Abstract
During seed germination, the transition from a quiescent metabolic state in a dry mature seed to a proliferative metabolic state in a vigorous seedling is crucial for plant propagation as well as for optimizing crop yield. This work provides a detailed description of the dynamics of protein synthesis during the time course of germination, demonstrating that mRNA translation is both sequential and selective during this process. The complete inhibition of the germination process in the presence of the translation inhibitor cycloheximide established that mRNA translation is critical for Arabidopsis seed germination. However, the dynamics of protein turnover and the selectivity of protein synthesis (mRNA translation) during Arabidopsis seed germination have not been addressed yet. Based on our detailed knowledge of the Arabidopsis seed proteome, we have deepened our understanding of seed mRNA translation during germination by combining two-dimensional gel-based proteomics with dynamic radiolabeled proteomics using a radiolabeled amino acid precursor, namely [35S]-methionine, in order to highlight de novo protein synthesis, stability, and turnover. Our data confirm that during early imbibition, the Arabidopsis translatome keeps reflecting an embryonic maturation program until a certain developmental checkpoint. Furthermore, by dividing the seed germination time lapse into discrete time windows, we highlight precise and specific patterns of protein synthesis. These data refine and deepen our knowledge of the three classical phases of seed germination based on seed water uptake during imbibition and reveal that selective mRNA translation is a key feature of seed germination. Beyond the quantitative control of translational activity, both the selectivity of mRNA translation and protein turnover appear as specific regulatory systems, critical for timing the molecular events leading to successful germination and seedling establishment.
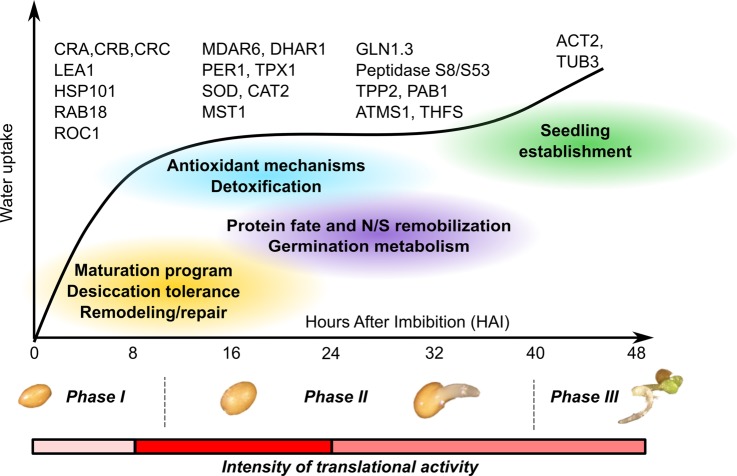
Seed germination is a vital stage in the plant life cycle during which embryo cells experience a programmed transition from a quiescent to a highly active metabolic state. Accordingly, seed quality in terms of germination vigor is of paramount importance for both ecological aspects and practical applications, as it influences the level, timing, and uniformity of seedling emergence in a wide range of environmental conditions (1, 2). In efforts to decipher the complex molecular mechanisms that control seed germination, genetic, genomic, and postgenomic analyses have been developed on the model plant Arabidopsis thaliana (3–6). Germination is classically described as a sequential time course divided into three major phases of seed water uptake (7). Phase I is characterized by rapid seed imbibition, which is crucial for the transition from the quiescent metabolic state of the dry seed to the high metabolic activity of the hydrated seed. Phase II corresponds to a period during which the seed imbibition level remains constant or slowly increases. Although this step occurs without visible morphological changes of the seed, it is characterized by germination-specific metabolism that prepares the seed for radicle protrusion and seedling growth. Phase II is achieved when the radicle (or a part that extends from the growing embryo) emerges through the seed coat thanks to cell elongation. Both Phase I and Phase II define germination sensu stricto, a developmental phase that occurs without cell division (8). Finally, Phase III corresponds to the resumption of water uptake when embryonic cells start to divide for seedling establishment. During the past decade, segregation-based genetic mapping approaches have enabled the characterization of several quantitative trait loci related to Arabidopsis seed germination quality (9–13). Also, numerous Arabidopsis mutant lines have been deeply exploited for the functional analysis of genes involved in seed germination (3–6). In addition, high-throughput functional genomic methods, such as transcriptomics and proteomics, uncovered key gene products involved in the germination process (3–6). Finally, thanks to a large set of transcriptomic data, a network model (SeedNet) describing global transcriptional regulations and interactions occurring in Arabidopsis seed germination has been created (14). This network model has proved helpful in efforts to understand how the transition from a quiescent to a germinated seed operates at the transcriptional level, and it also allowed the identification of positive and negative regulators of Arabidopsis seed germination.
A high proportion of the genome is already expressed in the mature dry Arabidopsis seed; over 10,000 transcripts have been detected (15–19). Proteomic studies disclosed that only protein translation is required for radicle protrusion, indicating that germination-specific proteins are translated from stored mRNAs (20, 21). Thus, these long-lived stored transcripts in mature dry seeds were proposed to play a critical role in seed germination, emphasizing the significance of post-transcriptional and translational controls (16, 20, 22, 23). Furthermore, proteomic approaches underlined the seed proteome's diversity as a result of numerous post-translational protein modifications (24). To date, differential transcriptomic and proteomic experiments have provided an overview of the accumulation patterns of mRNAs and proteins during the germination process, but without distinction between their stored and de novo synthesized counterparts. The present study highlights the dynamic regulation of protein translation and turnover during the time course of seed germination. To this end, we employed a combined approach involving two-dimensional gel-based differential proteomics and dynamic radiolabeled proteomics. The integration of these proteomic results with publicly available transcriptomic data (18) provides an original perspective on the contribution of dynamic protein turnover and selective mRNA translation in seed germination.
EXPERIMENTAL PROCEDURES
Plant Material and Germination Experiments
Nondormant seeds of Arabidopsis thaliana, accession Landsberg erecta (Ler), were used in all experiments. Germination assays were carried out at 25 °C, and seeds imbibed for 48 h under continuous light on three sheets of absorbent paper (Roundfilter paper circles, 45-mm diameter, Schleicher & Schuell, Dassel, Germany) and a black membrane filter with a white grid (ME 25/31, 45-mm diameter, Schleicher & Schuell) wetted with 1.3 ml of distilled water in covered plastic boxes (50-mm diameter).
Radiolabeling of Neosynthesized Proteins
To label neosynthesized proteins in vivo, six seed lots (50 mg; approximately 2,800 seeds; three biological replicates) were allowed to imbibe for, respectively, 8 h, 16 h, 24 h, 32 h, 40 h, and 48 h in the presence of [35S]-Met (1.85 MBq; MP Biomedicals SARL, Illkirch, France) for a period of 8 hours after imbibition (HAI)1 (i.e. from 0 to 8 h, 8 to 16 h, 16 to 24 h, 24 to 32 h, 32 to 40 h, and 40 to 48 h, respectively). Radioactive Met ([35S]-Met) has been used successfully to label neosynthesized proteins in germinating Arabidopsis seeds (20, 25, 26) and in other species (e.g. 27–29). Furthermore, there is ample circumstantial evidence that exogenously applied Met can easily penetrate into plant cells; indeed, altered plant phenotypes caused by the inhibition of Met biosynthesis and/or accumulation can be alleviated upon Met supplementation (30–34). Following incubation, total protein extracts were prepared as previously described (35). In order to prevent unspecific protein labeling by free [35S]-Met, seed protein extracts were precipitated by trichloroacetic acid/acetone before two-dimensional electrophoresis separation (35). The amount of protein synthesis was measured from trichloroacetic acid precipitation of aliquots of reaction mixtures spotted on Whatmann GF/C filters; after eight washing steps in cold 5% trichloroacetic acid and 0.04 m sodium pyrophosphate and two washing steps in absolute methanol, filters were dried and counted for radioactivity in a liquid scintillation counter (36). Protein extracts were also submitted to two-dimensional gel electrophoresis as described elsewhere (35).
Detection of Total Seed Proteins
Proteins on the two-dimensional gels were stained by silver nitrate (35). Then, stained two-dimensional gels were dried for 2 days at room temperature in a sandwich composed of, from bottom to top, one sheet of cellophane model Gel Dryer (Bio-Rad), two-dimensional gel, one sheet of Saran Wrap (VWR International SAS, Fontenay-sous-Bois, France), and one sheet of cellophane model Gel Dryer (Bio-Rad). After drying, silver-stained gels were scanned with a Sharp JX-330 scanner (Sharp Electronics, Roissy, France). Image analysis was carried out with ImageMaster 2D Elite version 4.01 software (Amersham Biosciences, Les Ulis, France).
Detection of Neosynthesized Proteins
The upper sheet of cellophane and the Saran Wrap sheet were peeled and exposed for 15 days to storage phosphor screens (GE Healthcare). These screens were submitted for phosphorimager analysis (Molecular Dynamics Storm 840 phosphorimager, Amersham Biosciences). Then, neosynthesized proteins revealed by autoradiography were matched with proteins detected on silver-stained two-dimensional gels according to both the position (molecular weight and pI) and the protein spot shape (supplemental Fig. S1). Proteins of interest were identified through comparison with the reference protein maps for the Arabidopsis seed proteome available online http://www.seed-proteome.com/. The amount of de novo synthesized proteins was calculated based on densitometric analyses of the spots on the autoradiography using ImageMaster 2D Elite version 4.01 software (Amersham Biosciences).
Protein Neosynthesis Clusters and Normalization
For protein neosynthesis clustering, the protein neosynthesis intensity of the different protein isoforms was first summed. Then, the protein neosynthesis amount of the resulting 158 nonredundant proteins was first normalized by their maximum value across the different time lapses. Finally, these protein-neosynthesis normalized intensities were clustered via the K-means method (eight clusters, 10 iterations) based on Pearson correlations using MeV software (37).
Functional Analysis of the Proteome Data
The Munich Information Center for Protein Sequence Functional Catalogue Database of the Arabidopsis genome (38) was used to annotate the 202 2DE protein spots characterized in the de novo synthesized proteome, as well as the 273 2DE protein spots characterized in the total proteome but not in the de novo synthesized set.
Heat Maps of Transcript Abundance, Protein Neosynthesis, and Protein Abundance
An Affymetrix ATH1 data set focused on Arabidopsis seed physiology between harvest and seedling establishment stages was retrieved (18). Microarray Analysis Suite 5.0 (Affymetrix, Santa Clara, CA) normalization was first performed on raw CEL files to determine present/absent/marginal calls for each probe set (39). Probe sets were considered present if they were detected in at least two replicates before GC-Robust Multiarray Average normalization was performed to obtain normalized intensities (39). From the 20, 24, 18, 20, 15, 10, 27, and 13 unambiguous Arabidopsis Gene Indexes (AGIs) of, respectively, Cluster 1 through Cluster 8 (supplemental Table S8), transcriptome data could be found for, respectively, 19, 22, 16, 17, 12, 6, 24, and 12 AGIs in the Arabidopsis transcriptome data set (18). For each gene, the corresponding normalized transcript value was divided by its maximum to reach a transcript abundance between 0 and 1. The probe intensities for GAPC1 (At3g04120) and ACT7 (At5g09810) in Cluster 8 were averaged. Similarly, the protein neosynthesis intensity and the total protein abundance of the corresponding AGI were also retrieved and normalized by their maximum intensity along the germination time course. Heat maps were built using R (40). All normalized data can be found in supplemental Table S11.
RESULTS
From the Full to the Neosynthesized Proteome
A proteomic analysis of Arabidopsis germination was carried out on nondormant seeds from the Ler accession displaying a high germination quality. Indeed, T1 and T50 values were achieved at 28 and 32 HAI, respectively (T1 and T50 correspond to the times at which, respectively, 1% and 50% of the seeds had germinated). In addition, the final percentage of germinated seeds reached 100%. Arabidopsis seed proteome analyses via 2DE revealed about 1,500 protein spots (2DE protein spots), of which 475 were identified via LC-MS/MS (supplemental Table S1 (41)). Because of mRNA processing, protein proteolysis, and chemical protein modification, one gene can produce many different protein species, resulting in wide proteome diversity (24). From one polypeptidic sequence corresponding to a single AGI, many different 2DE protein spots (protein isoforms) were visualized. Indeed, each protein isoform can display distinct electrophoretic properties as a result of variations in their molecular weight and/or isoelectric point (pI). These 475 2DE protein spots corresponded to 257 nonredundant proteins (241 unique AGIs and 16 multiple hits).
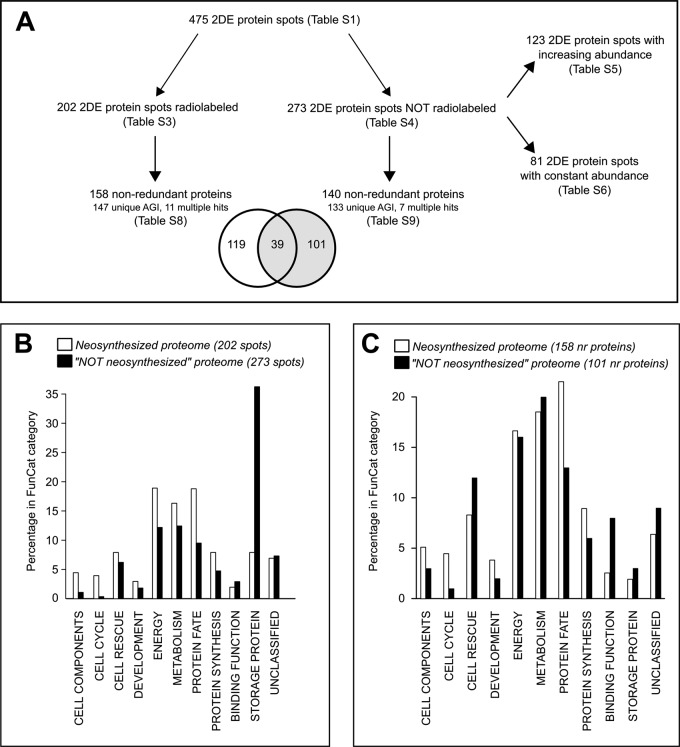
During the time course of seed germination, differential proteomics using silver-stained 2DE gels pointed out 241 and 220 2DE protein spots that were differentially accumulated (up or down) from 0 to 24 HAI (germination sensu stricto) and from 24 to 48 HAI (radicle protrusion), respectively (supplemental Table S2). Still, 147 2DE protein spots did not display abundance variations from dry seed up to radicle protrusion (supplemental Table S2). To our knowledge, no data are available regarding the turnover of these proteins. Thus, pulses of [35S]-Met were applied within six different 8-h time windows during the time course of seed germination (see “Experimental Procedures”). After each period of labeled precursor incorporation into newly synthesized proteins, total proteins were extracted and the amount of [35S]-Met incorporated into proteins was quantified (Table I). For each time window, the total proteins were then separated via 2DE and revealed by silver-staining, and the pattern of neosynthesized proteins was revealed by means of autoradiography (Fig. 1). We highlighted a de novo synthesis pattern for 202 2DE protein spots detected as radiolabeled during the germination process (Fig. 2A, supplemental Table S3). These 2DE protein spots contained some isoforms and corresponded to 158 nonredundant proteins (Fig. 2A, supplemental Table S3). It follows that out of the 475 2DE protein spots from the total proteome, 273 2DE protein spots were not detected as radiolabeled. Thus, the proteins corresponding to these 273 2DE protein spots can be considered as not de novo synthesized during germination (Fig. 2A, supplemental Table S4). Interestingly, among these 2DE protein spots, 123 were up-accumulated in the full proteome during seed germination (Fig. 2A, supplemental Table S5), a finding that emphasizes the importance of post-translational regulation of seed proteins in the germination process. Furthermore, 81 2DE protein spots showed neither de novo protein synthesis nor differential accumulation during Arabidopsis germination (Fig. 2A, supplemental Table S6). This suggests that the corresponding proteins had high stability and hence low turnover. Both the neosynthesized and the non-neosynthesized proteome appear to be complementary to ensure seed vigor.
Table I. Incorporation of [35S]-Met in neosynthesized proteins during the time course of Arabidopsis seed germination.
| Hours after imbibition (HAI) | 0–8 | 8–16 | 16–24 | 24–32 | 32–40 | 40–48 |
| cpm/μg protein | 48 ± 7 | 531 ± 43 | 498 ± 23 | 322 ± 19 | 353 ± 25 | 293 ± 24 |
Note: After each 8-h time window of [35S]-Met incorporation during seed germination, total proteins were extracted and the amount of [35S]-Met taken into proteins was quantified as counts per minute (cpm) per microgram (μg) of protein.
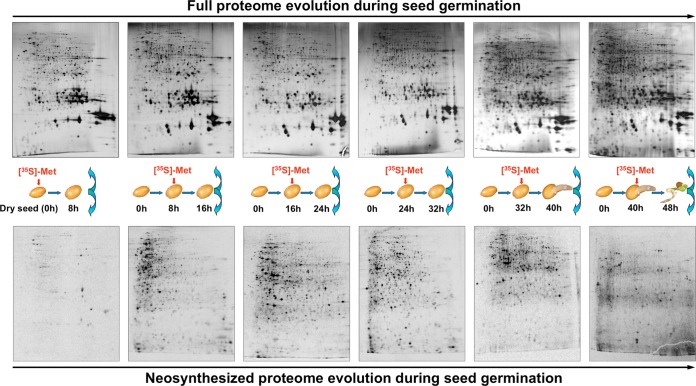
Fig. 1.
2DE patterns of total seed proteome (top row) compared with de novo synthesized proteome (bottom row) during Arabidopsis seed germination. Total proteins were separated via 2DE, and the patterns of the neosynthesized proteome were revealed by autoradiography of [35S]-Met pulse-chase time windows (0–8 HAI, 8–16 HAI, 16–24 HAI, 24–32 HAI, 32–40 HAI, and 40–48 HAI) during the time course of seed germination.
Fig. 2.
Functional analysis of the proteins neosynthesized or not neosynthesized in Arabidopsis germinating seeds. The analysis workflow applied to the present proteomic data is shown in A. The proteins corresponding to the 202 2DE radiolabeled protein spots and the 273 2DE protein spots detected as not radiolabeled in Arabidopsis germinating seeds were classified following FunCat classification (B). The same classification was then applied to the 158 and 101 nonredundant proteins neosynthesized or not during seed germination (C).
The frequency distribution of functional categories according to FunCat classification (38) was assessed for the 202 2DE protein spots that were radiolabeled and for the 273 2DE protein spots that were not radiolabeled (Fig. 2B, supplemental Table S7) during Arabidopsis seed germination. These 202 and 273 2DE protein spots corresponded to 158 and 140 nonredundant proteins, respectively (Fig. 2A). Among these nonredundant proteins, 39 were found in both de novo synthesized and non–de novo synthesized protein spots, and thus 101 proteins were never detected as de novo synthesized during seed germination (Fig. 2A). The frequency distribution of functional categories according to FunCat classification (38) was also assessed for the 158 nonredundant de novo synthesized proteins and the 101 nonredundant proteins detected as not de novo synthesized (Fig. 2C, supplemental Table S7). Most of these proteins were distributed among the six major following categories: “Cell Rescue, Defense and Virulence,” “Energy,” “Metabolism,” “Protein Fate,” “Protein Synthesis,” and “Storage Protein” (Fig. 2B). As expected, the “Storage Protein” category was more represented in the non-neosynthesized proteome than in the neosynthesized proteome of germinating seeds (Fig. 2B). The FunCat classification of the 202 radiolabeled protein spots revealed enrichment of the “Energy,” “Metabolism,” “Protein Fate,” “Protein Synthesis,” and “Storage Protein” categories while the “Energy” and “Storage Protein” categories were overrepresented for the 273 nonradiolabeled protein spots (Fig. 2B, supplemental Table S7). In the “Metabolism” category, the presence of several de novo synthesized proteins involved in sulfur amino acid metabolism (e.g. cysteine and methionine), which is crucial for seed germination, was evidenced (supplemental Table S3 (6, 34)).
De Novo Protein Synthesis Clustering
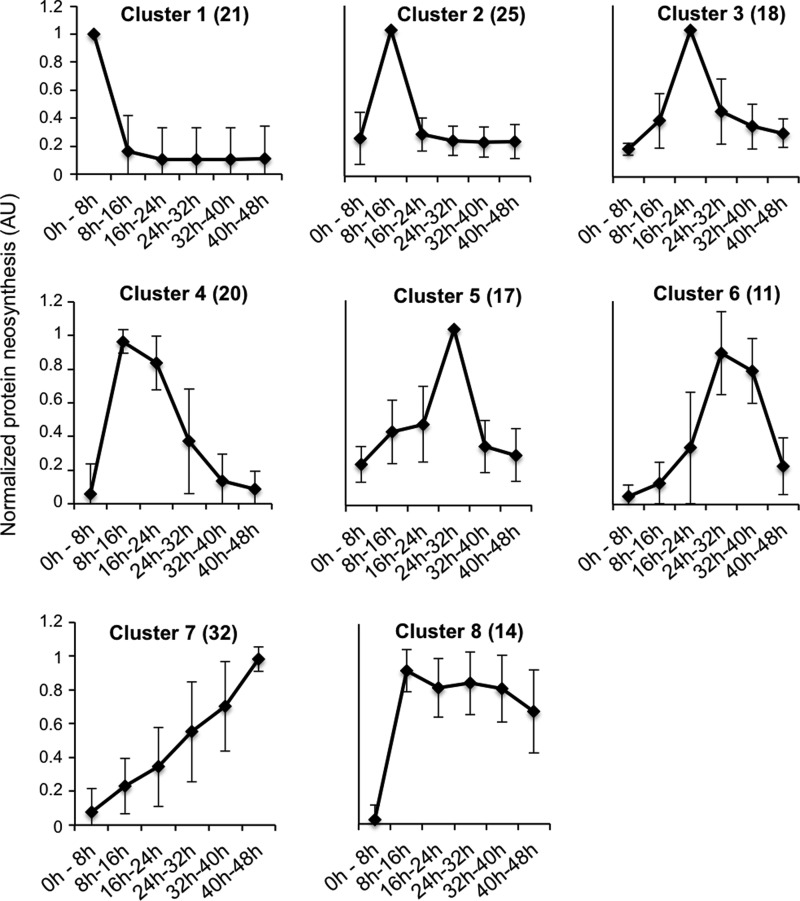
Stored mRNAs in dry seeds are accumulated during seed maturation, and some of them are required for germination completion. The application of cycloheximide, a known inhibitor of mRNA translation (supplemental Fig. S2), blocks seed germination (20). Thanks to the incorporation of [35S]-Met into newly synthesized proteins, the protein synthesis rate could be measured for the 158 nonredundant proteins. A K-means clustering analysis led to the identification of eight distinct kinetic patterns (Cluster 1 through Cluster 8) of de novo synthetized proteins (Fig. 3, supplemental Table S8).
Fig. 3.
K-means clustering of the neosynthesized proteins. Protein radiolabeled intensities corresponding to the same protein were summed when multiple protein isoforms were detected. This allowed us to obtain 158 nonredundant protein radiolabeled intensities. Then, for each protein, the protein neosynthesis intensity was normalized by its maximum value over the whole experiment to get a value between 0 and 1. Finally, a K-means clustering (eight clusters, 10 iterations) based on Pearson correlation was performed on the 158 normalized protein intensities using MEV software (37). The numbers in parentheses indicate the number of nonredundant neosynthesized proteins classified in each cluster.
Comparison of Transcriptome and Proteome Data from Arabidopsis Seed Germination
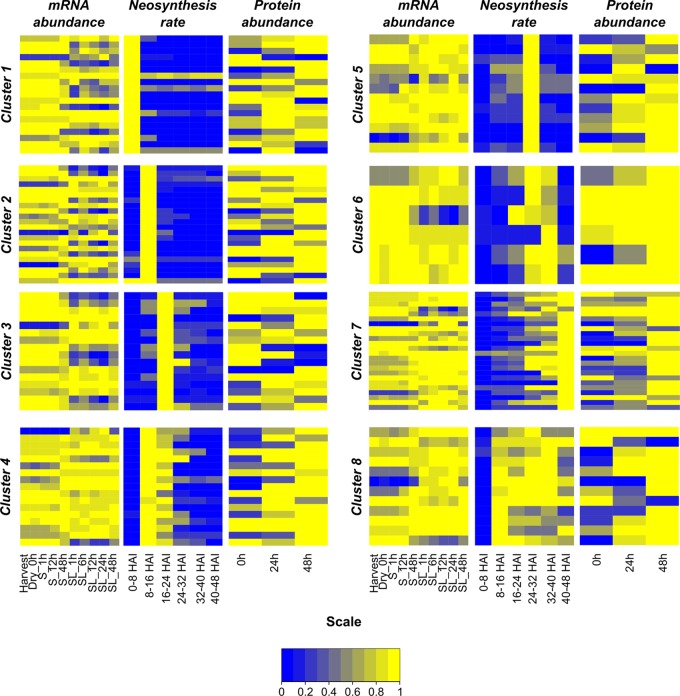
From the 147 unique AGIs, an analysis of mRNA abundances, protein neosynthesis rates, and protein abundances was carried out by comparing available transcriptome data (18) with the present proteomic results (Fig. 4, supplemental Table S11). In most cases, the heat map representation illustrated that the protein synthesis appeared to be independent of the RNA abundance, but the correlation with the protein abundance was higher.
Fig. 4.
Comparison of transcriptome and proteome data during Arabidopsis seed germination. From the 20, 24, 18, 20, 15, 10, 27, and 13 unambiguous AGIs of Cluster 1 to Cluster 8, respectively (supplemental Table S8), transcriptome data could be found for, respectively, 19, 22, 16, 17, 12, 6, 24, and 12 AGIs in a previously published data set (18). The raw Affymetrix CEL data were normalized using the GC-Robust Multiarray Average algorithm (39). For each gene, the corresponding normalized transcript value was divided by its maximum to obtain a transcript abundance between 0 and 1. The probe intensities for GAPC1 (At3g04120) and ACT7 (At5g09810) in Cluster 8 were averaged. Similarly, the protein neosynthesis intensity and the total protein abundance of the corresponding AGI were also retrieved and normalized to their maximum intensity along the germination time course. Heat maps were built using R (40). S: seeds stratified during the indicated number of hours; SL: seeds stratified and then germinated under continuous light for the indicated number of hours.
DISCUSSION
The present results shed new light on the mechanisms involved in the three classical phases of germination based upon water uptake by imbibing seeds (Fig. 5 (7)). The comparison of mRNA level, translational activity, and protein abundance emphasized that selective mRNA translation is a major regulatory mechanism of seed germination (Figs. 3 and 4). This process is both sequential and selective. Thus, it is tempting to speculate that a large set of seed-specific genes is regulated at the level of translation. In particular, these new data on protein turnover during Arabidopsis seed germination revealed contrasting protein behaviors (supplemental Tables S3–S6). This temporal profiling highlights that Arabidopsis seed germination consists of a series of sequential events that overlap the three canonical phases of seed germination (7). In the following, we discuss the observed eight different mRNA translation patterns in detail.
Fig. 5.
Metabolic transitions evidenced from Arabidopsis de novo synthesized proteins during the three canonical phases of seed germination. During Phase I, the hydrated seed translates stored mRNAs and restarts the late seed developmental program as exemplified by the synthesis of seed storage proteins (e.g. cruciferin (CRA)) and late embryogenesis abundant proteins (e.g. LEA1). The transition from Phase I to Phase II is characterized by the action of important remodeling/repair (e.g. rotamase cyclophilin (ROC1)), antioxidant (e.g. monodehydroascorbate reductase (MDAR6)), and detoxification mechanisms (e.g. mercaptopyruvate sulfurtransferase 1 (MST1)). During Phase II, seed storage and other proteins are degraded by the combined action of the proteasome (20S proteasome subunit PAB1) and peptidases (peptidase S8/S53, tripeptidyl peptidase II (TPPII)) that fuel amino-acid-incorporating metabolism (glutamine synthetase 1.3 (GLN1.3)). Immediately preceding Phase III, seedling establishment is prepared thanks to the mRNA translation cytoskeleton components (Actin2 (ACT2), Tubulin 3 (TUB3)). The vertical dashed lines distinguish the three phases of Arabidopsis seed germination based on water uptake kinetics. The red bar represents the mRNA translational activity based on radiolabeled Met incorporation (Table I).
Very Early Neosynthesized Proteins
Among the proteins synthesized in the 0–8 HAI time window, several proteins involved in desiccation tolerance were revealed, such as late embryogenesis-abundant proteins (At3g51810, At3g17520, At2g42560) and the RAB18 dehydrin (RAB18, At5g66400), all of which could help to maintain desiccation tolerance in seeds up to radicle protrusion (42–45). Late embryogenesis-abundant proteins are known to accumulate during seed maturation (acquisition of desiccation tolerance) and protect biomolecules (e.g. inner mitochondrial membrane) from dehydration-induced damages.
Seed Storage Proteins
Unexpectedly, several protein spots corresponding to seed storage proteins (SSPs) displayed a similar neosynthesis pattern. SSPs are proteins accumulated mainly during seed maturation and are typically described as a nitrogen and amino acid source for seed germination and seedling establishment. There are three 12S cruciferin annotated genes (CRA1, At5g44120; CRB, At1g03880; CRC, At4g28520) in the Arabidopsis Col-0 genome (46). Previously, it was shown that the mRNAs corresponding to the three 12S cruciferin genes in Arabidopsis are the most abundant in dry mature seeds (15, 16). It is worth noting that the Ler genome contains another copy of the CRA1 gene, called CRA2, which represents a tandem duplication (47). The present proteomic data did not allow us to differentiate CRA1 from CRA2, and therefore, in this study, the CRA1 and CRA2 protein data are grouped under the same AGI, namely, At5g44120. Interestingly, these four cruciferin genes generate more than 100 protein isoforms because of the high sensitivity of SSPs to post-translational modifications (24). During seed maturation, all four 12S cruciferin genes code for SSPs in the form of ∼50-kDa precursors. These precursor forms are then enzymatically processed into acidic α-subunits (∼30 kDa) and basic β-subunits (∼20 kDa) that remain linked together by a single interchain disulfide bond. The synthesis of the precursor forms encoded by the CRC (At4g28520) and CRB (At1g03880) genes has been detected only in the very early steps of seed germination (0–8 HAI), whereas the precursor forms encoded by the CRA1/CRA2 (At5g44120) gene are synthesized up to 16 HAI (supplemental Table S3). Radiolabeling of the acidic α-subunits and the basic β-subunits was also observed for all these genes up to 16 HAI (supplemental Table S3). This result highlights that the vacuolar-processing enzyme involved in post-translational processing of the precursor cruciferin forms into the corresponding acid and basic 12S cruciferin subunits is functional during the very early steps of seed germination. Vacuolar-processing enzyme is essential for the correct protein storage vacuole structure and compartmentalization of SSPs, and a seedling growth delay was reported in rice vpe mutants (48, 49). This indicates that SSP processing can affect early seedling development, presumably because such processing is required for the rapid use of these proteins as a source of nitrogen and amino acids. Moreover, SSP functions might not be limited to supplying the embryo with the nutrients required for germination. For example, the abundant 12S cruciferins were shown to display very high sensitivity to reactive oxygen species (ROS) (25, 26, 50) and could therefore represent a major and predominant ROS scavenger in seeds, an organ know to be continuously exposed to oxidative stress during desiccation, dormancy breaking, and germination (51). Thus, the rapid synthesis of SSPs upon imbibition could help the seed reconstitute protein reserves and cope with strong oxidative stress during natural imbibition/drying cycles in the soil.
Remodeling/Repair Mechanisms and Control of Water Uptake
Cluster 1 brings out proteins mostly neosynthesized during the first 8 HAI in the very early steps of seed germination (supplemental Table S8). Strikingly, most of these proteins are known to be associated with the seed maturation program. Our results are in accordance with those showing that Arabidopsis seed germination takes place in a short window of time after the start of imbibition during which the seed can still recruit its late maturation program (52). Such proteins are presumably translated from stored mRNAs (22), as this neosynthesized proteome is reminiscent of the reinduction of the maturation program as previously described in hydrated seeds in the presence of the transcriptional inhibitor α-amanitin (20). Notably, a peptidyl-prolyl cis-trans isomerase (cyclophilin family) named ROC1 (At4g38740) was specifically neosynthesized between 0 and 8 HAI, and its protein level subsequently remained abundant in the hydrated seeds up to radicle protrusion (supplemental Tables S1 and S7). Peptidyl-prolyl cis-trans isomerases catalyze the conversion of peptidyl-prolyl imide bonds in peptide and protein substrates. The Arabidopsis genome encodes at least five cytosolic cyclophilin family proteins (53). In plants, protein remodeling by proline cis-trans isomerization via ROC1 is emerging as a critical step in signaling cascades, given its role in stem elongation (54), seedling de-etiolation (55), and pollen germination (56). The present results suggest that ROC1 is involved in Arabidopsis seed germination.
Moreover, it is noteworthy that the subtilisin-like serine protease SBT1.7 (At5g67360), which is involved in the regulation of pectin methylesterification and therefore mucilage release upon imbibition (57), was also neosynthesized preferentially in the 0–8 HAI time window (supplemental Table S8). Mutants affected in mucilage production exhibit poor germination vigor when water availability is low (58). During the very first hours of seed imbibition, the released mucilage helps to control the speed of water uptake. Recent studies assumed involvement of the seed mucilage in assisting embryonic cells to maintain DNA integrity (59, 60). This supports the assumption that this mucilage contributes to seed environmental adaptation and plays an important role in successful germination.
Neosynthesized Proteins Associated with a Specific Germination Program
Clusters 2, 3, and 5 displayed proteins with a maximum of synthesis at, respectively, 8–16 HAI, 16–24 HAI, and 24–32 HAI, whereas Cluster 4 contained proteins highly neosynthesized from 8 to 24 HAI (supplemental Table S8). The period from 8 to 32 HAI corresponds to Phase II of seed germination, which leads to increased metabolic activity up to radicle protrusion.
Initiation of the Germination Program by Phosphoenolpyruvate/Oxaloacetate-related Metabolism
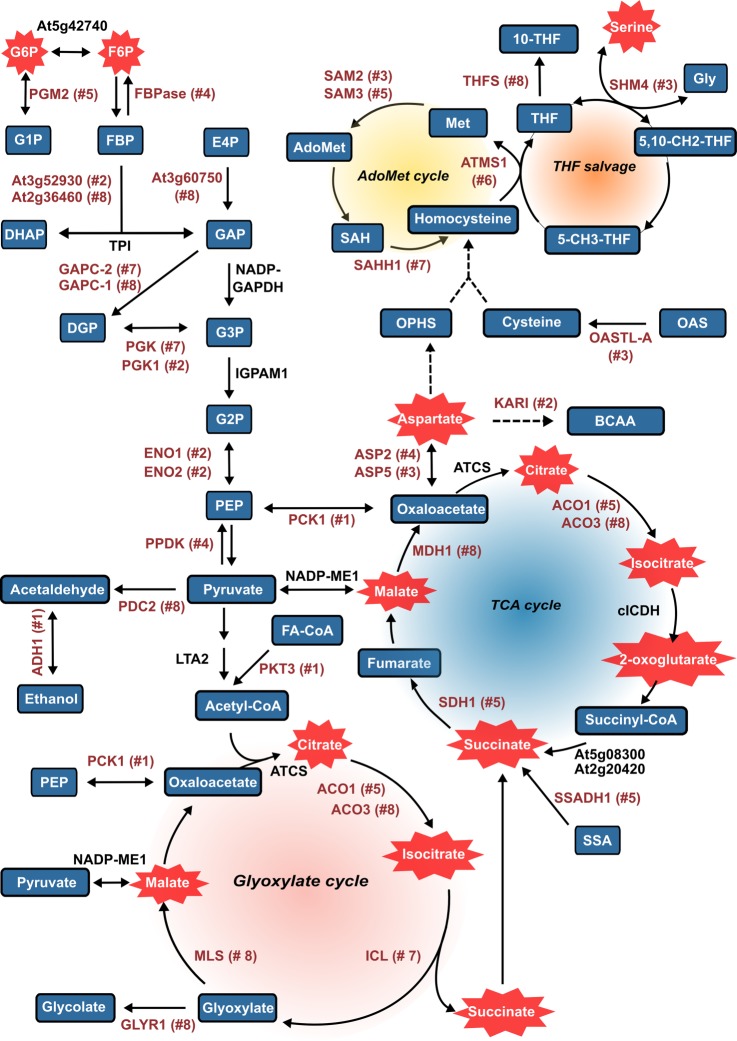
The early germination phase is defined as the sum of events occurring before testa rupture (61). Aerobic respiration is resumed rapidly upon imbibition and becomes the main energy source of the germinating seed. In the absence of carbon fixation via photosynthesis, the degradation of molecules for energy production (e.g. glycolysis, glyoxylate cycle, tricarboxylic acid (TCA) cycle) is presumed to be a key determinant of germination vigor. Accordingly, several enzymes involved in these energetic pathways were found neosynthesized during the early hours of the germination process. For instance, a peroxisomal 3-ketoacyl-CoA (PKT3/KAT2, At2g33150) involved in fatty acid β-oxidation was among the first proteins neosynthesized (Cluster 1). It has been reported that this protein positively regulates signaling via the phytohormone abscisic acid (62). In addition, de novo synthesis of key enzymes of the glyoxylate cycle, such as isocitrate lyase (At3g21720), aconitase 3 (At2g05710), and malate synthase (At5g03860), were pointed out from the early germination phase (Clusters 7 and 8), supporting early activation of lipid reserve remobilization upon seed imbibition. The concomitant neosynthesis of cytosolic malate dehydrogenase 1 (At1g04410, Cluster 8) suggests a possible conversion of the malate generated from both the glyoxylate and TCA cycles into oxaloacetate in the cytosol (Fig. 6). The generated oxaloacetate could then fuel amino acid synthesis pathways via aspartate aminotransferase activity (ASP5, At4g31990, Cluster 3; ASP2, At5g19550, Cluster 4) or be converted to phosphoenolpyruvate by phosphoenolpyruvate carboxykinase 1 (At4g37870), preferentially de novo synthesized between 0 and 8 HAI (Cluster 1). Indeed, phosphoenolpyruvate carboxykinase 1 was previously associated with the ability to mobilize storage lipids and proteins (63). The reversible nature of phosphoenolpyruvate carboxykinase 1 activity places this enzyme as a central element for the metabolic adaptation from germinating seeds to seedling establishment (64). Accordingly, de novo synthesis was observed for many enzymes involved in neoglucogenesis and/or glycolysis within the first 16 HAI (mainly in Clusters 2, 4, and 8; Fig. 6). For instance, two enolases (ENO1, At1g74030; ENO2, At2g36530) were present in Cluster 2 (Fig. 6, supplemental Table S8). These enzymes catalyze the reversible conversion of 2-phosphoglycerate into phosphoenolpyruvate in the glycolytic/neoglucogenic pathway. In developing Arabidopsis embryos, ENO1 and ENO2 are, respectively, responsible for most of the plastid and cytosolic enolase activity (65). The plastid-localized ENO1 has been identified and functionally characterized in Arabidopsis (66). The strong accumulation of fructose-6-P and glucose-6-P during the early steps of germination supports neoglucogenesis activity (67). Nonetheless, glycolysis is also presumed to occur, especially within plastids (23). Overall, the respective flux through these metabolic pathways seems to depend on the tissue and cell compartment considered but could also change along the time course of seed germination. Thus, several enzymes involved in reversible reactions of glycolysis/neoglucogenesis might be involved in the metabolic balance controlling germination vigor.
Fig. 6.
Neosynthesized proteins related to seed glycolysis/neoglucogenesis, energy, and amino acid metabolisms. The proteins neosynthesized (red) related to seed energy and neoglucogenesis metabolisms are displayed on their corresponding enzymatic reactions with their cluster number between brackets. The metabolites previously found to be up-accumulated during Arabidopsis seed germination are displayed within red stars (67). Proteins present in the total seed proteome but not neosynthesized during germination are indicated in black. 5,10-CH2-THF, 5,10-methylene-THF; ACO1/3, aconitase 1/3; ADH1, aldehyde dehydrogenase 1; ASP2/5, aspartate aminotransferase 2/5; ATCS, citrate synthase; ATMS1, cobalamin-independent methionine synthase 1; BCAA, branched-chain amino acids; CICDH, cytosolic NADP-dependent isocitrate dehydrogenase; DHAP, dihydroxyacetone phosphate; DGP, glycerate 1,3-diphosphate; E4P, erythrose-4-phosphate; ENO1/2, enolase 1/2; F6P, fructose-6-phosphate; FBP, fructose-1,6-bisphosphate; FBPase, fructose-1,6-bisphosphatase; G1P, glucose-1-phosphate; G6P, glucose-6-phosphate; G3P, glycerate-3-phosphate; G2P, glycerate-2-phosphate; GAP, glyceraldehyde-3-phosphate; GAPC-2, glyceraldehyde-3-phosphate dehydrogenase C2; Gly, glycine; GLYR1, glyoxylate reductase 1; ICL, isocitrate lyase; IGPAM1, 2,3-biphosphoglycerate-independent phosphoglycerate mutase 1; KARI, ketol-acid reductoisomerase I; LTA2, plastid E2 subunit of pyruvate decarboxylase; MDH1, malate dehydrogenase 1; MLS, malate synthase; NADP-GAPDH, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase; NADP-ME1, NADP-malic enzyme 1; OAS, O-acetylserine; OASTL-A, O-acetylserine (thiol) lyase cytosolic isoform A1; OPHS, O-phospho-l-homoserine; PCK1, phosphoenolpyruvate carboxykinase 1; PDC2, pyruvate decarboxylase 2; PGK, phosphoglycerate kinase; PGK2, phosphoglycerate kinase 1; PKT3, peroxisomal 3-ketoacyl-CoA thiolase 3; PGM2, phosphoglucomutase 2; PEP, phosphoenolpyruvate; PPDK, pyruvate orthophosphate dikinase; SAH, S-adenosyl-l-homocysteine; SAHH1, S-adenosyl-l-homocysteine hydrolase 1; SAM2/3, S-adenosylmethionine synthetase 2/3; SDH1, succinate dehydrogenase 1; SHM4, serine hydroxymethyltransferase 4; TCA cycle, tricarboxylic acid cycle; THF, tetrahydrofolate; THFS, 10-formyltetrahydrofolate (10-THF) synthetase; TPI, triose-phosphate isomerase; SSA, succinic semialdehyde; SSADH, succinic semialdehyde dehydrogenase.
Some of these enzymes also correspond to multifunctional proteins. For instance, ENO2 was described as a bifunctional enolase localized to the cytoplasm and to the nucleus. Indeed, the ENO2 protein sequence contains both DNA-binding and repression domains (68). It has been shown that Arabidopsis ENO2 can bind to the promoter of STZ/ZAT10 (At1g27730), a gene encoding a transcription factor with two Cys(2)/His(2)-type zinc-finger motifs that act as transcriptional repressors for oxidative stress responses, jasmonic acid biosynthesis, and cold responsive genes (68–71). ENO2 has a repressor role in STZ/ZAT10 expression, and transcriptome data show that the STZ/ZAT10 transcript is preferentially expressed at the end of seed maturation (Arabidopsis eFP Browser (72)). Furthermore, the STZ/ZAT10 transcript is strongly accumulated (9-fold) in 5-day-old wild-type seedlings treated with H2O2 and in seedlings subjected to drought stress (69, 73). Given all of its roles, the STZ/ZAT10 gene would be part of a late maturation program dedicated to seed drought resistance. Then, upon imbibition, the ENO2 protein that is preferentially neosynthesized between 8 and 16 HAI would repress STZ/ZAT10 expression to encourage the seed to exit its maturation program (loss of desiccation tolerance) and commit to seedling establishment.
Antioxidant Mechanisms and Detoxification
Five proteins related to ROS homeostasis—iron/manganese superoxide dismutase (At3g56350), 1-cysteine peroxiredoxin (At1g48130), catalase 2 (At4g35090), thioredoxin-dependent peroxidase 1 (At1g65980), and ferritin 2 (FER2, At3g11050)—are represented in the neosynthesized proteome. Successful seed germination has been suggested to occur within a certain oxidative window where ROS act as signaling molecules without deleterious consequences for biomolecules (51). Three proteins involved in ROS detoxification (iron/manganese superoxide dismutase, peroxiredoxin, and thioredoxin-dependent peroxidase 1) and one involved in free iron(II) sequestration (FER2) display maximum neosynthesis between 16 and 24 HAI (Cluster 3; supplemental Table S8). The present results indicate that the synthesis of ROS detoxifying enzymes occurs from 16 HAI in nondormant Arabidopsis seeds. Therefore, the above-mentioned oxidative window (51) would take place in the very early steps of Arabidopsis germination (before 16 HAI). Finally, the catalase 2 protein showed maximum neosynthesis between 24 and 32 HAI (Cluster 5; supplemental Table S8) in accordance with the significant production of H2O2 at 24 HAI in nondormant Arabidopsis seeds (50, 74). Free iron sequestration is also important in controlling ROS production caused by the Fenton reaction. Previously, FER2 was found to be the main ferritin isoform accumulated in dry seeds (75). In hydrated nondormant seeds, FER2 is translated not in the very early steps of seed germination but rather between 16 and 32 HAI, suggesting that this protein is involved in germination before radicle protrusion (supplemental Table S3). Interestingly, during germination, fer2 mutant seeds are sensitive to oxidative stress (76). It is important to underline that the FER1 (At5g01600) protein is accumulated in the early steps of seed development but not in dry seeds, as revealed by Western blot (76). However, we can detect the FER1 protein in the dry seed proteome with LC-MS/MS (supplemental Table S1). Remarkably, both FER1 and FER2 protein abundances strongly decreased during Arabidopsis germination to nearly undetectable levels (supplemental Table S1). Therefore, FER1 and FER2 are probably involved in both free iron and ROS homeostasis during seed germination. Interestingly, ferritins were described as classic examples of translationally regulated genes (77).
Ascorbate (vitamin C) is important for oxidative responses in both plants and animals. Yet the ascorbate pool appears to be relatively low in dry seeds (78). An intermediate during the oxidation process is a short-lived radical, monodehydroascorbate, which can be further enzymatically converted to ascorbate by monodehydroascorbate reductase (MDAR). The Arabidopsis genome contains five genes encoding MDARs (MDAR1, -2/3, -4, -5, and -6). These proteins are enzymatic components of the glutathione-ascorbate cycle, one of the major antioxidant systems in plants. In Arabidopsis, MDAR4 is required for seed storage lipid hydrolysis and postgerminative growth (79). Our results indicated that MDAR6 (At1g63940) is synthesized in the very early steps of seed germination (Cluster 1; supplemental Table S8), suggesting a rapid production of ascorbate from monodehydroascorbate during this process. The subcellular localization of MDAR6 has been assigned to the plastid compartment (80). Monodehydroascorbate can also be converted to a divalent oxidation product, dehydroascorbic acid, through spontaneous disproportionation or further oxidation (81). Fait et al. (67) described a significant increase in the level of dehydroascorbic acid during Arabidopsis seed germination. Dehydroascorbic acid can also be enzymatically reduced to ascorbate by dehydroascorbate reductase in a glutathione-dependent reaction. The enzyme dehydroascorbate reductase 1 (At1g19570) displayed a maximum of synthesis from 8 to 16 HAI in Arabidopsis seeds (Cluster 2; supplemental Table S8). Thus, MDAR6 and dehydroascorbate reductase 1 are key components of the ascorbate recycling system and would maintain significant levels of ascorbic acid in the very early stages of germination. Our results are in accordance with previous works indicating that ascorbate accumulation restarts very early (within a few hours) upon seed imbibition (82, 83). It is worth noting that the exogenous application of low concentrations of ascorbate can partially rescue rice seed germination from abscisic acid treatment (84). Moreover, an examination of the neosynthesized proteome highlights that GDP-d-mannose 3′,5′-epimerase (At5g28840), which converts GDP-d-mannose to GDP-l-galactose, exhibits a maximum of synthesis from 8 to 24 HAI in seeds (supplemental Table S8). Encoded by a single gene in the Arabidopsis genome, GDP-d-mannose 3′,5′-epimerase is a control point in the ascorbate biosynthetic process (85). Dehydroascorbic acid is derived from the antioxidant ascorbate, which is maintained at a constant steady-state level, apparently because it is replenished from the hexose phosphate pool (83). Taken together, our data indicate that the ascorbate recycling system would act upstream of the ascorbate biosynthetic process during seed germination.
Protein Fate (Folding)
Cluster 4 includes proteins highly neosynthesized from 8 to 24 HAI that are involved in protein fate, such as heat shock proteins (HSP91, At1g79930; HSP70, At3g12580; HSC70.5, At5g09590; HSP60.2, At2g33210) and protein disulfide isomerases (PDI4, At3g16110; PDI6, At1g77510) (supplemental Table S8). The Arabidopsis genome contains 12 PDI genes (86). PDIs are thiodisulfide oxidoreductases that catalyze disulfide bond formation and rearrangements in the substrate proteins, facilitating the folding of nascent polypeptides in the endoplasmic reticulum (ER). PDIs also function in the organelles outside the ER and play roles in redox signaling. However, little is known about their functions and their subcellular locations in Arabidopsis seeds. It has been reported that PDI5 (At1g21750) chaperones and inhibits cysteine proteases during trafficking to vacuoles prior to programmed cell death of the endothelium in developing seeds (86), and this protein is oxidized during Arabidopsis seed germination (50). In rice seeds, a PDI protein (LOC_Os11g09280) prevents proglutelin and prolamin from aggregating by chaperoning their segregation in the ER lumen (87, 88). In addition, PDI2 is highly expressed in Arabidopsis seeds; PDI2 accumulation is localized to both ER and nucleus, and a slight decrease in the germination rate of pdi2 mutant seeds relative to wild-type seeds has been observed (89). The strong expression of PDI2 in the outer cell layers of the seed and the PDI2 localization to the nucleus are intriguing. Notably, PDI2 interaction with Maternal Effect Embryo Arrest 8 (At1g25310) was reported, and Maternal Effect Embryo Arrest 8 is proposed to play a role in controlling the expression of genes in maternal tissue required for normal embryo and endosperm biogenesis (90). Moreover, our data disclosed that PDI11 (At2g47470) was also neosynthesized throughout germination until 40 HAI (Cluster 6). Thus, the present results suggest important roles for PDI4, PDI6, and PDI11 in Arabidopsis seed germination.
Preparation for Radicle Protrusion and Germination Sensu Stricto
Clusters 5 and 6 contained proteins that displayed maximum neosynthesis in the 24–32 HAI and 24–40 HAI time windows, respectively. Consequently, these proteins are presumably involved in the preparation of radicle protrusion.
Protein Degradation and Nitrogen Remobilization
Cytosolic glutamine synthetase 1–3 (At3g17820) belongs to Cluster 5, which coincides with protein degradation. Glutamine synthetase is a key enzyme for inorganic nitrogen assimilation and remobilization in plants. Indeed, glutamine synthetase catalyzes the ATP-dependent fixation of ammonium to the δ-carboxyl group of glutamate to form glutamine. Remarkably, genetic studies in maize showed the co-localization of a quantitative trait locus for high germination rates with the glutamine synthetase 1–3 gene, suggesting that high levels of glutamine synthetase activity lead to faster germination (91). Consequently, the release of amino acids from SSPs in germinating Arabidopsis seeds would start from 24 HAI, therefore anticipating radicle protrusion and seedling growth. This hypothesis is supported by the presence of two proteases, peptidase S8/S53 (At3g14067) and tripeptidyl peptidase II (TPPII) (At4g20850), in Clusters 5 and 6 that are highly neosynthesized from 24 HAI (supplemental Table S8). TPPII is the main exopeptidase involved in the processing of large proteasome products and is probably necessary for efficient protein turnover (92), especially contributing to the preferential catabolism of oxidized (carbonylated) proteins (93). Given that SSPs represent the major forms of carbonylated proteins in dry mature seeds (50), it seems conceivable that TPPII might contribute to the degradation of SSPs during seed germination via the 20S proteasome pathway. In agreement, three protein subunits of the 20S proteasome (PAB1, At1g16470; PAA1, At5g35590; PAE2, At3g14290 or At1g53850) were detected in the dry and germinating seed proteomes (supplemental Table S1). The 20S proteasome subunit PAE2 was constitutively neosynthesized from 8 to 48 HAI, whereas PAB1 was neosynthesized preferentially between 8 and 24 HAI (supplemental Table S8). It has been highlighted that the degradation of carbonylated proteins is partially mediated by the 20S proteasome in accordance with the favored detection of a 20S proteasome/TPPII couple in Arabidopsis seeds (93, 94).
Sulfur-containing Amino Acids (Cysteine and Methionine) and Folate Metabolism during Sensu Stricto Seed Germination
The de novo synthesis of several proteins related to sulfur-containing amino acids, the methionine cycle, and folate metabolism was highlighted in the present data (supplemental Table S3, Fig. 6). First, cytosolic O-acetylserine (thiol) lyase protein (At4g14880), contributing to cysteine synthesis (95, 96), was found in Cluster 3. In addition, the cobalamin-independent Met synthase (ATMS1, At5g17920) belongs to Cluster 6, which contains proteins with increasing de novo synthesis up to radicle protrusion (40 HAI) followed by a subsequent dramatic decline (supplemental Table S8). This enzyme, involved in the last step of methionine synthesis in plants (30, 97), plays a key role in seed germination because de novo Met synthesis is crucial to seed vigor and seedling establishment in Arabidopsis (6, 34). Moreover, Met is the precursor of S-adenosylmethionine (AdoMet), the universal methyl-group donor in biological reactions (30). The last step in the production of AdoMet is catalyzed by AdoMet synthetase, and the present data indicate that AdoMet synthetase 2 (SAM2, At4g01850) and AdoMet synthetase 3 (SAM3, At3g17390) are highly synthesized from 16 to 24 HAI (Cluster 3) and from 24 to 32 HAI (Cluster 5), respectively. This is in accordance with the characteristic accumulation of AdoMet synthetase just before radicle protrusion (34, 98). In addition, the presence of the S-adenosyl-homocysteine hydrolase 1 (At4g13940; Cluster 7) demonstrated the importance of AdoMet recycling, with very important neosynthesis starting from 24 HAI and continuing up to radicle protrusion. This enzyme is required for proper functioning of the methyl cycle to remove S-adenosyl-homocysteine, which is a potent inhibitor of the AdoMet-dependent methyltransferases (99). These results add a new line of evidence regarding the importance of methionine metabolism for seed germination sensu stricto (6).
Furthermore, of outstanding importance for seed germination, tetrahydrofolates (THFs) are essential co-factors mediating the transfer of one-carbon units (100). For instance, the reaction catalyzed by ATMS1 is a transfer of the methyl group from the 5-methyl-THF to homocysteine, yielding Met. In turn, in conditions where photorespiration is unlikely to take place, as in germinating seeds, 1-carbon formate could be incorporated into THF to form 10-formylTHF and, eventually, 5,10-methylene-THF (101). Finally, as we detected serine hydroxymethyltransferase 4 (At4g13930) in Cluster 4, the 5,10-methylene-THF could be converted to serine that accumulates during the first 24 h of germination (67). The origin of the necessary formate remains unclear, but it could be derived from oxalate metabolism.
Proteins Constitutively Neosynthesized in the Time Course of Seed Germination up to Seedling Establishment: Glucosinolate Remobilization
The proteins belonging to Cluster 7 displayed a progressive increase in protein neosynthesis toward radicle protrusion (supplemental Table S8). In this cluster, two proteins, nitrile-specifier protein 2 (At2g33070) and PYK10 protein (At3g09260), could be involved in glucosinolate catabolism. Glucosinolates are sulfur-rich plant secondary metabolites, and their overall content declines upon seed imbibition (102, 103). The Arabidopsis PYK10 protein (At3g09260) was described as a root- and hypocotyl-specific myrosinase (104) whose activity could release glucose and breakdown products, including defense compounds such as isothiocyanates. To date, the physiological role and natural substrate(s) of PYK10 are unknown, but functional β-glucosidase and β-fucosidase activities have been reported for PYK10 (105). In addition, nitrile-specifier protein 2 is abundant in both dry and germinating Arabidopsis seeds and is also progressively translated (supplemental Tables S1 and S7). Through in vitro assays on mature seed extracts, nitrile-specifier protein 2 was shown to preferentially promote nitrile production upon glucosinolate breakdown, thereby allowing efficient remobilization of the sulfur and nitrogen contents (106). Thus, glucosinolate degradation during Arabidopsis seed germination could represent a significant source of sulfur and nitrogen, thereby fueling the sulfur amino acid metabolism essential for germination (107, 108). These results provide further support for the crucial importance of sulfur metabolism in seed germination and vigor (6).
PYK10 is the major component of the ER bodies (109), and it has been shown that the PYK10 promoter mediates developmentally regulated gene expression that becomes specific to the root in advanced developmental stages and shows no activities in other parts of the mature plant (104). PYK10 is able to interact with PYK10-binding protein 1 (PBP1, At3g16420), which possesses two repeated regions, each of which is highly homologous to the α-chain of jacalin (plant-based lectin), a carbohydrate-binding protein (110). PBP1 was also detected as de novo synthesized during germination (Cluster 6), suggesting a marked necessity of the PYK10–PBP1 complex (β-glucosidase complex) before radicle protrusion. Indeed, PBP1 contributes to PYK10 activity and may act as a molecular chaperone that facilitates the correct polymerization of PYK10 when cells are exposed to stress. The PYK10 and PBP1 genes are both regulated by the NAI1 (At2g22770) basic-helix-loop-helix-type transcription factor (105). Moreover, it has also been demonstrated that NAI2 (At3g15950) deficiency in Arabidopsis specifically reduces the accumulation of PYK10 protein but does not affect the accumulation of PBP1 (111). nai1 and nai2 mutants display an absence of ER bodies, and the consequences of these mutations on seed biology would deserve to be studied in depth.
Cytoskeleton
Cytoskeletal elements involved in cytoplasmic organization, cell morphology, and motility should be essential to restart cellular metabolism during germination. This seems to be the case, as components of microtubules (β-tubulin 3, At5g62700; β-tubulin 8, At5g23860) and actin filaments (Actin 2, At3g18780) were detected as neosynthesized in Cluster 7 (supplemental Table S8). Furthermore, tubulins 3 and 4 (At5g19770; At1g04820) and actin 7 (At5g09810) were also neosynthesized and belonged to Cluster 4 and Cluster 8, respectively (supplemental Table S8). If the act2 mutation does not cause any obvious phenotypes except reduced fitness, the act7 mutation impairs germination and seedling and root growth (112, 113). These observations concur with previous works indicating that in germinating seeds, cytoskeleton reorganization is necessary for the large rates of cell elongation that precede radicle protrusion (114–116).
Constitutive de Novo Synthesis of Enzymes Involved in Energy and Neoglucogenesis Pathways
A large number of enzymes involved in the TCA cycle, the glyoxylate cycle, and glycolysis/neoglucogenesis were neosynthesized from the very early steps of seed germination up to seedling establishment (Fig. 6). Specifically, in Clusters 7 and 8, we found de novo synthesized proteins involved in energy production (TCA cycle, aconitase 3, malate dehydrogenase 1) or dedicated to the consumption of fatty acid (glyoxylate cycle, aconitase 3, isocitrate lyase, malate synthase). Most of these enzymes are important for seed vigor and seedling establishment (117–119). Cluster 8 contained proteins highly neosynthesized from 8 up to 48 HAI in a constant manner, with the exception of the very early steps of seed germination (supplemental Table S8). Interestingly, glyoxylate reductase 1 (GLYR1) (At3g25530) belongs to this cluster. This enzyme catalyzes the reduction of both glyoxylate and succinate semialdehyde, which are intermediates in the metabolism of glycolate and γ-aminobutyrate (GABA), respectively (120). Preferentially, GLYR1 catalyzes the NADPH-based reduction of glyoxylate into glycolate, and it has been proposed that glyoxylate detoxification by GLYR1 could provide an alternative sink for excess electrons during the recycling of reducing equivalents (121). Yet recently it was shown that the GLYR1 protein is exclusively present in the cytosol and is not found in the peroxisome (122). As GABA is converted to succinate semialdehyde in the mitochondria, GLYR1 could thus be involved in the detoxification of glyoxylate that accidently reaches the cytosol. In addition, the protein succinic semialdehyde dehydrogenase 1 (At1g79440) was found to be (i) abundant in dry seeds, (ii) highly de novo synthesized between 24 and 32 HAI, and (iii) accumulated to increasing amounts during the germination time course (supplemental Tables S1 and S7). The succinic semialdehyde dehydrogenase 1 enzyme can reduce succinate semialdehyde to γ-hydroxybutyrate. Together, these findings suggest that the detoxification of reactive aldehydes such as glyoxylate and succinate semialdehyde is very important for the metabolism of germinating seeds. The GABA shunt activity could also provide a major supply of succinate to the TCA cycle during germination. This could be related to the established role of the GABA shunt in regulating the C/N ratio in conditions where the carbon supply is limited (123).
Translational Machinery and Selective mRNA Translation
Translational control is exerted in all stages of plant development and affects a wide range of mRNAs (124, 125). Notably, protein synthesis is absolutely required for Arabidopsis seed germination (20), and translational activity is strongly reduced in aged seeds that exhibit depressed seed vigor (26, 119). Quantitatively, three distinct steps of mRNA translational activity are detected during seed germination (Table I). Around 10% (15/158 proteins) of the de novo synthesized proteins detected in this study are involved in mRNA translation (supplemental Table S10). These proteins were not part of Cluster 1, indicating that the stored translational machinery is already effective during the very first hours upon imbibition (0–8 HAI), as previously described for wheat germ extract (126). Thus, the low translational activity associated with the resumption of the late maturation program could be assimilated as an adaptive mechanism to protect the embryo in the very early steps of seed germination. Indeed, if the environmental conditions are not appropriate for germination and seedling establishment, this late maturation program might be extended, contributing as well to the maintenance of seed dormancy (23). From 8 HAI, both a qualitative and a quantitative failover of the translational program are observed in germinating seeds (Fig. 5, supplemental Table S8). The translational activity peaked between 8 HAI and 24 HAI before declining slightly and finally remaining stable until 48 HAI. The sequential neosynthesis of proteins involved in translational process (supplemental Table S10) strongly suggests an important modulation of mRNA translation. This finding supports previous results showing that a change in mRNA abundance for a given gene does not necessarily result in the expected change in the corresponding protein abundance (127–129). Based on the present results, the selective translation of mRNAs emerges as an important mechanism regulating molecular functions during Arabidopsis seed germination (Fig. 4). Moreover, regarding seed biology, the selective recruitment of either stored mRNAs or de novo synthetized mRNAs is also a likely mechanism. Indeed, the nuclear cap-binding complex (consisting of CBP20 and CBP80) plays a role in both seed germination and abscisic acid perception (130). The cap-binding complex binds co-transcriptionally to the 5′ cap of pre-mRNAs and is involved in the maturation, export, and degradation of nuclear mRNAs. In addition, in vitro experiments on maize seed germination showed that stored mRNAs are preferentially translated by the eIF(iso)4E isoform, a component of the cytoplasmic mRNA cap-binding complex (eIF4F) (124, 131). Finally, mRNA decay is of importance because impairment in the 3′-5′ exonuclease RRP41L delays germination and inhibits seedling growth (132). In the rrp41l knockout mutant, the mRNA levels of genes characteristic of the late maturation program (e.g. SSPs, 9-cis-epoxycarotenoid dioxygenases, oleosins, etc.) are considerably higher than in wild-type plants. Some of the identified checkpoints that regulate Arabidopsis seed germination through the modulation of mRNA transcription, selective recruitment of mRNA for translation, mRNA decay, protein translation, and protein activity are summarized in supplemental Fig. S3. As translational regulation is not dependent on mRNA half-lives, genes encoding stable mRNAs can still be dynamically regulated as long as their protein half-lives are short (129).
CONCLUSION AND PROSPECTS
Further research will address the question of the spatiotemporal control of mRNA translation during seed germination, notably concerning translational control by small RNAs (133–135) and mRNA oxidation (136). However, an unanswered question regarding oxidized mRNAs and small RNAs concerns the identity and determinants of mRNA decay and translational repression. In the present study, we opted for classic and efficient radiolabeling of proteins, compatible with a 2DE approach. However, technological improvements have recently allowed the use of stable isotopes (e.g. stable isotope labeling by amino acids in cell culture) to analyze the turnover of plant cellular proteomes. This strategy is combined with either 2DE gel-based proteome studies (137) or shotgun proteomic approaches (138). In the future it will be relevant to implement these novel approaches to obtain a more detailed picture of seed proteome turnover. The germinating seed is undoubtedly a relevant model for exploring the precise mechanisms of translational selectivity.
Supplementary Material
Footnotes
Author contributions: D.J. and L.R. designed research; R.H., G.C., and L.R. performed research; M.G., G.C., and L.R. contributed new reagents or analytic tools; M.G., R.H., E.A., and L.R. analyzed data; M.G., E.A., D.J., and L.R. wrote the paper.
* The French Ministry of Education and Research funded the Ph.D. fellowship awarded to E.A. The post-doctoral fellowships awarded to M.G. and E.A. were funded by the French Ministry of Industry (FUI, NUTRICE) and the European Commission's Seventh Framework Programme (KBBE, EcoSeed), respectively.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- HAI
- hours after imbibition
- 2DE
- two-dimensional gel electrophoresis
- AdoMet
- S-adenosylmethionine
- AGI
- Arabidopsis Gene Index
- ENO
- enolase
- ER
- endoplasmic reticulum
- FER
- ferritin
- GABA
- γ-aminobutyrate
- GLYR
- glyoxylate reductase
- Ler
- Landsberg erecta
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- MDAR
- monodehydroascorbate reductase
- PBP
- PYK10-binding protein
- PDI
- protein disulfide isomerase
- ROS
- reactive oxygen species
- SSP
- seed storage protein
- TCA
- tricarboxylic acid
- THF
- tetrahydrofolate
- TPP
- tripeptidyl peptidase.
REFERENCES
- 1. Finch-Savage W. E. (1995) Influence of seed quality on crop establishment, growth and yield. In Seed Quality: Basic Mechanisms and Agricultural Implications (Basra A. S., ed.), pp. 361–384, Food Product Press, New York [Google Scholar]
- 2. Donohue K., de Casas R. R., Burghardt L., Kovach K., Willis C. G. (2010) Germination, postgermination adaptation, and species ecological ranges. Annu. Rev. Ecol. Evol. Syst. 41, 293–319 [Google Scholar]
- 3. Holdsworth M. J., Finch-Savage W. E., Grappin P., Job D. (2008) Post-genomics dissection of seed dormancy and germination. Trends Plant Sci. 13, 7–13 [DOI] [PubMed] [Google Scholar]
- 4. North H., Baud S., Debeaujon I., Dubos C., Dubreucq B., Grappin P., Jullien M., Lepiniec L., Marion-Poll A., Miquel M., Rajjou L., Routaboul J. M., Caboche M. (2010) Arabidopsis seed secrets unravelled after a decade of genetic and omics-driven research. Plant J. 61, 971–981 [DOI] [PubMed] [Google Scholar]
- 5. Ligterink W., Joosen R. V. L., Hilhorst H. W. M. (2012) Unravelling the complex trait of seed quality: using natural variation through a combination of physiology, genetics and -omics technologies. Seed Sci. Res. 22, S45–S52 [Google Scholar]
- 6. Rajjou L., Duval M., Gallardo K., Catusse J., Bally J., Job C., Job D. (2012) Seed germination and vigor. Annu. Rev. Plant Biol. 63, 507–533 [DOI] [PubMed] [Google Scholar]
- 7. Bewley J. D. (1997) Seed germination and dormancy. Plant Cell. 9, 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sliwinska E., Bassel G. W., Bewley J. D. (2009) Germination of Arabidopsis thaliana seeds is not completed as a result of elongation of the radicle but of the adjacent transition zone and lower hypocotyl. J. Exp. Bot. 60, 3587–3594 [DOI] [PubMed] [Google Scholar]
- 9. Clerkx E. J., El-Lithy M. E., Vierling E., Ruys G. J., Blankestijn-De Vries H., Groot S. P., Vreugdenhil D., Koornneef M. (2004) Analysis of natural allelic variation of Arabidopsis seed germination and seed longevity traits between the accessions Landsberg erecta and Shakdara, using a new recombinant inbred line population. Plant Physiol. 135, 432–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Meng P. H., Macquet A., Loudet O., Marion-Poll A., North H. M. (2008) Analysis of natural allelic variation controlling Arabidopsis thaliana seed germinability in response to cold and dark: identification of three major quantitative trait loci. Mol. Plant. 1, 145–154 [DOI] [PubMed] [Google Scholar]
- 11. Galpaz N., Reymond M. (2010) Natural variation in Arabidopsis thaliana revealed a genetic network controlling germination under salt stress. PLoS One 5, e15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vallejo A. J., Yanovsky M. J., Botto J. F. (2010) Germination variation in Arabidopsis thaliana accessions under moderate osmotic and salt stresses. Ann. Bot. 106, 833–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. DeRose-Wilson L., Gaut B. S. (2011) Mapping salinity tolerance during Arabidopsis thaliana germination and seedling growth. PLoS One 6, e22832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bassel G. W., Glaab E., Marquez J., Holdsworth M. J., Bacardit J. (2011) Functional network construction in Arabidopsis using rule-based machine learning on large-scale data sets. Plant Cell. 23, 3101–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nakabayashi K., Okamoto M., Koshiba T., Kamiya Y., Nambara E. (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J. 41, 697–709 [DOI] [PubMed] [Google Scholar]
- 16. Kimura M., Nambara E. (2010) Stored and neosynthesized mRNA in Arabidopsis seeds: effects of cycloheximide and controlled deterioration treatment on the resumption of transcription during imbibition. Plant Mol. Biol. 73, 119–129 [DOI] [PubMed] [Google Scholar]
- 17. Okamoto M., Tatematsu K., Matsui A., Morosawa T., Ishida J., Tanaka M., Endo T. A., Mochizuki Y., Toyoda T., Kamiya Y., Shinozaki K., Nambara E., Seki M. (2010) Genome-wide analysis of endogenous abscisic acid-mediated transcription in dry and imbibed seeds of Arabidopsis using tiling arrays. Plant J. 62, 39–51 [DOI] [PubMed] [Google Scholar]
- 18. Narsai R., Law S. R., Carrie C., Xu L., Whelan J. (2011) In-depth temporal transcriptome profiling reveals a crucial developmental switch with roles for RNA processing and organelle metabolism that are essential for germination in Arabidopsis. Plant Physiol. 157, 1342–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dekkers B. J., Pearce S., van Bolderen-Veldkamp R. P., Marshall A., Widera P., Gilbert J., Drost H. G., Bassel G. W., Müller K., King J. R., Wood A. T., Grosse I., Quint M., Krasnogor N., Leubner-Metzger G., Holdsworth M. J., Bentsink L. (2013) Transcriptional dynamics of two seed compartments with opposing roles in Arabidopsis seed germination. Plant Physiol. 163, 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rajjou L., Gallardo K., Debeaujon I., Vandekerckhove J., Job C., Job D. (2004) The effect of alpha-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol. 134, 1598–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sano N., Permana H., Kumada R., Shinozaki Y., Tanabata T., Yamada T., Hirasawa T., Kanekatsu M. (2012) Proteomic analysis of embryonic proteins synthesized from long-lived mRNAs during germination of rice seeds. Plant Cell Physiol. 53, 687–698 [DOI] [PubMed] [Google Scholar]
- 22. Dure L., Waters L. (1965) Long-lived messenger RNA: evidence from cotton seed germination. Science 147, 410–412 [DOI] [PubMed] [Google Scholar]
- 23. Arc E., Chibani K., Grappin P., Jullien M., Godin B., Cueff G., Valot B., Balliau T., Job D., Rajjou L. (2012) Cold stratification and exogenous nitrates entail similar functional proteome adjustments during Arabidopsis seed dormancy release. J. Proteome Res. 11, 5418–5432 [DOI] [PubMed] [Google Scholar]
- 24. Arc E., Galland M., Cueff G., Godin B., Lounifi I., Job D., Rajjou L. (2011) Reboot the system thanks to protein post-translational modifications and proteome diversity: how quiescent seeds restart their metabolism to prepare seedling establishment. Proteomics 11, 1606–1618 [DOI] [PubMed] [Google Scholar]
- 25. Rajjou L., Belghazi M., Huguet R., Robin C., Moreau A., Job C., Job D. (2006) Proteomic investigation of the effect of salicylic acid on Arabidopsis seed germination and establishment of early defense mechanisms. Plant Physiol. 141, 910–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rajjou L., Lovigny Y., Groot S. P., Belghazi M., Job C., Job D. (2008) Proteome-wide characterization of seed aging in Arabidopsis: a comparison between artificial and natural aging protocols. Plant Physiol. 148, 620–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cuming A. C., Lane B. G. (1979) Protein synthesis in imbibing wheat embryos. Eur. J. Biochem. 99, 217–224 [DOI] [PubMed] [Google Scholar]
- 28. Walthall E. D., Brady T. (1986) The effect of the suspensor and gibberellic acid on Phaseolus vulgaris embryo protein synthesis. Cell Differ. 18, 37–44 [DOI] [PubMed] [Google Scholar]
- 29. Beltrán-Peña E., Ortíz-López A., Sánchez de Jiménez E. (1995) Synthesis of ribosomal proteins from stored mRNAs early in seed germination. Plant Mol. Biol. 28, 327–336 [DOI] [PubMed] [Google Scholar]
- 30. Ravanel S., Gakière B., Job D., Douce R. (1998) The specific features of methionine biosynthesis and metabolism in plants. Proc. Natl. Acad. Sci. U.S.A. 95, 7805–7812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maimann S., Wagner C., Kreft O., Zeh M., Willmitzer L., Höfgen R., Hesse H. (2000) Transgenic potato plants reveal the indispensable role of cystathionine beta-lyase in plant growth and development. Plant J. 23, 747–758 [DOI] [PubMed] [Google Scholar]
- 32. Kim J., Leustek T. (2000) Repression of cystathionine gamma-synthase in Arabidopsis thaliana produces partial methionine auxotrophy and developmental abnormalities. Plant Sci. 151, 9–18 [Google Scholar]
- 33. Gakière B., Ravanel S., Droux M., Douce R., Job D. (2000) Mechanisms to account for maintenance of the soluble methionine pool in transgenic Arabidopsis plants expressing antisense cystathionine gamma-synthase cDNA. C. R. Acad. Sci. III 323, 841–851 [DOI] [PubMed] [Google Scholar]
- 34. Gallardo K., Job C., Groot S. P., Puype M., Demol H., Vandekerckhove J., Job D. (2002) Importance of methionine biosynthesis for Arabidopsis seed germination and seedling growth. Physiol. Plant. 116, 238–247 [DOI] [PubMed] [Google Scholar]
- 35. Rajjou L., Belghazi M., Catusse J., Oge L., Arc E., Godin B., Chibani K., Ali-Rachedi S., Collet B., Grappin P., Jullien M., Gallardo K., Job C., Job D. (2011) Proteomics and posttranslational proteomics of seed dormancy and germination. Methods Mol. Biol. 773, 215–236 [DOI] [PubMed] [Google Scholar]
- 36. Dietrich J., Teissere M., Job C., Job D. (1985) Poly(dAT) dependent trinucleotide synthesis catalysed by wheat germ RNA polymerase II. Effects of nucleotide substrates and cordycepin triphosphate. Nucleic Acids Res. 13, 6155–6170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saeed A. I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., Sturn A., Snuffin M., Rezantsev A., Popov D., Ryltsov A., Kostukovich E., Borisovsky I., Liu Z., Vinsavich A., Trush V., Quackenbush J. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34, 374–378 [DOI] [PubMed] [Google Scholar]
- 38. Ruepp A., Zollner A., Maier D., Albermann K., Hani J., Mokrejs M., Tetko I., Guldener U., Mannhaupt G., Munsterkotter M., Mewes H. W. (2004) The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 32, 5539–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gautier L., Cope L., Bolstad B. M., Irizarry R. A. (2004) Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315 [DOI] [PubMed] [Google Scholar]
- 40. R Development Core Team (2012) R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 41. Galland M., Job D., Rajjou L. (2012) The seed proteome web portal. Front. Plant Sci. 3, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tolleter D., Jaquinod M., Mangavel C., Passirani C., Saulnier P., Manon S., Teyssier E., Payet N., Avelange-Macherel M. H., Macherel D. (2007) Structure and function of a mitochondrial late embryogenesis abundant protein are revealed by desiccation. Plant Cell. 19, 1580–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Olvera-Carrillo Y., Campos F., Reyes J. L., Garciarrubio A., Covarrubias A. A. (2010) Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis. Plant Physiol. 154, 373–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao P., Liu F., Ma M., Gong J., Wang Q., Jia P., Zheng G., Liu H. (2011) Overexpression of AtLEA3–3 confers resistance to cold stress in Escherichia coli and provides enhanced osmotic stress tolerance and ABA sensitivity in Arabidopsis thaliana. Mol. Biol. 45, 851–862 [PubMed] [Google Scholar]
- 45. Duan J., Cai W. (2012) OsLEA3–2, an abiotic stress induced gene of rice plays a key role in salt and drought tolerance. PLoS One 7, e45117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fujiwara T., Nambara E., Yamagishi K., Gotoc D. B., Naito S. (2002) Storage proteins. In The Arabidopsis Book (Somerville C. R., Meyerowitz E. M., eds.), pp. 1–12, American Society of Plant Biologists, Rockville, MD: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pang P. P., Pruitt R. E., Meyerowitz E. M. (1988) Molecular cloning, genomic organization, expression and evolution of 12S seed storage protein genes of Arabidopsis thaliana. Plant Mol. Biol. 11, 805–820 [DOI] [PubMed] [Google Scholar]
- 48. Gruis D., Schulze J., Jung R. (2004) Storage protein accumulation in the absence of the vacuolar processing enzyme family of cysteine proteases. Plant Cell. 16, 270–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kumamaru T., Uemura Y., Inoue Y., Takemoto Y., Siddiqui S. U., Ogawa M., Hara-Nishimura I., Satoh H. (2010) Vacuolar processing enzyme plays an essential role in the crystalline structure of glutelin in rice seed. Plant Cell Physiol. 51, 38–46 [DOI] [PubMed] [Google Scholar]
- 50. Job C., Rajjou L., Lovigny Y., Belghazi M., Job D. (2005) Patterns of protein oxidation in Arabidopsis seeds and during germination. Plant Physiol. 138, 790–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bailly C., El-Maarouf-Bouteau H., Corbineau F. (2008) From intracellular signaling networks to cell death: the dual role of reactive oxygen species in seed physiology. C. R. Biol. 331, 806–814 [DOI] [PubMed] [Google Scholar]
- 52. Lopez-Molina L., Mongrand S., Chua N. H. (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 98, 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chou I. T., Gasser C. S. (1997) Characterization of the cyclophilin gene family of Arabidopsis thaliana and phylogenetic analysis of known cyclophilin proteins. Plant Mol. Biol. 35, 873–892 [DOI] [PubMed] [Google Scholar]
- 54. Ma X., Song L., Yang Y., Liu D. (2013) A gain-of-function mutation in the ROC1 gene alters plant architecture in Arabidopsis. New Phytol. 197, 751–762 [DOI] [PubMed] [Google Scholar]
- 55. Trupkin S. A., Mora-Garcia S., Casal J. J. (2012) The cyclophilin ROC1 links phytochrome and cryptochrome to brassinosteroid sensitivity. Plant J. 71, 712–723 [DOI] [PubMed] [Google Scholar]
- 56. Ge W., Song Y., Zhang C., Zhang Y., Burlingame A. L., Guo Y. (2011) Proteomic analyses of apoplastic proteins from germinating Arabidopsis thaliana pollen. Biochim. Biophys. Acta 1814, 1964–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rautengarten C., Usadel B., Neumetzler L., Hartmann J., Bussis D., Altmann T. (2008) A subtilisin-like serine protease essential for mucilage release from Arabidopsis seed coats. Plant J. 54, 466–480 [DOI] [PubMed] [Google Scholar]
- 58. Penfield S., Meissner R. C., Shoue D. A., Carpita N. C., Bevan M. W. (2001) MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell. 13, 2777–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang X., Zhang W., Dong M., Boubriak I., Huang Z. (2011) The achene mucilage hydrated in desert dew assists seed cells in maintaining DNA integrity: adaptive strategy of desert plant Artemisia sphaerocephala. PLoS One 6, e24346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yang X., Baskin C. C., Baskin J. M., Zhang W., Huang Z. (2012) Degradation of seed mucilage by soil microflora promotes early seedling growth of a desert sand dune plant. Plant Cell Environ. 35, 872–883 [DOI] [PubMed] [Google Scholar]
- 61. Weitbrecht K., Muller K., Leubner-Metzger G. (2011) First off the mark: early seed germination. J. Exp. Bot. 62, 3289–3309 [DOI] [PubMed] [Google Scholar]
- 62. Jiang T., Zhang X. F., Wang X. F., Zhang D. P. (2011) Arabidopsis 3-ketoacyl-CoA thiolase-2 (KAT2), an enzyme of fatty acid beta-oxidation, is involved in ABA signal transduction. Plant Cell Physiol. 52, 528–538 [DOI] [PubMed] [Google Scholar]
- 63. Rylott E. L., Gilday A. D., Graham I. A. (2003) The gluconeogenic enzyme phosphoenolpyruvate carboxykinase in Arabidopsis is essential for seedling establishment. Plant Physiol. 131, 1834–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Penfield S., Rylott E. L., Gilday A. D., Graham S., Larson T. R., Graham I. A. (2004) Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid, and requires phosphoenolpyruvate carboxykinase 1. Plant Cell. 16, 2705–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Andriotis V. M., Kruger N. J., Pike M. J., Smith A. M. (2010) Plastidial glycolysis in developing Arabidopsis embryos. New Phytol. 185, 649–662 [DOI] [PubMed] [Google Scholar]
- 66. Prabhakar V., Lottgert T., Gigolashvili T., Bell K., Flugge U. I., Hausler R. E. (2009) Molecular and functional characterization of the plastid-localized phosphoenolpyruvate enolase (ENO1) from Arabidopsis thaliana. FEBS Lett. 583, 983–991 [DOI] [PubMed] [Google Scholar]
- 67. Fait A., Angelovici R., Less H., Ohad I., Urbanczyk-Wochniak E., Fernie A. R., Galili G. (2006) Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol. 142, 839–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lee H., Guo Y., Ohta M., Xiong L., Stevenson B., Zhu J. K. (2002) LOS2, a genetic locus required for cold-responsive gene transcription encodes a bi-functional enolase. EMBO J. 21, 2692–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sakamoto H., Maruyama K., Sakuma Y., Meshi T., Iwabuchi M., Shinozaki K., Yamaguchi-Shinozaki K. (2004) Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 136, 2734–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mittler R., Kim Y., Song L., Coutu J., Coutu A., Ciftci-Yilmaz S., Lee H., Stevenson B., Zhu J. K. (2006) Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 580, 6537–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Pauwels L., Morreel K., De Witte E., Lammertyn F., Van Montagu M., Boerjan W., Inze D., Goossens A. (2008) Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. U.S.A. 105, 1380–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Winter D., Vinegar B., Nahal H., Ammar R., Wilson G. V., Provart N. J. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Davletova S., Schlauch K., Coutu J., Mittler R. (2005) The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol. 139, 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Leymarie J., Vitkauskaite G., Hoang H. H., Gendreau E., Chazoule V., Meimoun P., Corbineau F., El-Maarouf-Bouteau H., Bailly C. (2012) Role of reactive oxygen species in the regulation of Arabidopsis seed dormancy. Plant Cell Physiol. 53, 96–106 [DOI] [PubMed] [Google Scholar]
- 75. Ravet K., Touraine B., Kim S. A., Cellier F., Thomine S., Guerinot M. L., Briat J. F., Gaymard F. (2009) Post-translational regulation of AtFER2 ferritin in response to intracellular iron trafficking during fruit development in Arabidopsis. Mol. Plant. 2, 1095–1106 [DOI] [PubMed] [Google Scholar]
- 76. Ravet K., Touraine B., Boucherez J., Briat J. F., Gaymard F., Cellier F. (2009) Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. Plant J. 57, 400–412 [DOI] [PubMed] [Google Scholar]
- 77. Eisenstein R. S., Munro H. N. (1990) Translational regulation of ferritin synthesis by iron. Enzyme. 44, 42–58 [DOI] [PubMed] [Google Scholar]
- 78. Bailly C., Leymarie J., Lehner A., Rousseau S., Come D., Corbineau F. (2004) Catalase activity and expression in developing sunflower seeds as related to drying. J. Exp. Bot. 55, 475–483 [DOI] [PubMed] [Google Scholar]
- 79. Eastmond P. J. (2007) Monodehyroascorbate reductase 4 is required for seed storage oil hydrolysis and postgerminative growth in Arabidopsis. Plant Cell. 19, 1376–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Obara K., Sumi K., Fukuda H. (2002) The use of multiple transcription starts causes the dual targeting of Arabidopsis putative monodehydroascorbate reductase to both mitochondria and chloroplasts. Plant Cell Physiol. 43, 697–705 [DOI] [PubMed] [Google Scholar]
- 81. Ishikawa T., Casini A. F., Nishikimi M. (1998) Molecular cloning and functional expression of rat liver glutathione-dependent dehydroascorbate reductase. J. Biol. Chem. 273, 28708–28712 [DOI] [PubMed] [Google Scholar]
- 82. Tommasi F., Paciolla C., Arrigoni O. (1999) The ascorbate system in recalcitrant and orthodox seeds. Physiol. Plant. 105, 193–198 [Google Scholar]
- 83. Arrigoni O., De Tullio M. C. (2002) Ascorbic acid: much more than just an antioxidant. Biochim. Biophys. Acta 1569, 1–9 [DOI] [PubMed] [Google Scholar]
- 84. Ye N., Zhu G., Liu Y., Zhang A., Li Y., Liu R., Shi L., Jia L., Zhang J. (2012) Ascorbic acid and reactive oxygen species are involved in the inhibition of seed germination by abscisic acid in rice seeds. J. Exp. Bot. 63, 1809–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Linster C. L., Clarke S. G. (2008) L-Ascorbate biosynthesis in higher plants: the role of VTC2. Trends Plant Sci. 13, 567–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Andème-Ondzighi C., Christopher D. A., Cho E. J., Chang S. C., Staehelin L. A. (2008) Arabidopsis protein disulfide isomerase-5 inhibits cysteine proteases during trafficking to vacuoles before programmed cell death of the endothelium in developing seeds. Plant Cell. 20, 2205–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Onda Y., Nagamine A., Sakurai M., Kumamaru T., Ogawa M., Kawagoe Y. (2011) Distinct roles of protein disulfide isomerase and P5 sulfhydryl oxidoreductases in multiple pathways for oxidation of structurally diverse storage proteins in rice. Plant Cell. 23, 210–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Takemoto Y., Coughlan S. J., Okita T. W., Satoh H., Ogawa M., Kumamaru T. (2002) The rice mutant esp2 greatly accumulates the glutelin precursor and deletes the protein disulfide isomerase. Plant Physiol. 128, 1212–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Cho E. J., Yuen C. Y., Kang B. H., Ondzighi C. A., Staehelin L. A., Christopher D. A. (2011) Protein disulfide isomerase-2 of Arabidopsis mediates protein folding and localizes to both the secretory pathway and nucleus, where it interacts with maternal effect embryo arrest factor. Mol. Cells 32, 459–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Pagnussat G. C., Yu H. J., Ngo Q. A., Rajani S., Mayalagu S., Johnson C. S., Capron A., Xie L. F., Ye D., Sundaresan V. (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132, 603–614 [DOI] [PubMed] [Google Scholar]
- 91. Limami A. M., Rouillon C., Glevarec G., Gallais A., Hirel B. (2002) Genetic and physiological analysis of germination efficiency in maize in relation to nitrogen metabolism reveals the importance of cytosolic glutamine synthetase. Plant Physiol. 130, 1860–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Tomkinson B., Lindas A. C. (2005) Tripeptidyl-peptidase II: a multi-purpose peptidase. Int. J. Biochem. Cell Biol. 37, 1933–1937 [DOI] [PubMed] [Google Scholar]
- 93. Polge C., Jaquinod M., Holzer F., Bourguignon J., Walling L., Brouquisse R. (2009) Evidence for the existence in Arabidopsis thaliana of the proteasome proteolytic pathway: activation in response to cadmium. J. Biol. Chem. 284, 35412–35424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Pickering A. M., Koop A. L., Teoh C. Y., Ermak G., Grune T., Davies K. J. (2010) The immunoproteasome, the 20S proteasome and the PA28αβ proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem. J. 432, 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Droux M., Ruffet M. L., Douce R., Job D. (1998) Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants: structural and kinetic properties of the free and bound enzymes. Eur. J. Biochem. 255, 235–245 [DOI] [PubMed] [Google Scholar]
- 96. Heeg C., Kruse C., Jost R., Gutensohn M., Ruppert T., Wirtz M., Hell R. (2008) Analysis of the Arabidopsis O-acetylserine(thiol)lyase gene family demonstrates compartment-specific differences in the regulation of cysteine synthesis. Plant Cell. 20, 168–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Gakière B., Job D., Douce R., Ravanel S. (1999) Characterization of the cDNA and gene for a cytosolic cobalamin-independent methionine synthase in Arabidopsis thaliana (Accession No. U97200). Plant Physiol. 120, 1206 [Google Scholar]
- 98. Li W., Han Y., Tao F., Chong K. (2011) Knockdown of SAMS genes encoding S-adenosyl-L-methionine synthetases causes methylation alterations of DNAs and histones and leads to late flowering in rice. J. Plant Physiol. 168, 1837–1843 [DOI] [PubMed] [Google Scholar]
- 99. Rocha P. S., Sheikh M., Melchiorre R., Fagard M., Boutet S., Loach R., Moffatt B., Wagner C., Vaucheret H., Furner I. (2005) The Arabidopsis homology-dependent gene silencing1 gene codes for an S-adenosyl-L-homocysteine hydrolase required for DNA methylation-dependent gene silencing. Plant Cell. 17, 404–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Jabrin S., Ravanel S., Gambonnet B., Douce R., Rebeillé F. (2003) One-carbon metabolism in plants. Regulation of tetrahydrofolate synthesis during germination and seedling development. Plant Physiol. 131, 1431–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Li R., Moore M., King J. (2003) Investigating the regulation of one-carbon metabolism in Arabidopsis thaliana. Plant Cell Physiol. 44, 233–241 [DOI] [PubMed] [Google Scholar]
- 102. Brown P. D., Tokuhisa J. G., Reichelt M., Gershenzon J. (2003) Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62, 471–481 [DOI] [PubMed] [Google Scholar]
- 103. Petersen B. L., Chen S., Hansen C. H., Olsen C. E., Halkier B. A. (2002) Composition and content of glucosinolates in developing Arabidopsis thaliana. Planta 214, 562–571 [DOI] [PubMed] [Google Scholar]
- 104. Nitz I., Berkefeld H., Puzio P. S., Grundler F. M. (2001) Pyk10, a seedling and root specific gene and promoter from Arabidopsis thaliana. Plant Sci. 161, 337–346 [DOI] [PubMed] [Google Scholar]
- 105. Matsushima R., Fukao Y., Nishimura M., Hara-Nishimura I. (2004) NAI1 gene encodes a basic-helix-loop-helix-type putative transcription factor that regulates the formation of an endoplasmic reticulum-derived structure, the ER body. Plant Cell. 16, 1536–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kissen R., Bones A. M. (2009) Nitrile-specifier proteins involved in glucosinolate hydrolysis in Arabidopsis thaliana. J. Biol. Chem. 284, 12057–12070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Janowitz T., Trompetter I., Piotrowski M. (2009) Evolution of nitrilases in glucosinolate-containing plants. Phytochemistry 70, 1680–1686 [DOI] [PubMed] [Google Scholar]
- 108. Zhang J., Sun X., Zhang Z., Ni Y., Zhang Q., Liang X., Xiao H., Chen J., Tokuhisa J. G. (2011) Metabolite profiling of Arabidopsis seedlings in response to exogenous sinalbin and sulfur deficiency. Phytochemistry 72, 1767–1778 [DOI] [PubMed] [Google Scholar]
- 109. Matsushima R., Kondo M., Nishimura M., Hara-Nishimura I. (2003) A novel ER-derived compartment, the ER body, selectively accumulates a beta-glucosidase with an ER-retention signal in Arabidopsis. Plant J. 33, 493–502 [DOI] [PubMed] [Google Scholar]
- 110. Yang H., Czapla T. H. (1993) Isolation and characterization of cDNA clones encoding jacalin isolectins. J. Biol. Chem. 268, 5905–5910 [PubMed] [Google Scholar]
- 111. Yamada K., Nagano A. J., Nishina M., Hara-Nishimura I., Nishimura M. (2008) NAI2 is an endoplasmic reticulum body component that enables ER body formation in Arabidopsis thaliana. Plant Cell. 20, 2529–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Gilliland L. U., Kandasamy M. K., Pawloski L. C., Meagher R. B. (2002) Both vegetative and reproductive actin isovariants complement the stunted root hair phenotype of the Arabidopsis act2–1 mutation. Plant Physiol. 130, 2199–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Gilliland L. U., Pawloski L. C., Kandasamy M. K., Meagher R. B. (2003) Arabidopsis actin gene ACT7 plays an essential role in germination and root growth. Plant J. 33, 319–328 [DOI] [PubMed] [Google Scholar]
- 114. de Castro R. D., van Lammeren A. A., Groot S. P., Bino R. J., Hilhorst H. W. (2000) Cell division and subsequent radicle protrusion in tomato seeds are inhibited by osmotic stress but DNA synthesis and formation of microtubular cytoskeleton are not. Plant Physiol. 122, 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Gallardo K., Job C., Groot S. P., Puype M., Demol H., Vandekerckhove J., Job D. (2001) Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiol. 126, 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Barroco R. M., Van Poucke K., Bergervoet J. H., De Veylder L., Groot S. P., Inze D., Engler G. (2005) The role of the cell cycle machinery in resumption of postembryonic development. Plant Physiol. 137, 127–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. de los Reyes B. G., Myers S. J., McGrath J. M. (2003) Differential induction of glyoxylate cycle enzymes by stress as a marker for seedling vigor in sugar beet (Beta vulgaris). Mol. Genet. Genomics 269, 692–698 [DOI] [PubMed] [Google Scholar]
- 118. Graham I. A. (2008) Seed storage oil mobilization. Annu. Rev. Plant Biol. 59, 115–142 [DOI] [PubMed] [Google Scholar]
- 119. Catusse J., Meinhard J., Job C., Strub J. M., Fischer U., Pestsova E., Westhoff P., Van Dorsselaer A., Job D. (2011) Proteomics reveals potential biomarkers of seed vigor in sugarbeet. Proteomics 11, 1569–1580 [DOI] [PubMed] [Google Scholar]
- 120. Simpson J. P., Di Leo R., Dhanoa P. K., Allan W. L., Makhmoudova A., Clark S. M., Hoover G. J., Mullen R. T., Shelp B. J. (2008) Identification and characterization of a plastid-localized Arabidopsis glyoxylate reductase isoform: comparison with a cytosolic isoform and implications for cellular redox homeostasis and aldehyde detoxification. J. Exp. Bot. 59, 2545–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Allan W. L., Clark S. M., Hoover G. J., Shelp B. J. (2009) Role of plant glyoxylate reductases during stress: a hypothesis. Biochem. J. 423, 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Ching S. L., Gidda S. K., Rochon A., van Cauwenberghe O. R., Shelp B. J., Mullen R. T. (2012) Glyoxylate reductase isoform 1 is localized in the cytosol and not peroxisomes in plant cells. J. Integr. Plant Biol. 54, 152–168 [DOI] [PubMed] [Google Scholar]
- 123. Michaeli S., Fait A., Lagor K., Nunes-Nesi A., Grillich N., Yellin A., Bar D., Khan M., Fernie A. R., Turano F. J., Fromm H. (2011) A mitochondrial GABA permease connects the GABA shunt and the TCA cycle, and is essential for normal carbon metabolism. Plant J. 67, 485–498 [DOI] [PubMed] [Google Scholar]
- 124. Muench D. G., Zhang C., Dahodwala M. (2012) Control of cytoplasmic translation in plants. Wiley Interdiscip. Rev. RNA 3, 178–194 [DOI] [PubMed] [Google Scholar]
- 125. Horiguchi G., Van Lijsebettens M., Candela H., Micol J. L., Tsukaya H. (2012) Ribosomes and translation in plant developmental control. Plant Sci. 191–192, 24–34 [DOI] [PubMed] [Google Scholar]
- 126. Takai K., Endo Y. (2010) The cell-free protein synthesis system from wheat germ. Methods Mol. Biol. 607, 23–30 [DOI] [PubMed] [Google Scholar]
- 127. Bärenfaller K., Grossmann J., Grobei M. A., Hull R., Hirsch-Hoffmann M., Yalovsky S., Zimmermann P., Grossniklaus U., Gruissem W., Baginsky S. (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320, 938–941 [DOI] [PubMed] [Google Scholar]
- 128. Piques M., Schulze W. X., Höhne M., Usadel B., Gibon Y., Rohwer J., Stitt M. (2009) Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Mol. Syst. Biol. 5, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 130. Hugouvieux V., Kwak J. M., Schroeder J. I. (2001) An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106, 477–487 [DOI] [PubMed] [Google Scholar]
- 131. Dinkova T. D., Márquez-Velázquez N. A., Aguilar R., Lázaro-Mixteco P. E., Sánchez de Jiménez E. (2011) Tight translational control by the initiation factors eIF4E and eIF(iso)4E is required for maize seed germination. Seed Sci. Res. 21, 85–93 [Google Scholar]
- 132. Yang M., Zhang B., Jia J., Yan C., Habaike A., Han Y. (2013) RRP41L, a putative core subunit of the exosome, plays an important role in seed germination and early seedling growth in Arabidopsis. Plant Physiol. 161, 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Liu P. P., Montgomery T. A., Fahlgren N., Kasschau K. D., Nonogaki H., Carrington J. C. (2007) Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 52, 133–146 [DOI] [PubMed] [Google Scholar]
- 134. Kim J. Y., Lee H. J., Jung H. J., Maruyama K., Suzuki N., Kang H. (2010) Overexpression of microRNA395c or 395e affects differently the seed germination of Arabidopsis thaliana under stress conditions. Planta 232, 1447–1454 [DOI] [PubMed] [Google Scholar]
- 135. Alonso-Peral M. M., Sun C., Millar A. A. (2012) MicroRNA159 can act as a switch or tuning microRNA independently of its abundance in Arabidopsis. PLoS One 7, e34751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Bazin J., Langlade N., Vincourt P., Arribat S., Balzergue S., El-Maarouf-Bouteau H., Bailly C. (2011) Targeted mRNA oxidation regulates sunflower seed dormancy alleviation during dry after-ripening. Plant Cell. 23, 2196–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Li L., Nelson C. J., Solheim C., Whelan J., Millar A. H. (2012) Determining degradation and synthesis rates of arabidopsis proteins using the kinetics of progressive 15N labeling of two-dimensional gel-separated protein spots. Mol. Cell. Proteomics 11, M111.010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Schütz W., Hausmann N., Krug K., Hampp R., Macek B. (2011) Extending SILAC to proteomics of plant cell lines. Plant Cell. 23, 1701–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.