Abstract
The association between NOD2 and tuberculosis (TB) risk has been reported widely, but the results of previous studies remained controversial and ambiguous. To assess the association between NOD2 polymorphisms and TB risk, a meta-analysis was performed. A literature search was conducted by using the PubMed, Ovid, ISI Web of Knowledge, Elsevier ScienceDirect, and Chinese National Knowledge Infrastructure (CNKI). We identified the data from all articles estimating the association between NOD2 polymorphisms and TB risk. In total, 2,215 cases and 1,491 controls in 7 case-control studies were included. In meta-analysis, we found significant association between the Arg702Trp polymorphism and TB risk (OR = 0.43, 95% CI = 0.20-0.90, P = 0.02). However, no significant association was found between the Arg587Arg (OR = 1.31, 95% CI = 0.83-2.07, P = 0.25) and Gly908Arg (OR = 0.78, 95% CI = 0.21-2.87, P = 0.71) polymorphisms and TB risk. The present meta-analysis suggested that NOD2 Arg702Trp polymorphism was likely to be a protective factor for TB. However, the Arg587Arg and Gly908Arg polymorphisms might not be the genetic risk factors for TB susceptibility.
Keywords: NOD2, Single-nucleotide polymorphism, meta-analysis, tuberculosis
Introduction
Tuberculosis (TB) is an infectious disease caused by various strains of mycobacteria, usually mycobacterium tuberculosis (Mtb) in humans. According to World Health Organization (WHO) report, about one-third of the current global population is infected asymptomatically with Mtb, of whom 10% will develop clinical disease during their lifetime 1. The difference between infection rate and incidence rate may be caused by smoking history, physical condition, genetic factor, socio-economic factor, and health resource allocation factor 2. Significant difference in the incidence of TB in different regions, ethnic groups, and populations also proved that genetic factor is responsible for TB susceptibility. The highest incidence of TB is in Asia (58%), followed by Africa (27%), the Eastern Mediterranean Region (8%), the European Region (4%), and the Region of the Americas (3%) 2. China has the world's second largest TB epidemic, and the epidemic in the Western China is higher than the national average 3. The active TB incidence of the Uygur, Kazak, and Mongolian populations in Western China was found to be 2.39 times higher than the Chinese Han in Eastern China. These facts together with other substantial evidence indicate that host genetic factors may play an important role in TB susceptibility, which contains single nucleotide polymorphisms (SNP) as a major factor. Numerous studies have been performed on the association of genetic variants with TB susceptibility including TLR 2 4, CTLA 4 5, MRC1 6, and PTPN22 7 SNPs.
Innate immunity is the first line of defense against Mtb. Pattern recognition receptors (PRRs) can recognize pathogen-associated molecular patterns of Mtb, causing macrophages activation and triggers an immune response. Toll-like receptors (TLR) and NOD-like receptors (NLR) are two main kinds of PRRs. NOD2 is a member of the NLR family located on chromosome 16q21. It has been shown to recognize muramyl dipeptide (MDP), a component of bacterial cell wall peptidoglycan (PGN), by Leucine-rich repeat (LRR) region. Upon recognition of agonist, NOD2 self-oligomerize to activate RICK via CARD-CARD interactions. RICK is then able to activate the IKK complex, which in turn activate NF-κB that ultimately induces pro-inflammatory cytokine transcription to clear intracellular bacteria 8. Austin et al. 9 sequenced NOD2 gene and found that Pro268Ser, Arg702Trp, and Ala725Gly were significantly associated with TB disease. Pro268Ser was found to be a susceptible SNP that can lead to nucleotide mutation resulting in amino acid changes, thereby changing the protein structure, thus affecting African Americans' TB incidence. But, no association between Arg702Trp, Gly908Arg and 3020insC of the NOD2 gene and TB was found in the Gambia population 10.
Many studies have reported the association between NOD2 polymorphisms and TB. However, the results are inconsistent and inconclusive due to limited sample sizes and different study populations. Therefore, in this article, we performed a systematic review and meta-analysis, based on literature identification until 17th July 2013 to summarize the associations between these polymorphisms and TB susceptibility.
Materials and Methods
Selection of Studies in Meta-analysis
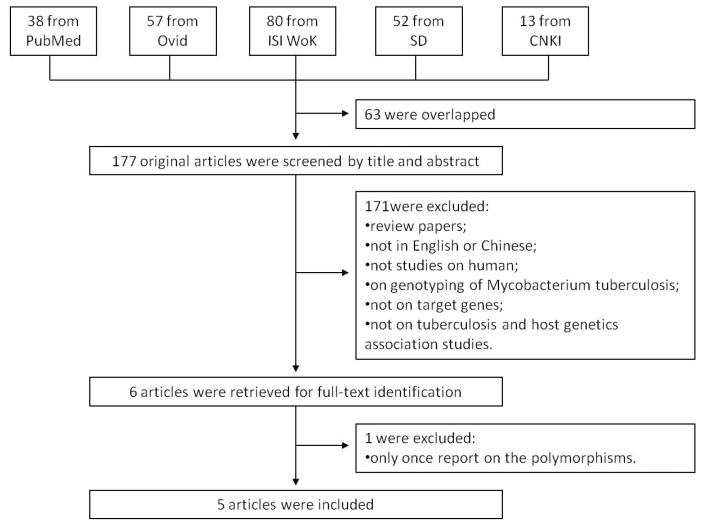
We searched the PubMed, Ovid, ISI Web of Knowledge, Elsevier ScienceDirect, and Chinese National Knowledge Infrastructure (CNKI) to identify studies of the association between TB susceptibility and NOD2 polymorphisms until 17th July 2013. The key words were: 'Mycobacterium tuberculosis' or 'tuberculosis' in combination with 'polymorphism' or 'variant' or 'genotype' or 'allele' or 'mutation', in combination with 'Nucleotide binding oligomerization domain containing 2' or 'NOD2' or 'CARD15' or 'CD' or 'ACUG' or 'BLAU' or 'IBD1' or 'NLRC2' or 'NOD2B' or 'CLR16.3' or 'PSORAS1'. The search results were limited to English and Chinese language articles. All retrieved titles and abstracts were examined for relevant studies. Inclusion criteria of our study were: 1) case control studies of unrelated individuals; 2) evaluation of NOD2 polymorphisms and TB susceptibility; 3) genotype distribution in both cases and controls were available. Exclusion criteria were: 1) study design based on family or sibling pairs; 2) genotype frequencies not reported; and 3) data from reviews and abstracts. Additional studies were also identified by hand searching reference lists of original studies and review articles including meta-analysis. The systematic review process is described in the flowchart of Figure 1.
Figure 1.
Flow chart showing the study selection procedure. Ovid: Ovid MEDLINE; ISI WoK: ISI Web of Knowledge; SD: Elsevier ScienceDirect; CNKI: China National Knowledge Infrastructure.
Data Extraction
All of the publications were assessed independently by two reviewers. For all studies, two reviewers extracted the following data from original publications: first author and year of publication; distribution of genotypes for each polymorphism among cases and controls; characteristics of the study design and the study population (study base, numbers and mean age of cases and controls, TB diagnosis, HIV status, source of controls, matching criteria and host ethnicity).
Bias Exclusion
Publication bias was evaluated by Begg's test and Egger's test 11, 12. Bias in location of studies, English language bias, and database bias was excluded by searching both English and Chinese database.
Statistical Analyses
Hardy-Weinberg Equilibrium (HWE) was examined in controls by asymptotic Pearson's Chi-square test for each polymorphism in each study. The association between polymorphism and TB was estimated by means of odds ratios (OR) and corresponding 95% confidence intervals (CI) comparing cases to controls. Study heterogeneity was tested by the Q test and I2 test 13, and the heterogeneity was considered significant if P-value was less than 0.05 or I2 > 50%. Fixed-effects models were adopted when P-value was more than 0.05; otherwise random-effects models were used. Statistical analyses were carried out using the Stata/SE 12.0 (College Station, TX, USA) and the Review Manager 5.1 software (Oxford, England).
Results
Characteristics of Included Studies
A total of 240 articles were achieved by literature search. As shown in Figure 1, after excluding those overlapped abstracts between the databases, 177 abstracts were retrieved for detailed evaluation. Six articles were left after removing 171 unsuitable articles, and one of them was excluded because its SNP was only once reported. Finally, 7 studies from 5 articles 9, 10, 14-16 were included in this review and meta-analysis. Among them, there were four studies for NOD2 Arg587Arg, seven studies for NOD2 Arg702Trp, and four studies for NOD2 Gly908Arg. Otherwise, there was only one study for SNPs such as Ala140Thr, Pro268Ser, Val955Ile, and 3020insC, which was eliminated. As shown in Table 1, the study participants were from diverse descents including American, African, and Asian. The pooled sample size was 3,706 (2,215 cases and 1,491 controls). The genotype and allele distributions of all the polymorphisms are shown in Table 2.
Table 1.
Characteristics of the 7 studies included in the meta-analysis.
| Author | Year | Country | Ethnicity | Age, years mean ± SD or mean (range) | Samples, n | Genotying method | ||
|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | |||||
| Austin et al | 2007 | United States | African Americans | 46.5 ± 12.9 | 45.4 ± 10.6 | 377 | 187 | PCR-sequencing |
| Möller et al | 2006 | South Africa | South Africans | 34 ± 14.8 | 27 ± 12.3 | 432 | 482 | Taq Man SNP genotyping assays |
| Singh et al | 2012 | Northern Indian states | Indians | 33.18 ± 13.29 | 36.85 ± 12.6 | 263 | 122 | PCR-RFLP |
| Stockton et al | 2004 | Gambia | Gambians | 36 (19-58) | 32 (18-50) | 320 | 320 | PCR-RFLP, PCR-FRET |
| Zhao et al_a | 2012 | China | Chinese Han | 36.47 ± 15.73 | 37.02 ± 15.29 | 219 | 215 | PCR-sequencing |
| Zhao et al_b | 2012 | China | Chinese Uygur | 28.8 ± 10.1 | 30.1 ± 8.9 | 86 | 72 | PCR-sequencing |
| Zhao et al_c | 2012 | China | Chinese Kazak | 28.8 ± 10.1 | 37.02 ± 15.29 | 120 | 93 | PCR-sequencing |
PCR = polymerase chain reaction; RFLP = restriction fragment length polymorphism; FRET = fluorescence resonance energy transfer.
Table 2.
Genotype and allele distribution of NOD2 polymorphisms in TB and controls.
| SNP | Study | Cases | Controls | HWE P | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TT | TG | GG | T | G | TT | TG | GG | T | G | |||
| NOD2 Arg587Arg | Austin et al | 377 | 0 | 0 | 754 | 0 | 187 | 0 | 0 | 374 | 0 | / |
| Zhao et al_a | 160 | 53 | 4 | 377 | 61 | 185 | 28 | 2 | 398 | 32 | 0.423 | |
| Zhao et al_b | 49 | 29 | 8 | 127 | 45 | 37 | 29 | 6 | 103 | 41 | 0.925 | |
| Zhao et al_c | 65 | 147 | 8 | 177 | 63 | 56 | 32 | 5 | 144 | 42 | 0.878 | |
| CC | CT | TT | C | T | CC | CT | TT | C | T | |||
| NOD2 Arg702Trp | Austin et al | 372 | 5 | 0 | 749 | 5 | 187 | 9 | 0 | 365 | 9 | 0.742 |
| rs2066844 | Möller et al | 421 | 6 | 0 | 848 | 6 | 468 | 11 | 0 | 947 | 11 | 0.799 |
| Singh et al | 263 | 0 | 0 | 526 | 0 | 122 | 0 | 0 | 244 | 0 | / | |
| Stockton et al | 192 | 0 | 0 | 384 | 0 | 192 | 0 | 0 | 384 | 0 | / | |
| Zhao et al_a | 217 | 0 | 0 | 434 | 0 | 215 | 0 | 0 | 430 | 0 | / | |
| Zhao et al_b | 86 | 0 | 0 | 172 | 0 | 72 | 0 | 0 | 144 | 0 | / | |
| Zhao et al_c | 120 | 0 | 0 | 240 | 0 | 93 | 0 | 0 | 186 | 0 | / | |
| GG | GC | CC | G | C | GG | GC | CC | G | C | |||
| NOD2 Gly908Arg | Austin et al | 377 | 0 | 0 | 754 | 0 | 187 | 0 | 0 | 374 | 0 | / |
| rs2066845 | Möller et al | 426 | 2 | 0 | 854 | 2 | 476 | 4 | 0 | 956 | 4 | 0.927 |
| Singh et al | 260 | 3 | 0 | 523 | 3 | 121 | 1 | 0 | 243 | 1 | 0.926 | |
| Stockton et al | 192 | 0 | 0 | 384 | 0 | 192 | 0 | 0 | 384 | 0 | / |
HWE: Hardy-Weinberg Equilibrium.
Quantitative Data Synthesis
NOD2 Arg587Arg polymorphism
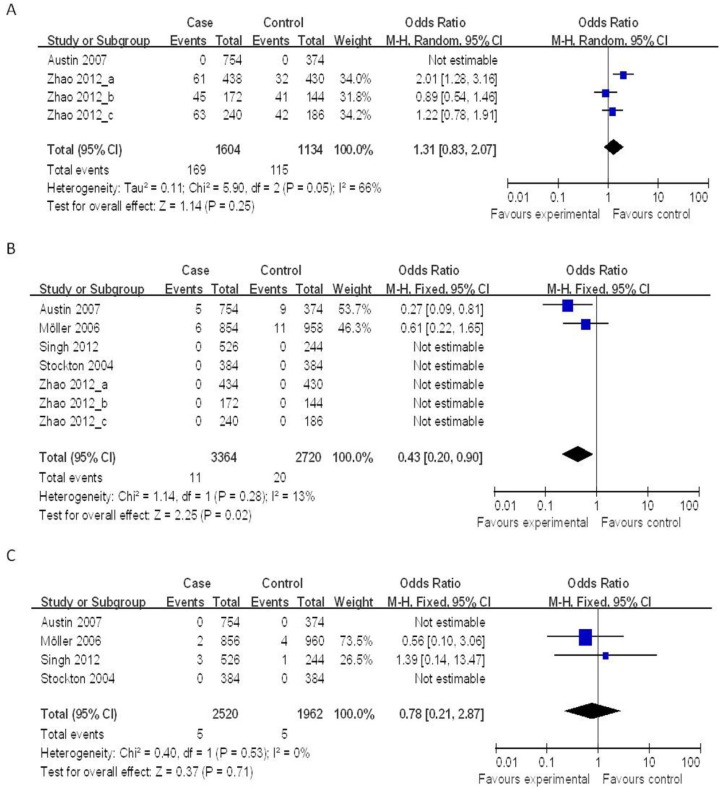
Four case-control studies (802 cases and 567 controls) on the relationship between the NOD2 Arg587Arg polymorphisms and the risk of TB were analyzed. We analyzed the heterogeneity of G vs. T for all studies and the value of χ2 was 5.90 with 2 degrees of freedom and P = 0.05. I2, another index of the test of heterogeneity, was 66%, suggesting a high heterogeneity. Thus, we chose the random-effect model to synthesize the data. Overall, no association was found in the allelic frequency with TB risk (OR = 1.31, 95% CI = 0.83-2.07, P = 0.25) (Figure 2A).
Figure 2.
Forrest plot of the association between NOD2 and TB risk in allele comparison. OR: odds ratio; CI: confidence interval; df: degrees of freedom. A: G vs. T of NOD2 Arg587Arg ; B: T vs. C of NOD2 Arg702Trp; C: C vs. G of NOD2 Gly908Arg.
NOD2 Arg702Trp polymorphism
Seven case-control studies (2,215 cases and 1,491 controls) were included in the meta-analysis on the relationship between the NOD2 Arg702Trp polymorphisms and the risk of TB. As shown in Figure 2B, the heterogeneity of T vs. C for all the studies was analyzed. The χ2 value was 1.14, with 1 df and P = 0.28, while I2 was 13%, suggesting a low heterogeneity. Thus, we chose the fixed-effect model, and found that the individuals who carried the T allele may have a decreased risk of TB (OR = 0.43, 95% CI = 0.20-0.90, P = 0.02) (Figure 2B).
NOD2 Gly908Arg polymorphism
Four case-control studies (1,392 cases and 1,111 controls) on the relationship between the NOD2 Gly908Arg polymorphisms and the risk of TB were included in the meta-analysis. The results on this polymorphism indicated that the C allele had no significant association to TB susceptibility as compared to the G allele under the fixed-effects models (OR = 0.78, 95% CI = 0.21-2.87, P = 0.71) (Figure 2C).
Publication Bias
Publication bias was analyzed by using the Begg's test and Egger's test. The publication bias by Begg's test showed no significant publication bias in NOD2 Arg587Arg, Arg702Trp, and Gly908Arg (P = 1.000, P = 1.000, P = 1.000). Then, Egger's test was also used. For NOD2 Arg587Arg polymorphism, the P value of Egger's test was 0.520. The results still did not suggest any evidence of publication bias.
Sensitivity Analysis
Sensitivity analysis was performed by sequentially excluding individual studies. After sequentially excluding each case-control study, statistically similar results were obtained for G vs. T of Arg587Arg, T vs. C of Arg702Trp, and C vs. G of Gly908Arg, indicating that our data were stability and liability in this meta-analysis.
Discussion
In this study, we found that NOD2 SNPs show different effects on the incidence of TB. The meta-analysis revealed an association between the Arg702Trp T allele and TB protection. At the same time, different ethnicity groups contribute to different TB susceptibility, such as Arg587Arg G allele is a TB risk SNP in Chinese Han while having no association with Kazak and Uygur. However, the Gly908Arg polymorphism did not show significant association with TB.
NOD2 is a kind of PRRs, which can identify Mtb invasion in cells. A recent report indicates that NOD2 and TLR pathways are non-redundant in the recognition of Mtb, but can synergize to induce a robust pro-inflammatory response 17. In addition, it has been shown in bone marrow-derived murine macrophages (BMMs) that RIP2 polyubiquitination occurs during Mtb incubation in a NOD2-dependent manner 18. Brooks et al. 19 also found that NOD2 in human macrophages can not only control the pro-inflammatory cytokine response, but also control the proliferation of Mtb. According to the importance of NOD2 in Mtb infection, we suggested that Arg587Arg, Arg702Trp, and Gly908Arg polymorphisms may be associated with TB susceptibility.
Arg587Arg is located on the winged helix and superhelical domain of the NOD2 protein coding by exon 4 and linked to the NACHT domain for synonymous mutation. Sato et al. 20 found that Arg587Arg G allele was associated with significantly better lung function in pulmonary sarcoidosis. In addition, Arg587Arg G allele has also been found to be a protective site of inflammatory bowel disease 21. Several studies have revealed a substantial contribution of synonymous SNPs to human disease risk 22. We suggested that Arg587Arg may be associated with susceptibility to TB in the Chinese Han population. However, in our meta-analysis, Arg587Arg GT genotype is a TB risk factor in Chinese Han, but not in Kazak and Uygur, with the fact that this SNP even do not exist in African Americans. The reasons for the difference were unclear, but the differences in the polymorphisms frequency and the differences in the infected Mtb strains might contribute to this heterogeneity.
Arg702Trp is also located in the exon 4, a missense mutation coding for LRR. Austin et al. 9 showed that the C to T change on Arg702Trp is a protective factor for African Americans, but, Möller et al. 15 indicated that the site has nothing to do with the incidence of TB in South Africa. In this meta-analysis, we found that Arg702Trp is a protective factor for the incidence of TB (OR = 0.43, 95% CI = 0.20-0.90, P = 0.02), although it appears only in the African population in limited articles. This SNP can lead to an amino acid changes, from a low hydrophobic arginine to a high hydrophobic tryptophan. Articles showed Arg702Trp did not change the membrane localization function of NOD2 protein 23, with the normal MDP-induced NF-κB activation 24. Because of the location of Arg702Trp, we speculated that Arg702Trp could reinforce the LRR combination with MDP, making it easier to clear intracellular Mtb.
Gly908Arg is also a missense mutation coding for LRR, which is located in the exon 8. Literature showed that Gly908Arg expressed in HEK293T cells reveals defects in NF-κB transcription 25, 26, leading to a broadly impaired transcriptional response and impaired cytokine, and co-stimulatory protein induction with MDP stimulation 27, 28, thus affecting bacterial diseases. Existing literature suggest that Gly908Arg is associated with Crohn's disease in the Dutch 29, Swedish 30, and Italian 31 populations. However, there was no such association with TB, which is reconfirmed by our meta-analysis.
However, there are still some limitations in our meta-analysis that should be considered when explaining the present results. Firstly, we did not track the unpublished articles to obtain data for analysis. The potential effect of this publication bias is unknown. Secondly, due to the limited number of studies, there are some SNPs such as 3020insC that were not included in this systematic analysis. Thirdly, the small sample sizes in some subgroup analyses may have limited statistical power to estimate the possible risk for NOD2 polymorphisms.
In conclusion, this systematic review summarized the association between NOD2 polymorphisms and TB susceptibility. Our results indicated that NOD2 Arg702Trp polymorphism is likely to be the protective allele. However, the Arg587Arg and Gly908Arg polymorphisms might not be the genetic risk factors for TB susceptibility, knowing the fact that Arg587Arg is only a risk factor in Chinese Han. More studies with a larger group of populations should be performed, especially in Caucasians.
Acknowledgments
This work was supported by grants from National Science and Technology Major Project of the Ministry of Science and Technology of China (No. 2012ZX10005001-006) and National Natural Science Foundation of China (No. 81072724, No.81273882).
References
- 1.World Health Organization, Fact sheet N°104. http://www.who.int/mediacentre/factsheets/fs104/en/index.html.
- 2.World Health Organization, Global Tuberculosis Report 2013. http://apps.who.int/iris/bitstream/10665/91355/1/9789241564656_eng.pdf.
- 3.Technical Guidance Group of the Fifth National TB Epidemiological Survey, The Office of the Fifth National TB Epidemiological Survey. The fifth national tuberculosis epidemiological survey in 2010. Chin J Antituber. 2012;34(8):485–508. [Google Scholar]
- 4.Zhang Y, Jiang T, Yang X. et al. Toll-Like Receptor -1, -2, and -6 Polymorphisms and Pulmonary Tuberculosis Susceptibility: A Systematic Review and Meta-Analysis. PLOS ONE. 2013;8(5):e63357. doi: 10.1371/journal.pone.0063357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang C, Jiang T, Wei L. et al. Association of CTLA4 Gene Polymorphisms with Susceptibility and Pathology Correlation to Pulmonary Tuberculosis in Southern Han Chinese. Int J Biol Sci. 2012;8(7):945–952. doi: 10.7150/ijbs.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X, Jiang F, Wei L. et al. Polymorphic Allele of Human MRC1 Confer Protection against Tuberculosis in a Chinese Population. Int J Biol Sci. 2012;8(3):375–82. doi: 10.7150/ijbs.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boechat AL, Ogusku MM, Sadahiro A. et al. Association between the PTPN22 1858C/T gene polymorphism and tuberculosis resistance. Infect Genet Evol. 2013;16:310–3. doi: 10.1016/j.meegid.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 8.Ting JP, Duncan JA, Lei Y. How the noninflammasome NLRs function in the innate immune system. Science. 2010;327:286–90. doi: 10.1126/science.1184004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Austin CM, Ma X, Graviss EA. Common nonsynonymous polymorphisms in the NOD2 gene are associated with resistance or susceptibility to tuberculosis disease in African Americans. J Infect Dis. 2008;197(12):1713–6. doi: 10.1086/588384. [DOI] [PubMed] [Google Scholar]
- 10.Stockton JC, Howson JM, Awomoyi AA. et al. Polymorphism in NOD2, Crohn's disease, and susceptibility to pulmonary tuberculosis. FEMS Immunol Med Microbiol. 2004;41(2):157–60. doi: 10.1016/j.femsim.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Egger M, Davey Smith G, Schneider M. et al. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Begg CB, Berlin JA. Publication bias and dissemination of clinical research. J Natl Cancer Inst. 1989;81:107–15. doi: 10.1093/jnci/81.2.107. [DOI] [PubMed] [Google Scholar]
- 13.Higgins JP, Thompson SG, Deeks JJ. et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh V, Gaur R, Mittal M. et al. Absence of nucleotide-binding oligomerization domain-containing protein 2 variants in patients with leprosy and tuberculosis. Int J Immunogenet. 2012;39(4):353–6. doi: 10.1111/j.1744-313X.2012.01085.x. [DOI] [PubMed] [Google Scholar]
- 15.Möller M, Nebel A, Kwiatkowski R. et al. Host susceptibility to tuberculosis: CARD15 polymorphisms in a South African population. Mol Cell Probes. 2007;21(2):148–51. doi: 10.1016/j.mcp.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Zhao M, Jiang F, Zhang W. et al. A novel single nucleotide polymorphism within the NOD2 gene is associated with pulmonary tuberculosis in the Chinese Han, Uygur and Kazak populations. BMC Infect Dis. 2012;12:91. doi: 10.1186/1471-2334-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferwerda G, Girardin SE, Kullberg BJ. et al. NOD2 and toll-like receptors are nonredundant recognition systems of Mycobacterium tuberculosis. PLoS Pathog. 2005;1:279–85. doi: 10.1371/journal.ppat.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y, Yin C, Pandey A. et al. NOD2 pathway activation by MDP or Mycobacterium tuberculosis infection involves the stable polyubiquitination of Rip2. J Biol Chem. 2007;282:36223–9. doi: 10.1074/jbc.M703079200. [DOI] [PubMed] [Google Scholar]
- 19.Brooks MN, Rajaram MV, Azad AK. et al. NOD2 controls the nature of the inflammatory response and subsequent fate of Mycobacterium tuberculosis and M. bovis BCG in human macrophages. Cell Microbiol. 2011;13(3):402–18. doi: 10.1111/j.1462-5822.2010.01544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sato H, Williams HR, Spagnolo P. et al. CARD15/NOD2 polymorphisms are associated with severe pulmonary sarcoidosis. Eur Respir J. 2010;35(2):324–30. doi: 10.1183/09031936.00010209. [DOI] [PubMed] [Google Scholar]
- 21.Lesage S, Zouali H, Cezard JP. et al. CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am J Hum Pharmacogenomics. 2007;8(8):1075–80. doi: 10.1086/339432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet. 2011;12(10):683–91. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- 23.Barnich N, Aguirre JE, Reinecker HC. et al. Membrane recruitment of NOD2 in intestinal epithelial cells is essential for nuclear factor-kB activation in muramyl dipeptide recognition. J Cell Biol. 2005;170:21–6. doi: 10.1083/jcb.200502153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDonald C, Chen FF, Ollendorff V. et al. A role for Erbin in the regulation of Nod2-dependent NF-kappaB signaling. J Biol Chem. 2005;280:40301–9. doi: 10.1074/jbc.M508538200. [DOI] [PubMed] [Google Scholar]
- 25.Inohara N, Ogura Y, Fontalba A. et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J Biol Chem. 2003;278:5509–12. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 26.Girardin SE, Boneca IG, Viala J. et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278:8869–72. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Moran T, Swanson E. et al. Regulation of IL-8 and IL-1beta expression in Crohn's disease associated NOD2/CARD15 mutations. Hum Mol Genet. 2004;13:1715–25. doi: 10.1093/hmg/ddh182. [DOI] [PubMed] [Google Scholar]
- 28.Netea MG, Ferwerda G, de Jong DJ. et al. Nucleotide-binding oligomerization domain-2 modulates specific TLR pathways for the induction of cytokine release. J Immunol. 2005;174:6518–23. doi: 10.4049/jimmunol.174.10.6518. [DOI] [PubMed] [Google Scholar]
- 29.van der Linde K, Boor PP, Houwing-Duistermaat JJ. et al. CARD15 mutations in Dutch familial and sporadic inflammatory bowel disease and an overview of European studies. Eur J Gastroenterol Hepatol. 2007;19(6):449–59. doi: 10.1097/01.meg.0000236887.44214.6a. [DOI] [PubMed] [Google Scholar]
- 30.Törkvist L, Noble CL, Lördal M. et al. Contribution of CARD15 variants in determining susceptibility to Crohn's disease in Sweden. Scand J Gastroenterol. 2006;41:700–5. doi: 10.1080/00365520500395245. [DOI] [PubMed] [Google Scholar]
- 31.Giachino D, van Duist MM, Regazzoni S. et al. Analysis of the CARD15 variants R702W, G908R and L1007fs in Italian IBD patients. Eur J Hum Genet. 2004;12(3):206–12. doi: 10.1038/sj.ejhg.5201130. [DOI] [PubMed] [Google Scholar]