbL2 mutant. For C and D, results are representative of two independent experiments and are expressed as means ± S.D. (error bars) of triplicates. E, percentages of mycobacteria-containing macrophages determined by counting infected cells under the microscope. Results are expressed as means ± S.E. (error bars) from three independent experiments.
To rule out the possibility that the differences in phagocytosis observed may result from mycobacterial aggregation, which would affect the cfu numbers, and to investigate whether the increased phagocytosis rate is associated with an increased number of infected cells, infected macrophages were directly counted under the microscope. For this purpose, several mutants were transformed with pMV261_mCherry and subsequently used to infect macrophages. The number of cells containing red fluorescent mycobacteria was then counted under a fluorescence microscope. As shown in Fig. 5E, the highest proportion of infected macrophages accounted for the papA4 mutant (40% of infected cells), and a proportion of 25% was found for the MMAR_2331 and wbbl2 mutants. In contrast, comparable proportions of infected cells were observed with the losA mutant and the wild-type strain (around 10%). Taken together, these data indicate that both class I and class II mutants are more efficiently engulfed by macrophages than class III mutants.
Altered LOS Production Increases Uptake of M. marinum by Amoebae
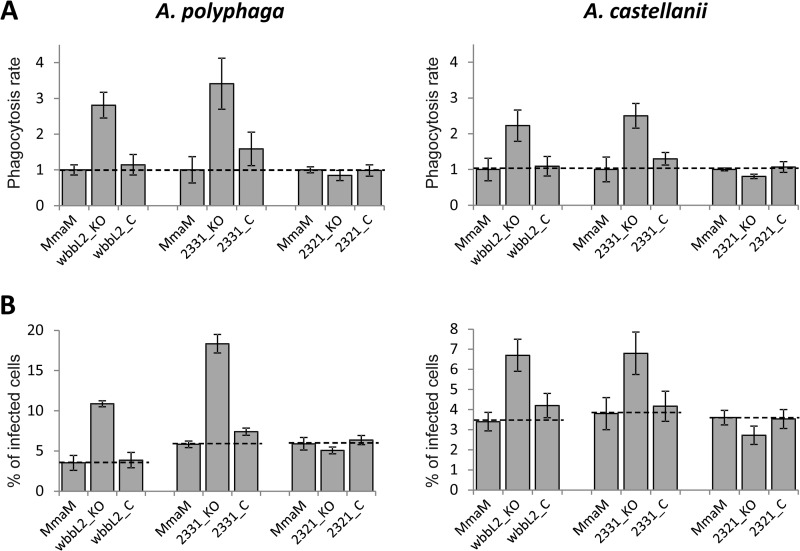
M. marinum is a waterborne pathogen that causes tuberculosis-like infection in ectotherms, such as frogs and fish (26). Free-living amoebae, such as Acanthamoeba, are professional phagocytes (27), and non-tuberculous mycobacteria, including M. marinum, have been recovered from samples of water colonized by free-living amoebae (28). Distinct mycobacterial species have been shown to resist destruction by amoebae (27, 29–31), which are thought to serve as a reservoir for non-tuberculous mycobacteria in the environment. Because M. marinum and amoebae share the same environmental niche, it is very likely that amoebae represent a natural host for M. marinum. A recent study demonstrated that the ESX-1 secretion system is required for M. marinum pathogenesis in A. castellanii and that M. marinum utilizes actin-based mobility in this host (32), indicating that these two virulence pathways used by M. marinum in macrophages are conserved during infection of amoebae. Therefore, we addressed whether, like in macrophages, alteration of the LOS profile of M. marinum influences the early interaction events with A. polyphaga and A. castellanii, which are both equally sensitive to gentamycin treatment used to eliminate extracellular mycobacteria. A. polyphaga were infected with class II and class III mutants. As shown in Fig. 6A (left panel), the uptake of the wbbL2 and MMAR_2331 mutants was significantly increased 3-fold with respect to the parental M. marinum strain M and was restored to wild-type levels in the corresponding complemented strains. In contrast, the phagocytosis rate of the MMAR_2321 mutant by A. polyphaga was comparable with that of M. marinum strain M (Fig. 6A, left panel).
FIGURE 6.
Phagocytosis of LOS mutants by Acanthamoeba. Amoebae were infected with M. marinum strains at an m.o.i. of 1. A, phagocytosis rates of the wbbL2, MMAR_2331, and the MMAR_2321 mutants and complemented (_C) strains by A. polyphaga (left panel) and A. castellanii (right panel) normalized to 1 for the wild-type strain. B, percentage of infected A. polyphaga (left panel) or A. castellanii (right panel) with the wbbL2, MMAR_2331, and MMAR_2321 mutants and complemented strains. Results are representative of three independent experiments and are expressed as means ± S.D. (error bars) of triplicates.
In an additional set of experiments, the percentage of infected amoebae was established after counting under the microscope. Whereas the percentage of infected A. polyphaga was increased by 2–4-fold with the wbbL2 and the MMAR_2331 mutants with respect to M. marinum strain M, no significant differences were observed with the MMAR_2321 mutant (Fig. 6B, left panel). The percentage of infected amoebae was restored to wild-type levels in both corresponding complemented strains. Moreover, infection studies performed in another amoebae species, A. castellanii, resulted in very similar effects characterized by an increase in phagocytosis of mycobacteria (Fig. 6A, right panel) and infected cells (Fig. 6B, right panel) in the case of wbbL2 and MMAR_2331 mutants. As for A. polyphaga, the MMAR_2321 mutant behaved like the parental M. marinum M strain in A. castellanii (Fig. 6, A and B, right panels). Overall, these findings suggest that early interactions and phagocytosis of Acanthamoeba are conditioned by the LOS pattern of M. marinum and that, as observed in macrophages, truncated LOS variants favor these early infection processes.
DISCUSSION
In this study, we identified six distinct LOS mutants among which three had been previously characterized. In agreement with earlier reports (10, 14–16), all mutants except MMAR_2321 exhibited a drier colony morphology, indicating a change in cell envelope structure. Because changes in the cell wall structure/composition are often correlated to modifications in membrane permeability and susceptibility to drugs (20, 33), we also examined the susceptibility of the papA4 and wbbL2 mutant strains to hydrophilic and hydrophobic antibiotics. Both mutants exhibited minimal inhibitory concentrations to isoniazid, streptomycin, rifampicin, and erythromycin that were comparable with those of the parental strain (data not shown), implying that altered LOS composition does not affect drug susceptibility.
Biochemical and structural analysis of the biosynthetic intermediates allowed us to precisely link the participation of the inactivated genes within the LOS pathway, extending our understanding of LOS biosynthesis. In particular, the absence of LOS-I to LOS-IV with the parallel increase of LOS-0, a new biosynthetic intermediate that had not been detected/described previously and corresponding to triacylated Glcp-(1–3)-Glcp-(1–4)-Glcp-(1–1)-Glcp, supposes that WbbL2 expresses rhamnosyltransferase activity. Although annotated as a conserved hypothetical protein, BLAST analyses revealed homology between MMAR_2331 and members of the HpcH/HpaI aldolase/citrate lyase family, suggesting that this enzyme may be implicated in the synthesis of the 4-C-branched substituent of caryophyllose. Accordingly, disruption of MMAR_2331 leads to a mutant unable to produce LOS-II to LOS-IV and that accumulates a LOS-II* lacking the terminal caryophyllose. MMAR_2321 presents 44% identity with a putative N-acyltransferase that catalyzes the transfer of a pyrrolidone derivative onto an α-4-amino-4,6-dideoxy-Galp of the polysaccharide from Vibrio cholerae O:5 (34). Therefore, we propose that MMAR_2321 encodes the N-acyltransferase involved in the linkage of the pyrrolidone cycle on the α-4-amino-4,6-dideoxy-Galp residue of the LOS-IV (12). Two-dimensional TLC and structural analyses indicated that disruption of MMAR_2321 led to a mutant lacking LOS-IV and demonstrated that the accumulating product was not a synthetic intermediate between LOS-III and LOS-IV but straight LOS-III. This suggests that the synthesis of a truncated 4-N-acylated-4,6-dideoxy-Galp prevents its block transfer onto LOS-III.
As a result of the generation of a wide panel of isogenic mutants, we were able to initiate structure-function relationship activities, which clearly pointed to an inverse correlation between LOS production and efficient uptake into professional phagocytic cells. This unexpected observation contrasts with a previous study in which LOS mutants demonstrated impaired cell entry efficiency (15). The discrepancies between the two studies may be related to different experimental procedures. First, Ren et al. (15) used an m.o.i. of 10 in their study as compared with an m.o.i. of 2 in our study. Second, mycobacterial cultures were performed using different media, which as reported previously affect the cell wall composition, including LOSs (10) and apolar glycolipids (35). In our study, all mutants were grown on Sauton's agar prior to infections as Sauton's agar has been shown to provide higher amounts of LOSs (10). Finally, the different phenotypes observed may also be dependent on the parental M. marinum strain used, the 1218R strain in the Ren et al. (15) study and the M strain in ours. Based on the assumption that LOSs could mask other cell wall determinants, it is possible that the two strains differ in the nature and composition of these factors, some of them being important for phagocytosis. Nevertheless, our study is based on a large panel of defined mutants that reproducibly shows an increase in macrophage entry efficiency for class I and class II mutants. This view is also further strengthened by very similar phenotypes observed in A. polyphaga and A. castellanii. It is also noteworthy that at later stages of infection all strains exhibited comparable intracellular growth rates (data not shown), suggesting that LOSs do not play a critical role in intramacrophage survival.
Previous infection studies with M. kansasii indicated that rough variants devoid of LOSs induce chronic infections, whereas smooth variants producing LOSs are rapidly eliminated (18), leading to the proposal that LOSs may be considered as an avirulence determinant capable of masking other cell wall-associated (glyco)lipids playing a role in virulence, such as lipoarabinomannan and phenolic glycolipids (17). Although the structures of LOSs in M. kansasii and M. marinum differ significantly, our findings support this “unmasking” hypothesis in M. marinum because early interactions with phagocytic cells, which represent the preferred residing niche of M. marinum, are amplified in mutants devoid of LOSs or defective in higher order LOS production. The reasons for the enhanced phagocytosis rate of these LOS mutants are currently not known, but it is very likely that the lack of LOSs results in the enhanced exposure of structures at the mycobacterial surface that will be recognized by specific macrophage receptors, eventually leading to a more virulent infection. In this context, it is worth mentioning that an LOS-IV mutant (wecE) has been shown to increase the degree of infection and formation of early granulomas in infected zebrafish embryos (14). Thus, these in vivo data suggest that LOSs can act as factors that suppress in the early bacterial infections, a view that is supported by our finding that LOS-deficient strains are more avidly phagocytosed by macrophages.
A similar unmasking hypothesis has recently emerged regarding glycopeptidolipids found in Mycobacterium smegmatis or Mycobacterium abscessus and known as metabolites required for sliding motility and biofilm formation (36). These glycolipids are also suspected to play a role in virulence by inhibiting phagocytosis (37). Importantly, the loss of glycopeptidolipid in M. abscessus resulted in the exposure of phosphatidyl-myo-inositol (38) and the increased expression and surface localization of lipoproteins (39) responsible for a strong proinflammatory response. This supposes that the outermost portion of the M. abscessus cell wall comprising glycopeptidolipids is masking underlying cell wall (glyco)lipids involved in stimulating the innate immune response, thereby affecting subsequent colonization.
Recent investigations demonstrated that LOS biosynthesis defects result in a tighter surface attachment of capsular proteins, including PE_PGRS proteins and EspE (14). Thus, whether the observed increased capacity of the LOS mutants of being phagocytosed is actually caused by the lack of higher order LOS production, the accumulation of lower order LOS structures, or the altered release of surface proteins, such as PE_PGRS, is not clear at this point. Interestingly, using an in vitro system, Stokes et al. (40) demonstrated that by removing the capsular material of M. tuberculosis there was a significant increase in phagocytosis by macrophages compared with the uptake of the wild-type unperturbed bacterium. Thus, by analogy with M. tuberculosis, it seems possible that removal of LOSs may perturb the outermost capsular layer of M. marinum, resulting in increased uptake by macrophages.
In M. tuberculosis, deficiency in diacyltrehalose and pentaacyltrehalose (PAT) affects the surface global composition of the mycobacterial cell envelope, improving the efficiency with which the bacilli bind to and enter phagocytic host cells, implying that PAT production affects early interaction between the bacilli and macrophages (41) similarly to LOSs in M. marinum. Because M. marinum does not produce diacyltrehalose/PAT and because of the structural relatedness between LOSs and diacyltrehalose/PAT (all are trehalose-based glycolipids), it can be inferred that LOS fulfills a function similar to that of diacyltrehalose/PAT, although the paradigm of the avirulence-promoting role of LOSs awaits further confirmation. LOSs are absent from M. tuberculosis, which may be explained by the fact that M. tuberculosis possesses fewer genes within the LOS cluster (15). However, LOSs are found in M. canettii, also known as the smooth tubercle bacilli (9), a species restricted to East Africa. With the recent completion of the M. canettii genome (42), it now becomes possible to compare its LOS loci with M. marinum and to generate isogenic mutants in this species to investigate the role of LOSs in phagocytosis and pathogenicity of M. canettii. Whether the presence of LOSs may be related to the decreased persistence and virulence of M. canettii compared with M. tuberculosis in mice remains an attractive hypothesis that needs to be addressed. Moreover, the recent observation that M. canettii can be ingested by amoebal trophozoites (31) implies that Acanthamoeba infection also represents an alternative means to dissect and address the role of LOSs in the uptake and virulence of this human pathogen.
As mentioned above, the profile and amount of the different LOS subspecies are dependent on the medium (10), indicating that M. marinum has the ability to modulate its LOS content. Therefore, one may speculate that the LOS profile varies under the different growth and environmental conditions encountered and/or during intracellular replication/survival of mycobacteria. That control/regulatory mechanisms of LOS production occur in the M. marinum-infected host would directly influence the early interactions with phagocytes and perhaps condition the outcome of the infection. In this context, a whiB4 (MMAR_5170) mutant showed a highly diminished LOS production, and WhiB4 was proposed to regulate the expression of various LOS biosynthetic genes (14). However, whether WhiB4 plays a role in regulating the LOS pathway during infection remains to be investigated.
There is now an increasing body of evidence suggesting that protozoa are a reservoir for environmental mycobacteria in the environment. In a recent study, Kennedy et al. (32) demonstrated that the requirement of both the ESX-1 secretion system and the actin-based motility of M. marinum in A. castellanii is similar to the situation encountered in macrophages (43). Our study shows that the impact of the LOS pattern of M. marinum was equivalent in macrophages and in Acanthamoeba. These findings not only contribute to the basic molecular understanding of the interactions between Acanthamoeba and M. marinum but also further support the view that the virulence pathways used by M. marinum in macrophage are conserved during Acanthamoeba infection.
Acknowledgments
We thank S. L. G. Cirillo and J. D. Cirillo for sharing the M. marinum Tn library.
This work was supported by French National Research Agency Grant ANR-10-MIDI-009 (ZebraFlam) (to L. K.), the InfectioPôle Sud Foundation (to V. K. S.), and the European Regional Development Fund under Operational Programme Innovative Economy Grant POIG.01.01.02-10-107/09 (to J. D. and J. P.).
- LOS
- lipooligosaccharide
- Rha
- rhamnose
- Me-Rha
- methylrhamnose
- Xyl
- xylose
- OADC
- oleic acid, albumin, dextrose, catalase
- SCO
- single crossover
- DCO
- double crossover
- mut
- mutated
- m.o.i.
- multiplicity of infection
- PYG
- peptone-yeast-glucose
- Hex
- hexose
- deHex
- deoxyhexose
- OS
- oligosaccharide
- Pent
- pentose
- Car
- caryophyllose
- PAT
- pentaacyltrehalose.
REFERENCES
- 1. Karakousis P. C., Bishai W. R., Dorman S. E. (2004) Mycobacterium tuberculosis cell envelope lipids and the host immune response. Cell. Microbiol. 6, 105–116 [DOI] [PubMed] [Google Scholar]
- 2. Kremer L., Baulard A. R., Besra G. S. (2000) in Molecular Genetics of Mycobacteria (Hatfull G. F., Jacobs W. R., Jr., eds) pp. 173–190, ASM Press, Washington, D. C [Google Scholar]
- 3. Brennan P. J., Nikaido H. (1995) The envelope of mycobacteria. Annu. Rev. Biochem. 64, 29–63 [DOI] [PubMed] [Google Scholar]
- 4. Neyrolles O., Guilhot C. (2011) Recent advances in deciphering the contribution of Mycobacterium tuberculosis lipids to pathogenesis. Tuberculosis 91, 187–195 [DOI] [PubMed] [Google Scholar]
- 5. Hunter S. W., Murphy R. C., Clay K., Goren M. B., Brennan P. J. (1983) Trehalose-containing lipooligosaccharides. A new class of species-specific antigens from Mycobacterium. J. Biol. Chem. 258, 10481–10487 [PubMed] [Google Scholar]
- 6. Hunter S. W., Jardine I., Yanagihara D. L., Brennan P. J. (1985) Trehalose-containing lipooligosaccharides from mycobacteria: structures of the oligosaccharide segments and recognition of a unique N-acylkanosamine-containing epitope. Biochemistry 24, 2798–2805 [DOI] [PubMed] [Google Scholar]
- 7. Gilleron M., Puzo G. (1995) Lipooligosaccharidic antigens from Mycobacterium kansasii and Mycobacterium gastri. Glycoconj. J. 12, 298–308 [DOI] [PubMed] [Google Scholar]
- 8. Hunter S. W., Barr V. L., McNeil M., Jardine I., Brennan P. J. (1988) Trehalose-containing lipooligosaccharide antigens of Mycobacterium sp.: presence of a mono-O-methyltri-O-acyltrehalose “core”. Biochemistry 27, 1549–1556 [DOI] [PubMed] [Google Scholar]
- 9. Daffe M., McNeil M., Brennan P. J. (1991) Novel type-specific lipooligosaccharides from Mycobacterium tuberculosis. Biochemistry 30, 378–388 [DOI] [PubMed] [Google Scholar]
- 10. Burguière A., Hitchen P. G., Dover L. G., Kremer L., Ridell M., Alexander D. C., Liu J., Morris H. R., Minnikin D. E., Dell A., Besra G. S. (2005) LosA, a key glycosyltransferase involved in the biosynthesis of a novel family of glycosylated acyltrehalose lipooligosaccharides from Mycobacterium marinum. J. Biol. Chem. 280, 42124–42133 [DOI] [PubMed] [Google Scholar]
- 11. Rombouts Y., Burguière A., Maes E., Coddeville B., Elass E., Guérardel Y., Kremer L. (2009) Mycobacterium marinum lipooligosaccharides are unique caryophyllose-containing cell wall glycolipids that inhibit tumor necrosis factor-alpha secretion in macrophages. J. Biol. Chem. 284, 20975–20988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rombouts Y., Elass E., Biot C., Maes E., Coddeville B., Burguière A., Tokarski C., Buisine E., Trivelli X., Kremer L., Guérardel Y. (2010) Structural analysis of an unusual bioactive N-acylated lipo-oligosaccharide LOS-IV in Mycobacterium marinum. J. Am. Chem. Soc. 132, 16073–16084 [DOI] [PubMed] [Google Scholar]
- 13. Rombouts Y., Alibaud L., Carrère-Kremer S., Maes E., Tokarski C., Elass E., Kremer L., Guérardel Y. (2011) Fatty acyl chains of Mycobacterium marinum lipooligosaccharides: structure, localization and acylation by PapA4 (MMAR_2343) protein. J. Biol. Chem. 286, 33678–33688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van der Woude A. D., Sarkar D., Bhatt A., Sparrius M., Raadsen S. A., Boon L., Geurtsen J., van der Sar A. M., Luirink J., Houben E. N., Besra G. S., Bitter W. (2012) Unexpected link between lipooligosaccharide biosynthesis and surface protein release in Mycobacterium marinum. J. Biol. Chem. 287, 20417–20429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ren H., Dover L. G., Islam S. T., Alexander D. C., Chen J. M., Besra G. S., Liu J. (2007) Identification of the lipooligosaccharide biosynthetic gene cluster from Mycobacterium marinum. Mol. Microbiol. 63, 1345–1359 [DOI] [PubMed] [Google Scholar]
- 16. Sarkar D., Sidhu M., Singh A., Chen J., Lammas D. A., van der Sar A. M., Besra G. S., Bhatt A. (2011) Identification of a glycosyltransferase from Mycobacterium marinum involved in addition of a caryophyllose moiety in lipooligosaccharides. J. Bacteriol. 193, 2336–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Belisle J. T., Brennan P. J. (1989) Chemical basis of rough and smooth variation in mycobacteria. J. Bacteriol. 171, 3465–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Collins F. M., Cunningham D. S. (1981) Systemic Mycobacterium kansasii infection and regulation of the alloantigenic response. Infect. Immun. 32, 614–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stinear T. P., Seemann T., Harrison P. F., Jenkin G. A., Davies J. K., Johnson P. D., Abdellah Z., Arrowsmith C., Chillingworth T., Churcher C., Clarke K., Cronin A., Davis P., Goodhead I., Holroyd N., Jagels K., Lord A., Moule S., Mungall K., Norbertczak H., Quail M. A., Rabbinowitsch E., Walker D., White B., Whitehead S., Small P. L., Brosch R., Ramakrishnan L., Fischbach M. A., Parkhill J., Cole S. T. (2008) Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 18, 729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alibaud L., Rombouts Y., Trivelli X., Burguière A., Cirillo S. L., Cirillo J. D., Dubremetz J. F., Guérardel Y., Lutfalla G., Kremer L. (2011) A Mycobacterium marinum TesA mutant defective for major cell wall-associated lipids is highly attenuated in Dictyostelium discoideum and zebrafish embryos. Mol. Microbiol. 80, 919–934 [DOI] [PubMed] [Google Scholar]
- 21. McAdam R. A., Quan S., Smith D. A., Bardarov S., Betts J. C., Cook F. C., Hooker E. U., Lewis A. P., Woollard P., Everett M. J., Lukey P. T., Bancroft G. J., Jacobs W. R., Jr., Duncan K. (2002) Characterization of a Mycobacterium tuberculosis H37Rv transposon library reveals insertions in 351 ORFs and mutants with altered virulence. Microbiology 148, 2975–2986 [DOI] [PubMed] [Google Scholar]
- 22. Parish T., Stoker N. G. (2000) Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146, 1969–1975 [DOI] [PubMed] [Google Scholar]
- 23. Ciucanu I., Kerek F. (1984) A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 131, 209–217 [Google Scholar]
- 24. Alexander D. C., Jones J. R., Tan T., Chen J. M., Liu J. (2004) PimF, a mannosyltransferase of mycobacteria, is involved in the biosynthesis of phosphatidylinositol mannosides and lipoarabinomannan. J. Biol. Chem. 279, 18824–18833 [DOI] [PubMed] [Google Scholar]
- 25. Mills J. A., Motichka K., Jucker M., Wu H. P., Uhlik B. C., Stern R. J., Scherman M. S., Vissa V. D., Pan F., Kundu M., Ma Y. F., McNeil M. (2004) Inactivation of the mycobacterial rhamnosyltransferase, which is needed for the formation of the arabinogalactan-peptidoglycan linker, leads to irreversible loss of viability. J. Biol. Chem. 279, 43540–43546 [DOI] [PubMed] [Google Scholar]
- 26. Tobin D. M., Ramakrishnan L. (2008) Comparative pathogenesis of Mycobacterium marinum and Mycobacterium tuberculosis. Cell Microbiol. 10, 1027–1039 [DOI] [PubMed] [Google Scholar]
- 27. Greub G., Raoult D. (2004) Microorganisms resistant to free-living amoebae. Clin. Microbiol. Rev. 17, 413–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thomas V., Herrera-Rimann K., Blanc D. S., Greub G. (2006) Biodiversity of amoebae and amoeba-resisting bacteria in a hospital water network. Appl. Environ. Microbiol. 72, 2428–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adékambi T., Ben Salah S., Khlif M., Raoult D., Drancourt M. (2006) Survival of environmental mycobacteria in Acanthamoeba polyphaga. Appl. Environ. Microbiol. 72, 5974–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cirillo J. D., Falkow S., Tompkins L. S., Bermudez L. E. (1997) Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect. Immun. 65, 3759–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mba Medie F., Ben Salah I., Henrissat B., Raoult D., Drancourt M. (2011) Mycobacterium tuberculosis complex mycobacteria as amoeba-resistant organisms. PLoS One 6, e20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kennedy G. M., Morisaki J. H., Champion P. A. (2012) Conserved mechanisms of Mycobacterium marinum pathogenesis within the environmental amoeba Acanthamoeba castellanii. Appl. Environ. Microbiol. 78, 2049–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chavadi S. S., Edupuganti U. R., Vergnolle O., Fatima I., Singh S. M., Soll C. E., Quadri L. E. (2011) Inactivation of tesA reduces cell wall lipid production and increases drug susceptibility in mycobacteria. J. Biol. Chem. 286, 24616–24625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hermansson K., Jansson P. E., Holme T., Gustavsson B. (1993) Structural studies of the Vibrio cholerae O:5 O-antigen polysaccharide. Carbohydr. Res. 248, 199–211 [DOI] [PubMed] [Google Scholar]
- 35. Rombouts Y., Brust B., Ojha A. K., Maes E., Coddeville B., Elass-Rochard E., Kremer L., Guerardel Y. (2012) Exposure of mycobacteria to cell wall-inhibitory drugs decreases production of arabinoglycerolipid related to mycolyl-arabinogalactan-peptidoglycan metabolism. J. Biol. Chem. 287, 11060–11069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nessar R., Reyrat J. M., Davidson L. B., Byrd T. F. (2011) Deletion of the mmpL4b gene in the Mycobacterium abscessus glycopeptidolipid biosynthetic pathway results in loss of surface colonization capability, but enhanced ability to replicate in human macrophages and stimulate their innate immune response. Microbiology 157, 1187–1195 [DOI] [PubMed] [Google Scholar]
- 37. Villeneuve C., Etienne G., Abadie V., Montrozier H., Bordier C., Laval F., Daffe M., Maridonneau-Parini I., Astarie-Dequeker C. (2003) Surface-exposed glycopeptidolipids of Mycobacterium smegmatis specifically inhibit the phagocytosis of mycobacteria by human macrophages. Identification of a novel family of glycopeptidolipids. J. Biol. Chem. 278, 51291–51300 [DOI] [PubMed] [Google Scholar]
- 38. Rhoades E. R., Archambault A. S., Greendyke R., Hsu F. F., Streeter C., Byrd T. F. (2009) Mycobacterium abscessus glycopeptidolipids mask underlying cell wall phosphatidyl-myo-inositol mannosides blocking induction of human macrophage TNF-α by preventing interaction with TLR2. J. Immunol. 183, 1997–2007 [DOI] [PubMed] [Google Scholar]
- 39. Roux A. L., Ray A., Pawlik A., Medjahed H., Etienne G., Rottman M., Catherinot E., Coppée J. Y., Chaoui K., Monsarrat B., Toubert A., Daffé M., Puzo G., Gaillard J. L., Brosch R., Dulphy N., Nigou J., Herrmann J. L. (2011) Overexpression of proinflammatory TLR-2-signalling lipoproteins in hypervirulent mycobacterial variants. Cell. Microbiol. 13, 692–704 [DOI] [PubMed] [Google Scholar]
- 40. Stokes R. W., Norris-Jones R., Brooks D. E., Beveridge T. J., Doxsee D., Thorson L. M. (2004) The glycan-rich outer layer of the cell wall of Mycobacterium tuberculosis acts as an antiphagocytic capsule limiting the association of the bacterium with macrophages. Infect. Immun. 72, 5676–5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rousseau C., Neyrolles O., Bordat Y., Giroux S., Sirakova T. D., Prevost M. C., Kolattukudy P. E., Gicquel B., Jackson M. (2003) Deficiency in mycolipenate- and mycosanoate-derived acyltrehaloses enhances early interactions of Mycobacterium tuberculosis with host cells. Cell. Microbiol. 5, 405–415 [DOI] [PubMed] [Google Scholar]
- 42. Supply P., Marceau M., Mangenot S., Roche D., Rouanet C., Khanna V., Majlessi L., Criscuolo A., Tap J., Pawlik A., Fiette L., Orgeur M., Fabre M., Parmentier C., Frigui W., Simeone R., Boritsch E. C., Debrie A. S., Willery E., Walker D., Quail M. A., Ma L., Bouchier C., Salvignol G., Sayes F., Cascioferro A., Seemann T., Barbe V., Locht C., Gutierrez M. C., Leclerc C., Bentley S. D., Stinear T. P., Brisse S., Médigue C., Parkhill J., Cruveiller S., Brosch R. (2013) Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat. Genet. 45, 172–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stamm L. M., Morisaki J. H., Gao L. Y., Jeng R. L., McDonald K. L., Roth R., Takeshita S., Heuser J., Welch M. D., Brown E. J. (2003) Mycobacterium marinum escapes from phagosomes and is propelled by actin-based motility. J. Exp. Med. 198, 1361–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stover C. K., de la Cruz V. F., Fuerst T. R., Burlein J. E., Benson L. A., Bennett L. T., Bansal G. P., Young J. F., Lee M. H., Hatfull G. F., Snapper S. B., Barletta R. G., Jacobs W. R., Jr., Bloom B. R. (1991) New use of BCG for recombinant vaccines. Nature 351, 456–460 [DOI] [PubMed] [Google Scholar]