Background: Cancerous inhibitor of protein phosphatase 2A (CIP2A) is overexpressed in most types of human cancer.
Results: Depletion of CIP2A prolongs cell division time and CIP2A interacts with NIMA-related kinase 2 (NEK2) during G2/M phase to facilitate centrosome separation.
Conclusion: CIP2A is involved in cell cycle progression through centrosome separation and mitotic spindle dynamics.
Significance: This provides a novel role for CIP2A in cell cycle progression.
Keywords: Cell Cycle, Centrosome, Mitosis, Mitotic Spindle, PP2A, CIP2A, Centrosome Separation, NEK2
Abstract
Cancerous inhibitor of protein phosphatase 2A (CIP2A) is overexpressed in most human cancers and has been described as being involved in the progression of several human malignancies via the inhibition of protein phosphatase 2A (PP2A) activity toward c-Myc. However, with the exception of this role, the cellular function of CIP2A remains poorly understood. On the basis of yeast two-hybrid and coimmunoprecipitation assays, we demonstrate here that NIMA (never in mitosis gene A)-related kinase 2 (NEK2) is a binding partner for CIP2A. CIP2A exhibited dynamic changes in distribution, including the cytoplasm and centrosome, depending on the cell cycle stage. When CIP2A was depleted, centrosome separation and the mitotic spindle dynamics were impaired, resulting in the activation of spindle assembly checkpoint signaling and, ultimately, extension of the cell division time. Our data imply that CIP2A strongly interacts with NEK2 during G2/M phase, thereby enhancing NEK2 kinase activity to facilitate centrosome separation in a PP1- and PP2A-independent manner. In conclusion, CIP2A is involved in cell cycle progression through centrosome separation and mitotic spindle dynamics.
Introduction
The human oncoprotein cancerous inhibitor of protein phosphatase 2A (CIP2A) is encoded by the KIAA1524 gene (1), and its overexpression has been reported in most human cancer types (2–25). In general, the PI3K-AKT and RAF-RAS-MEK signaling pathways are known to play a role in promoting malignant growth in human cancers. ETS1, one of the transcription factors activated by the RAS-MEK signaling pathway, increases the transcription of CIP2A mRNA (26). The CIP2A level is correlated with the proliferation of human cancer cells and with cancer progression in a large variety of human malignancies (5, 27), and CIP2A has a reported tumor-promoting role in several xenograft studies (5, 28–30). CIP2A performs its role in malignant cellular growth by inhibiting PP2A2 activity toward the oncogenic transcription factor c-Myc (28, 31). c-Myc expression rapidly induces the activation of cyclin E-cdk2 kinase activity, with the simultaneous release of p27Kip1 from cdk2 complexes, which is essential for G1/S transition and cell cycle progression (32, 33). Although CIP2A is known to cause c-Myc stabilization and is overexpressed in almost all human cancer types, with the exception of inhibiting PP2A activity toward c-Myc, the cellular function of CIP2A is poorly understood. Thus, the identification of a novel role for CIP2A in cell cycle progression would be essential.
NIMA-related kinase 2 (NEK2) is a serine/threonine kinase that participates in mitotic progression. NEK2 localizes to multiple sites, including the centrosome, nucleus, and cytoplasm. Moreover, NEK2 specifically associates with the chromosomes at M phase of the cell cycle (34), and its expression and activity peak in S and G2 phases to function as a core component of the human centrosome (35, 36). The role of NEK2 is to displace such centrosomal linker components as C-Nap1 and rootletin from the centrosomes by phosphorylation at the onset of mitosis. The disassembled linker proteins facilitate centrosome separation and the establishment of the mitotic spindle (35, 37–41). In addition to the role in centrosome separation, NEK2 is also known to be involved in meiotic events, including chromosomal condensation and segregation (42, 43), in microtubule stabilization through NIP2/centrobin (44, 45), and in kinetochore microtubule attachment stability through Hec1 phosphorylation (46–48).
In this study, we investigate for the first time the role of CIP2A in association with the cell cycle. CIP2A exhibited dynamic changes in localization, including the cytoplasm and centrosome, depending on the stage of the cell cycle. Additionally, the depletion of CIP2A resulted in the activation of SAC signaling because of impaired mitotic spindle dynamics, ultimately extending the duration of cell division. Our data indicate that CIP2A strongly interacts with NEK2 at G2/M phase, thereby enhancing NEK2 kinase activity to facilitate centrosome separation in a PP1- and PP2A-independent manner.
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid Screening
A cDNA encoding a dominant negative mutant of mouse PP2C ∈ (D302A) was cloned into pGBK-T7 to the produce “bait” vector, and this construct was used to screen a human brain cDNA library in the pACT2 vector. The dominant negative mutant was used because it was expected to associate with its substrate in a more stable manner than the wild-type protein in the cells. Five independent clones encoding human VAPA (vesicle-associated membrane protein-associated protein A) were isolated by screening 1 × 106 library clones.
Plasmids and Antibodies
Human cDNA NEK2A was purchased from OriGene. The full-length and truncated mutants of NEK2A were prepared by PCR and subcloned into the XhoI-BamHI sites of the pEGFP-C1 mammalian expression vector, which contains an N-terminal GFP epitope tag. The plasmid encoding CIP2A was a gift from Dr. Jukka Westermarck at the University of Turku and Abo Akademi University, Finland, and the plasmid encoding CIP2A #3 siRNA-resistant cDNA was provided by Jae-Sung Kim at the Korea Institute of Radiological and Medical Sciences. The full-length and truncated mutants of CIP2A were prepared by PCR and subcloned into the EcoRI-XhoI sites of the pCMVTag2B mammalian expression vector, which contains an N-terminal FLAG epitope tag.
The following commercial antibodies were used: anti-CIP2A, anti-β-actin, anti-cyclin A, and anti-cyclin B1 (Santa Cruz Biotechnology, San Diego, CA); anti-γ-tubulin and anti-FLAG antibody (M2) (Sigma); anti-α-tubulin and anti-lamin B1 (Abcam, Cambridge, MA), and anti-NEK2, anti-BubR1, and anti-GFP antibodies (BD Biosciences).
Cell Culture
HeLa and HEK293 cells were cultured using Dulbecco's modified Eagle's medium (Hyclone) supplemented with 10% FBS at 37 °C in a humidified atmosphere of 5% CO2 in air.
Transfection and Cell Synchronization
The transfection of siRNA into HeLa cells was performed using Dharmafect transfection reagent (Dharmacon, Lafayette, CO).
The following siRNA oligonucleotides were used to suppress CIP2A expression: GFP, 5′-GUU CAG CGU GUC CGG CGA GTT-3′; CIP2A #1, 5′-CUG UGG UUG UGU UUG CAC UTT-3′; CIP2A #2, 5′-GAC AGA AAC TCA CAC GAC TAT-3′; and CIP2A #3, 5′-GGA GUG GUU UGU CGG AGC A-3′. For NEK2 silencing, ON-TARGET plus SMART pool (Dharmacon, Lafayette, CO) siRNA oligonucleotides were used.
For the preparation of cells arrested in G1 phase (G0/G1 block), HeLa cells were serum-starved for 72 h in DMEM containing only 0.2% FBS. For synchronizing the cells in Gl/S phase, 2 μg/ml of aphidicolin was added to the culture medium for 16 h after cell starvation for 12 h. Early S phase-blocked cells were obtained by double thymidine block. S phase-blocked cells and mitotic cells were obtained by treatment with 100 μm 5-fluorodeoxyuridine for 24 h and 100 ng/ml nocodazole for 16 h, respectively. For G2/M phase arrest, HeLa cells were synchronized by treatment with 2 mm thymidine for 24 h, released for 3 h, and cultured in the presence of 100 ng/ml nocodazole for 12 h.
Immunoprecipitation
FLAG-tagged CIP2A and NEK2A were transfected into HEK293 cells using calcium phosphate. At 24 h after transfection, the cells were lysed in IP buffer (50 mm Tris-HCl (pH 8.0), 50 mm NaCl, 1 mm EDTA, 1% Triton X-100, and protease inhibitor mixture (Roche Diagnostics)) and homogenized by sonication for 3 s. Each cell extract was incubated with the indicated antibodies for 2 h, followed by an additional 2 h of incubation with protein A-agarose beads (Roche).
Western Blotting
Cell lysates were prepared using 5× SDS sample buffer and heating at 95 °C for 5 min. The proteins were separated electrophoretically on a 12% SDS-polyacrylamide gel and transferred onto a 0.45-μm pore size nitrocellulose membrane for 2 h. The membrane was incubated overnight with antibodies at 4 °C, followed by incubation with HRP-conjugated goat anti-mouse or anti-rabbit IgG (Fab) (Enzo Life Sciences, Farmingdale, NY) at room temperature for 2 h. The proteins were visualized with a PowerOpti-ECL Western blotting reagent (Animal Genetics, Yeongtong-gu, Gyeonggi-do, Korea) and analyzed using an LAS3000 luminescent image analyzer (Fuji Film, Tokyo, Japan).
Immunofluorescence and Microscopic Analysis
HeLa cells (1 × 105 cells) were seeded onto 0.1% gelatin-coated glass coverslips in 12-well plates. After 24 h, the cells were fixed with 70% methanol (−20 °C) for 10 min, washed with PBS, and then permeabilized with 0.1% Triton X-100 at room temperature for 10 min. After washing with PBS, the cells were blocked with 0.1% BSA at room temperature for 30 min and immunostained overnight with the following primary antibodies at 4 °C: 1:300 anti-CIP2A, 1:200 anti-Nek2, 1:500 anti-γ-tubulin, or 1:500 anti-α-tubulin. The secondary antibodies used were goat anti-mouse IgG, F(ab′)2-FITC, goat anti-mouse IgG, F(ab′)2-TRITC, goat anti-rabbit IgG, F(ab′)2-TRITC, goat anti-rabbit IgG, and F(ab′)2-FITC (Santa Cruz Biotechnology). Alternatively, cells were extracted with PHEMT buffer (60 mm Pipes, 25 mm Hepes (pH 6.9), 10 mm EGTA, 4 mm MgCl2, and 0.5% Triton X-100; Figs. 3, A and B, and 4, B and H, and supplemental Fig. S2) and then fixed with 4% paraformaldehyde for 15 min at room temperature. After washing with PBS, the cells were permeabilized and blocked with PBS-BT (PBS, 3% BSA, and 0.1% Triton X-100) for 30 min at room temperature. Coverslips were subsequently incubated in primary antibodies for 1 h, and FITC- or TRITC-coupled secondary antibodies diluted in PBS-BT. DNA was stained with 5 μg/ml Hoechst dye. Images were acquired with AxioVision 4.8.2 (Carl Zeiss) under a Zeiss Axiovert 200 m microscope using a 1.4 numerical aperture plan-Apo ×63 oil immersion lens and a HRm charge-coupled device camera. Deconvolved images were obtained using AutoDeblur v9.1 and AutoVisualizer v9.1 (AutoQuant Imaging). Some images were acquired under a confocal laser scanning microscope (FV 300, IX71, Olympus) using a ×40 dry lens and Fluoview v.5.0 software.
FIGURE 3.
CIP2A depletion impairs mitotic spindle dynamics. A–D and F, HeLa cells were transfected with siRNAs for 72 h, fixed, and stained with the indicated antibodies. Shown are the maximum projections from deconvolved z stacks of representative control or CIP2A-depleted HeLa cells stained for Mad2/BubR1 (A and B, red), α-tubulin (C–E, red) and DNA (blue). Scale bars = 5 μm. A, the Mad2 fluorescence-positive cells of the total metaphase cells were quantified (n = 102 cells for control, n = 112 for CIP2A-depleted cells). B, the BubR1 fluorescence-positive cells of the total metaphase cells were quantified. n = 100 cells for control, n = 98 for CIP2A-depleted cells. *, p < 0.05; **, p < 0.01; one-tailed Student's t test. C, HeLa cells stably expressing GFP-CENP-A were transfected with siRNAs and stained for α-tubulin (red) and DNA (blue). The images were acquired under a constant exposure time for each channel. The insets show the single focal planes of the boxed regions. Scale bar = 5 μm. D, total immunofluorescence intensity for tubulin on the metaphase spindle was quantified and plotted. In addition, siRNA-transfected cells were incubated at 4 °C for 15 min, and the microtubule fluorescence intensity of the metaphase cells was quantified and normalized to their respective control samples. n = 10 cells/condition. **, p < 0.01; ***, p < 0.001; one-tailed Student's t test. E, control, CIP2A-depleted or NEK2-depleted HeLa cells were treated with 0.1 μg/ml nocodazole for 10 min at 37 °C to completely depolymerize the mitotic spindle. The nocodazole-treated cells were washed twice with prewarmed PBS, released into fresh medium, and fixed 0 and 6 min after release. The images for tubulin were acquired under a constant exposure time, and the tubulin immunofluorescence intensity on the metaphase spindles was quantified and normalized to their respective control sample at the 0-min time point. n = 10 cells/condition. *, p < 0.05; ***, p < 0.001; one-tailed Student's t test. F, HeLa cells stably expressing GFP-α-tubulin were transfected with siRNAs. GFP fluorescence intensity was acquired every 0.5 s while a photobleaching laser was focused to a diffraction-limited spot in the cytoplasm away from the spindle. 10 half-spindles from 10 metaphase cells were quantified, and fluorescence signals for each half-spindle were normalized to their intensity at 0 s. The 10 half-spindles were then averaged at each time point to generate mean traces.
FIGURE 4.
Knockdown of CIP2A prolongs the duration of mitosis and NEK2A degradation. A, HeLa cells transfected with siRNAs for 24 h were plated, synchronized by DTB, released, and harvested at the indicated times. The expression of CIP2A, NEK2, cyclin A, and cyclin B1 were analyzed by Western blotting (WB). β-Actin served as the loading control. AS, asynchronized cells. B, HeLa cells were transfected with siRNAs for 72 h, fixed, and stained for NEK2 (red) and DNA (blue). Shown are the maximum projections from deconvolved z stacks of representative control cells or CIP2A-depleted cells. The NEK2 signals at the centrosomes were quantified. n = 10 cells/condition. **, p < 0.01, one-tailed Student's t test. Scale bar = 5 μm. C, HeLa cells were transfected with siRNAs and fixed and stained for NEK2 (red), γ-tubulin (green), and DNA (blue) after 72 h. The images were acquired under a constant exposure time for each channel. Shown are the maximum projections from deconvolved z stacks of the control and CIP2A-depleted cells. Scale bars = 5 μm. D, tubulin polymerization reactions were performed alone (Mock) or in the presence of 2.9 μg (high) or 0.58 μg of recombinant NEK2A protein or 10 μm paclitaxel. Polymerization was initiated by incubation at 37 °C and followed by absorbance readings at 340 nm.
Live Imaging
HeLa cells were transfected with siRNAs for 24 h and seeded (3 × 105) in a glass-bottom dish. The sequence of differential interference contrast/FITC images was acquired every 3 min for 24 h using an Olympus IX81 microscope. The experiments were performed in a temperature-controlled chamber that was maintained at 37 °C with a humidified atmosphere of 5% CO2 in air. Metamorph software (Molecular Devices, Dowingtown, PA) was used for acquisition and analysis.
Propidium Iodide Staining and Flow Cytometry
HeLa cells (1 × 106/100-mm dish) were synchronized by double thymidine block and released into fresh media for the indicated times. The cells were harvested at specific time points by trypsinization and fixed overnight with ice-cold 70% ethanol. After washing with PBS containing 0.25% Triton X-100, the cells were suspended in PBS containing 1% BSA, 20 μg/ml propidium iodide, and 10 μg/ml RNase A and incubated for 30 min at 37 °C in the dark. The samples were analyzed using a BD Biosciences FACScan, and the data of 20,000 events for each sample were plotted with CellQuest software (BD Biosciences).
Kinase Assays
The EGFP-NEK2A construct was transiently expressed for 48 h in HEK293 cells with the indicated siRNAs. The cells were scraped off into NEB lysis buffer (50 mm HEPES (pH 7.5), 100 mm NaCl, 10 mm MgCl2, 5 mm MnCl2, 5 mm KCl, 2 mm EDTA, 1 mm DTT, 0.1% Nonidet P-40, and protease inhibitor mixture) and lysed as described previously (49). The EGFP-NEK2A construct was immunoprecipitated using the GFP-Trap matrix (Chromotek, Martinsried, Germany). The purified kinases were washed three times in lysis buffer and twice in kinase buffer (25 mm MOPS (pH 7.2), 12.5 mm β-glycerophosphate, 25 mm MgCl2, 5 mm EGTA, 2 mm EDTA, and adding 0.25 mm DTT prior to use). The kinases bound to the GFP-Trap matrix were incubated with 1 μg of myelin basic protein substrate and a total ATP concentration of 100 μm ATP for 30 min at 30 °C. The reaction was stopped by adding ADP-Glo reagent. The in vitro kinase activity was measured using a ADP-Glo kinase assay kit (Promega, Madison, WI) following the protocol of the manufacturer. HEK293 cells were transfected with the pcDNA3.1 and CIP2A-FLAG plasmids for the in vitro kinase reaction using purified CIP2A. An empty pcDNA3.1 vector was used as the negative control. The cells were lysed, and the FLAG-tagged proteins were retrieved with 20 μl of anti-FLAG M2 affinity gel (Sigma) per each immunoprecipitation. The samples were rotated for 4 h at 4 °C. The resin was then washed three times with 1× wash buffer, and the proteins were eluted from the resin with elution buffer containing 100 μg/ml FLAG peptide for 30 min at 4 °C with gentle shaking. The supernatant was collected and stored at −20 °C until further analysis. For the kinase reaction, the eluted proteins were incubated with 0.5 μg of active recombinant NEK2 (Upstate), 50 μm of ATP, and 1 μg of myelin basic protein substrate.
Tubulin Polymerization Assay
The porcine brain tubulin polymerization assay was performed in 100-μl volumes at 37 °C using a tubulin polymerization assay kit (Cytoskeleton, Denver, CO). A340 was measured every 1 min for 30 min. The results are presented as the change in base line-corrected turbidity over time.
Fluorescence Loss in Photobleaching
HeLa cells stably expressing GFP-α-tubulin were transfected with a control or siCIP2A and placed in a sealed growth chamber heated to 37 °C. Cytoplasmic GFP-α-tubulin was photobleached with a fiberoptically pumped dye laser, and images were acquired at 0.5-s intervals for 300 s with ZEN 2011 software (LSM) under a confocal microscope (LSM 700, Carl Zeiss) with a ×40 objective. 10 half-spindles from 10 metaphase cells in each transfection were analyzed by measuring the absolute GFP-α-tubulin fluorescence intensity in a defined rectangle area contained entirely within each half-spindle. Fluorescence intensities for each half-spindle were normalized to their maximum intensity at the beginning of the time lapse, and the 10 normalized datasets were averaged to generate a single trace for each transfection.
RESULTS
CIP2A Localizes to Various Sites during the Cell Cycle
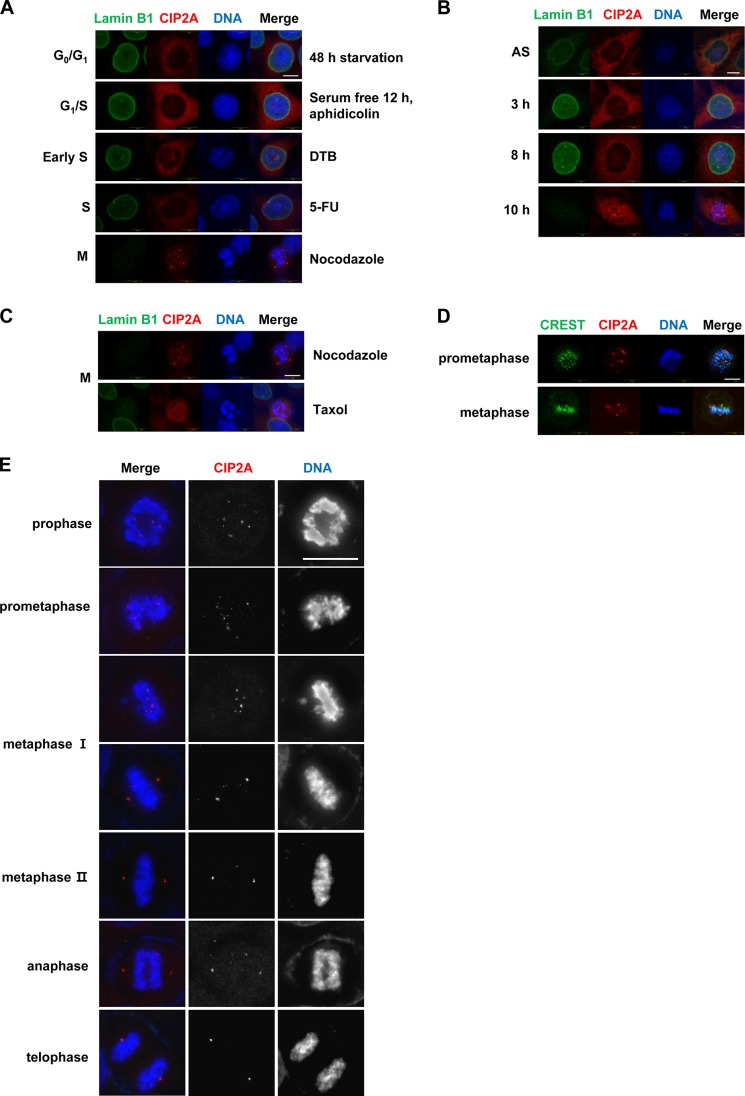
Although CIP2A is overexpressed in almost all types of human cancer and is known to inhibit PP2A activity toward c-Myc, the cellular function of CIP2A is poorly understood. A yeast two-hybrid assay was performed using CIP2A as the bait to identify novel binding partners that may provide clues to other cellular functions of CIP2A. One of the prey used was NEK2, which is known to be associated with centrosome splitting. Because it is known that CIP2A stimulates tumor cell proliferation, we decided to study role of CIP2A in the regulation of the cell cycle. To ascertain whether CIP2A subcellular localization changes during the cell cycle, HeLa cells were treated with chemical inhibitors to block the cell cycle at a specific stage. CIP2A was mainly localized in the cytosol, although a weak signal was observed in the nucleus at S phase and exhibited a multiple-focus shape in cells blocked at mitosis by nocodazole treatment (Fig. 1A). We also confirmed CIP2A localization using a double thymidine block (DTB). Consistent with nocodazole treatment, CIP2A was strongly localized as discrete foci at mitosis (Fig. 1B), implying that the CIP2A focal localization was not an artifact of the nocodazole treatment. This focal localization was also confirmed in mitotic arrest caused by nocodazole and taxol treatment (Fig. 1C). Because the centromere region appeared as multiple foci when immunostained, we examined whether CIP2A localized to the centromere region by coimmunostaining with CREST as a centromere marker. CIP2A did not colocalize with CREST at prometaphase and metaphase but displayed a centrosome localization pattern at metaphase (Fig. 1D).
FIGURE 1.
CIP2A localizes to various sites, including the centrosome, during the cell cycle. A–E, representative cells counterstained for DNA with Hoechst 33258. A and C, HeLa cells were starved or treated with the indicated drugs for synchronization in each phase. B, HeLa cells were synchronized by DTB and released into fresh medium at the indicated times. The cells were then immunostained with antibodies against lamin B1 (green) and CIP2A (red). AS, asynchronized cells; 5-FU, 5-fluorodeoxyuridine. D and E, HeLa cells were immunostained for CREST (green) and CIP2A (red). Scale bars = 10 μm.
CIP2A localization was observed in unperturbed cells to analyze the dynamics of localization during mitosis. CIP2A was detected as foci during prometaphase. After the completion of chromosome alignment, CIP2A was localized with centrosomes, and this was maintained until cytokinesis (Fig. 1E). Taken together, CIP2A is mainly localized in the cytosol yet moves to the centrosome after the completion of chromosome alignment until cytokinesis.
CIP2A Depletion Retards Centrosome Separation
Because centrosome separation occurs prior to mitosis and is a key step for the faithful segregation of chromosomes (50), and because the induction of active NEK2A induces premature centrosome splitting (38), the effect of CIP2A depletion on centrosome separation was examined. The protocol outlined in Fig. 2A was used to measure the distance between the centrosomes after treatment with monastrol to inhibit Eg5 activity. Three siRNAs against different regions of CIP2A were designed, and a CIP2A siRNA-resistant rescue vector (CIP2Arv) against the #3 siRNA was used in rescue experiments. The average distance between the centrosomes was 2.3 μm in the control cells, whereas the distance was reduced to 1.87 μm (mean) in the CIP2A-depleted cells. Importantly, the distance was restored by overexpression of CIP2Arv, excluding off-target effects of siRNA (Fig. 2B). This phenotype was further confirmed using HeLa cells stably expressing GFP-tubulin without synchronization and inhibition of the kinesin Eg5. Although the centrosomes were separated well at the beginning of cell division in the control cells, the GFP signal was barely separated in the CIP2A-depleted cells (Fig. 2C).
FIGURE 2.
Depletion of CIP2A retards centrosome separation and delays mitotic progression. A, schematic of the protocol used to treat the cells. HeLa cells were transfected with siRNAs or siRNAs with siRNA-resistant rescue vector (CIP2Arv), and the cells were plated onto glass coverslips for synchronization after 24 h. Following DTB, the cells were released into fresh medium. After 5 h, the cells were exposed to 100 μm monastrol for 4 h to inhibit Eg5-dependent centrosome splitting. The cells were then fixed and stained for γ-tubulin (red) and DNA (blue). B, the distance between the two centrosomes was analyzed (n = 15 cells/condition. The results of three independent experiments are shown. *, p < 0.05; **, p < 0.01; ***, p < 0.001; one-tailed Student's t test. Scale bar = 5 μm. C, GFP-tubulin stably expressing HeLa cells was transfected with siRNAs and plated onto a glass-bottom plate after 24 h. The distance between the two centrosomes was analyzed at the beginning of the cell division n = 67 cells for control, n = 54 for CIP2A-depleted cells. ***, p < 0.001, one-tailed Student's t test. Scale bar = 10 μm. D and F, HeLa cells stably expressing histone H2B-GFP were used to assess the time of mitosis and categorize the mitotic phases. D, cells were transfected with siRNAs, and mitotic progression was followed by time-lapse microscopy recording the DIC and GFP signals every 3 min. The arrowheads point to unaligned chromosomes. The mitosis time is given in minutes (n = 50 cells/condition). The results of three independent experiments are shown. *, p < 0.05, one-tailed Student's t test). Scale bars = 5 μm. E, HeLa cells transfected with siRNAs were synchronized with DTB and released into fresh medium. After the indicated times, the cells were fixed and stained with propidium iodide solution for DNA, followed by flow cytometry to analyze the DNA content. The distribution and percentage of cells in G0/G1 and G2/M phases of the cell cycle are indicated. AS, asynchronized cells. F, the percentages of mitotic cells with nuclear envelope breakdown to the formation of a bipolar spindle/metaphase plate with some unaligned chromosomes (prometaphase), the unaligned state to metaphase without unaligned chromosomes (metaphase I), metaphase without unaligned chromosomes to anaphase (metaphase II), anaphase/telophase and cytokinesis, and unaligned chromosomes over the total number of mitotic cells. n = 80 cells/condition. The results of four independent experiments are shown. *, p < 0.05; **, p < 0.01; ***, p < 0.001, one-tailed Student's t test.
Depletion of CIP2A Delays Mitotic Progression
Because CIP2A depletion reduces MDA-MB-231 cell proliferation (3, 5, 51) and is known to bind to NEK2, cell cycle progression in CIP2A-depleted cells was analyzed. HeLa cells stably expressing GFP-histone H2B were transfected with CIP2A siRNA, and cell division was observed using time-lapse microscopy. When the duration of cell division was measured by a count of more than 300 cells, most of the control cells were found to have completed cell division within 70 min. Conversely, the cell division time was extended in the CIP2A-depleted cells (Fig. 2D). To determine the effect of CIP2A depletion on cell cycle progression, CIP2A-depleted HeLa cells were synchronized using DTB and released from the G1/S boundary. The progression of the cell cycle was then analyzed using flow cytometry. The control and CIP2A-depleted cells progressed to G2/M phase by 8 h after DTB release. 23% of the control cells progressed from G2/M to G1 phase after 30 min, whereas only 9.5% of the CIP2A-depleted cells progressed to G1 phase. This delayed progression was sustained until 9.5 h (Fig. 2E). To determine which mitotic phase is linked to the delayed progression from G2/M to G1 phases in the CIP2A-depleted cells, we counted the CIP2A-depleted HeLa cells expressing GFP-histone H2B in each mitotic phase. CIP2A depletion increased the metaphase I population. Thus, the metaphase II population was decreased because of the extended time at metaphase I, and this phenotype was rescued by cotransfection with CIP2Arv (Fig. 2F). These results suggest that CIP2A may function as a cell cycle regulator during mitosis.
CIP2A Depletion Impairs Mitotic Spindle Dynamics
The SAC is a critical cellular mechanism that prevents chromosome segregation until the completion of chromosome congression. The SAC is activated when the kinetochores are not correctly attached to microtubules or when tension is not applied at the kinetochores. Because CIP2A depletion caused a delay in mitotic progression, we addressed whether CIP2A is associated with SAC activation by immunostaining Mad2 and BubR1 in CIP2A-depleted cells. Mad2 and BubR1 were well localized at the kinetochore in prometaphase of CIP2A knockdown cells, whereas Mad2 and BubR1 were still found at the kinetochore in metaphase cells. Only 18.1% of the metaphase control cells were Mad2-positive, and 28.8% of the metaphase CIP2A-depleted cells were recorded as positive for Mad2. Moreover, BubR1-positive metaphase cells increased from 31.1 to 41.2% for the control and CIP2A-depleted populations, respectively (Fig. 3, A and B). This phenotype was further confirmed using HeLa cells stably expressing GFP-CENP-A. Although the GFP-CENP-A signal was perfectly aligned along the metaphase plate in the control cells, the GFP signal was detected away from the metaphase plate in the CIP2A-depleted cells, and most of the unaligned centromeres were not attached to microtubules (Fig. 3C). This finding suggests that spindle stability or attachment is impaired in CIP2A-depleted cells. To determine whether SAC activation is associated with spindle formation and stability, the microtubule intensity was measured and was significantly reduced in the CIP2A-depleted cells prior to cold treatment and further decreased after cold treatment compared with the control cells (Fig. 3D). This phenotype was consistent with that of NEK2A-depleted cells. Next, the kinetics of microtubule repolymerization were examined. The cells were treated with nocodazole to completely depolymerize the microtubules and then released by a change to fresh medium. The rate of microtubule polymerization was then determined by measuring the microtubule intensity. The CIP2A- and NEK2A-depleted cells showed slower microtubule repolymerization than the control cells (Fig. 3E), indicating that CIP2A and NEK2A depletion are involved in the spindle dynamics. To further confirm this fact, the effect of CIP2A and NEK2A on spindle dynamics was directly analyzed with a fluorescence loss in photobleaching experiment that measured the turnover rate of α/β-tubulin heterodimers on the mitotic spindle. HeLa cells stably expressing GFP-α-tubulin were transfected with siCIP2A, siCIP2A with CIP2Arv, and siNEK2, and then the cytoplasms of metaphase cells were photobleached continuously while time-lapse images were captured every 0.5 s to record the decrease in GFP fluorescence on the spindle (Fig. 3F). The half-life of GFP-α-tubulin on the control metaphase spindle was 186 ± 47 s. This was decreased to 106 ± 44 s (mean) in siCIP2A metaphase spindles and rescued to 181 ± 49 s in siCIP2A- and CIP2Arv-transfected cells. It indicates that CIP2A depletion clearly impairs the rate of microtubule turnover on metaphase spindles. Collectively, CIP2A depletion impedes the dynamics of the mitotic spindle and proper centrosome separation.
Depletion of CIP2A Delays NEK2 Degradation
Because CIP2A binds to NEK2 and we found that they function in cell cycle progression, NEK2 levels were examined in CIP2A-depleted cells by immunoblot assay at the various time points after DTB release. NEK2 degradation was delayed significantly in CIP2A-depleted cells, which is consistent with mitotic delay in these cells. On the other hand, cyclin B degradation, which is one of the main events for mitotic exit, was also delayed in CIP2A-depleted cells (Fig. 4A). Because NEK2 is known to localize to the spindle pole, NEK2 localization and intensity were examined at the spindle poles of CIP2A-depleted cells. NEK2 localization was not influenced throughout the cell cycle (Fig. 4C). However, the fluorescence intensity of NEK2 at the centrosome of CIP2A-depleted cells increased more than 35% compared with controls, consistent with delayed NEK2 degradation (Fig. 4B). In addition, to establish whether the delayed NEK2 degradation affects microtubule polymerization, in vitro microtubule polymerization assays were performed. The recombinant NEK2A-added reaction mixture enhanced polymerization. However, when the NEK2A-added amount was high (2.9 μg of NEK2A), microtubule polymerization was decreased to mock control levels, whereas paclitaxel as a positive control showed the highest microtubule polymerization compared with mock controls (Fig. 4D).
CIP2A Interacts with and Affects NEK2A Kinase Activity
NEK2A was identified as a CIP2A-binding partner protein in a yeast two-hybrid screening. Thus, immunoprecipitation was performed to confirm whether NEK2A can form a complex with CIP2A in mammalian cells. Endogenous NEK2A and NEK2B clearly bound to CIP2A (Fig. 5A). Next, to determine the CIP2A region responsible for binding to NEK2A, CIP2A-truncated mutants were generated and tagged with FLAG. The deletion of the C-terminal portion of CIP2A (ΔC305), which includes the coiled-coil domain, resulted in a weak interaction with NEK2A but did not abolish their interaction. In contrast, an N-terminal deletion (ΔN600) removing the region containing the armadillo repeat sequence and leucine zipper domain did not affect NEK2A binding (Fig. 5B). Next, to determine the NEK2A region responsible for binding to CIP2A, NEK2A-truncated forms were generated and tagged with GFP. The removal of the C-terminal portion of NEK2A (ΔC40), which includes the coiled-coil domain, did not affect CIP2A binding, whereas deletion of the N-terminal portion of NEK2A (ΔN271), which covers the kinase domain, did abolish binding to CIP2A (Fig. 5C). These results indicate that the C-terminal coiled-coil domain of CIP2A and the N-terminal kinase domain of NEK2A are involved in their strong binding.
FIGURE 5.
CIP2A interacts with and affects NEK2A kinase activity. A–D, HEK293 cells were used for a coimmunoprecipitation analysis between CIP2A and NEK2. A, HEK293 cells lysates were immunoprecipitated (IP) with anti-CIP2A, and the immunoprecipitated proteins were analyzed by immunoblotting. B, FLAG-tagged CIP2A constructs were coexpressed with an NEK2A construct in HEK293 cells. The FLAG-tagged proteins were immunoprecipitated with anti-FLAG antibodies, and the coimmunoprecipitation of NEK2A was determined by immunoblotting (WB). C, HEK293 cells were transfected with plasmids expressing EGFP-NEK2A mutants and CIP2A-FLAG. The cell lysates were immunoprecipitated with anti-FLAG antibodies, followed by immunoblotting using the indicated antibodies. D, CIP2A was immunoprecipitated from asynchronized (AS), G1/S phase (DTB and released for 0 h), S phase (DTB 3 h), and G2/M phase (DTB 9 h) HeLa cells as indicated. The immunoprecipitate of CIP2A (IP) and the input whole-cell lysate was subjected to immunoblotting. E, NEK2A level was normalized by increasing the cell lysates obtained at 0 h and then immunoprecipitated with anti-CIP2A antibody. The immunoprecipitates and the input whole-cell lysate were subjected to immunoblotting. F, the EGFP-NEK2A construct was expressed with each siRNA in HEK293 cells. After 24 h, the cells were synchronized by DTB and harvested at 9 h after release. The NEK2 activity was measured using an in vitro kinase assay with EGFP-NEK2 immunoprecipitation. Data are mean ± S.D. n = 3. **, p < 0.01, one-tailed Student's t test. RLU, relative luminescence units. G, CIP2A-FLAG was purified from HEK293 cells transiently expressing CIP2A-FLAG using anti-FLAG M2 affinity gel. The NEK2A activity in the presence of CIP2A-FLAG was measured by an NEK2 kinase enzyme system using recombinant NEK2A and MBP protein as the substrate. pcDNA3.1 was used as a control. Data are mean ± S.D. n = 3. **, p < 0.01, one-tailed Student's t test. H, the EGFP-NEK2A construct was expressed with each siRNA in HEK293 cells. After 48 h, the cells were lysed, and immunoprecipitation was assessed using GFP-Trap. The immunoprecipitates and the whole-cell lysates (Input) were subjected to immunoblotting using anti-PP1α and γ isoform antibodies. I, the NEK2A-myc and NEK2A-FLAG constructs were expressed with each siRNA in HEK 293 cells. After 48 h, the cells were lysed, and immunoprecipitation was assessed using FLAG antibodies. The immunoprecipitates and the whole-cell lysates (Input) were subjected to immunoblotting using anti-myc, FLAG, and CIP2A antibodies. J, top panel, schematic of the protocol used to treat the cells. HeLa cells were transfected with siRNAs for 24 h and replated onto glass coverslips for synchronization. Following a double thymidine block, the cells were released into fresh medium. After 5 h, the cells were exposed to 100 μm monastrol for 4 h to inhibit Eg5-dependent centrosome splitting. Prior to fixation, the cells were treated with 1 nm of calyculin A or 0.1 nm okadaic acid. The cells were fixed and stained for γ-tubulin (red) and DNA (blue). Bottom panel, shown are the maximum projections from deconvolved z stacks of representative control cells or CIP2A-depleted cells. The distance between the two centrosomes was analyzed (n > 11 cells/condition. *, p < 0.05; **, p < 0.01; one-tailed Student's t test. Scale bar = 5 μm.
To identify the cell cycle stage at which CIP2A binds to NEK2A, cells were synchronized by DTB, released at the G1/S boundary, and then immunoprecipitated with anti-CIP2A antibody at the indicated time points. The binding of NEK2A and CIP2A markedly increased 9 h after release, which was the G2/M stage according to the cyclin A and B level (Fig. 5D). However, the increased NEK2A amount after release may be a cause of the enhanced binding between NEK2A and CIP2A at 9 h. To exclude this possibility, the NEK2A level was normalized by increasing amounts of cell lysates obtained at 0 h, and then immunoprecipitation analysis was performed. Although the NEK2A expression level was similar between the 0 and 9 h points, the binding of NEK2A and CIP2A was enhanced at 9 h (Fig. 5E). This indicates that CIP2A interacts with NEK2A specifically at the G2/M point. We next examined whether binding between CIP2A and NEK2 affects the function of NEK2. HEK293 cells were transfected with EGFP-NEK2A and CIP2A siRNA, and EGFP-NEK2A was immunoprecipitated. The NEK2A activity was then measured and found to have decreased by ∼22% in the CIP2A-depleted cells compared with the control cells (Fig. 5E).
There are four possible hypotheses to explain how CIP2A enhances NEK2A kinase activity. 1) CIP2A solely interacts with NEK2A, which leads to an enhancement of the kinase activity. 2) Because the formation of a PP1-NEK2A complex prevents NEK2A kinase activity, it is possible that CIP2A disrupts binding between NEK2A and PP1. 3) CIP2A-NEK2 binding may provide strong NEK2 dimerization, which is necessary for NEK2 kinase activity (52). 4) PP2A binding to CIP2A counteracts NEK2 kinase activity. To test the first hypothesis, CIP2A-FLAG was transfected into HEK293 cells, and the CIP2A-FLAG proteins were eluted from FLAG affinity-purifying agarose by competition with the FLAG peptide. NEK2A activity was increased when recombinant NEK2A was incubated with the purified CIP2A-FLAG protein (Fig. 5F). To test the second hypothesis, EGFP-NEK2A was cotransfected with siCIP2A in HEK293 cells and immunoprecipitated with EGFP-NEK2A. The level of endogenous PP1α and γ isoforms was then analyzed. The depletion of CIP2A did not affect the binding between NEK2A and PP1α and γ (Fig. 5G). For the third hypothesis, NEK2A-myc and NEK2A-FLAG were cotransfected with siCIP2A in HEK 293 cells, and NEK2A was immunoprecipitated with FLAG antibodies. The level of NEK2 dimerization was measured. However, the depletion of CIP2A did not affect the amount of NEK2 dimerization (Fig. 5H). To test the last hypothesis, the distance between prometaphase centrosomes was measured after treatment with a PP2A inhibitor. The defect of centrosome separation in the CIP2A-depleted cells was not rescued by treatment with calyculin A, an inhibitor of both PP1 and PP2A, or okadaic acid, a more specific PP2A inhibitor (Fig. 5I). These findings indicate that CIP2A enhances NEK2A activity in a PP1- and PP2A-independent manner.
DISCUSSION
The depletion of CIP2A in many tumors induces the inhibition of cell proliferation, whereas the overexpression of CIP2A with simultaneous RAS oncogene expression promotes the transformation of nonmalignant cells (28). CIP2A also positively regulates c-Myc expression, and c-Myc stimulates the G1/S transition by enhancing cyclin-dependent kinase (CDK) function via inhibition of the CDK inhibitors p27Kip1, p21Cip1, and p57Kip2 (33). Regardless, the molecular mechanism by which CIP2A increases cell proliferation has remained elusive to date. The cell cycle has been analyzed previously in an effort to identify the detailed molecular mechanism, although the CIP2A level showed no change with cell cycle progression (4). However, that study only employed a synchronization and release experiment. In this study, we specifically examined changes in the mitotic cell cycle using CIP2A-depleted cells. Although the CIP2A level was unchanged with cell cycle progression, consistent with previous results (Fig. 5D), CIP2A depletion delayed the mitotic cell cycle (Fig. 4E), indicating that CIP2A is indeed associated with mitotic cell cycle progression without fluctuations in its expression level. This finding provides direct evidence for a novel function of CIP2A in cell cycle progression.
NEK2A plays a critical role in centrosome separation by phosphorylating its target substrates at the beginning of mitosis (35, 38–40). In this study we showed that CIP2A stimulated NEK2A activity. Although the regulation mechanism by which CIP2A increases NEK2A activity should be elucidated in further studies, with these findings we can suggest the possible regulation mechanism. It is unlikely that CIP2A enhances NEK2A activity by stimulating NEK2A homodimerization, which is essential for its kinase activity (52), because CIP2A did not affect NEK2A dimerization (Fig. 5H). NEK2A activity is also negatively regulated by phosphatases (53–55), but we showed that CIP2A enhanced the endogenous NEK2 activity independently of PP1 and PP2A (Fig. 5I). On the other hand, because the catalytic domain of NEK2 has inhibitory autophosphorylation sites that negatively regulate NEK2 activity (56) and CIP2A bound with the catalytic domain of NEK2A, it is possible that the binding of CIP2A prevents inhibitory autophosphorylation. There is another explanation. The crystallographic analysis of NEK2 reveals that the active conformation and autoinhibitory conformation of NEK2 is interchangeable by binding of the regulatory molecule (57). Thus, the binding of CIP2A could change NEK2A conformation into an active form.
Additionally, NEK2 is mainly localized to the centrosome from G1 to G2 phases and begins to degrade at M phase (58). There is also a report that NEK2 is localized to both the nucleus and cytoplasm throughout the cell cycle and, moreover, associates with the chromosomes at M phase (34). CIP2A, however, was not localized to the centrosome at the G1 to G2 phases under our experimental conditions but was recruited to the centrosome at M phase. Hence, it is unlikely that the interaction between CIP2A and NEK2A functions at the G2 phase centrosome. There are three possibilities to explain this discrepancy. One possibility is that CIP2A may enhance NEK2A activity at the centrosome with CIP2A immediately dissociating, which would indicate that CIP2A and NEK2A binding is transient at the centrosome. Indeed, we did not observe CIP2A localization at the centrosome. Alternatively, although CIP2A and NEK2A may not interact at the centrosome, CIP2A and NEK2A can interact at other sites, such as the cytoplasm or chromosomes. Therefore, activated NEK2A could play a role at the centrosome. The last possibility is that the experimental conditions used may not have been optimal for observing CIP2A centrosome localization.
Similar to NEK2A, NEK2B is also localized at the centrosome, although the level of NEK2B is lower at S and G2 phases. However, the NEK2B level remains elevated, whereas the NEK2A level is reduced because of degradation at M phase (34, 58). A strong localization of CIP2A at the centrosome was observed after the completion of chromosome congression at M phase. Thus, CIP2A could interact with NEK2B at the late metaphase centrosome because CIP2A was also bound to NEK2B (Fig. 5A). Although several reports have revealed that the overexpression of NEK2B does not affect centrosome separation (58) and that NEK2B down-regulation leads to mitotic delay (59), little is known about the role of NEK2B. Further study of the CIP2A-NEK2B interaction will contribute to the understanding of the role of CIP2A and NEK2B in centrosome function and mitotic exit.
In addition to the delay of centrosome separation in CIP2A-depleted cells, the dynamics of the mitotic spindle were impaired, resulting in the activation of SAC signaling and the extension of the cell division time. The phenotypes presented by CIP2A-depleted cells can be understood in terms of the regulation of NEK2A-mediated Hec1 phosphorylation. Many studies have illustrated that NEK2A regulates the stability of kinetochore microtubule attachment through Hec1 Ser-165 phosphorylation and modulates chromosome alignment and SAC signaling (46–48). In this respect, it is conceivable that CIP2A-depleted cells show unstable kinetochore microtubule attachment and SAC activation because the NEK2A kinase activity is impaired. However, because cells with incomplete spindle pole separation exhibit higher rates of kinetochore chromosome misattachment (60) and centrosome separation is impaired in CIP2A-depleted cells, SAC activation and chromosome misalignment could occur in these cells.
It has been reported recently that NEK2 overexpression induces chromosomal instability in myeloma, head and neck squamous cell carcinoma, bladder carcinoma, glioblastoma, T cell acute lymphoblastic leukemia, colon carcinoma, and ovarian carcinoma by enhancing the expression of genes involved in cell cycle progression, DNA replication, and chromosome condensation (61). Because CIP2A has been reported as a gene that is highly correlated with NEK2 overexpression and CIP2A depletion impairs spindle dynamics and centrosome separation, the cooperation of NEK2 and CIP2A by enhancing NEK2 activity could be one of mechanisms by which chromosome instability is induced.
Structural maintenance of chromosomes (SMC) protein complexes play important roles in functional and structural chromosome organization, including chromosome condensation and sister chromatid cohesion processes (62–64). In addition to SMC1–6 proteins, several proteins containing SMC domains have been identified. Among these, the coiled-coil protein quantitatively enriched (Ccq1p) protein interacts with the centrosomal protein Pcp1p, a yeast homolog of pericentrin (65), and localizes to the telomere. Therefore, these two proteins link the centrosome and telomere, which is essential for meiosis. Ccq1p has an N-terminal armadillo (ARM) repeat and a C-terminal SMC domain/coiled-coil region that is involved in protein-protein interactions. CIP2A also has an N-terminal armadillo repeat and a C-terminal SMC domain/coiled-coil region. Moreover, CIP2A is strongly expressed in testes and promotes the proliferation of spermatogonial progenitor cells and spermatogenesis (66). Therefore, it is possible for CIP2A to play a critical role in meiosis by regulating NEK2 activity.
In summary, we demonstrate for the first time that CIP2A is associated with cell cycle regulation and exhibits dynamic changes in distribution, depending on the cell cycle stage. The depletion of CIP2A impairs cell cycle progression and leads to aberrations in centrosome separation, mitotic spindle dynamics, and SAC activation. We show that CIP2A is involved in centrosome separation through the activation of NEK2A activity. These results have important implications for an understanding of the role of CIP2A in cancer progression.
Acknowledgments
We thank K. Rhee, C. W. Lee, J. S. Kim, and K. Kim for material support. We also thank A.M. Fry for critical discussions.
This work was supported by National Research Foundation of Korea (NRF) Grants NRF-2012R1A2A1A03003709 and 2011-0030074 funded by the Korean government (Ministry of Science, ICT and Future Planning) and by Grant B110053-1101-0000200 of the Korea Health Industry Development Institute funded by the Ministry of Health and Welfare.
- PP2A
- protein phosphatase 2A
- SAC
- spindle assembly checkpoint
- TRITC
- tetramethylrhodamine isothiocyanate
- DTB
- double thymidine block
- SMC
- structural maintenance of chromosomes
- NIMA
- never in mitosis gene A.
REFERENCES
- 1. Kikuno R., Nagase T., Ishikawa K., Hirosawa M., Miyajima N., Tanaka A., Kotani H., Nomura N., Ohara O. (1999) Prediction of the coding sequences of unidentified human genes. XIV. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 6, 197–205 [DOI] [PubMed] [Google Scholar]
- 2. Wang L., Gu F., Ma N., Zhang L., Bian J. M., Cao H. Y. (2013) CIP2A expression is associated with altered expression of epithelial-mesenchymal transition markers and predictive of poor prognosis in pancreatic ductal adenocarcinoma. Tumour Biol. 34, 2309–2313 [DOI] [PubMed] [Google Scholar]
- 3. Li W., Ge Z., Liu C., Liu Z., Björkholm M., Jia J., Xu D. (2008) CIP2A is overexpressed in gastric cancer and its depletion leads to impaired clonogenicity, senescence, or differentiation of tumor cells. Clin. Cancer Res. 14, 3722–3728 [DOI] [PubMed] [Google Scholar]
- 4. Khanna A., Böckelman C., Hemmes A., Junttila M. R., Wiksten J. P., Lundin M., Junnila S., Murphy D. J., Evan G. I., Haglund C., Westermarck J., Ristimäki A. (2009) MYC-dependent regulation and prognostic role of CIP2A in gastric cancer. J. Natl. Cancer Inst. 101, 793–805 [DOI] [PubMed] [Google Scholar]
- 5. Côme C., Laine A., Chanrion M., Edgren H., Mattila E., Liu X., Jonkers J., Ivaska J., Isola J., Darbon J. M., Kallioniemi O., Thézenas S., Westermarck J. (2009) CIP2A is associated with human breast cancer aggressivity. Clin. Cancer Res. 15, 5092–5100 [DOI] [PubMed] [Google Scholar]
- 6. Niemelä M., Kauko O., Sihto H., Mpindi J. P., Nicorici D., Pernilä P., Kallioniemi O. P., Joensuu H., Hautaniemi S., Westermarck J. (2012) CIP2A signature reveals the MYC dependency of CIP2A-regulated phenotypes and its clinical association with breast cancer subtypes. Oncogene 31, 4266–4278 [DOI] [PubMed] [Google Scholar]
- 7. Huang L. P., Adelson M. E., Mordechai E., Trama J. P. (2010) CIP2A expression is elevated in cervical cancer. Cancer Biomark. 8, 309–317 [DOI] [PubMed] [Google Scholar]
- 8. Liu J., Wang X., Zhou G., Wang H., Xiang L., Cheng Y., Liu W., Wang Y., Jia J., Zhao W. (2011) Cancerous inhibitor of protein phosphatase 2A is overexpressed in cervical cancer and upregulated by human papillomavirus 16 E7 oncoprotein. Gynecol. Oncol. 122, 430–436 [DOI] [PubMed] [Google Scholar]
- 9. Dong Q. Z., Wang Y., Dong X. J., Li Z. X., Tang Z. P., Cui Q. Z., Wang E. H. (2011) CIP2A is overexpressed in non-small cell lung cancer and correlates with poor prognosis. Ann. Surg. Oncol. 18, 857–865 [DOI] [PubMed] [Google Scholar]
- 10. Katz J., Jakymiw A., Ducksworth M. K., Stewart C. M., Bhattacharyya I., Cha S., Chan E. K. (2010) CIP2A expression and localization in oral carcinoma and dysplasia. Cancer Biol. Ther. 10, 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vaarala M. H., Väisänen M. R., Ristimäki A. (2010) CIP2A expression is increased in prostate cancer. J. Exp. Clin. Cancer Res. 29, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qu W., Li W., Wei L., Xing L., Wang X., Yu J. (2012) CIP2A is overexpressed in esophageal squamous cell carcinoma. Med. Oncol 29, 113–118 [DOI] [PubMed] [Google Scholar]
- 13. Coenen E. A., Zwaan C. M., Meyer C., Marschalek R., Pieters R., van der Veken L. T., Beverloo H. B., van den Heuvel-Eibrink M. M. (2011) KIAA1524. A novel MLL translocation partner in acute myeloid leukemia. Leuk. Res. 35, 133–135 [DOI] [PubMed] [Google Scholar]
- 14. Wang J., Li W., Li L., Yu X., Jia J., Chen C. (2011) CIP2A is over-expressed in acute myeloid leukaemia and associated with HL60 cells proliferation and differentiation. Int. J. Lab. Hematol. 33, 290–298 [DOI] [PubMed] [Google Scholar]
- 15. Böckelman C., Lassus H., Hemmes A., Leminen A., Westermarck J., Haglund C., Bützow R., Ristimäki A. (2011) Prognostic role of CIP2A expression in serous ovarian cancer. Br. J. Cancer 105, 989–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Böckelman C., Hagström J., Mäkinen L. K., Keski-Säntti H., Häyry V., Lundin J., Atula T., Ristimäki A., Haglund C. (2011) High CIP2A immunoreactivity is an independent prognostic indicator in early-stage tongue cancer. Br. J. Cancer 104, 1890–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fang Y., Li Z., Wang X., Zhang S. (2012) CIP2A is overexpressed in human ovarian cancer and regulates cell proliferation and apoptosis. Tumour. Biol. 33, 2299–2306 [DOI] [PubMed] [Google Scholar]
- 18. Ren J., Li W., Yan L., Jiao W., Tian S., Li D., Tang Y., Gu G., Liu H., Xu Z. (2011) Expression of CIP2A in renal cell carcinomas correlates with tumour invasion, metastasis and patients' survival. Br. J. Cancer 105, 1905–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin Y. C., Chen K. C., Chen C. C., Cheng A. L., Chen K. F. (2012) CIP2A-mediated Akt activation plays a role in bortezomib-induced apoptosis in head and neck squamous cell carcinoma cells. Oral Oncol. 48, 585–593 [DOI] [PubMed] [Google Scholar]
- 20. Teng H. W., Yang S. H., Lin J. K., Chen W. S., Lin T. C., Jiang J. K., Yen C. C., Li A. F., Chen P. C., Lan Y. T., Lin C. C., Hsu Y. N., Wang H. W., Chen K. F. (2012) CIP2A is a predictor of poor prognosis in colon cancer. J. Gastrointest. Surg. 16, 1037–1047 [DOI] [PubMed] [Google Scholar]
- 21. He H., Wu G., Li W., Cao Y., Liu Y. (2012) CIP2A is highly expressed in hepatocellular carcinoma and predicts poor prognosis. Diagn. Mol. Pathol. 21, 143–149 [DOI] [PubMed] [Google Scholar]
- 22. Huang L. P., Savoly D., Sidi A. A., Adelson M. E., Mordechai E., Trama J. P. (2012) CIP2A protein expression in high-grade, high-stage bladder cancer. Cancer Med. 1, 76–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang P., Qiu J., You J., Hong J., Li B., Zhou K., Chen G., Yuan Y., Zou R. (2012) Expression and prognostic significance of CIP2A mRNA in hepatocellular carcinoma and nontumoral liver tissues. Biomarkers 17, 422–429 [DOI] [PubMed] [Google Scholar]
- 24. Xue Y., Wu G., Wang X., Zou X., Zhang G., Xiao R., Yuan Y., Long D., Yang J., Wu Y., Xu H., Liu F., Liu M. (2013) CIP2A is a predictor of survival and a novel therapeutic target in bladder urothelial cell carcinoma. Med. Oncol. 30, 406. [DOI] [PubMed] [Google Scholar]
- 25. Yi F., Ni W., Liu W., Bai J., Li W. (2013) Expression and biological role of CIP2A in human astrocytoma. Mol. Med. Rep. 7, 1376–1380 [DOI] [PubMed] [Google Scholar]
- 26. Khanna A., Okkeri J., Bilgen T., Tiirikka T., Vihinen M., Visakorpi T., Westermarck J. (2011) ETS1 mediates MEK1/2-dependent overexpression of cancerous inhibitor of protein phosphatase 2A (CIP2A) in human cancer cells. PLoS ONE 6, e17979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Laine A., Sihto H., Come C., Rosenfeldt M. T., Zwolinska A., Niemela M., Khanna A., Chan E. K., Kahari V. M., Kellokumpu-Lehtinen P. L., Sansom O. J., Evan G. I., Junttila M. R., Ryan K. M., Marine J. C., Joensuu H., Westermarck J. (2013) Senescence Sensitivity of Breast Cancer Cells Is Defined by Positive Feedback Loop between CIP2A and E2F1. Cancer Discov. 3, 182–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Junttila M. R., Puustinen P., Niemelä M., Ahola R., Arnold H., Böttzauw T., Ala-aho R., Nielsen C., Ivaska J., Taya Y., Lu S. L., Lin S., Chan E. K., Wang X. J., Grènman R., Kast J., Kallunki T., Sears R., Kähäri V. M., Westermarck J. (2007) CIP2A inhibits PP2A in human malignancies. Cell 130, 51–62 [DOI] [PubMed] [Google Scholar]
- 29. Ma L., Wen Z. S., Liu Z., Hu Z., Ma J., Chen X. Q., Liu Y. Q., Pu J. X., Xiao W. L., Sun H. D., Zhou G. B. (2011) Overexpression and small molecule-triggered downregulation of CIP2A in lung cancer. PLoS ONE 6, e20159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mathiasen D. P., Egebjerg C., Andersen S. H., Rafn B., Puustinen P., Khanna A., Daugaard M., Valo E., Tuomela S., Bøttzauw T., Nielsen C. F., Willumsen B. M., Hautaniemi S., Lahesmaa R., Westermarck J., Jäättelä M., Kallunki T. (2012) Identification of a c-Jun N-terminal kinase-2-dependent signal amplification cascade that regulates c-Myc levels in ras transformation. Oncogene 31, 390–401 [DOI] [PubMed] [Google Scholar]
- 31. Böckelman C., Koskensalo S., Hagström J., Lundin M., Ristimäki A., Haglund C. (2012) CIP2A overexpression is associated with c-Myc expression in colorectal cancer. Cancer Biol. Ther. 13, 289–295 [DOI] [PubMed] [Google Scholar]
- 32. Müller D., Bouchard C., Rudolph B., Steiner P., Stuckmann I., Saffrich R., Ansorge W., Huttner W., Eilers M. (1997) Cdk2-dependent phosphorylation of p27 facilitates its Myc-induced release from cyclin E/cdk2 complexes. Oncogene 15, 2561–2576 [DOI] [PubMed] [Google Scholar]
- 33. Pérez-Roger I., Solomon D. L., Sewing A., Land H. (1997) Myc activation of cyclin E/Cdk2 kinase involves induction of cyclin E gene transcription and inhibition of p27(Kip1) binding to newly formed complexes. Oncogene 14, 2373–2381 [DOI] [PubMed] [Google Scholar]
- 34. Ha Kim Y., Yeol Choi J., Jeong Y., Wolgemuth D. J., Rhee K. (2002) Nek2 localizes to multiple sites in mitotic cells, suggesting its involvement in multiple cellular functions during the cell cycle. Biochem. Biophys. Res. Commun. 290, 730–736 [DOI] [PubMed] [Google Scholar]
- 35. Fry A. M., Meraldi P., Nigg E. A. (1998) A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J. 17, 470–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Andersen J. S., Wilkinson C. J., Mayor T., Mortensen P., Nigg E. A., Mann M. (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426, 570–574 [DOI] [PubMed] [Google Scholar]
- 37. Fry A. M., Mayor T., Meraldi P., Stierhof Y. D., Tanaka K., Nigg E. A. (1998) C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol. 141, 1563–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Faragher A. J., Fry A. M. (2003) Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol. Biol. Cell 14, 2876–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mayor T., Hacker U., Stierhof Y. D., Nigg E. A. (2002) The mechanism regulating the dissociation of the centrosomal protein C-Nap1 from mitotic spindle poles. J. Cell Sci. 115, 3275–3284 [DOI] [PubMed] [Google Scholar]
- 40. Bahe S., Stierhof Y. D., Wilkinson C. J., Leiss F., Nigg E. A. (2005) Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J. Cell Biol. 171, 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bahmanyar S., Kaplan D. D., Deluca J. G., Giddings T. H., Jr., O'Toole E. T., Winey M., Salmon E. D., Casey P. J., Nelson W. J., Barth A. I. (2008) β-Catenin is a Nek2 substrate involved in centrosome separation. Genes Dev. 22, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rhee K., Wolgemuth D. J. (1997) The NIMA-related kinase 2, Nek2, is expressed in specific stages of the meiotic cell cycle and associates with meiotic chromosomes. Development 124, 2167–2177 [DOI] [PubMed] [Google Scholar]
- 43. Sonn S., Khang I., Kim K., Rhee K. (2004) Suppression of Nek2A in mouse early embryos confirms its requirement for chromosome segregation. J. Cell Sci. 117, 5557–5566 [DOI] [PubMed] [Google Scholar]
- 44. Jeong Y., Lee J., Kim K., Yoo J. C., Rhee K. (2007) Characterization of NIP2/centrobin, a novel substrate of Nek2, and its potential role in microtubule stabilization. J. Cell Sci. 120, 2106–2116 [DOI] [PubMed] [Google Scholar]
- 45. Park J., Rhee K. (2013) NEK2 phosphorylation antagonizes the microtubule stabilizing activity of centrobin. Biochem. Biophys. Res. Commun. 431, 302–308 [DOI] [PubMed] [Google Scholar]
- 46. Wei R., Ngo B., Wu G., Lee W. H. (2011) Phosphorylation of the Ndc80 complex protein, HEC1, by Nek2 kinase modulates chromosome alignment and signaling of the spindle assembly checkpoint. Mol. Biol. Cell 22, 3584–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen Y., Riley D. J., Zheng L., Chen P. L., Lee W. H. (2002) Phosphorylation of the mitotic regulator protein Hec1 by Nek2 kinase is essential for faithful chromosome segregation. J. Biol. Chem. 277, 49408–49416 [DOI] [PubMed] [Google Scholar]
- 48. Du J., Cai X., Yao J., Ding X., Wu Q., Pei S., Jiang K., Zhang Y., Wang W., Shi Y., Lai Y., Shen J., Teng M., Huang H., Fei Q., Reddy E. S., Zhu J., Jin C., Yao X. (2008) The mitotic checkpoint kinase NEK2A regulates kinetochore microtubule attachment stability. Oncogene 27, 4107–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mardin B. R., Lange C., Baxter J. E., Hardy T., Scholz S. R., Fry A. M., Schiebel E. (2010) Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nat. Cell Biol. 12, 1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mardin B. R., Isokane M., Cosenza M. R., Krämer A., Ellenberg J., Fry A. M., Schiebel E. (2013) EGF-induced centrosome separation promotes mitotic progression and cell survival. Dev. Cell 25, 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Choi Y. A., Park J. S., Park M. Y., Oh K. S., Lee M. S., Lim J. S., Kim K. I., Kim K. Y., Kwon J., Yoon do Y., Moon E. Y., Yang Y. (2011) Increase in CIP2A expression is associated with doxorubicin resistance. FEBS Lett. 585, 755–760 [DOI] [PubMed] [Google Scholar]
- 52. Fry A. M., Arnaud L., Nigg E. A. (1999) Activity of the human centrosomal kinase, Nek2, depends on an unusual leucine zipper dimerization motif. J. Biol. Chem. 274, 16304–16310 [DOI] [PubMed] [Google Scholar]
- 53. Helps N. R., Luo X., Barker H. M., Cohen P. T. (2000) NIMA-related kinase 2 (Nek2), a cell-cycle-regulated protein kinase localized to centrosomes, is complexed to protein phosphatase 1. Biochem. J. 349, 509–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Meraldi P., Nigg E. A. (2001) Centrosome cohesion is regulated by a balance of kinase and phosphatase activities. J. Cell Sci. 114, 3749–3757 [DOI] [PubMed] [Google Scholar]
- 55. Mi J., Guo C., Brautigan D. L., Larner J. M. (2007) Protein phosphatase-1α regulates centrosome splitting through Nek2. Cancer Res. 67, 1082–1089 [DOI] [PubMed] [Google Scholar]
- 56. Rellos P., Ivins F. J., Baxter J. E., Pike A., Nott T. J., Parkinson D. M., Das S., Howell S., Fedorov O., Shen Q. Y., Fry A. M., Knapp S., Smerdon S. J. (2007) Structure and regulation of the human Nek2 centrosomal kinase. J. Biol. Chem. 282, 6833–6842 [DOI] [PubMed] [Google Scholar]
- 57. Westwood I., Cheary D. M., Baxter J. E., Richards M. W., van Montfort R. L., Fry A. M., Bayliss R. (2009) Insights into the conformational variability and regulation of human Nek2 kinase. J. Mol. Biol. 386, 476–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hames R. S., Fry A. M. (2002) Alternative splice variants of the human centrosome kinase Nek2 exhibit distinct patterns of expression in mitosis. Biochem. J. 361, 77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fletcher L., Cerniglia G. J., Yen T. J., Muschel R. J. (2005) Live cell imaging reveals distinct roles in cell cycle regulation for Nek2A and Nek2B. Biochim. Biophys. Acta 1744, 89–92 [DOI] [PubMed] [Google Scholar]
- 60. Silkworth W. T., Nardi I. K., Paul R., Mogilner A., Cimini D. (2012) Timing of centrosome separation is important for accurate chromosome segregation. Mol. Biol. Cell 23, 401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhou W., Yang Y., Xia J., Wang H., Salama M. E., Xiong W., Xu H., Shetty S., Chen T., Zeng Z., Shi L., Zangari M., Miles R., Bearss D., Tricot G., Zhan F. (2013) NEK2 induces drug resistance mainly through activation of efflux drug pumps and is associated with poor prognosis in myeloma and other cancers. Cancer Cell 23, 48–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Haering C. H., Nasmyth K. (2003) Building and breaking bridges between sister chromatids. BioEssays 25, 1178–1191 [DOI] [PubMed] [Google Scholar]
- 63. Hirano T. (2006) At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 7, 311–322 [DOI] [PubMed] [Google Scholar]
- 64. Thadani R., Uhlmann F., Heeger S. (2012) Condensin, chromatin crossbarring and chromosome condensation. Curr. Biol. 22, R1012–1021 [DOI] [PubMed] [Google Scholar]
- 65. Flory M. R., Carson A. R., Muller E. G., Aebersold R. (2004) An SMC-domain protein in fission yeast links telomeres to the meiotic centrosome. Mol. Cell 16, 619–630 [DOI] [PubMed] [Google Scholar]
- 66. Ventelä S., Côme C., Mäkelä J. A., Hobbs R. M., Mannermaa L., Kallajoki M., Chan E. K., Pandolfi P. P., Toppari J., Westermarck J. (2012) CIP2A promotes proliferation of spermatogonial progenitor cells and spermatogenesis in mice. PLoS ONE 7, e33209. [DOI] [PMC free article] [PubMed] [Google Scholar]