Abstract
White spot syndrome virus (WSSV) is currently the most serious global threat for cultured shrimp production. Although its large, double-stranded DNA genome has been completely characterized, most putative protein functions remain obscure. To provide more informative knowledge about this virus, a proteomic-scale network of WSSV-WSSV protein interactions was carried out using a comprehensive yeast two-hybrid analysis. An array of yeast transformants containing each WSSV open reading frame fused with GAL4 DNA binding domain and GAL4 activation domain was constructed yielding 187 bait and 182 prey constructs, respectively. On screening of ∼28,000 pairwise combinations, 710 interactions were obtained from 143 baits. An independent coimmunoprecipitation assay (co-IP) was performed to validate the selected protein interaction pairs identified from the yeast two-hybrid approach. The program Cytoscape was employed to create a WSSV protein–protein interaction (PPI) network. The topology of the WSSV PPI network was based on the Barabási-Albert model and consisted of a scale-free network that resembled other established viral protein interaction networks. Using the RNA interference approach, knocking down either of two candidate hub proteins gave shrimp more protection against WSSV than knocking down a nonhub gene. The WSSV protein interaction map established in this study provides novel guidance for further studies on shrimp viral pathogenesis, host-viral protein interaction and potential targets for therapeutic and preventative antiviral strategies in shrimp aquaculture.
White spot syndrome virus (WSSV)1 is the causative agent of white spot disease (WSD) and is one of the most serious viral pathogens that threaten the shrimp culture industry worldwide. Because WSD causes rapid and high mortality up to 100% within 3–10 days after viral infection (1), it causes dramatic economic losses on farms. WSSV is a large enveloped, ovoid to bacilliform, double-stranded DNA (dsDNA) virus with a genome of ∼300 kb (See reviews in (2, 3)). The WSSV genome has been completely characterized for isolates from Thailand (GenBank accession number AF369029), China (accession number AF332093) and Taiwan (accession number AF440570). To expand its basic genetic information, various genomic and proteomic approaches have been applied to gain more insight into the molecular mechanisms of WSSV pathogenesis (See reviews in (2, 3)). However, the roles of most of the WSSV proteins still remain to be elucidated. This is due to the fact that many of its putative open reading frames (ORFs) lack homology to known proteins in the database. Protein–protein interaction studies can provide a valuable framework for understanding the roles of protein functions. Interaction studies of WSSV proteins have particularly focused on viral structural proteins (4–15). However, so far there has been no report on a protein–protein interaction (PPI) network for WSSV or any other crustacean virus. By contrast, several PPI networks for cellular organisms such as Saccharomyces cerevisiae (16, 17), Helicobacter pylori (18), Drosophila melanogaster (19), Caenarhabitis elegans (20), Plasmodium falciparum (21), and Homo sapiens (22, 23) and pathogens such as bacteriophage T7 (24), vaccinia virus (25), hepatitis C virus (26), and herpesviruses (27–29) have already been established. Therefore, the present study aimed to obtain a more fundamental understanding of WSSV protein interactions. A comprehensive yeast two-hybrid assay was employed to generate viral fusion proteins with DNA binding (BD) and activation (AD) domains in an array format that effectively allowed searching every possible binary interaction in WSSV. The interaction results from the yeast two-hybrid assays were subsequently validated by coimmunoprecipitation (co-IP). Topological properties of the WSSV PPI network were assessed and compared with previously published viral networks. Candidate viral hub proteins with high numbers of interacting partners were identified in this study and their significance was investigated using an RNA interference approach.
EXPERIMENTAL PROCEDURES
WSSV and Shrimp
The WSSV Taiwan isolate (WSSV-TW) originally isolated from a batch of WSSV-infected Penaeus monodon collected in Taiwan in 1994 (30, 31) was used as the template for WSSV ORFs amplification. To propagate the virus and use it in experimental challenges, a crude WSSV stock was prepared from moribund P. vannamei infected with WSSV-TW strain. Gill and stomach tissues were homogenized in TNE buffer (20 mm Tris-Hcl, pH 7.4, 400 mm NaCl, 1 mm EDTA) and clarified by centrifugation at 1500 × g for 10 min at 4 °C. The supernatant was then diluted in PBS at dilution of 1:100, filtered through 0.45 μm membrane filter and stored at −80 °C as a virus stock. SPF P. vannamei (5–6 g body weight) were kindly provided by Charoen Pokphand Company. The animals were acclimatized in a wet laboratory in 1000 L aquaria containing continuously aerated artificial seawater at 15 ppt and 26–28 °C for 2–3 days before experiments began.
Modification of pGBKT7 Plasmid
To enable simple homologous recombination, pGBKT7 or BD plasmid (Clontech) was modified to contain the multiplex cloning site (MCS) similar to pGADT7 or AD plasmid (Clontech). This was achieved by digestion of pGADT7 with NdeI and BamHI enzymes and use of the 48-bp released fragment for insertion into NdeI-BamHI digested pGBKT7. The sequence of the resulting plasmid named pGBKT7–1 was verified by DNA sequencing (Macrogen Co. Ltd., South Korea).
Selection and Amplification of WSSV Open Reading Frames (ORFs)
To construct WSSV protein arrays, criteria in selecting WSSV ORFs were as follows. First, all 180 ORF that encoding putative functional proteins predicted in the China isolate (GenBank accession number AF332093) (32) were chosen. All of these were also found in the Taiwan isolate (GenBank accession number AF440570). To enhance the integrity of the library, 22 predicted ORFs that appear in both the Thailand (GenBank accession number AF369029) and Taiwan isolates but are missing in the China isolate were included. Thus, a total of 202 putative ORFs were chosen for inclusion in this study. Among these, large ORFs with nucleotide sequences longer than 3135 bp were divided into sub-fragments to increase the success for PCR amplification and for protein expression in yeast cells. Thus, the total number of WSSV fragments required to be amplified was 246 (202 plus 44 fragments). Primers were designed in two sets to be used for two rounds of PCR amplification. The first primer set was designed based on the WSSV genome sequences to bind specifically to the selected fragments. Additionally, both upstream and downstream primers contained common sequences of 15–17 nucleotides that facilitated binding with the second primer. The second primer set (including BD and AD primer sub-sets) was designed to contain additional 34–35 nucleotide sequences that were homologous to sequences used in the yeast two-hybrid vectors. The primer set designs are shown in supplemental Table S1 and the sequences of the primers in supplemental Table S2.
A two-step PCR reaction was performed with primer set 1 in the first PCR reaction to amplify each WSSV fragment. In the second reaction WSSV fragments were amplified to contain flanking regions homologous to the yeast two-hybrid vectors, pGBKT7–1 and pGADT7. The first reaction was carried out in 25 μl containing 0.2 μm of each primer, 200 μm of dNTP mix, 1.5 mm of MgCl2, 2.5 U of TaqDNA polymerase (Invitrogen), 1× reaction buffer, and WSSV DNA template. The PCR protocol comprised initial activation at 94 °C for 3 min followed by 30 cycles at 94 °C for 30 s, annealing at 50 °C for 30 s, extension at 72 °C with time depending on the size of the DNA target (1 min per 1 kb) and then final extension at 72 °C for 5 min. After verification of the first PCR products by agarose gel electrophoresis, they were used as templates for the second PCR reaction with primer set 2. Each product was subjected to two more reactions (one with the BD primer set and another with the AD primer set). The second PCR reaction and protocol was the same as in the first reaction, except that the annealing step was at 65 °C instead of 50 °C. Finally, PCR products were purified using a PCR purification kit (Qiagen) before being cloned into pGBKT7–1 and pGADT7 vectors.
Generation of WSSV Protein Arrays
All PCR-generated WSSV DNA fragments were fused with GAL4 DNA binding domain (BD) and activation domain (AD) of pGBKT7–1 and pGADT7, respectively, by natural homologous recombination in yeast. The pGBKT7–1 and pGADT7 were linearlized by EcoRI and SmaI restriction enzymes. For bait construction, each amplified WSSV DNA fragment and linearized pGBKT7–1 vector were co-transformed into yeast Saccharomyces cerevisiae strain Y187 (Clontech). For prey construction, a recombination experiment was carried out using WSSV fragments and linearized pGADT7 in yeast strain AH109 (Clontech). High-throughput transformation was carried out as previously described (33). The experiment was conducted in 96-well plates so that each well contained one WSSV DNA fragment. After transformation, recombinant clones from each well were selected by growing on solid S.D. medium lacking tryptophan (S.D./-Trp) for baits or lacking leucine (S.D./-Leu) for prey selection. The transformants were then verified by being subjected to yeast colony PCR using vector primers (supplemental Table S2). The validated homologous recombinant clones (pools of 3–6 individual clones) were transferred into a 96-well plate containing appropriate selective media and the cultures were grown at 30 °C. The bait and prey constructs in 96-well plates were called as BD arrays or AD arrays, respectively.
High-throughput Screening of Protein Arrays by Yeast Mating
WSSV protein–protein interaction was sought using a yeast mating strategy in which each bait was used to screen for each prey in the AD arrays. Briefly, an individual bait was grown in 5 ml of S.D./-Trp at 30 °C for 24–48 h with shaking and then expanded by inoculation of 2 ml cultures into 30 ml of S.D./-Trp to be grown for a further 24–48 h. After yeast collection and resuspension in 15 ml of YPD containing 10% glucose, 2 μl of bait culture was dropped on the YPD agar containing 10% glucose, and yeast spots were allowed to dry at room temperature. For AD array preparation, 20 μl of each prey cultured in S.D./-Leu was inoculated into 80 μl of YPD containing 10% glucose and then grown at 30 °C overnight. Next, 3 μl of each prey culture was dropped on top of the bait spots and the resulting plate was incubated at 30 °C for 48 h. Diploid cells were then selected by replica-plating onto S.D./-Leu/-Trp followed by incubation for 48 h at 30 °C. To select positive interactions, the diploids were transferred to S.D. medium lacking adenine, histidine, leucine, and tryptophan (S.D./-Ade/-His/-Leu/-Trp) containing 40 μg/ml of X-α-gal followed by incubation at 30 °C for up to 2 weeks. Growth of blue yeast colonies on this selective medium was scored as a positive interaction. This screening for interactions was carried out independently at least twice. All baits used for screening were previously verified not to be auto-activated. This was done by mating all baits with yeast AH109 containing empty pGADT7 vector in the same manner as described above. Baits that grew and turned blue on selective media were not used for protein interaction screens.
Bioinformatic Data Analysis
Homology searches were performed using BLAST program in the NCBI database (http://www.ncbi.nlm.nih.gov/BLAST/). DNA and protein sequence analyses were carried out using the EXPASY server (http://au.expasy.org/). The protein–protein interaction networks were visualized using Cytoscape software (www.cytoscape.org). Network parameters were analyzed by the Cytoscape plug-in called Network analyzer (34).
Coimmunoprecipitation Assays
For validation of WSSV protein interactions from yeast two-hybrid assay, each WSSV gene fragment was cloned into heat inducible vectors pDHsp/V5 and pDHsp/FLAG containing a Drosophila heat shock protein 70 promoter (35) using the primers listed in supplemental Table S2. Additionally, to increase the molecular mass of small ORFs (coding a product less than 10 kDa), such WSSV genes were cloned into pDHsp/EGFP-V5 vector (14) for fusion with an EGFP (enhanced green fluorescent protein) tag at the C-terminal end. The FLAG-tagged and V5-tagged WSSV fusion plasmids were cotransfected pairwise into Sf-9 (Spodoptera frugiperda) cells. Cotransfection and coimmunoprecipitation followed methods previously described (35). The immunoprecipitated complexes were separated by SDS-PAGE before being subjected to immunoblot assay. The V5-tagged fusion proteins were detected with rabbit anti-V5 antibody (Sigma) and goat anti-rabbit IgG-HRP conjugate (Sigma). The FLAG-tagged fusion proteins were detected with mouse anti-FLAG antibody (Sigma) and goat anti-mouse IgG-HRP conjugate (Sigma).
Double-stranded RNA Production
Based on RNAi capacity prediction (36), 207-bp and 282-bp fragments of WSSV051 and WSSV517, respectively, were targeted for double-stranded RNA (dsRNA) production. The respective fragments were amplified by PCR using primer pairs as follows: WSSV051-RNAi-F and WSSV051-RNAi-R and WSSV517-RNAi-F and WSSV517-RNAi-R (Table I). The amplicons obtained were cloned into pDrive vector (Qiagen, Valencia, CA). Recombinant plasmids with opposite orientation of respective fragments were linearized with HindIII restriction enzyme to prepare sense and antisense templates. Single stranded RNAs were then generated by in vitro transcription using T7 RNA polymerase (Promega, Madison, WI). Equivalent amounts of sense and antisense RNA were mixed and annealed by incubation at 37 °C for 1 h. After DNA removal and purification, the dsRNA integrity was monitored in agarose electrophoresis gels and by RNase treatment with appropriate nucleases. The in vitro transcribed dsRNAs were subsequently quantified by spectrophotometer and freshly dissolved in 150 mm NaCl solution before use. Additionally, WSSV050 gene was selected as a nonhub gene and dsRNA-WSSV050 was prepared in the similar manner using specific primers listed in Table I. A nonrelated dsRNA used in control experiments was also designed from infectious myonecrosis virus (IMNV) (37).
Table I. Sequences and names of primers used.
| Names | Sequences (5′ to 3′) | Remarks |
|---|---|---|
| WSSV050-RNAi-F | TTAATTCGCTACTATTAATA | dsRNA production & transcription analysis |
| WSSV050-RNAi-R | TTTTATATCCTCAAGAG | |
| WSSV050-F | ATGCTTATTTTAATTAA | |
| WSSV051-RNAi-F | TTCAGGGCGGCTATCTTATG | dsRNA production & transcription analysis |
| WSSV051-RNAi-R | GCGGTAGCGTTCTCTTCATC | |
| WSSV051-F | TTCACGAACGGCTGCCATTT | |
| WSSV517-RNAi-F | AAGGAACGGAATGTCCACAA | dsRNA production & transcription analysis |
| WSSV517-RNAi-R | CCAGTGGTGCTGACGATG | |
| WSSV517-R | TCCCCGCGGTTTTGTTCCTTGTAATT | |
| ICP11-F | CGGATCCACCATGGCCACCTTCCAGACT | WSSV detection |
| ICP11-R | AGCGGCCGCCTTCTGTTGTTGGCACAAT | |
| Actin-F | AGGCTCCCCTCAACCCCAAGG | Internal control |
| Actin-R | GCAGTGATTCTGCATGCG |
Suppression of WSSV Hub Genes
Specific pathogen free (SPF) shrimp P. vannamei of average 5–6 g body weight (BW) were divided into four groups of 14 individuals each. Groups I, II, and III were injected with 50 μl of WSSV051, WSSV517, and IMNV dsRNAs, respectively (2.5 μg/g BW) at the lateral area of the fourth abdominal segment. Group IV was injected with 50 μl of 150 mm NaCl solution. At 2 days post dsRNA and NaCl administration, all groups were subsequently challenged with 50 μl of crude WSSV stock at a dilution of 5 × 10−5 that gave ∼50% shrimp death within 5 days (5-day LD50). At 3 days and 6 days post WSSV injection, four shrimp specimens from each group were sampled for RT-PCR and PCR analysis. Meanwhile, three shrimp specimens were kept for histopathology. Briefly, the cephalothorax region of samples was fixed in Davison's fixative solution and processed for standard paraffin sectioning (38). The sections were then stained with hematoxylin and eosin (H & E).
For the mortality test, a parallel experiment was carried out by dividing shrimp into four groups of 15 individuals each. All groups administrated dsRNAs or NaCl were then challenged with WSSV as described above. Shrimp mortality was monitored twice a day until 17 days post WSSV challenge. The mean time to death was calculated for the 15 shrimp in each group. Experiments were performed in duplicate. Statistical analysis was carried out using one-way analysis of variance with SigmaStat software followed by pairwise multiple comparison procedures (Holm-Sidak method) with differences considered to be statistically significant when p ≤ 0.05.
Comparison of Protective Effects Using dsRNAs From WSSV Hub and Nonhub Genes
To compare the protective effects of dsRNAs produced from WSSV hub and nonhub genes, a knockdown experiment was carried out as described above with some alternations. Briefly, whiteleg shrimp P. vannamei (average 5–6 g BW) were divided into four groups including group I (hub WSSV051 dsRNA injection), group II (hub WSSV517 dsRNA injection), group III (nonhub WSSV050 dsRNA injection), and group IV (NaCl injection control). Groups I to III had subgroups of two each for treatment with either 1 μg or 5 μg of dsRNA per gram BW. At 2 days post injection (dpi) of dsRNA or NaCl, shrimp in each group (24–34 individuals each) were challenged with crude WSSV stock at a 3-day LD50 dose. Shrimp mortality was monitored twice daily for 17 days. At days 3 and 6 of the experiment, shrimp specimens were sampled for investigation of WSSV transcripts by RT-PCR, as described above.
RT-PCR Analysis
Total RNA of shrimp specimens was isolated from gill tissues using TrizolTM (Invitrogen) as described in the manufacture's protocol. RNA quantity and quality were assessed by measuring absorbance at 260 and 280 nm. Semi-quantitative RT-PCR was employed to evaluate the expression profiles of hub genes WSSV051 and WSSV517 and nonhub gene WSSV050 after dsRNA treatement. WSSV partial sequences were amplified using respective primer pairs; WSSV051-F and WSSV051-RNAi-R for WSSV051, WSSV517-RNAi-F and WSSV517-R for WSSV517, and WSSV050-F and WSSV050-RNAi-R for WSSV050 (Table I). Partial P. vannamei β-actin (GenBank accession no. AF300705) amplified using primers described in Table I was used as a control. RT-PCR reactions were carried out in a 20 μl reaction solution containing of 400 ng of total RNA template, 0.2 μm of each forward and reverse primer, 0.8 μl of SuperScript One-Step RT/Platinum Taq mix (Invitrogen) and 1X reaction buffer. The reaction protocol comprised reverse transcription at 50 °C for 30 min followed by denaturation at 94 °C for 2 min followed by PCR cycling consisting of denaturation at 94 °C for 15 s, annealing at 55 °C for 30 s and extension at 68 °C for 30 s. The PCR cycling was optimized to detect each transcript as follows: 40 cycles for WSSV genes and 25 cycles for β-actin. RT-PCR products were analyzed by agarose gel electrophoresis.
WSSV Diagnosis by PCR
WSSV infection was evaluated by PCR amplification of ICP11 using specific primer pairs listed in Table I. DNA samples were prepared by homogenizing shrimp gill tissues in TF lysis buffer (50 mm Tris-HCl, pH 9, 50 mm NaCl, 100 mm EDTA, 2% SDS, 1 μg/ml proteinase K). After incubation of tissue lysates at 60 °C for 10 min, DNA was extracted using the standard phenol/chloroform protocol (39). DNA quantity and quality was measured by spectrophotometer. PCR reactions were carried out in a 20 μl reaction solution containing 500 ng of DNA sample, 0.2 μm of each forward and reverse primer, 200 μm of dNTP mix, 1.5 mm of MgCl2, 2.5 U of TaqDNA polymerase (Invitrogen), and 1× reaction buffer. The amplification protocol comprised first denaturation at 94 °C for 3 min followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s and extension at 72 °C for 30 s. Amplification of shrimp β-actin using primers listed in Table I and the same reaction components and thermal cycling protocol described for ICP11 above served as an internal control.
RESULTS
Construction of WSSV Protein Arrays
WSSV proteins fused with the GAL4 DNA-binding domain (BD) or activating domain (AD) were arranged in a matrix or array format to facilitate the search for pairwise protein interactions. WSSV ORFs predicted to encode functional proteins were amplified and revealed an ∼92% successful amplification rate. Of 246 fragments, 227 fragments were successfully prepared for homologous recombination with pGBKT7–1 (BD) plasmid whereas 226 fragments were prepared for pGADT7 (AD) plasmid. Amplification of most of the large ORFs of ∼3000–7000 bp failed. In the yeast homologous recombination steps, 187 (representing 170 ORFs) out of the starting 227 WSSV fragments were able to generate yeast transformants for bait arrays and 182 (representing 172 ORFs) out of the 226 WSSV fragments could produce transformed cells for prey arrays. Putative functions of WSSV ORFs in the arrays have been roughly classified into 6 categories; (1) DNA replication, transcription and regulation, (2) nucleotide metabolism, (3) structural proteins, (4) protein motifs and/or signatures, (5) protein modification, and (6) unknown (supplemental Table S3).
Identification of WSSV Protein–protein Interactions
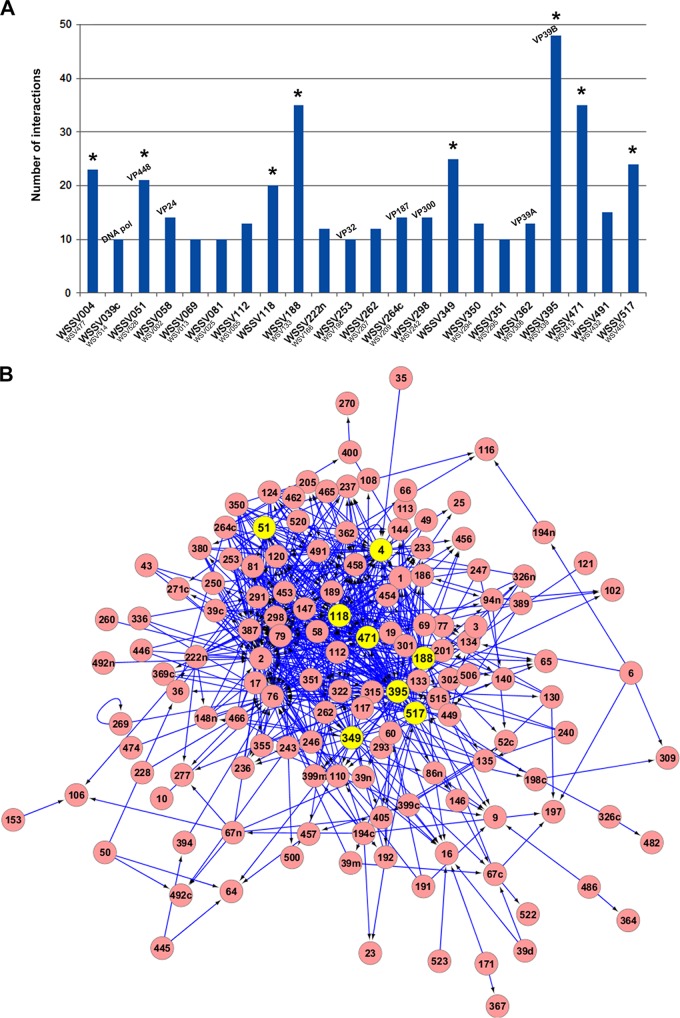
Before screening for protein interactions, the BD constructs that could autonomously induce reporter genes were screened out to eliminate false positive results. Of 187 baits, there were 31 baits that could activate HIS3, ADE2 and MEL1 (supplemental Table S4) reporter genes. Therefore, they were excluded from further screening. In the yeast two-hybrid assay, each bait was mated with every prey in the AD arrays, thereby generating 28,392 pairwise combinations of proteins (156 baits × 182 preys). After selecting the diploids, they were tested for reporter gene activation on S.D./-Ade/-His/-Leu/-Trp/X-α-gal plates. Growth of blue colonies representing potential positive interactions was scored. The interactions were independently performed at least twice. The reproducible results were scored as high confidence (H) results whereas the results with poor reproducibility were referred to as low confidence (L) results. During the mating experiments, it was found that 13 baits failed to generate diploid cells so they could not be used for further assays. The total interaction numbers from the remaining 143 baits were 710. Among these, 26 interaction pairs were detected in both orientations (i.e. bait-prey and prey-bait reciprocal interactions). It was also revealed that of the 143 baits, there were 42 baits that did not show interaction with any tested viral prey protein (i.e. 0 interactions). The distribution of the number of protein interaction partners of all tested baits was revealed to range from 0 to 48. The top 22 proteins with the highest number of interactions ranging from 10 to 48 were shown in Fig. 1A. The ratio of interactions to the total number of examined ORFs of WSSV was calculated as 4.97.
Fig. 1.
WSSV protein–protein interactions. A, Distribution of the number of protein interaction partners from the top 22 proteins with the highest connectivity. The x axis depicts the WSSV baits whereas the y axis represents the number of their interaction partners. The eight identified hubs are marked by asterisks. The ORF numbers for both Taiwan isolate (WSSV) and China isolate (WSV) are indicated. Known functions or predicted characteristic domains of some WSSV ORFs are indicated. B, A protein–protein interaction network of WSSV as generated by the Cytoscape program. WSSV proteins (nodes) are shown in circles and interactions (edges) are indicated by lines. The arrows point to target or prey proteins. The network consists of 142 nodes and 668 edges. Eight predicted hubs are indicated by yellow circles.
Building the WSSV Protein–Protein Interaction (PPI) Network
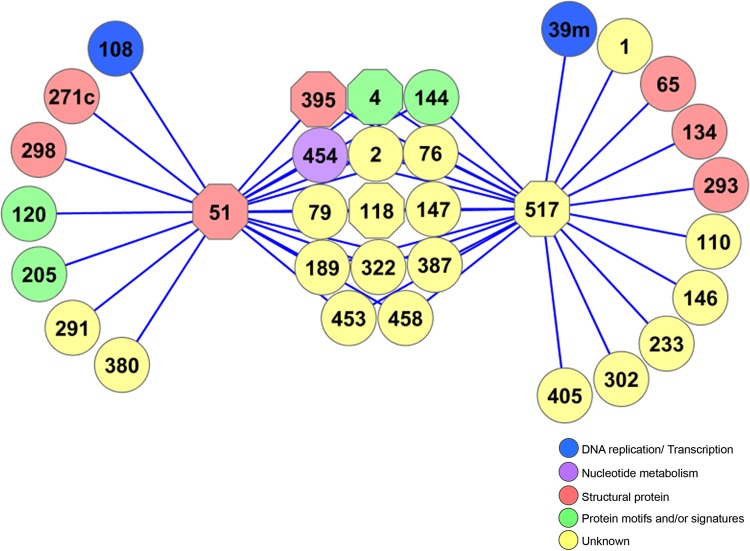
To build the WSSV protein–protein interaction (PPI) network, the open source software platform Cytoscape for visualization of complex-networks (40, 41) was employed. All the WSSV proteins that yielded interactions were set as input for the Cytoscape program. The results revealed that our WSSV PPI network comprised 668 interactions (edges) from 142 different proteins (nodes) (Fig. 1B). The 142 individual nodes were derived from 101 bait proteins and an additional 41 prey proteins. Proteins with a large number of interactions (usually called hubs) are likely to be more essential than proteins with a few links (42). However, there are many criteria to specify hub proteins (43). In this study, we defined the top 8 proteins with the highest number of interactions shown in Fig. 1 as hubs. Therefore, the hub proteins in the WSSV PPI network included WSSV004, WSSV051, WSSV118, WSSV188, WSSV349, WSSV395, WSSV471, and WSSV517 with the interaction partners ranging from 20–48. We focused primarily on two hubs, WSSV051 having 21 interacting proteins and WSSV517 with 24, for interaction validation and functional characterization. The local view of an interaction network through WSSV051 and WSSV517 hubs was then built to observe the relationship with their protein partners. It was shown in Fig. 2 that the network built based on the two hubs comprised 45 edges and 33 nodes. Three other hubs mentioned above were also present in this network.
Fig. 2.
The WSSV hub interactions. A PPI network was built based on protein interactions generated from two predicted hubs WSSV051 and WSSV517. Hub proteins present in this network are marked as octagons. The network represents 33 nodes and 45 edges. WSSV ORFs were assigned 5 different labeled colors according to their putative functions.
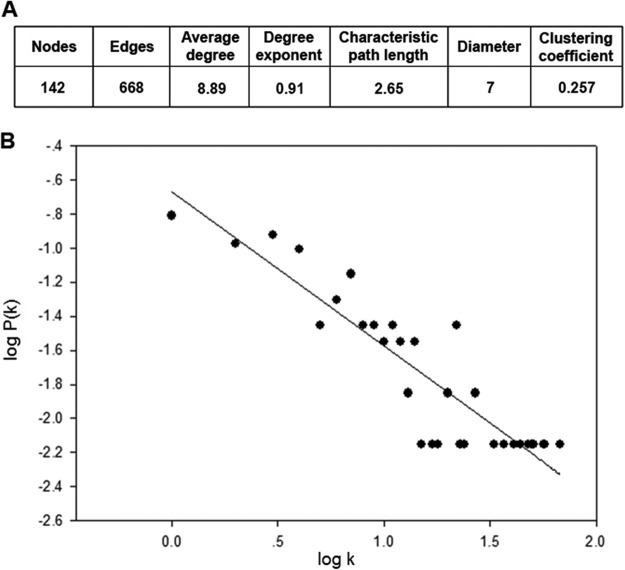
To characterize the WSSV PPI topological properties, the network parameters automatically obtained from the Cytoscape plug-in Network analyzer were evaluated based on the Barabási-Albert model (44). Complex networks can be classified into three different types as random, scale-free, and hierarchical networks (see review in (45)). One characterization parameter widely used to identify the type of complex network is the degree distribution, P(k), the probability that a selected node has k interactions. A logarithmic scale plotting of P(k) with k shown in Fig. 3 demonstrated that the degree distribution showed a linear regression with a slope of 0.91. This could be approximately fitted by a power law P(k)∼ k−γ, where γ is the degree exponent (i.e. the slope) (45). The significance of linear regression was therefore described by the equation of y = 30.38x−0.91 with R2 = 0.82. The degree exponent (0.91) of the WSSV network resembled exponents published for viral networks more than it resembled exponents published for cellular networks (supplemental Table S5). It also indicated that the WSSV PPI network was scale-free as has been found in most biological networks (44).
Fig. 3.
Properties of the WSSV PPI network. A, The network parameters of WSSV PPI were automatically calculated by the Network analyzer. B, Degree distribution of WSSV proteins (P(k)) and node degrees k were plotted on a log-log scale and fitted by linear regression. The distribution follows a power law of P(k)∼k−γ, where γ is the degree exponent (i.e. slope), indicating a characteristic of scale-free network where γ is 0.91.
WSSV PPI Confirmed by Coimmunoprecipitation
In this study, a coimmunoprecipitation assay (co-IP) was carried out to validate the quality of the hub interactions identified by the high-throughput yeast two-hybrid method. WSSV hub proteins WSSV051 and WSSV517 were constructed as FLAG fusion proteins whereas their interacting partners were constructed to have a V5 epitope tag. In this study, 62 pairs of interactions accounting for 34 ORFs were subjected to the co-IP assay (supplemental Fig. S1, S2, S3, and S4). Three randomly selected prey proteins that did not bind to WSSV hub proteins from the yeast two-hybrid approach were also negative by co-IP assays. A confirmation rate of ∼82% (31/38) was obtained for this assay. Some interaction pairs could not be confirmed because of relatively poor WSSV protein expression in the Sf-9 cell system. The interactions from the yeast two-hybrid study and the co-IP assay are summarized in Table II. In conclusion, the results demonstrated that representative WSSV protein interactions from the yeast two-hybrid data could be confirmed by independent methods.
Table II. Validation of yeast two-hybrid results by a co-IP assay. Y2H Yeast two-hybrid results; H High confidence interactions; L Low confidence interactions; − No interactions. + The Y2H results could be confirmed by co-IP assay; − The Y2H results could not be confirmed by co-IP assay; N Proteins not expressed in Sf-9; ND Not done.
| WSSV051 interacting protein |
WSSV517 interacting protein |
||||||
|---|---|---|---|---|---|---|---|
| Preya | Characteristic | Y2H | co-IP | Preya | Characteristic | Y2H | co-IP |
| WSSV002 | H | + | WSSV001 | H | + | ||
| WSSV004 (WSV477) | Cys2/Cys2-type Zinc finger | H | + | WSSV002 | H | + | |
| WSSV076 (WSV020) | H | − | WSSV004 (WSV477) | Cys2/Cys2-type Zinc finger | H | + | |
| WSSV079 (WSV023) | H | − | WSSV039m (WSV514) | DNA polymerase | H | ND | |
| WSSV108 (WSV051) | Immediate-early gene | H | − | WSSV065 (WSV009) | VP95 | H | + |
| WSSV118 | H | + | WSSV076 (WSV020) | H | − | ||
| WSSV120 (WSV063) | Zinc finger (RING finger) | H | N | WSSV079 (WSV023) | H | + | |
| WSSV144 | Ser-rich | H | + | WSSV110 (WSV053) | H | − | |
| WSSV147 | H | N | WSSV118 | H | + | ||
| WSSV189 (WSV134) | H | N | WSSV134 (WSV077) | VP36A | H | ND | |
| WSSV205 | His-rich | H | N | WSSV144 | Ser-rich | H | + |
| WSSV271c (WSV216) | VP124 | H | N | WSSV146 | H | ND | |
| WSSV291 (WSV235) | H | + | WSSV147 | H | N | ||
| WSSV298 (WSV242) | VP300/VP41B | H | + | WSSV189 (WSV134) | H | N | |
| WSSV322 (WSV267) | H | + | WSSV233 (WSV177) | H | ND | ||
| WSSV380 (WSV324) | H | + | WSSV293 (WSV237) | VP41A | H | N | |
| WSSV387 (WSV331) | H | + | WSSV302 | H | ND | ||
| WSSV395 (WSV339) | VP39B/VP39 | H | + | WSSV322 (WSV267) | H | + | |
| WSSV453 (WSV394) | H | N | WSSV387 (WSV331) | H | + | ||
| WSSV454 (WSV395) | Thymidine-Thymidylate kinase | H | + | WSSV395 (WSV339) | VP39B/VP39 | H | ND |
| WSSV458 (WSV399) | H | N | WSSV405 (WSV349) | H | + | ||
| WSSV113 (WSV056) | Cys2/His2-type Zinc finger | − | − | WSSV453 (WSV394) | H | N | |
| WSSV454 (WSV395) | Thymidine-Thymidylate kinase | H | + | ||||
| WSSV458 (WSV399) | H | N | |||||
| WSSV307 (WSV252) | Prenyl group binding site (CAAX box) (IPR001230) | − | − | ||||
| WSSV466 (WSV407) | Threonyl-tRNA synthetase (COG) | − | − | ||||
Protection of Shrimp Against WSSV by Knockdown of WSSV Hub Genes
The RNA interference (RNAi) approach has been employed to protect shrimp against viral pathogens including WSSV. However, the antiviral efficiency varied among the targeted genes. With respect to the WSSV PPI network described here, it was proposed that WSSV hubs which connect various proteins in the network might be ideal targets for antiviral strategies. Thus, the protective effects of dsRNA specific to the 2 WSSV hub genes WSSV051 and WSSV517 were examined. Groups injected with nonrelated dsRNA of IMNV (dsRNA-IMNV) and NaCl served as control groups. At 8 days post infection (dpi), the control shrimp groups injected with NaCl and dsRNA-IMNV showed 100 and 50% mortality, respectively, whereas shrimp injected with dsRNA-WSSV051 and dsRNA-WSSV517 revealed no mortality (Fig. 4). Although all shrimp died within 17 dpi, the mean time to death of groups dsRNA-WSSV051 (15 ± 0.44 days) and dsRNA-WSSV517 (16.13 ± 0.09 days) was about three times longer when compared with the NaCl control group (5.06 ± 0.25 days) and almost two times longer than the dsRNA-IMNV control group (9.38 ± 0.12 days) (Fig. 4). Statistical analysis revealed significant differences in mean time to death (p < 0.001) among all the groups, so the 1 day difference between the longest delay for dsRNA-WSSV517 and the second longest delay for dsRNA-WSSV051 was significant.
Fig. 4.
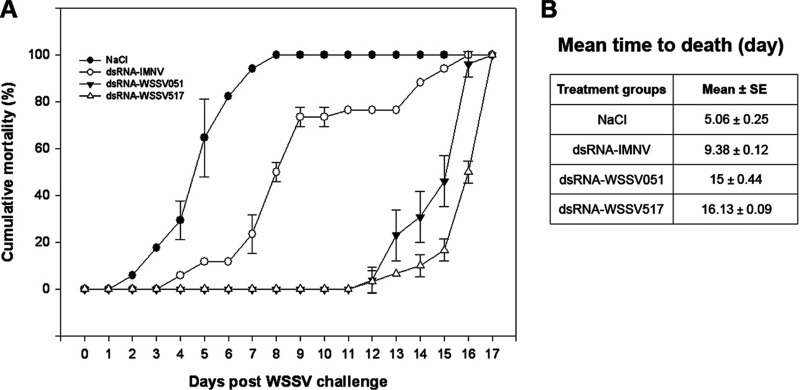
Cumulative mortality assay. A, Juvenile P. vannamei were injected intramuscularly with NaCl or dsRNAs (specific to IMNV, WSSV051 and WSSV517 genes) 2 days before WSSV challenge. There were 15 shrimp in each group and experiments were conducted for 17 days. The results are indicated as means with error bars of duplicate experiments. B, Mean time to death was calculated in each treatment group and is shown as mean with standard error (S.E.).
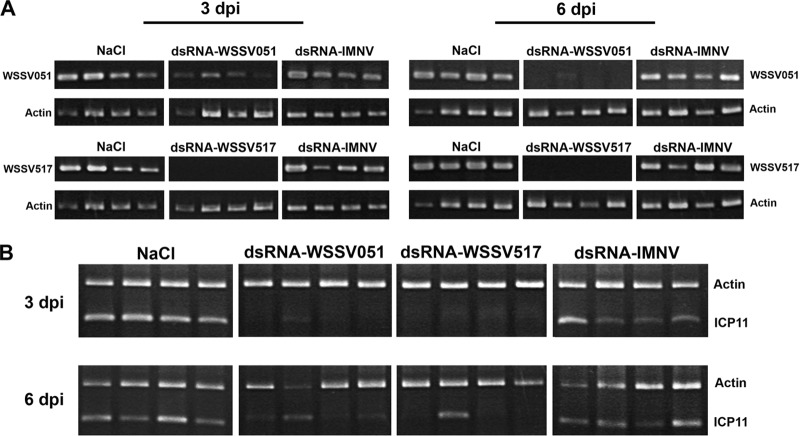
Knockdown Specificity of WSSV Hubs
To examine the specificity of dsRNAs mediated silencing of WSSV051 and WSSV517, RT-PCR analysis was conducted to measure respective viral transcripts using actin expression for comparison. At 3 dpi, WSSV051 transcripts were clearly reduced in shrimp treated with the dsRNA-WSSV051 when compared with control groups injected with NaCl and dsRNA-IMNV (Fig. 5A, top panel). Significant reduction of viral gene expression was also observed in the group injected with dsRNA-WSSV517 (Fig. 5A, bottom panel). The knockdown effect remained at 6 dpi (Fig. 5A, right panel). The results indicated that specific knockdown of hub genes was achieved at the transcription level.
Fig. 5.
Knockdown specificity of dsRNA for WSSV051 and WSSV517. A, RT-PCR analysis of WSSV051 and WSSV517 transcripts in gill tissues from shrimp from the knockdown experiments. Individual shrimp (4) from each treatment were collected at 3 and 6 days post injection (dpi). Shrimp actin transcripts served as the internal control. B, PCR analysis of ICP11 to monitor the viral loads from the shrimp specimens in (A). ICP11 and shrimp actin were separately amplified in each reaction but loaded in the same well.
The same shrimp specimens were also analyzed for the viral loads by PCR amplification using primers specific for WSSV gene ICP11. ICP11 (a DNA mimic protein) is the most highly expressed viral gene, making it a good indicator of WSSV infection (46, 47). As shown in Fig. 5B, at 3 dpi, ICP11 amplicons were barely detectable in shrimp treated with dsRNA-WSSV051 and dsRNA-WSSV517, whereas they were clearly detectable from the NaCl and dsRNA-IMNV groups. At 6 dpi, all the tested shrimp from the control groups clearly showed WSSV amplicons whereas they could be detected in only a few shrimp from the WSSV hub dsRNA treated-groups (Fig. 5B).
The significance of WSSV hub knockdown was further investigated histopathologically in shrimp tissues. The subcuticular epithelium of the shrimp stomach (a major target for WSSV) was examined by H&E staining. A representative photomicrograph at 3 dpi (Fig. 6) showed enlarged nuclei with basophilic intranuclear inclusions characteristic of WSSV infection in both control groups (bottom panel) but not in the group injected with dsRNA-WSSV051 or dsRNA-WSSV517 (top panel). The findings supported the results obtained from RT-PCR and PCR analysis as described above. Similar histopathological appearance was also observed in the 6 dpi samples except that one of the three shrimp in the dsRNA-WSSV517 group showed light WSSV lesions (data not shown) whereas all the shrimp examined from the control groups showed severe lesions. Taken together, the results demonstrated that the knockdown of WSSV hubs by dsRNA was specific and had consequences that were observed by a significantly longer mean time to death, lower viral loads and fewer WSSV lesions.
Fig. 6.
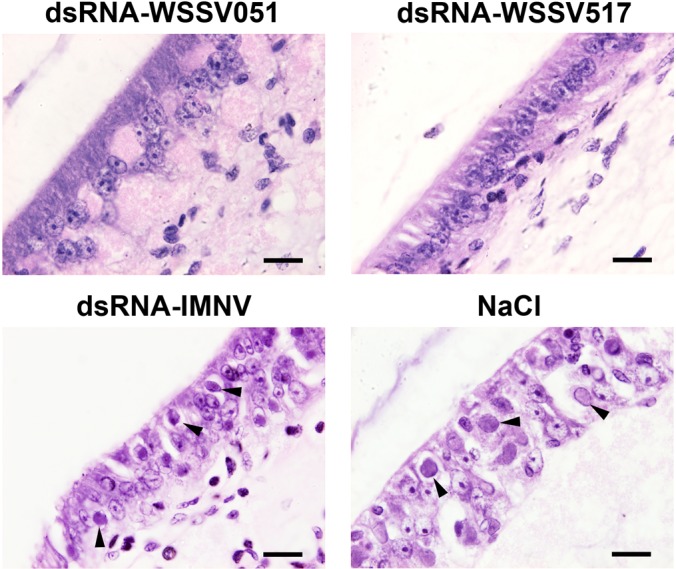
Histopathological examination for WSSV lesions in shrimp from the knockdown experiments. Hematoxylin and eosin stained sections of subcuticular epithelium of the stomach of shrimp from the knockdown experiments. Examples of tissue sections from 3 days post WSSV challenge are shown. Cells with basophilic intranuclear inclusions characteristic of WSSV infection (arrow heads) were revealed in the control groups but not observed from the groups injected with dsRNA specific for WSSV051 and WSSV517. Bar indicates 20 μm.
Suppression of a Nonhub Gene Yielded No Protection Against WSSV
To further examine the importance of WSSV hub genes, a preliminary experiment was carried out to test the protective effect of dsRNA against the arbitrarily chosen, nonhub gene WSSV050 (unknown function) with 3 binding partners (WSSV036, WSSV064 and WSSV492c). This knockdown experiment with either 1 or 5 μg/g BW dose of WSSV050 dsRNA resulted in 100% shrimp mortality within 6–10 days after WSSV challenge, the same as in the NaCl injected control group (Fig. 7). By contrast, protection was clearly seen when targeting hub genes WSSV051 and WSSV517, similar to the trends from the previous experiment. Curiously, the level of protection did not correlate with the dose of dsRNA used. Differences in level of protection between the two experiments probably related to differences in the batches of test shrimp used and the different challenge doses (higher in the second experiment). Specificity of WSSV gene knockdown was verified by RT-PCR of gill tissues collected at 3 and 6 days postchallenge (data not shown).
Fig. 7.
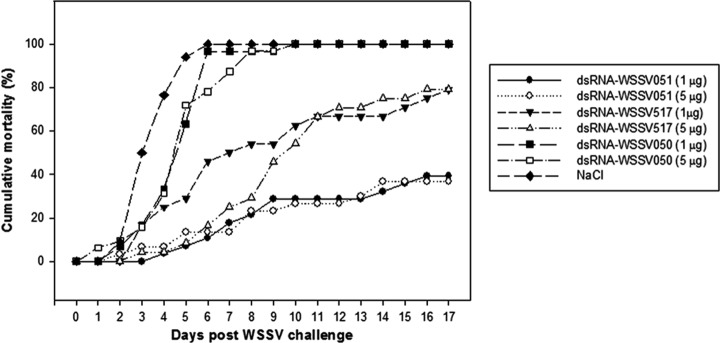
Comparison of protective effects between WSSV hub and nonhub genes. dsRNAs were prepared from WSSV hub genes (WSSV051 and WSSV517) and a nonhub gene (WSSV050). Two dosages of each dsRNA (1 μg and 5 μg dsRNA per g BW) were used for injection to P. vannamei 2 days before WSSV challenge. Control group was injected with NaCl solution followed by WSSV challenge. Shrimp mortality was recorded for 17 days after WSSV injection.
DISCUSSION
A WSSV PPI network has been successfully mapped using a comprehensive yeast two-hybrid assay to search every possible binary interaction in a Taiwan-isolated viral stock. On searching ∼28,000 possible pairwise protein interactions, 710 interactions were revealed. The explanation for these many interactions could be attributed to the high number of binding partners ranging from 20–48 for hub proteins identified in the assay. In addition, it was observed that some preys (WSSV002, WSSV076, WSSV118, WSSV120, and WSSV298) interacted with most of the baits, thereby generating a high number of interactions. Although the significance of these interactions needs to be further investigated, we speculated that they may act as adaptor proteins in certain viral pathways.
In our bait collections, there were 13 baits that failed to generate diploid cells. It might be because of instability of the plasmids or toxicity to yeast cells. Another 42 baits did not yield any interaction with WSSV protein preys. Those proteins may tend to interact with host proteins rather than viral proteins themselves and might therefore be considered to be involved in processes such as viral infection or host immune evasion. In addition, it was revealed that 31 baits (∼17% of the total constructed baits) had self-activation activity. This proportion was quite reasonable because self-activation is often observed in yeast two-hybrid assays. For example, investigation of a vaccinia virus PPI network revealed ∼9% and of a human PPI network ∼20% self-activating baits (23, 25). Interestingly, four (WSSV108, WSSV126, WSSV136 and WSSV156) out of the 31 auto-activating baits have been previously identified as WSSV immediate early (IE) genes with transcription activity (48), supporting our analysis. It was not known if the remaining self-activating baits (especially those with a zinc finger motif) found in this study might also have uninvestigated transcription functions, because most transcription factors contain zinc finger motifs that often mediate DNA binding activity (49). Additionally, several baits coding for WSSV structural proteins were autoactivators similar to results regarding virion protein interactions of herpes simplex virus type 1 that revealed ∼32% of viral structural proteins capable of auto-activation (50).
Excluding the baits that generated no diploid cells, yielded no interaction with WSSV proteins or caused auto-activation, 710 potential interactions among 101 baits were identified. Of these, only 26 could be supported by reciprocal bait-prey pairings (i.e. bidirectional). The low confirmation rate by reciprocal interaction was considered to result from characteristic technical limitations of the yeast two-hybrid system (27, 28). Similarly low bidirectionality has been previously reported for Kaposi's sarcoma-associated herpesvirus (KSHV) (27) (only 1 of 123), VZV (27) (only 10 of 173) and human cytomegalovirus (51) (only 11 of 79). Confirmation tests using an independent co-IP assay gave a validation rate of ∼82%. Lower confirmation rates have been reported in other PPI networks for humans (66%), (KSHV) (50%) and Epstein-Barr virus (EBV) (47%) (23, 27, 28). However, the number of WSSV protein interactions to be tested should be enlarged to increase reliability of our data. In addition, the new putative interactions need to be confirmed by other biological or biochemical assays. It is a common feature of the large-scale yeast two-hybrid screen to produce low coverage data, i.e. missing previously identified interactions (52). For example, minimal overlap interactions have been found for different high-throughput interaction datasets observed in Drosophila networks (19, 53, 54) and KSHV networks (27, 55). In addition, only six out of 43 interactions of the EBV proteins described by Calderwood et al. (28) were detected in the EBV network reported by Fossum et al. (29). Reasons for low coverage of our WSSV dataset might be because of incompleteness of the ORF arrays or to the fact that some interactions could not be detected because of limitations of the yeast two-hybrid assay. This was also evident, for example, when some interactions of KSHV structural proteins could not be found by yeast two-hybrid assay but could be detected by co-IP (55).
The topology of the WSSV PPI network followed the power law, indicating that it had scale-free properties similar to many cellular networks (44). A significant key of scale-free networks is hubs that hold the nodes together in the network. Nevertheless, the WSSV PPI network had an unusually small degree exponent (γ) different from those of known cellular networks but more closely related to those of viral PPI networks (supplemental Table S5). Uetz et al. (27) suggested that this unusual characteristic in the viral networks presumably reflected their incompleteness as stand-alone networks. They therefore modeled KSHV protein–human protein interactions using an orthologs-based approach which resulted in a complete change in network topology to that of a more typical scale-free network (27). A similar phenomenon might be expected for a WSSV protein-shrimp protein interaction network.
The putative WSSV hubs identified in this study were likely to play roles in many biological processes according to the putative functions of their targets such as nucleotide metabolism, DNA replication and virion assembly. WSSV051 has been identified as structural protein VP448 (also known as VP55) from purified WSSV virions using SDS-PAGE accompanied by mass spectrometry (56). A later study using a microarray approach also indicated that the time course expression pattern of WSSV051 clustered in the gene group related to WSSV envelope protein assembly (57). Interestingly, two structural proteins (WSSV298 and WSSV395) identified as WSSV051 interacting proteins in the present study were also classified in the same cluster of WSSV051 (57). This supports our interaction data and might suggest roles for these proteins in WSSV virion assembly. For WSSV517, the function remains unknown, but a previous report using a DNA microarray approach classified WSSV517 as an early gene because it was expressed at the early stage post WSSV infection (58). Interaction between WSSV517 and the middle part of DNA polymerase (WSSV039m) was revealed. Further study is required to explore possible roles of WSSV517 in viral DNA replication.
Explanations as to how one protein is capable of binding multiple partners have been recently described by means of intrinsic protein disorder (59–63). Intrinsically disordered proteins or intrinsically unstructured proteins are proteins characterized by lack of a well-defined structure even though they remain perfectly functional. There is bioinformatics and in vitro experimental evidence that intrinsic structural disorder is a distinctive and common characteristic of many hub proteins and that a disordered domain facilitates the different conformational requirements for binding to different partners (64–66).
RNAi is a powerful tool for gene silencing and holds promise for antiviral therapy in shrimp. This has drawn much attention for shrimp protection against viral diseases, and especially WSSV. Many WSSV genes such as VP15, VP19 VP24, VP26, VP28, VP281, protein kinase, DNA polymerase, ribonucleotide reductase large and small subunits (rr1 and rr2), immediate early genes (IE1 and IE3), and thymidine kinase and thymidylate kinase (tk-tmk) have been targeted using either dsRNA or small interfering RNA (siRNA) (67–72). The degree of protection has varied (e.g. ∼30–95% survival) and might be because of the different viral doses used in each experiment. It was also suggested that the protection effect depends on the function of the gene encoded proteins and their roles during the viral life cycle (71, 72). In this regard, we speculated that knockdown of the central hubs in WSSV network could provide shrimp protection against WSSV and prove their significances in viral processes. The importance of hub silencing has been previously described in the bacterial entry network of Chlamydia pneumoniae in which knockdown the hubs significantly inhibited C. pneumoniae infectivity in human cell lines (73). Our silencing of WSSV hubs by treating shrimp with hub dsRNAs significantly delayed mortality and improved survival in viral challenges when compared with untreated controls. Efficacy was supported by tissue examination that revealed WSSV lesions (hypertrophied nuclei) in every group except for the knockdown groups injected with WSSV051 and WSSV517 dsRNAs. In addition, examination revealed that expression of nontargeted WSSV genes such as the WSSV DNA polymerase was also suppressed (data not shown). We speculate that defects in DNA synthesis may have resulted from silencing of the two hub genes, leading to down-regulation of additional viral genes and to improved shrimp survival. Thus, the comparison of knocking down hub and nonhub genes supported the contention that WSSV hub-genes are important and may serve as potential targets for protection against shrimp mortality caused by WSSV. One requirement for reaching such a practical goal is a method to effectively deliver WSSV hub-specific dsRNAs to shrimp by oral administration.
WSSV has had an enormous impact on the global shrimp culture industry as a result of economic losses from severe mortality. Novel technology for virus control based on an improved understanding of WSSV is expected. Molecular studies have been carried out to gain insight into both shrimp immunity and WSSV pathology and have included both genomic and proteomic approaches. However, overall interactions (or interactomes) have not previously been reported and it is hoped that this first study of the WSSV PPI network will serve as the starting point for further work. For example, many WSSV proteins were identified as interesting candidates for testing in knockdown experiments similar to those we carried out for the hub genes WSSV051 and WSSV517. Another network topological feature called a bottleneck (a protein that connects more than two sub-networks) has also been proposed to be a good target for inhibition of protein interactions (74). Data mining and experimental validation for WSSV bottleneck proteins could be further investigated. Apart from a gene knockdown approach in disruption of protein interactions, other strategies have been previously proposed. These include, for example, development of specific antibodies and the use of small molecule inhibitors to selectively interfere with protein interactions (see reviews (75, 76)). The WSSV protein interaction data could then be employed as a fundamental resource to screen for such substances either by actual experiments, by computational based approaches or both.
Nevertheless, one dataset alone cannot completely elucidate protein functions or biological processes, and combination with other datasets may lead to significant new insights (77). For example, integration of the WSSV interaction dataset presented here with other high-throughput data sets such as WSSV transcriptional profiles may give a better understanding of the dynamic processes that occur during infection. Finally, the parallel development of a shrimp protein interaction network and a shrimp-virus interaction network, either by real experiment or by prediction from protein orthologs would allow for combination among networks and may lead to further insights on viral pathogenesis and the host response for the development of disease control strategies.
Supplementary Material
Acknowledgments
We thank Ms. C. Jaengsanong for skilled technical assistance.
Footnotes
Author contributions: P.S., S.S., W.H., T.W.F., and C.L. designed research; P.S., J.H., Y.C., K.P., P.L., W.M., and S.S. performed research; S.S., W.H., B.W., T.W.F., and C.L. contributed new reagents or analytic tools; P.S., S.S., W.H., T.W.F., and C.L. analyzed data; P.S., S.S., and T.W.F. wrote the paper.
* This work was supported by research grants from Mahidol University, L'Oreal Thailand ‘for Women in Science’ fellowship 2011, a National Taiwan University research grant (NTU-99R80830), National Science Council grants and National Cheng Kung University grants. P.S. was the recipient of a Royal Golden Jubilee Ph.D. scholarship (PHD/0231/2548) from the Thailand Research Fund. In addition, P.S. has been supported by a Mahidol University postdoctoral fellowship program.
 This article contains supplemental Figs. S1 to S4 and Tables S1 to S5.
This article contains supplemental Figs. S1 to S4 and Tables S1 to S5.
1 The abbreviations used are:
- WSSV
- white spot syndrome virus
- WSD
- white spot disease
- ORF
- open reading frame
- BD
- binding domain
- AD
- activation domain
- PPI
- protein–protein interaction
- IMNV
- infectious myonecrosis virus.
REFERENCES
- 1. Lightner D. V. (1996) A Handbook of Shrimp Pathology and Diagnostic Procedures for Diseases of Cultured Penaeid Shrimp. World Aquaculture Society, Baton Rouge, LA, U.S.A [Google Scholar]
- 2. Leu J. H., Yang F., Zhang X., Xu X., Kou G. H., Lo C. F. (2009) Whispovirus. Curr. Top. Microbiol. Immunol. 328, 197–227 [DOI] [PubMed] [Google Scholar]
- 3. Sánchez-Paz A. (2010) White spot syndrome virus: an overview on an emergent concern. Vet. Res. 41, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang X., Huang C., Tang X., Zhuang Y., Hew C. L. (2004) Identification of structural proteins from shrimp white spot syndrome virus (WSSV) by 2DE-MS. Proteins 55, 229–235 [DOI] [PubMed] [Google Scholar]
- 5. Witteveldt J., Vermeesch A. M., Langenhof M., de Lang A., Vlak J. M., van Hulten M. C. W. (2005) Nucleocapsid protein VP15 is the basic DNA binding protein of white spot syndrome virus of shrimp. Arch. Virol. 150, 1121–1133 [DOI] [PubMed] [Google Scholar]
- 6. Xie X., Yang F. (2006) White spot syndrome virus VP24 interacts with VP28 and is involved in virus infection. J. Gen. Virol. 87, 1903–1908 [DOI] [PubMed] [Google Scholar]
- 7. Chang Y. S., Liu W. J., Chou T. L., Lee Y. T., Lee T. L., Huang W. T., Kou G. H., Lo C. F. (2008) Characterization of white spot syndrome virus envelope protein VP51A and its interaction with viral tegument protein VP26. J. Virol. 82, 12555–12564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen J., Li Z., Hew C. L. (2007) Characterization of a novel envelope protein WSV010 of shrimp white spot syndrome virus and its interaction with a major viral structural protein VP24. Virology 364, 208–213 [DOI] [PubMed] [Google Scholar]
- 9. Jie Z., Xu L., Yang F. (2008) The C-terminal region of envelope protein VP38 from white spot syndrome virus is indispensable for interaction with VP24. Arch. Virol. 153, 2103–2106 [DOI] [PubMed] [Google Scholar]
- 10. Wan Q., Xu L., Yang F. (2008) VP26 of white spot syndrome virus functions as a linker protein between the envelope and nucleocapsid of virions by binding with VP51. J. Virol. 82, 12598–12601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu Q. H., Ma C. Y., Chen W. B., Zhang X. L., Liang Y., Dong S. L., Huang J. (2009) White spot syndrome virus VP37 interacts with VP28 and VP26. Dis. Aquat. Organ. 85, 23–30 [DOI] [PubMed] [Google Scholar]
- 12. Zhou Q., Xu L., Li H., Qi Y. P., Yang F. (2009) Four major envelope proteins of white spot syndrome virus bind to form a complex. J. Virol. 83, 4709–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lin Y., Xu L., Yang F. (2010) Tetramerization of white spot syndrome virus envelope protein VP33 and its interaction with VP24. Arch. Virol. 155, 833–838 [DOI] [PubMed] [Google Scholar]
- 14. Chang Y. S., Liu W. J., Lee C. C., Chou T. L., Lee Y. T., Wu T. S., Huang J. Y., Huang W. T., Lee T. L., Kou G. H., Wang A. H., Lo C. F. (2010) A 3D model of the membrane protein complex formed by the white spot syndrome virus structural proteins. PLoS ONE 5, e10718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li Z., Xu L., Li F., Zhou Q., Yang F. (2011) Analysis of white spot syndrome virus envelope protein complexome by two-dimensional blue native/SDS PAGE combined with mass spectrometry. Arch. Virol. 156, 1125–1135 [DOI] [PubMed] [Google Scholar]
- 16. Uetz P., Giot L., Cagney G., Mansfield T. A., Judson R. S., Knight J. R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Qureshi-Emili A., Li Y., Godwin B., Conover D., Kalbfleisch T., Vijayadamodar G., Yang M., Johnston M., Fields S., Rothberg J. M. (2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627 [DOI] [PubMed] [Google Scholar]
- 17. Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. U.S.A. 98, 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rain J. C., Selig L., De Reuse H., Battaglia V., Reverdy C., Simon S., Lenzen G., Petel F., Wojcik J., Schächter V., Chemama Y., Labigne A., Legrain P. (2001) The protein-protein interaction map of Helicobacter pylori. Nature 409, 211–215 [DOI] [PubMed] [Google Scholar]
- 19. Giot L., Bader J. S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y. L., Ooi C. E., Godwin B., Vitols E., Vijayadamodar G., Pochart P., Machineni H., Welsh M., Kong Y., Zerhusen B., Malcolm R., Varrone Z., Collis A., Minto M., Burgess S., McDaniel L., Stimpson E., Spriggs F., Williams J., Neurath K., Ioime N., Agee M., Voss E., Furtak K., Renzulli R., Aanensen N., Carrolla S., Bickelhaupt E., Lazovatsky Y., DaSilva A., Zhong J., Stanyon C. A., Finley R. L., White K. P., Braverman M., Jarvie T., Gold S., Leach M., Knight J., Shimkets R. A., McKenna M. P., Chant J., Rothberg J. M. (2003) A protein interaction map of Drosophila melanogaster. Science 302, 1727–1736 [DOI] [PubMed] [Google Scholar]
- 20. Li S., Armstrong C. M., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P.-O., Han J.-D. J., Chesneau A., Hao T., Goldberg D. S., Li N., Martinez M., Rual J.-F., Lamesch P., Xu L., Tewari M., Wong S. L., Zhang L. V., Berriz G. F., Jacotot L., Vaglio P., Reboul J., Hirozane-Kishikawa T., Li Q., Gabel H. W., Elewa A., Baumgartner B., Rose D. J., Yu H., Bosak S., Sequerra R., Fraser A., Mango S. E., Saxton W. M., Strome S., van den Heuvel S., Piano F., Vandenhaute J., Sardet C., Gerstein M., Doucette-Stamm L., Gunsalus K. C., Harper J. W., Cusick M. E., Roth F. P., Hill D. E., Vidal M. (2004) A map of the interactome network of the metazoan C. elegans. Science 303, 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. LaCount D. J., Vignali M., Chettier R., Phansalkar A., Bell R., Hesselberth J. R., Schoenfeld L. W., Ota I., Sahasrabudhe S., Kurschner C., Fields S., Hughes R. E. (2005) A protein interaction network of the malaria parasite Plasmodium falciparum. Nature 438, 103–107 [DOI] [PubMed] [Google Scholar]
- 22. Rual J. F., Venkatesan K., Hao T., Hirozane Kishikawa T., Dricot A., Li N., Berriz G. F., Gibbons F. D., Dreze M., Ayivi Guedehoussou N., Klitgord N., Simon C., Boxem M., Milstein S., Rosenberg J., Goldberg D. S., Zhang L. V., Wong S. L., Franklin G., Li S., Albala J. S., Lim J., Fraughton C., Llamosas E., Cevik S., Bex C., Lamesch P., Sikorski R. S., Vandenhaute J., Zoghbi H. Y., Smolyar A., Bosak S., Sequerra R., Doucette Stamm L., Cusick M. E., Hill D. E., Roth F. P., Vidal M. (2005) Towards a proteome-scale map of the human protein-protein interaction network. Nature 437, 1173–1178 [DOI] [PubMed] [Google Scholar]
- 23. Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F. H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Timm J., Mintzlaff S., Abraham C., Bock N., Kietzmann S., Goedde A., Toksöz E., Droege A., Krobitsch S., Korn B., Birchmeier W., Lehrach H., Wanker E. E. (2005) A human protein-protein interaction network: a resource for annotating the proteome. Cell 122, 957–968 [DOI] [PubMed] [Google Scholar]
- 24. Bartel P. L., Roecklein J. A., SenGupta D., Fields S. (1996) A protein linkage map of Escherichia coli bacteriophage T7. Nat. Genet. 12, 72–77 [DOI] [PubMed] [Google Scholar]
- 25. McCraith S., Holtzman T., Moss B., Fields S. (2000) Genome-wide analysis of vaccinia virus protein-protein interactions. Proc. Natl. Acad. Sci. U.S.A. 97, 4879–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Flajolet M., Rotondo G., Daviet L., Bergametti F., Inchauspé G., Tiollais P., Transy C., Legrain P. (2000) A genomic approach of the hepatitis C virus generates a protein interaction map. Gene 242, 369–379 [DOI] [PubMed] [Google Scholar]
- 27. Uetz P., Dong Y. A., Zeretzke C., Atzler C., Baiker A., Berger B., Rajagopala S. V., Roupelieva M., Rose D., Fossum E., Haas J. (2006) Herpesviral protein networks and their interaction with the human proteome. Science 311, 239–242 [DOI] [PubMed] [Google Scholar]
- 28. Calderwood M. A., Venkatesan K., Xing L., Chase M. R., Vazquez A., Holthaus A. M., Ewence A. E., Li N., Hirozane-Kishikawa T., Hill D. E., Vidal M., Kieff E., Johannsen E. (2007) Epstein-Barr virus and virus human protein interaction maps. Proc. Natl. Acad. Sci. U.S.A. 104, 7606–7611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fossum E., Friedel C. C., Rajagopala S. V., Titz B., Baiker A., Schmidt T., Kraus T., Stellberger T., Rutenberg C., Suthram S., Bandyopadhyay S., Rose D., von Brunn A., Uhlmann M., Zeretzke C., Dong Y. A., Boulet H., Koegl M., Bailer S. M., Koszinowski U., Ideker T., Uetz P., Zimmer R., Haas J. (2009) Evolutionarily conserved herpesviral protein interaction networks. PLoS Pathog. 5, e1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang C. H., Lo C. F., Leu J. H., Chou C. M., Yeh P. Y., Chou H. Y., Tung. M. C., Chang C. F., Su M. S., Kou G. H. (1995) Purification and genomic analysis of baculovirus associated with white spot syndrome (WSBV) of Penaeus monodon. Dis. Aquat. Organ. 23, 239–242 [Google Scholar]
- 31. Lo C. F., Hsu H. C., Tsai M. F., Ho C. H., Peng S. E., Kou G. H., Lightner D. V. (1999) Specific genomic DNA fragment analysis of different geographical clinical samples of shrimp white spot syndrome virus. Dis. Aquat. Organ. 35, 175–185 [Google Scholar]
- 32. Yang F., He J., Lin X., Li Q., Pan D., Zhang X., Xu X. (2001) Complete genome sequence of the shrimp white spot bacilliform virus. J. Virol. 75, 11811–11820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hazbun T. R., Miller J. P. (2005) Genome-wide analysis of protein-protein interactions using a two-hybrid array. Chapter 37, in Protein-Protein Interactions, 2nd edition (eds. Golemis, Adams). Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, U.S.A [Google Scholar]
- 34. Assenov Y., Ramírez F., Schelhorn S. E., Lengauer T., Albrecht M. (2008) Computing topological parameters of biological networks. Bioinformatics 24, 282–284 [DOI] [PubMed] [Google Scholar]
- 35. Leu J. H., Kuo Y. C., Kou G. H., Lo C. F. (2008) Molecular cloning and characterization of an inhibitor of apoptosis protein (IAP) from the tiger shrimp, Penaeus monodon. Dev. Comp. Immunol. 32, 121–133 [DOI] [PubMed] [Google Scholar]
- 36. Elbashir S. M., Lendeckel W., Tuschl T. (2001) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15, 188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Senapin S., Phiwsaiya K., Anantasomboon G., Sriphaijit T., Browdy C. L., Flegel T. W. (2010) Knocking down a Taura syndrome virus (TSV) binding protein Lamr is lethal for the whiteleg shrimp Penaeus vannamei. Fish Shellfish Immunol. 29, 422–429 [DOI] [PubMed] [Google Scholar]
- 38. Bell T. A., Lightner D. V. (1988) A handbook of normal shrimp histology. World Aquaculture Society, Baton Rouge, LA, U.S.A [Google Scholar]
- 39. Sambrook J., Russell D. W. (2001) Molecular cloning: a laboratory manual. 3 ed NY: Cold Spring Harbor [Google Scholar]
- 40. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cline M. S., Smoot M., Cerami E., Kuchinsky A., Landys N., Workman C., Christmas R., Avila-Campilo I., Creech M., Gross B., Hanspers K., Isserlin R., Kelley R., Killcoyne S., Lotia S., Maere S., Morris J., Ono K., Pavlovic V., Pico A. R., Vailaya A., Wang P. L., Adler A., Conklin B. R., Hood L., Kuiper M., Sander C., Schmulevich I., Schwikowski B., Warner G. J., Ideker T., Bader G. D. (2007) Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2, 2366–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jeong H., Mason S. P., Barabási A. L., Oltvai Z. N. (2001) Lethality and centrality in protein networks. Nature 411, 41–42 [DOI] [PubMed] [Google Scholar]
- 43. Vallabhajosyula R. R., Chakravarti D., Lutfeali S., Ray A., Raval A. (2009) Identifying hubs in protein interaction networks. PLoS ONE 4, e5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Barabási A. L., Albert R. (1999) Emergence of scaling in random networks. Science 286, 509–512 [DOI] [PubMed] [Google Scholar]
- 45. Barabási A. L., Oltvai Z. N. (2004) Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 5, 101–113 [DOI] [PubMed] [Google Scholar]
- 46. Wang H. C., Wang H. C., Kou G. H., Lo C. F., Huang W. P. (2007) Identification of icp11, the most highly expressed gene of shrimp white spot syndrome virus (WSSV). Dis. Aquat. Organ. 74, 179–189 [DOI] [PubMed] [Google Scholar]
- 47. Wang H. C., Wang H. C., Ko T. P., Lee Y. M., Leu J. H., Ho C. H., Huang W. P., Lo C. F., Wang A. H. (2008) White spot syndrome virus protein ICP11: a histone-binding DNA mimic that disrupts nucleosome assembly. Proc. Natl. Acad. Sci. U.S.A. 105, 20758–20763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li F., Li M., Ke W., Ji Y., Bian X., Yan X. (2009) Identification of the immediate-early genes of white spot syndrome virus. Virology 385, 267–274 [DOI] [PubMed] [Google Scholar]
- 49. Iuchi S. (2001) Three classes of C2H2 zinc finger proteins. Cell. Mol. Life Sci. 58, 625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee J. H., Vittone V., Diefenbach E., Cunningham A. L., Diefenbach R. J. (2008) Identification of structural protein-protein interactions of herpes simplex virus type 1. Virology 378, 347–354 [DOI] [PubMed] [Google Scholar]
- 51. To A., Bai Y., Shen A., Gong H., Umamoto S., Lu S., Liu F. (2011) Yeast two hybrid analyses reveal novel binary interactions between human cytomegalovirus-encoded virion proteins. PLoS ONE 6, e17796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Parrish J. R., Gulyas K. D., Finley R. L. (2006) Yeast two-hybrid contributions to interactome mapping. Curr. Opin. Biotechnol. 17, 387–393 [DOI] [PubMed] [Google Scholar]
- 53. Stanyon C. A., Liu G., Mangiola B. A., Patel N., Giot L., Kuang B., Zhang H., Zhong J., Finley R. L. (2004) A Drosophila protein-interaction map centered on cell-cycle regulators. Genome Biol. 5, R96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Formstecher E., Aresta S., Collura V., Hamburger A., Meil A., Trehin A., Reverdy C., Betin V., Maire S., Brun C., Jacq B., Arpin M., Bellaiche Y., Bellusci S., Benaroch P., Bornens M., Chanet R., Chavrier P., Delattre O., Doye V., Fehon R., Faye G., Galli T., Girault J.-A., Goud B., de Gunzburg J., Johannes L., Junier M.-P., Mirouse V., Mukherjee A., Papadopoulo D., Perez F., Plessis A., Rossé C., Saule S., Stoppa-Lyonnet D., Vincent A., White M., Legrain P., Wojcik J., Camonis J., Daviet L. (2005) Protein interaction mapping: a Drosophila case study. Genome Res. 15, 376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rozen R., Sathish N., Li Y., Yuan Y. (2008) Virion-wide protein interactions of Kaposi's sarcoma-associated herpesvirus. J. Virol. 82, 4742–4750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Huang C., Zhang X., Lin Q., Xu X., Hu Z., Hew C. L. (2002) Proteomic analysis of shrimp white spot syndrome viral proteins and characterization of a novel envelope protein VP466. Mol. Cell. Proteomics 1, 223–231 [DOI] [PubMed] [Google Scholar]
- 57. Tsai J. M., Wang H. C., Leu J. H., Hsiao H. H., Wang A. H., Kou G. H., Lo C. F. (2004) Genomic and proteomic analysis of thirty-nine structural proteins of shrimp white spot syndrome virus. J. Virol. 78, 11360–11370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lan Y., Xu X., Yang F., Zhang X. (2006) Transcriptional profile of shrimp white spot syndrome virus (WSSV) genes with DNA microarray. Arch. Virol. 151, 1723–1733 [DOI] [PubMed] [Google Scholar]
- 59. Kim P. M., Sboner A., Xia Y., Gerstein M. (2008) The role of disorder in interaction networks: a structural analysis. Mol. Syst. Biol. 4, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stein A., Pache R. A., Bernadó P., Pons M., Aloy P. (2009) Dynamic interactions of proteins in complex networks: a more structured view. FEBS J. 276, 5390–5405 [DOI] [PubMed] [Google Scholar]
- 61. Tyagi M., Shoemaker B. A., Bryant S. H., Panchenko A. R. (2009) Exploring functional roles of multibinding protein interfaces. Protein Sci. 18, 1674–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tsai C. J., Ma B., Nussinov R. (2009) Protein-protein interaction networks: how can a hub protein bind so many different partners? Trends Biochem. Sci. 34, 594–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dyson H. J. (2011) Expanding the proteome: disordered and alternatively folded proteins. Q. Rev. Biophys. 44, 467–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dunker A. K., Cortese M. S., Romero P., Iakoucheva L. M., Uversky V. N. (2005) Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 272, 5129–5148 [DOI] [PubMed] [Google Scholar]
- 65. Dosztányi Z., Chen J., Dunker A. K., Simon I., Tompa P. (2006) Disorder and sequence repeats in hub proteins and their implications for network evolution. J. Proteome. Res. 5, 2985–2995 [DOI] [PubMed] [Google Scholar]
- 66. Haynes C., Oldfield C. J., Ji F., Klitgord N., Cusick M. E., Radivojac P., Uversky V. N., Vidal M., Iakoucheva L. M. (2006) Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput. Biol. 2, e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Robalino J., Bartlett T., Shepard E., Prior S., Jaramillo G., Scura E., Chapman R. W., Gross P. S., Browdy C. L., Warr G. W. (2005) Double-stranded RNA induces sequence-specific antiviral silencing in addition to nonspecific immunity in a marine shrimp: convergence of RNA interference and innate immunity in the invertebrate antiviral response? J. Virol. 79, 13561–13571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Westenberg M., Heinhuis B., Zuidema D., Vlak J. M. (2005) siRNA injection induces sequence-independent protection in Penaeus monodon against white spot syndrome virus. Virus Res. 114, 133–139 [DOI] [PubMed] [Google Scholar]
- 69. Wu Y., Lü L., Weng S. P., Chan S. M., He J. G. (2007) Inhibition of white spot syndrome virus in Litopenaeus vannamei shrimp by sequence-specific siRNA. Aquaculture 271, 21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kim C. S., Kosuke Z., Nam Y. K., Kim S. K., Kim K. H. (2007) Protection of shrimp (Penaeus chinensis) against white spot syndrome virus (WSSV) challenge by double-stranded RNA. Fish Shellfish Immunol. 23, 242–246 [DOI] [PubMed] [Google Scholar]
- 71. Attasart P., Kaewkhaw R., Chimwai C., Kongphom U., Namramoon O., Panyim S. (2009) Inhibition of white spot syndrome virus replication in Penaeus monodon by combined silencing of viral rr2 and shrimp PmRab7. Virus Res. 145, 127–133 [DOI] [PubMed] [Google Scholar]
- 72. Sanjuktha M., Stalin Raj V., Aravindan K., Alavandi S. V., Poornima M., Santiago T. C. (2012) Comparative efficacy of double-stranded RNAs targeting WSSV structural and nonstructural genes in controlling viral multiplication in Penaeus monodon. Arch. Virol. 157, 993–998 [DOI] [PubMed] [Google Scholar]
- 73. Wang A., Johnston S. C., Chou J., Dean D. (2010) A systemic network for Chlamydia pneumoniae entry into human cells. J. Bacteriol. 192, 2809–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yu H., Kim P. M., Sprecher E., Trifonov V., Gerstein M. (2007) The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput. Biol. 3, e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zeng J., Zhang J., Tanaka T., Rabbitts T. H. (2013) Single domain antibody fragments as drug surrogates targeting protein-protein interactions inside cells. Antibodies 2, 306–320 [Google Scholar]
- 76. Metallo S. (2010) Intrinsically disordered proteins are potential drug targets. Curr. Opin. Chem. Biol. 14, 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ge H., Walhout A. J., Vidal M. (2003) Integrating ‘omic’ information: a bridge between genomics and systems biology. Trends Genet. 19, 551–560 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.