Abstract
Ubiquitination is a process that involves the covalent attachment of the 76-residue ubiquitin protein through its C-terminal di-glycine (GG) to lysine (K) residues on substrate proteins. This post-translational modification elicits a wide range of functional consequences including targeting proteins for proteasomal degradation, altering subcellular trafficking events, and facilitating protein-protein interactions. A number of methods exist for identifying the sites of ubiquitination on proteins of interest, including site-directed mutagenesis and affinity-purification mass spectrometry (AP-MS). Recent publications have also highlighted the use of peptide-level immunoaffinity enrichment of K-GG modified peptides from whole cell lysates for global characterization of ubiquitination sites. Here we investigated the utility of this technique for focused mapping of ubiquitination sites on individual proteins. For a series of membrane-associated and cytoplasmic substrates including erbB-2 (HER2), Dishevelled-2 (DVL2), and T cell receptor α (TCRα), we observed that K-GG peptide immunoaffinity enrichment consistently yielded additional ubiquitination sites beyond those identified in protein level AP-MS experiments. To assess this quantitatively, SILAC-labeled lysates were prepared and used to compare the abundances of individual K-GG peptides from samples prepared in parallel. Consistently, K-GG peptide immunoaffinity enrichment yielded greater than fourfold higher levels of modified peptides than AP-MS approaches. Using this approach, we went on to characterize inducible ubiquitination on multiple members of the T-cell receptor complex that are functionally affected by endoplasmic reticulum (ER) stress. Together, these data demonstrate the utility of immunoaffinity peptide enrichment for single protein ubiquitination site analysis and provide insights into the ubiquitination of HER2, DVL2, and proteins in the T-cell receptor complex.
Ubiquitin is a highly conserved, 8 kDa protein that can be covalently attached to substrate proteins, leading to changes in protein stability, subcellular localization, and pathway activation. Ubiquitination occurs primarily on lysine residues via a multistep process that requires the concerted action of three enzymes. First an E1 ubiquitin-activating enzyme uses ATP to form a high-energy thioester bond with ubiquitin. This charged E1 can subsequently interact with and transfer ubiquitin to an E2 ubiquitin-conjugating enzyme. E3 ubiquitin-ligases ultimately provide specificity to the reaction by facilitating the transfer of ubiquitin from a charged E2 to the substrate protein (1). As ubiquitination dictates the fate of modified proteins, characterizing the residues within specific proteins that can be modified by ubiquitin provides mechanistic insight into many biological processes.
Dysregulated ubiquitination of critical substrates has been associated with many human diseases including cancer and neurodegeneration (2–8). Currently, efforts are underway to gain a better understanding of factors modulating ubiquitination on a substrate by substrate basis. Because E3 ligases confer much of this specificity, many have become attractive as potential therapeutic targets (9). Understanding the precise targets of ubiquitination, and the stimuli that elicit this modification, will play a central role in validating these enzymes and their modulators as targets (3, 9–12).
A series of biochemical methods are available for detecting ubiquitination on both endogenous and overexpressed proteins. For endogenous proteins, a common diagnostic for ubiquitination involves protein-level immunoprecipitation followed by Western blot analysis using an antibody recognizing ubiquitin. Many ubiquitinated proteins have shorter half-lives and are present at lower levels than their unmodified counterparts. To overcome this challenge, cells overexpressing substrates of interest are often treated with proteasomal or lysosomal inhibitors to stabilize ubiquitinated proteins. Although this method is diagnostic for the presence of ubiquitination, it does not reveal the exact site(s) of ubiquitination. For this, site-directed mutagenesis is commonly employed to identify residues that may be ubiquitinated. Lysine residues are substituted with arginines (individually or in combination) and the mutant protein is examined by immunoprecipitation-Western blot analysis. In some cases, because of the number of lysine residues and the size of the protein, this task can be challenging. Functional redundancy can result in the ubiquitination of alternative lysines when preferred sites are mutated. Conversely, mutagenesis can inhibit ubiquitination by blocking the ligase-substrate interaction even when the substituted lysine was not the primary target of the modification.
Mass-spectrometry-based methods provide a means of generating direct evidence to demonstrate ubiquitination on a particular lysine. This can be achieved by immunoprecipitating the protein of interest, separating the captured proteins by SDS-PAGE, excising the high molecular weight modified protein, and performing in-gel tryptic digestion (referred to as the gel-based method). Tryptic digestion results in the generation of a di-glycine remnant that remains attached to ubiquitinated lysine residue. This remnant is derived from the C terminus of ubiquitin, and results in a mass shift of +114.0429 Da that can be detected by MS/MS. The use of multiple-reaction monitoring (MRM)-initiated detection has been reported to aid in identification of low abundance ubiquitinated peptides (13). Although gel-based methods have led to the successful identification of many ubiquitination sites (14–16), there are instances in which the sensitivity has not been sufficient to systematically define ubiquitination sites of interest.
Recent publications have demonstrated that peptide level immunoaffinity enrichment, which takes advantage of antibodies raised against the di-glycine remnant (K-GG)1 motif, is a powerful approach for cataloging ubiquitinated substrates from cell lysate (17–19). K-GG peptide immunoaffinity enrichment has been used to identify >5,000 sites from as little as 1 mg of input material in global profiling efforts (19) and to successfully map ubiquitination sites on the protein APOBEC3F (20). Considering the emerging role for this method in investigating ubiquitination on a global scale, we were interested in assessing its utility in the context of focused ubiquitination site mapping on individual proteins.
To investigate this, ubiquitination on three different proteins was characterized: the receptor tyrosine-protein kinase (RTK) erbB-2 (HER2), Dishevelled-2 (DVL2), and T cell receptor α (TCRα). For these three likely ubiquitinated substrates, efforts to map ubiquitination sites using protein level immunoprecipitation methods were each met with limited success. In contrast, K-GG peptide immunoaffinity enrichment using comparable amounts of starting material yielded multiple high confidence ubiquitination site identifications for each substrate. Our results demonstrate the effectiveness of K-GG peptide immunoaffinity enrichment at identifying ubiquitination sites not seen by other methods, while in parallel elucidating valuable information regarding the ubiquitination status in a global manner.
EXPERIMENTAL PROCEDURES
Cell Culture, Reagents, and Plasmids
For TCRα and DVL2 studies, HEK293T were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) and 100 μg/ml penicillin/streptomycin and transfected using Fugene HD (Roche) according to manufacturer's instructions. For HER2 studies, BT474 cells were maintained in RPMI supplemented with 10% FBS and Glutamax. For Jurkat studies, Jurkat cells were maintained in RPMI supplemented with 10% FBS.
Immunoprecipitations, Coomassies and Western Blots
For overexpressed TCRα ubiquitination site identification studies, cells were transfected 48 h before cell lysis with plasmids encoding C-terminally HA-tagged wild type TCRα (21, 22). 293T were treated with 10 μm MG132 for 3 h and lysed in RIPA buffer (50 mm Tris-HCl pH 8, 150 mm NaCl, 1% Nonidet P40, 0.5% sodium deoxycholate, and 0.1% SDS) supplemented with Protease inhibitor mixture EDTA-free (Roche Applied Science, Penzberg, Germany). For HER2 studies, BT474 cells were treated with 10 nm Trastuzumab (Genentech, Inc., South San Francisco, CA) for 2 h before lysis in RIPA buffer. Cellular lysates were quantified using the BCA Protein Assay kit (Pierce, Rockford, IL) according to manufacturer's protocol. For immunoprecipitation, 10 mg of protein was incubated with 3 μg of anti-HA (Santa Cruz Biotechnology, Santa Cruz, CA) antibody for TCRα or 3 μg anti-HER2 (7C2, Genentech, Inc.) for 1 h followed by addition of 100 μl Protein A/G agarose beads (Santa Cruz Biotechnology) overnight. TCRα was eluted using 50 μl HA peptide (1 mg/ml in TBS) for 30 min at room temperature; endogenous HER2 was eluted by boiling the beads in sample buffer.
For DVL2 ubiquitination site identification, DVL2-FLAG-Myc was transfected into HEK293T cells, along with HA-ubiquitin. At 48 h post-transfection, cells were treated with 25 μm MG132 (EMD Biosciences, Danvers, MA) for 2 h. Cells were lysed in Nonidet P-40 buffer (1% Nonidet P-40, 120 mm NaCl, 50 mm Tris-HCl pH 7.4, 1 mm EDTA), plus protease and phosphatase inhibitors. Denatured whole cell lysates were affinity purified with anti-FLAG M2 beads (SIGMA, St. Louis, MO) overnight, and washed in high salt buffer (20 mm HEPES pH 7.9, 1.5 mm MgCl2, 420 mm NaCl, 0.2 mm EDTA, 25% glycerol) for 10 min, then in low salt buffer (20 mm Tris-HCl pH 7.4, 300 mm NaCl, 0.2 mm EDTA, 20% glycerol, 0.1% Nonidet P-40) for 10 min (three repetitions) and overnight. DVL2 protein was then eluted with 500 μg/ml 3 × FLAG peptide and subjected to in-solution tryptic digestion or to K-GG peptide immunoaffinity enrichment.
Samples were separated on 4–12% Bis-Tris polyacrylamide gels and transferred to PVDF membrane (10% sample) or stained with SimplyBlue Safestain stain (Invitrogen, Carlsbad, CA) (90% sample). Western blots were performed to identify migration patterns of ubiquitin-modified protein (anti-ubiquitin antibody P4D1, Santa Cruz Biotechnology) and used to guide in-gel tryptic digestion.
In-solution and In-gel Tryptic Digests
For in-gel tryptic digests, gel pieces were diced into 1 mm cubes and destained in 50 mm Ambic in 50% ACN (v/v) for 15 min or until clear. Gel pieces were dehydrated with 30 μl of 100% ACN for 5 min, the liquid removed, and the gel pieces rehydrated in 5 mm DTT and incubated at 50 °C for 60 min. Gel pieces were again dehydrated in 100% ACN, liquid was removed and gel pieces were rehydrated with 12.5 mm iodoacetamide (IAA). Samples were incubated at room temperature, in the dark for 20 min. Gel pieces were washed with 50 mm Ambic and dehydrated with 100% ACN. Gel pieces were rehydrated with 10 ng/μl trypsin resuspended in 50 mm Ambic on ice for 1 h. Excess liquid was removed and gel pieces were digested with trypsin at 37 °C overnight. Peptides were extracted with 50% ACN/0.1% formic acid, followed by 100% ACN/0.1% formic acid. Peptides were dried to completion and resuspended in 2% ACN/0.1% formic acid.
For in-solution tryptic digestion, proteins were eluted from beads using 50% ACN/0.1% TFA and dried down in a SpeedVac. Proteins were then reconstituted in HEPES buffer (20 mm, pH 8) containing 4 m urea. Samples were reduced in the presence of 5 mm DTT at 60 °C for 30 min followed by alkylation with 12.5 mm IAA at room temperature, in the dark for 20 min. Samples were diluted with HEPES buffer to a final concentration of 2 m urea. Trypsin was added and digestion was performed at 37 °C overnight. The digestion was quenched with 20% TFA and STAGE tipped (21). A C18 STAGE tip was prepared using two wafers from 3 m Empore Disk C18, 47 mm. It was washed with 100 μl of 95% ACN/0.1% TFA and equilibrated with 3 × 100 μl of 0.1% TFA. The digest mixture was loaded onto the tip followed by 3 × 100 μl of 0.1% TFA washes. Peptides were then eluted with 2 × 50 μl of 60% ACN/0.1% TFA. A portion of the eluent (∼20%) was analyzed by LC-MS/MS and the remaining 80% was subjected to ubiquitin enrichment using K-GG peptide immunoaffinity enrichment.
K-GG Peptide Immunoaffinity Enrichment
Cells were subjected to immunoaffinity enrichment of ubiquitinated peptides using the PTMScan® protocol (Cell Signaling Technology, Danvers, MA), as previously described (22). Briefly, cells were lysed in urea lysis buffer, lysates were reduced with DTT, alkylated with IAA and digested with trypsin overnight at room temperature. Resultant peptides were desalted and dried. Peptides were resuspended in IAP buffer (Cell Signaling Technology) and K-GG peptides were immunoprecipitated using the K-GG motif antibody conjugated to agarose beads for 2 h at 4 °C. Peptides were eluted using 0.15% TFA, desalted using a STAGE tip, and analyzed by LC-MS/MS.
For endogenous TCRα ubiquitination studies, Jurkat cells were treated with vehicle, 5 μg/ml tunicamycin (48 h; SIGMA, St. Louis, MO), 10 μm MG132 (3 h), or tunicamycin and MG132 in combination. Protein lysate was collected from the four conditions and 35 mg of protein was subjected to the K-GG peptide immunoaffinity enrichment protocol (see above).
SILAC Analysis
Cells were cultured in SILAC RPMI (Pierce) supplemented with isotopically enriched forms of l-lysine (13C6,15N2 hydrochloride; 50 μg/ml) and l-arginine (13C6, 15N4 hydrochloride; 40 μg/ml) (Heavy) or the corresponding unlabeled form of each at the same concentration (Light). Both Heavy and Light forms of RPMI were further supplemented with l-Proline (200 μg/ml; to prevent metabolic conversion of Arg→Pro), l-glutamine (2 mm) and 10% dialyzed FBS (Pierce). After 1 week of metabolic labeling, cells were lysed, digested with trypsin, and analyzed by LC-MS/MS to ensure complete incorporation of the isotopic label. Light cell lysates (10 mg) were subjected to protein level enrichment, SDS-PAGE, followed by in-gel tryptic digestion. Heavy cell lysates (10 mg) were subjected to K-GG peptide immunoaffinity enrichment. Peptides resulting from either method were dried down separately, reconstituted in 10 μl 2% ACN/0.1% formic acid and combined together for LC-MS/MS analysis.
LC-MS/MS
Peptide mixtures were injected via an auto-sampler and loaded onto Symmetry® C18 column (1.7 μm BEH-130, 0.1 × 100 mm) at a flow rate of 1.5 μl/min using a NanoAcquity UPLC system (Waters, Milford, MA) as previously described (23). Peptides were separated at 1 μl/min using a gradient of 2% Solvent B to 25% Solvent B (where Solvent A is 0.1% Formic acid/2% ACN/water and Solvent B is 0.1% formic acid/2% water/ACN) applied over 35 or 60 min with a total analysis time of 60 or 90 min. Peptides were eluted directly into a Orbitrap XL (DVL2 studies) or Velos-Orbitrap-Elite mass spectrometer (all other studies) (ThermoFisher, San Jose, CA) at a spray voltage of 1.4 kV using an Advance CaptiveSpray ionization source (Michrom BioResources/Bruker, Auburn, CA). Precursor ions were analyzed in the FTMS at 60,000 resolution. MS/MS was performed in the LTQ with the instrument operated in data dependent mode whereby the top eight or top 15 most abundant ions were subjected for fragmentation on the Orbitrap XL or Orbitrap Elite, respectively.
For peptide identification, ReAdW (v.4.3.1) was used to generate peaks and MS/MS spectra were searched using Mascot (v.2.3.02) against a concatenated target-decoy database comprised of human protein sequences (UniProt Dec. 2011), known contaminants, epitope tagged-TCRα or -DVL2, and the reversed versions of each (119,247 total entries). For peptide identifications from Jurkat studies on ER stress, Jurkat specific sequences for TCRα (Gene AAA60626), TCRβ (Gene ADB80116), and TCRζ (Gene AAA60394) were appended to the concatenated target-decoy database using text editor. MS/MS spectra searched by Mascot using trypsin with up to two missed cleavages, a 50 ppm precursor ion tolerance, and 0.8 Da fragment ion tolerance. Variable modifications were permitted for methionine oxidation (+15.9949 Da), carbamidomethylation (+57.0215) on cysteine residues and ubiquitin di-glycine remnant (-GG signature) on lysine residues (+114.0429 Da). No more than three variable modifications were permitted in a given search. For SILAC experiments, heavy lysine (+8.0142) and heavy arginine (+10.0083) were also included. The data were also interrogated for the presence of C-terminal +114.0429 as a diagnostic for IAA artifacts (17).
Given that single protein analysis offers limited numbers of peptide spectral matches, peptide assignments were initially filtered to a 5% peptide level false discovery rate (FDR) using linear discriminant analysis (24). Following automated filtering, peptide spectral matches to substrate specific K-GG peptides of interest were manually validated and filtered data quantified at the peptide level using the VistaQuant algorithm either in direct or indirect cross-quantification (XQuant) mode (25, 26). Relative abundance ratios at the protein level were calculated from summed abundance measurements of peptides with VistaQuant Confidence Scores ≥ 83 (26, 27).
RESULTS
Methods of Ubiquitination Site Mapping on Individual Proteins
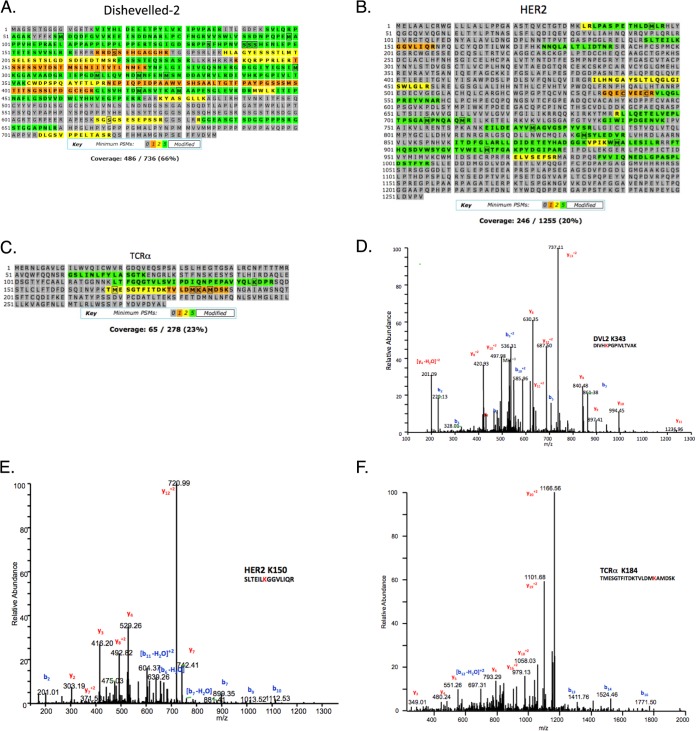
Dishevelled-2 (DVL2) is a cytoplasmic, multimodule adaptor protein in the Wnt signaling pathway (28) that is ubiquitinated by the KLHL12-cullin3-ubiquitin ligase (29). On Wnt stimulation, DVL2 is hyper-phosphorylated by CK1e, CK2, as well as RIPK4 (30–32). To profile DVL2 ubiquitination sites, FLAG-tagged DVL2 was overexpressed in HEK293T, purified by anti-FLAG immunoprecipitation, digested in-solution with trypsin, and analyzed by LC-MS/MS (Fig. 1B). Despite observing many unmodified DVL2 peptides (66% sequence coverage; Fig. 2A), no ubiquitinated DVL2 peptides were detected.
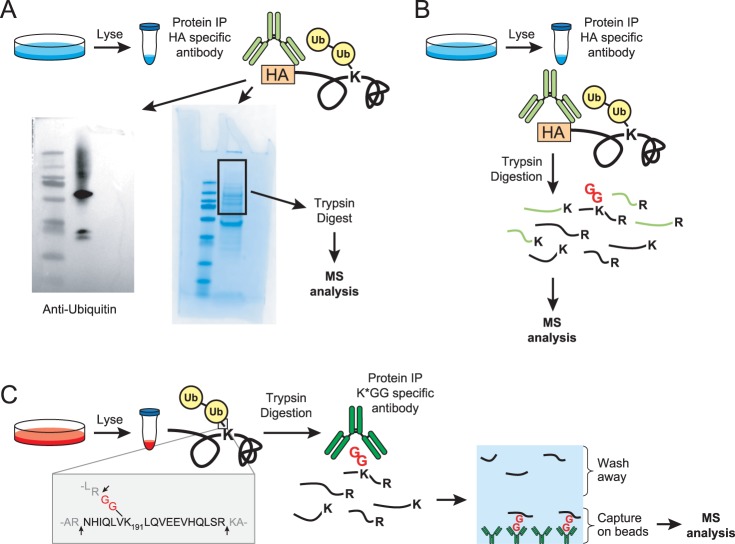
Fig. 1.
Methods to map ubiquitination sites. A, Schematic of the gel-based method for identifying ubiquitination sites on proteins of interest (POI). Proteins are immunoprecipitated using an immunoaffinity tag or with an antibody specific for a POI followed by SDS-PAGE analysis. Anti-ubiquitin Western blots are used to confirm the location of ubiquitinated POI and Coomassie stained gel bands are cut accordingly. In-gel tryptic digestions are performed, peptides are extracted, and then analyzed by LC-MS/MS. This example shows a representative Western blot and Coomassie from the HA-tagged TCRα studies. B, In solution digestion method on a POI. This is the same as (A) except trypsin digestions are performed immediately post-IP (in the presence of the IP antibody) and no SDS-PAGE analysis is performed. C, Schematic of the K-GG peptide immunoaffinity enrichment method for mapping ubiquitination sites on a POI. Whole cell lysates are digested with trypsin and the resultant peptides are immunoprecipitated with an antibody against the K-GG remnant left on ubiquitinated lysine residues. Peptides are eluted and analyzed by LC-MS/MS.
Fig. 2.
Ubiquitination site mapping on DVL2, HER2 and TCRα. Coverage reports obtained for (A) DVL2 using the in-solution digestion method, (B) HER2, and (C) TCRα using the gel-based method of ubiquitination site mapping. Representative spectra for ubiquitinated peptides from (D) DVL2, (E) HER2, and (F) TCRα using the K-GG peptide immunoaffinity enrichment method.
In parallel, we also investigated the ubiquitination status of the membrane-associated protein TCRα, a component of the T cell receptor (TCR). TCRα is a model substrate of the endoplasmic reticulum-associated degradation (ERAD) pathway that misfolds and is targeted for proteasomal degradation via the ERAD pathway when expressed in non-T cells lacking other members of the TCR complex (33, 34). Ubiquitination and degradation of this protein have been studied for nearly two decades, although the precise ubiquitination sites have only recently been proposed (35). We previously reported ubiquitination on three lysine residues (K118, K144, and K178) using a gel-based method; however, because TCRα is misfolded when ubiquitination occurs, we suspected additional lysine residues to also be modified. Using the previously described approach here (Fig. 1A), two ubiquitination sites on TCRα were observed in this study (K144 and K184) (Fig. 2C, Table I).
Table I. Identification of ubiquitination sites by various methods.
* Observed in a previous analysis.
We also investigated the ubiquitination status of the endogenous, membrane-associated protein HER2. HER2 is a member of the epidermal growth factor receptor family that plays an essential role in the pathogenesis and progression of certain aggressive types of breast cancer. HER2 is the target of the antibody-therapeutic trastuzumab (Herceptin®). It was previously reported that HER2 trafficking and degradation are associated with K48 and K63-linked poly-ubiquitination (36), although the precise ubiquitination sites on HER2 remain unknown. Additionally, inhibition of the chaperone HSP90 has been reported to induce ubiquitination and depletion of HER2 in a breast cancer cell line (37). We were interested in mapping endogenous HER2 ubiquitination sites in a breast cancer model and in characterizing the effect of trastuzumab treatment on HER2 ubiquitination patterns and protein stability. In this study, endogenous HER2 was immunoprecipitated from cellular lysates made from the BT474 HER2+ breast cancer cell line. Elutions were subjected to SDS-PAGE and the gel region from 175 kDa to the top of the gel, containing ubiquitin-modified HER2, was excised and subjected to in-gel tryptic digestion. Despite identification of many unmodified HER2 peptides, ubiquitination sites were not observed using this method (Fig. 2B, Table I).
In parallel with each of the DVL2, TCRα, and HER2 studies above, K-GG peptide immunoaffinity enrichment was performed. With DVL2, ubiquitination was observed on K343, K477, and K484 (nine unique peptides, 20 total PSMs, with mass errors ranging from two to eight ppm) (Fig. 2D, Table I, and supplemental Fig. S1). Two of these sites, K477 and K484, flank one of the RIPK4 phosphorylation sites in the DEP domain. Although further experiments are required to elucidate the coordination of phosphorylation and ubiquitination on DVL2, the proximity of these sites to one another is reminiscent of the phosphodegrons observed in certain cell cycle substrates and in growth factor receptor signaling (38, 39). For TCRα, K-GG peptide immunoaffinity enrichment led to the identification of ubiquitination on seven lysine residues: K74, K80, K86, K118, K144, K178, and K184 (30 unique peptides, 204 total PSMs, with mass errors that ranged from −8 to −2 ppm) (Fig. 2F, Table I, and supplemental Fig. S3). Similarly for HER2, K-GG peptide immunoaffinity enrichment of BT474 whole cell lysates yielded 11 ubiquitination sites: K150, K175, K716, K724, K736, K747, K753, K762, K765, K854, and K860 (17 unique peptides, 57 total PSMs, with mass errors that ranged from −8 to −2 ppm) (Fig. 2E, Table I, and supplemental Fig. S2).
Among the HER2 ubiquitination sites observed, five had not been previously reported (K716, K762, K765, K854, and K860), including four within the kinase domain. Ubiquitination was observed at these sites in untreated, dividing cells and remained unaltered by trastuzumab treatment (data not shown). The presence of ubiquitination sites within the kinase domain of an RTK has previously been shown to control agonist-dependent internalization and has been observed for many other kinases (18, 40–43). HER2 is known to form homo-dimers and hetero-dimers with the epidermal growth factor receptor (EGFR) and the structure of the HER2 kinase domain was recently solved (44). Nine of the ubiquitinated lysine residues identified in this study cluster on one side of the kinase domain (Fig. 3A, 3B) (18, 42). More experiments are required to determine how ubiquitination of the HER2 kinase domain affects protein dimerization and kinase activity as well as its impact on HER2-driven tumors and metastasis.
Fig. 3.
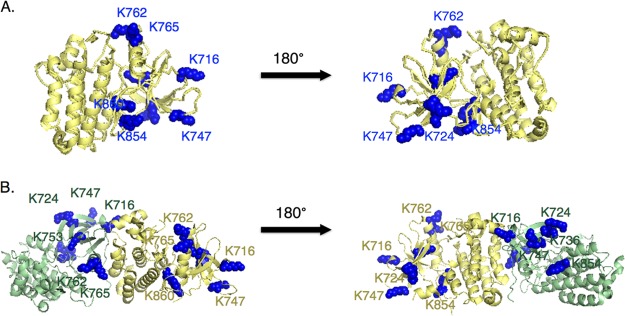
K-GG peptide immunoaffinity enrichment reveals ubiquitination sites on the kinase domain of HER2. A, The main structure of the kinase domain is labeled in yellow and lysine residues identified as ubiquitin carriers are labeled in blue in the left panel. The structure is rotated 180 degrees in the right panel. B, Same as A except the structure is shown in the context of the HER2 kinase domain homo-dimer. One monomer is labeled in yellow and one monomer is labeled in green for clarity.
Using Stable Isotope Labeling in Cell Culture (SILAC) to Compare Gel-based Methods of Ubiquitination Site Mapping to K-GG Peptide Immunoaffinity Enrichment
Given the consistent performance of K-GG peptide immunoaffinity enrichment relative to protein-level immunoprecipitation, we set up a SILAC experiment to directly compare the abundance of individual K-GG peptides between the two approaches. To assess the abundance of K-GG-peptides, cells were grown in either media containing isotopically heavy lysine (13C615N2) and arginine (13C6 15N4) or in isotopically light media. Cells were lysed and 10 mg of heavy protein lysate was subjected to K-GG peptide immunoaffinity enrichment. In parallel, 10 mg of light protein lysate was subjected to protein level immunoprecipitation, followed by SDS-PAGE, and in-gel tryptic digestion. Before LC-MS/MS analysis, peptide elutions from both methods were dried down separately and reconstituted in an equal volume. Samples were then combined together in a 1:1 ratio to compare the relative abundance of individual K-GG peptides derived from matching amounts of starting material (Fig. 4A). This analysis was carried out for both ectopically expressed TCRα in HEK293T cells and for endogenous HER2 from BT474 cells.
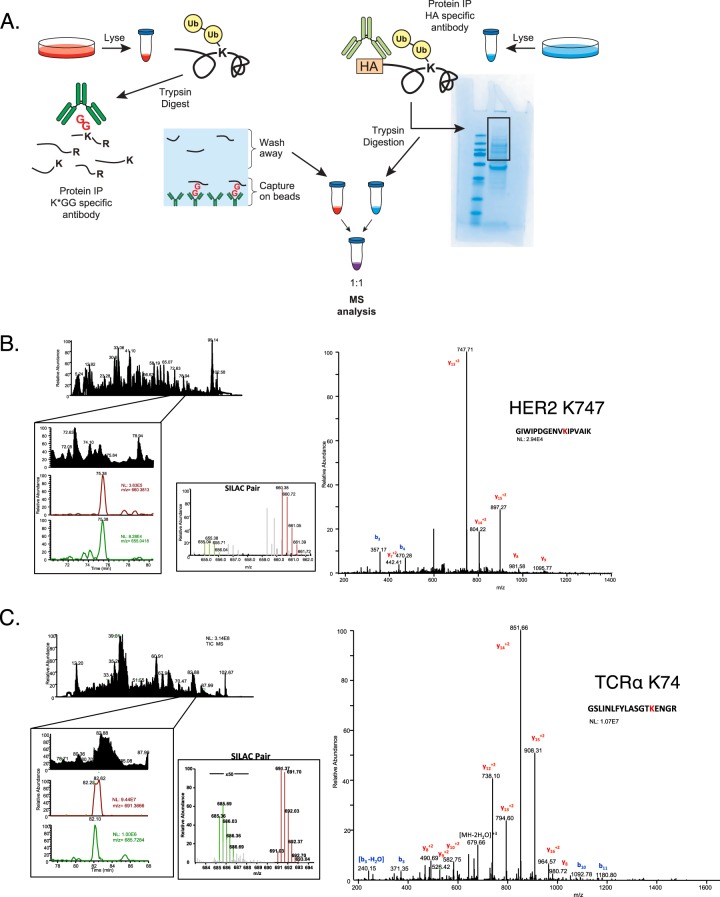
Fig. 4.
K-GG peptide immunoaffinity enrichment out-performs conventional methods of ubiquitination site mapping on single proteins. A, Cells grown in heavy media were lysed and 10 mg of protein was subjected to the K-GG peptide immunoaffinity enrichment method; cells grown in light media were lysed and 10 mg of protein was subjected to the gel-based method. Peptides eluted from each method were reconstituted separately in equal volumes. Samples were combined at a 1:1 ratio and examined by LC-MS/MS to compare the relative abundance of ubiquitinated peptides from our protein of interest isolated from each method. An example of a representative ubiquitinated peptide from (B) HER2 or (C) TCRα is shown. The top left panel shows the total ion chromatogram followed by the extracted ion chromatograms for ions corresponding to the heavy and light peptides (light ion on top, heavy ion on bottom). The middle panel is a narrow range full-MS scan depicting the relative intensity of the SILAC pair and the MS/MS spectrum for the ubiquitinated peptide is on the left panel. Heavy ions are labeled in green and light ions are labeled in red. b- and y- fragment ions are denoted by blue and red respectively. N.L. is the normalized level of the base peak for each ion.
In this SILAC approach, the identification of a single member of a SILAC pair is sufficient to quantify the abundance of a peptide in both conditions. Doing so permits the quantification of low abundance precursor ions that were present but did not trigger data-dependent MS/MS in one of the two conditions. For HER2, a series of heavy peptides stemming from K-GG peptide immunoaffinity enrichment were identified by MS/MS. Among these, the precursor ions for three out of the four remained undetectable in the light sample derived from protein level IP (Table II). For one ubiquitinated peptide detected by both methods (K747), the heavy ion precursor was four times more abundant in the heavy K-GG peptide immunoaffinity enrichment method than in the light gel-based method (Fig. 4B). Likewise, ubiquitinated peptides identified from TCRα were between 16 and 100 times more abundant in the heavy K-GG peptide immunoaffinity enriched samples than the light gel-based samples (Fig. 4C, Table III).
Table II. SILAC comparison of identification of HER2 ubiquitinated peptides. (SNR) is signal to noise ratio.
| Peptide | K# | Heavy SNR | Light SNR | Log2 (H-SNR/l-SNR) |
|---|---|---|---|---|
| R.SLTEILK*GGVLIQR.N | 150 | 3.36 | N.D. | N.Q. |
| K.VLGSGAFGTVYK*GIWIPDGENVKIPVAIK.V | 736 | 5.7 | N.D. | N.Q |
| K.GIWIPDGENVK*IPVAIK.V | 747 | 31.30 | 7.24 | 2.11 |
| R.NVLVK*SPNHVK.I | 854 | 2.46 | N.D. | N.Q. |
Table III. SILAC comparison of identification of TCRα ubiquitinated peptides; gel-based method vs. K-GG peptide immunoaffinity enrichment. (SNR) is signal to noise ratio.
| Peptides | K# | Heavy SNR | Light SNR | Log2 (H-SNR//l-SNR) |
|---|---|---|---|---|
| R.GSLINLFYLASGTK*ENGR.L | 74 | 1724.45 | 7.47 | 7.85 |
| R.LK*STFNSK.E | 80 | 731.98 | 1.42 | N.Q. |
| K.STFNSK*ESYSTLHIR.D | 86 | 814.43 | 9.75 | 6.38 |
| R.LKSTFNSK*ESYSTLHIR.D | 86 | 517.96 | 1.37 | N.Q. |
| R.ATGGNNK*LTFGQGTVLSVIPDIQNPEPAVYQLK.D | 118 | 117.81 | 2.08 | 5.82 |
| K.LTFGQGTVLSVIPDIQNPEPAVYQLK*DPR.S | 144 | 664.00 | 1.80 | N.Q. |
| K.TMESGTFITDK*TVLDMK.A | 178 | 103.55 | 4.05 | 4.66 |
| K.TMESGTFITDKTVLDMK*AMDSK.S | 184 | 43.98 | N.D. | N.Q. |
| K.TVLDMK*AMDSK.S | 184 | 632.88 | 0.77 | N.Q |
One consideration was that pre-enrichment at the protein level might improve the performance of K-GG peptide immunoaffinity enrichment by substantially reducing the number of other K-GG peptides competing for binding to the anti-K-GG antibody. To test this, a double IP strategy was implemented to examine the impact of protein pre-enrichment before K-GG peptide immunoaffinity enrichment. Elutions from HA-TCRα IPs were directly subjected to K-GG peptide immunoaffinity enrichment (isotopically light) and compared with K-GG peptide immunoaffinity enrichment alone (isotopically heavy). Again, performing the K-GG peptide immunoaffinity enrichment method in isolation yielded more ubiquitinated TCRα peptides. Nine ubiquitinated TCRα peptides were quantified from the K-GG peptide immunoaffinity enrichment, whereas, only five peptides from the double IP method had intensities great enough to quantify (Table IV). Of the peptides that were quantifiable from the double IP method, all were present at least fourfold higher in the K-GG peptide immunoaffinity enrichment method sample. This result suggests that pre-enrichment at the protein level provided no advantage and instead decreased the recovery of K-GG peptides for individual substrates of interest.
Table IV. SILAC comparison of identification of TCRα ubiquitinated peptides; double IP vs. K-GG peptide immunoaffinity enrichment. (SNR) is signal to noise ratio.
| Peptides | K# | Heavy SNR | Light SNR | Log2(H-SNR//l-SNR) |
|---|---|---|---|---|
| R.GSLINLFYLASGTK*ENGR.L | 74 | 1724.45 | 7.47 | 7.85 |
| R.LK*STFNSK.E | 80 | 731.98 | 1.42 | N.Q. |
| K.STFNSK*ESYSTLHIR.D | 86 | 814.43 | 9.75 | 6.38 |
| R.LKSTFNSK*ESYSTLHIR.D | 86 | 517.96 | 1.37 | N.Q. |
| R.ATGGNNK*LTFGQGTVLSVIPDIQNPEPAVYQLK.D | 118 | 117.81 | 2.08 | 5.82 |
| K.LTFGQGTVLSVIPDIQNPEPAVYQLK*DPR.S | 144 | 664.00 | 1.80 | N.Q. |
| K.TMESGTFITDK*TVLDMK.A | 178 | 103.55 | 4.05 | 4.66 |
| K.TMESGTFITDKTVLDMK*AMDSK.S | 184 | 43.98 | N.D. | N.Q. |
| K.TVLDMK*AMDSK.S | 184 | 632.88 | 0.77 | N.Q |
Detection of ER Stress-induced Ubiquitination Events Using K-GG Peptide Immunoaffinity Enrichment
Using K-GG peptide immunoaffinity enrichment, we set out to extend our findings from exogenously expressed TCRα and examine whether ER stress in T-cells elicited similar ubiquitination of the endogenously expressed TCRα protein. Protein level enrichment of endogenous TCRα is complicated by the fact that the human proteome contains an enormous level of sequence diversity within the TCRα protein (45). However, because K-GG peptide immunoaffinity enrichment relies on an antibody targeted against the K-GG epitope, it is possible to investigate ubiquitin modifications on a cell line-specific TCRα without the need to identify or produce an antibody that works for protein-level enrichment.
Jurkat T-cells were used to investigate the ubiquitination status of endogenous TCRα in response to chemically induced ER stress caused by tunicamycin. Tunicamycin is a nucleoside antibiotic that functions by blocking synthesis of N-linked glycosylation on proteins leading to an accumulation of misfolded proteins. Without glycosylation, misfolded proteins are shuttled through the ERAD pathway in a manner similar to exogenously expressed TCRα in non-T-cells. Jurkat cells were left untreated, treated for 48 h with 5 μg/ml tunicamycin, treated for 3 h with 10 μm MG132 to block the proteosome, or treated for 48 h with 5 μg/ml tunicamycin and 10 μm MG132 for the final 3 h to induce protein misfolding and prevent degradation of ubiquitinated proteins via the proteasome. Cells were lysed and subjected to K-GG peptide immunoaffinity enrichment and analyzed by LC-MS/MS. Fig. 5A illustrates the ubiquitination sites found on TCRα in the four different conditions. Although no peptide-spectral matches (PSMs) were detected in the untreated sample, levels of ubiquitinated peptides representing five distinct sites on TCRα were highly induced by treatment with the combination of tunicamycin and MG132. Only one PSM representing ubiquitinated TCRα was observed following tunicamycin treatment alone; likely because of the protein's short half-life (<30 min) when misfolded (33). As expected, detection of these K-GG peptides by LC-MS/MS required concurrent inhibition of the proteasome to stabilize ubiquitinated intermediates. After quantifying areas under the curve in cross-quant (XQuant) mode, low abundance ubiquitinated TCRα peptides were detected in both untreated and after MG132 treatment alone. XQuant mode infers the identity of unmatched peaks using retention times and high-resolution precursor ion masses from the list of confidently identified K-GG (48). This allows a more accurate comparison of the relative abundance of ubiquitinated peptides across multiple samples by including peptides that were not abundant enough to trigger MS/MS.
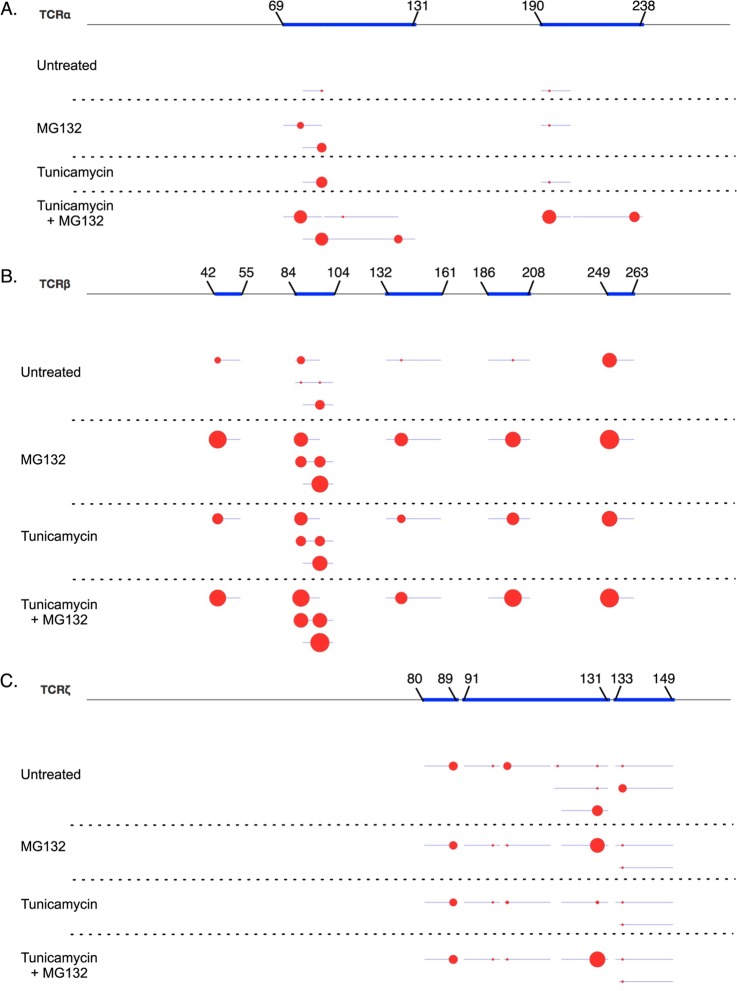
Fig. 5.
K-GG peptide immunoaffinity enrichment identifies ubiquitination sites on endogenous T-cell receptor components during ER stress. Jurkat T-cells were untreated, treated with tunicamycin, treated with MG132 or treated with a combination of tunicamycin and MG132. 35 mg of cellular lysate for each condition was digested with trypsin and subjected to K-GG peptide immunoaffinity enrichment. Area under the curve was quantified using the VistaQuant algorithm in XQuant mode. Protein coverage is represented by blue lines and individual ubiquitination sites for TCRα (A), TCRβ (B), or TCRζ (C) are represented as red circles on a linearized version of the protein. The area of the circle indicates the relative abundance among conditions on a logarithmic scale.
In addition to identifying ubiquitination sites on TCRα, we also identified ubiquitination of TCRβ on seven lysine residues in cells treated with tunicamycin and MG132 (Fig. 5B). However, similar levels of TCRβ ubiquitination was also observed in response to treatment with MG132 alone and at lower levels, in cells untreated or exposed to tunicamycin alone. These results suggest that TCRβ protein folding differs from TCRα and may be more sensitive to chemical perturbation. In contrast to TCRα and TCRβ, ubiquitination on TCRζ was relatively unchanged in response to treatment with MG132, tunicamycin, or the combination of both drugs (Fig. 5C). This result is expected because unlike TCRα and TCRβ, which are heavily glycosylated, TCRζ is not glycosylated and therefore should not be directly affected by treatment with tunicamycin.
Although our focused K-GG enrichment study revealed inducible ubiquitination of the T-cell receptor proteins of interest, additional information regarding the ubiquitination status of Caspase-3 and key mediators of the ER stress pathway (e.g. IRE1 and ATF6) was obtained (supplemental Fig. S4, supplemental Fig. S5, supplemental Table S1). These results demonstrate that the sensitivity of this method is sufficient to detect changes in ubiquitination levels across multiple samples and provides additional information regarding the ubiquitination status of other proteins that can be useful for interrogating pathway specific biology.
DISCUSSION
During the course of independent investigations of Wnt signaling, HER2 trafficking, and ERAD, we characterized ubiquitination on a series of overexpressed or endogenous proteins and were struck by the effectiveness of K-GG peptide immunoaffinity enrichment for characterizing individual proteins. Across three separate projects (DVL2, HER2, and TCRα), K-GG peptide immunoaffinity enrichment resulted in identification of more ubiquitination sites than protein level immunoaffinity enrichment. Using SILAC, a side-by-side comparison demonstrated that higher yields of ubiquitinated peptides for HER2 and TCRα were obtained using K-GG peptide immunoaffinity enrichment over methods involving protein level enrichment. This work was extended to T-cells where K-GG peptide immunoaffinity enrichment revealed that drug-induced changes in ubiquitination of the endogenous TCRα protein could be obtained as part of the global ubiquitination profile. The K-GG immunoaffinity enrichment strategy provided greater coverage of ubiquitination sites on substrates of interest in the absence of substrate specific pre-enrichment, despite the abundance of other K-GG peptides captured. These observations underscore difficulties associated with the presence of abundant, unmodified peptides, enrichment efficiency, and sample losses inherent to protein-level, gel based methods.
Gel-based methods have been a highly effective approach for identifying ubiquitination sites, owing to the separation of substoichiometric modified proteins from their unmodified counterparts. That said, these methods themselves have inherent limitations. One is that gel-based methods involve many steps where sample losses can occur. From the initial lysis step, methods involving protein level enrichment are at a disadvantage because most detergent based lysis buffers are only partially denaturing and therefore retain certain deubiquitinase (DUB) and protease activities (data not shown, (46)). For endogenous proteins, this can be compounded by the lack of available antibodies, especially for proteins like TCRα that have high variability across cell lines and samples. Furthermore, recovery of tryptic peptides from polyacrylamide gels can vary by peptide in ways that affect yield overall (46, 47).
These data suggest that K-GG peptide IPs are advantageous because they separate modified from unmodified peptides while avoiding the caveats associated with protein immunoprecipitation and gel based methods. One explanation may be that samples lysed in highly denaturing urea buffers display greater inhibited deubiquitinase activity. For three substrate proteins examined for this study, K-GG peptide immunoaffinity enrichment resulted in a greater depth-of-coverage from moderate amounts of starting material and minimal mass spectrometry instrument time. An additional benefit of this approach is that direct K-GG peptide immunoaffinity enrichment provides information regarding the ubiquitination status of proteins other than the protein of interest that can complement single substrate efforts, as was the case in our T-cell studies. That study identified a total of 896 proteins, including TCRα, that were inducibly ubiquitinated in response to ER stress (supplemental Figs. S4, S5, supplemental Table S1). This additional information provides context to interpret the results for TCRα, and serves as a valuable resource for other studies involving ubiquitination dynamics during ER stress. Although it remains challenging to capture low abundance, endogenous proteins, this is the case for both protein and peptide enrichment strategies and will likely require more extensive fractionation and enhanced LC-MS sensitivity to overcome.
This approach of employing motif-directed antibodies against post-translational modifications for single substrate studies may likewise be beneficial for other PTMs including phospho-Tyr, acetylation or methylation. Extending the use of this existing technique will be valuable as we seek both breadth and depth in our understanding of how modifications regulate proteins, pathways and cellular processes.
Supplementary Material
Acknowledgments
We thank members of the MPL group and Protein Chemistry department, including D. Bustos, L. Phu, J. Scheer, W. Sandoval, Q. Song, and C. Bakalarski for their helpful advice and suggestions. PTMScan studies performed at Genentech were carried out under a limited license from Cell Signaling Technology.
Footnotes
 This article contains supplemental Figs. S1 to S5 and Table S1.
This article contains supplemental Figs. S1 to S5 and Table S1.
1 The abbreviations used are:
- K-GG
- di-glycine remnant on ubiquitinated lysine residues after tryptic digestion
- SILAC
- stable isotope labeling by amino acids in cell culture
- PTM
- post-translational modification
- LC-MS/MS
- liquid chromatography followed by tandem MS analysis
- IP
- immunoprecipitation
- TCRα
- T cell receptor alpha
- TCRβ
- T cell receptor beta
- TCRζ
- T cell receptor zeta
- DVL2
- Dishevelled-2
- SNR
- signal to noise ratio
- POI
- protein of interest
- AP-MS
- affinity-purification mass spectrometry
- Ambic
- ammonium bicarbonate
- RTK
- receptor tyrosine-protein kinase
- ACN
- acetonitrile
- TFA
- Trifluoroacetic acid.
REFERENCES
- 1. Pickart C. M. (2001) Mechanisms underlying ubiquitination. Ann. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 2. Hoeller D., Dikic I. (2009) Targeting the ubiquitin system in cancer therapy. Nature 458, 438–444 [DOI] [PubMed] [Google Scholar]
- 3. Kong F., Zhang J., Li Y., Hao X., Ren X., Li H., Zhou P. (2013) Engineering a single ubiquitin ligase for the selective degradation of all activated ErbB receptor tyrosine kinases. Oncogene [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sharma V., Antonacopoulou A. G., Tanaka S., Panoutsopoulos A. A., Bravou V., Kalofonos H. P., Episkopou V. (2011) Enhancement of TGF-beta signaling responses by the E3 ubiquitin ligase Arkadia provides tumor suppression in colorectal cancer. Cancer Res. 71, 6438–6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ueki T., Park J. H., Nishidate T., Kijima K., Hirata K., Nakamura Y., Katagiri T. (2009) Ubiquitination and downregulation of BRCA1 by ubiquitin-conjugating enzyme E2T overexpression in human breast cancer cells. Cancer Res. 69, 8752–8760 [DOI] [PubMed] [Google Scholar]
- 6. Yang Y., Kitagaki J., Dai R. M., Tsai Y. C., Lorick K. L., Ludwig R. L., Pierre S. A., Jensen J. P., Davydov I. V., Oberoi P., Li C. C., Kenten J. H., Beutler J. A., Vousden K. H., Weissman A. M. (2007) Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res. 67, 9472–9481 [DOI] [PubMed] [Google Scholar]
- 7. Zhi X., Zhao D., Wang Z., Zhou Z., Wang C., Chen W., Liu R., Chen C. (2013) E3 ubiquitin ligase RNF126 promotes cancer cell proliferation by targeting the tumor suppressor p21 for ubiquitin-mediated degradation. Cancer Res. 73, 385–394 [DOI] [PubMed] [Google Scholar]
- 8. Balasubramanyam M., Sampathkumar R., Mohan V. (2005) Is insulin signaling molecules misguided in diabetes for ubiquitin-proteasome mediated degradation? Mol. Cell. Biochem. 275, 117–125 [DOI] [PubMed] [Google Scholar]
- 9. Cohen P., Tcherpakov M. (2010) Will the ubiquitin system furnish as many drug targets as protein kinases? Cell 143, 686–693 [DOI] [PubMed] [Google Scholar]
- 10. Sheng Z., Zhang S., Bustos D., Kleinheinz T., Le Pichon C. E., Dominguez S. L., Solanoy H. O., Drummond J., Zhang X., Ding X., Cai F., Song Q., Li X., Yue Z., van der Brug M. P., Burdick D. J., Gunzner-Toste J., Chen H., Liu X., Estrada A. A., Sweeney Z. K., Scearce-Levie K., Moffat J. G., Kirkpatrick D. S., Zhu H. (2012) Ser1292 autophosphorylation is an indicator of LRRK2 kinase activity and contributes to the cellular effects of PD mutations. Science Translational Med. 4, 164ra161. [DOI] [PubMed] [Google Scholar]
- 11. Dueber E. C., Schoeffler A. J., Lingel A., Elliott J. M., Fedorova A. V., Giannetti A. M., Zobel K., Maurer B., Varfolomeev E., Wu P., Wallweber H. J., Hymowitz S. G., Deshayes K., Vucic D., Fairbrother W. J. (2011) Antagonists induce a conformational change in cIAP1 that promotes autoubiquitination. Science 334, 376–380 [DOI] [PubMed] [Google Scholar]
- 12. Ernst A., Avvakumov G., Tong J., Fan Y., Zhao Y., Alberts P., Persaud A., Walker J. R., Neculai A. M., Neculai D., Vorobyov A., Garg P., Beatty L., Chan P. K., Juang Y. C., Landry M. C., Yeh C., Zeqiraj E., Karamboulas K., Allali-Hassani A., Vedadi M., Tyers M., Moffat J., Sicheri F., Pelletier L., Durocher D., Raught B., Rotin D., Yang J., Moran M. F., Dhe-Paganon S., Sidhu S. S. (2013) A strategy for modulation of enzymes in the ubiquitin system. Science 339, 590–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mollah S., Wertz I. E., Phung Q., Arnott D., Dixit V. M., Lill J. R. (2007) Targeted mass spectrometric strategy for global mapping of ubiquitination on proteins. Rapid Commun. Mass Spectrom. RCM 21, 3357–3364 [DOI] [PubMed] [Google Scholar]
- 14. Kirkpatrick D. S., Denison C., Gygi S. P. (2005) Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nat. Cell Biol. 7, 750–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003) A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21, 921–926 [DOI] [PubMed] [Google Scholar]
- 16. Danielsen J. M., Sylvestersen K. B., Bekker-Jensen S., Szklarczyk D., Poulsen J. W., Horn H., Jensen L. J., Mailand N., Nielsen M. L. (2011) Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol. Cell. Proteomics 10, M110 003590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bustos D., Bakalarski C. E., Yang Y., Peng J., Kirkpatrick D. S. (2012) Characterizing ubiquitination sites by peptide-based immunoaffinity enrichment. Mol. Cell. Proteomics 11, 1529–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., Harper J. W., Gygi S. P. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Udeshi N. D., Svinkina T., Mertins P., Kuhn E., Mani D. R., Qiao J. W., Carr S. A. (2013) Refined Preparation and Use of Anti-diglycine Remnant (K-{varepsilon}-GG) Antibody Enables Routine Quantification of 10,000s of Ubiquitination Sites in Single Proteomics Experiments. Mol. Cell. Proteomics 12, 825–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Albin J. S., Anderson J. S., Johnson J. R., Harjes E., Matsuo H., Krogan N. J., Harris R. S. (2013) Dispersed Sites of HIV Vif-Dependent Polyubiquitination in the DNA Deaminase APOBEC3F. J. Mol. Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rappsilber J., Mann M., Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protocols 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 22. Rush J., Moritz A., Lee K. A., Guo A., Goss V. L., Spek E. J., Zhang H., Zha X. M., Polakiewicz R. D., Comb M. J. (2005) Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 23, 94–101 [DOI] [PubMed] [Google Scholar]
- 23. Phu L., Izrael-Tomasevic A., Matsumoto M. L., Bustos D., Dynek J. N., Fedorova A. V., Bakalarski C. E., Arnott D., Deshayes K., Dixit V. M., Kelley R. F., Vucic D., Kirkpatrick D. S. (2011) Improved quantitative mass spectrometry methods for characterizing complex ubiquitin signals. Mol. Cell. Proteomics 10, M110 003756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villen J., Haas W., Sowa M. E., Gygi S. P. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 26. Bakalarski C. E., Elias J. E., Villen J., Haas W., Gerber S. A., Everley P. A., Gygi S. P. (2008) The impact of peptide abundance and dynamic range on stable-isotope-based quantitative proteomic analyses. J. Proteome Res. 7, 4756–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Elias J. E., Gygi S. P. (2010) Target-decoy search strategy for mass spectrometry-based proteomics. Meth. Mol. Biol. 604, 55–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Y., Appleton B. A., Wiesmann C., Lau T., Costa M., Hannoush R. N., Sidhu S. S. (2009) Inhibition of Wnt signaling by Dishevelled PDZ peptides. Nat. Chem. Biol. 5, 217–219 [DOI] [PubMed] [Google Scholar]
- 29. Angers S., Thorpe C. J., Biechele T. L., Goldenberg S. J., Zheng N., MacCoss M. J., Moon R. T. (2006) The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-beta-catenin pathway by targeting Dishevelled for degradation. Nat. Cell Biol. 8, 348–357 [DOI] [PubMed] [Google Scholar]
- 30. Peters J. M., McKay R. M., McKay J. P., Graff J. M. (1999) Casein kinase I transduces Wnt signals. Nature 401, 345–350 [DOI] [PubMed] [Google Scholar]
- 31. Willert K., Brink M., Wodarz A., Varmus H., Nusse R. (1997) Casein kinase 2 associates with and phosphorylates dishevelled. EMBO J. 16, 3089–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang X., McGann J. C., Liu B. Y., Hannoush R. N., Lill J. R., Pham V., Newton K., Kakunda M., Liu J., Yu C., Hymowitz S. G., Hongo J. A., Wynshaw-Boris A., Polakis P., Harland R. M., Dixit V. M. (2013) Phosphorylation of Dishevelled by Protein Kinase RIPK4 Regulates Wnt Signaling. Science 339, 1441–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu H., Kaung G., Kobayashi S., Kopito R. R. (1997) Cytosolic degradation of T-cell receptor alpha chains by the proteasome. J. Biol. Chem. 272, 20800–20804 [DOI] [PubMed] [Google Scholar]
- 34. Yu H., Kopito R. R. (1999) The role of multiubiquitination in dislocation and degradation of the alpha subunit of the T cell antigen receptor. J. Biol. Chem. 274, 36852–36858 [DOI] [PubMed] [Google Scholar]
- 35. Anania V. G., Bustos D. J., Lill J. R., Kirkpatrick D. S., Coscoy L. (2013) A Novel Peptide-Based SILAC Method to Identify the Posttranslational Modifications Provides Evidence for Unconventional Ubiquitination in the ER-Associated Degradation Pathway. Int. J. Proteomics 2013, 857918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marx C., Held J. M., Gibson B. W., Benz C. C. (2010) ErbB2 trafficking and degradation associated with K48 and K63 polyubiquitination. Cancer research 70, 3709–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mimnaugh E. G., Chavany C., Neckers L. (1996) Polyubiquitination and proteasomal degradation of the p185c-erbB-2 receptor protein-tyrosine kinase induced by geldanamycin. The Journal of biological chemistry 271, 22796–22801 [DOI] [PubMed] [Google Scholar]
- 38. Padron D., Sato M., Shay J. W., Gazdar A. F., Minna J. D., Roth M. G. (2007) Epidermal growth factor receptors with tyrosine kinase domain mutations exhibit reduced Cbl association, poor ubiquitylation, and down-regulation but are efficiently internalized. Cancer research 67, 7695–7702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Frescas D., Pagano M. (2008) Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nature reviews. Cancer 8, 438–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huang F., Kirkpatrick D., Jiang X., Gygi S., Sorkin A. (2006) Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Molecular cell 21, 737–748 [DOI] [PubMed] [Google Scholar]
- 41. Lopez-Otin C., Hunter T. (2010) The regulatory crosstalk between kinases and proteases in cancer. Nat. Rev. Cancer 10, 278–292 [DOI] [PubMed] [Google Scholar]
- 42. Wagner S. A., Beli P., Weinert B. T., Nielsen M. L., Cox J., Mann M., Choudhary C. (2011) A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics : MCP 10, M111 013284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huang F., Zeng X., Kim W., Balasubramani M., Fortian A., Gygi S. P., Yates N. A., Sorkin A. (2013) Lysine 63-linked polyubiquitination is required for EGF receptor degradation. Proc. Natl. Acad. Sci. U. S. A. 110, 15722–15727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Aertgeerts K., Skene R., Yano J., Sang B. C., Zou H., Snell G., Jennings A., Iwamoto K., Habuka N., Hirokawa A., Ishikawa T., Tanaka T., Miki H., Ohta Y., Sogabe S. (2011) Structural analysis of the mechanism of inhibition and allosteric activation of the kinase domain of HER2 protein. J. Biol. Chem. 286, 18756–18765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nikolich-Zugich J., Slifka M. K., Messaoudi I. (2004) The many important facets of T-cell repertoire diversity. Nat. Rev. Immunology 4, 123–132 [DOI] [PubMed] [Google Scholar]
- 46. Shuford C. M., Sederoff R. R., Chiang V. L., Muddiman D. C. (2012) Peptide production and decay rates affect the quantitative accuracy of protein cleavage isotope dilution mass spectrometry (PC-IDMS). Mol. Cell. Proteomics 11, 814–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Havlis J., Shevchenko A. (2004) Absolute quantification of proteins in solutions and in polyacrylamide gels by mass spectrometry. Anal. Chem. 76, 3029–3036 [DOI] [PubMed] [Google Scholar]
- 48. Kirkpatrick D. S., Bustos D. J., Dogan T., Chan J., Phu L., Young A., Friedman L. S., Belvin M., Song Q., Bakalarski C. E., Hoeflich K. P. (2013) Phosphoproteomic characterization of DNA damage response in melanoma cells following MEK/PI3K dual inhibition. Proceedings of the National Academy of Sciences of the United States of America 110, 19426–19431 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.