Background: Structural aspects of tristetraprolin (TTP) binding to mRNA are poorly understood.
Results: RNA binding can be differentially affected by sequence-equivalent residues within the tandem zinc fingers of TTP.
Conclusion: Residues at sequence-equivalent positions within each zinc finger may not be structurally or functionally equivalent.
Significance: Different TTP family members may have structural features that dictate unique modes of interacting with RNA.
Keywords: Cytokine, Inflammation, RNA-binding Protein, RNA Turnover, RNA-Protein Interaction, Tumor Necrosis Factor (TNF), AU-rich Elements
Abstract
Tristetraprolin (TTP), the best known member of a class of tandem (R/K)YKTELCX8CX5CX3H zinc finger proteins, can destabilize target mRNAs by first binding to AU-rich elements (AREs) in their 3′-untranslated regions (UTRs) and subsequently promoting deadenylation and ultimate destruction of those mRNAs. This study sought to determine the roles of selected amino acids in the RNA binding domain, known as the tandem zinc finger (TZF) domain, in the ability of the full-length protein to bind to AREs within the tumor necrosis factor α (TNF) mRNA 3′-UTR. Within the CX8C region of the TZF domain, mutation of some of the residues specific to TTP, not found in other members of the TTP protein family, resulted in decreased binding to RNA as well as inhibited mRNA deadenylation and decay. Evaluation of simulation solution models revealed a distinct structure in the second zinc finger of TTP that was induced by the presence of these TTP-specific residues. In addition, mutations within the lead-in sequences preceding the first C of highly conserved residues within the CX5C or CX3H regions or within the linker region between the two fingers also perturbed both RNA binding and the simulation model of the TZF domain in complex with RNA. We conclude that, although the majority of conserved residues within the TZF domain of TTP are required for productive binding, not all residues at sequence-equivalent positions in the two zinc fingers of the TZF domain of TTP are functionally equivalent.
Introduction
Humans express three members of the tristetraprolin (TTP)2 family of CCCH tandem zinc finger proteins: TTP (also known as ZFP36, Gos24, Tis11, and Nup475) (1–5); ZFP36L1 (cMG1, TIS11b, BRF1, and ERF1) (6, 7); and ZFP36L2 (TIS11d, BRF2, and ERF2) (7). A fourth family member, ZFP36L3 (8), has been identified only in rodents to date. Mammalian TTP family member proteins contain two CCCH motifs with characteristic internal spacing of CX8CX5CX3H; the two zinc fingers are separated by a 12-amino acid linker, and each is preceded by a lead-in sequence of (R/K)YKTE(L/P).
The physiological role of TTP began to be elucidated with the evaluation of TTP-knock-out (KO) mice, which developed an inflammatory syndrome that was eventually found to result from chronic excess of tumor necrosis factor α (TNF) (9–11). TTP was found to bind to AU-rich elements (AREs) within the 3′-untranslated region (3′-UTR) of TNF mRNA, leading to destabilization of the transcript (10, 12). Thus, the lack of TTP in macrophages derived from TTP-KO mice led to the increased stability of TNF mRNA and resulted in the excessive production of TNF protein (10). Additional mRNA targets, including GM-CSF, have been identified using cells from TTP-KO mice as well as a variety of other techniques (13–20).
Disruption of the genes encoding other TTP family members revealed other distinct phenotypes. For example, disruption of Zfp36l1 resulted in failure of chorio-allantoic fusion and embryonic lethality (21), whereas disruption of Zfp36l2 resulted in a severe hematopoietic defect and death within several weeks of birth (22). Both ZFP36L1 and ZFP36L2, as well as the more recently described rodent placental protein ZFP36L3, bind to similar ARE-containing sequences and promote the deadenylation and destabilization of ARE-containing transcripts in various assays (8, 23, 24).
The tandem zinc finger (TZF) domain of TTP has been established as the ARE binding domain (25–27). In particular, a previous study measured a Kd of 3.2 nm (at 24 °C) for a synthetic human TTP TZF domain peptide bound to a single binding site fluorescent RNA probe containing the optimum target sequence UUAUUUAUU (27). The integrity of the zinc fingers is critical for ARE binding because mutating any of the zinc-coordinating cysteines or histidines within either zinc finger of TTP leads to a complete loss of RNA binding activity (12, 25) and the subsequent inability to destabilize mRNA (28). The importance of various other amino acids within the TTP zinc fingers has also been demonstrated. For example, using the amino acid numbering for human TTP in GenBankTM accession number NP_003398.1,3 replacement of a single aromatic amino acid within the CX5C regions (Tyr120 in finger 1 or the equivalent residue Tyr158 in finger 2) or within the CX3H regions (Phe126 in finger 1 or the equivalent Phe164 in finger 2) with a non-aromatic amino acid resulted in the complete loss of TTP binding to RNA (25). Consistent with these findings, the NMR structure of the TZF domain of the TTP family member ZFP36L2 (PDB code 1RGO) (29), in complex with a nine-base core binding sequence, 1UUAUUUAUU9, highlighted the fact that the structural integrity of both CCCH zinc fingers is essential for ARE binding. In their structure, finger 1 interacted with the 6UAUU9 subsite, whereas finger 2 interacted with the 2UAUU5 subsite of a single RNA nonamer. The structure also revealed that the side chains of these aromatic residues within the TZF domain of ZFP36L2 stack with the RNA bases and explained the loss of binding observed upon mutation of aromatic residues in TTP. However, the importance of several other regions within the TZF domain of TTP to its RNA binding ability has yet to be determined.
The goal of this study was to elucidate some of the local structural aspects of the ability of TTP to bind to its RNA targets; this is the first, necessary step in ultimately modulating the stability of its target mRNAs. Sequence alignments of the TZF domains of TTP and related family members revealed that certain regions are highly conserved, such as the CX3H intervals within each finger and the lead-in sequences to each finger, whereas other positions can tolerate significant amino acid divergence. This is highlighted by the fact that the only existing structure for the TZF domain of a TTP family protein, that of human ZFP36L2 (PDB code 1RGO), contains several residues that differ from the analogous sequence positions within the TZF of TTP. Importantly, these amino acid differences occur in positions that have the potential to affect RNA binding and the local structure of the zinc fingers compared with that of ZFP36L2. Therefore, we have utilized sequence alignments and simulation solution models of the TZF domain of TTP in complex with RNA to guide a mutagenesis-based approach to evaluate the functional significance of the conserved and unique residues in TTP.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
Expression plasmids CMV.TTP.tag and its zinc finger mutants were constructed as described (12, 25). Unless otherwise specified, all TTP constructs and sequence numbers refer to human TTP; TTP mutants are numbered according to the GenBankTM RefSeq accession number for human TTP, NP_003398.1.3 The expression plasmid CMV.mTNFα(127–1325), and the cDNA transcript templates TNFα(1309–1332)(A)50/SK− and pA50/SK−, are described elsewhere (12, 28). The EGFP expression construct CMV.EGFP BGH3′ was made by releasing the HindIII-NotI fragment, containing the EGFP coding sequence, from pEGFP-N1 (Clontech) and ligating it with the HindIII and NotI-digested vector CMV.BGH3′/BS+ (12).
RNA Electrophoretic Mobility Shift Assay
The CMV.TTP.tag vector and its zinc finger mutants were expressed in HEK293 cells by transient transfection using the calcium phosphate precipitation method (25), and cytosolic extracts were prepared as described (28). Briefly, to each 100-mm plate of cells, 0.2 μg of CMV.TTP.tag or its zinc finger mutants and 4.8 μg of vector DNA (BS+) were added for a total amount of 5 μg of co-transfected DNA. Control extracts were prepared using 5 μg of vector (BS+) DNA/plate. Twenty-four h after removal of the transfection mixture, cytosolic extracts were prepared in a final concentration of 10 mm HEPES (pH 7.6), 40 mm KCl, and 20% glycerol and stored at −70 °C. Transfections were repeated several times to minimize variations in expression between different experiments. Extracts collected from each transfection were regarded as an individual set of samples. Western blotting was performed with an antibody directed at the HA epitope tag (HA-probe (F-7) HRP; Santa Cruz Biotechnology, Inc.). Within each independent set, relative protein expression from all the constructs was comparable, thus ensuring that the observed differences in binding did not arise due to differences in protein expression.
For the gel shift assays, a synthetic RNA oligonucleotide (probe TARE5, based on the AU-rich element within the mouse TNF mRNA 3′-UTR, GenBankTM accession number NM_013693.2, bp 1301–1332; see Fig. 3A for the sequence) was 3′-end-labeled with [32P]pCp and T4 RNA ligase. The labeled RNA (80 pmol) was eluted from a Quick Spin column (Roche Applied Science) and adjusted with labeling buffer to 0.8 pmol/μl. An initial set of assays to assess the apparent dissociation constant (Kd) of the probe at equilibrium was carried out using extracts from HEK293 cells transfected with vector alone (BS+) or CMV.TTP.tag (10 μg of protein/reaction) incubated with the labeled RNA probe (0.01–50 nm) in 10 mm HEPES (pH 7.6), 3 mm MgCl2, 40 mm KCl, and 5% (v/v) glycerol, in a final volume of 50 μl. After incubation at 20 °C for 30 min, the protein-RNA complexes formed were resolved on 8% non-denaturing acrylamide (37.5:1) gels. TTP-RNA complexes were quantitated using a PhosphorImager, and the probe-bound volume (volume quantitation by ImageQuant, Molecular Dynamics) was plotted against the concentrations of probe TARE5. Binding curves were generated using the GraphPad Prism (version 6) equation, Y = Bmax × Xh/(Kdh + Xh), where Bmax is the maximum specific binding, X is the probe concentration, Kd is the dissociation constant, and h is the Hill slope.
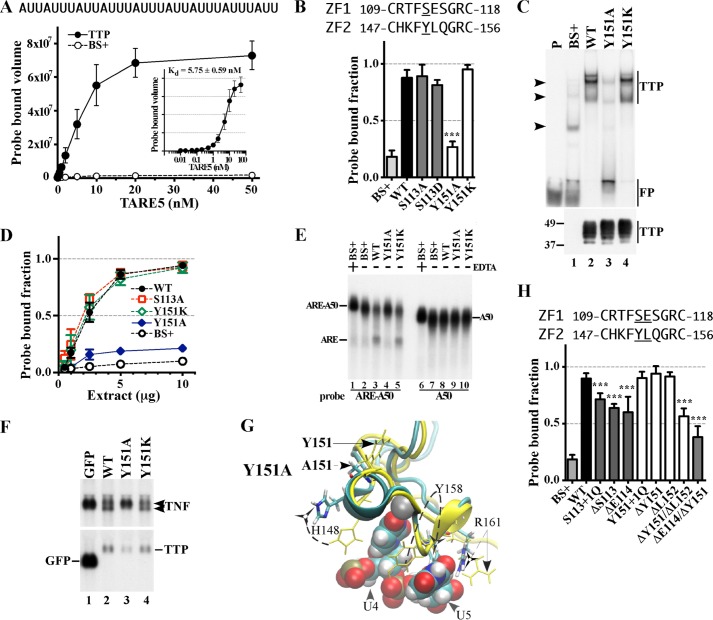
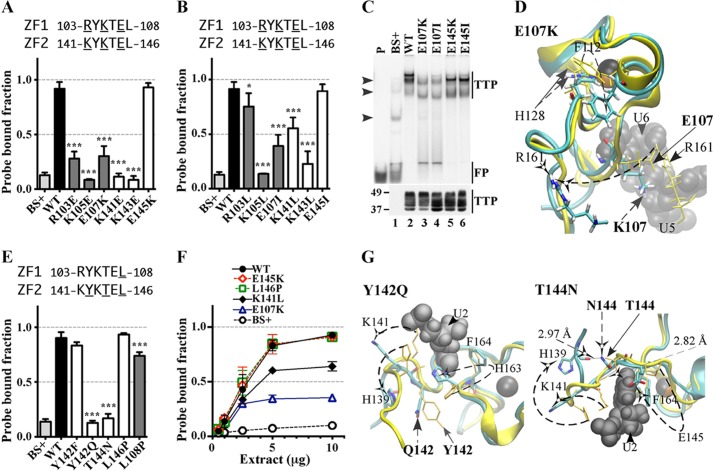
FIGURE 3.
Effect of mutated residues in the CX8C regions of the human TTP TZF domain on RNA binding and RNA stability. A, equilibrium binding of probe TARE5 (0.01–50 nm) to extracts of HEK293 cells transfected with TTP or vector (BS+) as described under “Experimental Procedures.” The sequence of the RNA probe is depicted above the graph. The inset shows the TTP probe binding curve, in which the x axis was transformed into antilog, and the apparent dissociation constant (Kd ± S.D.) was calculated from quantification of three independent experiments. B, the binding of protein to RNA is expressed as the probe-bound fraction. Above the graph are the amino acid sequences of the intervals. The underlined residues are those that were mutated and whose effects on binding are shown in the bar graph. The results from mutants of finger 1 are shown as gray columns, and those of finger 2 are shown as white columns. The results (mean ± S.D. (error bars)) are from three similar gel shift assays using 0.2 nm probe TARE5. C, the top panel shows representative gel shift assays using extracts from HEK293 cells transfected with vector alone (BS+), WT human TTP, or various mutant TTP constructs. Cytosolic extracts were incubated with 0.2 nm probe TARE5 as described under “Experimental Procedures” before loading on a non-denaturing acrylamide gel. A sample that contained probe alone in buffer was also loaded (lane P). The migration positions of the TTP-ARE complex (TTP) and the free probe (FP) are indicated to the right with vertical bars, and the three major endogenous protein-ARE complexes are labeled with arrowheads to the left. The bottom panel shows the relative amount of WT TTP protein and its mutants used in the binding reaction, as determined by Western blotting. D, binding assays with 0.2 nm TARE5 probe in serially diluted extract, where total protein ranges from 10 to 1 μg, were performed as described under “Experimental Procedures.” The probe-bound fractions (mean ± S.D.) are from three similar gel shift assays. E, shown is a deadenylation assay using these extracts (5 μg of protein/sample). In lanes 1 and 6, 20 mm EDTA was present during the incubation to inhibit the deadenylase activity. In lanes 1–5, the ARE-A50 probe was used. The full-length probe ARE-A50 and its deadenylated product ARE are indicated to the left. Probe A50 was used in the samples shown in lanes 6–10, and the migration of the probe is indicated to the right. F, Northern blots of a HEK293 cell co-transfection of CMV.TNF with control plasmid GFP (lane 1), WT TTP (lane 2), Y151A (lane 3), or Y151K (lane 4). Total cellular RNA was prepared from the cells, and 10 μg of RNA was used for each lane. The blots were hybridized with a TNF cDNA probe (top) or a mouse TTP cDNA probe and a GFP cDNA probe together (bottom). The migration positions of the TNF mRNA and the TTP mRNA are indicated to the right, and that of the GFP RNA is shown to the left. G, superposition of WT TTP finger 2 and its mutant Y151A structure ensembles. The wild-type TTP peptide backbone ribbon and side chains are in yellow, and the mutant Y151A peptide backbone ribbon is in cyan, with side chains in colors. Zinc atoms (superimposed) are shown as silver spheres. Also shown are ARE nucleosides U4 and U5 (RMSD value calculated from superposition of the TZF domains of Y151A with RNA-bound WT TTP is 8.98 Å). H, the binding of protein to RNA is expressed as the probe-bound fraction. The results (mean ± S.D.) are from three similar gel shift assays. The results from mutants of finger 1 are shown as gray columns, and those of finger 2 are shown as white columns. Other details are similar to those described in B.
In subsequent gel shift assays, extracts from HEK293 cells transfected with vector alone (BS+), CMV.TTP.tag, or zinc finger mutants (10 μg of protein/reaction, unless otherwise stated) were incubated with 0.2 nm TARE5 probe, and protein-RNA complexes were resolved and quantitated as described above. The initial probe input (0.2 nm) was also volume-quantitated. The results were expressed as probe-bound fractions (protein-RNA complex volume/input probe volume). The gel shift assays for vector alone, TTP, and mutants were performed several times using individual sets of extract samples collected independently. To compare the probe-bound fraction of each individual mutant to that of WT TTP (or of vector BS+), one-way analysis of variance, followed by a multiple-comparison post-test (Dunnett's test) were performed using GraphPad Prism (version 6). The results were expressed as average ± S.D. *, p = 0.01–0.05; **, p = 0.001–0.01; ***, p < 0.001. We also selected mutants with similar RNA-bound magnitudes as well as mutants with different RNA-bound magnitudes, compared with WT TTP, and performed gel shift assays with 0.2 nm TARE5 probe in serially diluted extract containing 1–10 μg of total protein.
Cell-free Deadenylation Assays
The templates for in vitro transcription were PCR-amplified from plasmids TNFα(1309–1332)(A)50/SK− or pA50/SK−, and the RNA probes were transcribed in the presence of [α-32P]UTP (800 Ci/mmol) and Ribo m7G Cap Analog (Promega), using the Promega Riboprobe in vitro transcription system protocol, as described (28). The in vitro transcribed products were purified from urea/TBE gels.
The in vitro deadenylation assays were performed as described (28). The reaction mixture, containing 5 μg of cell extract and 50,000 cpm of gel-purified RNA probe (2 nm per reaction) in 10 mm HEPES (pH 7.6), 3 mm MgCl2, 40 mm KCl, and 5% glycerol, in a final volume of 100 μl, was incubated at 37 °C for 60 min. The reaction was terminated by the addition of EDTA to achieve a final concentration of 20 mm. After phenol/chloroform extraction, 50 μl of the aqueous phase was added to 50 μl of 2× formamide stop solution and heated at 70 °C for 5 min. Aliquots of reaction products were analyzed on 6% acrylamide gels containing 7 m urea.
Northern Blotting
HEK293 cells were transiently transfected with CMV.mTNFα or other constructs in calcium phosphate precipitates as described previously (12). Each 100-mm dish of cells (0.8 × 106) was co-transfected with 1 μg of CMV.mTNFα and 10 ng of CMV.TTP.tag, zinc finger mutant, or control plasmid CMV.EGFP.BGH3′. Vector plasmid (BS+) was added to make the total amount of co-transfected DNA 5 μg for each plate. Twenty-four h after the removal of the transfection mixture, total cellular RNA was harvested from the HEK293 cells using the Illustra RNAspin mini-RNA isolation kit (GE Healthcare). Northern blots were hybridized to a random-primed, α-32P-labeled ∼1-kb NarI-BglII fragment of mTNF cDNA or a mouse TTP cDNA (1) together with the EGFP coding fragment.
Structural Models
Using molecular dynamics, simulation solution structures of the TTP TZF domain (in free and RNA-bound forms) and its mutants were generated. The initial structures were based on the RNA-bound structure of the TZF domain of ZFP36L2 (TIS11d) (PDB code 1RGO). Mutations were introduced into the sequence of ZFP36L2 using Coot (30) to generate a model of WT TTP in its RNA-bound form (with two CCCH-coordinated zinc ions) and energy-minimized using the program Amber, version 11 (31). The RNA TTP TZF domain complex was solvated in a box of water (about 9500 water molecules). A separate RNA-free WT TTP was also solvated in a box of water. Subsequently, the desired mutations in TTP were made, and the resulting mutant peptides were also solvated. Prior to equilibration, all systems were subjected to 1) 100-ps belly dynamics runs with fixed peptide, 2) minimization, 3) low temperature constant pressure dynamics at fixed protein to assure a reasonable starting density, 4) minimization, 5) stepwise heating molecular dynamics at constant volume, and 6) constant volume molecular dynamics for 1 ns. All final unconstrained trajectories were calculated at 300 K under constant volume (15 ns, time step 1 fs) using PMEMD (Amber version 11) to accommodate long range interactions (31). The parameters were taken from the FF03 force field (32) for the protein and the PARMBSC0 force field (33) for the RNA. The charge on zinc ions was +2, whereas cysteine coordinated with zinc carried a −1 charge. His coordinated to zinc remained neutral. The van der Waals parameters of zinc were adjusted to yield Zn–S and Zn–N distances of between 2.25 and 2.50 Å. No specific constraints were applied to maintain the zinc coordination. RMSD values were calculated using the ptraj module (Amber version 11). Images of molecular models were created using VMD.1.9.1 (34).
RESULTS
Sequence and Structural Features of the TTP TZF Domain
TTP family members are found from yeasts to humans. Although regions outside the TZF domain are highly variable, there is significant conservation of the TZF domain that most likely corresponds with its critical function as the binding partner for RNA. To gain insight into sequence features of human TTP that were identical to and distinct from those of other family members, we aligned the TZF domains from various TTP family members (Fig. 1). The numbering system used in this study is based on the human GenBankTM RefSeq accession number NP_003398.1.3
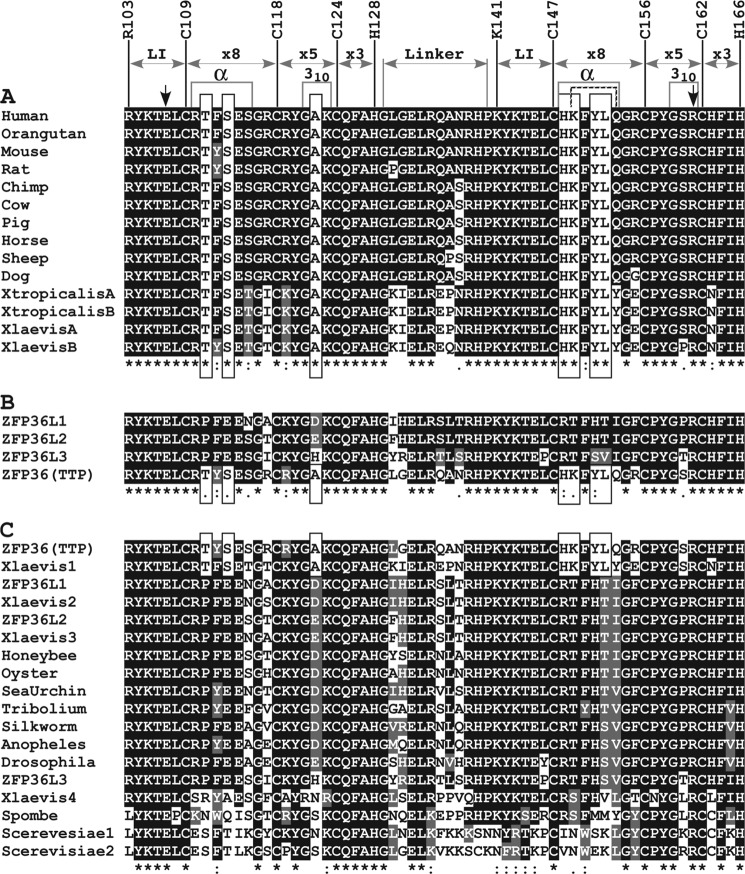
FIGURE 1.
Alignments of the tandem CCCH zinc fingers of TTP family members. A, an alignment of TZF domains from vertebrate TTP orthologues that were readily available using RefSeq proteins and expressed sequence tags. The numbers above the human TTP TZF sequence represent the residue position in NP_003398.1, and only the first residues of the lead-in sequence to each finger and the CCCH residues are indicated. The following are also indicated: the lead-in (LI) sequences to each finger; the interval distances within the CCCH motifs and the linker sequence; and the locations of those residues that form the helices (29). Note that in TTP the α-helix of finger 2 is indicated by the hatched lines. The two downward arrows indicate residues Glu107 and Arg161, whose ionic interaction stabilizes the two fingers (see Fig. 4B). The seven residues specific to TTP are highlighted in white background boxes. B, an alignment of the TZF domains of the four TTP family members known to exist in mice. C, an alignment of the TZF domains of TTP family members that occur in various more distant eukaryotes. The TTP family members from mouse and X. laevis were used as representatives of vertebrate family members; a sampling of yeasts, insects, and aquatic invertebrates was added. Alignments were generated using ClustalW and BoxShade. Asterisks, amino acid identity at a site; colons, a high degree of chemical conservation at that site; single dots, a lower degree of chemical conservation at that site. The GenBankTM accession numbers for sequences used in this figure are as follows: for TTP, human (NP_003398.1), orangutan (XP_002834549.2), mouse (NP_035886), rat (NP_579824), chimpanzee (XP_001136016), cow (NP_776918), pig (NP_001161891), horse (translation of CD536573.1), sheep (NP_001009765), dog (XP_541624), Silurana tropicalis (NP_001106542), X. laevis (NP_001081884); for other mouse TTP family members, ZFP36L1 (NP_031590), ZFP36L2 (NP_001001806), ZFP36L3 (NP_001009549); for other X. laevis TTP family members, ZFP36L1 (NP_001084214), ZFP36L2 (NP_001080610), C3H-4 (NP_001081889); for other organisms, oyster unnamed protein (translation of AAB69448.1), sea urchin unnamed protein (XP_782811.1), Schizosaccharomyces pombe Zfs1p (NP_596453), CTH1 and CTH2 from S. cerevisiae (AAB39897 and AAB39898, respectively); insect family members that are orthologues of the Drosophila melanogaster protein Tis11 (NP_511141), Apis mellifera (NP_001121248.1), Tribolium castaneum (XP_968440.1), Bombyx mori (translation of AK382012.1), Anopheles gambiae (XP_309752.3).
We then compared the sequence of the TTP TZF domain with its orthologues in various vertebrate species, including Xenopus laevis and Silurana tropicalis (Fig. 1A); compared the mouse TTP sequence with the other three family members from the same species (Fig. 1B); and compared mouse TTP with representative non-mammalian family members from species including yeasts (Fig. 1C). These alignments revealed several features that are highly conserved and many that were variable. Of particular interest were the seven residues that are only found in TTP and in general were found in all TTP orthologues. These TTP-unique residues are highlighted in Fig. 1A and include Thr111, Ser113, Ala122, His148, Lys149, Tyr151, and Leu152. The influences of some of these unique amino acids on binding to RNA and the predicted structure of the TZF domain are discussed further below.
Next, we generated a simulation solution structure model of TTP based on the only existing structure of ZFP36L2 (TIS11d) in complex with RNA (PDB code 1RGO). The TZF domains of TTP and ZFP36L2 are highly conserved (72% amino acid identity), including the CCCH motifs, linker spacing between the two fingers, and lead-in sequences, thus making ZFP36L2 a good candidate to use for our model generation. The RMSD values calculated from superposition of the ZFP36L2 NMR structure (R152-A218) with the final solution simulation model of the RNA-bound WT TTP TZF domain (Arg103–Pro168) and from superposition of finger 1 or finger 2 of the two proteins were 2.17, 0.82, and 1.19 Å, respectively. Values of RMSD from the similar superposition of the RNA-free TZF domain of TTP with the RNA-bound form were also calculated (2.59, 1.15, and 0.85 Å). These results imply that 1) the RNA-bound TZF domains of the ZFP36L2 NMR structure and the TTP model are similar; 2) the backbone structures that are already formed in the presence of zinc (29) are not much influenced by the presence/absence of RNA; and 3) the relatively small RMSDs are to be expected when only 28 residues (zinc finger 1 of TTP versus zinc finger1 of ZFP36L2 and zinc finger 2 of TTP versus zinc finger 2 of ZFP36L2) are compared because zinc and the coordinating residues are keeping the peptide backbone in place.
Superposition of the individual zinc finger domains from ZFP36L2 (Fig. 2A) and the simulation model of TTP (Fig. 2B) revealed that they share most structural features, including the following. 1) Pockets for RNA binding in TTP are largely analogous to those found in ZFP36L2 (Table 1); 2) the positions of the hydrophobic residues participating in stacking with RNA are strictly conserved (Table 1); 3) residues coordinating zinc are strictly conserved (Fig. 1); and 4) the orientation of the two fingers relative to one another appears to be stabilized in both proteins by a salt bridge between conserved residues Glu107 in TTP (Glu157 in ZFP36L2 (29) in the lead-in sequence to finger 1; Fig. 1A) and Arg161 in TTP (Arg211 in ZFP36L2, in the CX5C interval of finger 2; Fig. 1 and Table 1; see also Fig. 4).
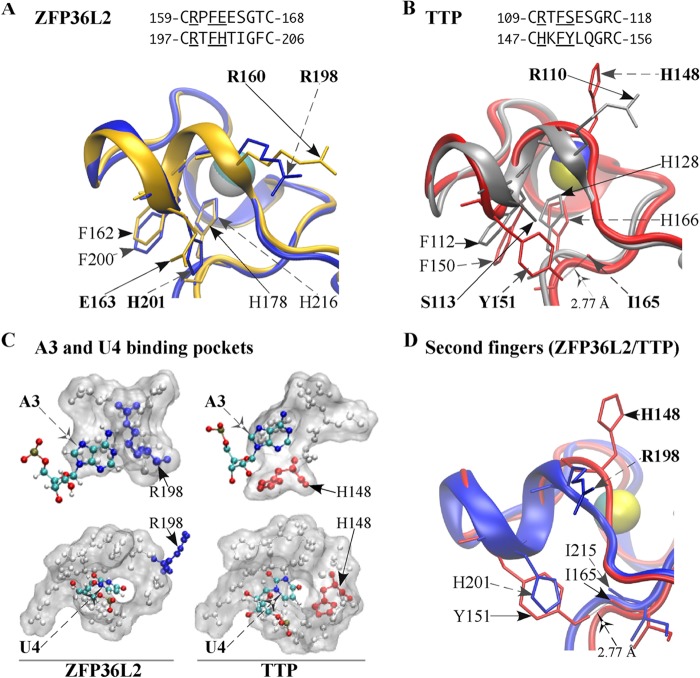
FIGURE 2.
Backbone superposition of TZF structural models of human ZFP36L2 (TIS11d) and TTP. The models shown were based on the initial RNA-bound structure of the TZF domain of ZFP36L2 (see “Experimental Procedures”). In A and B, finger 1 side chains are indicated by solid arrows, finger 2 side chains are indicated by dashed arrows, and zinc atoms are shown as spheres. A, superposition of the two fingers of ZFP36L2 (Arg153–Glu220). The ribbon diagram of the peptide backbone and selected side chains of finger 1 are depicted in gold, and those of finger 2 are in blue. Finger 1 side chains of Arg160 and Glu163, finger 2 side chains of Arg198 and His201, the zinc-coordinating residues His178/His216, and their respective stacking interacting residues Phe162/Phe200, are shown for both fingers. B, superposition of the two fingers of TTP (Arg103–Glu170). The ribbon diagram of the peptide backbone and selected side chains of finger 1 are depicted in silver, and those of finger 2 are in red. Finger 1 side chains of Arg110 and Ser113, finger 2 side chains of His148 and Tyr151, backbone of Ile165, the zinc-coordinating residues His128/His166, and their respective stacking interacting residues Phe112/Phe150 are shown for both fingers. The hydrogen bond formed by the side chain of Tyr151, and the backbone of Ile165 is also illustrated. C, surface representation of the binding pockets for the ARE bases A3 and U4. Nucleosides A3 and U4 are shown in colors. The side chain of Arg198 in finger 2 of ZFP36L2 is blue, and the side chain of His148 in finger 2 of TTP is red. Residues within the TZF domain of TTP or ZFP36L2 forming these binding pockets are listed in Table 1. D, Superposition of the second fingers of ZFP36L2 and TTP. The ribbon diagram of the peptide backbone and selected side chains of TTP finger 2 are shown in red, and those of ZFP36L2 are in blue. The hydrogen bond formed by the side chain of Tyr151 and the backbone of Ile165 is shown. Side chains of TTP shown are indicated by solid arrows, and side chains of ZFP36L2 are shown by dashed arrows. Zinc is displayed as spheres.
TABLE 1.
Residues that form the ARE base binding pockets in the TTP and ZFP36L2 TZF domains
The composition of the binding pockets for each RNA base of the RNA oligomer (1-UUAUUUAUU-9) was obtained from the TTP simulation solution model, with the equivalent residue in parentheses from the NMR structure of the TZF domain of ZFP36L2 (TIS11d; PDB code 1RGO) in complex with RNA. Binding pockets are made up largely of residues from equivalent sequence positions within the TZF domain of the two structures. The TZF domain of human TTP is comprised of residues 103–166 from GenBankTM accession number NP_003398.13; the corresponding residues of human ZFP36L2 (Tis11D) are 153–216 of NP_008818.3.
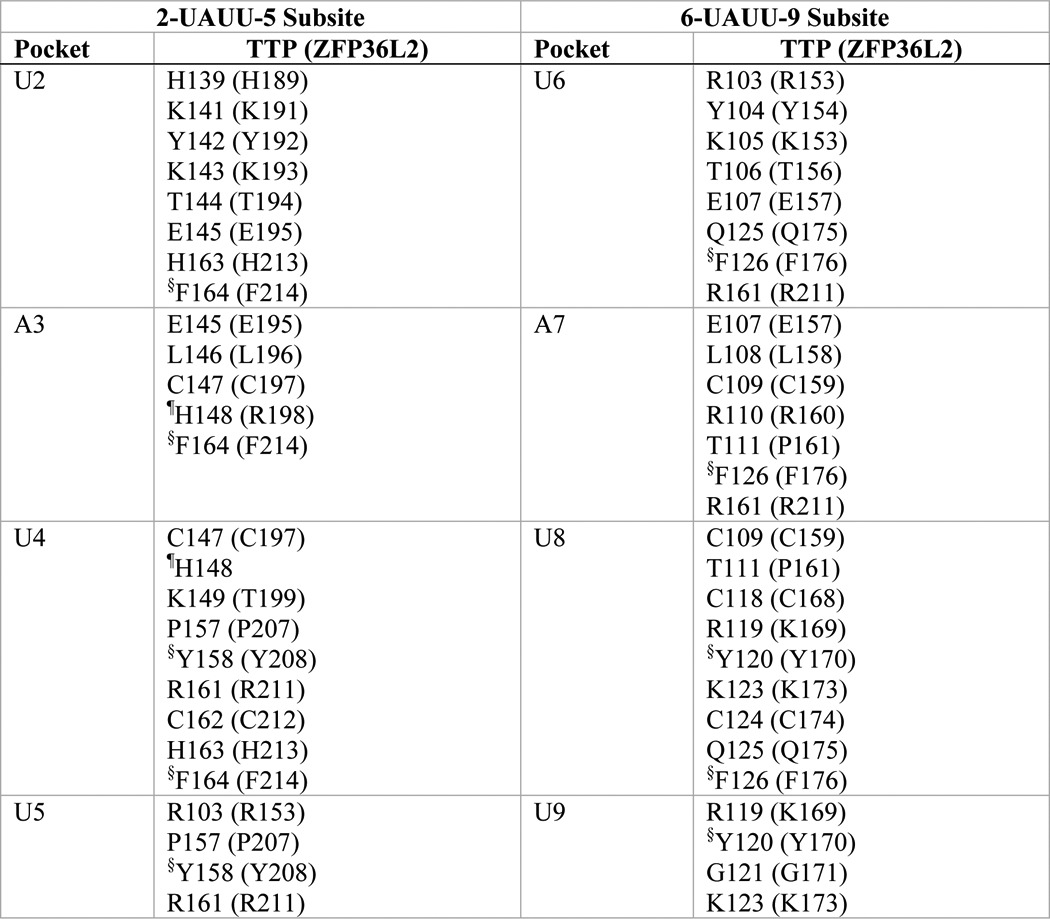
§ Side chain involved in stacking with the RNA bases: U2-Phe164-A3; U4-Tyr158-U5; U6-Phe126-A7; U8-Tyr120-U9 (in ZFP36L2: U2-Phe214-A3; U4-Tyr208-U5; U6-Phe176-A7; U8-Tyr170-U9).
¶ His148 of TTP is located at the rim of both A3 and U4 pockets. Arg198 of ZFP36L2 forms part of the wall of the pocket for A3.
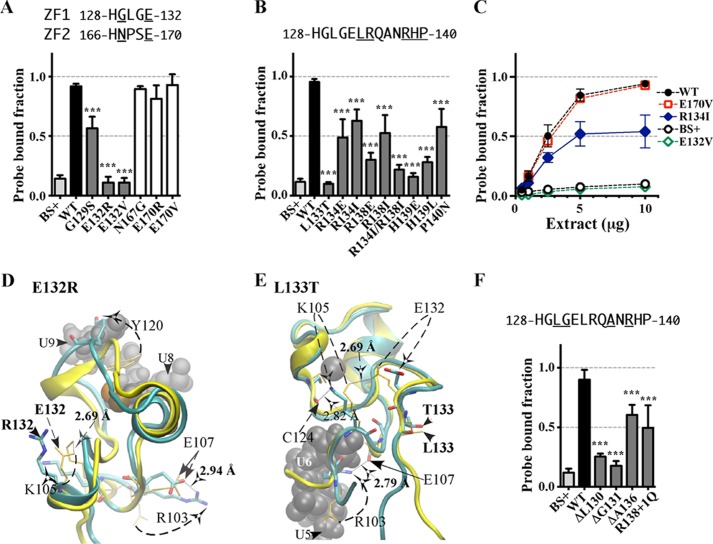
FIGURE 4.
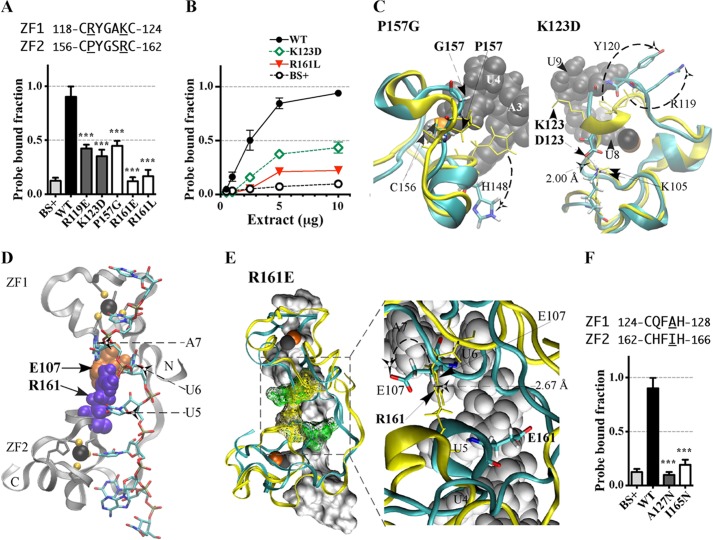
Effects of substitution mutants in the CX5CX3H intervals of the TTP TZF domain on RNA binding. The binding of protein to RNA is expressed as the probe-bound fraction. Above the graphs are the amino acid sequences of the intervals under study. The underlined residues are those that were mutated and whose effects on binding are shown in the bar graphs. The results from mutants of finger 1 are shown as gray columns, and those of finger 2 are shown as white columns. A, substitution mutants in the CX5C intervals. The results (mean ± S.D. (error bars)) are from four similar gel shift assays using 0.2 nm probe TARE5. B, binding assays with 0.2 nm TARE5 probe in serially diluted extracts, with total protein ranging from 10 to 1 μg. The probe-bound fractions (mean ± S.D.) are from four similar gel shift assays. C, superposition of the structural ensembles of finger 2 of WT TTP and mutant P157G (the RMSD is 10.37 Å; RMSD values were calculated from the superposition of the TZF domains of each mutant TTP with RNA-bound WT TTP) or finger 1 of WT TTP and mutant K123D (RMSD is 2.86 Å). The ribbon diagram of the WT TTP peptide backbone and side chains (sticks) are in yellow, the ribbon diagram of the mutant peptide backbone is in cyan, and side chains are shown in colors. RNA nucleosides are shown as gray spheres. Zinc atoms are shown as silver and copper (in P157G) or black and copper (in K123D) spheres. D, ribbon diagram of the peptide backbone of the human TTP TZF domain in complex with the ARE nonamer (sticks). The side chains of Glu107 (orange spheres, at the lead-in to finger 1) and Arg161 (purple spheres, at the C+5 position of the finger 2 CX5C region) are shown. Dashed arrows indicate nucleosides U5, U6, and A7 that interact with Glu107 and Arg161. Zinc atoms (black spheres) and the zinc-coordinating residues (ball and stick) of each finger are also displayed. E, superposition of the peptide backbone of the TZF domains of WT TTP (yellow ribbon) in complex with the ARE nonamer (surface representation) and the mutant R161E (cyan ribbon; RMSD is 4.66 Å). Zinc atoms are shown as black and copper spheres. Residues Glu107 and Arg161 (yellow mesh in the WT TZF domain) and Glu107 and Glu161 (green mesh in the mutant R161E) are shown (left). The right panel depicts the detailed view of the interaction between Arg161 and Glu107 (yellow sticks) in the WT and the non-interacting Glu107 and Glu161 (color sticks) in the mutant R161E. RNA nucleosides U4, U5, U6, and A7 (gray spheres) are indicated. F, substitution mutants in the CX3H intervals. The results (mean ± S.D.) are from four similar gel shift assays.
However, a comparison of these structures also indicates that the TZF domain of TTP would be expected to exhibit certain unique structural characteristics. For example, superposition of the two fingers of ZFP36L2 indicates that they adopt virtually identical folds (Fig. 2A), whereas in TTP, the peptide backbone conformation in finger 2 is not identical to that of finger 1 (Fig. 2B). Specifically, Arg160 and its counterpart Arg198 in the CX8C intervals within the two fingers of ZFP36L2 are confined to similar locations, as are the corresponding residues Glu163 and His201 (Fig. 2A), and are thus structurally identical (Fig. 2A). In contrast, in the CX8C intervals of fingers 1 and 2 of TTP, the residue pairs at the analogous sequence positions (Arg110/His148 and Ser113/Tyr151; Fig. 2B) conform differently from those found in ZFP36L2. Helicity differences at this region of the two proteins are probably responsible. In accordance with this observation, DSSP analysis (35) reveals that the ZFP36L2 finger 2 CX8C interval C+1 to C+6 segment forms two α-helical turns (backbone hydrogen bonds formed between Arg198 -CO and Thr202 -NH: 2.85 Å; between Thr199 -CO and Ile203 -NH: 2.79 Å). In contrast, DSSP analysis indicates that a hydrogen bond cannot be formed between the TTP finger 2 CX8C interval C+1 residue His148 backbone -CO and the C+5 residue Leu152 -NH because they are 5.92 Å apart. Thus, this region of TTP has only one α-helical turn, formed by the backbone hydrogen bond (2.90 Å) between C+2 and C+6 residues Lys149 and Gln153.
Not only are the two zinc fingers within TTP apparently structurally distinct from one another; there are notable differences between the second fingers of TTP and ZFP36L2. Specifically, His148, a residue that is unique to TTP, is present at the rim of both binding pockets for the RNA bases A3 and U4, whereas Arg198, the equivalent residue in ZFP36L2, is not a part of the pocket for U4 (Fig. 2C and Table 1) (also see TIS11d, PDB code 1RGO). In addition, a hydrogen bond formed between the TTP-unique residue, Tyr151, side chain -OH and the -CO backbone of Ile165 is absent in ZFP36L2 and may contribute to the difference in helical content of TTP finger 2 (Fig. 2D).
With the guidance of sequence alignments (Fig. 1) and structural models (Fig. 2), we made a panel of mutations to study the significance of residues within the TZF domain on the functional behavior of full-length TTP. Specifically, using cytosolic extracts prepared from HEK293 cells transiently transfected with these constructs, we examined the effects of these mutations on the ability of TTP to bind to an AU-rich RNA probe based on a bona fide TTP target mRNA, mouse TNF. From these studies, we consistently observed that loss of RNA binding directly correlated with the loss of the mutated proteins' ability to 1) promote deadenylation in a cell-free assay, 2) promote the decay of a probe based on the sequence of the TNF mRNA, and 3) destabilize the TNF mRNA in cells.
The CX8C Intervals
Before testing our panel of mutants, we first conducted three independent sets of equilibrium binding experiments with extracts of HEK293 cells, transfected with either full-length TTP or vector (BS+), in the presence of 12 increasing concentrations (0.01–50 nm) of the TARE5 probe (Fig. 3A). Fig. 3A demonstrates the ∼34-fold increase in probe-bound volume from expressed TTP compared with the endogenous proteins of the same approximate migration position on the gel (these cells are known not to express TTP or its other family members). The faint bands of endogenous protein-probe complexes can be seen in Fig. 3C (top and middle arrowheads). The apparent Kd of the probe for TTP under these conditions was 5.75 ± 0.58 nm. Based on these data, all subsequent gel shifts were performed using a 0.2 nm concentration of the TARE5 probe.
The sequence alignments shown in Fig. 1 illustrate that within the CX8C motifs of the TZF domain, the residues at the C+4 position, Ser113 (in finger 1) and Tyr151 (in finger 2), are unique to TTP and occur in all TTP proteins shown, from humans to frogs (Fig. 1A). Ser113 at the C+4 position in finger 1 is generally a glutamate in other family members, whereas Tyr151 at C+4 in finger 2 is a histidine in most other family members (Fig. 1). In addition, our simulation models suggest that these residues, although not components of the RNA base binding pockets (Table 1), are on the surface of the TTP TZF domain and have different relationships with regard to neighboring residues and thus may have the potential to affect RNA binding differently (Fig. 2). We therefore mutated Ser113 and Tyr151 to alanine as well as to similar residues at corresponding sequence positions within the TZF domain (aspartate in finger 1 or lysine in finger 2) of other TTP family members and evaluated the consequences on RNA binding using a gel shift assay. When assayed at a single extract protein concentration, mutation of Ser113 to either alanine or aspartate did not significantly affect TTP binding to the ARE probe (Fig. 3B). In contrast, the Y151A mutation exhibited markedly decreased TTP binding to the ARE probe, with results comparable with vector alone (Fig. 3, B and C (compare lane 3 with lane 2)). However, the Y151K mutant (Fig. 3, B and C (lane 4)) retained essentially normal ARE binding. The roughly equivalent expression of the mutant proteins in these experiments is documented in the Western blot shown in the bottom of Fig. 3C. Gel shift experiments using serial dilutions of these protein extracts showed that the levels of probe-protein binding were almost identical for WT TTP, S113A, and Y151K at all protein concentrations (Fig. 3D), consistent with a lack of effect of these mutations on RNA binding. Conversely, the mutant Y151A at all protein dilutions bound <20% of the input probe. These data suggest that the relative probe-bound fractions of the WT and various mutant TTP proteins remained remarkably constant at widely varying extract protein concentrations (Fig. 3D; also see Figs. 4B, 5F, and 6C).
FIGURE 5.
Effects of substitution mutants in the lead-in sequences to the zinc fingers on RNA binding. The binding of protein to RNA is expressed as the probe-bound fraction. Above the graphs are the amino acid sequences of the lead-in amino acids. The underlined residues are those that were mutated and whose effects on binding are shown in the bar graphs. The results from mutants of finger 1 are shown as gray columns, and those of finger 2 are shown as white columns. A and B, effects of mutant and WT TTP proteins, expressed as the probe-bound fraction. A, effects of charge reversal mutants; B, effects of neutral mutants. The results (mean ± S.D. (error bars)) are from three similar gel shift assays. C, the top panel shows representative gel shift assays using extracts from HEK293 cells transfected with vector alone (BS+), WT human TTP, or various mutant TTP constructs. Cytosolic extracts were incubated with 0.2 nm probe TARE5 as described under “Experimental Procedures” before loading on a non-denaturing acrylamide gel. A sample of probe alone in buffer was also loaded (lane P). Migration of the TTP-ARE complex (TTP) and the free probe (FP) are indicated on the right by vertical bars, and the three major endogenous protein-ARE complexes are labeled with arrowheads on the left. The bottom panel shows the relative amount of WT TTP protein and its mutants used in the binding reaction, as determined by Western blotting. D, superposition of WT TTP finger 1 and mutant E107K finger 1 structural ensembles (RMSD is 6.24 Å; RMSD values were calculated from superposition of the TZF domains of each mutant TTP with RNA-bound WT TTP). Wild-type TTP peptide backbone ribbon and side chains are in yellow, mutant E107K peptide backbone ribbon is in cyan, and side chains are in colors. RNA nucleosides U5 and U6 are shown as gray spheres. Zinc atoms are shown as black and copper spheres. E, comparisons in RNA binding are shown between mutant and WT TTP proteins; the results (mean ± S.D.) are from three similar gel shift assays. F, binding assays with 0.2 nm TARE5 probe in serially diluted extract with 1–10 μg of total protein. The probe-bound fractions (mean ± S.D.) are from three similar gel shift assays. G, superposition of WT TTP finger 2 and the Y142Q (RMSD value is 7.41 Å) or T144N (RMSD value is 10.66 Å) mutant model peptides. Color schemes are the same as in C, and RNA nucleoside U2 is shown. Zinc atoms are shown as silver and copper spheres.
FIGURE 6.
Effect of mutated residues or altered distances in the interfinger linker region of the TTP TZF domain on RNA binding. Above the graphs are the amino acid sequences of the intervals. The underlined residues are those that were mutated and whose effects on binding are shown in the bar graphs. A, comparisons between substitution mutants in the four-residue stretch after the CCCH motifs with WT TTP protein, expressed as probe-bound fraction. The results (mean ± S.D. (error bars)) are from three similar gel shift assays. The results from mutants of residues immediately after finger 1 are shown as gray columns, and results from those after finger 2 are shown as white columns. B, comparisons between substitution mutants in the linker region with WT TTP protein, expressed as probe-bound fraction. The results (mean ± S.D.) are from three similar gel shift assays. C, binding assays with 0.2 nm TARE5 probe in serially diluted extract with 1–10 μg of total protein. The probe-bound fractions (mean ± S.D.) are from three similar gel shift assays. Shown is superposition of the models of finger 1 of WT TTP and mutant peptides E132R (in D; RMSD value for the TZF domains from superposition with RNA-bound WT TTP is 5.27 Å) or L133T (in E; RMSD value is 6.93 Å). The WT TTP peptide backbone (ribbon) and side chains (sticks) are yellow, the mutant peptide backbone is cyan, and the side chains are in colors. Zinc atoms are shown as silver and copper spheres. RNA nucleosides (U9 and U8 in D; U6 and U5 in E) are shown as gray spheres. F, comparisons between deletion or addition mutants in the linker region with WT TTP protein, expressed as probe-bound fraction. The results (mean ± S.D.) are from three similar gel shift assays.
The finger 1 mutants S113A and S113D did not affect RNA binding; nor did they have any appreciable effect on deadenylation or TNF mRNA stability in cells (not shown). The greatly diminished RNA binding ability of the Y151A mutant was reflected in its inability to promote deadenylation of an ARE-containing polyadenylated probe (ARE-A50) (Fig. 3E, lane 4) compared with the WT protein (lane 3) and the Y151K mutant protein (lane 5); these effects were reflected in the decreased levels of the probe (ARE-A50) in response to WT TTP or the Y151K mutant and in the accumulation of the deadenylated probe fragment (ARE) (Fig. 3E, lanes 3 and 5). As expected, the probe (A50) that lacked the TTP binding site was not degraded by any of the extracts (Fig. 3E, lanes 6–10).
We next tested the ability of the mutant proteins to promote the instability of a TNF transcript in cells (12). In the presence of WT TTP or the Y151K mutant, the TNF transcript had the typical two-band appearance at steady state (Fig. 3F, lanes 2 and 4, top), with the decreased upper and increased lower bands corresponding to fully adenylated and deadenylated species, respectively (12). The total hybridizing TNF transcript (i.e. both upper and lower bands) with WT TTP was 47 ± 9% of control (p < 0.001), and the Y151K mutant TTP caused a decrease in transcript accumulation to 44 ± 9% of control that was similar to the effect with WT TTP (p = 0.71). However, the Y151A mutant produced steady state levels of the TNF transcript that were 76 ± 11% of control (p < 0.05) and exhibited a single band appearance similar to that seen in cells co-transfected with GFP alone (Fig. 3F). Taken together, it appeared that the Y151A mutant lost RNA binding activity and thus its ability to promote deadenylation in a cell-free assay and to promote TNF transcript instability in cells. Our subsequent mutational studies focused primarily on RNA binding of the mutant proteins, as assessed by gel shift assays.
To understand the mechanism of the decreased ability of the Y151A mutant to bind to RNA, we generated a simulation model of the TZF domain of the Y151A mutant and compared it with our simulation model of RNA-bound WT TTP (Fig. 3G). In the model of the WT protein, the -OH of Tyr151 forms a hydrogen bond with the backbone carbonyl O of Ile165 (Fig. 2B). The lack of this interaction in the Y151A mutant results in a slight overwinding of the helix that leads to the movement of His148 away from the rim of binding pocket for the U4 base of the RNA oligonucleotide. In addition, this mutation appears to disrupt the secondary structure and position of residues within the adjacent CX5C motif in finger 2 that may also influence RNA binding through 1) the loss of a 310 helix that distorts the binding pocket of U5, 2) movement of Tyr158 so that the aromatic ring can no longer participate in stacking with the RNA bases U4 and U5, and 3) failure of the side chain of Tyr158 to form hydrogen bonds with free oxygens of the phosphate backbone of U5 and U4. This is a striking example of different behaviors between the sequence-equivalent, TTP-specific amino acids within the two zinc fingers, in which the Ser113 mutation to alanine in the first zinc finger had no effect on TTP binding to RNA (Fig. 3B) or mRNA decay-promoting activity, as expected (not shown), whereas the sequence location-equivalent mutation, Y151A, in the second zinc finger essentially abrogated RNA binding and mRNA decay-promoting activities. This puzzling result can be explained by the models of the two mutant TZF domains; only Tyr151 in finger 2 orients neighboring residues that directly bind RNA, whereas Ser113, the analogous residue in finger 1, has no impact on RNA binding (Fig. 3G and Table 2).
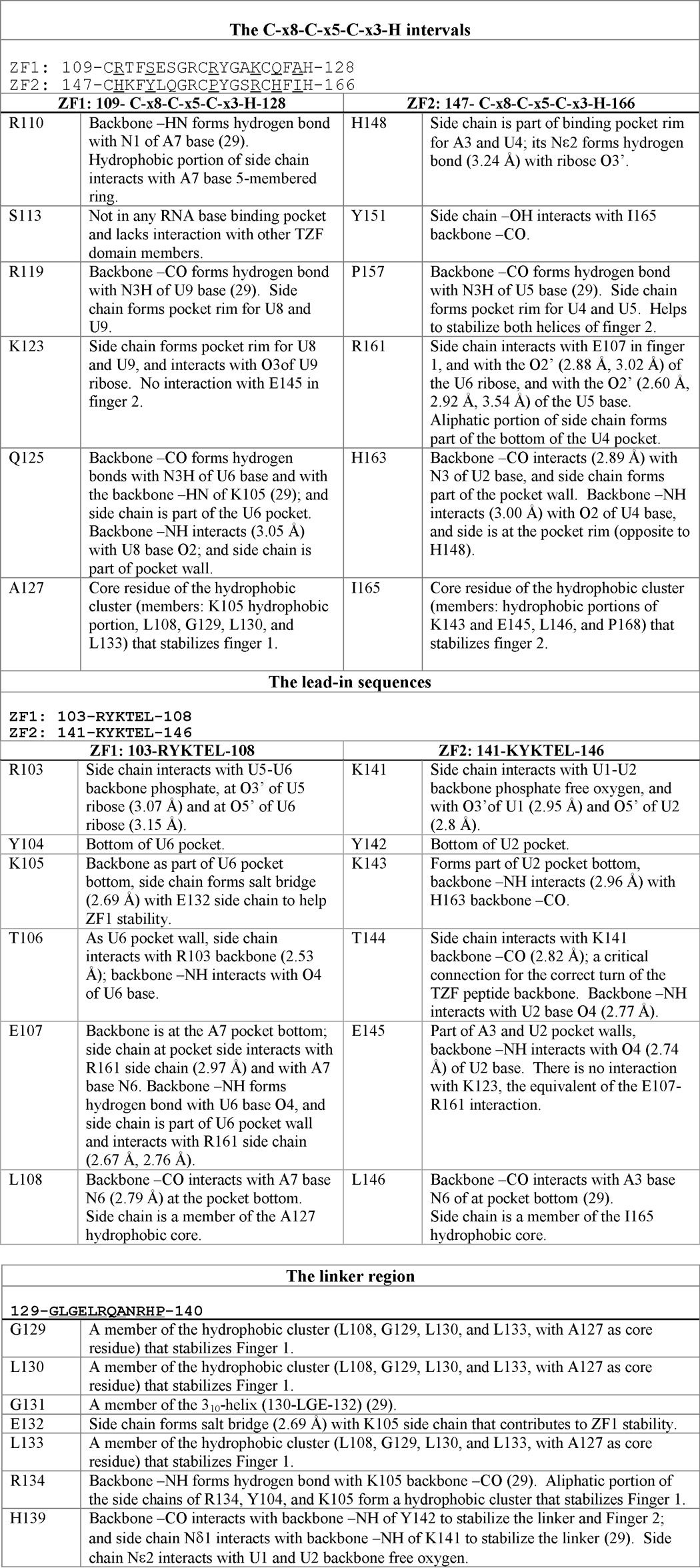
TABLE 2.
TTP TZF domain simulation solution model summary; interactions between TZF domain residues or between TZF domain amino acids and RNA bases
Results were obtained from the WT TTP TZF domain solution simulation model, as described under “Experimental Procedures” and in the legend for Table 1, in complex with the ARE RNA oligomer 1UUAUUUAUU9, ZF1 and ZF2, zinc fingers 1 and 2, respectively. In each section, the underlined residues are those whose mutations are investigated and/or discussed in this study.
We also investigated whether the RNA binding activity of TTP would be affected by changing the number of residues within the CX8C interval of either finger 1 or finger 2 because some TTP-like proteins, such as those in Caenorhabditis elegans, possess variations in the length of this region. The addition or deletion of a single residue in finger 1 significantly reduced RNA binding (Fig. 3H, S113 + 1Q, ΔS113, and ΔE114), whereas similar modifications in finger 2 had little or no effect (Fig. 3F, shown in white columns). Only when two or more (not shown) residues were deleted from finger 2 (ΔY151/ΔL152) was the reduction in RNA binding significant. Further, deletion of one residue from each finger (ΔE114/ΔY151) resulted in a reduction in RNA binding to the same extent as that of the single Glu114 deletion in finger 1. Thus, changes in length (by one residue) of the CX8C interval were tolerated in finger 2, but not in finger 1, and represent yet another example of how fingers 1 and 2 are likely to be functionally distinct in TTP.
The CX5C Intervals
The amino acid sequences within the CX5C intervals are highly conserved among the entire family of TTP proteins (Fig. 1, A–C). Our simulation model of TTP reveals that several of the residues within these intervals appear to interact directly with the RNA target (Tables 1 and 2). Changing the conserved residue Arg119 to glutamate in the C+1 position within the CX5C motif of finger 1 or reversing the charge of the conserved residue, Lys123, to aspartate, located at the C+5 position within the same interval, significantly decreased the ability of TTP to bind RNA compared with WT (Fig. 4A). Mutations at the sequence-equivalent positions in the CX5C interval of finger 2, P157G (C+1 position) and R161E (or R161L) (position C+5), both significantly reduced binding to the RNA probe compared with WT (Fig. 4A). In gel shift experiments using serial dilutions of these extracts, there were marked differences in magnitude of RNA bound when comparing WT TTP with the K123D and R161L mutations at all protein concentrations (Fig. 4B).
Examination of simulation models reveals the likely mechanism of the effects of mutations within the CX5C motifs (i.e. destabilization of RNA biding pockets). For instance, the R119E mutant model reveals that the side chain of this mutant turned away from the rim of both the U8 and U9 binding pockets (not shown). Mutation of the residue located at the corresponding position in finger 2, P157G, distorts the peptide backbone, resulting in disruption of the binding pockets for U4 and U5 bases as well as A3 (Fig. 4C and Tables 1 and 2).
Similarly, replacement of the conserved residue Lys123 in finger 1 with aspartate disrupts the binding pocket for bases U9 and U8. This mutation also alters the conformation of key residues, including that of Tyr120, so that it no longer participates in stacking with bases U9 and U8 (Fig. 4C and Tables 1 and 2). These direct and indirect changes should inhibit RNA binding.
Importantly, the models also indicate that the conserved residue Arg161 in finger 2 is critical in stabilizing the overall structure of the TZF domain, similar to Arg211 in finger 2 of ZFP36L2 (29). Not only does Arg161 participate in hydrogen bonds with the O2′ of the U5 base and the O2′ of the U6 ribose (heavy atom distances 2.88 and 3.02 Å; Table 2), it is also involved in an ionic interaction with Glu107 in finger 1 (Fig. 4D). This ionic interaction is likely to be critically important for orienting the two fingers relative to one another and for the overall stability of the TZF domain. Consistent with this observation, when the ionic interaction between Arg161 and Glu107 is disrupted by the R161E mutation, the proper orientation of the two fingers with respect to one other is severely compromised (Fig. 4E).
The CX3H Intervals
The CX3H intervals are also highly conserved among TTP family proteins. Within the CX3H motif, mutants Q125V (C+1 of finger 1) and H163L (C+1 of finger 2) did not significantly affect protein-RNA interactions (25). In contrast, A127N (C+3 of finger 1) and I165N (C+3 of finger 2) severely reduced RNA binding (Fig. 4F).
Ala127 (finger 1) and Ile165 (finger 2) are the core residues of the hydrophobic clusters of each finger, respectively (Table 2). As expected, our simulation models reveal that substituting either of these residues with polar amino acids disturbed these hydrophobic clusters and altered the helical conformation (not shown). In addition, the Ala127 to asparagine substitution indirectly alters the ionic interaction between Glu107 and Arg161, leading to the disruption of the proper orientation of the two fingers.
To evaluate if the CX5C and CX3H intervals within TTP could tolerate variations in length, we generated a number of deletion and insertion mutants. However, unlike the CX8C interval of finger 2 (Fig. 3H), our data indicate that the CX5C and CX3H intervals cannot tolerate changes in length (e.g. deletion mutants ΔA122 and ΔA127 and insertion mutant F164 + 1Q) (also data not shown).
The Lead-in Sequences
The sequences leading into both zinc fingers are highly conserved across the TTP family of proteins (Fig. 1, A–C). Our simulation models as well as the structure for ZFP36L2 described by Hudson et al. (29) indicate that the lead-in sequences are critical for RNA binding because there are several direct interactions between the positively charged residues within this region and the RNA backbone. For instance, our simulation model shows that Arg103 (finger 1) interacts with the O5′ of U6 (3.15 Å) and O3′ of U5 (3.07 Å). The equivalent residue in finger 2, Lys141, is predicted to interact with the O5′ of U2 (2.8 Å) and O3′of U1 (2.95 Å). Our model also indicates that residues within the lead-in sequences participate in ionic interactions that are critical for orienting the zinc fingers relative to one another (see Fig. 4D and Table 2).
To study these highly conserved lead-in sequences in more detail, we generated a panel of substitution mutants. Fig. 5A shows the results of charge reversal mutations on protein-RNA interactions. Changing the positively charged residues that directly interact with RNA, such as Arg103 (finger 1) or its sequence-equivalent Lys141 (finger 2) or Lys105 (finger 1) or its sequence-equivalent Lys143 (finger 2), to glutamate severely reduced protein-RNA binding (Fig. 5A).
Although reversing the charge of Glu107, in the lead-in sequence of finger 1, by substitution with a lysine led to a significant reduction in binding of the mutant protein to the RNA (Fig. 5, A and C (lane 3)), there was no change in RNA binding with the equivalent E145K mutant in finger 2 (Fig. 5, A and C (lane 5)). Furthermore, neutral substitutions of Glu107 decreased RNA binding to a similar extent as reversing the charge (Fig. 5, B and C (lane 4)), and the E145I mutation caused no binding reduction (Fig. 5, B and C (lane 6)). Statistical comparisons using analysis of variance with corrections for multiple comparisons showed that equivalent mutations within the two fingers, such as R103E versus K141E or K105E versus K141E, were not significantly different. However, as noted above, there were significant differences between the two fingers, with respect to RNA binding, between the charge reversal E107K and E145K and neutral mutants, E107I and E145I, R103L and K141L, and K105L and K143L.
As expected, the E107K mutant simulation model (Fig. 5D) reveals that this mutation disrupts the salt bridge with R161 (at the C+5 position within the CX5C region of the finger 2) that is present in the WT TTP simulation model (Figs. 4D and 5D). Loss of the ionic interaction disrupts the orientation of the two fingers relative to one another and disrupts the interaction between Arg161 and the RNA backbone, consistent with the significant decrease in RNA binding observed in our assays as well as the results of the R161E or R161L mutants (Fig. 4, A and E). In addition, the E107K mutant changes the helical content of the CX8C region of finger 1 and disrupts stacking between the Phe112 side chain and the zinc-coordinating residue, His128 (29), to further destabilize finger 1 (Fig. 5D).
The WT TTP simulation structure model indicates that the Glu107/Arg161 interaction is unique and that there is not an equivalent interaction between analogous residues Glu145 and Lys123 (Table 2). Our models indicate that Glu145 forms part of the pocket for the RNA bases A3 and U2, and its backbone interacts with the U2 base. In the E145K mutant, the long hydrophobic portion of the mutant side chain helps maintain the stability of these pockets (not shown).
In addition to our analysis of charged residues, we also mutated the neutral amino acids in the lead-in sequences of finger 1 (Leu108), and finger 2 (Tyr142, Thr144, or Leu146) (Fig. 5E). Substitution of Tyr142 with phenylalanine did not change binding of the mutant protein to the RNA probe, and there was no significant alteration in deadenylating activity or in the cell transfection TNF mRNA stability assay. In contrast, mutation to a non-aromatic residue, glutamine, significantly decreased binding to a level similar to that of extract from vector alone (BS+)-transfected cells.
Substitution of Thr144 with asparagine also reduced RNA binding to the level of extract from vector alone (BS+)-transfected cells. Mutation of Leu146 to proline (Fig. 5E) or serine (not shown) did not produce significant effects on the ability of TTP to bind RNA. The lack of effect of the proline mutation at this site is particularly relevant because of its natural occurrence in the rodent family member protein ZFP36L3 (8, 36) as well as in at least two family members from yeasts (37, 38) (Fig. 1C). In contrast, mutation of L108 (in finger 1) significantly reduced protein-RNA binding (Fig. 5E). The differences in RNA binding activities between these sequence-equivalent residues are probably due to destabilization of the A7 binding pocket introduced by the L108P mutant, where Arg110 moved away from the A7 base (Table 2) to interact with Glu114 (1.72 and 1.73 Å; not shown), whereas the L146P mutant led to structural rearrangements that preserved the A3 pocket (not shown). Consistent with these observations, there were no observable differences between WT TTP and the E145K or L146P mutant in RNA binding gel shift experiments using serial dilutions of the protein extracts (Fig. 5F); however, there were magnitude differences in RNA bound between WT and mutants K141L and E107K.
The WT simulation model shows that the backbones of Tyr142 and Thr144 in the lead-in sequence to the second zinc finger form part of the binding pocket for U2 (Tables 1 and 2). Consistent with the significant decrease we observed in our protein-RNA binding assay, the Y142Q and T144N mutants change the binding pocket such that residues His139 and Lys141 no longer interact with the U1-U2 backbone (Fig. 5G). The T144N mutant also resulted in the loss of a hydrogen bond (2.82 Å) between Thr144 and Lys141, which is crucial for the correct turn of the peptide backbone. These alterations resulted in a change in the orientation of the fingers relative to one another, apparently making the TZF domain-RNA association impossible.
The Linker Region and Residues after the C-terminal End of Zinc Finger 2
The linker region in TTP family member proteins serves as a bridge that connects the two zinc fingers and contains both conserved and non-conserved residues (Fig. 1). Because the linker region has the potential to influence the positioning of the fingers relative to one another, we sought to understand the contribution of individual amino acids as well as the length requirements to TTP-RNA association.
We generated single point mutants of the most highly conserved residues within the linker region, encompassing positions 129, 132–134, and 138–140. All single point mutants in this region significantly reduced binding to the RNA probe compared with that of WT TTP. However, the most dramatic effects were observed upon mutation of Glu132, Leu133, and His139, which reduced binding to RNA to levels similar to extracts from vector-transfected cells (Fig. 6, A and B). Our simulation models showed that the single mutations E132R and L133T (Fig. 6, D and E) result in the loss of the ionic interaction between the side chains of Lys105 and Glu132 (2.69 Å), an important interaction that contributes to stabilizing finger 1. Further, all four mutant peptides, E132R, L133T, R134E, and R138E (not shown), result in a newly formed ionic interaction between Arg103 and Glu107 that renders the crucial interaction between Glu107 (finger 1) and Arg161 (finger 2) impossible, an interaction earlier established as critical for the overall structural stability of the TZF domain of TTP (Fig. 4D).
In contrast to the linker region, mutation of the conserved residues Asn167 or Glu170 (Fig. 6A), which are at sequence-equivalent positions in finger 2 to Gly129 and Glu132 in finger 1, did not significantly affect TTP-RNA binding). Gly129 is part of the hydrophobic cluster (Ala127 as the core residue), but Asn167 seems not to be part of the hydrophobic cluster in which Ile165 is the core (Table 2). The side chains of Glu132 and Lys105 (lead-in to finger 1; see Fig. 6D) form a hydrogen bond to stabilize finger 1, but there is no obvious connection between Glu170 and Lys143.
Other structural changes in TTP also are predicted to occur as a result of mutating the conserved residues within the linker region. Besides mutants E132R and L133T (Fig. 6, D and E), the charge reversal substitution R134E results in the formation of a new salt bridge between Glu134 and Arg138 that is predicted to drastically alter the conformation of surrounding residues, thus destabilizing the TZF structure (Table 2). The ZFP36L2 NMR structure (29) also predicts that His139 is involved in stabilizing the structure of the second zinc finger. In this prediction, His139 would contribute to RNA binding by forming part of the binding pocket for U2 and interacting with the backbone free oxygen of U2 and U1. Consistent with this prediction, mutation of this residue had dramatic effects on the ability of TTP to bind RNA (Fig. 6B and Tables 1 and 2). In addition, mutation of His139 led to rearrangement of Lys141 such that its interactions with the phosphate backbones of U2 and U1 were eliminated (Table 2). In fact, any mutation in the TZF domain that affected the position of His139 would lead to a great reduction in TTP association with the RNA (see Fig. 5, E and G).
Fig. 6C shows that RNA binding using serial dilutions of extracts from cells expressing WT TTP was similar to that seen with an E170V mutant. However, the R134I mutation inhibited binding by ∼50% at 10 or 5 μg of protein, and the E132V mutation eliminated binding entirely.
To study the length requirements of the linker on the ability of TTP to bind RNA, we made a series of single deletions within the linker region. We first deleted three less well conserved residues in the linker region, Leu130, Gly131, or Ala136 (Fig. 1). Deletion of Leu130 or Gly131 significantly decreased RNA binding (Fig. 6F). In contrast, deleting the equivalent residues following the second zinc finger, Pro168 and Ser169, had no detectable effect on RNA binding (not shown). Deleting a residue in the middle of the linker, Ala136, resulted in a significant although less severe decrease in RNA binding (Fig. 6F). When the distance between the two fingers was increased by inserting a glutamine after Arg138, the binding of the mutant to the ARE probe was significantly reduced (Fig. 6E). Thus, reduction in length may be moderately tolerated in the middle but not at the beginning of the linker, whereas the effect was relatively moderate when the length of the linker was increased by one residue. Further expansions of the linker were not compatible with productive RNA binding (not shown).
DISCUSSION
In the present study, we created a panel of TZF domain mutants within the context of full-length human TTP and made a series of simulation solution models of the normal and mutant TZF domain to better understand unique features of RNA binding by TTP. We identified several key features of the TTP-mRNA interaction, including the following. 1) TTP-specific residues lead to important predicted structural differences between the two zinc fingers of TTP as well as differences between TTP and its related family member, ZFP36L2, bound to the same mRNA target; 2) not all residues at analogous sequence positions within the two fingers are functionally equivalent; and 3) the majority of the conserved residues within the TZF domain are required for high affinity RNA binding, including an interaction between Arg161 (finger 2) and Glu107 (finger 1), which is critically important for properly orienting each finger of TTP to the other.
Another key finding is that, whereas the two fingers in ZFP36L2 were reported to be nearly identical (29), finger 2 of TTP is predicted to be structurally distinct from finger 1 (see Fig. 2B). The structural differences between the two fingers of TTP were mainly observed in the CX8C intervals. It is noteworthy that six of the seven TTP-specific residues (see Fig. 1) are located in these intervals. A portion of this segment (finger 1 residues C+1 to C+6; finger 2 residues C+2 to C+6) forms an α-helix in each finger. However, the two fingers in TTP differ in their helical content, probably due to the interaction of a TTP-specific residue within the CX8C interval of finger 2, Tyr151, with Ile165 as well as a single helical turn in finger 2, contrasting with two turns in finger 1. As a consequence, the comparatively relaxed helical segment in finger 2 (see Fig. 2B) is more tolerant of expansion or deletion by a single residue than the corresponding segment of TTP finger 1. Further, the α-helix of finger 2 of TTP (only one turn and involving three TTP-specific residues) is more relaxed compared with that of ZFP36L2 (two turns; see Fig. 2D). In contrast, the α-helix in finger 1 of TTP (two TTP-unique residues) is quite similar to that of ZFP36L2 (not shown).
Consistent with these proposed structural differences, mutants in the CX8C interval of finger 2 of TTP are functionally distinct in that they behave very differently when compared with mutants at equivalent sequence positions of finger 1. Certain residues in this segment of both fingers appear to be directly involved in RNA binding (residues C+1, C+2, and C+3; see Tables 1 and 2). However, as shown in the simulation models, the TTP-specific residues at the C+4 position, Ser113 in finger 1 and Tyr151 in finger 2, make different contributions to RNA binding despite their sequence positional equivalency (see Table 2). In addition, they have different spatial orientations, as compared with the analogous residue pairs in ZFP36L2 (Glu163 and His201) that occupy the same space with similar steric boundaries (see Fig. 2A). Mutating Ser113 (to alanine or aspartate) in TTP finger 1 did not alter RNA binding (see Fig. 3) or affect the helicity of finger 1 (not shown). In finger 2, a Y151K mutation that has similar steric boundaries did not change TTP binding to RNA; however, the Y151A mutant significantly affected RNA binding as well as the helical content of finger 2 (see Fig. 3E), probably through altering the position of neighboring residues involved in direct contacts with RNA.
Comparison of two additional residues within the CX8C region of TTP, His148 (C+1, finger 2) and Arg110 (C+1, finger 1), also supports the proposal that similar positional residues within this region are not necessarily functionally equivalent, although these residues are physio-chemically similar. We previously observed that RNA binding was decreased when Arg110 was mutated (25). Although the TTP-specific residue His148 participates in both the A3 and U4 binding pockets, association of TTP with the TNF ARE was not affected when His148 was mutated (H148E or H148L). However, the conformation of the His148 aromatic ring was altered away from the binding pocket in certain TTP TZF mutants in which there was severe loss of RNA binding (e.g. Y151A (see Fig. 3, B and E), P157G (see Fig. 4A), and I165N (see Fig. 4F), due to apparent overwinding of the helix in the finger 2 CX8C region. The end result is the destabilization of the binding pocket for U4 or A3, as shown in the simulation mutant structures (see Figs. 3G and 4C). Also noteworthy is that whereas His148 within TTP participates in both A3 and U4 binding, the corresponding Arg198 residue of ZFP36L2 is only involved in A3 binding. Thus, His148 at this region of finger 2 appears to play a unique role in TTP RNA binding; whether it is functionally distinct from the analogous residue in other family members remains to be determined.
Besides the corresponding pairs in the CX8C intervals (C+1, Arg110/His148; C+4, Ser113/Tyr151) that are neither functionally nor structurally equivalent, there are other sequence-equivalent but functionally different pairs: Glu107/Glu145 and Leu108/Leu146 in the lead-in sequences (the fifth and sixth positions; see Fig. 5). Thus, the two fingers of TTP contain structurally and functionally distinct residues.
In addition, the conserved sequence-equivalent residues Lys123/Arg161 (C+5 in the CX5C intervals of finger 1 and finger 2, respectively) show differences in their contribution to the TTP TZF domain structure. Arginine 161, in finger 2, interacts with Glu107 of the lead-in to finger 1, thus stabilizing the orientation of the two fingers. The Arg161/Glu107 interaction serves as one of the key elements in the stability of the zinc fingers and, consequently, for the association with RNA (see Fig. 4 and Table 2). This is demonstrated in several mutants (e.g. E107I, E107K, E132R, E132V, L133T, R134E, or R138E) that disrupt the Glu107/Arg161 interaction, resulting in loss of RNA binding ability. In contrast to Arg161 (finger 2), which stabilizes the overall structure of the TZF domain, our simulation structures revealed that the sequence-equivalent residue in finger 1, Lys123, directly interacts with the U9 ribose (see Table 2). Thus, this observation highlights the potential importance of U9 in the interaction between full-length TTP and the RNA nonamer.
We also investigated some of the highly conserved residues in the TZF domain within the mammalian TTP-like family members using other assays. As expected, we found that mutants unable to bind RNA in the cell-free assay were unable to deadenylate the ARE-A50 probe in a cell-free assay or to destabilize mRNA in intact cells. These data strongly support the concept that high affinity and high capacity RNA binding is a necessary requirement for the subsequent physiological activity of TTP to promote mRNA decay.
The lead-in sequences ((R/K)YKTEL) to both fingers are also highly conserved across the TTP family of proteins and have been suggested in the ZFP36L2-RNA complex to make several direct contacts with RNA (29). The severe loss of RNA binding resulting from mutation of the positively charged residues in the lead-in sequences of both fingers supports their role for productive RNA binding.
It is also interesting to note that mutations of the conserved residues within the lead-in sequences, Leu108 (finger 1) and Leu146 (finger 2), resulted in different outcomes with respect to RNA binding (see Fig. 5E) and thus represent another example of a clear distinction between the functional roles of sequence-equivalent residues. Specifically, L108P resulted in a significant loss of RNA binding, probably due to perturbations in the peptide backbone, whereas the L146P did not affect RNA binding.
We have also found that conserved residues within the linker region between the two fingers are critical for the TTP-mRNA interaction. The highly conserved residue Glu132, located at the fourth position after the zinc-coordinating histidine of finger 1 (see Fig. 1), is crucial to TTP-RNA association, as shown by the fact that both the E132R and E132V mutants were detrimental to TTP-ARE binding (Fig. 6). The simulation model indicates that a salt bridge between Glu132 and Lys105 at the lead-in to finger 1 may be a significant interaction in the TTP-RNA association (see Table 2). Interestingly, the glutamate (Glu170) at the fourth position after the zinc-coordinating histidine of finger 2 is also highly conserved among the mammalian TTP family proteins, but the mutant E170R or E170V had no negative effect on RNA binding (see Fig. 6). It is difficult, however, to determine whether these two highly conserved glutamates located at the fourth positions after each CCCH motif are structurally equivalent, due to the fact that our model template, the TZF-ARE NMR structure (29), ended at the glutamate of the fourth position after the second CCCH motif. Whether protein sequences beyond the zinc fingers influence RNA binding remains to be determined.
There are many types of CCCH zinc finger proteins (39, 40), and there is a group of such proteins in C. elegans that contain TTP family protein-like lead-in sequences to their two CCCH zinc fingers (41) but exhibit various lengths between the first and the second Cs within the zinc fingers and in the linker between the two fingers. These CCCH-type zinc finger proteins in C. elegans have been reported to bind RNA (42–45). For instance, the TZF domain of the C. elegans protein MEX-5 has been shown to bind the TNF ARE with high affinity (43), although the two CCCH motifs of MEX-5 are separated by 23 amino acids, five more than the TTP family proteins. In contrast, as shown here, the insertion of a single residue in the linker region of TTP (e.g. R138R + 1Q; see Fig. 6C) was not tolerated, although the linker region of ZFP36L2 has been suggested to be a relatively relaxed segment (29).
Using a systematic approach to mutate the TZF domain of TTP, followed by functional evaluation and structural modeling, we have shown the importance of each region of the TZF domain working in concert with other regions, leading to the ultimate association with ARE-containing RNA. Our present data, as well as previous studies (10, 23, 25), indicate that for the optimal recognition of the UUAUUUAUU sequence, the TZF domain needs to have almost all of the following consensus characteristics: the (R/K)YKTEL lead-in, the exact CX5CX3H interval size in both fingers, and 18 residues between the two CCCH motifs, as well as the highly conserved residues in these regions. Thus, it seems likely that the other CCCH-type zinc fingers, with varying sequence characteristics, may bind distinct RNA elements.
Many other questions remain about this protein-RNA interaction, such as whether residues in the TZF domain specific to a given TTP family member would act in concert with other domains of the same protein to modulate the target mRNA because the sequences are so different outside each member's TZF domain. Thus, to fully delineate how TTP binds to RNA and initiates the process leading to subsequent RNA destabilization would require the use of full-length TTP and longer RNA target sequences. It is interesting to note that we have found little or no detectable TNF ARE binding when the TTP TZF domain is embedded in the middle of an unrelated protein (MARCKSL1 or GFP),4 although binding to an ARE target sequence can be readily demonstrated using a synthetic or recombinant TZF domain peptide (26, 27) or a fusion version of the TZF domain located at the C- or the N-terminal end of an unrelated protein (46).
The activities of TTP in cells are regulated by a multitude of factors, including phosphorylation (47). Whether any of the TZF domain residues are phosphorylated in intact cells, either in the basal state or after stimulation and whether this phosphorylation would affect the TZF domain binding to RNA are not known. However, the present study suggests that phosphorylation is unlikely to play a role in the formation of the TZF-RNA complex, based on the following considerations concerning the potentially “phosphorylatable” residues Ser113, Thr144, and Tyr151: 1) although the side chain of Ser113 does not interact with neighboring residues or with RNA, and thus has the potential to be phosphorylated, binding of TTP to RNA was not affected by mutating Ser113 to either alanine or aspartate; and 2) simulation models indicated that the side chain -OH of Thr144 and Tyr151 participate in hydrogen bonding with the backbone carbonyl of other residues (Thr144 with Lys141, Tyr151 with Ile165), thus making it unlikely that these two sites are accessible by protein kinases.
It is also not clear how TTP family member proteins interact with other proteins and complexes in regulating the stability or translation of their target mRNAs in response to stimuli, in different phases of development, or in specific cells and tissues. Nevertheless, some insight has been gained through a recent study in which a direct interaction between the conserved C-terminal domain of human TTP and the human NOT1 protein was demonstrated (48). Furthermore, disrupting this interaction with TTP C-terminal mutants interfered with TTP-dependent deadenylation of a target transcript in cells. Although much remains unknown about this process, the increased understanding of the relationship between the TZF domain and the ARE presented here is an important step that should aid us in the necessary future studies.
Acknowledgments
We thank Drs. Lars Pedersen and R. Scott Williams for careful reading of the manuscript and helpful discussions. We also thank the members of the NIEHS, National Institutes of Health, DNA Sequencing Core and Dr. Gerald Wilson for useful discussions.
This work was supported by the National Institutes of Health, NIEHS, Division of Intramural Research.
To be consistent with our previous publications, we have used the NCBI Reference Sequence NP_003398.1 to identify the human TTP protein residues. Recently, GenBankTM updated a version of the protein sequence (NP_003398.2) to include six additional residues at the N terminus.
W. S. Lai and P. J. Blackshear, unpublished observations.
- TTP
- tristetraprolin
- ARE
- AU-rich element
- TZF
- tandem zinc finger
- RMSD
- root mean square deviation
- PDB
- Protein Data Bank.
REFERENCES
- 1. Lai W. S., Stumpo D. J., Blackshear P. J. (1990) Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. J. Biol. Chem. 265, 16556–16563 [PubMed] [Google Scholar]
- 2. Taylor G. A., Lai W. S., Oakey R. J., Seldin M. F., Shows T. B., Eddy R. L., Jr., Blackshear P. J. (1991) The human TTP protein. Sequence, alignment with related proteins, and chromosomal localization of the mouse and human genes. Nucleic Acids Res. 19, 3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Heximer S. P., Forsdyke D. R. (1993) A human putative lymphocyte G0/G1 switch gene homologous to a rodent gene encoding a zinc-binding potential transcription factor. DNA Cell Biol. 12, 73–88 [DOI] [PubMed] [Google Scholar]
- 4. Ma Q., Herschman H. R. (1991) A corrected sequence for the predicted protein from the mitogen-inducible TIS11 primary response gene. Oncogene 6, 1277–1278 [PubMed] [Google Scholar]
- 5. DuBois R. N., McLane M. W., Ryder K., Lau L. F., Nathans D. (1990) A growth factor-inducible nuclear protein with a novel cysteine/histidine repetitive sequence. J. Biol. Chem. 265, 19185–19191 [PubMed] [Google Scholar]
- 6. Gomperts M., Pascall J. C., Brown K. D. (1990) The nucleotide sequence of a cDNA encoding an EGF-inducible gene indicates the existence of a new family of mitogen-induced genes. Oncogene 5, 1081–1083 [PubMed] [Google Scholar]
- 7. Varnum B. C., Ma Q. F., Chi T. H., Fletcher B., Herschman H. R. (1991) The TIS11 primary response gene is a member of a gene family that encodes proteins with a highly conserved sequence containing an unusual Cys-His repeat. Mol. Cell. Biol. 11, 1754–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blackshear P. J., Phillips R. S., Ghosh S., Ramos S. V., Richfield E. K., Lai W. S. (2005) Zfp36l3, a rodent X chromosome gene encoding a placenta-specific member of the Tristetraprolin family of CCCH tandem zinc finger proteins. Biol. Reprod. 73, 297–307 [DOI] [PubMed] [Google Scholar]
- 9. Taylor G. A., Carballo E., Lee D. M., Lai W. S., Thompson M. J., Patel D. D., Schenkman D. I., Gilkeson G. S., Broxmeyer H. E., Haynes B. F., Blackshear P. J. (1996) A pathogenetic role for TNF α in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4, 445–454 [DOI] [PubMed] [Google Scholar]
- 10. Carballo E., Lai W. S., Blackshear P. J. (1998) Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science 281, 1001–1005 [DOI] [PubMed] [Google Scholar]
- 11. Carballo E., Blackshear P. J. (2001) Roles of tumor necrosis factor-α receptor subtypes in the pathogenesis of the tristetraprolin-deficiency syndrome. Blood 98, 2389–2395 [DOI] [PubMed] [Google Scholar]
- 12. Lai W. S., Carballo E., Strum J. R., Kennington E. A., Phillips R. S., Blackshear P. J. (1999) Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor α mRNA. Mol. Cell. Biol. 19, 4311–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carballo E., Lai W. S., Blackshear P. J. (2000) Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood 95, 1891–1899 [PubMed] [Google Scholar]
- 14. Ogilvie R. L., Abelson M., Hau H. H., Vlasova I., Blackshear P. J., Bohjanen P. R. (2005) Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. J. Immunol. 174, 953–961 [DOI] [PubMed] [Google Scholar]
- 15. Lai W. S., Parker J. S., Grissom S. F., Stumpo D. J., Blackshear P. J. (2006) Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol. Cell. Biol. 26, 9196–9208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Datta S., Biswas R., Novotny M., Pavicic P. G., Jr., Herjan T., Mandal P., Hamilton T. A. (2008) Tristetraprolin regulates CXCL1 (KC) mRNA stability. J. Immunol. 180, 2545–2552 [DOI] [PubMed] [Google Scholar]
- 17. Stoecklin G., Tenenbaum S. A., Mayo T., Chittur S. V., George A. D., Baroni T. E., Blackshear P. J., Anderson P. (2008) Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. J. Biol. Chem. 283, 11689–11699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Horner T. J., Lai W. S., Stumpo D. J., Blackshear P. J. (2009) Stimulation of polo-like kinase 3 mRNA decay by tristetraprolin. Mol. Cell. Biol. 29, 1999–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ogilvie R. L., Sternjohn J. R., Rattenbacher B., Vlasova I. A., Williams D. A., Hau H. H., Blackshear P. J., Bohjanen P. R. (2009) Tristetraprolin mediates interferon-γ mRNA decay. J. Biol. Chem. 284, 11216–11223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tudor C., Marchese F. P., Hitti E., Aubareda A., Rawlinson L., Gaestel M., Blackshear P. J., Clark A. R., Saklatvala J., Dean J. L. (2009) The p38 MAPK pathway inhibits tristetraprolin-directed decay of interleukin-10 and pro-inflammatory mediator mRNAs in murine macrophages. FEBS Lett. 583, 1933–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stumpo D. J., Byrd N. A., Phillips R. S., Ghosh S., Maronpot R. R., Castranio T., Meyers E. N., Mishina Y., Blackshear P. J. (2004) Chorioallantoic fusion defects and embryonic lethality resulting from disruption of Zfp36L1, a gene encoding a CCCH tandem zinc finger protein of the Tristetraprolin family. Mol. Cell. Biol. 24, 6445–6455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stumpo D. J., Broxmeyer H. E., Ward T., Cooper S., Hangoc G., Chung Y. J., Shelley W. C., Richfield E. K., Ray M. K., Yoder M. C., Aplan P. D., Blackshear P. J. (2009) Targeted disruption of Zfp36l2, encoding a CCCH tandem zinc finger RNA-binding protein, results in defective hematopoiesis. Blood 114, 2401–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lai W. S., Carballo E., Thorn J. M., Kennington E. A., Blackshear P. J. (2000) Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J. Biol. Chem. 275, 17827–17837 [DOI] [PubMed] [Google Scholar]
- 24. Carrick D. M., Lai W. S., Blackshear P. J. (2004) The tandem CCCH zinc finger protein tristetraprolin and its relevance to cytokine mRNA turnover and arthritis. Arthritis Res. Ther. 6, 248–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lai W. S., Kennington E. A., Blackshear P. J. (2002) Interactions of CCCH zinc finger proteins with mRNA. Non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. J. Biol. Chem. 277, 9606–9613 [DOI] [PubMed] [Google Scholar]
- 26. Blackshear P. J., Lai W. S., Kennington E. A., Brewer G., Wilson G. M., Guan X., Zhou P. (2003) Characteristics of the interaction of a synthetic human tristetraprolin tandem zinc finger peptide with AU-rich element-containing RNA substrates. J. Biol. Chem. 278, 19947–19955 [DOI] [PubMed] [Google Scholar]
- 27. Brewer B. Y., Malicka J., Blackshear P. J., Wilson G. M. (2004) RNA sequence elements required for high affinity binding by the zinc finger domain of tristetraprolin. Conformational changes coupled to the bipartite nature of Au-rich MRNA-destabilizing motifs. J. Biol. Chem. 279, 27870–27877 [DOI] [PubMed] [Google Scholar]
- 28. Lai W. S., Kennington E. A., Blackshear P. J. (2003) Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol. Cell. Biol. 23, 3798–3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hudson B. P., Martinez-Yamout M. A., Dyson H. J., Wright P. E. (2004) Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat. Struct. Mol. Biol. 11, 257–264 [DOI] [PubMed] [Google Scholar]
- 30. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Case D. A., Darden T. A., Cheatham I., T. E., Simmerling C. L., Wang J., Duke R. E., Luo R., Walker R. C., Zhang W., Merz K. M., Roberts B., Wang B., Hayik S., Roitberg A., Seabra G., Kolossváry I., Wong K. F., Paesani F., Vanicek J., Wu X., Brozell S. R., Steinbrecher T., Gohlke H., Cai Q., Ye X., Wang J., Hsieh M.-J., Cui G., Roe D. R., Mathews D. H., Seetin M. G., Sagui C., Babin V., Luchko T., Gusarov S., Kovalenko A., Kollman P. A. (2010) AMBER, version 11, University of California, San Francisco [Google Scholar]
- 32. Duan Y., Wu C., Chowdhury S., Lee M. C., Xiong G., Zhang W., Yang R., Cieplak P., Luo R., Lee T., Caldwell J., Wang J., Kollman P. (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 24, 1999–2012 [DOI] [PubMed] [Google Scholar]
- 33. Pérez A., Marchán I., Svozil D., Sponer J., Cheatham T. E., 3rd, Laughton C. A., Orozco M. (2007) Refinement of the AMBER force field for nucleic acids. Improving the description of α/γ conformers. Biophys. J. 92, 3817–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Humphrey W., Dalke A., Schulten K. (1996) VMD. Visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–38 [DOI] [PubMed] [Google Scholar]
- 35. Kabsch W., Sander C. (1983) Dictionary of protein secondary structure. Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 [DOI] [PubMed] [Google Scholar]
- 36. Frederick E. D., Ramos S. B., Blackshear P. J. (2008) A unique C-terminal repeat domain maintains the cytosolic localization of the placenta-specific tristetraprolin family member ZFP36L3. J. Biol. Chem. 283, 14792–14800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thompson M. J., Lai W. S., Taylor G. A., Blackshear P. J. (1996) Cloning and characterization of two yeast genes encoding members of the CCCH class of zinc finger proteins. Zinc finger-mediated impairment of cell growth. Gene 174, 225–233 [DOI] [PubMed] [Google Scholar]
- 38. Cuthbertson B. J., Liao Y., Birnbaumer L., Blackshear P. J. (2008) Characterization of zfs1 as an mRNA-binding and -destabilizing protein in Schizosaccharomyces pombe. J. Biol. Chem. 283, 2586–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liang J., Song W., Tromp G., Kolattukudy P. E., Fu M. (2008) Genome-wide survey and expression profiling of CCCH-zinc finger family reveals a functional module in macrophage activation. PLoS One 3, e2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang D., Guo Y., Wu C., Yang G., Li Y., Zheng C. (2008) Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genomics 9, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shimada M., Kawahara H., Doi H. (2002) Novel family of CCCH-type zinc-finger proteins, MOE-1, -2 and -3, participates in C. elegans oocyte maturation. Genes Cells 7, 933–947 [DOI] [PubMed] [Google Scholar]
- 42. Tabara H., Hill R. J., Mello C. C., Priess J. R., Kohara Y. (1999) pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development 126, 1–11 [DOI] [PubMed] [Google Scholar]
- 43. Pagano J. M., Farley B. M., McCoig L. M., Ryder S. P. (2007) Molecular basis of RNA recognition by the embryonic polarity determinant MEX-5. J. Biol. Chem. 282, 8883–8894 [DOI] [PubMed] [Google Scholar]
- 44. Jadhav S., Rana M., Subramaniam K. (2008) Multiple maternal proteins coordinate to restrict the translation of C. elegans nanos-2 to primordial germ cells. Development 135, 1803–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Farley B. M., Pagano J. M., Ryder S. P. (2008) RNA target specificity of the embryonic cell fate determinant POS-1. RNA 14, 2685–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kedar V. P., Zucconi B. E., Wilson G. M., Blackshear P. J. (2012) Direct binding of specific AUF1 isoforms to tandem zinc finger domains of tristetraprolin (TTP) family proteins. J. Biol. Chem. 287, 5459–5471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brooks S. A., Blackshear P. J. (2013) Tristetraprolin (TTP). Interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochim. Biophys. Acta 1829, 666–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fabian M. R., Frank F., Rouya C., Siddiqui N., Lai W. S., Karetnikov A., Blackshear P. J., Nagar B., Sonenberg N. (2013) Structural basis for the recruitment of the human CCR4-NOT deadenylase complex by tristetraprolin. Nat. Struct. Mol. Biol. 20, 735–739 [DOI] [PMC free article] [PubMed] [Google Scholar]