Abstract
Many pathological processes are not directly correlated to dramatic alterations in protein levels. The changes in local concentrations of important proteins in a subset of cells or at specific loci are likely to play a significant role in disease etiologies, but the precise location might be unknown, or the concentration might be too small to be adequately sampled for traditional proteomic techniques. Matrix-assisted laser desorption ionization (MALDI) imaging mass spectrometry (IMS) is a unique analytical method that combines analysis of multiple molecular species and of their distribution in a single platform. As reproducibility is essential for successful biomarker discovery, it is important to systematically assess data quality in biologically relevant MALDI IMS experiments. In the present study, we applied four simple tools to study the reproducibility for individual sections, within-group variation, and between-group variation of data acquired from brain sections of 21 animals divided into three treatment groups. We also characterized protein changes in distinct regions of the striatum from six-month-old rats treated neonatally (postnatal days 9–10) with the cyanobacterial toxin β-N-methylamino-l-alanine (BMAA), which has been implicated in neurodegenerative diseases. The results showed that optimized experimental settings can yield high-quality MALDI IMS data with relatively low variation (14% to 15% coefficient of variance) that allow the characterization of subtle changes in protein expression in various subregions of the brain. This was further exemplified by the dose-dependent reduction of myelin basic protein in the caudate putamen and the nucleus accumbens of adult rats neonatally treated with BMAA (150 and 460 mg/kg). The reduction in myelin basic protein was confirmed through immunohistochemistry and indicates that developmental exposure to BMAA may induce structural effects on axonal growth and/or directly on the proliferation of oligodendrocytes and myelination, which might be important for the previously shown BMAA-induced long-term cognitive impairments.
Many disease processes are characterized not by large changes in the transcriptome or proteome, but by discrete changes in a subset of cells or at specific loci. This may be particularly true for many pathophysiological processes underlying different brain disorders. The brain is highly organized in topographic maps and functionally related circuitries that might not be easily recognized or isolated for traditional proteomic analysis. In many cases the precise localization of molecular correlates to a disease remains unknown. For example, in the current study, rats treated neonatally (postnatal days (PND)1 9–10) with the cyanobacterial toxin β-N-methylamino-l-alanine (BMAA) developed transient behavioral changes such as disturbed motor function and hyperactivity in neonates (1), as well as long-term cognitive impairments in adults (2, 3). Acute neurodegeneration has been observed in several brain regions, including retrosplenial and cingulate cortices, the dentate gyrus, and CA1, after a high dose of BMAA (460 mg/kg). However, lower doses of BMAA induce cognitive impairments without any acute or long-term cell death (2, 3), and there is a need to further pinpoint brain areas with molecular changes that might be correlated with the behavioral abnormalities in the animals.
For exploratory proteomic analysis with retained spatial information, mass spectrometry imaging (or imaging mass spectrometry (IMS)) is one of the most effective methods. With the use of different sample preparation protocols, MALDI IMS can be utilized to analyze peptides and proteins as well as lipids, pharmaceuticals, and metabolites directly on tissues (4). It is possible to quantify drugs in tissue using MALDI IMS (4–6), but the protocol requires standards deposited onto tissue for reference and does not translate well to simultaneous exploratory studies of many peptides or proteins (7). The application of MALDI IMS to projects of biological and clinical significance has increased since the introduction of the technology (8). However, many papers on IMS are still of a methodological character and are based on sections from one or a few subjects, although the ultimate goal is to apply MALDI IMS in biologically relevant experiments using a large set of subjects. Whether the samples are derived from clinical cohorts or from animal models of diseases, MALDI IMS requires good reproducibility and tools to assess data quality.
The experimental sources of variation in IMS are similar across mass spectrometry (MS) platforms, including histology-directed tissue profiling, protein/serum profiling, and Surface enhanced laser desorption/ionisation time of flight (SELDI-TOF) profiling (9–11). The spectral quality depends generally on factors such as tissue/sample preparation, matrix deposition, MS acquisition, and MS processing (4, 12, 13). For example, the tissue surface needs to be carefully prepared to ensure reproducible and high-quality imaging without compromising the original spatial distribution of the molecules of interest. The choice of washing protocol, matrix, solvent composition, and deposition method will influence the outcome of the study, and each combination should be carefully chosen and optimized for each type of tissue and IMS experiment (4, 14, 15). Sensitivity can easily be assessed by plotting the signal-to-noise ratio, peak heights, or peak areas as a function of the amount of matrix (16). In addition to manual sprayers, new automated matrix deposition devices using nebulization, vibration, sublimation, and automated spotters have been developed to ensure standardized procedures (4). In the first description of matrix deposited by an acoustic spotter for tissue-profiling MS, the reproducibility of the spot-to-spot peak intensity of microdispensed matrix droplets displayed a coefficient of variance (CV) of 43%, which represented a significant improvement relative to manual matrix deposition via capillary (CV = 77% (17)). To obtain high signal quality and reproducible IMS results, we have used robotic spotting in several studies (2, 18–21). Although this strategy is time consuming and the image resolution is limited to the size and spacing of the matrix deposits (usually 200 to 300 μm), we have repeatedly observed low variance in peak intensities from several brain areas. Typically, a lower %CV is observed in the analysis of small proteins using sinapinic acid as a matrix (15% CV, compared with 30% CV using 2,5-dihydroxybenzoic acid for neuropeptide analysis) (2, 18–21). This is comparable to results for histology-directed profiling MALDI IMS (34% CV (9)).
MALDI MS data preprocessing algorithms for baseline correction, spectral noise removal, and normalization commonly used with profiling MALDI MS have been applied to imaging MS data of peptides and small proteins. Visual improvements were observed in postprocessed ion images, suggesting that this preprocessing strategy is appropriate, and it provides the basis upon which the present study was designed (22–25). In one experiment on two serial rat brain sections 72 μm apart, the molecular ion images were co-registered and the correlation coefficients of six peaks were calculated and ranged from 0.51 to 0.89, revealing relatively good section-to-section variability (25).
Some studies retrieve statistical information on different treatment groups directly from imaging data (2, 19–21, 26–31). However, most IMS studies rely on a histo-directed profiling experiment to generate data for statistical analysis, followed by a single IMS experiment of one or two sections to study the localization of interesting peptides and proteins (for example, see Refs. 22 and 32–35). This is a sensible rationale when the regions of interest are known prior to the experiment. By contrast, we have used imaging as an exploratory tool to localize affected areas and then proceeded to use histology-directed profiling to obtain data for the two following technical repeats (21).
As reproducibility is imperative for successful biomarker discovery and image analysis, it is important to systematically assess data quality in biologically relevant MALDI IMS experiments. In the present study, we applied four simple tools for assessment of the effects of sample preparation and preprocessing algorithms including the normalization of MALDI IMS data on brain tissue sections from a relatively large experiment on 21 animals divided into three treatment groups. To our knowledge, no similar strategy has been presented previously, even though the individual tools are well known. We also used MALDI IMS as a discovery tool in a non-hypothesis-driven study to pinpoint brain areas of interest and characterize long-term protein changes in adult rats neonatally treated with BMAA.
MATERIALS AND METHODS
Chemicals
Unless otherwise stated, all chemicals including BMAA hydrochloride (≥97%) were obtained from Sigma-Aldrich Co. (St. Louis, MO).
Experimental Design
The experimental design and the housing conditions were identical to those previously reported to induce acute motor disturbance in neonates and cognitive changes in adult animals (1–3). In short, pregnant outbred Wistar rats were obtained from Scanbur BK AB (Sollentuna, Sweden), and the litters were cross-fostered at the day of birth (PND 0). The male pups were given one daily subcutaneous injection (20 μl/g) of BMAA at 150 mg/kg (corresponding to 200 mg/kg BMAA HCl; n = 8) or 460 mg/kg (corresponding to 600 mg/kg BMAA HCl; n = 7) freshly dissolved in Hanks' balanced salt solution, or vehicle (n = 7), for 2 days on PND 9 to 10. After weaning on PND 22 and onward, two to four male rats were housed together in standard macrolon cages (59 cm × 38 cm × 20 cm) in their respective treatment groups. All animal experiments were approved by the Uppsala Ethical Committee on Animal Experiments and followed the guidelines of Swedish legislation on animal experimentation (Animal Welfare Act SFS1998:56) and European Union legislation (Convention ETS123 and Directive 86/609/EEC).
MALDI IMS
The animals were killed by decapitation at 23 weeks of age, and the brains were immediately frozen in dry ice and stored at −80 °C. The left hemispheres of the brains from three control animals and three animals from the high-dose group were cut in sagittal sections at a level (lateral 2.62 mm (36)) that included brain areas such as hippocampus, striatum, thalamus, amygdala, and substantia nigra for initial screening (Fig. 1A). For the full-scale experiment, coronal cryosections were obtained from the right hemispheres of all animals at the level of the striatum (2.28 mm relative to the bregma (36)) (Figs. 1B and 1C). The brain sections (12 μm) were thaw mounted on conductive glass slides suitable for MALDI IMS analysis (Bruker Daltonics, Bremen, Germany). The sections were dried under vacuum and stored at −80 °C. In total, brain sections from 21 animals were analyzed (vehicle-control, n = 6; 150 mg/kg BMAA, n = 8; 460 mg/kg BMAA, n = 7). Due to technical error in the processing of the glass slides, tissue from one control animal could not be analyzed.
Fig. 1.
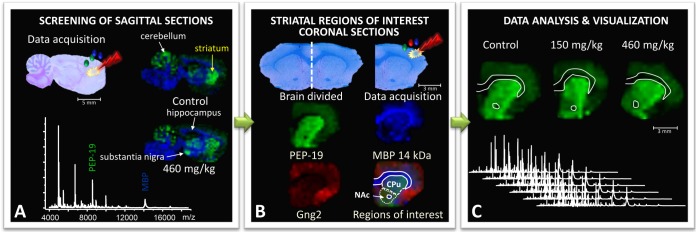
Neonatal rats were treated with BMAA (150 mg/kg and 460 mg/kg) or vehicle, and their brains were dissected and processed for MALDI IMS at adult age. A, to identify brain regions affected by BMAA, an initial screening of sagittal brain sections from the left hemispheres of animals in the high-treatment group and vehicle-treated animals was performed. Several areas displayed BMAA-induced changes in peak intensities, including the striatum and hippocampus. B, the striatum was selected for further studies in the full-scale experiment, and coronal striatal cryosections were obtained from the right hemispheres of all 21 animals (control, n = 6; BMAA 150 mg/kg, n = 8; 460 mg/kg, n = 7). For statistical analysis, distinct regions of interest (ROIs) were determined. Histology and ion distribution images of several proteins were used to define the striatum, divided into caudate putamen (CPu) and nucleus accumbens (NAc). The anterior commissure was excluded from the NAc ROIs. C, the spectra from each ROI were exported and analyzed. The peak intensity distribution of individual peaks is visualized in user-defined colors, and the maximum peak intensity in each image was set at 100% (m/z 7725 red Gng2; m/z 6718 green PEP-19; m/z 14,144 blue MBP).
To fix tissue and remove excessive salts and lipids, the cryosections were washed in 70% (30 s) and 95% (2 × 30 s) ethanol as previously described (2, 37). The MALDI matrix (10 mg/ml sinapinic acid mixed with 5 mg/ml 2′,4′-dihydroxyacetophenone in 60% ethanol, 10% ammonium acetate, and 0.3% trifluoroacetic acid) was applied using a chemical inkjet printer (CHIP-1000, Shimadzu, Kyoto, Japan) as previously described (2). Droplets of 100 pl sinapinic acid/2′,4′-dihydroxyacetophenone were placed in arrays (15 drops per pass for 24 passes) over the sections at a spatial resolution of 300 μm by 300 μm. Mass spectra were immediately acquired using an Ultraflex II (Bruker Daltonics, Bremen, Germany) equipped with smart beam technology, operating in linear positive mode. Prior to image analysis, each MALDI plate was externally calibrated using a standard protein mix (Protein Calibration Standard I, m/z range 3–25 kDa, Bruker Daltonics, Bremen, Germany). A mass range of 3.5–22.3 kDa was analyzed with a laser frequency of 100 Hz accumulating 300 laser shots per spot. Data sequence preparation, MS acquisition, and visualization were performed using the FlexImaging and FlexControl software (v 3.0, Bruker Daltonics, Bremen, Germany). Data postprocessing by means of baseline reduction, normalization by total ion current, and data reduction was performed as previously described (2, 16, 18–20). The total ion current sums before normalization were compared and found to display the characteristic normal distribution within each group and no statistical differences between groups, thus minimizing the risk of introducing artificial errors.
Regions of Interest and Data Analysis
For statistical analysis of BMAA-induced changes of peak intensities, regions of interest (ROIs) were defined (Fig. 1B). Histology together with ion-distribution images of several proteins were used to define the striatum, divided into the caudate putamen (CPu) and the nucleus accumbens (NAc). The 14-kDa ion of myelin basic protein (MBP) is associated with white matter fiber tracts and is clearly localized to the corpus callosum and the anterior commissure (38). PEP-19, which is abundant in the striatum (39), and Gng2, which is highly expressed in the adjacent subcortical structure claustrum (40), were found to have distinct localizations at the studied brain level and were therefore used to aid in the definition of the medial and lateral borders of the ROIs (Fig. 1B). The anterior commissure was excluded from the NAc ROIs (Fig. 1B). The spectra from each ROI were exported as ascii files and analyzed using Origin software (Fig. 1C). Peaks were automatically detected in each spectrum, and peak borders were determined using pBin as previously described (18). A peak was included only if it was present in at least 10% of all spectra, corresponding to about 78 out of a total 786 spectra for CPu and 23 out of 233 for NAc. In total, the peak limits were calculated for 1160 peaks for CPu and 1075 peaks in NAc, and peak area integration within the corresponding limits was performed with a self-written R script (18). The data were not log transformed in any step of the analysis.
Protein Identification and Validation via On-tissue Digestion Combined with MALDI IMS
Protein peaks were identified by means of HPLC separation followed by trypsinization and LC-MS/MS analysis as described before (2), as well as through comparisons of mass and tissue distribution previously studied via MALDI IMS (2, 16, 37, 39–41). For some proteins, including MBP, the identities were further validated by visualization of the distribution of tryptic peptides using on-tissue digestion combined with MALDI IMS as previously described (2, 38).
Immunofluorescence Analysis of MBP
Coronal cryosections from the right hemisphere of the brain were used for antigen fluorescent immunohistochemistry using mouse anti-MBP (1:1000, SMI-94, Calbiochem, Darmstadt, Germany) as the primary antibody. The epitope is within amino acids 70–89, and the antibody could therefore recognize several isoforms of MBP, including the 21.5- and 17.2-kDa forms. The fluorescent secondary antibody anti-mouse-Alexa Fluor 488 (1:500, Invitrogen, Carlsbad, CA) was used for visualization. Cell nuclei were stained by DAPI. Sections incubated without primary antibody (only the antibody diluent, 2.5% horse serum in phosphate buffered saline, in the primary antibody step) served as negative controls.
Statistical Analysis
Intergroup comparisons of protein peaks were tested using nonparametric Kruskal–Wallis analysis of variance followed by post hoc analysis using the Mann–Whitney U test. The Pearson product-moment correlation coefficient (R) was used to measure the strength of the linear dependence of intensity-intensity plots. Analysis of variance and Student's t tests were used for intergroup comparisons of the total ion current and MBP immunoreactivity. Differences were considered statistically significant at p < 0.05.
RESULTS AND DISCUSSION
Experimental Setup
In the present study, several strategies were employed to ensure signal reproducibility and data quality. Sample collection (post-mortem times of around 45 s) and the sample washing protocol were standardized. Matrix composition and application via robotic spotting were optimized in a pilot study to ensure reproducibility and high-quality images (data not shown). The sections from different treatment groups were mounted on MALDI-compatible slides in a randomized fashion to avoid the systematic experimental errors that may occur over time as the matrix is printed or spectra are acquired. To further reduce the experimental variation, all brain sections were analyzed on the same day.
Ion Distribution Maps
The first step in assessing imaging quality compared the ion distribution maps of 15 known proteins across all animals (Fig. 2). The proteins were selected among previously identified proteins with known distributions in rat and mouse brain (2, 16, 37, 39–41). Imaging and histology-directed tissue-profiling MALDI MS are characterized by identical sources of variation; however, there are undeniable benefits to the evaluation of MS data quality when it comes to imaging MS, as the evaluation of multiple ion distribution images quickly reveals overnormalization effects and defects in the experimental data (18). For instance, microtears in tissue commonly cause dispersion of the matrix and cause changes in the ratio of analyte to matrix concentrations, as well as impair crystallization into deposits that can be accurately targeted during MS acquisition. This will result in a loss of sensitivity, and the ion density maps will consistently display reduced peak intensity across several molecular ions (see inset in Fig. 2). For single ion distribution images it is possible to analyze the variation of pixel intensities, thereby obtaining an overall index of experimental success, given that the anatomical localization of the analyte is evenly distributed throughout the tissue or ROI (42). Similarly, digital image correlation analysis can be used to both measure variance and detect differences in MALDI imaging datasets (43). However, an approach incorporating the visualization and inspection of ion distribution maps as shown in Fig. 2 is a relatively fast and sensitive way to evaluate an imaging experiment. In the current experiment, a high degree of concordance was observed across treatment groups, and no cases of overnormalization effects or loss of signal sensitivity could be detected in the ROIs, the CPu and NAc (Fig. 2).
Fig. 2.
Ion distribution maps of known proteins in the rat striatum and cortex show a high degree of concordant appearance between groups (A, control group; B, 150 mg/kg BMAA group; and C, 460 mg/kg BMAA group). Each row represents the molecular images from one animal. The inset shows the effect of microtears in tissue, which result in a loss of sensitivity and ion density maps that consistently display reduced peak intensity across several molecular ions. The maximum peak intensity in each image was set at 100%. ThB10 = thymosin beta-10, ThB4 = thymosin beta-4, COX VIIa = cytochrome c oxidase subunit 7A2, PEP19 = Purkinje cell protein 4, Gng7 = guanine nucleotide-binding protein subunit gamma-7, NG = neurogranin, Gng2 = guanine nucleotide-binding protein subunit gamma-2, UBC = ubiquitin, ACBP = acyl-CoA-binding protein, CytOx = cytochrome oxidase, H4 = histone H4, Cyt C = cytochrome c somatic, MIF = macrophage migration inhibitory factor, H2b = histone H2B, and MBP 14 kDa = myelin basic protein. The proteins were selected from previously identified proteins with known distributions in rat and mouse brain (2, 16, 37, 39–41).
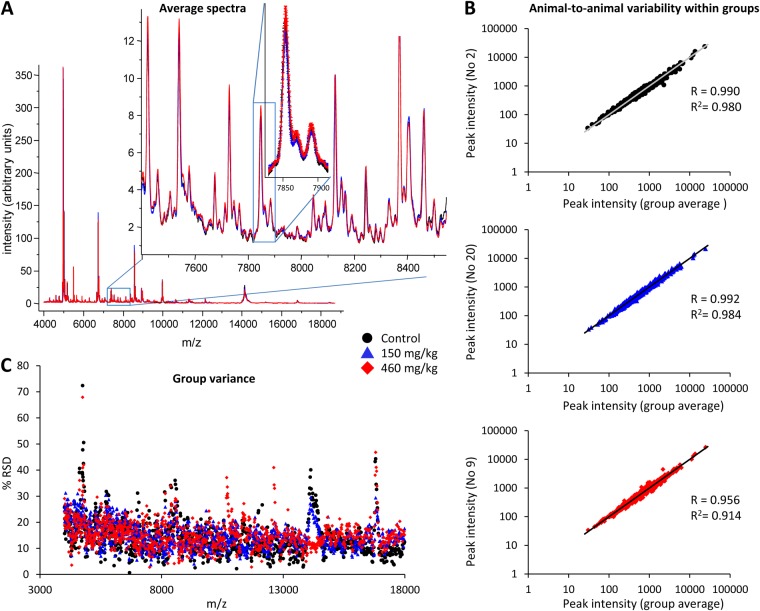
Average MS Traces
Plotting the average spectra for each ROI and group revealed a high number of peaks (Fig. 3A). About a thousand peaks were detected using Origin, and although a large number of these were peak shoulders that often are the result of oxidation and neutral losses, these were also labeled as peaks. Plotting the group average mass spectra and standard error indicated a high degree of overlap between the groups (see inset in Fig. 3A; n = 6 to 8 per group).
Fig. 3.
Assessment of experimental variability. A, average striatal mass spectra from BMAA- and vehicle-treated rats. Over 1000 peaks in the mass range of 3.5–20 kDa were detected and analyzed via MALDI IMS, including many low-intensity peaks (inset). The spectra displayed a high degree of overlap of both peaks and baseline between the treatment groups, as well as a high degree of overlap in signal variation demonstrating good IMS reproducibility (second inset, mean intensity + S.E., n = 6 to 8 per group). B, intensity-intensity plots of peak intensities in the striatum from one representative animal of each group (animals 2, 20, and 9) versus their respective group average. Each dot represents one peak. A linear trend line with a slope of 1 was added for comparison, and Pearson's coefficient R was calculated (values ranged from 0.95 to 0.99 in the experiment). C, the spread of peak measurements can be visualized by plotting the percent relative standard deviation (%RSD) of each peak area over m/z. A low %RSD of peak measurements was seen in the CPu spectra of rats treated with BMAA or vehicle. The group variances were largely overlapping and were not significantly different from each other. Control, n = 6; BMAA 150 mg/kg, n = 8; 460 mg/kg, n = 7.
Intensity-Intensity Plots for Analysis of Animal-to-animal Variability within Groups
After peak detection and selection of the peaks of interest (here, a signal-to-noise ratio of >3 and present in at least 10% of all spectra), a matrix consisting of peak intensities per ROI and animal was created, and the average peak intensity for each group and m/z was calculated. In general, plotting all peak intensities for each animal in a dataset against its own group average is one of the most effective ways to quickly reveal high animal-to-animal variation within groups. These intensity-intensity plots assume a small variation in peak intensity for each specified peak in all animals within the same treatment group. In the most successful experiment, all peaks will display a linear relationship with a slope of 1 and a Pearson's correlation coefficient close to 1 (R ≈ 1), whereas an unsuccessful experiment will not yield a linear relationship. In the present experiment, all intensity-intensity plots displayed a high degree of linearity, with R ranging from 0.95–0.99 and having a mean of 0.986 (representative plots for three animals are shown in Fig. 3B). This approach has previously been used for evaluating histology-directed profiling MS experiments (22, 34). All intensity-intensity plots for every animal are shown in supplemental Fig. S1.
Analysis of Group Variance
The data distribution for each peak can also give an estimate of the experimental variability (Fig. 3C). Because of the large differences between peak intensities within a spectrum, ranging over 3 orders of magnitude, the mean percent relative standard deviation (%RSD = S.D./mean × 100, also known as %CV) was calculated for each peak and treatment group. The mean %RSD ranged from 14.3% for the control group to 15.4% for the group that received 460 mg/kg BMAA (Fig. 3C; peak intensities were not log transformed and did not differ significantly between groups). Plotting the %RSD as a function of peak m/z can also reveal abnormalities introduced by, for example, overnormalization or data preprocessing errors (18). The %RSD plots will also show whether one group displays a higher degree of variability than the control group, indicating that the statistical analysis may be skewed.
Together, these quality measures show that the data from this relatively large MALDI IMS experiment with optimized experimental settings are of high quality with fairly low variation, which allows the characterization of subtle changes in protein expression ranging, here from 9% to 24% relative to control levels.
Effects of BMAA on Protein Expression in Striatum Determined via MALDI IMS
To identify brain regions affected by the environmental neurotoxin BMAA, an initial screening of sagittal brain sections from animals in the high-treatment group (460 mg/kg) and from vehicle-treated animals was performed. Hundreds of small proteins could be detected in the mass range from 4000 to 20,000 Da (Fig. 1A). Several areas displayed BMAA-induced changes in peak intensities, including the striatum, which was divided into CPu and NAc sections and further studied in the full-scale experiment (Fig. 1B). Our previous pathological study of the brain from the same animals showed that the high dose of BMAA could induce neuronal degeneration, calcification, and astrogliosis in the CA1 segment of the adult hippocampus that could be detected via MALDI IMS (2). However, no acute or long-term histopathological changes have so far been observed in the striatum with any dose (2, 44). In the present study, BMAA (150 mg/kg and 460 mg/kg) caused discreet long-term changes in protein intensities in both CPu and NAc of adult (neonatally treated) rats. The regional analysis of BMAA-induced changes revealed 10 and 18 statistically significantly changed protein peaks in the CPu and the NAc of both high- and low-dose groups relative to controls.
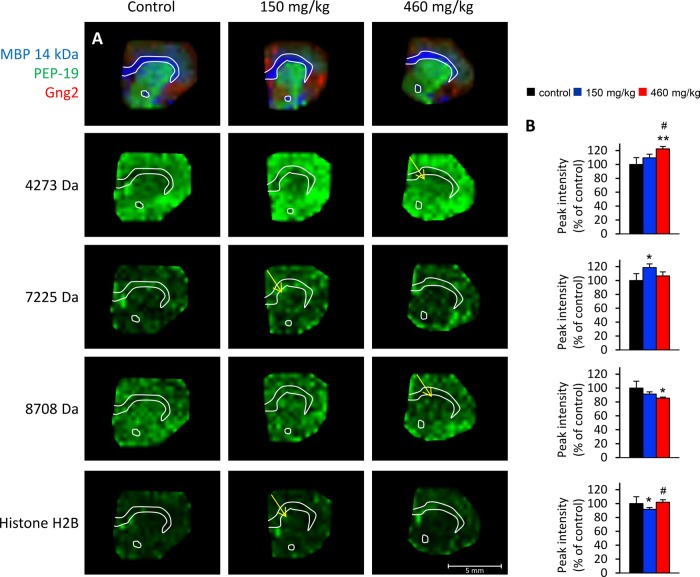
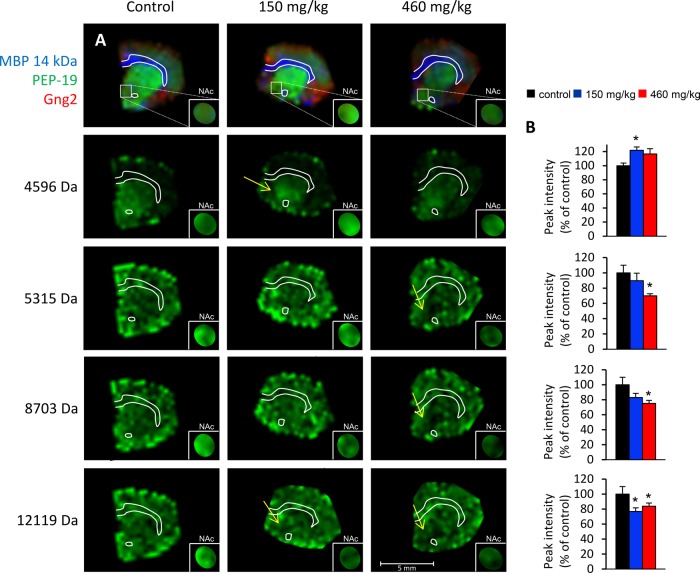
The identities of three proteins, histone H2B, MBP 14kDa, and MBP 18.5 kDa, which were affected in BMAA-treated animals, were determined as described in materials and methods. Several peaks (2 in CPu and 10 in NAc) were post-translational modifications of the MBP proteins (see below). Other affected ion peaks could not be conclusively identified (Figs. 4 and 5; supplemental Table S1). The levels of histone H2B were found to be decreased in CPu of the low-treatment group (10% decrease; Fig. 4). This could indicate effects of BMAA treatment on chromatin regulation and structure. However, no changes in post-translational modifications or in other known histones such as H2A, H3, or H4 were detected. In the NAc, most effects were localized to the dorsomedial area of the NAc, encompassing the NAc core more than the NAc shell territories (see arrows and insets in Fig. 5).
Fig. 4.
MALDI ion distribution maps of BMAA-induced long-term changes of peak intensities in the CPu of adult rats. A, four selected proteins are shown: m/z 4273, m/z 7225, m/z 8708, and m/z 13,778 histone H2B. The regional location of reduced protein expression is indicated by arrows. Proteins of interest are displayed in green and visualized at a fixed absolute intensity threshold. The upper panel shows the outline of the section and the distribution of three common proteins as shown in Fig. 1. B, the diagrams show the peak intensity (percentage of control ± S.E.) for the selected proteins in CPu. Vehicle (black), BMAA 150 mg/kg (blue), and BMAA 460 mg/kg (red). *p < 0.05 and **p < 0.01 relative to vehicle control animals. #p < 0.05 relative to 150 mg/kg (Kruskal–Wallis test and Mann–Whitney U test). Control, n = 6; BMAA 150 mg/kg, n = 8; 460 mg/kg, n = 7.
Fig. 5.
MALDI imaging of affected peak intensities of unknown identity in the NAc of adult rats neonatally treated on PND 9–10 with BMAA or vehicle. A, four selected proteins are shown: m/z 4596, m/z 5315, m/z 8703, and m/z 12,119. The regional location of reduced protein expression is indicated by arrows. Proteins of interest are highlighted in green and visualized at a fixed absolute intensity threshold. The small insets are magnifications (1.5×) of NAc with the protein of interest displayed at a higher color intensity to make interpretation of the results easier. A mask has been applied to block surrounding signal outside the NAc. The upper panel shows the outline of the section and the distribution of three common proteins, as shown in Fig. 1. B, the diagrams show the peak intensity (percentage of control ± S.E.) for the selected proteins in NAc. Vehicle (black), BMAA 150 mg/kg (blue), and BMAA 460 mg/kg (red). *p < 0.05 relative to vehicle control animals (Kruskal–Wallis test and Mann–Whitney U test). Control, n = 6; BMAA 150 mg/kg, n = 8; 460 mg/kg, n = 7.
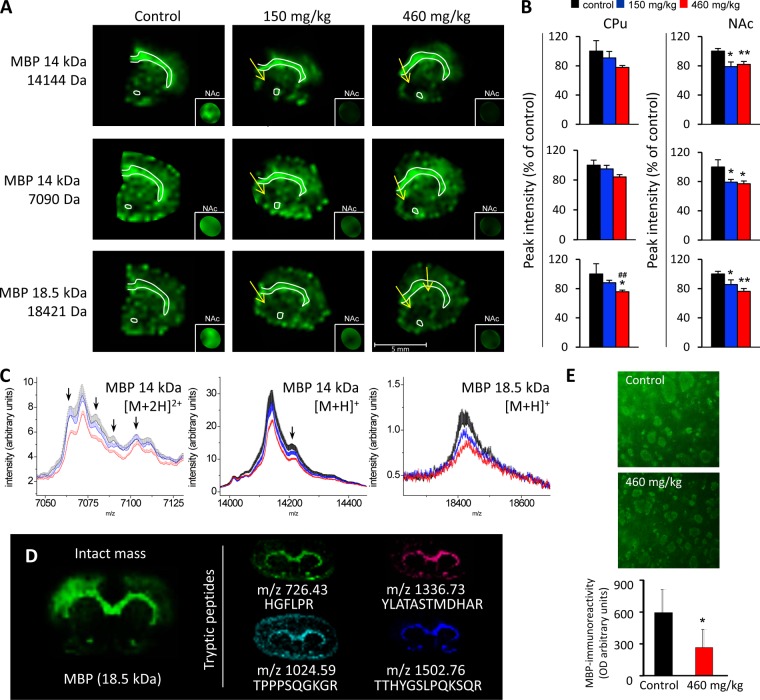
Dose-dependent Decrease of MBP in Striatum
MBP is a major structural protein in myelin that is essential for the formation and maintenance of myelin in the CNS. Knockout of MBP suggests that this protein, like many myelin-associated proteins, is probably a negative regulator of axonogenesis (45). Roles in signaling and in interactions with the cytoskeleton are examples of other suggested functions (46). Several isoforms of MBP are formed by alternative splicing of the mRNA transcript—21.5, 20.2, 18.5, 17.24, 17.22, and 14 kDa in the mouse and rat and 21.5, 20.2, 18.5, and 17.2 kDa in humans. The isoforms are differentially expressed during development, but the predominant isoform in the adult human CNS is the 18.5-kDa MBP, and the 14-kDa isoform is predominant in adult mice and rats (46). MALDI IMS can be used to analyze the levels and the localization of the different isoforms at the same time, through study of their specific intact masses. In the present study, neonatal exposure to 460 mg/kg BMAA resulted in a 25% decrease of the peak intensity of striatal 18.5-kDa MBP isoform at adult age relative to the vehicle control group. A small, nonsignificant decrease of about 10% was detected in group receiving the lower dose (150 mg/kg), indicating a dose-dependent decrease of the level of 18.5-kDa MBP in the CPu (Figs. 6A–6C). A similar result was observed for the 14-kDa MBP isoform, which was detected as both singly and doubly charged ions (Figs. 6A–6C). Most striking, a BMAA-induced reduction of both the 18.5-kDa and the 14-kDa MBP isoform occurred dorsally of the anterior commissure in the NAc, including the medial parts of the NAc core and shell (arrows and inset magnifications in Fig. 6A). The statistical analysis revealed that several post-translational modifications of both isoforms were reduced in BMAA-treated animals. A similar trend was observed in the CPu (arrows in spectra in Fig. 6C). The identity of MBP was validated by visualization of tryptic peptides using on-tissue digestion (Fig. 6D). The image analysis of immunofluorescence-stained brain sections showed a decrease in MBP optical density in the high-dose group (p < 0.05), which confirmed the MALDI IMS analysis (Fig. 6E). There was a trend toward both a lower number of small white matter fiber bundles (p = 0.12) and a reduction in bundle size (p = 0.10) in high-dose animals. Interestingly, no changes in MBP levels were detected in the hippocampus of the same animals in our previous MALDI IMS study (2).
Fig. 6.
The MALDI IMS analysis revealed a dose-dependent decrease of MBP levels in the CPu and NAc of adult rats neonatally treated on PND 9–10 with BMAA or vehicle. A, the 14-kDa (both singly and doubly charged) and 18.5-kDa MBP isomers are shown in green. The regional location of reduced protein expression in BMAA-treated animals is indicated by arrows. The small insets are magnifications (1.5×) of NAc with the protein of interest displayed at a higher color intensity to make the interpretation of the results easier. A mask has been applied to block surrounding signal outside the NAc. All proteins were visualized at the same fixed absolute intensity threshold. B, the bar histograms show the peak intensity (percentage of control ± S.E.) for the selected proteins in CPu and NAc. Vehicle (black), BMAA 150 mg/kg (blue), and BMAA 460 mg/kg (red). *p < 0.05 and **p < 0.01 relative to vehicle control animals. ##p < 0.01 relative to 150 mg/kg (Kruskal–Wallis test and Mann–Whitney U test). Control, n = 6; BMAA 150 mg/kg, n = 8; 460 mg/kg, n = 7. C, the corresponding average spectra (± S.E. bars) from CPu show the protein of interest as well as several possible post-translational modifications that also are reduced in BMAA-treated animals. Arrows point to peak maxima that have the same tissue distribution as the main MBP peak. Vehicle (black), BMAA 150 mg/kg (blue), and BMAA 460 mg/kg (red). D, the identity of MBP was validated using on-tissue tryptic digestion MALDI IMS by comparing the distribution of tryptic peptides with the distribution of the intact mass of the protein (18.5-kDa isoform is shown) on an adjacent section. The small images show tryptic peptides generated from the digestion of all main isoforms of MBP. E, immunofluorescence showing the expression of MBP in the NAc of rats treated with BMAA (460 mg/kg) or vehicle. A distinct decrease in the intensity and area of fiber bundles was seen in the NAc of a BMAA-treated rat relative to the control. The image analyses revealed a decrease in MBP expression in the high-dose group and confirmed the MALDI IMS analysis. Control, n = 6; BMAA 460 mg/kg, n = 7.
The dose-dependent reduction of MBP in CPu and NAc appears not to be isoform specific, but a general reduction of MBP may reflect lower protein expression levels, a reduced number of axons, or changed myelination of the existing axons in the striatum. This could indicate BMAA-induced effects on neurodevelopmental processes such as axonal growth and/or direct effects on the proliferation of oligodendrocytes and myelination. The time window for exposure of the rat pups (PND 9 and 10) corresponds to a phase of fast growth of the brain. During this period, rapid neurodevelopmental changes such as axonal and dendritic growth, establishment of neuronal connections, synaptogenesis, apoptosis, and proliferation of oligodendrocytes followed by myelination take place in a strictly controlled and region-specific way (47). The glutamate receptor agonist BMAA may affect these neurodevelopmental processes directly, as the glutamatergic system is important for the modulation of these events (48). Peptidomic analyses of the neonatal striatum have also revealed BMAA-induced changes in neuropeptides such as enkephalins and VGF nerve growth factor–derived peptides that are critical for brain development (49). Myelination of axons by oligodendrocytes is necessary for the rapid conduction of impulses and accelerates the communication between neurons (50, 51). Myelin integrity is therefore important for functional neurocircuitry, and demyelination and the loss/dysfunction of oligodendrocytes are features of disease states such as multiple sclerosis and Alzheimer disease (52) and may contribute to cognitive deficits associated with aging (53). BMAA may affect the myelination directly, as the glutamatergic system stimulates oligodendrocyte proliferation and promotes myelination and the overstimulation of glutamate receptors kills these myelin-producing glial cells via an excitotoxic mechanism (54, 55). This is further supported by a recent study that showed BMAA to be gliotoxic (56). The hippocampus has direct projections to the striatum (57, 58), so it is also possible that the decreased levels of MBP are secondary to the neuronal loss previously observed in the hippocampus of some animals (2), as neonatal brain lesions have been observed to induce a loss of myelination in the projection areas of adult rats (59). However, neuronal cell loss was detected in only four of the animals in the high-dose group. Furthermore, none of the animals in the lower dose group had any observable neuronal lesions in the hippocampus or in any other studied brain areas. The observed decrease in MBP could contribute to the learning and memory impairments seen in the animals, as the striatum is important for several cognitive processes and cognitive tasks, especially procedural learning (60–62). The finding may also be of importance for the suggested linkage of cyanobacteria, BMAA, and avian vacuolar myelinopathy (63, 64).
CONCLUSION
Reproducibility is essential for successful biomarker discovery, and much effort has previously been put into assessing and improving data quality for MALDI IMS. In the present study we applied methods for data assessment and the reduction of experimental variability in a relatively large MALDI IMS experiment with brain sections from 21 animals. The study shows that optimized experimental settings can yield high-quality IMS data with relatively low variation (here 14% to 15% CV) that allow the characterization of subtle changes in protein expression (here 9% to 24% of the control group) in various subregions of the brain. Through the use of MALDI IMS as a discovery tool in a non-hypothesis-driven approach, rats neonatally treated with BMAA (150 mg/kg and 460 mg/kg) were shown to have experienced a dose-dependent reduction of MBP in the CPu and the NAc six months after exposure. The reduction in MBP was confirmed with immunohistochemistry and may reflect lower protein expression or a reduced number of myelinated axons in the striatum. This could be of importance for the explanation of the previously reported cognitive impairments in the animals.
Supplementary Material
Acknowledgments
Ms. Raili Engdahl and Professor Eva Brittebo are gratefully acknowledged for technical assistance and for valuable comments during the preparation of the manuscript, respectively.
Footnotes
* Financial support was given by the Swedish Research Council FORMAS and the Swedish Research Council (Grant Nos. 522-2006-6414 and 521-2007-5407).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- PND
- postnatal day
- BMAA
- β-N-methylamino-l-alanine
- CPu
- caudate putamen
- CV
- coefficient of variance
- IMS
- imaging mass spectrometry
- MALDI
- matrix-assisted laser desorption ionization
- MBP
- myelin basic protein
- NAc
- nucleus accumbens
- ROI
- region of interest
- %RSD
- percent relative standard deviation.
REFERENCES
- 1. Karlsson O., Lindquist N. G., Brittebo E. B., Roman E. (2009) Selective brain uptake and behavioral effects of the cyanobacterial toxin BMAA (beta-N-methylamino-L-alanine) following neonatal administration to rodents. Toxicol. Sci. 109, 286–295 [DOI] [PubMed] [Google Scholar]
- 2. Karlsson O., Berg A. L., Lindstrom A. K., Hanrieder J., Arnerup G., Roman. E., Bergquist J., Lindquist N. G., Brittebo E. B., Andersson M. (2012) Neonatal exposure to the cyanobacterial toxin BMAA induces changes in protein expression and neurodegeneration in adult hippocampus. Toxicol. Sci. 130, 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Karlsson O., Roman E., Brittebo E. B. (2009) Long-term cognitive impairments in adult rats treated neonatally with beta-N-methylamino-L-alanine. Toxicol. Sci. 112, 185–195 [DOI] [PubMed] [Google Scholar]
- 4. Norris J. L., Caprioli R. M. (2013) Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chem. Rev. 113, 2309–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stoeckli M., Staab D., Schweitzer A. (2007) Compound and metabolite distribution measured by MALDI mass spectrometric imaging in whole-body tissue sections. Int. J. Mass Spectrom. 260, 195–202 [Google Scholar]
- 6. Nilsson A., Fehniger T. E., Gustavsson L., Andersson M., Kenne K., Marko-Varga G., Andren P. E. (2010) Fine mapping the spatial distribution and concentration of unlabeled drugs within tissue micro-compartments using imaging mass spectrometry. PLoS One 5, e11411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pan S., Aebersold R., Chen R., Rush J., Goodlett D. R., McIntosh M. W., Zhang J., Brentnall T. A. (2009) Mass spectrometry based targeted protein quantification: methods and applications. J. Proteome Res. 8, 787–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Caprioli R. M., Farmer T. B., Gile J. (1997) Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 69, 4751–4760 [DOI] [PubMed] [Google Scholar]
- 9. Cornett D. S., Mobley J. A., Dias E. C., Andersson M., Arteaga C. L., Sanders M. E., Caprioli R. M. (2006) A novel histology-directed strategy for MALDI-MS tissue profiling that improves throughput and cellular specificity in human breast cancer. Mol. Cell. Proteomics 5, 1975–1983 [DOI] [PubMed] [Google Scholar]
- 10. Dijkstra M., Vonk R. J., Jansen R. C. (2007) SELDI-TOF mass spectra: a view on sources of variation. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 847, 12–23 [DOI] [PubMed] [Google Scholar]
- 11. Villanueva J., Philip J., Chaparro C. A., Li Y., Toledo-Crow R., DeNoyer L., Fleisher M., Robbins R. J., Tempst P. (2005) Correcting common errors in identifying cancer-specific serum peptide signatures. J. Proteome Res. 4, 1060–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Albrethsen J. (2007) Reproducibility in protein profiling by MALDI-TOF mass spectrometry. Clin. Chem. 53, 852–858 [DOI] [PubMed] [Google Scholar]
- 13. Hilario M., Kalousis A., Pellegrini C., Muller M. (2006) Processing and classification of protein mass spectra. Mass Spectrom. Rev. 25, 409–449 [DOI] [PubMed] [Google Scholar]
- 14. Schwartz S. A., Reyzer M. L., Caprioli R. M. (2003) Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J. Mass Spectrom. 38, 699–708 [DOI] [PubMed] [Google Scholar]
- 15. Heeren R. M. A., Kükrer-Kaletaş B., Taban I. M., MacAleese L., McDonnell L. A. (2008) Quality of surface: the influence of sample preparation on MS-based biomolecular tissue imaging with MALDI-MS and (ME-)SIMS. Appl. Surf. Sci. 255, 1289–1297 [Google Scholar]
- 16. Hanrieder J., Wicher G., Bergquist J., Andersson M., Fex-Svenningsen A. (2011) MALDI mass spectrometry based molecular phenotyping of CNS glial cells for prediction in mammalian brain tissue. Anal. Bioanal. Chem. 401, 135–147 [DOI] [PubMed] [Google Scholar]
- 17. Aerni H. R., Cornett D. S., Caprioli R. M. (2006) Automated acoustic matrix deposition for MALDI sample preparation. Anal. Chem. 78, 827–834 [DOI] [PubMed] [Google Scholar]
- 18. Hanrieder J., Ljungdahl A., Andersson M. (2012) MALDI imaging mass spectrometry of neuropeptides in Parkinson's disease. J. Vis. Exp. 60, 3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hanrieder J., Ljungdahl A., Falth M., Mammo S. E., Bergquist J., Andersson M. (2011) L-DOPA-induced dyskinesia is associated with regional increase of striatal dynorphin peptides as elucidated by imaging mass spectrometry. Mol. Cell. Proteomics 10, M111.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ljungdahl A., Hanrieder J., Falth M., Bergquist J., Andersson M. (2011) Imaging mass spectrometry reveals elevated nigral levels of dynorphin neuropeptides in L-DOPA-induced dyskinesia in rat model of Parkinson's disease. PLoS One 6, e25653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hanrieder J., Ekegren T., Andersson M., Bergquist J. (2013) MALDI imaging of post-mortem human spinal cord in amyotrophic lateral sclerosis. J. Neurochem. 124, 695–707 [DOI] [PubMed] [Google Scholar]
- 22. Norris J. L., Cornett D. S., Mobley J. A., Andersson M., Seeley E. H., Chaurand P., Caprioli R. M. (2007) Processing MALDI mass spectra to improve mass spectral direct tissue analysis. Int. J. Mass Spectrom. 260, 212–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Williams B., Cornett S., Dawant B., Crecelius A., Bodenheimer B., Caprioli R. (2005) An algorithm for baseline correction of MALDI mass spectra. In Proceedings of the 43rd Annual Southeast Regional Conference, Vol. 1, pages 137–142 Editor: Mario Guimaraes ACM Press, Kennesaw, GA [Google Scholar]
- 24. Deininger S. O., Cornett D. S., Paape R., Becker M., Pineau C., Rauser S., Walch A., Wolski E. (2011) Normalization in MALDI-TOF imaging datasets of proteins: practical considerations. Anal. Bioanal. Chem. 401, 167–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McDonnell L. A., van Remoortere A., van Zeijl R. J., Deelder A. M. (2008) Mass spectrometry image correlation: quantifying colocalization. J. Proteome Res. 7, 3619–3627 [DOI] [PubMed] [Google Scholar]
- 26. Oppenheimer S. R., Mi D., Sanders M. E., Caprioli R. M. (2010) Molecular analysis of tumor margins by MALDI mass spectrometry in renal carcinoma. J. Proteome Res. 9, 2182–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Willems S. M., van Remoortere A., van Zeijl R., Deelder A. M., McDonnell L. A., Hogendoorn P. C. (2010) Imaging mass spectrometry of myxoid sarcomas identifies proteins and lipids specific to tumour type and grade, and reveals biochemical intratumour heterogeneity. J. Pathol. 222, 400–409 [DOI] [PubMed] [Google Scholar]
- 28. Jones E. A., van Remoortere A., van Zeijl R. J., Hogendoorn P. C., Bovee J. V., Deelder A. M., McDonnell L. A. (2011) Multiple statistical analysis techniques corroborate intratumor heterogeneity in imaging mass spectrometry datasets of myxofibrosarcoma. PLoS One 6, e24913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Onishi S., Tatsumi Y., Wada K., Yang H. J., Sugiura Y., Setou M., Yoshikawa H. (2013) Sulfatide accumulation in the dystrophic terminals of gracile axonal dystrophy mice: lipid analysis using matrix-assisted laser desorption/ionization imaging mass spectrometry. Med. Mol. Morphol. 46, 160–165 [DOI] [PubMed] [Google Scholar]
- 30. Meistermann H., Norris J. L., Aerni H. R., Cornett D. S., Friedlein A., Erskine A. R., Augustin A., De Vera Mudry M. C., Ruepp S., Suter L., Langen H., Caprioli R. M., Ducret A. (2006) Biomarker discovery by imaging mass spectrometry: transthyretin is a biomarker for gentamicin-induced nephrotoxicity in rat. Mol. Cell. Proteomics 5, 1876–1886 [DOI] [PubMed] [Google Scholar]
- 31. Kriegsmann M., Seeley E. H., Schwarting A., Kriegsmann J., Otto M., Thabe H., Dierkes B., Biehl C., Sack U., Wellmann A., Kahaly G. J., Schwamborn K., Caprioli R. M. (2012) MALDI MS imaging as a powerful tool for investigating synovial tissue. Scand. J. Rheumatol. 41, 305–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Uys J. D., Grey A. C., Wiggins A., Schwacke J. H., Schey K. L., Kalivas P. W. (2010) Matrix-assisted laser desorption/ionization tissue profiling of secretoneurin in the nucleus accumbens shell from cocaine-sensitized rats. J. Mass Spectrom. 45, 97–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Burnum K. E., Cornett D. S., Puolitaival S. M., Milne S. B., Myers D. S., Tranguch S., Brown H. A., Dey S. K., Caprioli R. M. (2009) Spatial and temporal alterations of phospholipids determined by mass spectrometry during mouse embryo implantation. J. Lipid Res. 50, 2290–2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yanagisawa K., Shyr Y., Xu B. J., Massion P. P., Larsen P. H., White B. C., Roberts J. R., Edgerton M., Gonzalez A., Nadaf S., Moore J. H., Caprioli R. M., Carbone D. P. (2003) Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet 362, 433–439 [DOI] [PubMed] [Google Scholar]
- 35. Stauber J., Lemaire R., Franck J., Bonnel D., Croix D., Day R., Wisztorski M., Fournier I., Salzet M. (2008) MALDI imaging of formalin-fixed paraffin-embedded tissues: application to model animals of Parkinson disease for biomarker hunting. J. Proteome Res. 7, 969–978 [DOI] [PubMed] [Google Scholar]
- 36. Paxinos G., Watson C. (2007) The Rat Brain in Stereotaxic Coordinates, 6th ed., Academic Press, Oxford [Google Scholar]
- 37. Andersson M., Groseclose M. R., Deutch A. Y., Caprioli R. M. (2008) Imaging mass spectrometry of proteins and peptides: 3D volume reconstruction. Nat. Methods 5, 101–108 [DOI] [PubMed] [Google Scholar]
- 38. Groseclose M. R., Andersson M., Hardesty W. M., Caprioli R. M. (2007) Identification of proteins directly from tissue: in situ tryptic digestions coupled with imaging mass spectrometry. J. Mass Spectrom. 42, 254–262 [DOI] [PubMed] [Google Scholar]
- 39. Skold K., Svensson M., Nilsson A., Zhang X., Nydahl K., Caprioli R. M., Svenningsson P., Andren P. E. (2006) Decreased striatal levels of PEP-19 following MPTP lesion in the mouse. J. Proteome Res. 5, 262–269 [DOI] [PubMed] [Google Scholar]
- 40. Mathur B. N., Caprioli R. M., Deutch A. Y. (2009) Proteomic analysis illuminates a novel structural definition of the claustrum and insula. Cereb. Cortex 19, 2372–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pierson J., Norris J. L., Aerni H. R., Svenningsson P., Caprioli R. M., Andren P. E. (2004) Molecular profiling of experimental Parkinson's disease: direct analysis of peptides and proteins on brain tissue sections by MALDI mass spectrometry. J. Proteome Res. 3, 289–295 [DOI] [PubMed] [Google Scholar]
- 42. Kallback P., Shariatgorji M., Nilsson A., Andren P. E. (2012) Novel mass spectrometry imaging software assisting labeled normalization and quantitation of drugs and neuropeptides directly in tissue sections. J. Proteomics 75, 4941–4951 [DOI] [PubMed] [Google Scholar]
- 43. Thiele H., Heldmann S., Trede D., Strehlow J., Wirtz S., Dreher W., Berger J., Oetjen J., Kobarg J. H., Fischer B., Maass P. (2013) 2D and 3D MALDI-imaging: conceptual strategies for visualization and data mining. Biochim. Biophys. Acta March 4 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 44. Karlsson O., Roman E., Berg A. L., Brittebo E. B. (2011) Early hippocampal cell death, and late learning and memory deficits in rats exposed to the environmental toxin BMAA (beta-N-methylamino-L-alanine) during the neonatal period. Behav. Brain Res. 219, 310–320 [DOI] [PubMed] [Google Scholar]
- 45. Phokeo V., Kwiecien J. M., Ball A. K. (2002) Characterization of the optic nerve and retinal ganglion cell layer in the dysmyelinated adult Long Evans Shaker rat: evidence for axonal sprouting. J. Comp. Neurol. 451, 213–224 [DOI] [PubMed] [Google Scholar]
- 46. Boggs J. M. (2006) Myelin basic protein: a multifunctional protein. Cell. Mol. Life Sci. 63, 1945–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Davison A. N., Dobbing J. (1968) Applied Neurochemistry, pp. 253–316, Blackwell Scientific Publications, Oxford, UK [Google Scholar]
- 48. Ruediger T., Bolz J. (2007) Neurotransmitters and the development of neuronal circuits. Adv. Exp. Med. Biol. 621, 104–115 [DOI] [PubMed] [Google Scholar]
- 49. Karlsson O., Kultima K., Wadensten H., Nilsson A., Roman E., Andren P. E., Brittebo E. B. (2013) Neurotoxin-induced neuropeptide perturbations in striatum of neonatal rats. J. Proteome Res. 12, 1678–1690 [DOI] [PubMed] [Google Scholar]
- 50. Guerri C., Pascual M. (2010) Mechanisms involved in the neurotoxic, cognitive, and neurobehavioral effects of alcohol consumption during adolescence. Alcohol 44, 15–26 [DOI] [PubMed] [Google Scholar]
- 51. Rodier P. M. (1995) Developing brain as a target of toxicity. Environ. Health Perspect. 103, 73–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McTigue D. M., Tripathi R. B. (2008) The life, death, and replacement of oligodendrocytes in the adult CNS. J. Neurochem. 107, 1–19 [DOI] [PubMed] [Google Scholar]
- 53. Albert M. (1993) Neuropsychological and neurophysiological changes in healthy adult humans across the age range. Neurobiol. Aging 14, 623–625 [DOI] [PubMed] [Google Scholar]
- 54. Cavaliere F., Urra O., Alberdi E., Matute C. (2012) Oligodendrocyte differentiation from adult multipotent stem cells is modulated by glutamate. Cell Death Dis. 3, e268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Matute C., Alberdi E., Domercq M., Sanchez-Gomez M. V., Perez-Samartin A., Rodriguez-Antiguedad A., Perez-Cerda F. (2007) Excitotoxic damage to white matter. J. Anat. 210, 693–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chiu A. S., Gehringer M. M., Braidy N., Guillemin G. J., Welch J. H., Neilan B. A. (2013) Gliotoxicity of the cyanotoxin, beta-methyl-amino-L-alanine (BMAA). Sci. Rep. 3, 1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cohen M. X., Schoene-Bake J. C., Elger C. E., Weber B. (2009) Connectivity-based segregation of the human striatum predicts personality characteristics. Nat. Neurosci. 12, 32–34 [DOI] [PubMed] [Google Scholar]
- 58. Friedman D. P., Aggleton J. P., Saunders R. C. (2002) Comparison of hippocampal, amygdala, and perirhinal projections to the nucleus accumbens: combined anterograde and retrograde tracing study in the Macaque brain. J. Comp. Neurol. 450, 345–365 [DOI] [PubMed] [Google Scholar]
- 59. Schneider M., Koch M. (2005) Behavioral and morphological alterations following neonatal excitotoxic lesions of the medial prefrontal cortex in rats. Exp. Neurol. 195, 185–198 [DOI] [PubMed] [Google Scholar]
- 60. Devan B. D., Hong N. S., McDonald R. J. (2011) Parallel associative processing in the dorsal striatum: segregation of stimulus-response and cognitive control subregions. Neurobiol. Learn Mem. 96, 95–120 [DOI] [PubMed] [Google Scholar]
- 61. Myhrer T. (2003) Neurotransmitter systems involved in learning and memory in the rat: a meta-analysis based on studies of four behavioral tasks. Brain Res. Brain Res. Rev. 41, 268–287 [DOI] [PubMed] [Google Scholar]
- 62. Knowlton B. J., Mangels J. A., Squire L. R. (1996) A neostriatal habit learning system in humans. Science 273, 1399–1402 [DOI] [PubMed] [Google Scholar]
- 63. Bidigare R. R., Christensen S. J., Wilde S. B., Banack S. A. (2009) Cyanobacteria and BMAA: possible linkage with avian vacuolar myelinopathy (AVM) in the south-eastern United States. Amyotroph. Lateral Scler. 10 Suppl 2, 71–73 [DOI] [PubMed] [Google Scholar]
- 64. Wiley F. E., Wilde S. B., Birrenkott A. H., Williams S. K., Murphy T. M., Hope C. P., Bowerman W. W., Fischer J. R. (2007) Investigation of the link between avian vacuolar myelinopathy and a novel species of cyanobacteria through laboratory feeding trials. J. Wildl. Dis. 43, 337–344 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.