Background: Polysaccharide lyases degrade anionic polysaccharides and are important in bacterial biofilm formation and host-pathogen interactions.
Results: Putative alginate lyase (Smlt1473) from Stenotrophomonas maltophillia displays activity toward multiple polysaccharides in a pH-regulated manner.
Conclusion: Smlt1473 is a unique polysaccharide lyase with pH-dependent activity toward mammalian, plant, and microbial substrates.
Significance: Characterization of Smlt1473 allows for an understanding of possible roles in biofilm formation and pathogenesis.
Keywords: Biofilm, Host-Pathogen Interactions, Hyaluronate, Multifunctional Enzymes, Polysaccharide
Abstract
Polysaccharide lyases (PLs) catalyze the depolymerization of anionic polysaccharides via a β-elimination mechanism. PLs also play important roles in microbial pathogenesis, participating in bacterial invasion and toxin spread into the host tissue via degradation of the host extracellular matrix, or in microbial biofilm formation often associated with enhanced drug resistance. Stenotrophomonas maltophilia is a Gram-negative bacterium that is among the emerging multidrug-resistant organisms associated with chronic lung infections as well as with cystic fibrosis patients. A putative alginate lyase (Smlt1473) from S. maltophilia was heterologously expressed in Escherichia coli, purified in a one-step fashion via affinity chromatography, and activity as well as specificity determined for a range of polysaccharides. Interestingly, Smlt1473 catalyzed the degradation of not only alginate, but poly-β-d-glucuronic acid and hyaluronic acid as well. Furthermore, the pH optimum for enzymatic activity is substrate-dependent, with optimal hyaluronic acid degradation at pH 5, poly-β-d-glucuronic acid degradation at pH 7, and alginate degradation at pH 9. Analysis of the degradation products revealed that each substrate was cleaved endolytically into oligomers comprised predominantly of even numbers of sugar groups, with lower accumulation of trimers and pentamers. Collectively, these results imply that Smlt1473 is a multifunctional PL that exhibits broad substrate specificity, but utilizes pH as a mechanism to achieve selectivity.
Introduction
Polysaccharides play critical structural and signaling roles in a diverse range of biological structures including plant and mammalian extracellular matrix as well as microbial biofilm. Many biologically important polysaccharides contain uronic acids, including hyaluronic acid (HA),2 a linear polysaccharide comprised of repeating disaccharide units of d-glucuronic acid (GlcA) and N-acetyl-d-glucosamine (GlcNAc) (1). HA is a major component of the extracellular matrix in eukaryotes and plays numerous roles in cell motility, inflammation, and angiogenesis (2, 3). Other polyuronides are major components of plant and algal cell walls, such as alginate in brown algae (4) and poly-β-GlcA in green algae (5). Alginate is a linear copolymer comprised of d-mannuronic acid (ManA) and l-guluronic acid (GulA) arranged in repeating poly-β-ManA blocks, poly-α-GulA blocks, or alternating poly-MG blocks (6). Bacteria such as Pseudomonas aeruginosa also synthesize and secrete alginate as a major component of microbial biofilms. In the case of pathogens such as P. aeruginosa, biofilm formation is often correlated with enhanced adhesion to mammalian cells or inert surfaces as well as increased resistance to antimicrobials (7). Thus, polysaccharides play several key roles in the organization and function of multicellular mammalian and microbial structures.
Polysaccharide lyases (PLs) are a class of enzymes that degrade polyuronides via a β-elimination mechanism, resulting in the formation of unsaturated products containing 4-deoxy-l-erythro-hex-4-enopyranosyluronic acid at the non-reducing end. The β-elimination mechanism proceeds in three major steps (Fig. 1): (i) neutralization of the negative charge of the C5 carboxylate group of the +1 sugar ring, reducing the pKa of the C5 proton and thereby assisting in its abstraction, (ii) formation of enolate anion intermediate by removal of C5 proton by a Brønsted base, and (iii) concomitant formation of a double bond between C4 and C5 and cleavage of the C4-O-1 glycosidic bond via electron transfer (8). Although several crystallographic and biochemical studies have confirmed the 3-step β-elimination mechanism, identifying which catalytic residues are responsible for each step is still an area of active research (9–12). In particular, whereas a conserved tyrosine and histidine present in numerous classes of PL are both known to play an important role in the catalytic mechanism, it is unclear whether the histidine acts as the proton donor and tyrosine as the proton acceptor, a mechanism proposed for the HA lyase of Streptococcus sp. (11, 12) or if tyrosine acts as proton donor and acceptor and histidine stabilizes an intermediate product, a mechanism proposed for A1-III alginate lyase of Sphingomonas sp. (9). Thus, defining the exact proton donor-acceptor pair for catalysis in PLs remains an area of active research.
FIGURE 1.
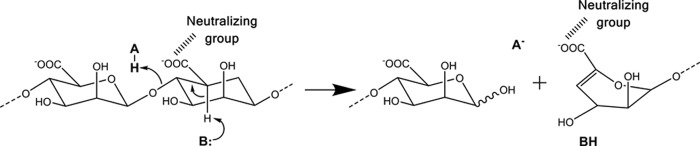
Schematic of β-elimination reaction mechanism.
Biologically, microbial PLs play diverse roles in biofilm formation as well as in host-pathogen interactions. In particular, HA lyases secreted by Group A Streptococcus are thought to act as spreading factors, enabling bacteria and toxins to disseminate throughout the host by degrading the high molecular weight HA present in the extracellular matrix (13). In Mycobacterium tuberculosis, secreted HA lyases generate low molecular weight sugars from HA that serve as carbon source for bacteria to replicate in deep tissue infections (14). In the case of P. aeruginosa biofilm formation, the periplasmic alginate lyase AlgL regulates the chain length and concentration of alginate in the periplasm during secretion (15). Loss of AlgL results in cell lysis for mucoid strains of P. aeruginosa due to alginate accumulation in the periplasm (16), and AlgL overexpression leads to truncation of alginate to short-chain polysaccharides unable to form extended extracellular aggregates. Addition of AlgL to mucoid P. aeruginosa in vitro has been demonstrated to enhance antimicrobial killing efficiency, and has attracted interest as a possible adjuvant for inhaled antimicrobial therapies (17). Thus, understanding the underlying structural basis for polysaccharide turnover by PLs is important not only in terms of resolving the basis of substrate specificity, but also in targeting microbial PLs that may contribute to pathogenicity.
To understand the role that PLs may play in microbial biofilm formation as well as virulence, we characterized a predicted alginate lyase (Smlt1473) from Stenotrophomonas maltophilia strain k279a. S. maltophilia is an emerging, multidrug-resistant organism often associated with chronic lung infections, especially in cystic fibrosis patients. Of particular note, the prevalence of biofilm-producing S. maltophilia strains isolated from the lungs of cystic fibrosis patients and from contaminated medical equipment has implicated biofilm formation as a mechanism contributing to S. maltophilia infection and multidrug resistance (18). We find that Smlt1473 is secreted when overexpressed in Escherichia coli, and secretion depends on a predicted lipoprotein secretion signal sequence. Strikingly, Smlt1473 is not only active against alginate, but also demonstrates significant activity toward poly-GlcA and HA in a pH-regulated manner; optimal enzymatic activity toward HA is observed at pH 5, poly-GlcA at pH 7, and alginate at pH 9. The oligosaccharide products formed by Smlt1473 cleavage are comprised mainly of disaccharides, with odd numbered products such as trimers and pentamers produced in lower abundance. Collectively our results indicate Smlt1473 is a novel PL with broad, but pH-regulated, substrate specificity, which has implications for both biofilm formation and bacterial pathogenesis.
EXPERIMENT PROCEDURES
Subcloning, Expression, and Purification
Unless otherwise stated, standard molecular biology techniques were used for subcloning and site-directed mutagenesis (19). An E. coli codon-optimized nucleotide sequence of smlt1473 (corresponding to GenBankTM protein accession number CAQ45011) was subcloned into pET28a(+) (Invitrogen) as a BamHI-XhoI insert. Mutagenic primers were designed via PrimerX (bioinformatics.org/primerX) and point mutations were generated via the QuikChange II Site-directed Mutagenesis kit (Agilent Technologies). Nucleotide sequences containing point mutations were confirmed by DNA sequencing (GeneWiz).
For expression, constructs were electroporated into E. coli BL21(DE3) cells and plated on LB agar plates containing 50 μg/ml of kanamycin. An individual colony was cultured in 5 ml of LB medium supplemented with 50 μg/ml of kanamycin for 16 h at 37 °C, 200 rpm. Then 2 ml of saturated culture was added to 200 ml of LB and incubated for 16 h at 18 °C, 200 rpm. The culture was then diluted to an A600 of 0.8 in 800 ml of LB and grown for 1 h at 18 °C, 200 rpm. After 1 h, protein expression was induced by the addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside and the culture was incubated at 18 °C, 200 rpm, for another 16–20 h.
For purification, cells were harvested at 8,000 × g for 15 min at 4 °C, washed once in 20 ml of ice-cold PBS, resuspended in 15 ml of lysis buffer (100 mm HEPES, 500 mm NaCl, 10% w/v glycerol, 10 mm imidazole), and sonicated at 15 watts, 50% duty, for 15 min total processing time. The soluble cell lysate was clarified by centrifugation at 17,000 × g for 20 min at 4 °C and His6-tagged Smlt1473 was purified from the sample by immobilized metal ion affinity chromatography (IMAC). Cell lysate was passed over a column containing 15 ml of Ni2+-bound Chelating Sepharose Fast Flow resin (GE Healthcare) pre-equilibrated in IMAC Buffer A (20 mm HEPES, 500 mm NaCl, 10% (w/v) glycerol, 10 mm imidazole) at a flow rate of 1.8 ml/min via a BioLogic LP chromatography system (Bio-Rad) with a fraction collector. The column was washed for 70 min with IMAC Buffer A to remove any unbound proteins, before applying a gradient from 0 to 100% IMAC Buffer B (20 mm HEPES, 500 mm NaCl, 10% (w/v) glycerol, 500 mm imidazole) over the course of 200 min. Fractions were assayed for protein content via Bradford reagent (Bio-Rad) and samples containing purified Smlt1473 were pooled together and dialyzed against 4 liters of 20 mm sodium phosphate buffer (pH 8) for 40 h at 4 °C with one buffer exchange. To confirm that recombinant Smlt1473 was responsible for the observed lyase activity and not a protein contaminant present in E. coli lysate, a control IMAC purification was run using BL21 cells transformed with the empty parent vector, pET28a(+). The lyase activity of fractions collected at low (∼10 mm), medium (∼85 mm), and high (∼200 mm) imidazole concentrations was tested by adding 2 μl of each fraction to 18 μl of 1 mg/ml of HA, poly-GlcA, or poly-ManA buffered at pH 5, 7, or 9, respectively. Reactions were incubated for 10 min are room temperature before measuring the concentration of 4-deoxy-l-erythro-hex-4-enopyranosyluronic acid via the thiobarbituric acid (TBA) method as described previously (20).
Purity of Smlt1473 was also evaluated by SDS-PAGE, Western blotting, and MALDI-TOF. For MALDI-TOF, 1 μl of protein sample was mixed with 1 μl of sinapinic acid matrix, spotted onto a MSP 96 target ground steel plate (Bruker), and allowed to air dry. Samples were then analyzed via a Microflex mass spectrometer (Bruker Daltonics).
Preparation of Whole Cell Lysate and Subcellular Fractions
Two 1-ml samples of cultures induced for 12 h were harvested by centrifugation at 8,000 × g for 10 min at 4 °C. One pellet was resuspended in 200 μl of 4 m urea and designated the whole cell lysate sample. The second pellet was resuspended in 200 μl of ice-cold sucrose buffer (20 mm HEPES, 1 mm EDTA, 20% (w/v) sucrose), incubated on ice for 15 min, centrifuged, resuspended in 200 μl of ice-cold 5 mm MgSO4, and incubated on ice for 15 min. The resulting supernatant containing the periplasmic proteins was collected by centrifugation at 17,000 × g for 10 min at 4 °C.
SDS-PAGE and Immunoblotting
Protein samples (40 μl) were mixed with 10 μl of 5× Lammeli sample buffer and heated for 5 min at 90 °C before loading 25 μl onto a 4% stacking, 12% separating acrylamide gel with MES running buffer. Precision Plus Protein All Blue Standard (Bio-Rad) was used as a molecular weight standard. Samples were run at 100 V and transferred to a nitrocellulose membrane (Amersham Biosciences Hybond ECL) for 90 min at 17 V at 4 °C. Transfer was confirmed by staining the membrane with 0.1% Ponceau S. The membrane was then blocked overnight at 4 °C with 5% fat-free milk in TBST, washed once with TBST, incubated for 1 h at room temperature with mouse monoclonal anti-His6 tag primary antibody (Cell Signaling) diluted 3000-fold in 5% fat-free milk, washed five times with TBST, incubated for 1 h at room temperature with horseradish peroxidase-conjugated anti-mouse IgG secondary antibody (Cell Signaling) diluted 3000-fold in 5% fat-free milk, washed five times with TBST, and then twice with TBS. The membrane was developed with ECL plus Western blotting detection reagents (RPN2132, GE Healthcare), a chemiluminescent horseradish peroxidase substrate.
Plate Assay to Detect Extracellular Lyase Activity
Induced culture (5 μl) was spotted onto LB plates solidified with 1% agarose and supplemented with 50 μg/ml of kanamycin and 1 mg/ml of poly-ManA or poly-GlcA. Plates were incubated at room temperature for 12 h, gently flooded with 10% cetyl pyridinium chloride, and incubated at room temperature for 30 min. A clearing zone surrounding the area treated with culture was indicative of extracellular lyase activity (21).
Polysaccharide Sample Preparation
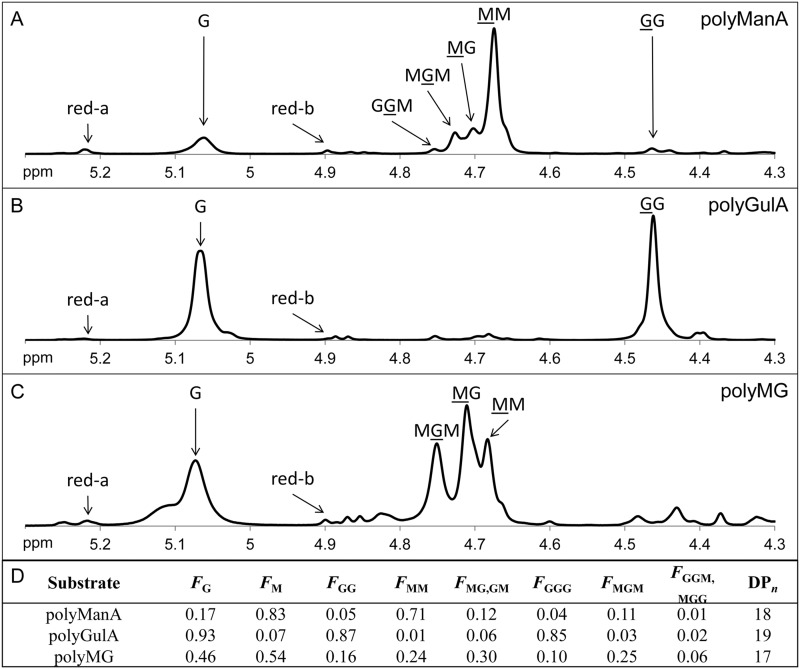
Sodium alginate, medium viscosity, was obtained from MP Biomedicals. Hyaluronic acid potassium salt from human umbilical cord was a kind gift of Dr. Ellen Pure (The Wistar Institute). Poly-GlcA was prepared from Avicel PH-105 NF (FMC Biopolymer) by first converting the microcrystalline cellulose to regenerated amorphous cellulose by dissolution in 86.2% H3PO4 (22) and then converting the regenerated amorphous cellulose to poly-GlcA via 2,2,6,6-tetramethylpipelidine-1-oxyl radical-NaBr-NaClO-mediated oxidation (23). Poly-GlcA samples were precipitated with ethanol, dried, resuspended in ddH2O, and dialyzed against 4 liters of ddH2O for 40 h at 4 °C with one buffer exchange. Total carbohydrate concentration was measured via the phenol-sulfuric acid method (24) and uronic acid concentration was measured via the carbazole method (25). The percent oxidation of the poly-GlcA sample was determined by comparing the total sugar concentration of the sample against the uronic acid concentration. d-Glucuronic acid (Acros Organics) was used to generate standard curves for each assay. 100% oxidation would correspond to an equal concentration of carbohydrates and uronic acids (i.e. all carbohydrate in the sample is oxidized to uronic acid). The poly-GlcA sample prepared from Avicel PH-150 NF was found to be 99.48 ± 4.64% oxidized (data not shown). The composition of the poly-GlcA sample was further confirmed by proton nuclear magnetic resonance (1H NMR) spectroscopy (data not shown). The 1H NMR spectrum matched spectrum obtained from other poly-GlcA samples, with two doublets at 4.42 and 3.75 ppm corresponding to H1 and H5 of the glucuronic ring and three triplets at 3.55, 3.50, and 3.25 ppm corresponding to H4, H3, and H2, respectively (26). Poly-ManA, poly-GulA, and poly-MG block structures were prepared from sodium alginate by partial acid hydrolysis as described by previously (6). Each fraction was resuspended in ddH2O and dialyzed against 4 liters of ddH2O for 40 h at 4 °C with one buffer exchange. The MG ratio of each alginate block was determined via 1H NMR according to the standard test method ASTM F2259 (27) (Fig. 2, A–C). Fig. 2D depicts the molar fractions of monomers (FG and FM), dimers (FGG, FMM, FGM, and FMG), and trimers (FGGG, FMGM, FGGM, and FMGG) of each alginate block structure as well as the average degree of polymerization (DPn). Briefly, poly-ManA contained 83% ManA, poly-GulA contained 7% ManA, and poly-MG contained 54% ManA. Additionally, 71% of the dimers in poly-ManA were ManA-ManA, 87% of the dimers in poly-GulA were GulA-GulA, and 60% of the dimers in poly-MG were ManA-GulA or GulA-ManA (Fig. 2D). All 1H NMR measurements were carried out using a 500 MHz Bruker Avance II spectrometer (Bruker). Polysaccharide samples were lyophilized, resuspended in D2O, lyophilized again, and resuspended in D2O at a concentration of 10 mg/ml. 20 μl of 0.3 m 3-(trimethylsilyl)propionate-2,2,3,3-d4 acid in D2O was added to the NMR sample tube as an internal standard. Data were collected at 80 °C with sample spinning, proton spectral width, number of scans, relaxation delay, proton pulse angle, and acquisition time at 20 Hz, −0.5 → 9.5 ppm, 64, 2 s, 90°, and 4.096 s, respectively.
FIGURE 2.
Composition of alginate blocks. 1H NMR spectrum of poly-ManA (A), poly-GulA (B), and poly-MG (C) at 10 mg/ml in D2O. Signals relevant to the determination of chemical structure are indicated with an arrow and labeled with the type of residue generating the signal. For dimer or trimer sequences the underlined residue contains the hydrogen responsible for the signal. Red a and red b correspond to α and β reducing ends, respectively, and were used to determine the average degree of polymerization. D, the molar fractions of monomers (FG and FM), dimers (FGG, FMM, FMG, and FGM), and trimers (FGGG, FMGM, FGGM, and FMMG) in each substrate were calculated according to the standard test method ASTM F2259 (27). DPn indicates average degree of polymerization.
Enzyme Activity Assays
The β-elimination mechanism of PLs results in the formation of a double bond whose accumulation was monitored by measuring the change in absorbance at 235 nm (28). Absorbance measurements were made in 1-s intervals using an Ultrospec 3300 pro UV-visible spectrophotometer with a detection limit of 0.001 absorbance units at 235 nm/min. One unit of enzyme activity was defined as an increase in absorbance at 235 nm of 1.0/min at 25 °C (1 unit = 1 ΔA235 nm min−1) (29–31). Purified Smlt1473 (17.5 μg) was added to a final volume of 350 μl for reactions containing alginate, poly-ManA, poly-GulA, poly-MG, or HA. Due to the higher activity against poly-GlcA versus all other substrates, 1.75 μg of protein was added to reactions containing poly-GlcA. The pH of all reactions was maintained by using a specific buffer for a given pH (acetate for pH 4–6, phosphate for pH 6–8, Tris for pH 6.5–9, and glycine for pH 9–10) at a total ionic strength of 30 mm. For the determination of optimal pH and buffer conditions, the enzyme was incubated in a given buffer without substrate for 10 min before being added to a solution containing a final polysaccharide concentration of 1 mg/ml in the same buffer. For the determination of Michaelis constant (Km) and maximum velocity (Vmax) for each substrate, the polysaccharide concentration (S) was varied from 0.0625 to 2 mg/ml and initial rates (vi) were fitted to the Michaelis-Menten equation, v = VmaxS/(Km + S), with a generalized reduced gradient (GRG2) nonlinear optimization program (32). For all substrates, R-squared and correlation values were greater than 0.965 and 0.985, respectively. Lyase activity was also confirmed via the TBA method (20).
Circular Dichroism (CD)
Proteins samples (300 μl) at 300 μg/ml in 20 mm sodium phosphate buffer (pH 8) were added to a 1-mm path length quartz cuvette (Starna) and ellipticity was measured from 190 to 260 nm in a J-815 circular dichroism spectrometer (JASCO). The scan speed was 200 nm/min with three accumulations per sample.
Electrospray Ionization Mass Spectrometry (ESI-MS) of Oligosaccharide Products
Reaction mixtures containing 5 mg of digested oligosaccharides were desalted by size exclusion chromatography via a Bio-Gel P-2 column (1.5 cm by 45 cm) equilibrated with ddH2O (Bio-Rad). The flow rate was set to 0.1 ml/min and fractions containing uronic acids were pooled together, lyophilized, and resuspended in 1 ml of 1:1 (v/v) methanol:water. Oligosaccharide samples were delivered to a Thermo LTQ mass spectrometer with HESI-II probe (Thermo) by a syringe pump at a flow rate of 5 μl/min. MS was run in negative mode with a mass scan range of 150 to 2000 Da, capillary temperature of 250 °C, electrospray needle voltage of 3.5 kV, and nitrogen sheath gas at 8 p.s.i.
High-performance Liquid Chromatography (HPLC) of Oligosaccharide Products
HPLC experiments were run on an Agilent 1100 series HPLC value system equipped with a 96-well autosampler and UV-visible detector. Smlt1473 at a final concentration of 50 μg/ml (5 μg/ml for poly-GlcA samples) was added to poly-ManA (3 mg/ml, 30 mm Tris, pH 9), poly-GlcA (3 mg/ml, 30 mm Tris, pH 7), or HA (1 mg/ml, 30 mm acetate, pH 5) at the final concentrations and buffers indicated in parentheses. The reaction mixture (15 μl) was injected after 24 h onto a TSKgel SuperOligoPW column (Tosoh) equipped with corresponding guard column. The mobile phase was 20 mm sodium phosphate buffer (pH 8) plus 250 mm NaCl and the flow rate was 0.275 ml/min. Unsaturated uronic acid products were detected by absorbance at 235 nm. A series of polyethylene glycol standards (molecular weight 200, 600, 1000, 1500, and 2000; Alfa Aesar) were used to generate a calibration curve.
Sequence Alignment
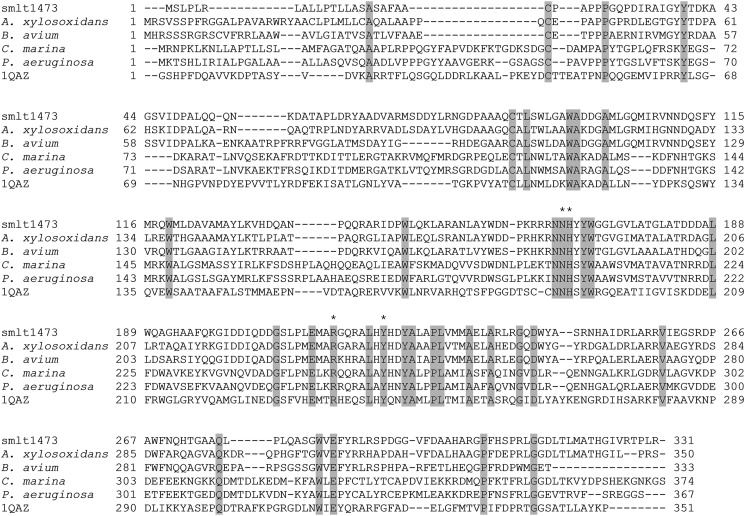
According to the Carbohydrate Active Enzymes (CAZy) database (33), Smlt1473 belongs to polysaccharide lyase family 5 (PL-5). The amino acid sequences of PL-5 lyases from Achromobacter xylosoxidans (Swiss-Prot F7SXN4), Bordetella avium (34), Cobetia marina (Swiss-Prot Q9ZNB7), P. aeruginosa (35), and Spingomonas sp. A1 (PDB 1QAZ) (36) were aligned with Smlt1473 using COBALT (37).
Homology Modeling
The alginate lyase A1-III (Protein Data Bank 1QAZ) from Sphingomonas sp. A1 was chosen as a template for homology modeling based on having the highest sequence identity (30%) among polysaccharide lyases with solved crystal structures (36). Despite the low sequence identity between the two PLs (30%), which can limit the accuracy of homology models, comparative analysis of high-resolution PL crystal structures revealed similar folds in many cases despite having low overall sequence identity. Other groups have also noted alginate lyases share overall low sequence identity, even within specific structural subfamilies, but have been able to use highly conserved regions important for enzymatic activity to generate accurate homology models consistent with biochemical characterization. We used SWISS-MODEL to build a model for Smlt1473 based on Protein Data Bank code 1QAZ (38–40). All images based on the resultant model were generated using PyMOL (41).
RESULTS
Heterologous Expression and One-step Purification of Smlt1473 via IMAC
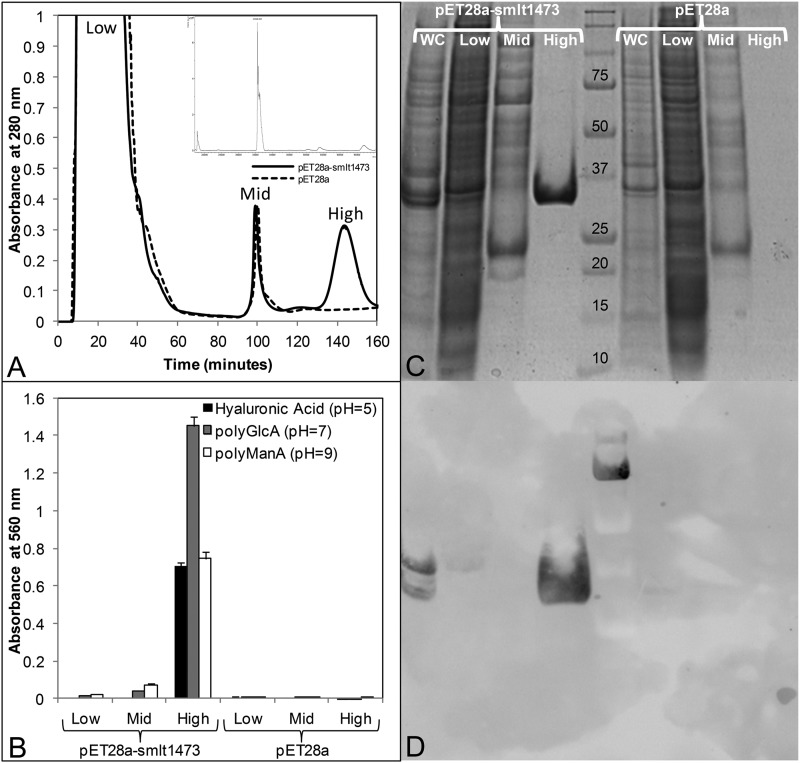
An E. coli codon-optimized nucleotide sequence of smlt1473 was subcloned into pET28a(+) as a BamHI-XhoI insert without a stop codon, resulting in the recombinant lyase containing a C-terminal His6 tag. Expression was carried out at 18 °C, 200 rpm, and soluble cell lysate was prepared via sonication. Smlt1473 was purified by passing the cell lysate over a Ni2+-bound chelating Sepharose column as described under “Experimental Procedures.” At least 50 mg of enzyme was purified per liter of the induced culture. The molecular mass of Smlt1473 without the signaling peptide was determined to be 35586.9 Da by MALDI-TOF, in close agreement with the predicted molecular mass of 35550.9 Da (Fig. 3A, inset). A control purification experiment was performed with lysate prepared from BL21 cells containing the pET28a vector alone. Fig. 3A depicts a chromatogram of each purification experiment. Cell lysate was added at t = 0 min and the column was washed with buffer containing 10 mm imidazole to remove unbound proteins. At t = 70 min an imidazole gradient was initiated, which increased the imidazole concentration by 2.5 mm per minute. At t = 100 min (∼85 mm imidazole) nonspecifically bound proteins eluted from the column. At t = 145 min (∼200 mm imidazole) recombinant Smlt1473 eluted from the column. Fractions collected at low (10 mm), mid (85 mm), and high (200 mm) imidazole concentrations were analyzed for lyase activity by mixing 2 μl of each fraction with 18 μl of 1 mg/ml of HA, poly-GlcA, or poly-ManA in 30 mm buffer at pH 5, 7, or 9, respectively. After 10 min the samples were analyzed for lyase activity by the TBA method (20) (Fig. 3B). Additionally each fraction was analyzed via SDS-PAGE (Fig. 3C) and immunoblotting (Fig. 3D). The high imidazole pET28a-smlt1473 fraction exhibited lyase activity against HA, poly-GlcA, and poly-ManA (Fig. 3B) and contained a single protein with a molecular mass of ∼35.5 kDa (Fig. 3C), which appeared on an anti-His6 tag Western blot (Fig. 3D). Conversely, the high imidazole pET28a fraction exhibited no lyase activity (Fig. 3B), contained no proteins visible by SDS-PAGE (Fig. 3C) nor anti-His6 tag Western blot (Fig. 3D). Therefore, recombinant Smlt1473 was purified in a one-step fashion via IMAC and was responsible for the lyase activity against each polysaccharide substrate.
FIGURE 3.
Heterologous expression and one-step purification of Smlt1473 by immobilized metal ion affinity chromatography. A, chromatogram of IMAC purification. Cell lysates prepared from E. coli BL21 cells transformed with pET28a-smlt1473 or pET28a vector and induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside at 18 °C, 200 rpm, were added to Ni2+-bound chelating Sepharose column at t = 0 min and a flow rate of 1.8 ml/min. At t = 70 min a linear imidazole gradient was applied, increasing the imidazole concentration 2.5 mm per minute. Low (∼10 mm), mid (∼85 mm), and high (∼200 mm) imidazole fractions were collected for both pET28a-smlt1473 and pET28a samples. B, fractions were analyzed for lyase activity by mixing 2 μl of sample with 18 μl of 1 mg/ml of HA, poly-GlcA, or poly-ManA in 30 mm buffer at pH 5, 7, or 9, respectively. Oligosaccharide products formed via lyase enzymatic activity were detected by the TBA method (20). All reactions were performed in triplicate and error is reported as S.D. C, SDS-PAGE analysis of each fraction. WC corresponds to whole cell lysate prior to IMAC purification. D, anti-His6 tag Western blot of each fraction.
Heterologous Secretion of Smlt1473 Is Dependent on a Predicted Lipoprotein Signal
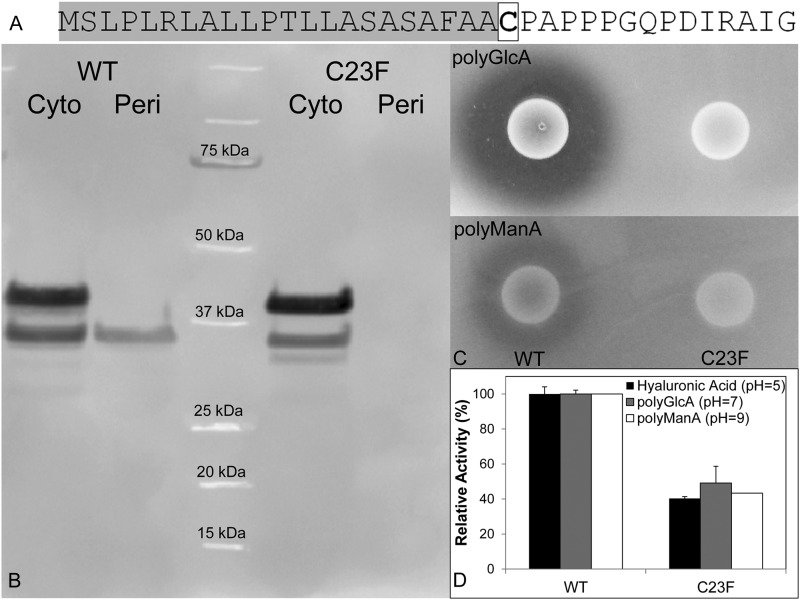
The primary sequence for Smlt1473 is predicted to contain a N-terminal signal peptide (SignalP) that includes an Ala-Ala-Cys motif at positions 21–23, which is consistent with a consensus sequence for bacterial lipoprotein secretion (42). Secretion of lipoproteins requires the addition of a N-acyl-S-diacylglyceryl moiety to the cysteine residue in the +1 position (boxed in Fig. 4A) (43). To determine whether Smlt1473 is a secreted lipoprotein when expressed heterologously in E. coli, we compared the subcellular distribution and lyase activity from BL21(DE3) cells expressing wild-type (WT) Smlt1473 and a mutant (C23F) where the predicted lipid-modified cysteine had been replaced with phenylalanine. Cultures expressing both WT and C23F were separated into whole cell lysate and periplasmic fractions, and immunoblotting revealed that although WT could traffic to the periplasmic space, C23F was completely retained in the cytosol (Fig. 4B). Comparing purified Smlt1473 from the periplasmic fractions and whole cell lysates, both proteins migrate at their predicted molecular masses (35.5 kDa without signaling peptide, 41.2 kDa with signaling peptide, Fig. 4A); this indicates that recognition and processing of Smlt1473 is dependent on the predicted lipoprotein secretion signal. Additionally, cell suspensions expressing WT and C23F were spotted onto LB-agarose plates containing 1 mg/ml of poly-GlcA or poly-ManA and extracellular lyase activity was visualized by staining with 10% cetylpyridinium chloride. As indicated in Fig. 4C, a clearing zone is present surrounding BL21(DE3) cells expressing WT, but not for C23F. Additionally, the C23F mutant was purified via IMAC and its activity was compared with WT Smlt1473. The C23F mutant retained between 40 and 50% of WT activity (Fig. 4D), suggesting that signaling and secretion may play a role in activating Smlt1473. Thus, Smlt1473 is a secreted lipoprotein with extracellular activity when overexpressed in E. coli, and proper processing and secretion is dependent upon a predicted N-terminal lipoprotein secretion signal.
FIGURE 4.
Trafficking of WT Smlt1473 and a C23F mutant. A, sequence of N-terminal signaling peptide of Smlt1473 highlighted in gray and the lipid-modified cysteine 23 residue boxed. A C23F mutant was prepared via site-directed mutagenesis. B, anti-His6 tag Western blot of E. coli BL21 cultures expressing wild-type Smlt1473 and C23F mutant separated into whole cell lysate and periplasmic fractions. C, E. coli BL21 cultures expressing wild-type Smlt1473 and C23F mutant dotted onto LB plates solidified with 1% agarose, and supplemented with 50 μg/ml of kanamycin and 1 mg/ml of poly-GlcA (top) or poly-ManA (bottom). After a 12-h incubation at 25 °C, the plates were treated with 10% cetylpyridinium chloride. Clearing zone indicates degradation of polysaccharide in surrounding media. D, activity of purified Smlt1473 WT and C23F. Enzyme activity was monitored by absorbance at 235 nm. All reactions were performed in triplicate and error is reported as S.D.
Substrate Specificity Analysis Revealed a pH-sensitive Mechanism
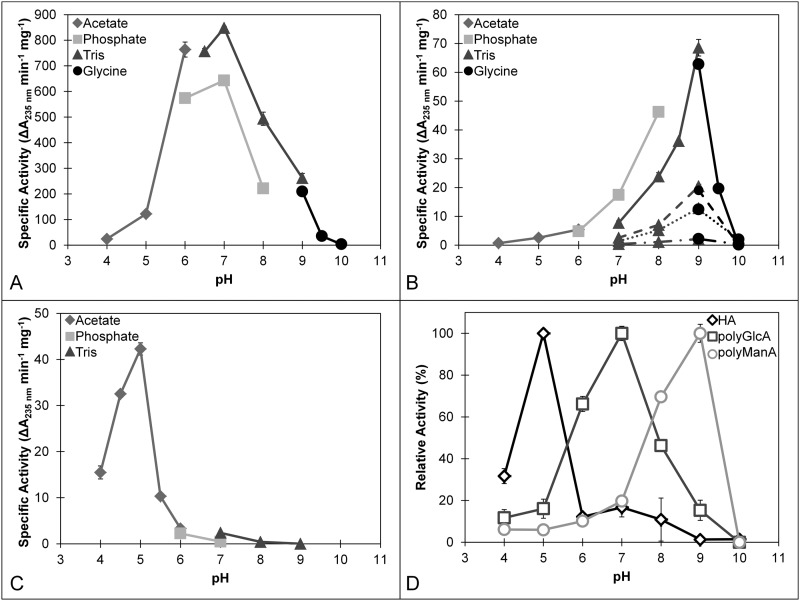
The enzymatic activity of purified Smlt1473 was tested against the following nine polyuronides: alginate (MP Biomedicals), poly-ManA, poly-GulA, poly-MG, poly-GlcA, poly-α-d-galacturonic acid (poly-GalA; Alfa Aesar), HA (MP Biomedicals), heparin (Sigma), and heparin sulfate (Sagent Pharmaceuticals). Of these substrates, significant activity in terms of increased absorbance at 235 nm was measured for poly-ManA, poly-GlcA, and HA. This is illustrated in Table 1, in which the initial rates for each substrate (1 mg/ml) are given at their respective optimal pH values. Of particular note, specific activities for each of the three most active substrates (HA, poly-GlcA, and poly-ManA) were found to be strongly dependent on pH, with optimal activity for HA at pH 5, poly-GlcA at pH 7, and poly-ManA at pH 9 (Fig. 5). The highest overall specific activity (848.3 ± 6.3 units/mg at pH 7) was for poly-GlcA (Fig. 5A), which is among the highest reported for poly-GlcA-specific lyases (30, 44, 45). For poly-ManA the specific activity (68.5 ± 2.9 units/mg at pH 9) was also significant (Fig. 5B), being within an order of magnitude to the reported values for poly-ManA-specific lyases (31, 46, 47). Likewise, whereas the specific activity for HA (42.3 ± 1.3 units/mg at pH 5) was the lowest for the three major substrates (Fig. 5C), it is within an order of magnitude of the reported values for bacterial hyaluronidases (48). Optimal pH values for each substrate were independently confirmed by the TBA method (Fig. 5D). Kinetic parameters for each of the three substrates were also determined according to the Michaelis-Menten equation (Table 2), with Vmax values similar in magnitude and trend to the specific activities for each of the three main substrates (poly-ManA, poly-GlcA, and HA); Vmax for poly-GlcA is ∼10-fold greater versus poly-ManA or HA. The Km values for the three substrates also follow a similar trend to Vmax and specific activity, with a 5-fold larger Km for HA versus poly-GlcA (30, 47). Thus, based on the highest specific activity for poly-GlcA (Fig. 5A), one could tentatively conclude that Smlt1473 is a poly-GlcA-specific lyase. However, the multiple pH optima exhibited by Smlt1473 for maximal HA, poly-GlcA, and poly-ManA cleavage (Fig. 5), as well as comparable kinetic parameters and specific activities to published values for lyases specific to each of these substrates (Table 2), indicate that Smlt1473 is a multifunctional PL with potent activity against poly-ManA, poly-GlcA, and HA.
TABLE 1.
Substrate specificity of Smlt1473
17.5 μg of Smlt1473 was added to 1 mg/ml of each substrate in 30 mm buffer at various pH. Enzymatic activity was monitored by absorbance at 235 nm. One unit of activity (U) was defined as an increase in absorbance at 235 nm of 1.0 per minute at 25 °C.
| Substrate | Source | pH | Specific activity |
|---|---|---|---|
| Units/mg | |||
| Alginate | MP Biomedicals | 9 | 20.4 ± 0.7 |
| Poly-ManA | Prepared from alginate | 9 | 68.5 ± 2.9 |
| Poly-GulA | Prepared from alginate | 9 | 2.1 ± 0.2 |
| Poly-MG | Prepared from alginate | 9 | 12.8 ± 0.4 |
| Poly-GlcA | Prepared from Avicel PH-105 NF | 7 | 848.3 ± 6.3 |
| Poly-GalA | Alfa Aesar | 5,7,9 | NDa |
| Hyaluronan | MP Biomedicals | 5 | 42.3 ± 1.3 |
| Heparin | Sigma | 5,7,9 | ND |
| Heparin sulfate | Sagent Pharmaceuticals | 5,7,9 | ND |
a No detectable activity (detection limit: 0.001 absorbance units at 235 nm per minute).
FIGURE 5.
Optimal pH of Smlt1473 for poly-GlcA (A), alginate-based substrates (B), and HA (C). Enzyme activity was monitored by absorbance at 235 nm (A–C) and TBA method (D). Various buffer systems were utilized: acetate (pH 4 to 6), phosphate (pH 6 to 8), Tris (pH 6.5 to 9), and glycine (pH 9 to 10). B, various alginate block types were tested. Solid line, poly-ManA; dashed line, alginate; dotted line, poly-MG; dash dot, poly-GulA. D, TBA confirmation of optimal pH for HA, poly-GlcA, and poly-ManA. 100% activity was taken as the activity at the optimal pH for a given substrate. All reactions were performed in triplicate and error is reported as S.D.
TABLE 2.
Kinetic parameters of Smlt1473
| Substrate | Buffer | Km | Vmax |
|---|---|---|---|
| mg ml−1 | ΔA235 nm min−1 mg−1 | ||
| Poly-GlcA | 30 mm Tris, pH 7 | 0.14 | 940.0 |
| Poly-ManA | 30 mm Tris, pH 9 | 0.26 | 91.6 |
| Hyaluronan | 30 mm Acetate, pH 5 | 0.55 | 71.5 |
Mono- and Divalent Cations as Well as l-Ascorbic Acid 6-Hexadecanoate Inhibit Lyase Activity
Several polysaccharide lyases are reported to have enhanced activity in response to addition of divalent cations such as calcium or magnesium as well as at elevated NaCl concentrations (31, 44, 49). In the case of Smlt1473, however, activity was significantly inhibited by the presence of both mono- and divalent cations (Table 3). At 50 mm NaCl, Smlt1473 is essentially inactive against HA activity, and retains ∼30% of its optimal activity against poly-ManA and poly-GlcA; at 200 mm NaCl, activity is reduced to below 10% optimal activity. For CaCl2 the inhibitory effects are even more pronounced; 2 mm CaCl2 reduces HA activity by nearly half, and reduces poly-ManA as well as poly-GlcA activity to nearly 30% of optimal activity. The monosaccharide derivative l-ascorbic acid 6-hexadecanoate (Vcpal), which has been shown previously to act as a specific inhibitor of bacterial hyaluronan lyases (50), also acts as a specific inhibitor for Smlt1473, with HA activity reduced by about 50% and poly-GlcA as well as poly-ManA activity reduced to 30% of optimal activity at a concentration of 100 μm Vcpal (Table 3).
TABLE 3.
Effect of various chemicals on Smlt1743 lyase activity
Enzyme was incubated in 30 mm buffer containing NaCl, CaCl2, or Vcpal at the indicated concentrations for 10 min before adding 17.5 μg (for poly-ManA and HA) or 1.75 μg (for poly-GlcA) to 1 mg/ml substrate in 30 mm buffer with the equivalent concentration of chemical. Enzyme activity was monitored by change in absorbance at 235 nm. Activity in the absence of any chemicals was taken to be 100%.
| Chemical | Concentration | Relative activity |
||
|---|---|---|---|---|
| Poly-GlcA (pH 7) | Poly-ManA (pH 9) | HA (pH 5) | ||
| mm | % | |||
| None | 100 ± 0.7 | 100 ± 4.2 | 100 ± 3.2 | |
| NaCl | 10 | 79.9 ± 5.6 | 104.0 ± 2.0 | 75.6 ± 0.2 |
| 50 | 29.6 ± 1.4 | 26.5 ± 1.0 | 0.5 ± 0.2 | |
| 200 | 9.0 ± 0.5 | 3.2 ± 0.3 | NDa | |
| CaCl2 | 1 | 68.5 ± 0.6 | 70.7 ± 8.8 | 73.5 ± 3.0 |
| 2 | 32.9 ± 0.8 | 27.1 ± 4.9 | 58.9 ± 2.3 | |
| Vcpal | 0.05 | 59.1 ± 3.7 | 64.5 ± 1.5 | 72.7 ± 5.0 |
| 0.1 | 29.9 ± 2.1 | 31.4 ± 1.1 | 45.6 ± 3.1 | |
a No detectable activity (detection limit: 0.001 absorbance units at 235 nm per minute).
Identification of Conserved, Catalytic Residues Determining Lyase Activity
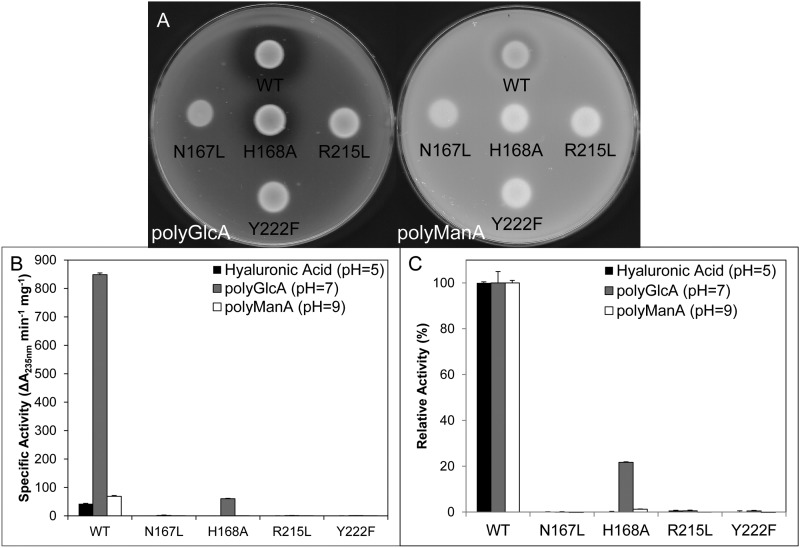
As stated previously, the β-elimination mechanism of PLs requires a neutralizing group, proton acceptor, and protein donor (8). Despite diverse substrate specificity, secondary structure content, and tertiary folds, the residues responsible for each of the aforementioned roles appears to be highly conserved across many polysaccharide lyase families and fall into two general categories: (i) lyases that require a divalent cation (usually Ca2+) for activity due to its role as the neutralizing group, with lysine or arginine acting as the base and water as the acid, and (ii) lyases that do not require a divalent cation and instead neutralize the C5 carboxylate group via interactions with nearby arginine, asparagine, or glutamine residues, with tyrosine or histidine acting as the base and tyrosine as the acid (51). For example, the crystal structure of Streptococcus pneumoniae hyaluronate lyase in complex with substrate revealed that the amide group of Asn329 acts as the neutralizing group by interacting directly with the C5 carboxylate in a bidentate fashion, with Tyr408/His399 acting as the acid/base pair (12). Similarly, the crystal structure of Sphingomonas sp. A1-III alginate lyase in complex with substrate revealed that in addition to Asn191, the guanidinium group of Arg239 interacts with the C5 carboxylate, with Tyr246 acting as both acid and base (47). Due to the fact that Smlt1473 activity is strongly inhibited by Ca2+ (Table 3), we focused on the Arg/Asn-Tyr/His neutralization-acid/base mechanism. As stated previously, Smlt1473 belongs to PL-5 CAZy family. A multiple sequence alignment of Smlt1473 with four other PL-5 lyases, including A1-III alginate lyase from Sphingomonas sp. (47), was performed with COBALT (Fig. 6) (37). Based on this alignment, four conserved, putative active-site residues in Smlt1473 were identified (Asn167, His168, Arg215, and Tyr222) and predicted inactivating mutants (N167L, H168A, R215L, and Y222F) were expressed, purified, and characterized. Enzymatic activity of each mutant was observed directly from a plate assay with poly-GlcA and poly-ManA as the substrates (Fig. 7A), and quantified in terms of the change in absorbance at 235 nm (Fig. 7B) and the TBA method (Fig. 7C). All four mutations (N167L, H168A, R215L, and Y222F) were found to significantly diminish activity (less than 0.5% of WT) against all three substrates (poly-GlcA, poly-ManA, and HA) with the exception of H168A, which retained low activity (10% of WT) against poly-GlcA at pH 7. Interestingly, whereas the H168A mutant retained ∼10% of WT activity against poly-GlcA, its specific activity (59.9 ± 1.8 units/mg at pH 7) was comparable with optimal, WT Smlt1473 activity against poly-ManA (68.5 ± 2.9 units/mg at pH 9) and HA (42.3 ± 1.3 units/mg at pH 5) (Table 1 and Fig. 5). To confirm that the mutant lyases retained secondary structure equivalent to wild-type, CD spectra were collected for wild-type and each mutant (N167L, H168A, R215L, and Y222F). For WT and mutant Smlt1473, a predominantly α-helical secondary structure was observed in terms of specific minima at 208 and 222 nm. The calculated percent helicity for WT was 48.3% (52), with less than 5% change in percent helicity for each mutant, indicating the overall secondary structure was not perturbed significantly by the mutations (data not shown). Therefore, the catalytic mechanism of Smlt1473 appears to be dependent on four highly conserved residues (Asn167, His168, Arg215, and Tyr222) that are consistent with an Asn/Arg-Tyr/His neutralization-acid/base mechanism.
FIGURE 6.
Sequence alignment of Smlt1473 with polysaccharide lyases belonging to PL-5. Identical residues are highlighted in gray. Residues predicted to participate in the catalytic mechanism of Smlt1473 are marked by asterisks.
FIGURE 7.
Mutagenesis of predicted catalytic residues. A, E. coli BL21 cultures expressing Smlt1473 WT, N167L, H168A, R215L, and Y222F were dotted onto LB plates solidified with 1% agarose and supplemented with 50 μg/ml of kanamycin and 1 mg/ml of poly-GlcA (left) or poly-ManA (right). A clearing zone is indicative of lyase activity. B, specific activity of Smlt1473 WT and catalytic mutants against HA, poly-GlcA, and poly-ManA at their respective optimal pH values. Enzyme activity was monitored by absorbance at 235 nm. C, TBA confirmation of activity of Smlt1473 WT and putative catalytic mutants against HA, poly-GlcA, and poly-ManA at their respective optimal pH values. 100% activity was taken as the activity of WT against each substrate. All reactions were performed in triplicate and error is reported as S.D.
Analysis of Oligosaccharides Products formed by Smlt1473
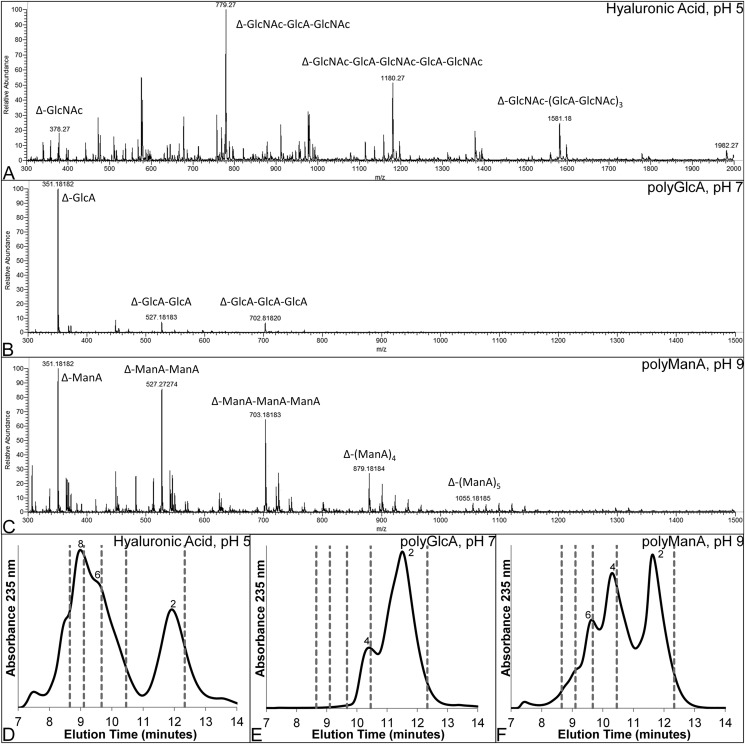
To gain further insight into the mechanism by which unsaturated products are generated by Smlt1473, enzymatic cleavage products of HA, poly-GlcA, and poly-ManA were monitored by ESI-MS and HPLC. In general, for all three substrates, the products observed by ESI-MS after 24 h are low molecular weight polysaccharides spanning from dimers to octamers (Fig. 8, A–C). For poly-GlcA, which is the substrate with the highest Vmax (Table 2), the products are dimer, trimer, and tetramer, whereas for poly-ManA and HA, products include dimers through octamers. Using HPLC to determine the relative abundance of each of these products for a given substrate, the major product formed from poly-GlcA degradation after 24 h was dimer (Fig. 8D), whereas poly-ManA and HA yielded dimers, tetramers, hexamers, and octamers as the major products (Fig. 8, E and F). The presence of multiple higher order oligmers implies Smlt1473 cleaves its substrates in an endolytic manner with the highest conversion to dimer, the terminal product, observed for the substrate with the highest specific activity (poly-GlcA). The predominance of dimers and tetramers as end products, with small amounts of trimer and pentamer, is consistent with cleavage patterns observed for other polysaccharide lyases that process substrates in an endolytic manner (44, 53).
FIGURE 8.
Analysis of oligosaccharide products formed by Smlt1473 digestion. A–C, ESI-MS of oligosaccharide products formed by Smlt1473 enzymatic cleavage of HA at pH 5 (A), poly-GlcA at pH 7 (B), and poly-ManA at pH 9 (C). Each relevant peak is labeled with a mass and oligosaccharide sequence. “Δ” designates 4-deoxy-l-erythro-hex-4-enopyranosyluronic acid, the unsaturated residue at the non-reducing end. D–F, HPLC chromatogram of products formed after 24 h for HA (D), poly-GlcA (E), and poly-ManA (F). Numbers near peaks indicate the degree of polymerization for each major product. Gray dashed lines indicate the elution time of polyethylene glycol standards (2000, 1500, 1000, 600, and 200 daltons from left to right).
DISCUSSION
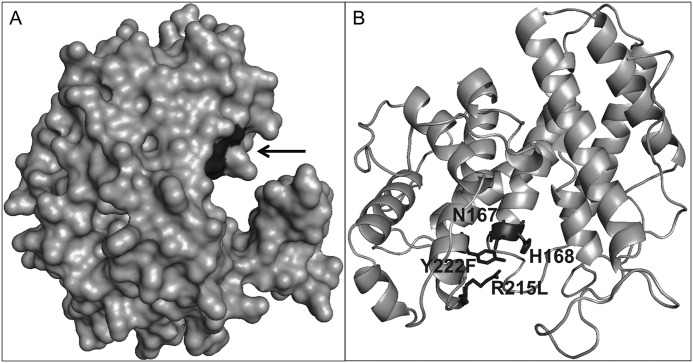
The catalytically important residues for Smlt1473 were identified through site-directed mutagenesis as Asn167, His168, Arg215, and Tyr222, with inactivating mutations for each of these residues resulting in nearly complete loss of Smlt1473 activity toward each of the three most active substrates (poly-GlcA, poly-ManA, HA) (Table 1 and Fig. 5). Thus, our results are most consistent with a mechanism in which (i) the C5 carboxylate group is neutralized by arginine and asparagine and (ii) tyrosine or histidine acts as the base and tyrosine as the acid. This is illustrated further in the homology model generated for Smlt1473 (Fig. 9) based on A1-III alginate lyase from Sphingomonas sp. A1-III alginate lyase (PDB 1QAZ) (36). In the model, Asn167 and Arg215 cluster in a deep cleft along with putative acid/base catalytic residues His168 and Tyr222, with their overall proximity within the putative active site consistent with their ability to neutralize the C5 carboxylate of the substrate. Inactivating mutants (N167L, R215L) also diminished activity to below 0.5% of WT independent of substrate type (Fig. 7). Corresponding asparagine and arginine residues have been shown to be important in the catalytic mechanism of alginate lyases (47), hyaluronate lyases (12), xanthan lyases (10), and chondroitin AC lyases (54). As stated previously, crystal structures of S. pneumoniae hyaluronate lyase and Sphingomonas sp. A1-III alginate lyase in complex with substrates have shown that the corresponding arginine and asparagine residues are within bonding distance (∼3Å) of the C5 carboxylate of the +1 sugar ring (9, 12, 47). Thus, our results point to Asn167 and Arg215 likely acting to neutralize the negative charge of the C5 carboxylate, thereby reducing the pKa of C5 hydrogen on the +1 sugar and enabling its abstraction.
FIGURE 9.
Homology model of Smlt1473. Homology model built using SWISS-MODEL and Protein Data Bank file 1QAZ as a template. Images were generated in PyMOL. A, surface model of Smlt1473. Black arrow indicates location of the putative catalytic residues in evident enzymatic cleft. B, tentative catalytic residues Asn167, His168, Arg215, and Tyr222 were found to be located in close proximity to each other.
In terms of the key residues responsible for general acid/base catalysis for Smlt1473, the unique pH optima for each of the key substrates suggests multiple, substrate-dependent mechanisms may occur (Table 1 and Fig. 5). Evidence for two general mechanisms exist (9–12, 55, 56), in which (i) tyrosine acts as both the base and acid and (ii) histidine acts as the base and tyrosine acts as the acid. Tyrosine is proposed to act as both proton acceptor (base) and donor (acid) in A1-II (56) and A1-III (9) alginate lyase from Sphingomonas sp. A1, as well as xanthan lyase from Bacillis sp. (10) and chondroitin AC lyase from Arthrobacter aurescens (57), based on crystal structures in complex with the corresponding substrate and the distance of the conserved catalytic histidine from the C5 hydrogen of the +1 sugar. The authors also propose that the histidine, rather than acting as a proton donor or acceptor, instead plays a role in either neutralizing the negative charge of the C5 carboxylate, stabilizing the enolate intermediate formed between proton abstraction and donation by tyrosine, or assisting in the deprotonation of the hydroxyl group of the catalytic tyrosine, enabling the tyrosine to act as the base. In contrast, histidine is proposed to act as the proton acceptor and tyrosine as the proton donor in the alginate lyase Atu3025 from Agrobacterium tumefaciens (55), as well as the HA lyases of Streptococcus sp. (11, 12), again based on their positioning relative to the C5 hydrogen on the +1 sugar. Given the optimal activity against HA at acidic pH 5 versus poly-GlcA at pH 7 and poly-ManA at pH 9 (Table 1 and Fig. 5), our results are most consistent with a model in which His168 plays the role of proton acceptor at acidic pH for optimal HA activity, similar to the bacterial HA lyases of Streptococcus sp., which have optimal activity at acidic pH (11, 12), and His168 or Tyr222 plays the role of the proton acceptor at neutral or basic pH for optimal poly-GlcA and poly-ManA activity, similar to the alginate lyases of Sphingomonas sp., which have optimal activity at neutral or basic pH (9, 56).
To provide further insight into the possible mechanisms by which multiple, divergent substrates could be processed by Smlt1473 (Fig. 5), we estimated pKa values for each of the active site residues using PROPKA 3.1 (58–61). Based on the homology model of Smlt1473 (Fig. 9), the predicted pKa of His168 was shifted to 2.6, consistent with the measured HA activity at acidic pH 5, whereas Tyr222 was predicted to have a pKa of 9.8, thus unlikely to act as a proton acceptor at acidic pH. At neutral or basic pH, however, the roles of His168 and Tyr222 become less clear, with the possibility of His168 acting as the proton acceptor and Tyr222 acting as the donor, or Tyr222 acting as both. It is interesting to note that the H168A mutant retains 10% activity against poly-GlcA at pH 7 (Fig. 7), implying that it is not absolutely necessary for catalysis. Although poly-GlcA activity was significantly diminished by the H168A mutation, the specific rate of H168A against poly-GlcA was comparable with the rate of WT Smlt1473 against poly-ManA and HA at their respective optimal pH values. Given that Smlt1473 activity toward poly-ManA and HA is comparable with other lyases specific to these substrates (47, 48), the residual activity of H168A is significant and suggests the possibility of Tyr222 acting as both proton acceptor and donor. Based on the homology model for Smlt1473 (Fig. 9) and PROPKA calculations, no other residues present within the predicted active site are likely to act as a replacement to His168 as putative proton donors or acceptors, consistent with the idea that Tyr222 acts as proton donor and acceptor at pH 7 for poly-GlcA. Thus, our overall results are most consistent with a model in which His168 acts as a specific proton acceptor at acidic pH to selectively cleave HA, based on predicted pKa values of His168 and Tyr222, whereas at neutral (poly-GlcA) or basic (poly-ManA) pH Tyr222 acts as both proton acceptor and donor.
It is also interesting to consider the pH-regulated substrate cleavage for Smlt1473 in the context mechanisms proposed for other polysaccharide eliminases. In particular, Shaya et al. (62) proposed a mechanism for heparinase II in which His202 acted as proton acceptor in the cleavage of heparin and Tyr257 acted as proton acceptor in the cleavage of heparan sulfate. Heparin contains mostly GlcNAc and GlcA, whereas heparin sulfate contains N-sulfoglucosamine (GlcNS) and iduronic acid, the C5 epimer of GlcA. The heparin activity of His202 was attributed to the fact that the H5 proton of GlcA (found in heparin) points in the opposite direction of Tyr257, therefore preventing Tyr257 from acting as a proton acceptor in GlcA containing substrates. Conversely the H5 proton of iduronic acid (found in heparan sulfate) points toward Tyr257, allowing the tyrosine to act as both proton acceptor and donor (62). In the case of Smlt1473, all three of the most active substrates contain GlcA (poly-GlcA and HA) or its C2 epimer ManA (poly-ManA), however, only HA contains GlcNAc. GlcNAc contains a C2 acetylamino group and lacks the negatively charged C6 carboxyl indicative of uronic acids. Additionally, only HA contains both (1,4) and (1,3) O-glycosidic bonds, resulting in a distinct macromolecular conformation. The aromatic and polar residues surrounding the active site of PLs have been shown to interact with the −2, −1, +1, and +2 sugar residues of the substrate via hydrogen bonding and π-stacking interactions (4). Therefore the structural and chemical differences between HA and poly-GlcA/poly-ManA could result in a unique binding site for HA in the active site cleft of Smlt1473 and thus a unique positioning of His168 and Tyr222 with respect to H5 proton of the +1 GlcA residue in HA. Further structural and biochemical studies are currently underway to resolve which catalytic mechanism occurs at acidic pH for HA versus neutral and basic pH for poly-GlcA and poly-ManA, as well as identify potential substrate binding residues that assist in the docking and positioning of all three substrates.
PLs can act on polysaccharides endolytically, exolytically, or a combination of both. Endolytic enzymes bind to an internal site on the polysaccharide, cleave one glycosidic bond, and detach from the substrate, resulting in the simultaneous accumulation of multiple unsaturated products of various sizes (44, 45, 63, 64). Exolytic enzymes bind to the non-reducing end of the polysaccharide and removes mono-, di-, or trisaccharides from the end until the entire macromolecule has been processed, resulting in the accumulation of predominantly one unsaturated product, with little to no accumulation of differently sized oligosaccharides (46, 64–66). The HA lyases of Streptococcus sp. exhibit a hybrid mechanism, in which HA is digested via an initial random endolytic cleavage and subsequent exolytic processing into disaccharides (12). Analysis of the unsaturated polysaccharide products formed by Smlt1473 cleavage revealed the enzyme digested the substrates into oligomers of various sizes. The major products consisted of multiples of a disaccharide repeat, with residual trimers and pentamers present as well (Fig. 8). The reduction in higher order oligomers also correlates with the specific activity for a given substrate (Table 1 and Fig. 5); the predominant end product is a dimer for poly-GlcA, whereas a broad mixture of monomer, dimer, and higher-order oligomers are observed for poly-ManA and HA. The rapid production of multiple higher order unsaturated products suggests an endolytic digestion of each substrate (Fig. 8), similar to the endolytic alginate lyases A1-I, A1-II, and A1-III of Sphingomonas sp. (63), as well as the endolytic poly-GulA-specific lyases of Sinorhizobium meliloti M5N1CS (45) and Trichoderma reesei (44). Each of the aforementioned endolytic lyases produce products that vary in size by one sugar ring, including dimers, trimers, tetramers, and pentamers, which is indicative of a random endolytic mechanism in which the enzyme exhibits no bias for the initial size of the substrate, assuming it is larger than the minimum substrate size (64, 67). The ability of Smlt1473 to process substrates with divergent chemical structures such as HA and poly-ManA is also unique among bacterial lyases; most reported bacterial HA lyases show activity toward related substrates such as chondroitin (3), but to our knowledge no reported bacterial lyase is capable of processing both non-acetylglucosamine containing polysaccharides such as poly-ManA as well as acetylglucosamine containing heteropolysaccharides such as HA.
Overall, it is interesting to consider the possible biological roles Smlt1473 may play in S. maltophilia physiology as well as possible uses in polysaccharide processing. Our results demonstrate that Smlt1473 is secreted when overexpressed heterologously in E. coli, and secretion depends on a predicted lipoprotein signaling sequence (Fig. 4). As mentioned before, secreted bacterial hyaluronan lyases have been shown to act as spreading factors to enhance invasion through degrading the host extracellular matrix as well as act as metabolic enzymes capable of generating sugars from host HA to promote bacterial growth during deep tissue infections (14). Alternatively, one recent study of the biofilm composition of two mucoid S. maltophilia strains isolated from cystic fibrosis patients identified a unique structure comprised of d-lactate-substituted, O-acetylated GalA and GlcA blocks (68). Given high Smlt1473 activity against both HA and poly-GlcA (Table 1 and Fig. 5), it is possible that Smlt1473 participates in biofilm formation similar to AlgL in P. aeruginosa, acts as a spreading factor similar to other bacterial hyaluronidases, or potentially both (13, 15).
Acknowledgments
We thank Dr. Robert Ryan (University College Cork) for providing S. maltophilia strain k279a, and Dr. Emily Wong (Lehigh Valley Hospital) for helpful discussions and advice.
This work was supported by Lehigh University, Lehigh Valley Hospital, and a Faculty Innovation Grant (FIG).
- HA
- hyaluronic acid
- GlcA
- d-glucuronic acid
- GlcNAc
- N-acetyl-d-glucosamine
- ManA
- d-mannuronic acid
- GulA
- l-guluronic acid
- PL
- polysaccharide lyase
- IMAC
- immobilized metal ion affinity chromatography
- TBA
- thiobarbituric acid
- DPn
- degree of polymerization
- ESI-MS
- electrospray ionization mass spectrometry
- PL-5
- CAZy polysaccharide lyase family 5
- GalA
- d-galacturonic acid
- Vcpal
- l-ascorbic acid 6-hexadecanoate.
REFERENCES
- 1. Laurent T. C., Fraser J. R. (1992) Hyaluronan. FASEB J. 6, 2397–2404 [PubMed] [Google Scholar]
- 2. Girish K. S., Kemparaju K. (2007) The magic glue hyaluronan and its eraser hyaluronidase. A biological overview. Life Sci. 80, 1921–1943 [DOI] [PubMed] [Google Scholar]
- 3. Stern R., Jedrzejas M. J. (2006) Hyaluronidases. Their genomics, structures, and mechanisms of action. Chem. Rev. 106, 818–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wong T. Y., Preston L. A., Schiller N. L. (2000) ALGINATE LYASE. Review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu. Rev. Microbiol. 54, 289–340 [DOI] [PubMed] [Google Scholar]
- 5. Ray B. (2006) Polysaccharides from Enteromorpha compressa. Isolation, purification and structural features. Carbohydr. Polym. 66, 408–416 [Google Scholar]
- 6. Haug A., Larsen B., Smidsrod O. (1967) Studies on the sequence of uronic acid residues in alginic acid. Acta Chem. 21, 691–704 [Google Scholar]
- 7. Hentzer M., Teitzel G. M., Balzer G. J., Heydorn A., Molin S., Givskov M., Parsek M. R. (2001) Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183, 5395–5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gacesa P. (1987) Alginate-modifying enzymes. A proposed unified mechanism of action for the lyases and epimerases. FEBS Lett. 212, 199–202 [Google Scholar]
- 9. Yoon H. J., Hashimoto W., Miyake O., Murata K., Mikami B. (2001) Crystal structure of alginate lyase A1-III complexed with trisaccharide product at 2.0 Å resolution. J. Mol. Biol. 307, 9–16 [DOI] [PubMed] [Google Scholar]
- 10. Maruyama Y., Hashimoto W., Mikami B., Murata K. (2005) Crystal structure of Bacillus sp. GL1 xanthan lyase complexed with a substrate. Insights into the enzyme reaction mechanism. J. Mol. Biol. 350, 974–986 [DOI] [PubMed] [Google Scholar]
- 11. Mello L. V., De Groot B. L., Li S., Jedrzejas M. J. (2002) Structure and flexibility of Streptococcus agalactiae hyaluronate lyase complex with its substrate. Insights into the mechanism of processive degradation of hyaluronan. J. Biol. Chem. 277, 36678–36688 [DOI] [PubMed] [Google Scholar]
- 12. Jedrzejas M. J., Mello L. V., de Groot B. L., Li S. (2002) Mechanism of hyaluronan degradation by Streptococcus pneumoniae hyaluronate lyase. Structures of complexes with the substrate. J. Biol. Chem. 277, 28287–28297 [DOI] [PubMed] [Google Scholar]
- 13. Hynes W. L., Walton S. L. (2000) Hyaluronidases of Gram-positive bacteria. FEMS Microbiol. Lett. 183, 201–207 [DOI] [PubMed] [Google Scholar]
- 14. Hirayama Y., Yoshimura M., Ozeki Y., Sugawara I., Udagawa T., Mizuno S., Itano N., Kimata K., Tamaru A., Ogura H., Kobayashi K., Matsumoto S. (2009) Mycobacteria exploit host hyaluronan for efficient extracellular replication. PLoS Pathog. 5, e1000643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bakkevig K., Sletta H., Gimmestad M., Aune R., Ertesvåg H., Degnes K., Christensen B. E., Ellingsen T. E., Valla S. (2005) Role of the Pseudomonas fluorescens alginate lyase (AlgL) in clearing the periplasm of alginates not exported to the extracellular environment. J. Bacteriol. 187, 8375–8384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jain S., Ohman D. E. (2005) Role of an alginate lyase for alginate transport in mucoid Pseudomonas aeruginosa. Infect. Immun. 73, 6429–6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alkawash M. A., Soothill J. S., Schiller N. L. (2006) Alginate lyase enhances antibiotic killing of mucoid Pseudomonas aeruginosa in biofilms. APMIS 114, 131–138 [DOI] [PubMed] [Google Scholar]
- 18. Pompilio A., Crocetta V., Confalone P., Nicoletti M., Petrucca A., Guarnieri S., Fiscarelli E., Savini V., Piccolomini R., Bonaventura G. D. (2010) Adhesion to and biofilm formation on IB3–1 bronchial cells by Stenotrophomonas maltophilia isolates from cystic fibrosis patients. BMC Microbiol. 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sambrook J., Russell D. (2001) Molecular Cloning: A Laboratory Manual, 3 Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 20. Weissbach A., Hurwitz J. (1959) The formation of 2-keto-3-deoxyheptonic acid in extracts of Escherichia coli B. I. Identification. J. Biol. Chem. 234, 705–709 [PubMed] [Google Scholar]
- 21. Gacesa P., Wusteman F. S. (1990) Plate assay for simultaneous detection of alginate lyases and determination of substrate specificity. Appl. Environ Microbiol. 56, 2265–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Y.-H., Cui J., Lynd L. R., Kuang L. R. (2006) A transition from cellulose swelling to cellulose dissolution by o-phosphoric acid. Evidence from enzymatic hydrolysis and supramolecular structure. Biomacromolecules 7, 644–648 [DOI] [PubMed] [Google Scholar]
- 23. Isogai A., Kato Y. (1998) Preparation of polyuronic acid from cellulose by TEMPO-mediated oxidation. Cellulose 5, 153–164 [Google Scholar]
- 24. Taylor K. A. C. C. (1995) A modification of the phenol/sulfuric acid assay for total carbohydrates giving more comparable absorbances. Appl. Biochem. Biotechnol. 53, 207–211 [Google Scholar]
- 25. Bitter T., Muir H. M. (1962) A modified uronic acid carbazole reaction. Anal. Biochem. 4, 330–334 [DOI] [PubMed] [Google Scholar]
- 26. Tahiri C., Vignon M. R. (2000) TEMPO-oxidation of cellulose. Synthesis and characterization of polyglucuronans. Cellulose 7, 177–188 [Google Scholar]
- 27. ASTM Standard F2259–10, 2012e1 (2012) “Standard Test Method for Determining the Chemical Composition and Sequence in Alginate by Proton Nuclear Magnetic Resonance (1H NMR) Spectroscopy,” ASTM International, West Conshohocken, PA, DOI 10.1520/F2259-10R12E01 [DOI] [Google Scholar]
- 28. Preiss J., Ashwell G. (1962) Alginic acid metabolism in bacteria. I. Enzymatic formation of unsaturated oligosac-charides and 4-deoxy-l-erythro-5-hexoseulose uronic acid. J. Biol. Chem. 237, 309–316 [PubMed] [Google Scholar]
- 29. Tøndervik A., Klinkenberg G., Aarstad O. A., Drabløs F., Ertesvåg H., Ellingsen T. E., Skjåk-Bræk G., Valla S., Sletta H. (2010) Isolation of mutant alginate lyases with cleavage specificity for di-guluronic acid linkages. J. Biol. Chem. 285, 35284–35292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Konno N., Habu N., Maeda I., Azuma N., Isogai A. (2005) Celluronate (β-1,4-linked polyglucuronate) lyase from Brevundimonas sp. SH203. Purification and characterization. Carbohydr. Polym. 64, 589–596 [Google Scholar]
- 31. Xiao L., Han F., Yang Z., Lu X.-z., Yu W.-g. (2006) A novel alginate lyase with high activity on acetylated alginate of Pseudomonas aeruginosa FRD1 from Pseudomonas sp. QD03. World J. Micro. Biot. 22, 81–88 [Google Scholar]
- 32. Fylstra D., Lasdon L., Watson J., Waren A. (1998) Design and use of the microsoft Excel solver. Interfaces 28, 29–55 [Google Scholar]
- 33. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy). An expert resource for glycogenomics. Nucleic Acids Res. 37, D233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sebaihia M., Preston A., Maskell D. J., Kuzmiak H., Connell T. D., King N. D., Orndorff P. E., Miyamoto D. M., Thomson N. R., Harris D., Goble A., Lord A., Murphy L., Quail M. A., Rutter S., Squares R., Squares S., Woodward J., Parkhill J., Temple L. M. (2006) Comparison of the genome sequence of the poultry pathogen Bordetella avium with those of B. bronchiseptica, B. pertussis, and B. parapertussis reveals extensive diversity in surface structures associated with host interaction. J. Bacteriol. 188, 6002–6015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boyd A., Ghosh M., May T. B., Shinabarger D., Keogh R., Chakrabarty A. M. (1993) Sequence of the algL gene of Pseudomonas aeruginosa and purification of its alginate lyase product. Gene 131, 1–8 [DOI] [PubMed] [Google Scholar]
- 36. Yoon H. J., Mikami B., Hashimoto W., Murata K. (1999) Crystal structure of alginate lyase A1-III from Sphingomonas species A1 at 1.78 Å resolution. J. Mol. Biol. 290, 505–514 [DOI] [PubMed] [Google Scholar]
- 37. Papadopoulos J. S., Agarwala R. (2007) COBALT. Constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079 [DOI] [PubMed] [Google Scholar]
- 38. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL workspace. A web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 39. Bordoli L., Kiefer F., Arnold K., Benkert P., Battey J., Schwede T. (2009) Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 4, 1–13 [DOI] [PubMed] [Google Scholar]
- 40. Kiefer F., Arnold K., Künzli M., Bordoli L., Schwede T. (2009) The SWISS-MODEL repository and associated resources. Nucleic Acids Res. 37, D387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. DeLano W. L. (2010) The PyMOL Molecular Graphics System, Version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 42. Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP and releated tools. Nat. Protoc. 2, 953–971 [DOI] [PubMed] [Google Scholar]
- 43. von Heijne G. (1989) The structure of signal peptides from bacterial lipoproteins. Protein Eng. 2, 531–534 [DOI] [PubMed] [Google Scholar]
- 44. Konno N., Igarashi K., Habu N., Samejima M., Isogai A. (2009) Cloning of the Trichoderma reesei cDNA encoding a glucuronan lyase belonging to a novel polysaccharide lyase family. Appl. Environ. Microbiol. 75, 101–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Da Costa A., Michaud P., Petit E., Heyraud A., Colin-Morel P., Courtois B., Courtois J. (2001) Purification and properties of a glucuronan lyase from Sinorhizobium meliloti M5N1CS (NCIMB 40472). Appl. Environ. Microbiol. 67, 5197–5203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Farrell E. K., Tipton P. A. (2012) Functional characterization of AlgL, an alginate lyase from Pseudomonas aeruginosa. Biochemistry 51, 10259–10266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mikami B., Ban M., Suzuki S., Yoon H. J., Miyake O., Yamasaki M., Ogura K., Maruyama Y., Hashimoto W., Murata K. (2012) Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III. Acta Crystallogr. D Biol. Crystallogr. 68, 1207–1216 [DOI] [PubMed] [Google Scholar]
- 48. Gase K., Ozegowski J., Malke H. (1998) The Streptococcus agalactiae hylB gene encoding hyaluronate lyase. Completion of the sequence and expression analysis. Biochim. Biophys. Acta 1398, 86–98 [DOI] [PubMed] [Google Scholar]
- 49. Lee S. I., Choi S. H., Lee E. Y., Kim H. S. (2012) Molecular cloning, purification, and characterization of a novel poly-MG-specific alginate lyase responsible for alginate MG block degradation in Stenotrophomas maltophilia KJ-2. Appl. Microbiol. Biotechnol. 95, 1643–1653 [DOI] [PubMed] [Google Scholar]
- 50. Botzki A., Rigden D. J., Braun S., Nukui M., Salmen S., Hoechstetter J., Bernhardt G., Dove S., Jedrzejas M. J., Buschauer A. (2004) l-Ascorbic acid 6-hexadecanoate, a potent hyaluronidase inhibitor. X-ray structure and molecular modeling of enzyme-inhibitor complexes. J. Biol. Chem. 279, 45990–45997 [DOI] [PubMed] [Google Scholar]
- 51. Garron M. L., Cygler M. (2010) Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology 20, 1547–1573 [DOI] [PubMed] [Google Scholar]
- 52. Louis-Jeune C., Andrade-Navarro M. A., Perez-Iratxeta C. (2012) Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins 80, 374–381 [DOI] [PubMed] [Google Scholar]
- 53. Heyraud A., Colin-Morel P., Girond S., Richard C., Kloareg B. (1996) HPLC analysis of saturated or unsaturated oligoguluronates and oligomannuronates. Application to the determination of the action pattern of Haliotis tuberculata alginate lyase. Carbohydr. Res. 291, 115–126 [DOI] [PubMed] [Google Scholar]
- 54. Huang W., Boju L., Tkalec L., Su H., Yang H. O., Gunay N. S., Linhardt R. J., Kim Y. S., Matte A., Cygler M. (2001) Active site of chondroitin AC lyase revealed by the structure of enzyme-oligosaccharide complexes and mutagenesis. Biochemistry 40, 2359–2372 [DOI] [PubMed] [Google Scholar]
- 55. Ochiai A., Yamasaki M., Mikami B., Hashimoto W., Murata K. (2010) Crystal structure of exotype alginate lyase Atu3025 from Agrobacterium tumefaciens. J. Biol. Chem. 285, 24519–24528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yamasaki M., Ogura K., Hashimoto W., Mikami B., Murata K. (2005) A structural basis for depolymerization of alginate by polysaccharide lyase family-7. J. Mol. Biol. 352, 11–21 [DOI] [PubMed] [Google Scholar]
- 57. Lunin V. V., Li Y., Linhardt R. J., Miyazono H., Kyogashima M., Kaneko T., Bell A. W., Cygler M. (2004) High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism. J. Mol. Biol. 337, 367–386 [DOI] [PubMed] [Google Scholar]
- 58. Li H., Robertson A. D., Jensen J. H. (2005) Very fast empirical prediction and rationalization of protein pKa values. Proteins 61, 704–721 [DOI] [PubMed] [Google Scholar]
- 59. Bas D. C., Rogers D. M., Jensen J. H. (2008) Very fast prediction and rationalization of pKa values for protein-ligand complexes. Proteins 73, 765–783 [DOI] [PubMed] [Google Scholar]
- 60. Olsson M. H. M., Sondergard C. R., Rostkowski M., Jensen J. H. (2011) PROPKA3. Consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 7, 525–537 [DOI] [PubMed] [Google Scholar]
- 61. Sondergard C. R., Olsson M. H. M., Rostkowski M., Jensen J. H. (2011) Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J. Chem. Theory Comput. 7, 2284–2295 [DOI] [PubMed] [Google Scholar]
- 62. Shaya D., Zhao W., Garron M. L., Xiao Z., Cui Q., Zhang Z., Sulea T., Linhardt R. J., Cygler M. (2010) Catalytic mechanism of heparinase II investigated by site-directed mutagenesis and the crystal structure with its substrate. J. Biol. Chem. 285, 20051–20061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yoon H. J., Hashimoto W., Miyake O., Okamoto M., Mikami B., Murata K. (2000) Overexpression in Escherichia coli, purification, and characterization of Sphingomonas sp. A1 alginate lyases. Protein Expr. Purif. 19, 84–90 [DOI] [PubMed] [Google Scholar]
- 64. Preston J. F., 3rd, Rice J. D., Ingram L. O., Keen N. T. (1992) Differential depolymerization mechanisms of pectate lyases secreted by Erwinia chrysanthemi EC16. J. Bacteriol. 174, 2039–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Miyake O., Hashimoto W., Murata K. (2003) An exotype alginate lyase in Sphingomonas sp. A1. Overexpression in Escherichia coli, purification, and characterization of alginate lyase IV (A1-IV). Protein Expr. Purif. 29, 33–41 [DOI] [PubMed] [Google Scholar]
- 66. Konno N., Habu N., Iihashi N., Isogai A. (2008) Purification and characterization of exo-type cellouronate lyase. Cellulose 15, 453–463 [Google Scholar]
- 67. Jandik K. A., Gu K., Linhardt R. J. (1994) Action pattern of polysaccharide lyases on glycosaminoglycans. Glycobiology 4, 289–296 [DOI] [PubMed] [Google Scholar]
- 68. Cescutti P., Cuzzi B., Liut G., Segonds C., Di Bonaventura G., Rizzo R. (2011) A novel highly charged exopolysaccharide produced by two strains of Stenotrophomonas maltophilia recovered from patients with cystic fibrosis. Carbohydr. Res. 346, 1916–1923 [DOI] [PubMed] [Google Scholar]