Background: Peroxisome import receptor Pex5 harbors multiple WXXX(F/Y) sequences for Pex14 binding.
Results: Pex5 contains a novel essential Pex14-binding site with faster binding kinetics than the canonical WXXX(F/Y).
Conclusion: The new Pex14-binding site is critical for matrix protein import due to fast dissociation rates.
Significance: Peroxisomal protein import requires sequential Pex14 binding of Pex5, which may trigger docking and formation of the translocation pore.
Keywords: Peroxisomes, Protein Dynamics, Protein Motifs, Protein Structure, Protein Targeting, Protein-Protein Interactions, Subcellular Organelles
Abstract
Protein import into peroxisomes relies on the import receptor Pex5, which recognizes proteins with a peroxisomal targeting signal 1 (PTS1) in the cytosol and directs them to a docking complex at the peroxisomal membrane. Receptor-cargo docking occurs at the membrane-associated protein Pex14. In human cells, this interaction is mediated by seven conserved diaromatic penta-peptide motifs (WXXX(F/Y) motifs) in the N-terminal half of Pex5 and the N-terminal domain of Pex14. A systematic screening of a Pex5 peptide library by ligand blot analysis revealed a novel Pex5-Pex14 interaction site of Pex5. The novel motif composes the sequence LVAEF with the evolutionarily conserved consensus sequence LVXEF. Replacement of the amino acid LVAEF sequence by alanines strongly affects matrix protein import into peroxisomes in vivo. The NMR structure of a complex of Pex5-(57–71) with the Pex14-N-terminal domain showed that the novel motif binds in a similar α-helical orientation as the WXXX(F/Y) motif but that the tryptophan pocket is now occupied by a leucine residue. Surface plasmon resonance analyses revealed 33 times faster dissociation rates for the LVXEF ligand when compared with a WXXX(F/Y) motif. Surprisingly, substitution of the novel motif with the higher affinity WXXX(F/Y) motif impairs protein import into peroxisomes. These data indicate that the distinct kinetic properties of the novel Pex14-binding site in Pex5 are important for processing of the peroxisomal targeting signal 1 receptor at the peroxisomal membrane. The novel Pex14-binding site may represent the initial tethering site of Pex5 from which the cargo-loaded receptor is further processed in a sequential manner.
Introduction
Import of peroxisomal matrix proteins is a cycling multistep process comprising the post-translational recognition of fully synthesized and folded peroxisomal proteins in the cytosol, transport to the peroxisome, docking of the receptor cargo complex at the organelle surface, and receptor-dependent formation of a transient protein-conducting channel (1, 2). After translocation of the matrix proteins across the membrane, the receptors are released into the cytosol to initiate a further round of import (3).
For the cytosolic cargo recognition, folded and even oligomerized peroxisomal matrix proteins interact with the peroxisome targeting signal (PTS)5 receptors Pex5 or Pex7 via cognate targeting sequences PTS1 or PTS2, respectively (4). Only a minor class of peroxisomal proteins contains the PTS2 signal, which corresponds to a nonapeptide sequence near the N terminus. The majority of matrix proteins harbor PTS1 motifs that consist of the tripeptide –SKL (or a conserved variant thereof) at the extreme C terminus of the protein. The PTS1 motif interacts with the tetratricopeptide repeat domain in the C-terminal half of Pex5. The intrinsically disordered N-terminal half of Pex5 per se is capable of performing all transport steps of the receptor cycle, including docking and pore formation and dislocation from the peroxisomal membrane (5, 6).
A peroxisomal membrane complex consisting of Pex13 and Pex14 (and Pex17 in yeast) provides the primary docking site for the cargo-loaded receptors (7, 8). Pex14 fulfills additional important functions in cargo translocation across the peroxisomal membrane. For the model organism Saccharomyces cerevisiae, it was shown that Pex14 together with Pex5 constitutes a large dynamic channel at the peroxisomal membrane, which is supposed to act as a protein conducting pore (9). Recent data suggest that the Pex14-Pex5 interaction also plays a critical role for release of the cargo protein into the lumen of the peroxisome (10).
Pex5 interacts with Pex14 via diaromatic penta-peptide motifs that were originally defined by the signature WXXX(F/Y) and are located in the N-terminal halves of all Pex5 proteins. The number of the conserved WXXX(F/Y) motifs varies depending on the species with, for example, two in yeast or seven in human Pex5 (11). It was shown that each of the seven WXXX(F/Y) motifs of the human PTS1 receptor can interact with the highly conserved N-terminal domain (NTD) of Pex14 with equilibrium dissociation constants in the nanomolar range (12, 13). A recent biophysical study showed that the presence of multiple WXXX(F/Y) motifs in the N terminus of Pex5 allows the formation of higher order complexes with the Pex14-NTD in vitro, although it is not clear whether this also reflects the situation in peroxisomes in vivo (14, 15). Interestingly, human Pex19, which is supposed to act as an import receptor/chaperone for peroxisomal membrane proteins, binds competitively to the same surface in Pex14-NTD via an amphipathic helix-forming penta-peptide sequence (16). In contrast to typical Pex5 WXXX(F/Y) motifs, the identified FFXXXF of Pex19 binds with opposite directionality. The mode of binding is essentially similar as the ligand adopts an amphipathic α-helical conformation from which the protruding conserved aromatic side chains are positioned into hydrophobic pockets formed by Pex14-NTD.
A crystallographic study (17) and NMR analysis (16) of the free Pex14-NTD revealed that the Pex5 binding domain of Pex14 adopts a three helical fold. S. cerevisiae Pex5 binds the N-terminal Pex14 domain via a short segment containing the reverse motif FXXXW (18), whereas the Pex14-binding site of Leishmania Pex5 has been mapped to a region without any sequence similarity to WXXX(F/Y) motifs (19). Recently, it was reported that Pex14 interacts with tubulin and that this interaction also depends on the Pex14-NTD (20).
Taken together, these data demonstrate that the Pex14-NTD is a small structured domain that mediates versatile protein-protein interactions linked to peroxisomal function. For binding to the N-terminal Pex14 domain, ligands harbor the well studied WXXX(F/Y) motif or variants thereof. However, not all conserved WXXX(F/Y) motifs form stable complexes with Pex14, indicating that the consensus sequence might be essential but not sufficient for Pex14 binding. Prominent examples are two typical WXXX(F/Y) motifs of S. cerevisiae Pex5 (18, 21) or one of the three WXXX(F/Y) motifs of Trypanosoma brucei Pex5 (22). These sequences may mediate the interaction with other peroxins, as shown for human and yeast Pex5 motifs, which both can interact with the Src homology 3 domain of Pex13 (8, 13, 24, 25).
In this study, we identified a novel LVXEF-binding motif for human Pex14 in the N-terminal half of human Pex5. The new motif binds to the Pex14-NTD with a comparable affinity but with notably distinct binding kinetics than the well characterized diaromatic WXXX(F/Y) motifs. NMR data and structural analysis revealed that the novel motif binds in a similar orientation as the WXXX(F/Y) motif but that the tryptophan pocket is now occupied by a Leu residue. Interestingly, replacement of the new motif by a canonical WXXX(F/Y) motif with different binding properties impairs Pex5-dependent protein import. Our data suggest that the novel Pex14-binding site represents the initial tethering site of Pex5 from which the cargo-loaded receptor is further processed in a sequential manner.
EXPERIMENTAL PROCEDURES
Construction of Pex5 Fragment Expression Vectors
For DNA cloning, Escherichia coli strain DH5α was used. The human His-tagged Pex5 fragments Pex5-(1–113) and Pex5-(1–131) were amplified by PCR from pET9d-His6-Pex5L (11) using the forward T7 promotor primer (Novagen) and the reverse primers KU582 (Pex5-(1–131)) and KU644 (Pex5-(1–113)) and subcloned into NcoI/SalI-digested pET9d-His6-Pex5L. The essential tryptophan in the first WXXX(F/Y) motif (Trp-118) was replaced by an alanine using the QuikChange XL site-directed mutagenesis kit (Stratagene) using primer pair KU690/KU1278. Pex5-(1–110) was amplified by PCR from pET9d-His6-Pex5L (11) using primer pair 1–110 forward and 1–110 reverse. The resulting PCR fragment was subcloned into a pETM11 vector (EMBL, Heidelberg, Germany) using NcoI and Acc65I restriction sites. Mutagenic disruption of the novel Pex14-binding site was achieved by substitution of the amino acid sequence LVAEF by AAAAA. Mutagenesis was carried out by overlapping PCR using the primer pairs (forward, T7 promotor primer/reverse; KU1102 and KU1101/reverse T7 termination). As templates both pET9d-His6-Pex5L and pET9d-His6-Pex5-(1–131) were used.
Amplification products were subcloned in a modified E. coli expression plasmid pET24d, allowing N-terminal fusion of a His6 tag (modified by G. Stier, EMBL) by using XbaI and NotI restriction endonuclease recognition sites. In addition, the full-length PCR product was restricted with NcoI and blunted before it was digested by NotI for subcloning into EcoRV/NotI-digested mammalian expression vector pcDNA3.1-zeo+. All point mutations were verified by DNA sequencing. The sequences of the primers used are listed in Table 1.
TABLE 1.
Oligonucleotides used
| Designation | Sequence (5′ to 3′) |
|---|---|
| T7 promotor | TAATACGACTCACTATAG |
| T7 termination | GCTAGTTATTGCTCAGCGG |
| KU582 | CTAGGTCGACTATACATCCACAGCATCTCCAGC |
| KU644 | CTAGGTCGACCTAGGCCAAGTCTGCCACACC |
| KU690 | TGGCCTTGTCTGAGAATGCGGCCCAGGAGTTTCTT |
| KU1101 | GGAGTAGCTTCTGAAGATGAGGCGGCGGCGGCGGCGCTGCAGGACCAGAATGCACCC |
| KU1102 | GGGTGCATTCTGGTCCTGCAGCGCCGCCGCCGCCGCCTCATCTTCAGAAGCTACTCC |
| KU1255 | TATAATGAGACTGACGCGTCCCAAGAATTCATCTCTG |
| KU1278 | AAGAAACTCCTGGGCCGCATTCTCAGACAAGGCCA |
| KU1279 | CAGAGATGAATTCTTGGGACGCGTCAGTCTCATTATA |
| RE2142 | GAGTAGCTTCTGAAGATGAGTGGGCGCAGGAGTTCCTGCAGGACCAGAATGC |
| RE2143 | GCATTCTGGTCCTGCAGGAACTCCTGCGCCCACTCATCTTCAGAAGCTACTC |
| RE3533 | GATCAGATCTATGGCAATGCGGGAGCTGG |
| RE3534 | GATCGTCGACTCACTGGGGCAGGCCAAAC |
| 1–110_F | TAATCCATGGCAATGCGGGAGCTGGTGG |
| 1–110_R | TATTGGTACCTTATGCCACACCAGGGGCTCTCTGG |
Expression and Purification of Recombinant Proteins
The genes coding for recombinant proteins were all expressed in E. coli strain BL21(DE3). Expression of the genes encoding His6-tagged Pex5 proteins and GST-Pex14-(1–78) were carried out as described previously (11). Purifications of His6-tagged Pex5 fragments using nickel-nitrilotriacetic acid-agarose and ResourceQ (GE Healthcare) anion exchange chromatography and GST-Pex14-(1–78) were carried out as described previously (11).
Pex14-(16–80) and Pex5-(1–110) for NMR studies were expressed as fusion proteins with a His6 tag followed by a tobacco etch virus cleavage site as described previously for Pex14-(16–80) (16). 13C,15N- or 15N-labeled proteins were expressed in minimal M9 medium supplemented with 2 g/liter [13C]glucose and/or 1 g/liter [15N]ammonium chloride, respectively. Protein samples for NMR studies were purified by Ni2+ affinity chromatography followed by tobacco etch virus cleavage and removal of the His6 tag by a second Ni2+ affinity chromatography step. The final step of purification consisted of size exclusion chromatography on a HiLoad 16/60 Superdex75 column (GE Healthcare) in 20 mm sodium/phosphate buffer, pH 6.5, 100 mm NaCl, and 1 mm DTT.
Peptide Binding
Synthetic peptides were purchased from Peptide Specialty Laboratories (Heidelberg, Germany) and dialyzed extensively against water before use in NMR studies. Immobilized peptides were synthesized by the Fmoc (N-(9-fluorenyl)methoxycarbonyl) method of solid phase peptide chemistry on a Milligen/Biosearch 9050 Pep-synthesizer. Cleavage of the peptides from the resin and removal of the protecting groups were achieved by treating the peptide with 90% trifluoroacetic acid, 2.5% phenol, 2.5% 1,2-ethanedithiol, 1% triisopropylsilane (all from Sigma), and 5% water (26). A peptide library of the protein sequences of human Pex5-(1–343) and a combinatorial library to analyze amino acid substitutions of Pex5-(59–67) were generated by the Spots technique according to the manufacturer's protocol (ABIMED, Auto-Spot-Robot ASP 222) as described previously (26). The library representing Pex5-(1–343) consisted of 15-mer peptides with a 13-amino acid overlapping region that were immobilized on a cellulose membrane. The library was incubated with purified His6-tagged Pex14-(1–78) as described previously (16). Bound Pex14-(1–78) was immunodetected using monoclonal anti-His antibodies (Qiagen).
NMR Spectroscopy
NMR experiments were recorded at 25 °C on 900, 600, or 500 MHz Bruker Avance NMR spectrometers equipped with cryogenic triple resonance gradient probes. All spectra were processed with NMRPipe (27) and analyzed in Sparky (33). NMR spectra for assignment and structural determination of the Pex14-(16–80)·Pex5-(57–71) complex were acquired on a sample containing 13C,15N-labeled Pex14-(16–80) bound to unlabeled Pex5-(57–71). Protein backbone and side chain assignments were obtained using standard triple-resonance experiments (28).1H chemical shifts of unlabeled Pex5-(57–71) in the bound form were assigned based on ω1-filtered TOCSY and NOESY spectra. Intermolecular NOEs between Pex14-(16–80) and Pex5-(57–71) were obtained from a ω1-filtered NOESY spectrum (28) with a mixing time of 80 ms. Paramagnetic relaxation enhancements (PREs) were recorded on Pex14-(16–80) in complex with either of two 16-mer Pex5 peptides (CASEDELVAEFLQDQN or ASEDELVAEFLQDQNC). The additional cysteine residue at the N or C terminus of each peptide was used for covalent attachment of the iodoacetamido-proxyl spin label. PRE data were recorded and analyzed as described elsewhere (30).
Structure Determination
The solution structure of the Pex14-(16–80)·Pex5-(57–71) complex was calculated with the previously established protocol using a modified version of Aria1.2/CNS (30). The structure of free Pex14-(16–80) (Protein Data Bank code 3FF5) (17) was used as input for structure calculation and maintained using noncrystallographic symmetry restraints. Distance restraints were derived from intermolecular (Pex14-Pex5) and intramolecular (Pex5) NOEs. Backbone torsion angle restraints were obtained from chemical shifts of Pex5-(1–110) in complex with Pex14-(16–80) using TALOS+ (31). All restraints were employed during a molecular dynamics and simulated annealing run. The final structures were refined in a shell of water molecules (32) and validated using the iCing web interface. Molecular images were generated with PyMOL (Schrödinger, LLC).
Pex5-Pex14 Interaction Assays
Isothermal titration calorimetry (ITC) measurements were carried out at 25 °C using an iTC200 calorimeter (GE Healthcare). Pex14-(16–80) at a concentration of ∼600 μm in buffer A (20 mm sodium phosphate, 100 mm NaCl, pH 6.5) was injected in the cell containing Pex5-(1–110) at a concentration of ∼35 μm in buffer A. After correction for heat of dilution, the data were fitted to a one-site binding model using the Microcal Origin 7.0 software.
The interaction between Pex5 proteins and GST-Pex14-(1–78) was also studied using surface plasmon resonance spectroscopy with a BIAcore 2000 instrument (BIAcore AB) as described previously (11). Data were evaluated with the BIAevaluation software version 4.1.1 (BIAcore AB) using the 1:1 Langmuir binding fit. Interaction assays using size exclusion chromatography were performed on a Superose 6 PC 3.2/30 column (GE Healthcare) as described previously (11).
In Vivo Studies
Pex14-binding sites in full-length Pex5 were disrupted either individually or simultaneously by substituting amino acids 62–66 by penta-alanine (LVAEF → AAAAA) or by replacing essential tryptophans of respective WXXX(F/Y) motifs, Trp-118 (W118A) and Trp-140 (W140A) by alanine. Mutagenesis of LVAEF was carried out by overlapping PCR using mutagenic primers KU1101 and KU1102 and pcDNA3-PEX5S (23) as a template. Substitution of the tryptophans of the first and the second WXXX(F/Y) motif with alanines was achieved by using QuikChange XL site-directed mutagenesis kit (Stratagene) and primer pairs KU690/KU1278 and KU1255/KU1279, respectively. For substitution of LVAEF by WAQEF, primers RE2142/RE2143 were used. A bicistronic expression plasmid coding for EGFP-SKL and nontagged Pex14 (pMF120 (29)) was kindly provided by Marc Fransen (Leuven, Belgium). pIRES2-EGFP-SKL was constructed by replacing the open reading frame of PEX14 with the original multiple cloning site from pIRES2-EGFP. The PEX5S open reading frames with and without mutations were amplified using primer pair RE3533/3534 and subcloned into pIRES-EGFP-SKL using BglII and SalI restriction sites. Primer sequences are shown in Table 1.
The bicistronic expression vectors coding for Pex5 variants and EGFP-PTS1 were transfected into the human cell line ΔPEX5T, which was derived from Pex5-deficient Zellweger patient fibroblasts (20). Forty-eight hours after transfection, cells were subjected to immunofluorescence microscopy using polyclonal rabbit antiserum against human Pex14 and monoclonal antibody against AFP (MP Biomedicals, Heidelberg, Germany). For quantitative analysis, three to six independent transfections of each pIRES-Pex5-EGFP-SKL expression plasmid were monitored. Based on the appearance of EGFP-SKL fluorescence pattern, about 100 cells of each experiment were visually categorized in full or partial complementation of peroxisomal import as visualized by punctate staining pattern or no complementation indicated by diffuse nonpunctate cytosolic background staining. All micrographs were recorded on a Zeiss Axioplan 2 microscope with a Zeiss Plan-Apochromat 63 ×/1.4 oil objective and an Axiocam MR digital camera and were processed with AxioVision 4.6 software (Zeiss, Jena, Germany). The steady-state level of Pex5 expression was assessed by immunoblot analyses of transfected cells using polyclonal rabbit antibodies against HsPex5. Anti-prohibitin antibodies (Abcam) were used as loading control.
RESULTS
Identification of a Novel Pex14-binding Site in the N-terminal Domain of Human Pex5
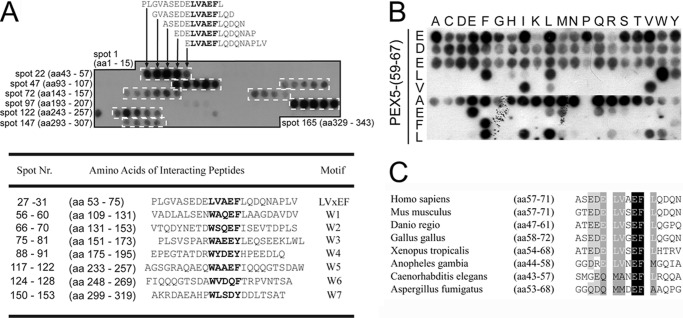
To identify the Pex14-binding sites of human Pex5, we performed a peptide scan analysis. The membrane array consisted of synthetic 15-mer peptides that sequentially overlap by 13 residues and represented Pex5-(1–343) composing the entire sequence of the Pex14-interacting region (11). The cellulose-bound peptides were incubated with His-tagged Pex14-NTD, and bound protein was visualized by immunodetection with anti-His antibodies. Peptides containing one of the seven known WXXX(F/Y) motifs did bind the Pex14 probe resulting in a staining pattern of clusters of four to six successive peptides (Fig. 1A, white boxes). In accordance with previously reported dissociation constants (KD) for WXXX(F/Y) binding (12), the strongest immunological labeling was obtained with peptides containing either the first WXXX(F/Y) sequence (amino acids 118–122, spots 56–60) or the fifth WXXX(F/Y) motif (amino acids 243–247, spots 117–122). Surprisingly, the peptide scan revealed, in addition to the seven known Pex14-binding sites, staining for five consecutive peptides that do not contain a WXXX(F/Y) motif (Fig. 1). The overlapping region of the new binding site represents 7 amino acids with the sequence ELVAEFL. The core sequence LVAEF resembles the Pex5 WXXX(F/Y) motifs with the major difference that tryptophan is replaced by the hydrophobic amino acid leucine (Fig. 1A).
FIGURE 1.
Identification of a novel Pex14-binding motif in the N-terminal domain of the human PTS1 receptor. A, peptide scan analysis of Pex5 for Pex14-binding sites. 15-mer peptides with two amino acid (aa) shifts between successive peptides representing amino acids 1–343 of Pex5 were synthesized on a cellulose membrane. The first spot of each row is indicated by numbers. After incubation with purified His-tagged Pex14-NTD, interacting peptide spots were detected with anti-His antibodies. Sequences of interacting peptide arrays (dashed boxes), including the novel binding motif (spots 27–31), are shown, and the overlapping amino acids of all peptides within each array are highlighted in gray, and core sequences with highest structural similarity are indicated by bold letters. B, substitution analysis of the conserved residues of the novel Pex14-binding region of human Pex5. Each row represents a variant of the human Pex5-(59–67) sequence, EDELVAEFL, in which the indicated core amino acid was replaced with any of the 20 proteinogenic amino acids. His-tagged Pex14-NTD interacting peptides were detected by immunoblot analysis using anti-His antibodies. C, LVXEF motifs within the N-terminal domain of Pex5p are conserved among several groups of metazoans. Representative sequences are shown. Similar sequences with the consensus MXXEF were detected in Pex5 sequences of C. elegans and filamentous fungi represented here by A. fumigatus.
To further characterize the sequence requirements of the novel Pex14-binding motif, we carried out a peptide scan substitution analysis. To this end, 15-mer peptides corresponding to the Pex5 sequence 59EDELVAEFL67 were synthesized to harbor a single amino acid exchange at each position of the minimal binding sequence (Fig. 1B). The major difference between the novel LVAEF and canonical WXXX(F/Y) motifs concerns the two first residues. Whereas in WXXX(F/Y) motifs only the tryptophan is invariant for interaction with Pex14 (12, 16), in the monoaromatic 62LVAEF66 binding sequence, two hydrophobic residues Leu-62 and Val-63 are critical for Pex14 binding. However, the leucine residue in the new motif is interchangeable with the aromatic residues tryptophan, phenylalanine, and tyrosine (Fig. 1B). Interestingly, the peptide containing tryptophan instead of leucine seems to bind to Pex14-NTD with a higher affinity than the original sequence. Common to all Pex14-binding motifs of human Pex5 is the invariant position of an aromatic residue, either phenylalanine or tyrosine at position 5 of the penta-peptide as well as the preference for a glutamate in position 4.
Sequence alignment of known PTS1 receptors revealed that an LVXEF sequence motif preceding the first canonical WXXX(F/Y) motif is conserved in many metazoa (Fig. 1C). In Caenorhabditis elegans, the residues at positions 1 and 2 of the corresponding sequence are methionine and alanine, respectively, both of which would interfere with binding of the peptide to human Pex14. Remarkably, the LVXEF sequence motif is absent in Pex5 from plants and yeasts but similar sequences were found in Pex5 of several filamentous fungi, including Aspergillus fumigatus.
NMR Analysis and Structure of the Complex of Pex14-NTD with the Novel Pex5 LVXEF Motif
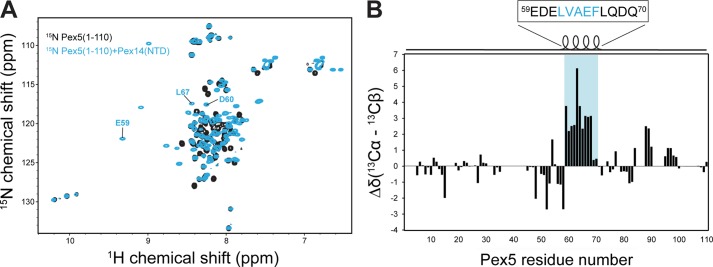
To further characterize the binding of Pex5 to Pex14-NTD using NMR spectroscopy, we studied a construct comprising only the first 110 residues of human Pex5, which lacks any of the known WXXX(F/Y) motifs. NMR fingerprint spectra of 15N-labeled Pex5-(1–110) show very little dispersion, characteristic of an unstructured protein. However, upon addition of unlabeled Pex14-NTD, the spectrum changes significantly, indicating a strong interaction between Pex5-(1–110) and Pex14-NTD (Fig. 2A). Analysis of 13Cα/β secondary chemical shifts revealed that the region including residues Glu-59 to Gln-70 of human Pex5 (59EDELVAEFLQDQ70) forms an α-helix upon interacting with Pex14-NTD (Fig. 2B). The identified α-helix contains the core sequence LVAEF, matching the sequence obtained from our peptide scan analysis.
FIGURE 2.
Residues Glu-59 to Gln-70 of Pex5 form an α-helix upon interacting with Pex14-NTD. A, 1H,15N HSQC spectra of 15N-Pex5(1–110), free (black) and in complex with Pex14-NTD (cyan). Few residues undergoing large chemical shift perturbations are labeled. B, plot of 13Cα/β secondary chemical shifts of Pex5-(1–110) in complex with Pex14. Positive values indicate an α-helical conformation for residues Glu-59 to Gln-70 of Pex5 that harbor the novel LVXEF motif (highlighted in cyan).
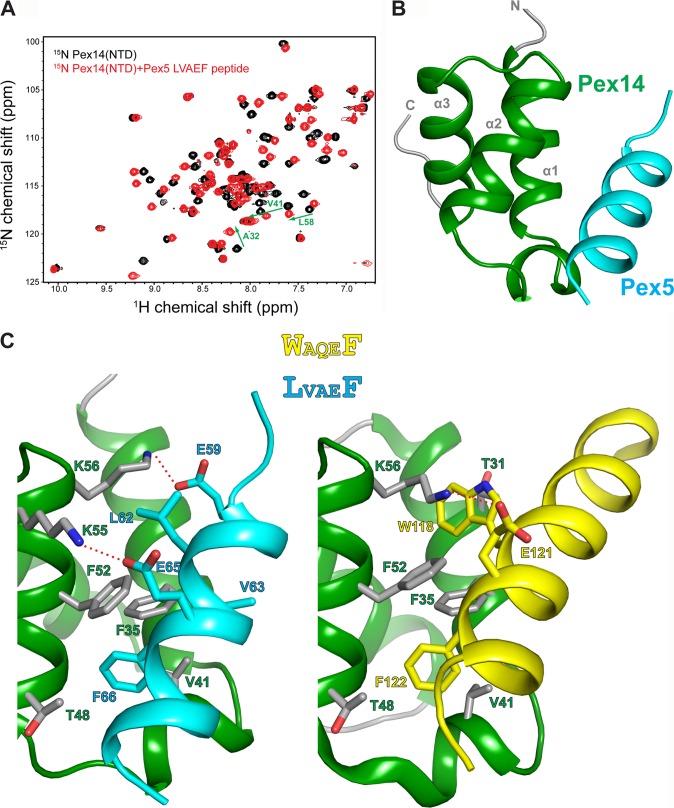
The binding site of the LVXEF motif on the structure of Pex14-NTD was mapped based on chemical shift perturbations seen upon addition of a 16-mer Pex5 peptide containing the LVAEF sequence to 15N-labeled Pex14-NTD (Fig. 3A). The chemical shift perturbations (data not shown) involve the same set of residues that have been previously shown to bind to a WXXX(F/Y) motif of Pex5, indicating that the new motif interacts with the same surface of Pex14-NTD (16). To elucidate molecular details for the recognition of the novel Pex5 motif by Pex14-NTD, we determined the solution structure of the complex. The structure of the complex was determined based on the previously reported structure of Pex14-NTD (16, 17), using a protocol that we have recently established (30), and included 47 intermolecular NOE-derived distance restraints between Pex14-NTD and Pex5-(57–71) (Table 2). The Pex14-NTD adopts a three-helical bundle in which helices α1 and α2 constitute the main binding interface. The Pex5 LVXEF motif binds in an α-helical conformation to the hydrophobic surface of Pex14-NTD (Fig. 3B). Three conserved residues (Leu-62, Val-63, and Phe-66) of the LVXEF motif are involved in hydrophobic interactions with Pex14-NTD. Phe-66 is inserted into a hydrophobic pocket in Pex14 and engages contacts with Pex14 Phe-35 and Phe-52 on helices α1 and α2, respectively. Leu-62 is not deeply buried in the second hydrophobic pocket of Pex14 but interacts with the aliphatic side chain of the conserved Lys-56 in Pex14. In addition, Pex5 Val-63 is involved in hydrophobic interactions with Pex14 Val-41 and Phe-35. The conserved Val-41 is centrally located in the short 310-helix of Pex14-NTD between helices α1 and α2 and makes contact with both Val-63 and Phe-66 in the LVXEF motif of Pex5 (Fig. 3C). Thus, similar to the previously reported structures, where hydrophobic pockets in Pex14-NTD accommodate aromatic side chains of WXXX(F/Y) motifs or the inversely oriented FFXXXF motif in Pex5 and Pex19, respectively (16), the LVXEF motif uses the same pocket for recognition of phenylalanine. In contrast, recognition of the LV sequence in the newly identified LVXEF motif is distinct from the interactions with Phe or Trp in canonical WXXX(F/Y) motifs (Fig. 3C).
FIGURE 3.
NMR structure of the Pex14-NTD-Pex5-(57–71) complex. A, 1H,15N HSQC spectra of 15N-Pex14-NTD, free (black), and titrated with 2-fold excess of Pex5-(57–71) (red). B, ribbon representation of the lowest energy NMR structure of the Pex14-NTD·Pex5-(57–71) complex. C, structural comparison of Pex14 interaction with LVAEF (left) and WAQEF (right) peptide ligands of Pex5. Side chains of residues involved in intermolecular interactions are shown as sticks. The recognition of Phe (in LVAEF and WAQEF) by Pex14 is similar in the two structures; however, the main difference lies in the recognition of LV (in LVAEF) and Trp (in WAQEF) residues. Plausible salt bridges between Glu and Lys side chains are indicated by red dashed lines in both structures.
TABLE 2.
NMR and structural statistics
Statistics are reported for the 10 lowest energy structures from a total of 100 calculated. r.m.s.d. is root mean square deviation.
| NMR restraints | |
| Total NOE distance restraints | 173 |
| Inter(Pex14-Pex5 peptide) | 47 |
| Intra(Pex5 peptide) | 126 |
| PRE-derived distance restraints | 113 |
| Hydrogen-bonds | 16 |
| Φ + ψ torsion angle restraintsa | 30 |
| Restraint violations | |
| Distance violations >0.3 | 0.0 |
| Dihedral angle violations >5° | 0.0 |
| r.m.s.d. from experimental distance restraints | 0.031 ± 0.007 |
| r.m.s.d. from experimental torsion angle restraints | 0.698 ± 0.273° |
| Deviations from idealized geometry | |
| Bonds | 0.005 ± 0.000 |
| Angles | 0.668 ± 0.007° |
| Impropers | 0.965 ± 0.083° |
| Coordinate precision r.m.s.d.b | |
| Backbone | 0.56 ± 0.16 |
| Heavy atom | 1.17 ± 0.07 |
| Ramachandran plot analysisb | |
| Residues in most favored regions | 97.5% |
| Residues in additionally allowed regions | 2.5% |
| Residues in generously allowed regions | 0% |
| Residues in disallowed regions | 0% |
a Dihedral angle restraints were derived from TALOS+.
b This applies to residues 25–70 of Pex14 and 57–71 of Pex5.
Electrostatic interactions also play an important role in ligand binding by Pex14-NTD. As described previously, the binding interface of Pex14-NTD forms a highly positively charged surface complementary to the negatively charged Pex5 and Pex19 ligands (16). The structure of the Pex14-Pex5 LVXEF motif suggests that Lys-55 and Lys-56 of Pex14 form salt bridges with Glu-65 and Glu-59 of Pex5, respectively (Fig. 3C). According to our peptide scan analysis, Glu-65 in the Pex5 LVXEF motif is critical for Pex14 binding. Furthermore, charge-reversal substitution of Glu-59 located C-terminally to the LVXEF motif leads to loss of Pex14 binding (Fig. 1B). Thus, electrostatic interactions make important contributions to the recognition of the Pex5 LVXEF motif by Pex14.
Our previous structural studies have shown that Pex14-NTD can interact with helical motifs of Pex5 and Pex19 in opposite orientations. The Pex5 LVAEF peptide adopts the same directionality as the previously reported Pex5 WXXX(F/Y) peptide (16). To further validate the orientation of the novel Pex5 LVXEF motif helix, we used PRE data for two spin-labeled peptide ligands with the spin label attached either to the N or C terminus. Addition of a spin-labeled peptide to Pex14-NTD leads to line broadening of amide protons that are in the vicinity of the spin label. Analysis of the PRE data for both peptides shows that experimentally observed line broadening effects are in agreement with the theoretically calculated ones based on our NMR structure (Fig. 4). Thus, the PRE data independently confirm that the Pex5 LVXEF motif binds to Pex14 in the same orientation as the Pex5 WXXX(F/Y) peptide.
FIGURE 4.
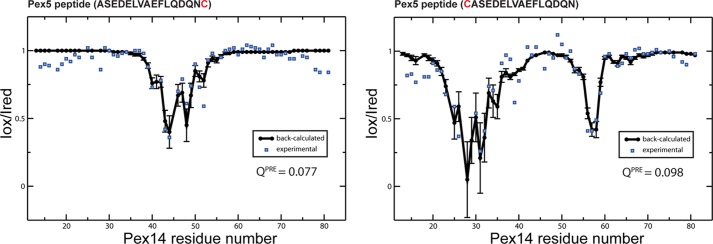
PRE experiments confirm the orientation of the Pex5-(57–71) peptide bound to Pex14-NTD observed in the NMR structure. Comparison of experimental (blue squares) and back-calculated PRE data (black circles, error bars indicate variations across the NMR ensemble) obtained for Pex14-NTD in complex with C-terminally (left panel) and N-terminally (right panel) spin-labeled Pex5 peptides. In both cases, there is excellent agreement between experimental and back-calculated values, as judged from the low value of the quality factor QPRE. The paramagnetic spin label was attached to a terminal cysteine residue added to the Pex5 peptide (shown in red).
Equilibrium and Kinetic Binding Properties of Pex14-NTD·Pex5 Complexes
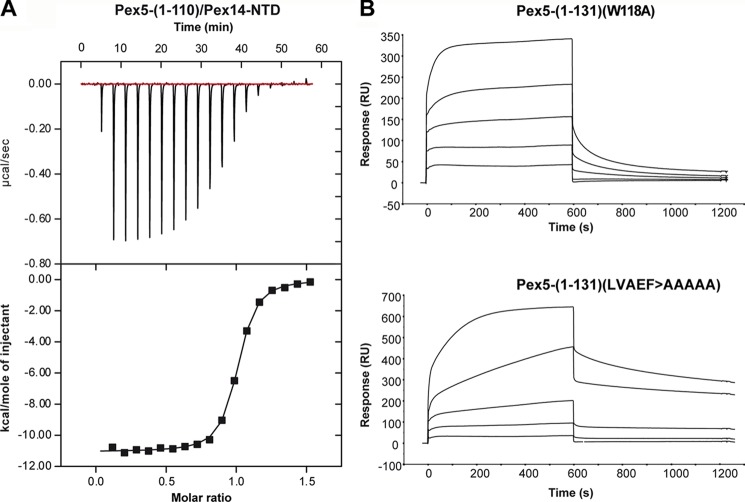
We determined the binding affinities and kinetic rate constants for the complex of Pex14-NTD with the novel Pex5 motif using isothermal titration calorimetry and surface plasmon resonance experiments. The ITC data show that Pex5-(1–110) binds to Pex14-NTD with a dissociation constant (KD) of 157 ± 9 nm (Fig. 5A), demonstrating high affinity binding, similar to the WXXX(F/Y) motifs (12, 16). The stoichiometry of binding was 1:1, confirming that the identified binding sequence represents the only accessible interacting site within the N-terminal 110 residues of Pex5.
FIGURE 5.
Rate and equilibrium binding constants of the interaction of regions of Pex5 and Pex14. A, ITC curve of the interaction between Pex5-(1–110) and the N-terminal domain of Pex14 (Pex14-NTD). Pex14-NTD was injected into the cell, containing Pex5-(1–110), and the heat of binding was measured per mol of injectant. The raw data were fitted to a one-site binding model, which yielded a KD of 157 ± 9 nm with a stoichiometry of 0.97 ± 0.002. B, for surface plasmon resonance spectroscopy analyses, 700 response units (RU) of purified GST-Pex14-NTD were immobilized on an anti-GST sensor chip surface, and varying concentrations of purified Pex5-(1–131)(W118A) and Pex5-(1–131)(LVAEF > AAAAA) were applied. The concentrations used for both Pex5-(1–131) constructs were as follows: 1, 4, 15, 60, and 240 nm (graphs from bottom to top). Analyses were performed as described under “Experimental Procedures,” and the results are summarized in Table 3.
For analysis by surface plasmon resonance experiments, GST-Pex14-NTD was coupled to the surface of sensor chips and incubated with increasing concentrations of the N-terminal Pex5 fragment. The LVXEF containing Pex5-(1–117) interacted with Pex14-NTD with a KD of 87 nm (Table 3). Interestingly, a longer N-terminal fragment, Pex5-(1–131), which harbors both the LVXEF and an additional WXXX(F/Y) motif (amino acids 118–122), exhibited a much higher binding affinity with a KD of 4.6 nm.
TABLE 3.
Rate and equilibrium binding constants of the interaction of immobilized GST-Pex14-NTD and Pex5 proteins
Association rate constants, kon, dissociation rate constants, koff, and equilibrium binding constants, KD, were determined by surface plasmon resonance spectroscopy using GST-Pex14-NTD immobilized on an anti-GST surface as ligand and the various Pex5 proteins as analytes. Disruption of Pex14-binding sites, either of the new motif (LVAEF) or the N-terminal WXXX(F/Y) motifs (Trp-118 and Trp-140), by alanine replacements are indicated. For each Pex5 construct, the number of remaining Pex14-binding sites is shown (Pex14-BS(n)).
| Pex5- | Pex14-BS | kon | koff | KD |
|---|---|---|---|---|
| n | m−1s−1 | s−1 | m | |
| 1–117 | 1 | 7.0 × 104 | 6.1 × 10−3 | 87 × 10−9 |
| 1–131 (W118A) | 1 | 7.5 × 104 | 6.6 × 10−3 | 87 × 10−9 |
| 1–131 (LVAEF → AAAAA) | 1 | 2.6 × 104 | 0.2 × 10−3 | 9.2 × 10−9 |
| 1–131 | 2 | 31 × 104 | 1.4 × 10−3 | 4.5 × 10−9 |
| Full length | 8 | 55 × 104 | 1.2 × 10−3 | 2.2 × 10−9 |
| Full-length (LVAEF → AAAAA) | 7 | 67 × 104 | 1.7 × 10−3 | 2.5 × 10−9 |
| Full-length (LVAEF → AAAAA, W118A, W140A) | 5 | 65 × 104 | 2.8 × 10−3 | 4.3 × 10−9 |
To analyze and compare the kinetic properties for the interaction of the novel and canonical Pex14-binding motifs, fragments containing mutations disrupting either the novel Pex14-binding site or the WXXX(F/Y) motif (WAQEF) were analyzed (Fig. 5B). The kinetic binding constants of monovalent complex formation were assessed by fitting the surface plasmon resonance binding curves assuming the 1:1 Langmuir binding model.
Inactivation of the Pex14-binding W1 motif was achieved by substitution of the tryptophan by alanine (12). The LVXEF-containing Pex5-(1–131) (with inactive W1) exhibited similar binding kinetics as the shorter fragment Pex5-(1–117) (Table 3). To study the kinetics of W1-containing Pex5-(1–131), the newly identified LVXEF motif was replaced by penta-alanine. As shown by in vitro binding assays, this mutation abolishes the monovalent interaction between N-terminal fragments of Pex5 and Pex14 (Fig. 6).
FIGURE 6.
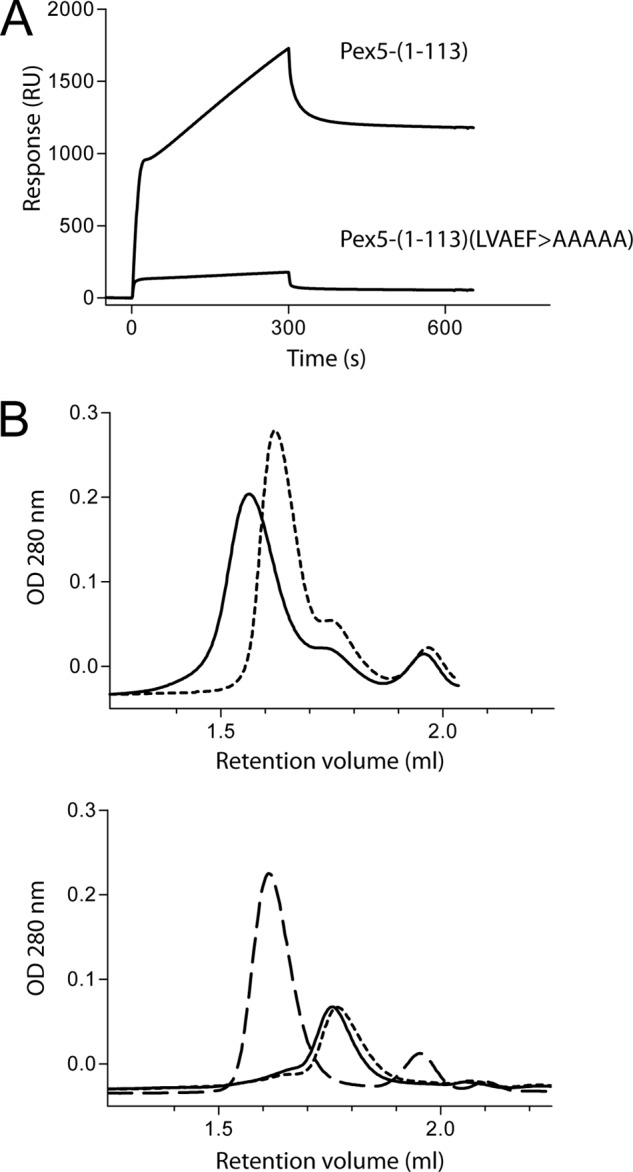
Substitution of LVXEF motif with AAAAA inhibits in vitro binding of an N-terminal fragment of Pex5 to the N-terminal domain of Pex14. A, surface plasmon resonance spectroscopy shows that Pex5-(1–113) binds to immobilized GST-tagged N-terminal domain of Pex14 (GST-Pex14-NTD), although Pex5-(1–113)(LVAEF > AAAAA) lacking the LVXEF motif does not. For the binding experiments, the concentration of the Pex5 fragments was 50 nm. B, isolated N-terminal domain of Pex14 (Pex14-NTD) was preincubated with wild type or the mutant Pex5-(1–113), which lacks the LVXEF motif, and the mixtures were analyzed by size exclusion chromatography. The curves in the upper panel were recorded after applying mixtures of GST-Pex14-NTD either with Pex5-(1–113) (solid line) or with Pex5-(1–113)( LVAEF > AAAAA) (dotted line). The data show that incubation of GST-Pex14-NTD with the Pex5-(1–113) results in a shift to higher molecular weight, indicative of complex formation. This shift is not observed with the mutated Pex5 fragment Pex5-(1–113)(LVAEF > AAAAA). As control, the lower panel displays gel filtration profiles of the purified fragments in the absence of binding partners: GST-Pex14-NTD (dashed line), Pex5-(1–113) (solid line), and Pex5-(1–113)(LVAEF > AAAAA) (dotted line). Optical density, OD, of the column effluent was monitored at 280 nm.
The fragment containing the functional WXXX(F/Y) motif displayed a 3-fold slower association rate (kon) and a 33-fold reduced dissociation rate (koff) compared with the same fragment containing only the functional LVXEF motif (Table 3). Thus, the novel LVXEF motif exhibits significantly faster binding kinetics with Pex14 than the adjacent WXXX(F/Y) motif.
Functional Analysis of the Novel Pex14-binding Site
To study the physiological relevance of the LVXEF motif, we replaced these amino acids with five alanines in the context of full-length Pex5 and tested its ability to rescue the peroxisome import defect of Pex5-deficient fibroblast cells. For this purpose, a pIRES vector was constructed, which encodes a bicistronic mRNA for full-length Pex5 harboring the respective point mutations and EGFP fused with the peroxisomal matrix protein targeting sequence –SKL (EGFP-PTS1) at its C terminus. 48 h after transfection, cells were inspected by fluorescence microscopy. A peroxisomal punctate pattern indicates either partial or full complementation of the import phenotype (Fig. 7A). Wild-type Pex5 rescued the PTS1 import defect of most of the transfected cells (82%), whereas the LVAEF → AAAAA mutant restored peroxisomal protein import in only 43% of the cells. Thus, most cells exhibited no punctate staining, indicative of a complete mislocalization of the peroxisomal marker protein to the cytosol and thus a severe import defect. Moreover, almost all cells that still exhibited a punctate staining upon expression of the LVAEF → AAAAA mutant displayed a partial import defect with strong cytosolic background (Fig. 7A, lower panel). The data indicate that inactivation of the novel Pex14-binding motif strongly affects the efficiency of peroxisomal protein. Importantly, the inactivation of the LVXEF motif (LVAEF → AAAAA) showed a more severe effect on matrix protein import than mutations of the adjacent first WXXX(F/Y) motif (W118A) or the second WXXX(F/Y) motif (W140A). When these mutations were combined with mutation of the LVAEF sequence (LVAEF → AAAAA, W118A, W140A), the complementing activity of the triple mutant protein was almost completely abolished (Fig. 7A). Remarkably, the severe import defect of the triple mutation does not correlate with a notable loss of affinity with Pex14 as indicated by surface plasmon resonance binding experiments with purified proteins (Table 3). Fig. 6B shows that the different complementing activity of wild-type and mutant Pex5 is not due to different steady-state levels of the proteins.
FIGURE 7.
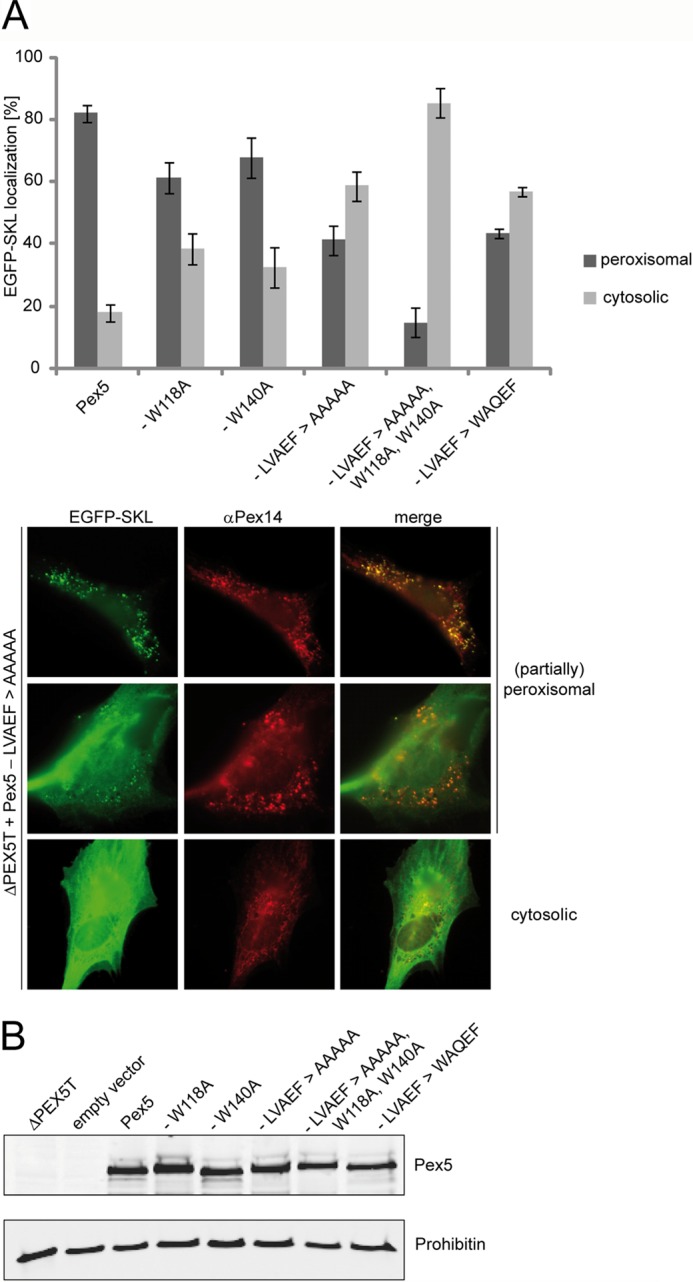
Functional complementation of PEX5-deficient fibroblasts by Pex5 carrying mutations in distinct Pex14-binding sites. Full-length Pex5 was mutagenized either in single Pex14-binding sites by alanine substitution of the LVXEF motif (LVAEF > AAAAA), the first (W118A), or the second (W140A) WXXX(F/Y) motif, or in combination (LVAEF > AAAAA, W118A,W140A). Furthermore, the LVXEF motif was substituted by the first WXXX(F/Y) motif, indicated by LVAEF > WAQEF. All constructs were expressed in Pex5-deficient fibroblasts (ΔPEX5T) from bicistronic expression vectors coding for the full-length Pex5 variants as indicated and EGFP fused to a PTS1 (EGFP-SKL). A, localization of the peroxisomal matrix marker protein EGFP-SKL was monitored by immunofluorescence microscopy using antibodies against EGFP. Peroxisomes were labeled in red by immunofluorescence microscopy of the peroxisomal membrane marker Pex14. Representative cells expressing the Pex5-LVAEF > AAAAA mutant are shown in the lower panel. A congruent punctate red and green fluorescent pattern indicates partial or complete restoration of peroxisomal protein transport, whereas diffuse cytosolic staining of EGFP-SKL shows that the protein is at least partially mislocalized to the cytosol, indicating a low complementation activity of transfected Pex5 variant. The graph shows a quantitative analysis of mutant phenotype complementation by the individual Pex5 variants. Values were obtained from three or six independent transfection experiments. For each experiment, 100 transfected cells were categorized into those enabling EGFP-PTS1 import (peroxisomal) and those without complementation of the import defect (cytosolic). B, immunoblot analysis shows that all Pex5 variants are present at the same steady-state level as the plasmid-encoded full-length wild-type Pex5. Negative controls show lysates of cells either nontransfected or transfected with empty vector (pIRES2). The analysis of prohibitin served as loading control.
We next asked whether a canonical WXXX(F/Y) sequence can functionally replace the novel monoaromatic LVXEF motif. To this end, the residues Leu-62, Val-63, and Ala-64 of the LVAEF sequence were substituted by tryptophan, alanine, and glutamine. The corresponding penta-peptide sequence (62WAQEF66) matches exactly the sequence of the N-terminal WXXX(F/Y) motif. Remarkably, this mutant displays a similar phenotype as the Pex5 variant that lacks a functional Pex14-binding site at this position (Fig. 7A). Most of the cells transfected with the substitution mutant (LVAEF → WAQEF) exhibit a severe import defect. Apparently, a WXXX(F/Y) motif cannot fully account for the LVXEF motif, indicating that not only the presence of a Pex14-binding site at this N-terminal position determines import efficiency but that also the kinetic properties of the binding site play a critical role. Replacement of the first WXXX(F/Y) motif by LVAEF did not affect the complementing activity of full-length Pex5 or of the variants Pex5-(LVAEF → AAAAA) and Pex5-(LVAEF → WAQEF) (data not shown). This further supports the view that the fast on/off rates of Pex14 binding are only critical at the N-terminal position.
DISCUSSION
The highly conserved N-terminal domain of Pex14 possesses a much broader binding specificity for peptide ligands as anticipated. Originally, the sequence signature of the core binding region has been defined as WXX(QED)(FY) based on the analysis of human Pex5-Pex14 interaction (12). However, the existence of 2–9 WXXX(F/Y) motifs in each of the known Pex5 sequences can neither predict that the regions containing the consensus sequences indeed interact with Pex14 nor does it exclude that additional Pex14-binding sites exist. By using a peptide scan approach, we here identified all linear Pex14-binding sites of human Pex5 experimentally. Besides the seven WXXX(F/Y) motifs, we detected a novel site that includes Pex5 residues 62–66 with the sequence LVAEF. NMR analysis showed that similar to WXXX(F/Y) motifs, the novel penta-peptide motif adopts an amphipathic α-helix where the phenylalanine residue is recognized by one of the two hydrophobic pockets in Pex14-NTD. However, compared with canonical WXXX(F/Y) ligands, the Leu and Val residues do not completely occupy the corresponding pocket but form alternative hydrophobic interactions with residues in the proximity. Furthermore, our structural and mutational analyses suggest that electrostatic interactions may play a more important role in binding of Pex14 to LVXEF compared with the canonical WXXX(F/Y) motif. The length and helical conformation show that the novel LVXEF motif is a variant of the WXXX(F/Y) motif that binds to the same surface of Pex14. The LVXEF ligand has a comparable affinity to human Pex14 (KD around 100 nm) as the seven WXXX(F/Y) motifs of Pex5 (KD values ranging from 3 to 150 nm) (12, 16).
Interestingly, comparison of the newly identified LVXEF motif with the first WXXX(F/Y) motif at amino acids positions 118–122 of human Pex5 revealed distinct binding kinetics. Most remarkably, the LVXEF motif exhibits a 33 times higher dissociation rate when compared with a diaromatic Pex14 ligand (Fig. 5B and Table 3). One possible explanation for these differences could be that the hydrophobic pocket in the binding surface of Pex14 is better suited to accommodate a single tryptophan residue as in the WXXX(F/Y) motif than the two amino acids Leu and Val of the LVAEF sequence.
Alanine substitution of the hydrophobic residues or the whole penta-peptide sequence of the LVXEF motif disrupts interaction with Pex14-NTD. However, site-directed mutagenesis of the new motif in full-length constructs of Pex5 does not significantly disturb Pex14-NTD interaction in terms of affinity and kinetic properties (Table 3). These in vitro results could be expected because each of the remaining WXXX(F/Y) motifs per se can act as high affinity ligands for Pex14. Thus, a triple mutant of full-length Pex5 lacking the novel motif and two adjacent WXXX(F/Y) motifs retains full affinity with Pex14-NTD (Table 3). In contrast, the same triple mutant expressed in Pex5-deficient cells almost completely failed to rescue the import defect (Fig. 7A). In fact, comparison of the complementing activities of single site mutants revealed that mutagenesis of the LVXEF motif has the most severe effect on the protein import into peroxisomes.
The in vivo results indicate a critical role for the new Pex14-binding site, which cannot be efficiently substituted by other Pex5 WXXX(F/Y) motifs even with higher affinity for Pex14. A possible mechanistic role for the new motif could be derived from the fast kinetics of LVXEF interaction with Pex14. In comparison with WXXX(F/Y) motifs, the LVXEF ligand associates three times faster, and the dissociation rate is even 33 times faster. Accordingly, it is tempting to speculate that the novel Pex14-interacting site in vivo also establishes the first contact between the PTS1 receptor and its docking protein Pex14. On the basis of the relatively high dissociation rate of the LVXEF·Pex14 complex, it seems possible that after initial contact, Pex14 is then transferred to other WXXX(F/Y) motifs with higher affinity for further processing of the PTS1 receptor at the peroxisomal membrane. The hypothesis that the LVXEF sequence serves as the initial docking and loading site is supported by in vivo studies. Intriguingly, replacement of the novel motif in human Pex5 with a high affinity WXXX(F/Y) motif impairs the function of Pex5 in the same way as its substitution with a sequence that does not interact with Pex14 at all. This further suggests that the structural and kinetic features of the LVXEF motif but not simply the presence of a Pex14-binding site at the N terminus of Pex5 are important for the function of the receptor in matrix protein import.
In conclusion, our data clearly demonstrate that human Pex5 contains eight Pex14-binding sites, and each of these motifs per se is a high affinity ligand for the peroxisomal docking protein Pex14. An intriguing question is whether these eight motifs can interact with Pex14 in a sequential manner or whether simultaneous binding of these motifs would form higher order complexes of Pex14 and Pex5. The presence of multiple, partially redundant binding sites could be advantageous for docking of the PTS1 receptor by increasing the avidity of Pex5-Pex14 interaction. However, this work and other recent data suggest distinct roles of the different Pex14-binding sites in Pex5 during protein import.
Although still speculative, we propose a mechanistic model enabling sequential binding of the receptor during the protein import cascade (Fig. 8). In this model, the new motif serves as a preferred docking site for cargo-loaded PTS1 receptor. Dissociation and sliding along the Pex5 polypeptide chain could initiate further binding of Pex14 to WXXX(F/Y) motifs 1–5 resulting in the formation of a multimeric Pex5-Pex14 complex that presumably represents the protein-conducting channel enabling matrix protein translocation across the peroxisomal membrane. Interestingly, a stable subcomplex consisting of Pex5 and Pex14 with a 1:5 stoichiometry could be isolated from rat liver peroxisomes (6). Finally, Pex14 binding to the downstream-located WXXX(F/Y) motifs six and seven triggers cargo release (10). Pex13 might also play a critical role in pore assembly or disassembly because it was shown that WXXX(F/Y) motifs 2–4 of Pex5 are able to interact with this peroxin (13). To test the model, future studies need to be initiated to characterize the membrane topology of Pex5 mutants defective in one or more Pex14-binding sites.
FIGURE 8.
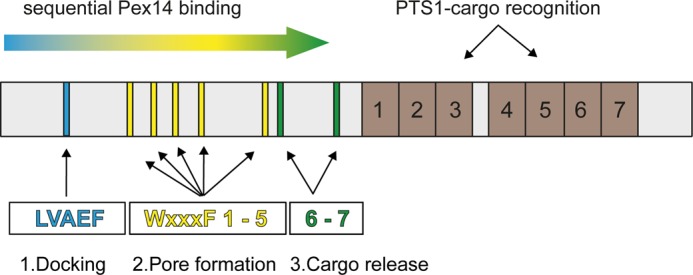
Sliding model of the Pex5-Pex14 interaction. Although PTS1-containing peroxisomal matrix proteins bind to the C-terminal seven tetratricopeptide repeat domains of Pex5 (brown boxes, 1–7), Pex14 interacts via its N-terminal domain with eight penta-peptide motifs located in the N-terminal half of the long isoform of human Pex5 (cyan, yellow and green bars). 1, N-terminal LVXEF motif (LVAEF, cyan) is supposed to make the first contact to Pex14 and serves as the initial membrane docking site of the receptor-cargo complex. 2, upon dissociation, Pex14 can associate with one of the adjacent WXXX(F/Y) motifs W1 to W5, and the novel LVXEF motif can recruit another Pex14. Sequential uploading of W1 to W5 correlates with structural changes of the Pex5-cargo·Pex14 complex and may result in the formation of a transient protein conducting channel in the peroxisomal membrane. 3, finally, interaction of the WXXX(F/Y) motifs W6 and W7 with Pex14-NTD enables cargo release into the peroxisomal matrix.
Acknowledgments
We thank Elisabeth Becker for technical assistance and Philipp Wortmann (Technische Universität München) for NMR data analysis.
This work was supported in part by Deutsche Forschungsgemeinschaft Grants FOR1905 (to R. E. and M. S.), GRK1721 (to M. S.), and SFB594 (to M. S.) and European Commission Grant FP7 NMI3, 226507 (to M. S.).
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (code 4BXU) have been deposited in the Protein Data Bank (http://wwpdb.org/).
The NMR chemical shifts and restraints have been deposited in the Biological Magnetic Resonance Data Bank under accession number 19368.
- PTS
- peroxisomal targeting signal
- NTD
- N-terminal domain
- PRE
- paramagnetic relaxation enhancement
- ITC
- isothermal titration calorimetry
- EGFP
- enhanced GFP.
REFERENCES
- 1. Ma C., Agrawal G., Subramani S. (2011) Peroxisome assembly: matrix and membrane protein biogenesis. J. Cell Biol. 193, 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nuttall J. M., Motley A., Hettema E. H. (2011) Peroxisome biogenesis: recent advances. Curr. Opin. Cell Biol. 23, 421–426 [DOI] [PubMed] [Google Scholar]
- 3. Platta H. W., Hagen S., Erdmann R. (2013) The exportomer: the peroxisomal receptor export machinery. Cell. Mol. Life Sci. 70, 1393–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rucktäschel R., Girzalsky W., Erdmann R. (2011) Protein import machineries of peroxisomes. Biochim. Biophys. Acta 1808, 892–900 [DOI] [PubMed] [Google Scholar]
- 5. Schäfer A., Kerssen D., Veenhuis M., Kunau W. H., Schliebs W. (2004) Functional similarity between the peroxisomal PTS2 receptor binding protein Pex18p and the N-terminal half of the PTS1 receptor Pex5p. Mol. Cell. Biol. 24, 8895–8906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gouveia A. M., Guimarães C. P., Oliveira M. E., Sá-Miranda C., Azevedo J. E. (2003) Insertion of Pex5p into the peroxisomal membrane is cargo protein-dependent. J. Biol. Chem. 278, 4389–4392 [DOI] [PubMed] [Google Scholar]
- 7. Albertini M., Rehling P., Erdmann R., Girzalsky W., Kiel J. A., Veenhuis M., Kunau W.-H. (1997) Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell 89, 83–92 [DOI] [PubMed] [Google Scholar]
- 8. Bottger G., Barnett P., Klein A. T., Kragt A., Tabak H. F., Distel B. (2000) Saccharomyces cerevisiae PTS1 receptor Pex5p interacts with the SH3 domain of the peroxisomal membrane protein Pex13p in an unconventional, non-PXXP-related manner. Mol. Biol. Cell 11, 3963–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meinecke M., Cizmowski C., Schliebs W., Krüger V., Beck S., Wagner R., Erdmann R. (2010) The peroxisomal importomer constitutes a large and highly dynamic pore. Nat. Cell Biol. 12, 273–277 [DOI] [PubMed] [Google Scholar]
- 10. Freitas M. O., Francisco T., Rodrigues T. A., Alencastre I. S., Pinto M. P., Grou C. P., Carvalho A. F., Fransen M., Sá-Miranda C., Azevedo J. E. (2011) PEX5 protein binds monomeric catalase blocking its tetramerization and releases it upon binding the N-terminal domain of PEX14. J. Biol. Chem. 286, 40509–40519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schliebs W., Saidowsky J., Agianian B., Dodt G., Herberg F. W., Kunau W. H. (1999) Recombinant human peroxisomal targeting signal receptor PEX5. Structural basis for interaction of PEX5 with PEX14. J. Biol. Chem. 274, 5666–5673 [DOI] [PubMed] [Google Scholar]
- 12. Saidowsky J., Dodt G., Kirchberg K., Wegner A., Nastainczyk W., Kunau W. H., Schliebs W. (2001) The di-aromatic penta-peptide repeats of the human peroxisome import receptor PEX5 are separate high affinity binding sites for the peroxisomal membrane protein PEX14. J. Biol. Chem. 276, 34524–34529 [DOI] [PubMed] [Google Scholar]
- 13. Otera H., Setoguchi K., Hamasaki M., Kumashiro T., Shimizu N., Fujiki Y. (2002) Peroxisomal targeting signal receptor Pex5p interacts with cargoes and import machinery components in a spatiotemporally differentiated manner: Conserved Pex5p WXXXF/Y motifs are critical for matrix protein import. Mol. Cell. Biol. 22, 1639–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stanley W. A., Sokolova A., Brown A., Clarke D. T., Wilmanns M., Svergun D. I. (2004) Synergistic use of synchrotron radiation techniques for biological samples in solution: a case study on protein-ligand recognition by the peroxisomal import receptor Pex5p. J. Synchrotron Radiat. 11, 490–496 [DOI] [PubMed] [Google Scholar]
- 15. Shiozawa K., Konarev P. V., Neufeld C., Wilmanns M., Svergun D. I. (2009) Solution structure of human Pex5·Pex14·PTS1 protein complexes obtained by small angle x-ray scattering. J. Biol. Chem. 284, 25334–25342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Neufeld C., Filipp F. V., Simon B., Neuhaus A., Schüller N., David C., Kooshapur H., Madl T., Erdmann R., Schliebs W., Wilmanns M., Sattler M. (2009) Structural basis for competitive interactions of Pex14 with the import receptors Pex5 and Pex19. EMBO J. 28, 745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Su J. R., Takeda K., Tamura S., Fujiki Y., Miki K. (2009) Crystal structure of the conserved N-terminal domain of the peroxisomal matrix protein import receptor, Pex14p. Proc. Natl. Acad. Sci. U.S.A. 106, 417–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kerssen D., Hambruch E., Klaas W., Platta H. W., de Kruijff B., Erdmann R., Kunau W. H., Schliebs W. (2006) Membrane association of the cycling peroxisome import receptor Pex5p. J. Biol. Chem. 281, 27003–27015 [DOI] [PubMed] [Google Scholar]
- 19. Madrid K. P., Jardim A. (2005) Peroxin 5-peroxin 14 association in the protozoan Leishmania donovani involves a novel protein-protein interaction motif. Biochem. J. 391, 105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bharti P., Schliebs W., Schievelbusch T., Neuhaus A., David C., Kock K., Herrmann C., Meyer H. E., Wiese S., Warscheid B., Theiss C., Erdmann R. (2011) PEX14 is required for microtubule-based peroxisome motility in human cells. J. Cell Sci. 124, 1759–1768 [DOI] [PubMed] [Google Scholar]
- 21. Williams C., van den Berg M., Distel B. (2005) Saccharomyces cerevisiae Pex14p contains two independent Pex5p binding sites, which are both essential for PTS1 protein import. FEBS Lett. 579, 3416–3420 [DOI] [PubMed] [Google Scholar]
- 22. Choe J., Moyersoen J., Roach C., Carter T. L., Fan E., Michels P. A., Hol W. G. (2003) Analysis of the sequence motifs responsible for the interactions of peroxins 14 and 5, which are involved in glycosome biogenesis in Trypanosoma brucei. Biochemistry 42, 10915–10922 [DOI] [PubMed] [Google Scholar]
- 23. Dodt G., Braverman N., Wong C., Moser A., Moser H. W., Watkins P., Valle D., Gould S. J. (1995) Mutations in the PTS1 receptor gene, PXR1, define complementation group 2 of the peroxisome biogenesis disorders. Nat. Genet. 9, 115–125 [DOI] [PubMed] [Google Scholar]
- 24. Pires J. R., Hong X., Brockmann C., Volkmer-Engert R., Schneider-Mergener J., Oschkinat H., Erdmann R. (2003) The ScPex13p SH3 domain exposes two distinct binding sites for Pex5p and Pex14p. J. Mol. Biol. 326, 1427–1435 [DOI] [PubMed] [Google Scholar]
- 25. Douangamath A., Filipp F. V., Klein A. T., Barnett P., Zou P., Voorn-Brouwer T., Vega M. C., Mayans O. M., Sattler M., Distel B., Wilmanns M. (2002) Topography for independent binding of α-helical and PPII-helical ligands to a peroxisomal SH3 domain. Mol. Cell 10, 1007–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hilpert K., Winkler D. F., Hancock R. E. (2007) Peptide arrays on cellulose support: SPOT synthesis, a time and cost efficient method for synthesis of large numbers of peptides in a parallel and addressable fashion. Nat. Protoc. 2, 1333–1349 [DOI] [PubMed] [Google Scholar]
- 27. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 28. Sattler M., Schleucher J., Griesinger C. (1999) Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. NMR Spectrosc. 34, 93–158 [Google Scholar]
- 29. Huybrechts S. J., Van Veldhoven P. P., Hoffman I., Zeevaert R., de Vos R., Demaerel P., Brams M., Jaeken J., Fransen M., Cassiman D. (2008) Identification of a novel PEX14 mutation in Zellweger syndrome. J. Med. Genet. 45, 376–383 [DOI] [PubMed] [Google Scholar]
- 30. Simon B., Madl T., Mackereth C. D., Nilges M., Sattler M. (2010) An efficient protocol for NMR spectroscopy-based structure determination of protein complexes in solution. Angew. Chem. Int. Ed. Engl. 49, 1967–1970 [DOI] [PubMed] [Google Scholar]
- 31. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Linge J. P., Williams M. A., Spronk C. A., Bonvin A. M., Nilges M. (2003) Refinement of protein structures in explicit solvent. Proteins 50, 496–506 [DOI] [PubMed] [Google Scholar]
- 33. Goddard. T. D., Kneller D. G. (2007) SPARKY, Version 3, University of California, San Francisco [Google Scholar]