Abstract
The Hippo pathway, which is conserved from Drosophila to mammals, has been recognized as a tumor suppressor signaling pathway governing cell proliferation and apoptosis, two key events involved in organ size control and tumorigenesis. Although several upstream regulators, the conserved kinase cascade and key downstream effectors including nuclear transcriptional factors have been defined, the global organization of this signaling pathway is not been fully understood. Thus, we conducted a proteomic analysis of human Hippo pathway, which revealed the involvement of an extensive protein–protein interaction network in this pathway. The mass spectrometry data were deposited to ProteomeXchange with identifier PXD000415. Our data suggest that 550 interactions within 343 unique protein components constitute the central protein–protein interaction landscape of human Hippo pathway. Our study provides a glimpse into the global organization of Hippo pathway, reveals previously unknown interactions within this pathway, and uncovers new potential components involved in the regulation of this pathway. Understanding these interactions will help us further dissect the Hippo signaling-pathway and extend our knowledge of organ size control.
The Hippo pathway has been extensively studied in the past two decades and is known to play crucial roles in regulating cell proliferation and apoptosis, thus contributing to organ size control, organism development, and cancer formation (1–3). The Hippo pathway was initially identified in the Drosophila system through genetic mosaic screening for tumor suppressor genes. Mutation in Hippo components, such as Hippo (4–8), Salvador (9), Warts (10, 11), and Mats (12), results in tissue overgrowth phenotypes in Drosophila. The Hippo and Warts proteins, which associate with their respective adaptor proteins Salvador and Mats, constitute a kinase cascade (1–3) and phosphorylate the key downstream target protein Yorkie (13). Yorkie is a transcriptional co-activator that controls the transcriptional program downstream of the Hippo pathway through its interaction with transcription factor Scalloped (14–17) in the nucleus; it promotes transcription of a number of downstream genes involved in cell proliferation and anti-apoptosis. The critical event in the Hippo pathway is regulation of the nuclear localization of Yorkie. Activation of Hippo pathway leads to Hippo/Warts-dependent phosphorylation of Yorkie. 14-3-3 proteins recognize phosphorylated Yorkie, preventing its nuclear localization (18) and thus suppressing its nuclear transcriptional co-activator function and inhibiting cell proliferation.
The Hippo pathway is highly conserved from Drosophila to mammals, as almost all the components in Drosophila have recognizable orthologs in mammals. In mammalian systems, MST1/2 (Hippo orthologs) and the adaptor protein SAV1 (Salvador orthologs) form the kinase cascade with LATS1/2 (Warts orthologs) and adaptor proteins MOB1A/B (Mats orthologs). YAP1 and TAZ (Yorkie orthologs) function together with TEAD1/2/3/4 (Scalloped orthologs) in the nucleus and regulate the transcription of downstream genes. Similar to the situation in Drosophila, activation of the Hippo pathway in mammalian systems leads to LATS1/2-dependent phosphorylation of YAP1/TAZ, which prevents YAP1/TAZ nuclear localization via binding to 14-3-3 in cytoplasm and also initiates the degradation of YAP1/TAZ by β-TRCP E3 ligase (19). Furthermore, WWC1 (Kibra orthologs) (20–22), NF2 (Merlin orthologs) (23–25) and FRMD6 (Expanded orthologs) (26) have been identified as upstream regulators for the core kinase cascade. Although the upstream receptors are still elusive, recent studies suggest that GPCR (27) and protease-activated receptors (28, 29) may be the upstream components that link extracellular signals to activation of the Hippo pathway in mammals.
Besides the core components in the Hippo pathway, some new regulators have recently been identified through different methods, such as a genetic screen in Drosophila, a short-hairpin RNA (shRNA)1-mediated loss-of-function screen, and proteomic studies for new interacting proteins. A number of these newly identified regulators control YAP1/TAZ phosphorylation and localization through regulation of the core kinase cascade in the Hippo pathway. For example, the centrosome protein NPHP4 was shown to negatively regulate of the Hippo pathway by directly interacting with LATS1 and diminishing its ability to phosphorylate YAP1 or TAZ (30). The sterile 20 kinase TAOK1 was discovered to positively regulate the Hippo pathway by phosphorylating MST1 kinase, a component of the Hippo kinase cascade (31, 32). LIM-domain-containing proteins such as Ajuba suppress YAP1 phosphorylation by binding to LATS1 kinase and SAV1 adaptor protein (33) and mediate the crosstalk between EGFR-MAPK signaling and the Hippo pathway (34). In addition, RASSF proteins antagonize Hippo signaling through their association with MST1/2 and SAV1 (35, 36).
Another group of newly identified regulators of the Hippo pathway act by directly suppressing YAP1/TAZ oncogenic activities via physical protein–protein interaction, which is independent of the canonical kinase cascade. For example, we and others identified the angiomotin protein family (AMOT, AMOTL1, and AMOTL2) as the major binding partners of YAP1 or TAZ (37–39). Angiomotin proteins facilitate the translocation of both wild-type and phosphorylated YAP1 or TAZ from nucleus to cytoplasm, thereby suppressing the oncogenic functions of YAP1 in the nucleus. In addition, non-receptor protein tyrosine phosphatase 14 (PTPN14) suppresses YAP1 nuclear localization in a density-dependent manner (40, 41), which requires a direct protein–protein interaction between PTPN14 and YAP1, but not the tyrosine phosphatase activity of PTPN14 (40, 42). In the context of tissue homeostasis and cancer development, polarity determinant protein SCRIB was found to translocate TAZ to the cell membrane and the translocation is crucial for self-renewal and tumor-initiation capacities of breast cancer stem cells (43). Adhesion junction protein α-catenin is also required for homeostasis in skin by controlling YAP1 activity (44, 45). Loss of α-catenin in mice leads to over proliferation of epidermal stem cells and the development of skin squamous cell carcinoma (44).
Together, these newly identified regulators include (1) upstream regulators (NF2, FRMD6, WWC1, RASSF1–8, and TAO1), (2) adhesion junction proteins (AMOT, AMOTL1, AMOTL2, PTPN14, β-catenin, and α-catenin), (3) tight junction proteins (PAR3, PAR6, and ZO2), (4) septate junction complex proteins (SCRIB, LLGL1/2, DLG1, and CRB3), and (5) others (ASSP, NPHP4, JUP1, and LIMD1). Although more and more proteins have been demonstrated to regulate the Hippo pathway, the dissection of cell context-dependent connections among these regulators is lagging behind. The limited understanding of the connections among these newly identified Hippo regulators and the canonical Hippo pathway prompted us to initiate a proteomic analysis to establish the protein–protein interaction network of the Hippo pathway. We reasoned that such a study will help to elucidate the biological functions and cross-talk involving this important signaling pathway. Moreover, identification of additional new associated proteins will provide further insights into the possible biological functions, regulatory mechanisms, and cellular signaling network associated with the Hippo pathway.
To elucidate the interactome involving the Hippo pathway, we took advantage of tandem affinity purification followed by mass spectrometry analysis for the identification of associated proteins. We employed an unbiased methodology, SAINT (Significance Analysis of INTeractome), to identify high-confidence candidate interacting proteins (HCIPs) for all of the known major Hippo components as well as regulators involved in this pathway. This study led to the discovery of many new components of the Hippo pathway, expanded the roles of the Hippo pathway in multiple biological processes, and established a core protein–protein interaction network that is valuable resource for further mechanistic studies of this pathway.
EXPERIMENTAL PROCEDURES
Antibodies
Anti-YAP1 antibody for immunoprecipitation was raised by immunizing rabbit with bacterially expressed and purified GST-fused full-length human YAP1 protein (Cocalico Biologicals, Inc., Reamstown, PA). Anti-YAP1 and phosphor-YAP1 (S127) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and Cell Signaling Technology (Danvers, MA) respectively. Anti-α-tubulin and anti-Flag (M2) monoclonal antibodies and anti-Flag polyclonal antibody were obtained from Sigma-Aldrich. Anti-Myc and anti-GFP monoclonal antibodies were purchased from Santa Cruz Biotechnology. Anti-LATS1 and anti-phoshor-LATS1 (Ser909) polyclonal antibodies were purchased from Cell Signaling Technology. Anti-CIT monoclonal antibody was obtained from Millipore.
Constructs and Viruses
The plasmids encoding Hippo pathway components (FRMD6, JUB, LATS1, LLGL1, LLGL2, MOB1A, MOB1B, MST1, MST2, NF2, NT-AMOT, p130AMOT, p80AMOT, PTPN14, RASSF1, RHOA, SAV1, SCRIB, SMAD7, TAZ, TEAD2, TEAD4, WWC1, and YAP1) were purchased from Harvard Plasmids Resource and Open Biosystems. AMOTL1, AMOTL2, and DLG1 plasmids were kindly provided by Dr. Anthony P. Schmitt (Pennsylvania State University), Dr. Anming Meng (Tsinghua University), and Dr. Alexandra Newton (University of California at San Diego), respectively. LATS2 and its kinase dead mutant LATS2KR were kindly provided by Dr. Moshe Oren (Weizmann Institute). LIMD1and NPHP4 plasmids were kindly provided by Dr. Greg Longmore (Washington University) and Dr. Bernhard Schermer (University of Cologne), respectively. All constructs were generated by polymerase chain reaction (PCR) and subcloned into pDONOR201 vector using Gateway Technology (Invitrogen, Carlsbad, CA) as the entry clones. For tandem affinity purification (TAP) analysis, all the entry clones were subsequently recombined into lentiviral-gateway-compatible destination vector for the expression of C-terminal triple (S tag-Flag tag-SBP tag, or SFB) tagged fusion proteins.
Gateway-compatible destination vectors with indicated SFB tag, GST tag, Myc tag, or GFP tag were used to express various fusion proteins for the WWC1/CIT and YAP1/CCDC85C studies. For CIT truncation mutants, residues 1∼360, 1∼1297, 1∼1700, 1412∼2027, 1564∼2027, or 1882∼2027 were deleted in D1 to D6 mutants, respectively. PCR-mediated site-directed mutagenesis was used to generate point mutations or deletions. CIT PY motif PPTY (residues 1926∼1929) was either mutated to PPTA and referred as CITmPY, or deleted and referred as PY motif deletion mutant CITdPY. Two WW domains (WW1: residues12∼34; WW2: residues 59∼88) located at the N terminus of WWC1 were deleted as single-deletion mutants WWC1dWW1and WWC1dWW2 or double-deletion mutant WWC1dWW1/2. The CCDC85C PY motif (residues 316∼319) was deleted in the CCDC85CdPY mutant. The two WW domains of YAP1 (WW1: residues 130∼152; WW2: residues 236∼258) were either single deleted as YAP1dWW1 and YAP1dWW2 mutants, or double deleted as YAP1dWW1/2.
Two individual pGIPZ lentiviral shRNAs targeting CIT were obtained from the shRNA and ORFeome core facility at The University of Texas MD Anderson Cancer Center. The shRNA sequences were as follows:
CIT shRNA-1# (V3LHS_366009): 5′-AGCGACAGAATGTCAGCAT-3′;
CIT shRNA-2# (V3LHS_366011): 5′-ACGATGAGCTGCTAGAAAA-3′;
Control shRNA: 5′-TCTCGCTTGGGCGAGAGTAAG-3′.
All lentiviral supernatants were generated by transient transfection of 293T cells with helper plasmids pSPAX2 and pMD2G (kindly provided by Dr. Zhou Songyang, Baylor College of Medicine) and harvested 48 h later. Supernatants were passed through a 0.45-μm filter and used to infect MCF10A cells with the addition of 8 μg/ml polybrene.
Cell Culture and Transfection
293T and HeLa cells were purchased from American Type Culture Collection and maintained in Dulbecco modified essential medium (DMEM) supplemented with 10% fetal bovine serum at 37 °C in 5% CO2 (v/v). MCF10A cells were kindly provided by Dr. Dihua Yu (MD Anderson Cancer Center). MCF10A cells were maintained in DMEM/F12 medium supplemented with 5% horse serum, 200 ng/ml epidermal growth factor, 500 ng/ml hydrocortisone, 100 ng/ml cholera toxin, and 10 μg/ml insulin at 37 °C in 5% CO2 (v/v). All culture media contained 1% penicillin and streptomycin antibiotics. Plasmid transfection was performed using the polyethylenimine reagent.
Tandem Affinity Purification of SFB-tagged Protein Complexes
HEK293T cells were infected twice with lentivirus encoding SFB-fused Hippo group or control group proteins. Cells stably expressing the SFB-tagged Hippo group or control group proteins were selected by culturing in medium containing 2 μg/ml puromycin and confirmed by immunostaining and Western blotting. For affinity purification, 293T cells were subjected to lysis in NETN buffer (with protease inhibitors) at 4 °C for 20 min. Crude lysates were subjected to centrifugation at 4 °C and 14,000 rpm for 15 min. Supernatants were incubated with streptavidin-conjugated beads (Amersham Biosciences) for 1 h at 4 °C. The beads were washed three times with NETN buffer, and bounded proteins were eluted with NETN buffer containing 2 mg/ml biotin (Sigma) nearly 90 min at 4 °C. The eluates were incubated with S protein beads (Novagen, Madison, WI) for 1 h. The beads were washed three times with NETN buffer and subjected to SDS-PAGE. Protein bands were excised and subjected to mass spectrometry analysis (performed by Taplin Mass Spectrometry Facility, Harvard Medical School).
Data Analysis and Bioinformatics Analysis
The prey and bait protein sequences were downloaded from the UniProt Consortium (46). The whole Hippo pathway and individual component interactomes were generated by Cytoscape (47) and Ingenuity pathway software (Ingenuity Systems, www.ingenuity.com). The function annotations were also analyzed by Ingenuity pathway software. The heatmap for the hierarchical clustering was generated by MeV-4.8.1 and Heatmap Builder software.
For the evaluation of potential protein–protein interactions, raw data from the mass spectrometry analysis were subjected to assessment by the SAINT methodology (48, 49). The spectra counts from Hippo group and control group proteins were assembled as a matrix for all of the bait and prey proteins. In total, 9239 binary interactions were identified in 64 experiments carried out with 32 Hippo group purifications, 31 control group purifications, and a vector control.
According to the SAINT algorithms, 20,000 simulation runs were performed for Gibbs sampling. The total spectra count (TSC) was fixed for each bait, and these spectra counts were assembled together as a pool of all the bait spectra counts. Then we randomly drew spectra from the pool and randomly assigned them to the baits until the number of spectra reached the number in the real data. We simulated 100,000 such runs to get the distribution of SAINT probability scores by random chance. The separation of positive and negative distributions was considered but not for the impact of extremely high-count interactions on the scoring of low-count interactions or for division of spectra counts by the total spectra counts of each purification. According to the SAINT methodology, the interactions with over 0.8 probability score were kept for the following analysis; 1415 interactions passed this filtration.
We used the semi-supervised mixture model to further eliminate type II errors. The probability distributions P(Xij True) and P(Xij False) of individual preys were also used to calculate the probability of true interaction. We filtered out preys with P(Xij False) > P(Xij True). Common contaminants and abundant proteins were removed by this method; 1081 interactions passed this filtration. In total, 550 interactions passed both filtrations and were designated as HCIPs.
GST Pull-down Assay
GST-fused YAP1 was expressed and purified in Escherichia coli BL21 cells. GST-YAP1 protein (2 μg) was immobilized on GST-Sepharose 4B beads and incubated with various cell lysates for 2 h at 4 °C. Beads were washed three times. Proteins bound to beads were eluted and subjected to SDS-PAGE and Western blotting analysis.
Immunofluorescent Staining
Immunofluorescent staining was performed as described previously (50). Briefly, cells cultured on coverslips were fixed with 4% paraformaldehyde for 10 min at room temperature and then cells were extracted with 0.5% Triton X-100 solution for 5 min. After blocking with TBST containing 1% BSA, cells were incubated with indicated primary antibodies for 1 h at room temperature. After that, cells were washed and incubated with FITC or Rhodamine-conjugated second primary antibodies for 1 h. Cells were counterstained with 100 ng/ml 4′,6-diamidino-2-phenylindole (DAPI) for 2 min to visualize nuclear DNA. The cover slips were mounted onto glass slides with anti-fade solution and visualized under a Nikon ECLIPSE E800 fluorescence microscope with a Nikon Plan Fluor 60× oil objective lens (NA 1.30).
Luciferase Assay
For luciferase reporter assay, 293T cells were seeded in 12-well plates the day before transfection. 5×UAS luciferase reporter plasmid, TEAD4-Gal4 (kindly provided by Dr. Kun-Liang Guan), PRL-SV40, and indicated plasmids were co-transfected into 293T cells. Twenty-four hours later, cells were subjected to lysis, and luciferase activities were assayed in the lysates by Dual-Luciferase Reporter Assay System kit (Promega, Madison, WI) following protocols provided by the manufacturer. The measured luciferase activities were normalized to the Renilla activity.
Three-dimensional Culture of MCF10A Cells for Acini Formation
MCF10A cells (5 × 103) stably transfected or infected with indicated plasmids were grown in growth factor-reduced BD Matrigel matrix (BD Biosciences) within the eight-well chamber slide system (Fisher Scientific). Cultured cells were analyzed for invasive acini formation after 7 days of growth in Matrigel. At least three replicates (∼100 GFP-positive acini counted per replicate) were performed in one experiment.
RESULTS
Proteomic Analysis of the Human Hippo Pathway
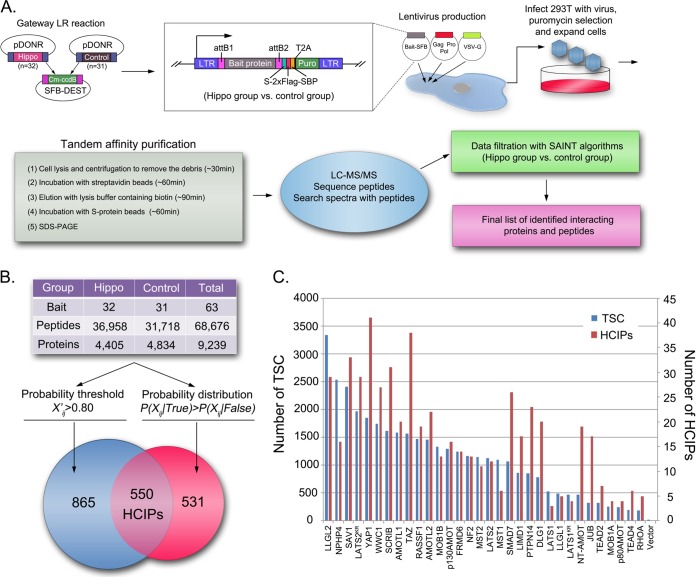
To establish the protein–protein interaction network of the human Hippo pathway, we conducted proteomic analysis using tandem affinity purification followed by mass spectrometry analysis. Thirty-two human proteins linked to Hippo pathway functions (Hippo group), as well as 31 unrelated proteins (control group), were stably expressed as the SFB triple-tagged fusion proteins in 293T cells. Western blotting and immunostaining were carried out to validate the correct protein expression and cellular localization for each stable cell line (data not shown). After two rounds of affinity purifications, proteins in the final eluate were identified by LC-MS/MS analysis (Fig. 1A). The complete protein and peptide identification lists are shown in supplemental Table S1. The relative raw data and mass spectrometry searching results can be found in proteomeXchange (PXD000415). Total spectra accounts for 4405 unique proteins in the Hippo group and 4834 unique proteins in the control group were subjected to SAINT, an unbiased filtration methodology, for identification of HCIPs.
Fig. 1.
Proteomic analysis of the human Hippo pathway. A, Schematic illustration of major steps involved in the tandem affinity purification-mass spectrometry analysis of the human Hippo pathway. Thirty-two Hippo pathway relevant proteins, together with 31 unrelated control proteins and control vector, were constructed into C-terminal SFB tag fused lentiviral vector through gateway technology. 293T cells stably expressing each bait protein were generated by lentivirus infection and puromycin selection. Through the standard tandem affinity purification steps, purified protein complexes were identified by mass spectrometry analysis and final interactive proteins were generated by SAINT algorithms based filtration. B, The total peptide and protein numbers obtained from mass spectrometry analysis are listed. The probability threshold (X'ij) > 0.80 was used as the cutoff to identify HCIPs, as suggested by the SAINT method. We also applied another filtration using the prey information in 31control purifications to remove the nonspecific bindings or contaminants. The numbers of HCIPs remaining after these two filtrations are shown here. C, The total spectral counts (TSC; blue) and corresponding number of HCIPs (red) for each Hippo bait protein are shown together.
Using two-pool analysis, a matrix with all the prey proteins and bait proteins was assembled for Hippo and control groups that included 64 experiments and 9239 proteins. As suggested by SAINT algorithms, we performed the first filtration by choosing (X'ij) > 0.80 as the probability threshold cutoff to identify HCIPs. The second filtration was used to remove the nonspecific binding proteins according to probability distribution P(Xij True) > P(Xij False) analysis based on the prey information of the Hippo group and the control group. Collectively, we identified 550 HCIPs involved in the Hippo pathway (Fig. 1B and supplemental Table S2). Given the fact that the 293T cells were mostly around 80∼90% confluent under normal growth conditions when they collected for this study, the total spectra counts and HCIPs for each Hippo bait protein identified here (Fig. 1C) were considered the basal state interactome of the Hippo pathway (supplemental Figs. S1 and S2).
Overview of the Interaction Network in the Human Hippo Pathway
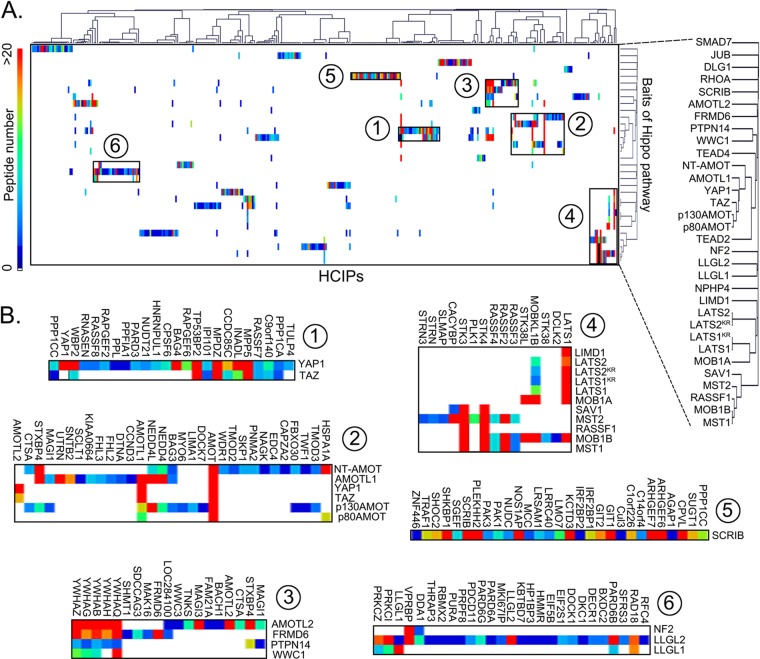
To analyze the similarity of prey-bait interactions among the Hippo pathway, an unbiased hierarchical clustering of all the HCIPs and bait proteins in the Hippo group was used to assemble the interaction data into a coherent network (Fig. 2A). Two prominent clusters were identified around YAP1 and TAZ (clusters 1 and 2 in Fig. 2B), confirming the central roles of these two proteins in the Hippo pathway (supplemental Fig. S3A). Cluster 1 validated previously reported interactions for YAP1 or TAZ, which include PPP1CA (51), WBP2 (52), and TP53BP2 (53). Several uncharacterized preys in this cluster, such as CCDC85C and C9orf140, were considered as potential regulators of YAP1 or TAZ. Cluster 2 confirmed that the angiomotin family proteins are the major associated proteins for YAP1 and TAZ (supplemental Fig. S3C). Cluster 3, which grouped two Hippo upstream regulators (WWC1 and FRMD6) and two-YAP1-associated proteins (AMOTL2 and PTPN14) together, which revealed a previously uncharacterized crosstalk between WWC1/FRMD6 and AMOTL2/PTPN14 in the regulation of the Hippo pathway. Cluster 4 defined the classic kinase cascade in the Hippo pathway, which includes the LAST1/2 and MST1/2 kinases and their adaptor proteins MOB1A/B and SAV1 (supplemental Fig. S3B). It includes other proteins, such as Ajuba LIM protein, LIMD1 (33), and RASSF1 (36), that have also been characterized previously as regulators of these kinase complexes. Cluster 5 concentrated on the bait protein SCRIB, which has been shown to restrain TAZ's activities in breast cancer stem cells (43) and regulate Warts kinase activation in Drosophila (54). The clustered preys consisted mostly of other signaling pathway regulators, such as PAK1, PAK3, SHOC2, IRFBP1, IRFBP2, GIT1, and GIT2, which indicated that SCRIB functions as a scaffold protein, mediating the crosstalk between various signaling pathways and the Hippo pathway. Cluster 6 grouped three bait proteins, LLGL1, LLGL2, and NF2, which are all membrane-associated proteins that mediate the upstream signals to regulate the Hippo pathway (23, 55–57).
Fig. 2.
Hierarchical cluster analysis of HCIPs in the Hippo pathway identified by SAINT algorithms. A, A heatmap was generated from hierarchical clustering of 550 HCIPs for 32 Hippo bait proteins. Six prominent HCIP clusters were manually selected and enlarged below (B). The color of squares in the heat map represents the number of identified HCIP peptides for each bait protein (see scale at left of panel A).
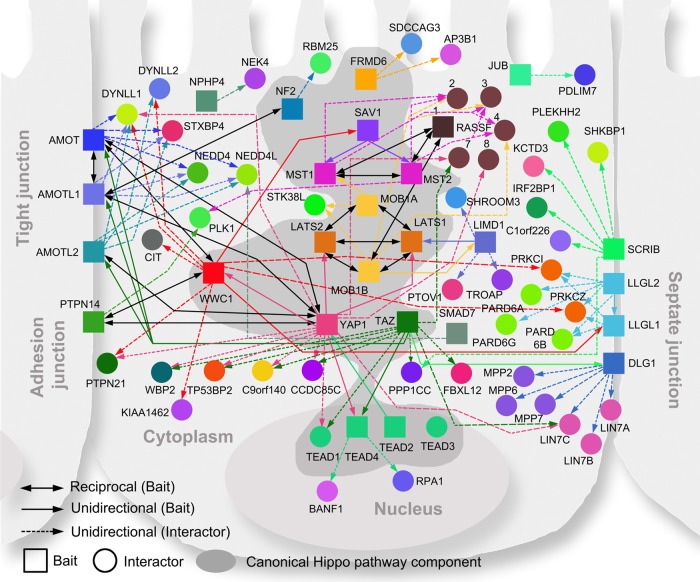
The current understanding of the Hippo pathway is that the core kinase complexes in cytoplasm receive the signals from upstream regulators, which localize in cytoplasm and the cell membrane, and then transduce the signals to YAP1 or TAZ via phosphorylation. YAP1 and TAZ shuttle between cytoplasm and nucleus. Their nuclear binding partners TEAD1/2/3/4 control the transcription of downstream genes involved in cell proliferation and anti-apoptosis. This general scheme of Hippo pathway regulation was confirmed by our proteomic analysis. As shown in Fig. 3, the identified regulators of the Hippo pathway come from various cellular compartments, including cell membrane, cell-cell junctions, centrosome, cytoplasm, and nucleus, indicating that many signals can be involved in the regulation of the Hippo pathway (supplemental Table S3).
Fig. 3.
Protein–protein interaction network of the Hippo pathway. Canonical Hippo pathway components are highlighted with a dark gray background. The unidirectional (solid colored and single arrows) and reciprocal (solid black and double arrows) interactions between baits (squares) are shown. Each bait protein is rendered in a unique color and linked to its major HCIPs (circles) by a dashed line with a single arrow in the corresponding color.
To characterize these Hippo pathway and associated proteins in the context of biological processes, we carried out Gene Ontology (GO) analysis to identify the GO process for each bait-associated HCIP (Fig. S4). GO process analysis linked the Hippo pathway to a wide variety of cellular functions, while mostly focusing on cell signaling, cell cycle, cell death and survival, cell development, cell growth and proliferation, and cell–cell interactions, all of which are fundamental functions of the Hippo pathway (3). These major cellular functions contribute to the systematic functions of the Hippo pathway in tissue development, nervous system development, embryonic development, and tissue morphology, which clearly are linked to the known roles of the Hippo pathway in organ development and organ size control. The disease and disorder GO analysis linked the Hippo pathway mostly to cancer, developmental disorders, and hereditary disorders, which have already been widely studied for the Hippo pathway activity.
Proteomic Data Validation
To verify our proteomic data, all the HCIPs were searched in various protein–protein interaction databases, including BioGrid, STRING, BIND, DIP, and HPRD. 162 interactions among the 550 HCIPs (∼29.45%) have been previously reported (supplemental Table S2).
We also performed co-immunoprecipitation (co-IP) experiments to validate the identified HCIPs. As shown in Table S4, HCIPs in WWC1 and YAP1 interactomes were chosen for the validation of protein–protein interaction. 15 HCIPs were shown to interact with WWC1, which represents 75% positive rate in 20 co-IP experiments we performed (Table S4, Fig. 4, and supplemental Fig. S5A). 24 YAP1 HCIPs among a total of 29 co-IP experiments were confirmed to associate with YAP1, which indicates a positive rate of 82.7% (Table S4 and supplemental Fig. S5B). Although we could not obtain all of the Table S4 constructs encoding WWC1 and YAP1 HCIPs for these co-IP experiments, the current positive interaction rates of all of the HCIPs for both WWC1 and YAP1 are over 55% (Table S4). These results demonstrate the efficacy of our proteomic methodology and the ability to identify bona fide interacting proteins in HCIPs.
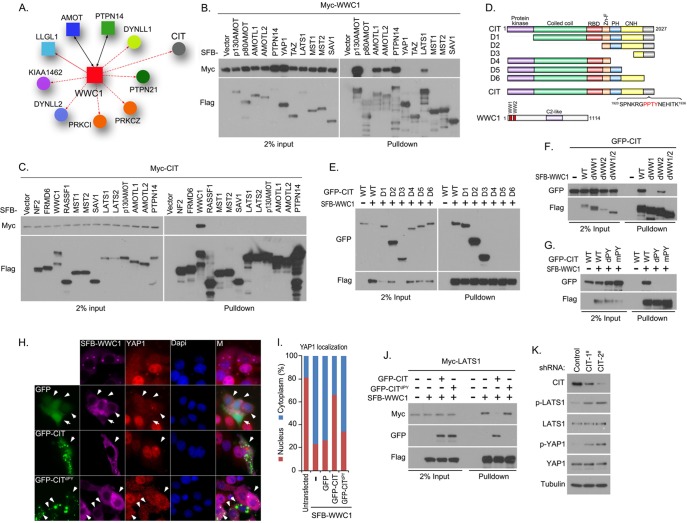
Fig. 4.
Proteomic analysis and validation of the WWC1 interactome. A, Schematic illustration of major WWC1-interacting proteins. WWC1 was also reverse-identified in bait protein AMOT- or PTPN14-containing protein complexes, which are indicated by solid black lines with double arrows. Bait protein LLGL1 in the WWC1 interactome is indicated by a solid line with a single arrow, whereas other prey proteins are linked to WWC1 via a dashed line with a single arrow. B, WWC1 associated with a group of YAP1-binding proteins, including AMOT, AMOTL1, AMOTL2, and PTPN14. Myc-tagged WWC1 was co-expressed with the SFB-tagged Hippo pathway proteins indicated in 293T cells. Pull-down experiments were carried out with S protein beads, and immunoblotting was performed with anti-Myc and anti-Flag antibodies. C, Identification of CIT as a new WWC1-associated protein. Myc-tagged CIT was co-expressed with the SFB-tagged Hippo pathway proteins indicated in 293T cells. Pull-down experiments were carried out with S-protein beads, and immunoblotting was performed with anti-Myc and anti-Flag antibodies. D, Schematic illustration of truncation mutants of CIT used in this study. The PY motif of CIT and two WW domains of WWC1 are indicated. E, The C-terminal region of CIT was required for its binding to WWC1. 293T cells were transfected with constructs encoding wild-type or the indicated CIT truncation mutant together with a construct encoding SFB-WWC1. Pull-down experiments were carried out with S-protein beads, and the precipitates were immunoblotted with antibodies recognizing GFP or Flag-tag. F, Two WW domains of WWC1 were required for its interaction with CIT. 293T cells were transfected with constructs encoding GFP-tagged CIT together with a construct encoding SFB-tagged wild-type or indicated WW domain-deletion mutants of WWC1. Pull-down experiments were carried out using S-protein beads, and immunoblotting was performed with indicated antibodies recognizing GFP or Flag-tag. G, The PY motif located within the C-terminal region of CIT was required for its interaction with WWC1. 293T cells were transfected with a construct encoding wild-type or indicated CIT PY motif mutation or deletion mutant together with construct encoding SFB-WWC1. Pull-down experiments were carried out using S-protein beads, and immunoblotting was performed with indicated antibodies recognizing GFP or Flag-tag. H, I, CIT negatively regulated WWC1-mediated YAP1 nucleus-to-cytoplasm translocation. The localization of endogenous YAP1 was detected by anti-YAP1 antibody in HeLa cells expressing indicated SFB-tagged WWC1 together with GFP-tagged CIT or its PY motif deleted mutant. The localizations of WWC1 and CIT were visualized with anti-Flag antibody and GFP fluorescence. Nuclei were stained by DAPI. M stands for merged. I, The percentages of nuclear and cytoplasmic localized YAP1 were quantified. J, CIT competed with LATS1 for binding to WWC1. 293T cells were transfected with a construct encoding GFP-tagged wild-type or CIT PY motif-deleted mutant together with a construct encoding SFB-WWC1 and Myc-LATS1. Pull-down experiments were carried out using S-protein beads, and immunoblotting was performed with indicated antibodies recognizing Myc, GFP or Flag-tag. K, YAP1 and LATS1 phosphorylation decreased in cells in which CIT was down-regulated. MCF10A cells transfected with CIT shRNAs were analyzed for indicated total proteins or phosphorylated proteins by Western blotting.
Functional Validation of WWC1 Interactome Revealed CIT as a Negative Regulator of the Hippo Pathway
From our proteomic data set, we found that an upstream regulator of the Hippo pathway, WWC1 (Kibra), forms complexes with angiomotin and PTPN14, two proteins that are known regulators of YAP1 (Fig. 4A). In Drosophila, Kibra functions together with Merlin and Expanded in a protein complex localized at the apical region of epithelial cells, which act as upstream regulators of the Salvador/Warts/Hippo/Mats kinase complex. In mammalian cells, WWC1 restrains YAP1's activity through LATS1/2 kinases but not MST1/2 kinases (58, 59), which differs from its functions in Drosophila. In human cells, the detailed mechanism for WWC1-dependent regulation of the Hippo pathway is still unclear. To confirm our proteomic data, we checked the interaction between WWC1 and some Hippo pathway components, including angiomotin family proteins, PTPN14, YAP1, and TAZ as well as LATS1 and MST1/2 kinases. As reported, WWC1 associates with LATS1 kinase, but not with YAP1, TAZ, or MST1/2 kinases (Fig. 4B). Moreover, robust interactions of p130AMOT, AMOTL1, AMOTL2, and PTPN14 with WWC1 were detected, indicating a potential crosstalk between upstream regulators of the Hippo pathway and these newly identified YAP1-associated proteins (supplemental Fig. S3D). Indeed, the Drosophila ortholog of PTPN14, Pez, has been shown to interact with Kibra and function as an upstream regulator of the Hippo pathway (60). Interestingly, WWC1 was also identified in angiomotin and PTPN14 reciprocal purifications (supplemental Table S2). Together, our proteomic data suggest that WWC1 is likely a signal integrator in the human Hippo pathway and reveal potential crosstalk between WWC1 and angiomotin protein family and PTPN14 in the regulation of the Hippo pathway.
Besides angiomotin proteins and PTPN14, CIT (the rho-interacting, serine/threonine kinase 21) was identified as a major WWC1-associated protein (Fig. 4A). CIT has been found to localize at midbody and control RhoA localization and activity during cytokinesis (61–65). We found that CIT specifically interacted with WWC1, but not with a series of other Hippo pathway components or regulators (Fig. 4C). To identify the region of CIT required for the WWC1 binding, several CIT truncations were generated (Fig. 4D). As shown in Fig. 4E, the C-terminal region of CIT was required for its association with WWC1. Because WWC1 harbors two WW domains at its N terminus that bridge the interaction with LATS1/2 kinases, and most regulating events in the Hippo pathway are mediated by WW domain-PY motif-based interactions, we analyzed the C-terminal amino acid sequence of CIT and discovered one PY motif in this region (Fig. 4D). Mutating (mPY) or deleting (dPY) this PY motif disrupted its association with WWC1 (Fig. 4G). Consistent with these findings were our findings that both WW domains of WWC1 were required for its binding to CIT and that the first WW domain contributed more to the interaction with CIT (Fig. 4F). These data suggest that the interaction between CIT and WWC1 was mediated by interaction of the PY motif at the C-terminus of CIT with the two WW domains on WWC1.
As WWC1 has been shown to be a positive regulator of the Hippo pathway, overexpression of WWC1 enhanced the activation of LATS1/2 kinases and YAP1 phosphorylation, which was reflected by YAP1 nucleus-to-cytoplasm translocation (Fig. 4H). To evaluate the cellular functions of CIT, YAP1 cellular localization was determined in cells where WWC1 was co-expressed with CIT. As shown in Fig. 4H and 4I, wild-type CIT, but not the PY motif-deleted CIT (CITdPY) or GFP control, reversed WWC1-mediated nucleus-to-cytoplasm translocation of YAP1. These data indicate that CIT may suppress the activation of the Hippo pathway via its interaction with WWC1.
In human cells, WWC1 activates the Hippo pathway through its binding to the LATS1/2 kinases, which increases the activation and phosphorylation of these kinases. Interestingly, the association of WWC1 with these kinases is also mediated by the two WW domains, and this association is required for the activation of these kinases. As CIT and LATS1/2 kinases bind to WWC1 through the same region on WWC1, we tested the hypothesis that CIT could compete with LATS1/2 kinases for the binding to WWC1. Indeed, overexpression of wild-type CIT, but not the PY motif-deleted mutant of CIT (CITdPY), attenuated the interaction between WWC1 and LATS1 (Fig. 4J). These data suggest that CIT may inhibit the activation of the Hippo pathway by suppressing formation of the LATS-WWC1 complex. To test this hypothesis, we determined the phosphorylation levels of LATS1 kinase and YAP1 in CIT knockdown cells. As shown in Fig. 4K, both LATS1 and YAP1 phosphorylation increased in cells transduced with either of two different CIT shRNAs. These data support a potential role for CIT as a negative regulator of the Hippo pathway through its interaction with WWC1.
On the basis of the potential crosstalk between WWC1 with PTPN14 and angiomotin family proteins and the findings that CIT protein is associated with WWC1 and negatively regulates Hippo signaling, we propose that WWC1 is a critical node in the Hippo pathway.
CCDC85C is a New YAP1-interacting Protein
YAP1 is the key downstream effector for the Hippo pathway; all signaling events in the Hippo pathway finally converge on YAP1, which determines transcription of the downstream targeted genes. Because of that, we took a close look at the YAP1 interactome with the objective of identifying any new regulators of YAP1 from our proteomic data.
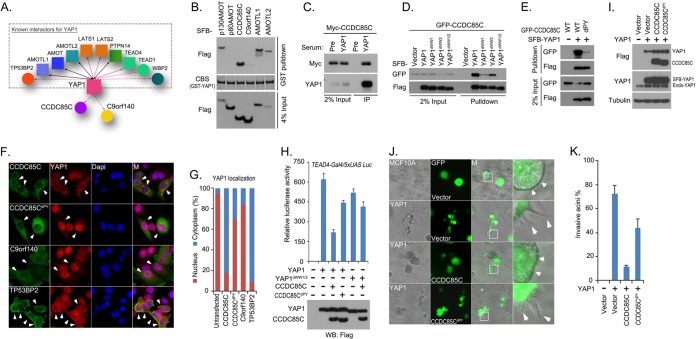
In the YAP1 interactome, we identified many previously reported YAP1-associated proteins, such as p130AMOT, AMOTL1, AMOTL2, PTPN14, LATS1, LATS2, TEAD4, TP53BP2, and WBP2, validating our proteomic studies (Fig. 5A). We also identified two unknown proteins, coiled-coil domain-containing protein 85C (CCDC85C) and chromosome 9 open reading frame 140 (C9orf140), in the YAP1 prey list (Fig. 5A). Bacterially purified GST-YAP1 was able to pull-down SFB-tagged CCDC85C (Fig. 5B). The binding of CCDC85C to YAP1 was equivalent to that of other known YAP1-associated proteins such as p130AMOT, AMOTL1, and AMOTL2 (Fig. 5B). However, we detected only weak interaction between C9orf140 and YAP1 (Fig. 5B), suggesting that C9orf140 may not bind directly to YAP1. Furthermore, endogenous YAP1 could immunoprecipitate Myc-tagged CCDC85C (Fig. 5C). These data indicate that CCDC85C is a new YAP1-binding protein.
Fig. 5.
Analysis of YAP1 interactome suggests that CCDC85C is a new regulator of YAP1. A, Schematic illustration of major YAP1-interacting proteins. YAP1 was also identified in the bait protein AMOT-, AMOTL1-, AMOTL2- or PTPN14-containing protein complexes and therefore they are indicated by solid black lines with double arrows. Bait proteins TEAD4, LATS1, and LATS2 are indicated in the YAP1 interactome by a solid line with a single arrow, while other prey proteins are linked to YAP1 via a dashed line with a single arrow. The reported YAP1-binding proteins in this interactome are grouped in a dashed square; two previously unknown prey proteins, CCDC85C and C9orf140, were left outside of the square. B, CCDC85C is a newly discovered YAP-associated protein. Bacterially expressed and purified GST-YAP1 fusion protein was used to pull down exogenously expressed SFB-tagged p130AMOT, p80AMOT, CCDC85C, C9orf140, AMOTL1, and AMOTL2 in vitro. CBS, coomassie blue stain. C, Association of endogenous YAP1and Myc-tagged CCDC85C was confirmed by a co-immunoprecipitation experiment. Immunoprecipitation (IP) assay was performed using 293T cell extracts and anti-YAP1 serum. Pre-immune serum was used as the control. Immunoblotting was performed with indicated anti-YAP1 anti-serum or anti-Myc antibody. D, Two WW domains of YAP1 were required for its interaction with CCDC85C. 293T cells were transfected with constructs encoding GFP-tagged CCDC85C together with constructs encoding SFB-tagged wild-type or indicated WW domain-deletion mutants of YAP1. Pull-down experiments were carried out using S-protein beads, and immunoblotting was performed with indicated antibodies recognizing GFP or Flag-tag. E, The PY motif of CCDC85C was required for its interaction with YAP1. 293T cells were transfected with a construct encoding GFP-tagged wild-type or indicated CCDC85C PY motif-deleted mutant together with a construct encoding SFB-YAP1. Pull-down experiments were carried out using S-protein beads and immunoblotting was performed with indicated antibodies recognizing GFP or Flag-tag. F, G, CCDC85C translocated YAP1 from nucleus into cytoplasm. The localization of endogenous YAP1 was detected by anti-YAP1 antibody in HeLa cells expressing indicated SFB-tagged wild-type or PY motif deleted CCDC85C mutant, C9orf140, or TP53BP2. The localization of indicated SFB-tagged proteins was detected with anti-Flag antibody. Nuclei were stained by DAPI. M stands for merged. The percentages of nuclear and cytoplasmic YAP1 localization are quantified in (G). H, CCDC85C inhibited YAP1 transactivation activity in a luciferase reporter assay. Luciferase reporter assay was performed by cotransfecting indicated YAP1 or YAP1 two WW domain-deleted mutant with indicated CCDC85C or its PY motif-deleted mutant in 293T cells. Firefly Renilla was used as the internal control. Data are presented as mean ± s.d. from three different experiments. The transfected proteins were detected by Western blotting as shown at the bottom of the panel. I, Overexpression of CCDC85C or its PY motif -deleted mutant in MCF10A cells overexpressing YAP1. MCF10A cells expressing SFB-YAP1 were transduced with viral particles encoding SFB-tagged CCDC85C or its PY motif-deleted mutant. This lentiviral vector contains a separate promoter that controls the expression of GFP. Exogenously expressed YAP1 and CCDC85C were detected by anti-Flag antibody, whereas endogenous and exogenous YAP1 were detected by anti-YAP1 antibody. J, CCDC85C suppressed invasive acini formation by MCF10A cells overexpressing YAP1. MCF10A cells in (I) were subjected to 3D culture in Matrigel. GFP indicated cells positively transduced with control virus or viral particles expressing wild-type CCDC85C or its PY motif-deleted mutant. White squares are enlarged in the far right column. Small white arrows indicate normal acini morphology and large white arrows indicate invasive acini morphology. K, The percentages of invasive acini shown in (J) were quantified.
A majority of YAP1-interacting proteins associate with YAP1 through the binding of their PY motif/motifs (PPXY) with the WW domains on YAP1. Thus, we first tested and confirmed that the two WW domains of YAP1 are required for its interaction with CCDC85C (Fig. 5D). We found one PY motif (PPSY) located in the middle region of CCDC85C. The interaction between YAP1 and CCDC85C was dramatically decreased when this PY motif of CCDC85C was deleted (Fig. 5E). These data suggest that the interaction between YAP1 and CCDC85C requires the two WW domains of YAP1 and the PY motif of CCDC85C.
The key regulating event in the Hippo pathway is the subcellular localization of YAP1. In the canonical Hippo pathway, LATS1/2 kinases phosphorylate YAP1, which consequently provides a docking site for 14-3-3 binding and therefore retains YAP1 in cytosol. Another group of proteins, which includes AMOT, AMOTL1, AMOTL2, and PTPN14, prevents YAP1 nuclear localization through direct protein–protein interaction. Because the association of CCDC85C with YAP1 is similar to that of angiomotin family proteins or PTPN14, we tested whether CCDC85C could also translocate YAP1 from nucleus to cytoplasm. Exogenously expressed CCDC85C mostly localized in the cytoplasm and we observed a dramatic translocation of endogenous YAP1 from nucleus to cytoplasm in cells expressing CCDC85C (Figs. 5F and 5G). Moreover, the PY motif-deleted mutant of CCDC85C (CCDC85CdPY), which lost its affinity for YAP1, failed to translocate YAP1 to cytoplasm (Figs. 5F and 5G). As a control, C9orf140 did not bind directly to YAP1 (Fig. 5B) and thus could not promote YAP1 translocation to cytoplasm (Figs. 5F and 5G). On the other hand, TP53BP2 is a known YAP1 binding protein (53) that was able to facilitate the translocation of YAP1 to cytoplasm and cell membrane (Figs. 5F and 5G). Together, these findings indicate that CCDC85C is a previously unrecognized negative regulator of YAP1 and that physical interaction between CCDC85C and YAP1 can induce the translocation of YAP1 from nucleus into cytoplasm.
Because CCDC85C inhibits YAP1 nuclear localization, we asked whether CCDC85C could negatively regulate the transactivation activity and oncogenic function of YAP1. Indeed, we found that the expression of CCDC85C suppressed YAP1-dependent transactivation activity, which requires a physical interaction between the two proteins (Fig. 5H). Overexpression of YAP1 can lead to the transformation of immortalized human mammary epithelial MCF10A cells (66), which in 3-dimentional culture displayed invasive acini morphology featuring with branch-like morphology (Figs. 5I and 5J). When wild-type CCDC85C or its PY motif-deleted mutant were introduced into YAP1-overexpressing MCF10A cells (Fig. 5I), wild-type CCDC85C, but not CCDC85CdPY, reduced invasive acini formation in 3D culture (Fig. 5J and 5K). These results suggest that CCDC85C functions as a negative regulator of YAP1, promoting nuclear-to-cytoplasm translocation of YAP1 via direct protein–protein interaction.
DISCUSSION
The Hippo pathway plays crucial roles in a variety of cellular functions, including cell proliferation, cell death, stem cell self-renewal, embryonic development, and cancer formation (3). In this study, we performed high-content proteomics studies and established an interaction landscape for this pathway (Fig. 3). The identification of 550 HCIPs greatly expanded our current understanding of the Hippo pathway. These newly discovered HCIPs will provide directions for future functional analysis, helping us to achieve a comprehensive understanding of the networks that connect and function together with the Hippo pathway.
In this study, we took advantage of our modified tandem affinity purification (TAP) method. Our choice of this method was based on its several features that are advantageous for the purification of protein complexes: (1) it allows purification of protein complexes within 3∼4 h after cell lysis, greatly reducing the risk of protein degradation during purification (Fig. 1A); (2) only one buffer is used from the beginning to the end, avoiding any potential disruption of protein complexes due to changes of buffer conditions; and (3) more importantly, since this approach does not use an antibody-based affinity column, it circumvents the common problems of antibody leakage during purification and antibody cross-reactions. Compared with one-step tag- and antibody-based purification methods, this modified tandem affinity purification method increases the likelihood of identifying genuine binding proteins and decreases the amount of nonspecific contaminants. We have used this method extensively in recent years and successfully identified many important regulators in the DNA damage pathway (67–69) and other cancer related pathways (70–73).
Our proteomic data and hierarchical clustering analysis confirm the essential roles of YAP1/TAZ and the core kinase complexes in the Hippo pathway, as most identified HCIPs connect to these complexes. In addition, there may be crosstalk between several known upstream components (FRMD6 and WWC1) and the recently identified components (PTPN14, AMOT, AMOTL1, and AMOTL2) involved in regulation of the Hippo pathway, since they cluster together (Fig. 2) and physically bind to each other (Fig. 4). Moreover, we also identified two new components of the Hippo pathway, the WWC1-interacting protein CIT and the YAP1-associated protein CCDC85C.
As an effector of Rho GTPase, CIT is known to play critical roles in cell proliferation and cell cycle progress, mostly from its functions in cytokinesis (61–65). In this study, we identified CIT as a negative regulator of the Hippo pathway, which indicates that CIT may function as an oncogene. Indeed, CIT gene expression was elevated in various human tumors (Oncomine: https://www.oncomine.org/resource/login.html) and correlated with poor survival in breast cancer patients (PrognoScan: http://www.prognoscan.org/) (data not shown). It remains to be determined whether this potential oncogenic activity of CIT is linked with its role in the Hippo pathway, its involvement in cytokinesis, or both. Interestingly, there seems to be a connection between mitosis and Hippo pathway, because several known components of Hippo pathway, including LATS1 (74–77), LATS2 (78), MST2 (79), and SAV1 (79), also play important functions in the regulation of mitosis. It remains to be resolved whether and how the Hippo pathway and mitosis are interconnected, especially how the dual functions of CIT in these processes are regulated.
CCDC85C is another YAP1-associated protein identified by our proteomic study. CCDC85C associates with YAP1 through the WW domains on YAP1, which are also binding sites for many other YAP1-interacting proteins including AMOT, AMOTL1, AMOTL2, and PTPN14. It is still unknown exactly how these proteins coordinate and participate in the regulation of YAP1. Nevertheless, together with the CIT-WWC1 interaction, the CCDC85C-YAP1 interaction further supports the importance of “WW domain-PY motif” in signal transduction in the Hippo pathway (80).
In summary, our study provides the first picture of the protein–protein interaction network involved in the human Hippo pathway. Of course, this represents only a static snapshot of this network. It is highly likely that this protein–protein interaction network will change under specific growth conditions or stimuli, and these changes will provide the directions for future proteomic studies.
Supplementary Material
Acknowledgments
We thank all our colleagues in Dr. Chen's laboratory for insightful discussion and technical assistance, especially Jingsong Yuan. We thank Dr. Jae-il Park and Dr. Li Ma for the insightful discussion and advice. We thank Drs. Xin Wang, HaeYun Jung, Dahu Chen, Jinsong Zhang, Hailong Piao, and Peijing Zhang for technical assistance. We thank Drs. Susan Tucker and Shelley Herbrich (Department of Bioinformatics and Computational Biology, MD Anderson Cancer Center) as well as Dr. Rudy Guerra (Rice University) for bioinformatics discussion and assistance. We also want to thank Drs. Steven Gygi and Ross Tomaino (Taplin Mass Spectrometry Facility, Harvard Medical School) for their help with mass spectrometry analysis and providing raw data for the submission of this manuscript. We want to thank the PRIDE Team for their help uploading data to ProteomeXchang database. We also want to thank Dr. Yutong Sun and Shan Shao (shRNA and ORFeome core facility, MD Anderson Cancer Center) for the ORFs and shRNAs.
Footnotes
* This work was supported in part by the Department of Defense Era of Hope research scholar award to J.C. (W81XWH-09-1-0409). J.C. is also a recipient of an Era of Hope Scholar award from the Department of Defense (W81XWH-05-1-0470) and a member of M.D. Anderson Cancer Center (CA016672). X.L. is a recipient of a Computational Cancer Biology Training Program Felloship supported by the Cancer Prevention and Research Institutes of Texas and a Jeffery Lee Cousins Fellowship in Lung Cancer Research.
 This article contains supplemental Tables S1 to S4 and Figs. S1 to S5.
This article contains supplemental Tables S1 to S4 and Figs. S1 to S5.
1 The abbreviations used are:
- shRNA
- short-hairpin RNA
- SAINT
- Significance Analysis of INTeactome
- HCIP
- high-confidence candidate interacting proteins
- SFB
- S tag-Flag tag-SBP
- DAPI
- 4′, 6-diamidino-2-phenylindole
- GO
- gene ontology.
REFERENCES
- 1. Pan D. (2010) The hippo signaling pathway in development and cancer. Dev. Cell 19, 491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhao B., Li L., Lei Q., Guan K. L. (2010) The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 24, 862–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yu F. X., Guan K. L. (2013) The Hippo pathway: regulators and regulations. Genes Dev. 27, 355–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harvey K. F., Pfleger C. M., Hariharan I. K. (2003) The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114, 457–467 [DOI] [PubMed] [Google Scholar]
- 5. Jia J., Zhang W., Wang B., Trinko R., Jiang J. (2003) The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 17, 2514–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pantalacci S., Tapon N., Léopold P. (2003) The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat. Cell Biol. 5, 921–927 [DOI] [PubMed] [Google Scholar]
- 7. Udan R. S., Kango-Singh M., Nolo R., Tao C., Halder G. (2003) Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 5, 914–920 [DOI] [PubMed] [Google Scholar]
- 8. Wu S., Huang J., Dong J., Pan D. (2003) hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114, 445–456 [DOI] [PubMed] [Google Scholar]
- 9. Tapon N., Harvey K. F., Bell D. W., Wahrer D. C., Schiripo T. A., Haber D., Hariharan I. K. (2002) salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110, 467–478 [DOI] [PubMed] [Google Scholar]
- 10. Justice R. W., Zilian O., Woods D. F., Noll M., Bryant P. J. (1995) The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 9, 534–546 [DOI] [PubMed] [Google Scholar]
- 11. Xu T., Wang W., Zhang S., Stewart R. A., Yu W. (1995) Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121, 1053–1063 [DOI] [PubMed] [Google Scholar]
- 12. Lai Z. C., Wei X., Shimizu T., Ramos E., Rohrbaugh M., Nikolaidis N., Ho L. L., Li Y. (2005) Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell 120, 675–685 [DOI] [PubMed] [Google Scholar]
- 13. Huang J., Wu S., Barrera J., Matthews K., Pan D. (2005) The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122, 421–434 [DOI] [PubMed] [Google Scholar]
- 14. Goulev Y., Fauny J. D., Gonzalez-Marti B., Flagiello D., Silber J., Zider A. (2008) SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr. Biol. 18, 435–441 [DOI] [PubMed] [Google Scholar]
- 15. Wu S., Liu Y., Zheng Y., Dong J., Pan D. (2008) The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev. Cell 14, 388–398 [DOI] [PubMed] [Google Scholar]
- 16. Zhang L., Ren F., Zhang Q., Chen Y., Wang B., Jiang J. (2008) The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev. Cell 14, 377–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhao B., Ye X., Yu J., Li L., Li W., Li S., Yu J., Lin J. D., Wang C. Y., Chinnaiyan A. M., Lai Z. C., Guan K. L. (2008) TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao B., Wei X., Li W., Udan R. S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L., Zheng P., Ye K., Chinnaiyan A., Halder G., Lai Z. C., Guan K. L. (2007) Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao B., Li L., Tumaneng K., Wang C. Y., Guan K. L. (2010) A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Genes Dev. 24, 72–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baumgartner R., Poernbacher I., Buser N., Hafen E., Stocker H. (2010) The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev. Cell 18, 309–316 [DOI] [PubMed] [Google Scholar]
- 21. Genevet A., Wehr M. C., Brain R., Thompson B. J., Tapon N. (2010) Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev. Cell 18, 300–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu J., Zheng Y., Dong J., Klusza S., Deng W. M., Pan D. (2010) Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev. Cell 18, 288–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang N., Bai H., David K. K., Dong J., Zheng Y., Cai J., Giovannini M., Liu P., Anders R. A., Pan D. (2010) The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev. Cell 19, 27–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pellock B. J., Buff E., White K., Hariharan I. K. (2007) The Drosophila tumor suppressors Expanded and Merlin differentially regulate cell cycle exit, apoptosis, and Wingless signaling. Dev. Biol. 304, 102–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hamaratoglu F., Willecke M., Kango-Singh M., Nolo R., Hyun E., Tao C., Jafar-Nejad H., Halder G. (2006) The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 8, 27–36 [DOI] [PubMed] [Google Scholar]
- 26. Badouel C., Gardano L., Amin N., Garg A., Rosenfeld R., Le Bihan T., McNeill H. (2009) The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev. Cell 16, 411–420 [DOI] [PubMed] [Google Scholar]
- 27. Yu F. X., Zhao B., Panupinthu N., Jewell J. L., Lian I., Wang L. H., Zhao J., Yuan H., Tumaneng K., Li H., Fu X. D., Mills G. B., Guan K. L. (2012) Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 150, 780–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mo J. S., Yu F. X., Gong R., Brown J. H., Guan K. L. (2012) Regulation of the Hippo-YAP pathway by protease-activated receptors (PARs). Genes Dev. 26, 2138–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yu F. X., Mo J. S., Guan K. L. (2012) Upstream regulators of the Hippo pathway. Cell Cycle 11, 4097–4098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Habbig S., Bartram M. P., Müller R. U., Schwarz R., Andriopoulos N., Chen S., Sägmüller J. G., Hoehne M., Burst V., Liebau M. C., Reinhardt H. C., Benzing T., Schermer B. (2011) NPHP4, a cilia-associated protein, negatively regulates the Hippo pathway. J. Cell Biol. 193, 633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boggiano J. C., Vanderzalm P. J., Fehon R. G. (2011) Tao-1 phosphorylates Hippo/MST kinases to regulate the Hippo-Salvador-Warts tumor suppressor pathway. Dev. Cell 21, 888–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Poon C. L., Lin J. I., Zhang X., Harvey K. F. (2011) The sterile 20-like kinase Tao-1 controls tissue growth by regulating the Salvador-Warts-Hippo pathway. Dev. Cell 21, 896–906 [DOI] [PubMed] [Google Scholar]
- 33. Das Thakur M., Feng Y., Jagannathan R., Seppa M. J., Skeath J. B., Longmore G. D. (2010) Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr. Biol. 20, 657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reddy B. V., Irvine K. D. (2013) Regulation of Hippo Signaling by EGFR-MAPK Signaling through Ajuba Family Proteins. Dev. Cell 24, 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hwang E., Ryu K. S., Pääkkönen K., Guntert P., Cheong H. K., Lim D. S., Lee J. O., Jeon Y. H., Cheong C. (2007) Structural insight into dimeric interaction of the SARAH domains from Mst1 and RASSF family proteins in the apoptosis pathway. Proc. Natl. Acad. Sci. U.S.A. 104, 9236–9241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Polesello C., Huelsmann S., Brown N. H., Tapon N. (2006) The Drosophila RASSF homolog antagonizes the hippo pathway. Curr. Biol. 16, 2459–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chan S. W., Lim C. J., Chong Y. F., Pobbati A. V., Huang C., Hong W. (2011) Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J. Biol. Chem. 286, 7018–7026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao B., Li L., Lu Q., Wang L. H., Liu C. Y., Lei Q., Guan K. L. (2011) Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 25, 51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang W., Huang J., Chen J. (2011) Angiomotin-like proteins associate with and negatively regulate YAP1. J. Biol. Chem. 286, 4364–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang W., Huang J., Wang X., Yuan J., Li X., Feng L., Park J. I., Chen J. (2012) PTPN14 is required for the density-dependent control of YAP1. Genes Dev. 26, 1959–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang J. M., Nagatomo I., Suzuki E., Mizuno T., Kumagai T., Berezov A., Zhang H., Karlan B., Greene M. I., Wang Q. (2013) YAP modifies cancer cell sensitivity to EGFR and survivin inhibitors and is negatively regulated by the non-receptor type protein tyrosine phosphatase 14. Oncogene 32, 2220–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lin J. I., Poon C. L., Harvey K. F. (2013) The Hippo size control pathway–ever expanding. Sci. Signal 6, pe4. [DOI] [PubMed] [Google Scholar]
- 43. Cordenonsi M., Zanconato F., Azzolin L., Forcato M., Rosato A., Frasson C., Inui M., Montagner M., Parenti A. R., Poletti A., Daidone M. G., Dupont S., Basso G., Bicciato S., Piccolo S. (2011) The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 147, 759–772 [DOI] [PubMed] [Google Scholar]
- 44. Silvis M. R., Kreger B. T., Lien W. H., Klezovitch O., Rudakova G. M., Camargo F. D., Lantz D. M., Seykora J. T., Vasioukhin V. (2011) alpha-catenin is a tumor suppressor that controls cell accumulation by regulating the localization and activity of the transcriptional coactivator Yap1. Sci. Signal 4, ra33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schlegelmilch K., Mohseni M., Kirak O., Pruszak J., Rodriguez J. R., Zhou D., Kreger B. T., Vasioukhin V., Avruch J., Brummelkamp T. R., Camargo F. D. (2011) Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell 144, 782–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. The-UniProt-Consortium (2012) Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res. 40, D71–D75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Smoot M. E., Ono K., Ruscheinski J., Wang P. L., Ideker T. (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Breitkreutz A., Choi H., Sharom J. R., Boucher L., Neduva V., Larsen B., Lin Z. Y., Breitkreutz B. J., Stark C., Liu G., Ahn J., Dewar-Darch D., Reguly T., Tang X., Almeida R., Qin Z. S., Pawson T., Gingras A. C., Nesvizhskii A. I., Tyers M. (2010) A global protein kinase and phosphatase interaction network in yeast. Science 328, 1043–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Choi H., Larsen B., Lin Z. Y., Breitkreutz A., Mellacheruvu D., Fermin D., Qin Z. S., Tyers M., Gingras A. C., Nesvizhskii A. I. (2011) SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods 8, 70–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang W., Chen L., Ding Y., Jin J., Liao K. (2008) Centrosome separation driven by actin-microfilaments during mitosis is mediated by centrosome-associated tyrosine-phosphorylated cortactin. J. Cell Sci. 121, 1334–1343 [DOI] [PubMed] [Google Scholar]
- 51. Wang P., Bai Y., Song B., Wang Y., Liu D., Lai Y., Bi X., Yuan Z. (2011) PP1A-mediated dephosphorylation positively regulates YAP2 activity. PLoS One 6, e24288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chan S. W., Lim C. J., Huang C., Chong Y. F., Gunaratne H. J., Hogue K. A., Blackstock W. P., Harvey K. F., Hong W. (2011) WW domain-mediated interaction with Wbp2 is important for the oncogenic property of TAZ. Oncogene 30, 600–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Espanel X., Sudol M. (2001) Yes-associated protein and p53-binding protein-2 interact through their WW and SH3 domains. J. Biol. Chem. 276, 14514–14523 [DOI] [PubMed] [Google Scholar]
- 54. Verghese S., Waghmare I., Kwon H., Hanes K., Kango-Singh M. (2012) Scribble acts in the Drosophila fat-hippo pathway to regulate warts activity. PLoS One 7, e47173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jukam D., Desplan C. (2011) Binary regulation of Hippo pathway by Merlin/NF2, Kibra, Lgl, and Melted specifies and maintains postmitotic neuronal fate. Dev. Cell 21, 874–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Parsons L. M., Grzeschik N. A., Allott M. L., Richardson H. E. (2010) Lgl/aPKC and Crb regulate the Salvador/Warts/Hippo pathway. Fly 4, 288–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Grzeschik N. A., Parsons L. M., Allott M. L., Harvey K. F., Richardson H. E. (2010) Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr. Biol. 20, 573–581 [DOI] [PubMed] [Google Scholar]
- 58. Moleirinho S., Chang N., Sims A. H., Tilston-Lünel A. M., Angus L., Steele A., Boswell V., Barnett S. C., Ormandy C., Faratian D., Gunn-Moore F. J., Reynolds P. A. (2013) KIBRA exhibits MST-independent functional regulation of the Hippo signaling pathway in mammals. Oncogene 32, 1821–1830 [DOI] [PubMed] [Google Scholar]
- 59. Xiao L., Chen Y., Ji M., Dong J. (2011) KIBRA regulates Hippo signaling activity via interactions with large tumor suppressor kinases. J. Biol. Chem. 286, 7788–7796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Poernbacher I., Baumgartner R., Marada S. K., Edwards K., Stocker H. (2012) Drosophila Pez acts in Hippo signaling to restrict intestinal stem cell proliferation. Curr. Biol. 22, 389–396 [DOI] [PubMed] [Google Scholar]
- 61. Watanabe S., De Zan T., Ishizaki T., Narumiya S. (2013) Citron-kinase mediates transition from constriction to abscission through its coiled-coil domain. J. Cell Sci. 126, 1773–1784 [DOI] [PubMed] [Google Scholar]
- 62. Bassi Z. I., Verbrugghe K. J., Capalbo L., Gregory S., Montembault E., Glover D. M., D'Avino P. P. (2011) Sticky/Citron kinase maintains proper RhoA localization at the cleavage site during cytokinesis. J. Cell Biol. 195, 595–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gai M., Camera P., Dema A., Bianchi F., Berto G., Scarpa E., Germena G., Di Cunto F. (2011) Citron kinase controls abscission through RhoA and anillin. Mol. Biol. Cell 22, 3768–3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Eda M., Yonemura S., Kato T., Watanabe N., Ishizaki T., Madaule P., Narumiya S. (2001) Rho-dependent transfer of Citron-kinase to the cleavage furrow of dividing cells. J. Cell Sci. 114, 3273–3284 [DOI] [PubMed] [Google Scholar]
- 65. Madaule P., Eda M., Watanabe N., Fujisawa K., Matsuoka T., Bito H., Ishizaki T., Narumiya S. (1998) Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature 394, 491–494 [DOI] [PubMed] [Google Scholar]
- 66. Overholtzer M., Zhang J., Smolen G. A., Muir B., Li W., Sgroi D. C., Deng C. X., Brugge J. S., Haber D. A. (2006) Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc. Natl. Acad. Sci. U.S.A. 103, 12405–12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lou Z., Minter-Dykhouse K., Wu X., Chen J. (2003) MDC1 is coupled to activated CHK2 in mammalian DNA damage response pathways. Nature 421, 957–961 [DOI] [PubMed] [Google Scholar]
- 68. Liu T., Ghosal G., Yuan J., Chen J., Huang J. (2010) FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science 329, 693–696 [DOI] [PubMed] [Google Scholar]
- 69. Feng L., Huang J., Chen J. (2009) MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev. 23, 719–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fu Z., Malureanu L., Huang J., Wang W., Li H., van Deursen J. M., Tindall D. J., Chen J. (2008) Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat. Cell Biol. 10, 1076–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kim J. E., Chen J., Lou Z. (2008) DBC1 is a negative regulator of SIRT1. Nature 451, 583–586 [DOI] [PubMed] [Google Scholar]
- 72. Maddika S., Chen J. (2009) Protein kinase DYRK2 is a scaffold that facilitates assembly of an E3 ligase. Nat. Cell Biol. 11, 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Maddika S., Kavela S., Rani N., Palicharla V. R., Pokorny J. L., Sarkaria J. N., Chen J. (2011) WWP2 is an E3 ubiquitin ligase for PTEN. Nat. Cell Biol. 13, 728–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tao W., Zhang S., Turenchalk G. S., Stewart R. A., St John M. A., Chen W., Xu T. (1999) Human homologue of the Drosophila melanogaster lats tumour suppressor modulates CDC2 activity. Nat. Genet. 21, 177–181 [DOI] [PubMed] [Google Scholar]
- 75. Hirota T., Morisaki T., Nishiyama Y., Marumoto T., Tada K., Hara T., Masuko N., Inagaki M., Hatakeyama K., Saya H. (2000) Zyxin, a regulator of actin filament assembly, targets the mitotic apparatus by interacting with h-warts/LATS1 tumor suppressor. J. Cell Biol. 149, 1073–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bothos J., Tuttle R. L., Ottey M., Luca F. C., Halazonetis T. D. (2005) Human LATS1 is a mitotic exit network kinase. Cancer Res. 65, 6568–6575 [DOI] [PubMed] [Google Scholar]
- 77. Chiyoda T., Sugiyama N., Shimizu T., Naoe H., Kobayashi Y., Ishizawa J., Arima Y., Tsuda H., Ito M., Kaibuchi K., Aoki D., Ishihama Y., Saya H., Kuninaka S. (2012) LATS1/WARTS phosphorylates MYPT1 to counteract PLK1 and regulate mammalian mitotic progression. J. Cell Biol. 197, 625–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yabuta N., Mukai S., Okada N., Aylon Y., Nojima H. (2011) The tumor suppressor Lats2 is pivotal in Aurora A and Aurora B signaling during mitosis. Cell Cycle 10, 2724–2736 [DOI] [PubMed] [Google Scholar]
- 79. Mardin B. R., Lange C., Baxter J. E., Hardy T., Scholz S. R., Fry A. M., Schiebel E. (2010) Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nat. Cell Biol. 12, 1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Salah Z., Aqeilan R. I. (2011) WW domain interactions regulate the Hippo tumor suppressor pathway. Cell Death Dis. 2, e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.