Abstract
MicroRNAs are a novel class of powerful endogenous regulators of gene expression. MiR-378 and miR-378* are localized in the first intron of the Ppargc1b gene that codes the transcriptional co-activator PGC-1β. The latter regulates energy expenditure as well as mitochondrial biogenesis. The miR-378:miR-378* hairpin is highly expressed in cardiac cells. To better assess their role in cardiomyocytes, we identified miR-378 and miR-378* targets via a proteomic screen. We established H9c2 cellular models of overexpression of miR-378 and miR-378* and identified a total of 86 down-regulated proteins in the presence of either one of these miRs. Functional annotation clustering showed that miR-378 and miR-378* regulate related pathways in cardiomyocytes, including energy metabolism, notably glycolysis, cytoskeleton, notably actin filaments and muscle contraction. Using bioinformatics algorithms we found that 20 proteins were predicted as direct targets of the miRs. We validated eight of these targets by quantitative RT-PCR and luciferase reporter assay. We found that miR-378 targets lactate dehydrogenase A and impacts on cell proliferation and survival whereas miR-378* targets cytoskeleton proteins actin and vimentin. Proteins involved in endoplasmic reticulum stress response such as chaperone and/or calcium buffering proteins GRP78, PPIA (cyclophilin A), calumenin, and GMMPA involved in glycosylation are repressed by these miRs. Our results show that the miR-378/378* hairpin establishes a connection among energy metabolism, cytoskeleton remodeling, and endoplasmic reticulum function through post-transcriptional regulation of key proteins involved in theses pathways.
MicroRNAs (miRs)1 are 18–25-nucleotide noncoding RNAs that are known to regulate gene expression in a sequence-specific manner. They bind to mRNAs causing their degradation or translational inhibition (1). MiRs are transcribed as primary transcripts that fold to form miRNA:miRNA* stem loop duplexes (2). In the main processing pathway, the primary transcripts are cleaved in the nucleus by the Drosha/Dgcr8 microprocessor complex into 70 nucleotide precursor miRNA hairpins and then transported into the cytoplasm for further processing by Dicer before the strand of the stem-loop duplex with the lower stability at the 5′ end is incorporated into the RISC complex. The other strand, called miRNA*, is often rapidly degraded (2). However, alternative noncanonical pathways exists in the cell and the expression of one of the two strands is sometimes tissue specific, i.e. the miRNA* can be matured and expressed in some cell types (3). MiRNAs can also derive from introns of coding genes in which case they are called mirtrons that are spliced into pre-miRNA hairpin mimics then cleaved by Dicer into functional miRs (4).
MiRs have been described as key players in signaling and transcriptional pathways modulating cardiac development, function and disease (5). MiR-378 and miR-378* are mirtrons derived from a single hairpin located in the first intron of the Peroxisome proliferator-activated receptor gamma coactivator 1-beta (Ppargc1b) gene that encodes the PGC-1β protein. The latter is a powerful transcriptional coactivator of nuclear respiratory factor-1 (NRF-1) and ERRs (estrogen receptor-related receptors), which are key factors in the expression of many mitochondrial genes and mitochondrial biogenesis (6, 7). PGC-1β is necessary for adaptive thermogenesis to cold and exertional exercise (8). In conjunction with PGC-1α, PGC-1β is important for perinatal cardiac development (8). In the adult heart, PGC-1β maintains mitochondrial and contractile function following pressure overload hypertrophy by preserving glucose metabolism and preventing oxidative stress (9).
MiR-378 and 378* are expressed in different cell types. MiR-378 has been mostly described as an oncogene-like microRNA in different cancer cell lines (10–12). MiR-378 stimulates tumor growth and cell survival via its repressive effect on tumor supressors Sufu and Fus-1 (13). In breast cancer cells infected with an adenovirus overexpressing the miR-378:378* hairpin, miR-378* was found to be more abundant than miR-378 and responsible for a shift from oxidative phosphorylation to a high-rate glycolytic metabolism associated with increased lactate production, a phenomenon known as the Warburg effect (14). The expression of miR-378 and miR-378* is associated with increased lipogenesis in adipocytes (15, 16). Knock-out of miR-378 and miR-378* in mice leads to a resistance to high-fat diet-induced obesity, enhanced mitochondrial fatty acid metabolism, and elevated oxidative capacity of insulin-target tissues (17).
The miR-378:miR-378* hairpin is highly expressed in cardiac cells. Deep sequencing data showed that miR-378 is among the 20 most abundant microRNAs in the heart in different species (18, 19). However, miR-378 and miR-378* functions in the heart have not been described so far. Somewhat contradictory reports have been published concerning the role of miR-378 in cardiac cell survival in vitro. In serum deprived rat H9c2 cardioblast cells exposed to hypoxia, miR-378 overexpression enhanced cell viability through the inhibition of caspase 3 expression (20) whereas in primary cultures of neonatal rat cardiomyocytes it enhanced H2O2 induced cell death by direct targeting of IGF1R (21). More recently, miR-378 was shown to block cardiac hypertrophy by targeting Grb2, an upstream component of Ras-signaling (22). To better assess the role of miR-378 in cardiac cells, we decided to identify miR-378 (miR-378-3p) and miR-378* (miR-378-5p) targets via a proteomic screen. We included miR-378* in our study because, on the contrary to most miRs, the two strands of the miR-378:miR-378* duplex have been detected in several studies. In a deep sequencing study performed in an HL-1 murine cardiac cell line, miR-378 and 378* were found to represent 1.6% and 0.1% respectively of all miRs detected in the heart, putting both of them in the top 10% of total miR expression (23). In a study performed by Vacchi-Suzzi et al. in rat adult heart, miR-378* represented in average 0.003% of total miR expression in the myocardium still placing it in the top 20% of all microRNAs expressed in the heart (19).
Here, we identified several new targets of miR-378 and 378* in an H9c2 cell line derived from rat fetal atrial cardiomyocytes. We show that these two microRNAs differentially regulate lactate dehydrogenase expression and play a role in cytoskeleton remodeling and the expression of chaperones and calcium buffering proteins in the endoplasmic reticulum (ER). All targets were highly relevant for cardiac cell differentiation and metabolism.
EXPERIMENTAL PROCEDURES
Cell Culture
We used the H9c2 cardiac cell line derive from rat fetal atria (ATCC® Number: CRL-1446 ™). Cells were cultured in Dulbecco's modified Eagles medium (LONZA, 4.5g/l glucose, with pyruvate) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. Cells were passaged every 72 h. Twenty-four hours before transfection experiments, cells were seeded at a density of 75,000 cells/ml in antibiotics free medium.
MiR Precursor Transfection
H9c2 cells were cultured as described above and transfected 24 h after plating with a range of 0.06 to 25 nm for microRNA Mimics (Ambion, Invitrogen) or 50 nm microRNA power-inhibitor (Exiqon) using lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA) transfection agent (1:200 final concentration) as suggested by the manufacturer. Cells were harvested 72 h after transfection.
Two-dimensional Gel Electrophoresis, Gel Staining, Image Acquisition
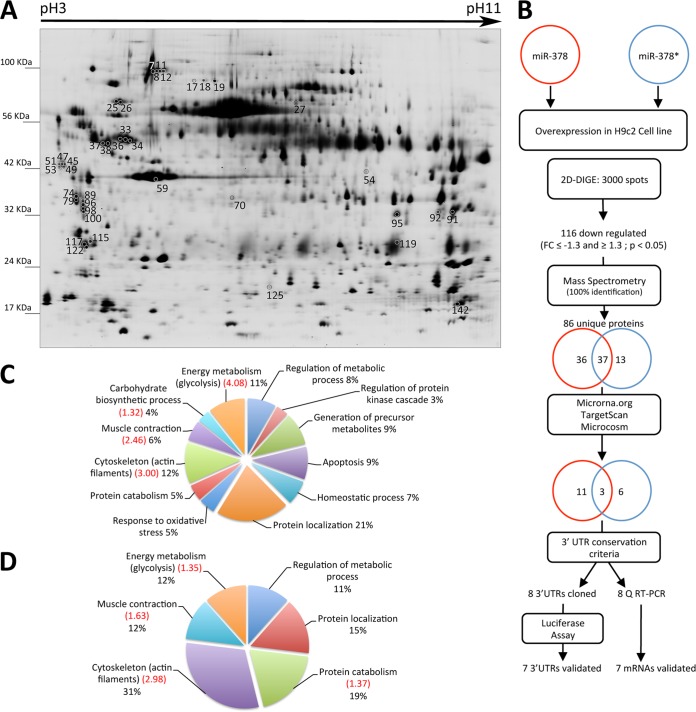
Transfected cells (6 cm Petri dishes) were washed twice with PBS and proteins were extracted in UTC lysis buffer (8 m urea, 2 m thiourea, 4% CHAPS, 50 mm dithiothreitol (DTT)). The crude extract was homogenized using a syringe and needle (21 gauge) and centrifuged at 20,000 × g for 30 min at 4 °C. The supernatant was precipitated using a two-dimensional clean-up kit (GE Healthcare) following the manufacturer's recommendations. Protein pellets were dissolved in lysis buffer without DTT. Protein concentration was determined using Bradford assay (Bio-Rad, Hercules, CA). To screen for protein amount differences and obtain quantitative protein ratios, Two-dimensional differential in gel electrophoresis (DIGE) was performed on four samples for each group, that is, four nontransfected extracts (RNAiMAX only), four transfected with miR-378, and four with miR-378* (12 nm each). Control and transfected protein extracts (50 μg) were labeled with the CyDye DIGE Fluor minimal labeling kit (GE Healthcare) following the manufacturer's recommendations. A dye swap (Cy3 and Cy5) was used to normalize for potential differences in labeling efficiencies. An internal standard was made by mixing equal amounts of proteins from each sample and labeled with Cy2. For analytical gels, 24-cm Immobiline DryStrip gels (pH 3–11NL (nonlinear)) were loaded with a total protein amount of 150 μg (50 μg of two independent groups and 50 μg of internal standard). For preparative gels, we used 400 μg of pooled control unlabeled protein extracts. Strips were rehydrated overnight in the presence of proteins in a volume of 350 μl of rehydration solution (8 m urea, 2 m thiourea, 2% CHAPS, 1.2% DeStreak Reagent, 0.5% IPG buffer pH 3–11NL). Isoelectric focusing was performed with an Ettan IPGphor2 apparatus (GE Healthcare) at 150 V for 2 h, followed by stepwise application of 200, 1000, and 8000 V for a total of 40,000 V-h. After focusing and equilibration, strips were applied to 12.5% gradient SDS-PAGE gels and sealed with 0.6% agarose, containing bromphenol blue. Electrophoresis was performed at 15 °C in an Ettan DALT-six tank with electrophoresis buffer (25 mm Tris, pH 8.3, 192 mm glycine, and 0.2% SDS) for a total of 2000 V-h. Analytical gels were imaged using an Ettan DIGE Imager (GE Healthcare) scanner. Data were analyzed with DeCyder 7.0 software (GE Healthcare) using the batch process mode. The fold change is the ratio of the mean spot value from a transfected condition (n = 4) on the control nontransfected cells (n = 4). An unpaired Student's t test was used to determine the statistical difference of the mean ratio between control and transfected groups. We chose 2 criteria for spot selection: a threshold fold change ≤-1.3 or ≥ 1.3 and a p value <0.05. Unlabeled preparative gels were fixed and stained with Coomassie Brilliant Blue G-250 for 5 days and scanned with the Ettan DIGE imager using the Cy5 channel (Ex: 635/30 nm, Em: 680/30 nm). Matching the protein spots of interest between analytical and preparative gels was performed manually.
Protein Identification by Mass Spectrometry (MS) and Database Searching
In-gel digestion was carried out with trypsin as described by Shevchenko et al. (24) with minor modifications and using for all steps a Freedom EVO 100 digester/spotter robot (Tecan, Switzerland). Spots were first destained two times with a mixture of 100 mm ammonium bicarbonate (ABC) and 50% (v/v) acetonitrile (ACN) for 45 min at 22 °C and then dried using 100% ACN for 15 min. Protein spots were then reduced with 25 mm ABC containing 10 mm DTT for 1 h at 60 °C and then alkylated with 55 mm iodoacetamide in 25 mm ABC for 30 min in the dark at 22 °C. Gels pieces were washed twice with 25 mm ABC and finally shrunk two times with 100% ACN for 15 min and dried using 100% ACN for 10 min. Bands were finally completely dehydrated after 1 h at 60 °C. Gel pieces were incubated with 13 μl of sequencing grade modified trypsin (Promega, USA; 12.5 μg/ml in 40 mm ABC with 10% ACN, pH 8.0) overnight at 40 °C. After digestion, peptides were washed with 30 μl of 25 mm ABC, shrunk with 100% ACN and extracted twice with a mixture of 50% ACN-5% formic acid (FA) and dried.
For MS and MS/MS MALDI analysis, peptides were redissolved in 4 μl of alpha-CHCA (2,5 mg/ml in 70% ACN-0.1% TFA). An aliquot of 1.5 μl of each sample was spotted directly onto a dry MALDI plate (ABSciex, Foster City, CA). Peptides on MALDI plate were then desalted with a cold solution of ammonium phosphate 10 mm–0.1% TFA. The analysis of samples was performed using a MALDI-TOF-TOF 4800 mass spectrometer (ABSciex). Spectra acquisition and processing was performed using the 4000 series explorer software (ABSciex) version 3.5.28193 in positive reflectron mode at fixed laser fluency with low mass gate and delayed extraction. External plate calibration was performed using four calibration points spotted onto the four corners of the plate using a mixture of five external standards (PepMix 1, LaserBio Labs, Sophia Antipolis, France). Peptide masses were acquired by steps of 50 spectra for the range of 900 to 4000Da. MS spectra were summed from 500 laser shots from an Nd-YAG laser operating at 355 nm and 200Hz. After filtering tryptic-, keratin- and matrix-contaminant peaks up to 15 parent ions were selected for subsequent MS/MS fragmentation according to mass range, signal intensity, signal to noise ratio, and absence of neighboring masses in the MS spectrum. MS/MS spectra were acquired in 1 kV positive mode and 1000 shots were summed by increments of 50. Database searching was carried out using Mascot version 2.2 (MatrixScience, London, UK) via GPS explorer software (ABSciex) version 3.6 combining MS and MS/MS interrogations on rattus from SwissProt data bank (7620 sequences, May 2011, www.expasy.org). A second search was performed on mus and homo sapiens SwissProt databanks that are better annotated than the rattus to provide alternative identifications for some proteins with low score. The search parameters were as follows: carbamidomethylation as a variable modification for cysteins and oxidation as a variable modification for methionines. Up to 1 missed tryptic cleavage was permitted and mass accuracy tolerance of 30 ppm for precursors and 0.3 Da for fragments were used for all tryptic mass searches. Positive identification was based on a Mascot score above the significance level (i.e. <5%). The reported proteins were those with the highest number of peptide matches.
MS and MS/MS ORBITRAP analyses were realized using an Ultimate 3000 Rapid Separation Liquid Chromatographic (RSLC) system (Thermo Fisher Scientific) online with a hybrid LTQ-Orbitrap-Velos mass spectrometer (Thermo Fisher Scientific). Briefly, peptides were loaded and washed on a C18 reverse phase precolumn (3 μm particle size, 100 Å pore size, 150 μm i.d., 1 cm length). The loading buffer contains 98% H2O, 2% ACN and 0.1% TFA. Peptides were then separated on a C18 reverse phase resin (2 μm particle size, 100 Å pore size, 75 μm i.d., 15 cm length) with a 4 min “effective gradient” from 100% A (0.1% FA and 99.9% H2O) to 50% B (80% ACN, 0.085% FA and 19.915% H2O).
The Linear Trap Quadrupole Orbitrap mass spectrometer acquired data throughout the elution process and operated in a data dependent scheme with full MS scans acquired with the Orbitrap, followed by up to 20 LTQ MS/MS CID spectra on the most abundant ions detected in the MS scan. Mass spectrometer settings were: full MS (AGC: 1*106, resolution: 6*104, m/z range 400–2000, maximum ion injection time: 500 ms); MS/MS (AGC: 5*103, maximum injection time: 50 ms, minimum signal threshold: 500, isolation width: 2Da, dynamic exclusion time setting: 15 s). The fragmentation was permitted of precursor with a charge state of 2, 3, 4, and up. For the spectral processing, the software used to generate .mgf files is Proteome discoverer 1.2. The threshold of Signal to Noise for extraction values is 3. Database searching parameters were the same as MALDI except the precursor mass tolerance that was set to 5 ppm and the fragment mass tolerance to 0.45 Da.
RNA Extraction and Quantification
Cells were scraped in RNA NOW reagent (Biogentex, League City, TX) according to the manufacturer's instructions. To analyze microRNA abundance, 400 ng of RNA were reverse transcribed using miScript Reverse Transcription Kit (Qiagen, Hilden, Germany). For mRNA abundance assessment, 1 μg of RNA was reverse transcribed using M-MuLV Reverse Transcriptase (Fermentas/Thermo Fisher Scientific, Burlington, Canada). Quantitative PCR was performed with LightCycler® 480 SYBR Green I Master (Roche, Basel, Switzerland) for mRNA and miScript SYBR Green PCR Kit (Qiagen) for microRNA analysis. HPRT and miR-16 were used as a reference for mRNA and miRNA respectively. Primer sequences are available in the supplementary data.
3′UTR Cloning into psiCheck2 Vector
The 3′UTRs of potential miRNA target genes were PCR amplified with Phusion high fidelity polymerase (Finnzymes, Espoo, Finland) during 30 amplification cycles (denaturation 94 °C, annealing and elongation temperature 60 °C, 1min 30). The obtained double stranded DNA fragments were introduced into the XhoI and NotI sites at the multiple cloning site of the psiCHECKTM-2 Vector (Promega, Madison, WI) downstream of the Renilla luciferase reporter gene. In addition, a Firefly luciferase gene under the control of an independent HSV-TK promoter is present in the vector and provides an internal normalization signal. The sequence of the 3′UTR inserts in the psiCheck vector was verified by sequencing (Beckmann Coulter Genomics, Brea, CA). Primer sequences are available in the supplementary data.
Cotransfection and Dual Luciferase Assay
Cells were cotransfected with 200 ng 3′UTR containing plasmid and 12 nm miR mimic or a negative control miR (AM17110, Ambion, Applied Biosystems). Transfection was performed according to the manufacturer's instructions using Lipofectamine 2000 reagent (Invitrogen) in sextuplicates for each experimental condition. Experiments were repeated at least twice. Forty-eight hours later, cells were harvested and luciferase activities were measured with the Luciferase Dual-Reporter Kit (Promega) as suggested by the manufacturer. Renilla/Firefly ratio represents normalized, relative 3′UTR-reporter activity.
Western Blot Analysis
One-dimensional PAGE was carried out using the NuPAGE® Novex® 4–12% Bis-Tris Gel from Invitrogen according to the manufacturer's instructions. Proteins were transferred onto nitrocellulose membranes (Hybond C, Amersham Biosciences) and the blots were saturated in TBS-T buffer (20 mm Tris, pH 7.5, 136.8 mm NaCl, and 0.1% (v/v) Tween 20) containing 5% (w/v) BSA. The following primary antibodies were used for Western blot: rabbit monoclonal anti-Lactate Dehydrogenase (EP15666) ab52488 (1:100000, Abcam, Cambridge, MA), mouse monoclonal anti-Vimentin V5255 (1:200, Sigma), rabbit polyclonal anti-Grp78 (BiP) #3183 (1:250, Cell Signaling, Danvers, MA), rabbit polyclonal anti-Cyclophilin A 07–313 (1:1000, Merck Millipore), rabbit polyclonal anti-Grp94 #2104 (1:1000, Cell Signaling), and mouse monoclonal anti-α-Tubulin T5168 (1:2000, Sigma). HRP-conjugated secondary anti-rabbit antibody BI2407 (1:1000, P.A.R.I.S Biotech) and anti-mouse IgM antibody sc-2973 (1:1000, SantaCruz, Santa Cruz, CA) were used to detect the proteins by ECL. Alternitavely, IRDye® 800CW Goat (polyclonal) Anti-Rabbit IgG 926–32211 (1:2500, Li-Cor) and IRDye® 800CW Goat (polyclonal) Anti-Mouse IgG 926–32210 (1:2500, Li-Cor) were used to detect the proteins by infrared imaging using the Li-Cor Odyssey system. Proteins were extracted in RIPA buffer (50 mm Tris, 150 mm NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0,1% SDS, 1 mm EDTA, pH 7.4, protease inhibitor mixture (Roche Applied Science)) incubated for 30 min on ice, and then centrifuged for 30 min at 20,000 × g. The supernatant was recovered and quantified using the Bradford Assay (Bio-Rad).
Phalloidin Staining and Image Quantification
Actin was stained with TRITC-labeled phalloidin and 10 images were taken for each condition on a ZEISS microscope at 10×. Quantification was done with imageJ on 16-bit images by measuring the integrated density of the whole photographed section.
LDH Activity Assay
Lactate dehydrogenase converts pyruvate to lactate while transforming one NADH to NAD+. We measure NADH extinction at 340 nm in a buffer containing 50 mm imidazole, 4 mm magnesium acetate, 50 mm KCl and 0.712 mg/ml BSA, 0.5 mm beta-NADH and 2 mm Na-Pyruvate. Two micrograms of proteins were used for each sample. We performed the measurements on six samples per condition each in duplicate.
Live and Dead Assay
Living and dead cell number was assayed using the Live and Dead Assay from Promokine (PK-CA707–30002) as suggested by the manufacturer. A calibration curve was realized by seeding incremental number of cells and counting viable cells after 96 h. For the dead cell calibration, 100% dead cells were obtained after 30 min ethanol treatment. For the experiment, cells were seeded at a density of 6000 cells/well in 96-well plates in quintuplicate. Cells were transfected 24 h after plating and total live and dead cell number was quantified 72 h after transfection (96h total) on a TECAN Infinite F500 plate reader at 515 nm (green-fluorescent Calcein AM) for live cells and 620 nm (red-fluorescent ethidium homodimer III) excited respectively at 490 and 530 nm.
Functional Annotation Clustering
We performed functional annotation clustering with the DAVID Bioinformatics Resources 6.7 (25, 26). We used Uniprot IDs to generate gene lists and selected the rat genome as background. We selected Biological processes (BP_FAT) and KEGG pathways to perform the functional annotation clustering using the default parameters suggested by the DAVID algorithm. Functional annotation clustering is a method that minimizes redundant annotations in the report by grouping similar annotations together using the Kappa statistics technique to measure the degree of the common genes between two annotations. It is followed by fuzzy heuristic clustering, to classify the groups of similar annotations according to kappa values. We used medium stringency parameters for the clustering (i.e. a value of 3 for the similarity term overlap, a minimal kappa value of 0.50 for the similarity threshold, a value of 3 for the initial and final group membership and 0.50 for the multiple linkage threshold). The group enrichment score is the geometric mean (in -log scale) of member's functional annotation p values (EASE-modified Fisher Exact test) in a corresponding annotation cluster. Annotation clusters with enrichment scores > 1.3 (i.e. p value <0.05) were considered significant.
Statistical Analysis
The data are expressed as mean ± S.E. An unpaired Student's t test was used to determine the probability value (p value). p values of p < 0.001(***), p < 0.01 (**), and p < 0.05 (*) were considered statistically significant.
RESULTS
Proteomic Changes in miR-378 and 378* H9C2 Transfected Cells
To identify differentially expressed proteins we prepared whole protein extracts from H9c2 cells transfected with miR-378, miR-378* and a control group (Fig. 1A). The 2D-DIGE experiment was analyzed with DeCyder software and revealed an average of 3000 unique protein spots per gel. Principal component analysis using all matched spots separated the control, the miR-378 transfected sample and miR-378* transfected sample (supplemental Fig. S1). For quantitative analysis we used as selection criteria a p value ≤ 0.05 and a fold change (FC) ≥ 1.3 or ≤-1.3. We identified 116 protein spots that were significantly altered by either miR-378 or miR-378* (supplemental Table S1). Strikingly, all were down-regulated with miR overexpression in agreement with the mainly repressive function of miRs. We identified 114 spots by MALDI-TOF and ORBITRAP (supplemental Table S2) corresponding to 86 unique proteins, 73 of which were down-regulated following miR-378 transfection and 50 following miR-378* transfection. There was an overlap of 37 proteins repressed in both conditions (Fig. 1B). We then performed functional annotation clustering using the DAVID Bioinformatics Resources 6.7 (25, 26). The algorithm recognized 68 proteins or ‘DAVID IDs’ out of 73 for miR-378 and 46 out of 50 for miR-378*, the majority of which were clustered. Interestingly, the significantly enriched clusters (Enrichment score ≥ 1.30) were the same in both groups. These were cytoskeleton (including several components of the actin thin filaments), muscle contraction and energy metabolism (Figs. 1C and 1D). The most significant cluster was glycolysis (Enrichment score: 4.08) in the miR-378 group and cytoskeleton (Enrichment score: 2.98) in the miR-378* group. The complete list of clusters is provided in supplemental Table S3. Among the KEGG pathways, the dilated cardiomyopathy pathway and the regulation of actin cytoskeleton were significantly enriched in the miR-378* group (Enrichment score: 3.3 and 1.75 respectively). To identify the direct targets of each miR, we searched for the Ensembl transcript IDs of the 86 repressed proteins using the BioMart portal system. We looked for potential miR-378 or miR-378* binding sites in their 3′UTR using three different miR target prediction algorithms: Microrna.org (27), TargetScan (28) and Microcosm (29). We grouped the data from these three databases and found 11 potential targets for miR-378, six for miR-378*, and three in common for the two miRs (Table I).
Fig. 1.
Two-dimensional DIGE analysis of H9c2 proteins from controls, miR-378 and miR-378* transfected cells. A, Scan of master gel used for spot comparison and selection. Whole protein preparations were differentially labeled with Cydyes and separated by isoelectric focusing (Immobiline DryStrip gels pH3–11NL) and apparent molecular weight (nominal range 15,000 to 100,000). Shown spots correspond to proteins coded by mRNA predicted as targets of miR-378 and/or miR-378* (listed under Table I). B, Workflow for identification of miR-378/378* targets and validation strategy. Numbers inside the circles represent proteins down-regulated with miR-378 (red) and miR-378* (blue) as well as those predicted as targets by different algorithms. C–D, Graphical representation of DAVID functional annotation clustering of the identified proteins down-regulated in the presence of miR-378 (C) and miR-378* (D). Enrichment scores higher than 1.3 (= -log10 0.05) are indicated in red between brackets.
Table I. List of proteins coded by mRNA predicted as targets of miR-378 and/or miR-378*.
| miR | Spot numbera | Accessionb | Protein nameb | mirSVRc | miRandac | Context+c | Consd | Functione |
|---|---|---|---|---|---|---|---|---|
| Target of miR-378 | 8, 11, 12 | ACTN4_RAT | Alpha-actinin-4 | −0.5918 | M, H | Actin crosslink formation | ||
| 17, 18, 19 | GELS_RAT | Gelsolin | 16.0005 | M | Actin filament capping | |||
| 27, 36 | DPYL3_RAT | Dihydropyrimidinase-related protein 3 | −0.04 | Actin crosslink formation | ||||
| 54 | GMPPA_RAT | Mannose-1-phosphate guanyltransferase a | −0.0115 | M, H | GDP-mannose biosynthetic process | |||
| 70 | DDAH1_RAT | Dimethylarginine dimethylaminohydrolase 1 | 0.09 | M, H | Arginine metabolic process | |||
| 91, 92 | LDHA_RAT | l-lactate dehydrogenase A chain | −0.0181 | M | Glycolysis, lactate metabolic process | |||
| 95 | CNN3_RAT | Calponin-3 | −0.1071 | M | Actomyosin structure organization | |||
| 115 | 1433G_RAT | 14-3-3 protein gamma | −0.0009 | Signal transduction | ||||
| 117 | 1433T_RAT | 14-3-3 protein theta | −0.0649 | M | Protein binding | |||
| 119 | CYBP_RAT | Calcyclin-binding protein | −0.27 | Cardiac muscle cell differentiation | ||||
| 122 | 1433Z_RAT | 14-3-3 protein zeta/delta | −0.0182 | Protein targeting to mitochondrion | ||||
| Target of miR-378* | 33, 34 | VIME_RAT | Vimentin | −0.0401 | M, H | Intermediate filament organization | ||
| 45, 47, 49, 51, 53 | CALU_RAT | Calumenin | −0.0270 | M, H | Response to organic cyclic compound | |||
| 59 | ACTS_RAT | Actin, alpha skeletal muscle | −0.0106 | M, H | Heart contraction | |||
| 74, 79 | TPM2_RAT | Tropomyosin beta chain | 16.3864 | Regulation of ATPase activity | ||||
| 89, 96, 98, 100 | TPM1_RAT | Tropomyosin alpha-1 chain | −0.0083 | Sarcomere organization | ||||
| 125 | PARK7_RAT | Protein DJ-1 | −0.2971 | Negative regulation of cell death | ||||
| Targets of both miRs | 25.26 | GRP78_RAT | 78 kDa glucose-regulated protein | −0.2755* | 18.8279* | −0.16 | M, H | ER response, response to glucose starvation |
| 37, 38 | TBB5_RAT | Tubulin beta-5 chain | −0.0908/−0.0041* | Microtubule-based movement | ||||
| 142 | PPIA_RAT | Peptidyl-prolyl cis trans isomerase A | −0.4677* | 17.3748 | M, H | Protein folding |
a Spot number refers to Fig. 1.
b Uniprot accession ID and protein name.
c MiR target scores. Microrna.org: mirSVR, microcosm: miRanda, Targetscan: context + *, score for miR-378* when mRNA is predicted as a target for both miRs.
d Conservation of the rat binding site with mouse (M) and human (H).
e Biological function indicated by GO database.
Target Validation
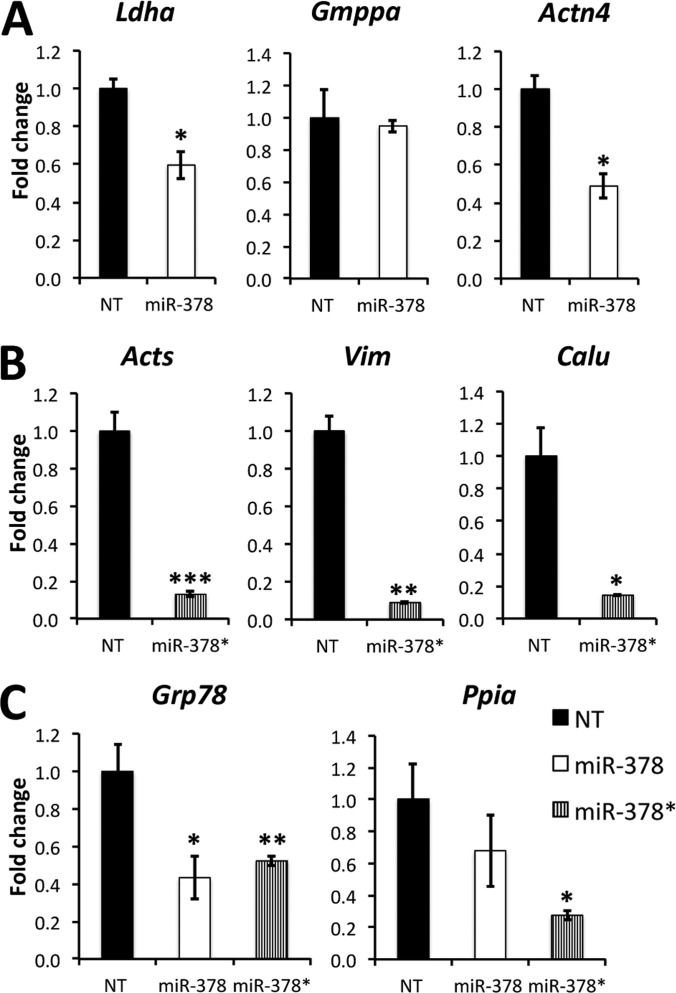
To validate the microRNA targets, we selected genes of interest based on seed sequence binding site conservation among rat, human, and mouse (supplemental Fig. S2). The list includes mannose-1-phosphate guanyltransferase α (GMPPA, FC = −1.49, p = 0.0108), α-actinin 4 (ACTN4, FC = −2.34, p = 0.00032), and Lactate Dehydrogenase A (LDHA, FC = −1.62, p = 0.002) as miR-378 targets, skeletal alpha-actin (ACTS, FC = −1.56, p = 0.0016), vimentin (VIM, FC = −1.7, p = 0.000056), and calumenin (CALU, FC = −2.33, p = 0.00031) as miR-378* targets and Glucose-regulated protein 78 (GRP78, FC = −1.67 p = 0.003 for miR-378; FC = −1.12 p = 0.17 for miR-378*) and Peptidyl-prolyl cis-trans isomerase A (PPIA, FC = −1.72, p = 0.006 for miR-378; FC = −1.23, p = 0.083 for miR-378*) as targets for the two microRNAs.
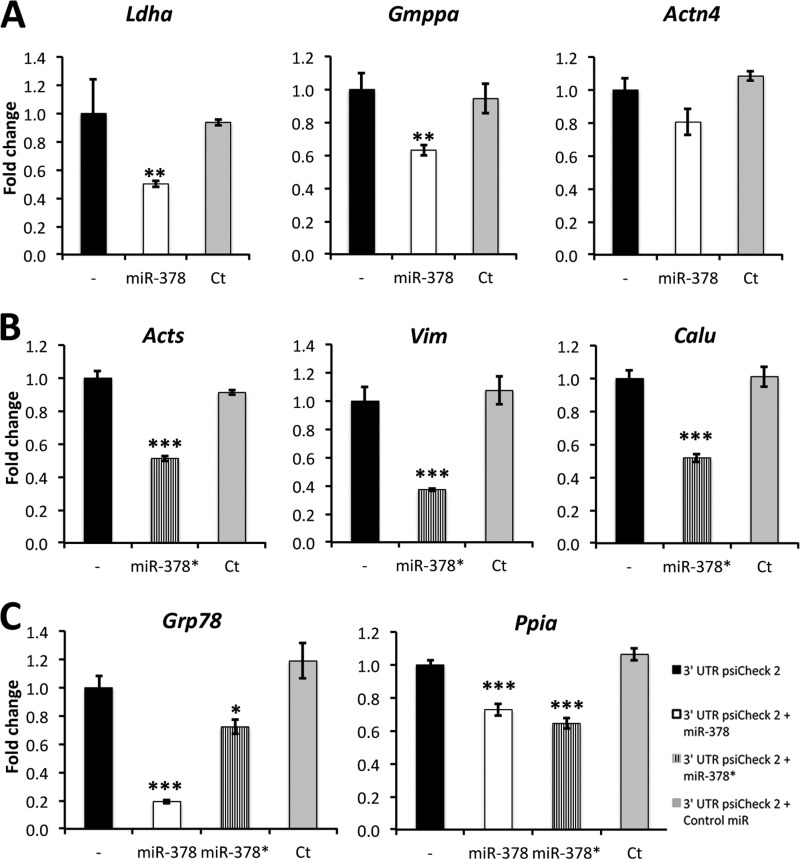
We first validated the eight chosen targets by Q RT-PCR and found that the mRNAs encoding these proteins were down-regulated in agreement with our proteomic screen, except for two miR-378 targets: Gmppa and Ppia (Fig. 2). To further assess and confirm these targets we performed Luciferase reporter assays where a portion of the 3′UTR of each gene was cloned downstream from a Renilla Luciferase reporter gene in PSICHECK-2 vectors and transfected with and without miR-378 or 378* (Fig. 3). All targets were significantly decreased except for Actn4. A negative control miR had no effect on the validated targets. Our results suggest that miR-378 and 378* intervene in different cellular functions.
Fig. 2.
Analysis of mRNA expression by Q RT-PCR. A, miR-378 targets. B, miR-378* targets. C, Targets of both miRs. Whole RNA was extracted from H9c2 cells transfected with miR-378 or miR-378*. We performed mRNA reverse transcription followed by semi-quantitative real time PCR using hprt as a reference gene. Ratios are expressed as fold change over nontransfected (NT) cells. Data are expressed as mean ± S.E. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 miR versus control. n = 3 for each group.
Fig. 3.
Validation of microRNA targets by Luciferase assay. A, miR-378 targets. B, miR-378* targets. C, Targets of both miRs. H9c2 cells were cotransfected with PsiCHECK-2 vectors containing the 3′UTR of indicated mRNAs downstream of Firefly luciferase alone (-) or with premiR-378, premiR-378* or a negative control premiR (Ct). Normalized Firefly/Renilla (internal control) luciferase values are presented as a fold change over the mean value (set to 1) obtained for the same construct without miRNA transfection. Data are expressed as mean ± S.E. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 miR versus control. The data were collected from two independent experiments, which were run with sextuplicates.
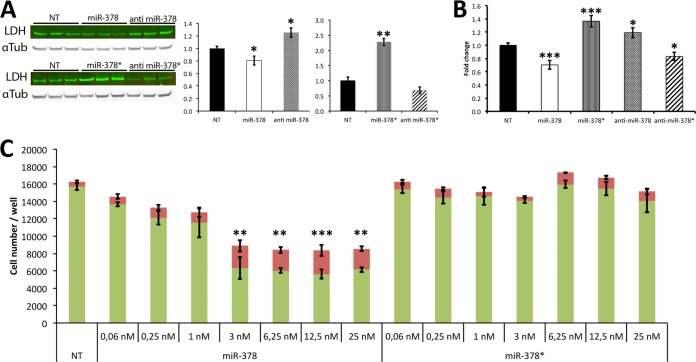
MiR-378 and 378* Differentially Regulate LDHA
MiR-378 and 378* had opposite effects on LDHA. The protein level was significantly down-regulated with miR-378 in our proteomic screen (FC = −1.62, p = 0.002) and was slightly up-regulated with miR-378* (FC = 1.27, p = 0.130). To further evaluate this differential regulation we performed Western blots. LDHA expression was 20% lower in miR-378 transfected cells than in control cells and 60% higher after miR-378* transfection (Fig. 4). When we transfected the microRNA inhibitors, we found LDHA to be 30% over expressed with anti miR-378 and 30% under expressed with anti miR-378*. We also assayed LDH activity and found it to be 30% decreased in the presence of miR-378 when compared with control and 40% increased in the presence of miR-378* (Fig. 4). Conversely, inhibiting miR-378 caused an increase in LDH activity whereas inhibiting miR-378* caused a decrease in LDH activity. Our results confirm LDHA as a direct target of miR-378. Because lactate production by LDHA is associated with higher glycolytic rates and increased cell proliferation (30), we looked at the impact of miR-378 and miR-378* on cell proliferation and survival in H9c2 cells. Using the live and dead assay, we found that miR-378 repressed cell growth and increased cell death in a dose dependent manner whereas miR-378* did not affect these parameters (Fig. 4).
Fig. 4.
LDH regulation by miR-378 and miR-378*. A, Western blots: LDH expression following transfection of miRs and antimiRs. Alpha-Tubulin is used as loading control. Graphs on the right: quantification of band intensities. Normalized LDH/tubulin values are presented as fold change over nontransfected cells (NT), B, Total LDH activity following transfection of miRs and antimiRs. Data are expressed as fold change over nontransfected cells (NT). C, Live/Dead assay measuring the total number of live (green) and dead (red) cells per well for each condition. Cells were seeded at 6000 cells/well in 96-well plates in quintuplicate and analyzed 72 h after transfection. Data are expressed as mean ± S.E. **p ≤ 0.01, ***p ≤ 0.001 miR versus control for both live and dead values.
MiR-378* Regulates Cytoskeleton
Among validated miR-378* targets, we found skeletal alpha-actin and vimentin, a type III intermediate filament. To further assess the effects of miR-378* on these cytoskeleton proteins, we performed Western blots and immunostaining that confirmed our results. Vimentin expression was repressed by 50% in the presence of miR-378* (Fig. 5A). F-actin staining with fluorescent phalloidin revealed a significant decrease in signal in the presence of miR-378* when compared with controls or a negative control miR. The use of an anti miR-378* on the other hand, caused an increase in the level of F-actin (Fig. 5B). Our results clearly show an important role of miR-378* in repressing actin and vimentin expression.
Fig. 5.
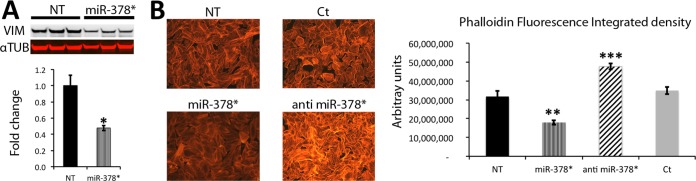
Cytoskeleton modulation by miR-378*. A, Western blot showing repressed levels of vimentin in the presence of miR-378*. Alpha-Tubulin is used as loading control. B, F-actin staining with TRITC-labeled phalloidin in H9c2 cells transfected with: miR-378*, anti miR-378*, miR negative control (Ct) and nontransfected (NT). Graph on the right: Image J quantification of Phalloidin signal intensity measured from the whole field. The total signal is decreased in the presence of miR-378* and increased in the presence of anti miR-378*. Data are expressed as mean ± S.E. **p ≤ 0.01, ***p ≤ 0.001 miR versus control. n = 6 for each group.
MiR-378 and miR-378* Regulate Proteins Linked to Endoplasmic Reticulum Functions
Our proteomic screen also revealed calumenin as a miR-378 target and GRP78 and PPIA as targets of the two miRs. Calumenin is found in the lumen of the endoplasmic and sarcoplasmic reticulum, it has Ca2+ binding motifs and can bind to SERCAs (Sarco/Endoplasmic Reticulum Ca2+-ATPase) (31). GRP78 also known as HSPA5 or BiP, is a multifunctional chaperone located in the ER where it acts as a negative regulator of signaling mediators involved in the unfolded protein response (UPR) but also regulates the flow of ER Ca2+ to preserve cytoplasmic calcium homeostasis (32). PPIA, also known as cyclophilin A, a target of the immunosuppressant drug cyclosporin A, is involved in the maturation and folding of native proteins (33). To further explore the impact of the two miRs on these proteins, we performed Western blots to validate our previous results. GRP78 was significantly down-regulated especially in the presence of miR-378, validating our proteomics screen findings as well as our Q-PCR and luciferase validations (Fig. 6). PPIA was significantly down-regulated in the presence of miR-378* but not with miR-378 also validating our Q-PCR and luciferase results where we found miR-378* to be a more efficient inhibitor of PPIA expression. Although not a direct target of either miRs, GRP94, also known as endoplasmin (ENPL) is an important player of the UPR response along with GRP78 (34). Interestingly, GRP94 expression was also down-regulated in the presence of either microRNAs (supplemental Table S1). Western blot analysis confirmed this result (Fig. 6).
Fig. 6.
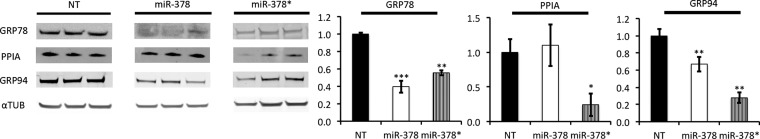
Repression of chaperone proteins by miR-378 and miR-378*. Western blot analysis showed that Grp78 was repressed in the presence of miR-378 and to a lesser extent in the presence of miR-378*. PPIA was not repressed by miR-378 but only by miR-378*. Grp94 expression pattern was also repressed by the two miRs. Data are expressed as mean ± S.E. **p ≤ 0.01, ***p ≤ 0.001 miR versus control. n = 3 for each group.
DISCUSSION
In this study we sought to improve our understanding of the repertoire of genes regulated by miR-378 and miR-378* in cardiac cells. Identification of miRNA targets is central to understanding the biological roles of miRNAs. We overexpressed miR-378 and miR-378* in H9c2 cells, a rat embryonic cardiac myoblast cell line to investigate proteome modulation by these microRNAs.
We performed DIGE experiments in combination with mass spectrometry identification, bioinformatics predictions and functional assays to identify direct targets for miR-378 and miR-378*.
With our proteomic approach, we identified a total of 73 proteins down-regulated with miR-378 overexpression and 50 down-regulated with miR-378* overexpression. Thirty-seven proteins were down-regulated when either microRNA was transfected. This result shows that these two miRs arising from the same hairpin converge in part on similar pathways despite different seed sequences. This was further exemplified by the gene ontology clustering analysis showing several identical enriched biological functions between the two sets of repressed proteins. MiR-378 and miR-378* modulate a common subset of biological pathways in cardiac cells: glycolysis, cytoskeleton, and ER Ca2+ buffering and chaperone proteins (Fig. 7). However, identification of a set of proteins repressed by a miRNA is susceptible to detection of indirect targets that are repressed through secondary effects, and experimental validation of the targets is required. We cross referenced our target list to online databases (microcosm, microrna.org, targetscan) and found 11 potential targets for miR-378, six for miR-378* and three potentially targeted by the two miRs. We validated and confirmed eight mRNA targets by Q-PCR and luciferase assay.
Fig. 7.
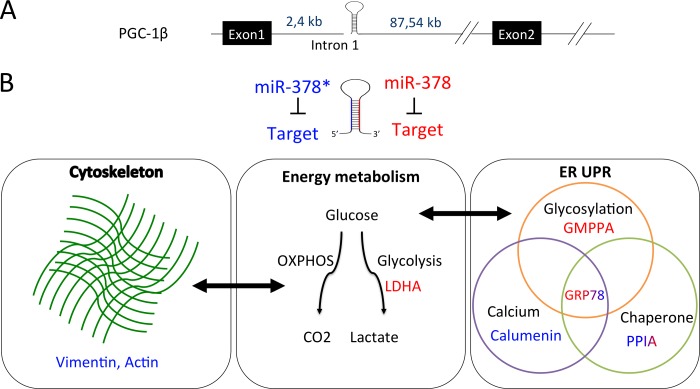
Biological functions regulated by miR-378 and miR-378*. A, MiR-378* and miR-378 are produced from a mirtron located in the first intron of the PGC-1β gene. B, MiR-378* targets are shown in blue and miR-378 targets in red. Targets of both miRs are shown in a shade of blue and red. Mir-378* represses cytoskeleton proteins that are involved in a cross-talk with OXPHOS energetic pathway. MiR-378 represses LDHA. This enzyme, involved in the last step of glycolysis, is required for cell proliferation. Both miRs repress ER proteins involved in glycosylation, Ca2+ buffering and chaperone activity. These pathways play a major role in the unfolded protein response (UPR). GRP78 is more repressed by miR-378 than by miR-378* and vice versa for PPIA. Glucose deprivation is known to induce GRP78 and alter glycosylation.
Role of the miR-378/miR-378* Hairpin in the Balance Between Oxidative Phosphorylation and Glycolysis
We found that miR-378 inhibits LDHA expression whereas miR-378* indirectly activates its expression in H9c2 cells. The latter is in agreement with a study by Eichner et al. where overexpression of miR-378* in breast cancer cells inhibited transcriptional regulators ERRγ and GABPA reducing oxidative phosphorylation (OXPHOS), which indirectly caused an increase in the rate of glycolysis and LDHA activity. This metabolic shift, known as the Warburg effect, is characteristic of cancer cells (14). Our results, showing an antagonistic role for miR-378 and miR-378* on LDHA expression in H9c2 cells, further confirm an important role of the miR-378 hairpin in the balance between glycolysis and lactate production and OXPHOS. In the heart, the expression of the miR-378/378* hairpin increases progressively after birth and miR-378 is more abundant than miR-378* (21). Cardiac cells have a high oxidative capacity and fulfill their energetic requirements mainly via the OXPHOS pathway. PGC-1β is a prominent activator of this pathway via co-activation of the transcription factors NRF-1 and 2 that regulate genes required for mitochondrial oxidative phosphorylation (35). Our results suggest that miR-378-mediated inhibition of LDHA could complement PGC-1β function. Indeed, miR-378 inhibition of LDHA will limit the conversion of pyruvate into lactate, thus favoring pyruvate use in oxidative phosphorylation to produce higher quantities of ATP per mole of glucose. Glycolysis produces much less ATP but is thought to facilitate the incorporation of carbon into biomass by allowing some glucose molecules to be diverted into the pentose phosphate pathway and macromolecular precursors required for lipids, amino acids and nucleotides biosynthesis (36). LDHA inhibition by siRNA has been reported to block cell proliferation (30). The LDH activity that eliminates excess carbon may facilitate high glycolytic rate and the diverse use of glucose required for proliferation. Our findings showing that miR-378 overexpression represses LDHA expression and blocks cell proliferation is in agreement with this hypothesis.
Mir-378* Impact on Vimentin and Actin Cytoskeleton and Their Link With Mitochondrial Activity
It has been shown that a deficiency in OXPHOS causes alterations in protein expression in the distribution of cytoskeleton components and mitochondria (37). Mitochondria support in the cell is under the control of cytoskeletal proteins, including microtubules, actin microfilaments and intermediate filaments such as vimentin and desmin (37–39). We have previously shown that cytoskeleton disorganization correlates with energy metabolism defects during heart failure (40). Tracking mitochondrial movement in cells in which vimentin has been disrupted showed that their motility is highly increased whereas at the basal state the majority of mitochondria are anchored at sites in the cytoplasm to supply local ATP (41). Conversely, inhibition of mitochondria complex IV causes a collapse of the vimentin network around the nucleus (37). Actin polymerization is also important for mitochondrial fission (42). Hence, direct targeting of vimentin and actin by miR-378* could contribute to its known impact on OXPHOS metabolism (14).
Regulation of Chaperones and Endoplasmic Reticulum Proteins by miR-378 and miR-378*
We found several proteins related to Ca2+ buffering (calumenin, GRP78) and chaperone activity (PPIA, GRP78) to be repressed by the two miRs. Low glucose levels in the cell induces synthesis of GRP78 (43) a chaperone protein, whose increase is a marker of ER stress (32). GRP78 also preserves cytoplasmic calcium homeostasis by regulating the Ca2+ flow of the ER (32). This Ca2+ flow plays an important role in the regulation of mitochondria activity. Drugs altering the homeostasis of calcium in ER such as thapsigargin, an inhibitor of Serca2 pumps, are strong inducers of ER stress. Cyclophilin A inhibition is associated with the UPR and an induction of ER stress associated proteins Grp78 and CHOP (44). GMPPA is the alpha subunit of the GMPP enzyme that catalyzes the reaction converting mannose-1-phosphate or glucose-1-phosphate and GTP to GDP-mannose or GDP-glucose, respectively (45). GDP-mannose plays a key role in the biosynthetic pathway that produces the N-linked oligosaccharides of many membrane and secretory glycoproteins in the ER of eukaryotic cells. Interference with the glycosylation process triggers the UPR (46).
Altogether, these data and our results suggest a tight entanglement of these pathways through their regulation by miR-378 and miR-378*. The first strongly repressed GRP78 whereas the second is a strong inhibitor of PPIA expression. Moreover, GRP78 is involved in mitogenesis and cellular proliferation (32), which suggests that its repression by miR-378 along with LDHA inhibition blocks cellular proliferation.
In the present study, we report the novel observation that miR-378 and miR-378* regulate different but related pathways in cardiomyocytes. An elegant study recently identified the mRNAs that are differentially regulated following miR-378:miR-378* hairpin overexpression in the mouse heart (47). In agreement with our results, at least three putative targets in their data set match our list of direct miR-378 or miR-378* targets including alpha skeletal actin mRNA that is strongly repressed in Matkovich's data set (FC = −8.24), Lactate dehydrogenase A and Grp78. As highlighted in this study and others (18, 19, 23, 47), both miR-378 and miR-378* are co-expressed in cardiac cells. Interestingly, the ratio of each strand of the hairpin may vary according to the stage of cardiomyocyte differentiation. For instance, we found that the Q-PCR miR-378/378* ratio is 2.3 under baseline conditions in H9c2 cells but increases to 10.17 in neonatal rat cardiomyocytes (data not shown). The latter are much less proliferative than H9c2 cells and develop a higher oxidative metabolism in agreement with an increased impact of miR-378. Our study suggests that the balance in the ratio between the two strands of the hairpin may have an impact on the metabolism and differentiation state of cardiac cells.
In conclusion, our results show that miR-378 and miR-378* establish a connection among energy metabolism, cytoskeleton remodeling, and ER function through post-transcriptional regulation of key proteins involved in theses pathways. It is important to note that, because the two-dimensional DIGE approach mainly detects quantitative differences among abundant cellular proteins, our observations provide proof of concept rather than a thorough accounting of the proteomic changes under the control of these miRs. Further investigation and the use of high throughput deep proteomics techniques will be required to fully understand the role of these microRNAs in the regulation of cardiac functions.
Supplementary Material
Acknowledgments
We thank Prof. Denise Paulin for valued scientific training. We thank Renée Ventura-Clapier, Anne Garnier and colleagues from INSERM U-769 for valuable scientific advice and kind gift of antibodies. Protein identifications were made at the 3P5 proteomic facility, Université Paris Descartes, Institut Cochin, Paris. We thank Cedric Broussard for expert analysis of mass spectrometry data.
Footnotes
* This work was supported by the French Agence Nationale pour la Recherche (M.M., Z.L., ANR-08-GENOPATH-038) and the Association Française contre les Myopathies (AFM). Y.M. and E.T. were supported by PhD fellowships from the Lebanese National Council for Scientific Research and the French Ministry of Research, respectively.
 This article contains supplemental Figs. S1 and S2 and Tables S1 to S3.
This article contains supplemental Figs. S1 and S2 and Tables S1 to S3.
1 The abbreviations used are:
- miR
- microRNA
- ER
- endoplasmic reticulum
- OXPHOS
- oxidative phosphorylation.
REFERENCES
- 1. Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. 9, 102–114 [DOI] [PubMed] [Google Scholar]
- 2. Lau P. W., MacRae I. J. (2009) The molecular machines that mediate microRNA maturation. J. Cell. Mol. Med. 13, 54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miyoshi K., Miyoshi T., Siomi H. (2010) Many ways to generate microRNA-like small RNAs: non-canonical pathways for microRNA production. Mol. Genet. Genomics 284, 95–103 [DOI] [PubMed] [Google Scholar]
- 4. Berezikov E., Chung W. J., Willis J., Cuppen E., Lai E. C. (2007) Mammalian mirtron genes. Mol. Cell 28, 328–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Small E. M., Frost R. J., Olson E. N. (2010) MicroRNAs add a new dimension to cardiovascular disease. Circulation 121, 1022–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lin J., Puigserver P., Donovan J., Tarr P., Spiegelman B. M. (2002) Peroxisome proliferator-activated receptor gamma coactivator 1beta (PGC-1beta), a novel PGC-1-related transcription coactivator associated with host cell factor. J. Biol. Chem. 277, 1645–1648 [DOI] [PubMed] [Google Scholar]
- 7. Kamei Y., Ohizumi H., Fujitani Y., Nemoto T., Tanaka T., Takahashi N., Kawada T., Miyoshi M., Ezaki O., Kakizuka A. (2003) PPARgamma coactivator 1beta/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc. Natl. Acad. Sci. U.S.A. 100, 12378–12383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lai L., Leone T. C., Zechner C., Schaeffer P. J., Kelly S. M., Flanagan D. P., Medeiros D. M., Kovacs A., Kelly D. P. (2008) Transcriptional coactivators PGC-1alpha and PGC-lbeta control overlapping programs required for perinatal maturation of the heart. Genes Dev. 22, 1948–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Riehle C., Wende A. R., Zaha V. G., Pires K. M., Wayment B., Olsen C., Bugger H., Buchanan J., Wang X., Moreira A. B., Doenst T., Medina-Gomez G., Litwin S. E., Lelliott C. J., Vidal-Puig A., Abel E. D. (2011) PGC-1beta deficiency accelerates the transition to heart failure in pressure overload hypertrophy. Circ. Res. 109, 783–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Redova M., Poprach A., Nekvindova J., Iliev R., Radova L., Lakomy R., Svoboda M., Vyzula R., Slaby O. (2012) Circulating miR-378 and miR-451 in serum are potential biomarkers for renal cell carcinoma. J. Translat. Med. 10, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xie P., Xu F., Cheng W., Gao J., Zhang Z., Ge J., Wei Z., Xu X., Liu Y. (2012) Infiltration related miRNAs in bladder urothelial carcinoma. J. Huazhong Univ. Sci. Technol. 32, 576–580 [DOI] [PubMed] [Google Scholar]
- 12. Liu H., Zhu L., Liu B., Yang L., Meng X., Zhang W., Ma Y., Xiao H. (2012) Genome-wide microRNA profiles identify miR-378 as a serum biomarker for early detection of gastric cancer. Cancer Lett. 316, 196–203 [DOI] [PubMed] [Google Scholar]
- 13. Lee D. Y., Deng Z., Wang C. H., Yang B. B. (2007) MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc. Natl. Acad. Sci. U.S.A. 104, 20350–20355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eichner L. J., Perry M. C., Dufour C. R., Bertos N., Park M., St-Pierre J., Giguère V. (2010) miR-378(*) mediates metabolic shift in breast cancer cells via the PGC-1beta/ERRgamma transcriptional pathway. Cell Metab. 12, 352–361 [DOI] [PubMed] [Google Scholar]
- 15. Gerin I., Bommer G. T., McCoin C. S., Sousa K. M., Krishnan V., MacDougald O. A. (2010) Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am. J. Physiol. Endocrinol. Metab. 299, E198–E206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sacco J., Adeli K. (2012) MicroRNAs: emerging roles in lipid and lipoprotein metabolism. Current Opinion Lipidol. 23, 220–225 [DOI] [PubMed] [Google Scholar]
- 17. Carrer M., Liu N., Grueter C. E., Williams A. H., Frisard M. I., Hulver M. W., Bassel-Duby R., Olson E. N. (2012) Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378*. Proc. Natl. Acad. Sci. U.S.A. 109, 15330–15335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rao P. K., Toyama Y., Chiang H. R., Gupta S., Bauer M., Medvid R., Reinhardt F., Liao R., Krieger M., Jaenisch R., Lodish H. F., Blelloch R. (2009) Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ. Res. 105, 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vacchi-Suzzi C., Hahne F., Scheubel P., Marcellin M., Dubost V., Westphal M., Boeglen C., Büchmann-Møller S., Cheung M. S., Cordier A., De Benedetto C., Deurinck M., Frei M., Moulin P., Oakeley E., Grenet O., Grevot A., Stull R., Theil D., Moggs J. G., Marrer E., Couttet P. (2013) Heart structure-specific transcriptomic atlas reveals conserved microRNA-mRNA interactions. PloS One 8, e52442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fang J., Song X. W., Tian J., Chen H. Y., Li D. F., Wang J. F., Ren A. J., Yuan W. J., Lin L. (2012) Overexpression of microRNA-378 attenuates ischemia-induced apoptosis by inhibiting caspase-3 expression in cardiac myocytes. Apoptosis 17, 410–423 [DOI] [PubMed] [Google Scholar]
- 21. Knezevic I., Patel A., Sundaresan N. R., Gupta M. P., Solaro R. J., Nagalingam R. S., Gupta M. (2012) A novel cardiomyocyte-enriched microRNA, miR-378, targets insulin-like growth factor 1 receptor: implications in postnatal cardiac remodeling and cell survival. J. Biol. Chem. 287, 12913–12926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nagalingam R. S., Sundaresan N. R., Gupta M. P., Geenen D., Solaro R. J., Gupta M. (2013) A cardiac enriched microRNA, miR-378 blocks cardiac hypertrophy by targeting Ras-signaling. J. Biol. Chem. 288, 11216–11232 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23. Humphreys D. T., Hynes C. J., Patel H. R., Wei G. H., Cannon L., Fatkin D., Suter C. M., Clancy J. L., Preiss T. (2012) Complexity of murine cardiomyocyte miRNA biogenesis, sequence variant expression and function. PloS One 7, e30933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shevchenko A., Loboda A., Ens W., Schraven B., Standing K. G., Shevchenko A. (2001) Archived polyacrylamide gels as a resource for proteome characterization by mass spectrometry. Electrophoresis 22, 1194–1203 [DOI] [PubMed] [Google Scholar]
- 25. Huang da W., Sherman B. T., Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang D. W., Sherman B. T., Lempicki R. A. (2008) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 27. Betel D., Wilson M., Gabow A., Marks D. S., Sander C. (2008) The microRNA.org resource: targets and expression. Nucleic Acids Res. 36, D149–D153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grimson A., Farh K. K., Johnston W. K., Garrett-Engele P., Lim L. P., Bartel D. P. (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Griffiths-Jones S., Grocock R. J., van Dongen S., Bateman A., Enright A. J. (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34, D140–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fantin V. R., St-Pierre J., Leder P. (2006) Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell 9, 425–434 [DOI] [PubMed] [Google Scholar]
- 31. Sahoo S. K., Kim T., Kang G. B., Lee J. G., Eom S. H., Kim do H. (2009). Characterization of calumenin-SERCA2 interaction in mouse cardiac sarcoplasmic reticulum. J. Biol. Chem. 284, 31109–31121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Avila M. F., Cabezas R., Torrente D., Gonzalez J., Morales L., Alvarez L., Capani F., Barreto G. E. (2013) Novel interactions of GRP78: UPR and estrogen responses in the brain. Cell Biol. Int. 37, 521–532 [DOI] [PubMed] [Google Scholar]
- 33. Wang P., Heitman J. (2005) The cyclophilins. Genome Biol. 6, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marzec M., Eletto D., Argon Y. (2012) GRP94: An HSP90-like protein specialized for protein folding and quality control in the endoplasmic reticulum. Biochim. Biophys. Acta 1823, 774–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Riehle C., Abel E. D. (2012) PGC-1 proteins and heart failure. Trends Cardiovasc. Med. 22, 98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vander Heiden M. G., Cantley L. C., Thompson C. B. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Annunen-Rasila J., Ohlmeier S., Tuokko H., Veijola J., Majamaa K. (2007) Proteome and cytoskeleton responses in osteosarcoma cells with reduced OXPHOS activity. Proteomics 7, 2189–2200 [DOI] [PubMed] [Google Scholar]
- 38. Tang H. L., Lung H. L., Wu K. C., Le A. H., Tang H. M., Fung M. C. (2008) Vimentin supports mitochondrial morphology and organization. Biochem. J. 410, 141–146 [DOI] [PubMed] [Google Scholar]
- 39. Guzun R., Gonzalez-Granillo M., Karu-Varikmaa M., Grichine A., Usson Y., Kaambre T., Guerrero-Roesch K., Kuznetsov A., Schlattner U., Saks V. (2012) Regulation of respiration in muscle cells in vivo by VDAC through interaction with the cytoskeleton and MtCK within Mitochondrial Interactosome. Biochim. Biophys. Acta 1818, 1545–1554 [DOI] [PubMed] [Google Scholar]
- 40. Diguet N., Mallat Y., Ladouce R., Clodic G., Prola A., Tritsch E., Blanc J., Larcher J. C., Delcayre C., Samuel J. L., Friguet B., Bolbach G., Li Z., Mericskay M. (2011) Muscle creatine kinase deficiency triggers both actin depolymerization and desmin disorganization by advanced glycation end products in dilated cardiomyopathy. J. Biol. Chem. 286, 35007–35019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nekrasova O. E., Mendez M. G., Chernoivanenko I. S., Tyurin-Kuzmin P. A., Kuczmarski E. R., Gelfand V. I., Goldman R. D., Minin A. A. (2011) Vimentin intermediate filaments modulate the motility of mitochondria. Mol. Biol. Cell 22, 2282–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Korobova F., Ramabhadran V., Higgs H. N. (2013) An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 339, 464–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Munro S., Pelham H. R. (1986) An Hsp70-like protein in the ER: identity with the 78 kd glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell 46, 291–300 [DOI] [PubMed] [Google Scholar]
- 44. Pallet N., Bouvier N., Bendjallabah A., Rabant M., Flinois J. P., Hertig A., Legendre C., Beaune P., Thervet E., Anglicheau D. (2008) Cyclosporine-induced endoplasmic reticulum stress triggers tubular phenotypic changes and death. Am. J. Transplantation 8, 2283–2296 [DOI] [PubMed] [Google Scholar]
- 45. Ning B., Elbein A. D. (2000) Cloning, expression and characterization of the pig liver GDP-mannose pyrophosphorylase. Evidence that GDP-mannose and GDP-Glc pyrophosphorylases are different proteins. Eur. J. Biochem. 267, 6866–6874 [DOI] [PubMed] [Google Scholar]
- 46. Chakrabarti A., Chen A. W., Varner J. D. (2011) A review of the mammalian unfolded protein response. Biotechnol. Bioengineer. 108, 2777–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Matkovich S. J., Hu Y., Dorn G. W., 2nd (2013) Regulation of cardiac microRNAs by cardiac microRNAs. Circ. Res. 113, 62–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.