Abstract
This review focuses on chaperone-mediated autophagy (CMA), one of the proteolytic systems that contributes to degradation of intracellular proteins in lysosomes. CMA substrate proteins are selectively targeted to lysosomes and translocated into the lysosomal lumen through the coordinated action of chaperones located at both sides of the membrane and a dedicated protein translocation complex. The selectivity of CMA permits timed degradation of specific proteins with regulatory purposes supporting a modulatory role for CMA in enzymatic metabolic processes and subsets of the cellular transcriptional program. In addition, CMA contributes to cellular quality control through the removal of damaged or malfunctioning proteins. Here, we describe recent advances in the understanding of the molecular dynamics, regulation and physiology of CMA, and discuss the evidence in support of the contribution of CMA dysfunction to severe human disorders such as neurodegeneration and cancer.
Keywords: chaperone-mediated autophagy, neurodegeneration, cancer, aging
Introduction
Intracellular proteins are subjected to continuous turn-over through coordinated synthesis, degradation and recycling of their component amino acids. This constant renewal of the proteome assures its proper functioning and permits a tight control of intracellular levels of proteins as a way to modulate multiple intracellular processes1,2. Proteins can undergo degradation by the proteasome or by lysosomes. Delivery of proteins to lysosomes for degradation, or autophagy, can occur through different mechanisms3. In some instances, such as macroautophagy, proteins are sequestered in vesicles that form in the cytosol and then fuse with lysosomes to transfer their contents for degradation. In other cases, such as microautophagy, proteins are trapped inside vesicles that form directly through the invagination of the lysosomal membrane. These vesicles then pinch off into the lysosomal lumen and are degraded by the proteases inside lysosomes. However, not all lysosomal delivery involves vesicles. Proteins can be targeted from the cytosol to the lysosomal membrane and then gain access to the lumen of this organelle by directly crossing its membrane. This process is known as chaperone-mediated autophagy (CMA) and constitutes the focus of this review4.
One of the distinctive features of CMA is that proteins that undergo degradation by this autophagic pathway are selected individually through a recognition motif in their amino acid sequences5. This allows for the removal of specific proteins without disturbance of neighboring ones and makes CMA an efficient system for degradation of damaged or abnormal proteins, and surplus subunits of multi-protein complexes. In addition, this selectivity permits CMA to play a regulatory role in multiple cellular processes by contributing to modulate intracellular levels of enzymes, transcription factors and cell maintenance proteins4.
In this review, we first summarize the main steps and components involved in CMA and the diversity of physiological functions attributed to this autophagy pathway, and then we discuss the consequences of CMA malfunctioning in the context of disease and aging.
How was CMA discovered?
Even in the early years, when it was widely accepted that autophagy was a non-selective in bulk process, some studies started to propose that selective degradation by lysosomes could be a possibility. This notion originated from the observation that cytosolic proteins underwent degradation in lysosomes at different rates6. This was further supported experimentally by the fact that removal of nutrients from the culture media accelerated the lysosomal degradation of some proteins microinjected into the cytosol of cultured fibroblasts, but it did not affect degradation of other proteins delivered by the same procedure7. Enzymatic fragmentation of one of the proteins susceptible to starvation-induced lysosomal degradation led to the identification of a pentapeptide motif in that protein that was necessary8,9 and sufficient10 to mediate its lysosomal degradation. Shortly after, this pentapetide11 was identified as a binding site for a cytosolic chaperone, hsc70, and this binding was proven necessary for the lysosomal degradation of the substrate protein12.
Selectivity was not the only unexpected feature of this new form of lysosomal degradation. In fact, the characteristic that still makes this type of autophagy, nowadays known as CMA, very distinct from other autophagic processes is the fact that the substrate proteins seem to cross the lysosomal membrane directly to reach the lumen. In contrast to macroautophagy, where intermediate cytosolic vesicles (autophagosomes) are responsible for trapping the cargo protein and delivering it to lysosomes, two independent groups were able to reproduce in vitro this chaperone-dependent uptake and degradation of cytosolic proteins by lysosomes isolated either from fibroblast or from rat liver13,14. This transport of substrate was also very different from microautophagy because arrival of substrates to the lysosomal lumen did not require the formation of the characteristic invaginations of the lysosomal membrane that trap cytosolic substrates in the case of microautophagy. Furthermore, the in vitro studies demonstrated that the chaperone-dependent lysosomal degradation was saturable at the level of lysosomal binding and uptake, and required the presence of some specific proteins at the lysosomal membrane because partial degradation of lysosomal surface proteins was sufficient to block both binding and translocation of substrates13,15.
The molecular dissection of this process using the in vitro system with isolated lysosomes, cells in culture and different organs from rodents led to the identification of the subset of lysosomal proteins that mediate substrate binding and uptake. Along with integral membrane proteins, these studies demonstrated that specific chaperones were required at both sides of the lysosomal membrane to complete substrate translocation. The dependence on chaperones was the reason that motivated the naming of this process as CMA in 200016.
How does CMA work?
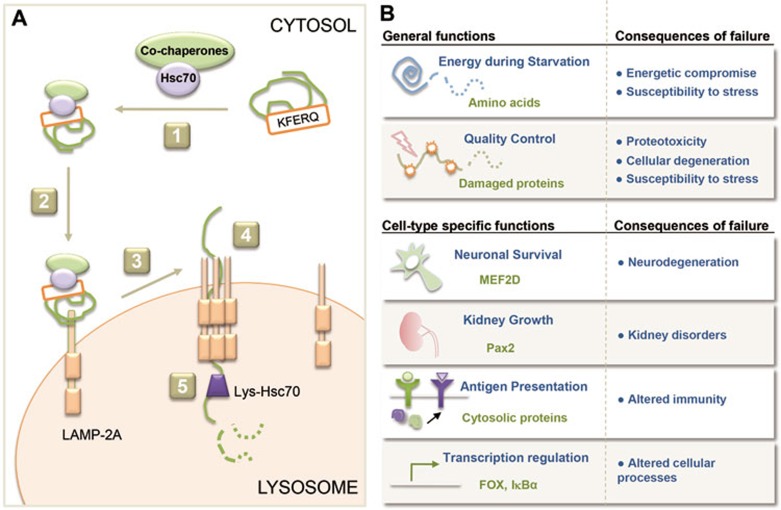
CMA is a multi-step process that involves: (I) substrate recognition and lysosomal targeting; (II) substrate binding and unfolding; (III) substrate translocation and (IV) substrate degradation in the lysosomal lumen (Figure 1A).
Figure 1.
Steps and physiological functions of CMA. (A) Proteins degraded by CMA are identified in the cytosol by a chaperone complex that, upon binding to the targeting motif in the substrate protein (1), brings it to the surface of lysosomes (2). Binding of the substrate to the cytosolic tail of the receptor protein LAMP-2A promotes LAMP-2A multimerization to form a translocation complex (3). Upon unfolding, sustrate proteins cross the lysosomal membrane (4) assisted by a luminal chaperone and reach the lysosomal matrix where they undergo complete degradation (5). (B) General and cell-type specific functions of CMA and consequences of CMA failure in different organs and systems.
Recognition of substrate proteins takes place in the cytosol through the binding of a constitutive chaperone, the heat shock-cognate protein of 70 KDa (hsc70), to a pentapeptide motif present in the amino acid sequences of all CMA substrates12. This motif consists of an invariant amino acid, a glutamine (Q) residue, at the beginning or end of the sequence, one of the two positively charged amino acids, lysine (K) or arginine (R), one of the four hydrophobic amino acids, phenylalanine (F), valine (V), leucine (L) or isoleucine (I) and one of the two negatively charged amino acids, glutamic acid (E) or aspartic acid (D)5. The fifth amino acid in the sequence can be one of the indicated positive or hydrophobic residues. Motifs can become accessible for chaperone recognition after protein unfolding in the case of motifs buried in the core of the protein; after proteins disassemble from multiprotein complexes if the motif was hidden in the regions of protein-protein interaction; or when proteins are released from the subcellular membranes in those instances where the motif is in the region of binding to the membrane. The fact that the CMA motif is based on the charge of the amino acids makes it possible to create a motif out of an incomplete four-amino acid motif through post-translational modifications such as phosphorylation or acetylation. For example, phosphorylation of a cysteine (C), serine (S) or tyrosine (Y) residue can provide the negative charge missing in some incomplete motifs. In addition, acetylation of a K residue makes it comparable to the Q missing in some partial motifs, which explains the recent discovery that acetylation contributes to the targeting of some glycolytic enzymes17 or even of pathogenic proteins such as huntingtin18 to lysosomes for degradation via CMA. Although still not demonstrated experimentally, it is also plausible that in those motifs where the positive charge is contributed by a K residue, acetylation of this residue or even ubiquitination may prevent recognition and binding by hsc70, and reduce CMA degradation of the cognate substrates when they are post-translationally modified.
Once bound to the chaperone, the substrate is targeted to the surface of the lysosomes where it interacts with the cytosolic tail of the single-span membrane protein lysosome-associated membrane protein type 2A (LAMP-2A19) (Figure 1A). This protein is one of the three variants that originate from the alternative splicing of a single gene (lamp2) and that have different cytosolic and transmembrane regions, but share a common luminal domain. LAMP-2A is present at the lysosomal membrane as monomers and in association with other proteins to form a multi-protein complex required for substrate translocation20. The assembly of LAMP-2A into this complex is dynamic and is driven by the binding of the substrate to this receptor protein. During the transition from monomer to multimer, the stability of LAMP-2A is maintained through its interaction with a form of hsp90 located at the luminal side of the lysosomal membrane20. The substrate can bind to the receptor while still in a folded state, but in order to cross the lysosomal membrane, the substrate needs to undergo unfolding21. This process is likely mediated by hsc70 and some of its cochaperones detected at the lysosomal membrane, and is completed before the LAMP-2A complex is fully assembled (Figure 1A).
The mechanism of substrate internalization and the way in which multimerization of LAMP-2A contributes to this process are not fully understood. Multimerization of LAMP-2A requires a GXXG motif present in the transmembrane region of this protein. Interestingly, mutations in this region that prevent multimerization block substrate translocation but do not affect binding of substrates to the cytosolic tail of LAMP-2A20. However, under experimental conditions in which LAMP-2A is locked in the multimeric conformation, binding of substrates to the cytosolic tail is no longer possible. These findings disproved our initial prediction that a complex of LAMP-2A with CMA-related chaperones was stably present at the lysosomal membrane to mediate substrate translocation. On the contrary, later experiments revealed that the multimerization of LAMP-2A is transient and that as soon as the substrate is translocated into the lumen, LAMP-2A disassembles from this complex in an hsc70-dependent manner20. The multimeric complex that mediates substrate translocation is of a defined size (700 kDa) and is composed mainly of LAMP-2A molecules but at least two other proteins can also be found associated to the complex22,23 (Kon et al., unpublished). Interestingly, these additional components seem important to regulate the stability of the multimeric complex but not essential for translocation, as inserting LAMP-2A in proteoliposomes in the presence of hsc70 is sufficient to mediate substrate translocation, although understandably at a lower efficiency than lysosomes (Cuervo AM and Dice JF, unpublished).
Translocation of the substrate protein across the lysosomal membrane requires the presence of a form of hsc70 (lys-hsc70) normally resident in lysosomes8. Although the specific way in which this chaperone contributes to translocation has not been elucidated yet, it has been proposed that it may function actively by pulling the substrate proteins in a ratchet-like manner, or alternatively may hold onto the substrate passively to prevent its return to the cytosol. Hsc70 bears in its amino acid sequence two targeting motifs for CMA and can indeed behave as a substrate protein24. However, it is unlikely that lys-hsc70 reaches the lysosomal lumen through CMA since experimental blockage of CMA does not affect the content of hsc70 in lysosomes25. Likewise, blockage of macroautophagy does not reduce the luminal content of hsc70 either, discarding that the chaperone could be delivered upon cytosolic sequestration through autophagosome-lysosome fusion26. It is possible that hsc70 is internalized as multivesicular bodies from the surface of late endosomes and reaches the lysosomal compartment through endosomal-lysosomal fusion. Once in the lumen, hsc70 stability is highly dependent on the lysosomal pH, where small increases in pH are sufficient to promote its degradation in this compartment24.
After the substrate is translocated into the lysosomal lumen, LAMP-2A is rapidly dissembled from the translocation complex into monomers where substrates can bind again20. Binding and translocation of substrates by CMA are coordinated steps but they can be dissociated under conditions that compromise assembly of LAMP-2A into the translocation complex. Consequently, the rate of CMA can be modulated by the rate of assembly/disassembly of the translocation complex20. Multiple factors may participate in the regulation of this process, for example, changes in the fluidity of the lysosomal membrane that affect lateral mobility or changes in protein density of this membrane27. To date, a pair of proteins, GFAP and EF1α, have been the first one described to specifically modulate LAMP-2A assembly/disassembly in a GTP-dependent manner (since EF1α is a GTP-binding protein)22. Association of GFAP to the translocation complex contributes to its stabilization. Once the substrate has passed through the complex, disassembly occurs by the mobilization of GFAP from the complex to bind phosphorylated forms of GFAP resident in the membrane and normally caped by EF1α.
Rates of CMA are also directly dependent on the content of LAMP-2A at the lysosomal membrane. Levels of LAMP-2A can be regulated through transcriptional upregulation, as in the case of oxidative stress28, or through changes in the degradation rate of LAMP-2A at the lysosomal membrane, as occurs when CMA is upregulated during prolonged starvation29,30. Although there is still relatively limited information about the signaling pathways that contribute to modulating CMA activity, a negative regulatory effect of the nuclear receptor, retinoic acid receptor α, on CMA has been recently described31. Signaling through this receptor inhibits, among others, LAMP-2A transcription and this blockage is released when CMA upregulation is required.
Validation of a protein as a CMA substrate requires more than the mere identification of a putative CMA targeting motif in its amino acid sequence, as this motif has also been recently shown to be utilized for targeting of proteins to late endosomes for endosomal microautophagy32. Although both CMA and endosomal microautophagy share hsc70 as the targeting chaperone, the dependence on LAMP-2A is exclusive for CMA. Ultimately, the best way to confirm that a protein is a CMA substrate is to reproduce its binding and translocation across the membrane of isolated lysosomes33.
What are the physiological functions of CMA?
The first role proposed for CMA was to contribute to amino acid recycling during prolonged starvation, a condition in which CMA is maximally activated15 (Figure 1B). Although some levels of basal CMA activity can be detected in almost all cells, starvation has been one of the best characterized stimuli for CMA. In contrast to macroautophagy that becomes activated shortly after starvation and reaches its maximal peak, at least for protein degradation, around 4-6 h, CMA is gradually activated after 8-10 h of starvation and persists at maximal activity for up to three days15,25. Proteins degraded under these conditions were thought to be those proteins no longer necessary for cells under starvation, which could be broken down for amino acid recycling to sustain synthesis in the absence of nutrients. However, it is also possible that this form of regulated degradation under prolonged starvation allows for changes in the proteome aimed at adapting the cell to the new conditions. For example, degradation of inhibitors of transcription factors by CMA during starvation has already been demonstrated for specific transcription programs34. Likewise, degradation of regulatory metabolic enzymes by CMA has been shown to contribute to the metabolic changes that allow, for example, for cancer cells to adapt to low nutrient conditions17,35.
The other important function that CMA fulfills in cells is quality control, directly linked to the ability of this pathway to selectively remove single proteins from the cytosol (Figure 1B). CMA is upregulated during oxidative stress, where it contributes to the degradation of oxidized proteins28. In fact, inability to upregulate CMA under those conditions results in marked accumulation of oxidative damage and reduced cellular viability25. It is also possible that CMA contributes to quality control of cytosolic-assembled protein complexes by eliminating excess subunits. For example, different subunits of the catalytic core of the proteasome bear CMA-targeting motifs and have been shown to be selectively degraded in lysosomes by CMA36. Whether their degradation is a pre-assembly event or whether subunits that already formed part of the proteasome are actively disassembled and targeted to CMA as a way to reduce overall proteasome activity, requires future investigation.
CMA is also upregulated in other conditions that lead to protein damage such as exposure to denaturing toxic compounds15. Activation of CMA has also been shown to support survival of retinal cells upon activation of a pro-apoptotic program in those cells37. Recent studies have demonstrated that CMA is activated during hypoxia and this activation is required for cell survival38. Although the specific substrates degraded by CMA under these conditions are not fully elucidated, the fact that the hypoxia-inducible factor 1 has been confirmed as a CMA substrate39 supports a possible regulatory effect of CMA on the intensity and duration of the cellular response to hypoxia.
In addition to these general functions of CMA, there are also other instances in which activation of CMA contributes to modulate cell type-specific functions (Figure 1B). For example, degradation of the transcription factor Pax2 by CMA in kidney is important to control tubular cell growth40, and explains why in conditions such as diabetes when CMA is compromised, kidneys undergo pronounced hypertrophy41. Selective degradation of a neuronal survival factor, transcription factor myocyte enhancer factor 2D, has been shown to occur, at least in part by CMA, and this is essential to assure proper neuronal response to injury42. In this case, intuitively a reduction of CMA activity could be considered beneficial as it should contribute to increasing cellular levels of myocyte enhancer factor 2D. However, the myocyte enhancer factor 2D removed by CMA is no longer the functional form, which will compromise function of the active form if remained in neurons. This selective turnover of the inactive forms of proteins by CMA has also been shown in the case of specific cancer cells for the PMK2 enzyme, which, once inactive, is targeted for removal by CMA upon acetylation17. In this case too, removal of the non-functional enzyme is required to maintain the functional pool.
Although the purpose of CMA in most cells is the complete degradation of proteins into their constituent amino acids, in some specialized cells such as professional antigen presenting cells, a role for CMA in antigen presentation has been proposed43. Future studies are needed to determine if partial degradation of CMA substrates can also occur in other cell types. Study of the consequences of CMA blockage in different cells types could help to identify additional cell-specific functions. To date, the best way to block this autophagic pathway is through knock-down or knock-out of LAMP-2A, because manipulation of any of the other CMA components (hsc70, hsp90, GFAP or EF1α), is usually more difficult to interpret due to the additional cellular functions of these proteins. Although chemical activators of CMA are now available31, considerably less progress has been made in the identification of specific inhibitors of this pathway. Unfortunately, some of the small molecules initially described to modulate CMA44 have proven to be rather non-specific and to affect the activity of other cellular quality-control mechanisms, again making interpretation of results obtained with these drugs difficult.
How do alterations in CMA contribute to disease?
Increasing evidence demonstrates that malfunction of CMA plays a key role in the pathogenesis of severe human disorders45,46,47. Often, the mechanisms underlying the alterations of CMA in these pathologies involve perturbations in the functioning of the CMA translocation complex. Both diminished and enhanced CMA activities have been shown to associate with diseases, which highlights the importance of a tight regulation of CMA activity. In this review, we have selected neurodegenerative diseases as an example of pathologies associated with reduced CMA activity, and oncongenic processes as an example of pathological conditions in which enhanced CMA activity facilitates disease progression.
Reduced CMA and neurodegeneration
A common theme unifying different neurodegenerative pathologies is the failure of the proteolytic systems to adequately dispose of deleterious proteins2,48. Such mishandling of aberrant proteins alters proteostasis and often leads to the precipitation of protein aggregates that contribute to neuronal demise49. The involvement of CMA in neurodegeneration is two-fold, as it contributes to the elimination of pathogenic proteins, but also often, becomes victim of the toxic effect of these aberrant proteins50,51,52,53. This dual role of CMA in neurodegenerative disorders makes it necessary to analyze the status of this autophagy pathway in disease in order to determine whether interventions aimed at enhancing CMA activity could be of potential value or not, depending on the degree of compromise of this pathway and the reversibility of the CMA blockage.
Parkinson's disease (PD)
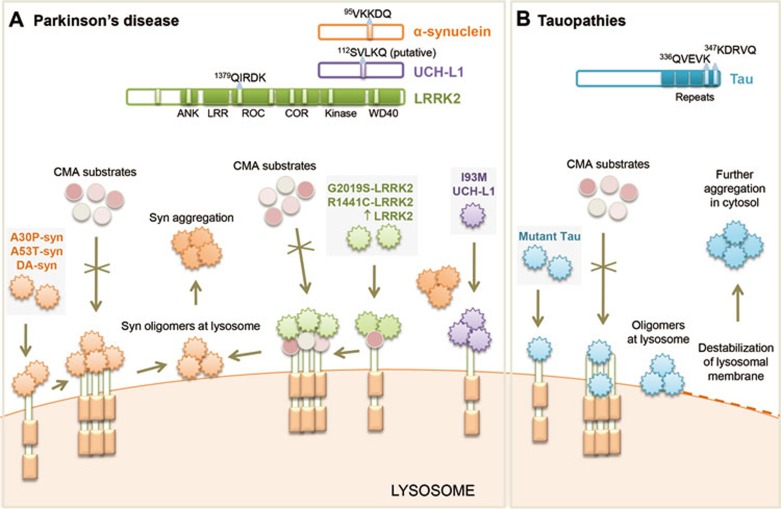
Impairment of CMA is intimately linked to the pathogenesis of Parkinson's disease (PD) - the most prevalent neurodegenerative movement disorder that is characterized by the selective loss of dopaminergic neurons and subsequent motor deficits. Dysfunction in CMA has been described in both familial50,52,54,55 and sporadic51,52 PD. In the case of familial PD, sequence analysis reveals the presence of CMA-targeting motifs in the majority of PD-related proteins, supporting an important role for CMA in the control of their intracellular levels (Figure 2A). Indeed, the two most commonly mutated proteins in patients with familial PD, α-synuclein and leucine-rich repeat kinase 2 (LRRK2), have both been demonstrated to undergo degradation in lysosomes via CMA using various experimental systems such as isolated lysosomes, primary mouse neuronal cultures, mouse models of PD and even neuronal-differentiated induced pluripotent stem cells and brains from familial50,52,54,55 and sporadic51,52 PD patients. In contrast, pathogenic mutant variants of α-synuclein (for example, A30P and A53T mutants) and of LRRK2 (for example, G2019S and R1441C mutants), despite being recognized by cytosolic hsc70 and successfully delivered to the lysosomal membrane, fail to reach the lysosomal lumen to be degraded by CMA50,52 (Figure 2A). Internalization of α-synuclein and LRRK2 mutants into lysosomes is obstructed due to aberrant interactions of these toxic proteins with LAMP-2A. Pathogenic α-synuclein mutants bind to LAMP-2A with abnormally high affinity thus preventing their translocation across the lysosomal membrane50 (Figure 2A). Similarly, LRRK2 mutant proteins show enhanced lysosomal binding in the presence of other CMA substrates, which in the process interfere with the proper organization of the active CMA translocon52 (Figure 2A). The toxic interactions of α-synuclein and LRRK2 mutants with the CMA transporter preclude not only their own degradation, but also inhibit the degradation of other CMA substrates50,52.
Figure 2.
Impairment of CMA by pathogenic proteins contributes to neurodegeneration. (A) Mechanisms of CMA failure in Parkinson's disease (PD). Many PD-related proteins bear CMA-targeting motifs (α-synuclein, UCH-LI and LRRK2 shown here) (top). LRRK2 has eight CMA-targeting motifs but only the sequence of the most commonly used is shown. Both wild-type α-synuclein and LRRK2 are degraded by CMA. Mutant forms of these proteins and of UCH-L1 bind abnormally to LAMP-2A, albeit via different mechanisms, leading to blockage of their own degradation as well as degradation of other CMA substrates. Dopamine-modified α-synuclein and abnormally high levels of wild-type LRRK2 also impair CMA. Failure of CMA causes accumulation and aggregation of these toxic proteins that could contribute to Lewy body formation in PD. Alterations of CMA by mutant LRRK2 and UCH-L1 show converging toxic effects on α-synuclein aggregation. (B) Perturbation of CMA by mutant tau in tauopathies. wild-type tau protein is a bona fide CMA substrate carrying two CMA-targeting motifs (top). Pathogenic variants of tau fail to translocate fully into the lysosomal lumen. Such inefficient translocation promotes partial cleavage of tau and formation of tau oligomers at the lysosomal membrane resulting in destabilization of lysosomal membrane and lysosomal leakage. Release of lysosomal tau oligomers into the cytosol may act as a precursor for further tau aggregation.
Particularly noteworthy is that mutations in one of the PD-related proteins also interfere with the degradation of other pathogenic proteins by CMA52. For example, mutant LRRK2 exacerbates the intracellular accumulation of α-synuclein, in part by preventing its clearance through CMA. Although α-synuclein is still delivered to lysosomes, it fails to translocate into the lumen due to the disruption of the CMA translocation complex by LRRK2 (Figure 2A). Interestingly, the persistence of α-synuclein bound to the lysosomal membrane promotes its multimerization into toxic oligomers that further compromise CMA activity, and could, in principle, contribute to the seeding of protein aggregates characteristic of this disorder. These dual pathogenic effects of reduced elimination of the pathogenic protein and additional CMA blockage should considerably trigger a more severe PD pathology in LRRK2-mediated PD cases. Similar abnormal interaction with CMA components has also been described for another PD-associated protein, the ubiquitin C-terminal hydrolase L1 (UCH-L1). Wild-type UCH-L1 interacts with CMA-related chaperones and LAMP-2A, but the levels of these interactions are abnormally increased by the PD-linked I93M mutation in UCH-L1 leading to blockage of α-synuclein degradation by CMA56 (Figure 2A).
Alterations of CMA have also been implicated in sporadic PD, which accounts for the majority of the PD cases51,52. Perturbation of α-synuclein degradation as a consequence of unfavorable post-translational modifications caused by environmental or cellular stressors (for example, pesticides, oxidative stress, so on) is a key event in the pathogenesis of sporadic PD and various synucleinopathies57. Among the different post-translational modifications described for the α-synuclein proteins that accumulate in inclusions in PD neurons, dopamine-modified α-synuclein has shown reduced susceptibility to CMA degradation in a manner similar to that of familial α-synuclein mutants51. Like α-synuclein mutants, the tight binding yet inefficient translocation of dopamine-modified α-synuclein by the CMA translocation complex inhibits the degradation of dopamine-modified α-synuclein as well as other CMA substrates. Similar to the case of the mutant variants, the persistence of the post-translationally modified forms of α-synuclein bound to the lysosomal membrane, and likely the arrival of other CMA substrates at the lysosomal surface, promote the formation of highly toxic α-synuclein oligomers or protofibrils at the lysosomal membrane51. Recent studies support that even in the absence of noticeable post-translational modifications, an increase in the cellular levels of either α-synuclein50 or LRRK252 beyond a tolerable threshold, has very similar inhibitory effects on CMA activity. In fact, these two proteins seem to potentiate each other's toxic effect on CMA52.
The proposed involvement of CMA malfunction in PD pathogenesis is further supported by the overt changes in the CMA components seen in the post-mortem brain samples from PD patients. Increased levels of LAMP-2A have been observed in the early stages of PD both in mouse models54 and in brains of PD patients52. However, in advanced stages, reduced levels of LAMP-2A and hsc70 have been detected instead in the dopaminergic neurons of human brain regions58. In fact, there seems to be a good correlation between regional deficiency in LAMP-2A, used as a surrogate marker for CMA function, and the selective vulnerability of the brain regions to α-synuclein aggregation59. Although, as described in later sections, age-related changes in the LAMP-2A protein itself could be behind the gradual reduction in CMA and subsequent acceleration of the disease in the older patients; a recent study shows that deregulation of microRNAs in PD brains may underlie the down-regulation of some CMA components in the affected neurons60. In addition, a sequence variation in the promoter region of LAMP-2 identified recently in a PD patient61, opens up the possibility that alterations in CMA components may be behind some forms of PD. The fact that both chemical31 and genetic62 upregulation of CMA have been shown to be capable of alleviating cellular toxicity associated with pathogenic forms of α-synuclein supports that the changes in CMA observed in PD are not a mere consequence of the disease, but that rather they contribute to pathogenesis.
Alzheimer's disease
The compromise of CMA function as a consequence of blockage of this autophagic pathway by the pathogenic variants of proteins that normally undergo degradation by CMA is not limited to PD. In fact, CMA blockage also plays a role in influencing the levels of neurofibrillary tangles arising from the aggregation of mutant tau proteins associated with Alzheimer's disease and tauopathies53. As in the case of the PD-related proteins, normal tau undergoes degradation by CMA upon hsc70 recognition of one of the two targeting motifs in its C-terminus53 (Figure 2B). In contrast, mutant tau variants exhibit peculiar lysosomal processing by CMA. The mutant tau protein, once bound to LAMP-2A, is only partially internalized and the portion of the protein that gains entry into the lysosomal lumen is trimmed, resulting in the formation of smaller amyloidogenic tau fragments at the lysosomal membrane53. These fragments oligomerize directly at the surface of lysosomes, resulting in disruption of the lysosomal membrane integrity and blockage of normal CMA function. Release of these toxic mutant tau oligomers from the lysosomes upon membrane rupture could further seed tau aggregation in the cytosol by acting as a nucleating center (Figure 2B). Besides tau, a connection between CMA and a second Alzheimer's disease-related protein, the regulator of calcineurin 1 (RCAN1) has also been established. CMA degrades RCAN1, a protein whose high expression in Alzheimer's disease brains has been linked to neuronal demise63. The possible contribution of the blockage of CMA by pathogenic tau to the elevation of RCAN1 levels in the affected neurons awaits evaluation. In light of the described interplay between different PD-pathogenic proteins in the context of CMA, it is plausible that the pathogenic effect of mutant tau and the aberrant levels of RCAN1 are interconnected and both contribute to determining the severity of the Alzheimer's disease phenotype.
Abnormally enhanced CMA and cancer
While CMA deficiency characterizes many neurodegenerative pathologies, upregulation of CMA has been linked to the survival and proliferation of cancer cells17,35,64. Examination of CMA activity in a wide array of cancer cell lines and human tumor biopsies has demonstrated a consistent increase in basal CMA activity35. Activation of CMA is mostly due to an increase in the LAMP-2A levels in these cancer cells and tumors. Genetic knock-down of LAMP-2A in cancer cells helped to establish that CMA is required for cancer cell proliferation, optimal tumor growth and metastasis35.
The beneficial effect associated with upregulated CMA may be different depending on the cell type. The enhanced CMA has been shown to be necessary to sustain enhanced glycolysis to meet the bioenergetic demand of rapid cancer cell growth and proliferation. Selective blockage of CMA in cancer cells results in transcriptional attenuation of several rate-limiting glycolytic enzymes, and the subsequent reduction in glycolysis and ATP production35. In other cancers, the decrease in glycolysis observed upon CMA blockage is not due to changes at the transcriptional level, but rather at the protein level. As mentioned in previous sections, the inactive forms of PKM2, one of the rate-limiting glycolytic enzymes, are eliminated via CMA17. Blockage of CMA in cancer cells leads to the accumulation of these non-functional forms of PKM2 and the subsequent decrease in glycolysis and energetic compromise. Lastly, it is possible that in other tumors, part of the negative consequences of CMA blockage are due to reduced quality control. While in many cancer cell types, prolonged blockage of CMA elicits upregulation of the ubiquitin proteasome system (UPS), thus preventing accumulation of damaged substrates normally degraded by CMA35, in other types of cancers, accumulation of oxidized and misfolded proteins has been proposed to underlie the toxic effect of CMA blockage23,64. The upregulated CMA in these latter cells helps to ameliorate intracellular stress and promotes the activation of oncogenic survival pathways23. The mechanism behind induction of CMA in tumorigenesis is still unclear, although it is tempting to postulate that microRNA deregulation may underlie the cancer-related increase in LAMP-2A expression as seen in the aberrant microRNA-mediated regulation of CMA components in neurodegeneration60.
Although the dependence of cancer cells on CMA suggests a pro-oncogenic function for CMA, the effect of CMA in normal cells seems to be quite the opposite, as it protects cells from the damage caused by extracellular and intracellular injuries, which, if allowed to accumulate could facilitate oncogenesis. In addition, a recent study shows that CMA takes on an anti-oncogenic role in non-proliferating tumor cells by reducing the cellular levels of mutant p53 through CMA degradation65. Furthermore, CMA has also been shown to control proteolysis of another pro-oncogenic protein, the epidermal growth factor receptor pathway substrate 8 (Eps8) implicated in solid malignancies66.
In summary, a complex relationship exists between CMA and cancer biology which warrants further studies to better understand the multifaceted roles of CMA in tumorigenesis. Nevertheless, from the therapeutic point of view, manipulation of CMA is highly promising based on the fact that blockage of CMA in human tumor explants in mice through knock-down of LAMP-2A has proven effective in not only reducing tumor growth and metastasis but also in inducing tumor shrinkage through cancer cell necrotic death35.
How does aging affect CMA?
Alterations in CMA are not limited only to pathological conditions. Functional decline in CMA also occurs with physiological aging. Reduced CMA activity has been observed in many cell types and tissues of old rodents, as well as in cells derived from aged individuals16,67. Age-dependent decay in CMA appears to be caused by age-related changes in the lipid constituents of lysosomal membrane that alter the dynamics and stability of LAMP-2A in the lysosomes of old organisms27,67,68. While the transcription, synthesis and lysosomal targeting of the LAMP-2A protein during lysosome biogenesis remain unchanged from young to old individuals, the stability of LAMP-2A at the lysosomal membrane is greatly compromised with increasing age67. Undesired changes in the lipid composition of the lysosomal membrane abnormally enhance the degradation of LAMP-2A in the lysosomal lumen, and as a result, the binding and translocation of substrate proteins by lysosomes are markedly reduced in older organisms68. Interestingly, similar lipid changes can be induced at the lysosomal membrane through diets with high lipidic content, thus underscoring the importance of the diet in the control of this autophagic pathway and the possible acceleration of its decline with age.
Experimental blockage of CMA activity in cultured cells suggests that a direct consequence of the age-dependent failure in CMA is the loss of CMA-mediated homeostasis such as the removal of oxidatively damaged proteins and the ability to respond to stressors69. Consequently, age-dependent decline in CMA could be a major aggravating factor in accelerating the pathological changes in many age-related disorders. Genetic manipulation to preserve CMA function in old rodents by expressing an exogenous copy of LAMP-2A in mouse liver has proven effective in improving the healthspan of aged animals70. Restored CMA functions in the LAMP-2A transgenic animals result in improved cellular homeostasis, enhanced resistance to different stressors and preservation of organ functions. Such pronounced beneficial effects in prolonging healthspan underscore CMA as an important anti-aging mechanism.
Does CMA communicate with other proteolytic systems to maintain proteostasis?
An important aspect to keep in mind when thinking about the contribution of CMA to cellular physiology and disease is the fact that CMA does not function in isolation or completely independent of other degradative systems. On the contrary, CMA activity is tightly coordinated not only with that of other forms of autophagy but also with the UPS (reviewed in71,72). The points of interaction between CMA and these systems are multiple and seem to function in different directions.
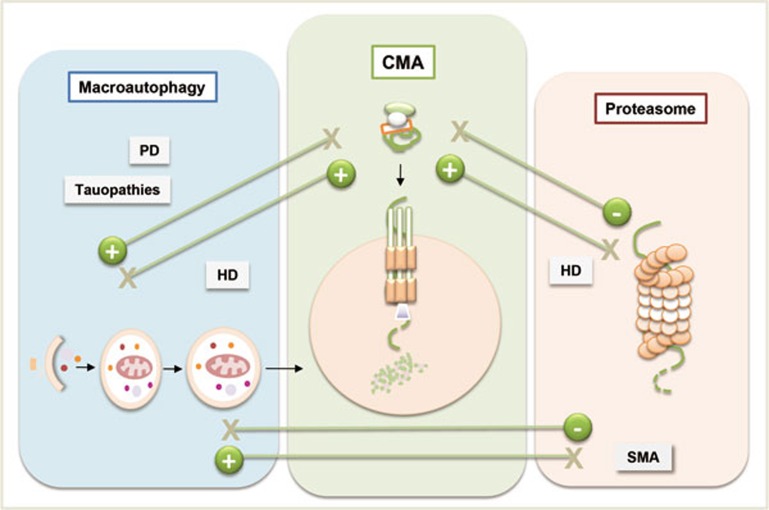
Compensatory upregulation of CMA has been described to limit the severity of damages caused by primary defects in macroautophagy and UPS in cultured cells (Figure 3). Cross talk between CMA and macroautophagy is exemplified by the observed constitutive activation of CMA in cells deficient in macroautophagy73. Similarly, many cells respond to chemical blockage of the proteasome by upregulating CMA74. Conversely, macroautophagy is highly induced in response to CMA blockage25. Although the functions of macroautophagy and CMA are distinct and non-redundant, they can still compensate for each other to sustain cell survival under normal conditions. However, in the event of stresses, this compensatory response may not suffice to allow cells to cope well. Additionally, compromised CMA perturbs functioning of the UPS, at least during the early stages of acute CMA blockage75, likely by affecting the turnover of specific proteasome subunits76, which in turn may alter proteasome assembly. Interestingly, the maintenance of efficient CMA functions in old rodents has been shown to also positively preserve UPS activity70.
Figure 3.
Cross talk between different proteolytic systems. Increasing evidence shows that different proteolytic systems are wired through multi-levels of interactions to maintain cellular homeostasis. Examples of cross talk implicated in neurodegenerative diseases are highlighted in grey boxes. PD, Parkinson's disease; HD, Huntington's disease; SMA, Spinal muscular atrophy.
This coordinated nature of the proteolytic responses could explain the late onset of many age-related disorders that originate from loss of proteostasis. For example, recent studies have revealed that constitutive upregulation of CMA77 compensates for the dual failure of macroautophagy78,79 and UPS80 seen in mouse models of Huntington's disease (HD). Activation of CMA in HD is achieved through both enhanced transcription and increased stability of LAMP-2A in the affected cells77. However, the ability of CMA to compensate for the severe proteolytic deficiency in HD cells is limited by the progressive functional decline in CMA with age (Figure 3).
Synergy between macroautophagy and CMA is also crucial in PD and in certain tauopathies where the blockage of CMA is often compensated by activation of macroautophagy53,81,82,83. This upregulation of macroautophagy is crucial for removing the toxic α-synuclein and tau oligomers to alleviate these conditions81,82,83.
Understanding the molecular determinants that efficiently reroute a protein substrate from one degradative pathway to another or upregulate the activity of one pathway over the other would allow us to better manipulate the various proteolytic systems to efficiently correct proteostasis deficiencies and, in this way, slow down disease progression. Molecules such as ubiquitin could become interesting therapeutic targets in lieu of their putative important role in the cross talk among autophagic pathways. For example, adding to the already well-characterized role of ubiquitin in proteasome-mediated degradation, cells utilize specific ubiquitin topology to promote aggregation of pathogenic proteins in transitory aggresomal structure for removal by macroautophagy84. The recent identification of enhancers that promote this type of aggrephagy of pathogenic proteins and of determinants that differentiate their degradation by basal or inducible macroautophagy82, may allow, in the future, to upregulate specific stress-coping mechanisms to prevent overloading and subsequent blockage of other components of the proteostasis system.
Concluding remarks
In recent years, the identification of a plethora of new CMA substrates and of deficiencies in CMA associated with diverse human pathologies has expanded our understanding of the importance of CMA in multiple cellular functions. These findings have also provided a wider perspective of the repercussion that CMA deregulation has on cellular and organismal homeostasis and function. The growing number of connections between CMA and human diseases has generated interest in modulating CMA activity for therapeutic purposes. Genetic manipulation to enhance CMA has proved useful in mitigating mutant α-synuclein-induced neurodegeneration in mouse models of PD62. Similarly, experimental upregulation of CMA also attenuates the toxicity associated with HD in brain slice cultures18 and interventions that enhance targeting of the HD toxic protein to CMA have also succeeded in slowing down neurodegeneration in HD mouse models85. Interestingly, rather than targeting the pathogenic proteins that induce CMA blockage in each of these diseases, the promising results from attempting to slow down the decline of CMA with age by preserving normal levels of the CMA receptor until late in life, support that restoration of CMA in old organisms could be of value to prevent a myriad of age-related diseases.
While genetic interventions may be unfeasible or highly challenging in the elderly, recent studies have provided more amenable options through the modulation of dietary lipid intake and the development of retinoic acid derivatives that specifically regulate CMA without affecting other autophagic pathways31,68. Inverse interventions may be considered under those instances such as cancer in which blockage of CMA may be useful. However, the challenge remains in administrating selective blockage of CMA only in cancer cells without compromising normal CMA functions in healthy cells. Also of potential therapeutic value are interventions targeting the cross talk among different autophagic pathways, although additional research in this area is required before these types of interventions can be implemented. Thus far, little is known about the molecular modulators of this cross talk and the universality of these processes. For example, while studies in fibroblasts, hepatocytes and neurons support that compensation between CMA and macroautophagy in these cells is bidirectional, recent studies show that retinal cells can respond to the blockage of macroautophagy by upregulating CMA, although the compensation does not work in the opposite direction86, highlighting a critical role of CMA in the maintenance of retinal homeostasis.
Acknowledgments
We thank Dr Samantha Orenstein-Hutten (Albert Einstein College of Medicine, USA) for critically reviewing this manuscript. Work in our laboratories is supported by grants from the NIH/NIA, NIH/NINDS, the P. Glenn Foundation and a generous gift from R&R Belfer (to AMC) and by an ARF Tier 1 M4010961 (to EW).
References
- van Oosten-Hawle P, Porter RS, Morimoto RI. Regulation of organismal proteostasis by transcellular chaperone signaling. Cell. 2013;153:1366–1378. doi: 10.1016/j.cell.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth DM, Balch WE. Modeling general proteostasis: proteome balance in health and disease. Curr Opin Cell Biol. 2011;23:126–134. doi: 10.1016/j.ceb.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- Dice JF, Walker CD, Byrne B, Cardiel A. General characteristics of protein degradation in diabetes and starvation. Proc Natl Acad Sci USA. 1978;75:2093–2097. doi: 10.1073/pnas.75.5.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auteri JS, Okada A, Bochaki V, Dice JF. Regulation of intracellular protein degradation in IMR-90 human diploid fibroblasts. J Cell Physiol. 1983;115:159–166. doi: 10.1002/jcp.1041150210. [DOI] [PubMed] [Google Scholar]
- Agarraberes FA, Dice JF. A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J Cell Sci. 2001;114:2491–2499. doi: 10.1242/jcs.114.13.2491. [DOI] [PubMed] [Google Scholar]
- Backer JM, Bourret L, Dice JF. Regulation of catabolism of microinjected ribonuclease A requires the amino-terminal 20 amino acids. Proc Natl Acad Sci USA. 1983;80:2166–2170. doi: 10.1073/pnas.80.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer J, Dice J. Covalent linkage of ribonuclease S-peptide to microinjected proteins causes their intracellular degradation to be enhanced by serum withdrawal. Proc Natl Acad Sci USA. 1986;83:5830–5834. doi: 10.1073/pnas.83.16.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HI, Dice JF. Peptide sequences that target proteins for enhanced degradation during serum withdrawal. J Biol Chem. 1988;263:6797–6803. [PubMed] [Google Scholar]
- Chiang H, Terlecky S, Plant C, Dice JF. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989;246:382–385. doi: 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Terlecky SR, Dice JF, Knecht E. Selective binding and uptake of ribonuclease A and glyceraldehyde-3-phosphate dehydrogenase by isolated rat liver lysosomes. J Biol Chem. 1994;269:26374–26380. [PubMed] [Google Scholar]
- Terlecky SR, Chiang H-L, Olson TS, Dice JF. Protein and peptide binding and stimulation of in vitro lysosomal proteolysis by the 73-kDa heat shock cognate protein. J Biol Chem. 1992;267:9202–9209. [PubMed] [Google Scholar]
- Cuervo AM, Knecht E, Terlecky SR, Dice JF. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol. 1995;269:C1200–C1208. doi: 10.1152/ajpcell.1995.269.5.C1200. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275:31505–31513. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- Lv L, Li D, Zhao D, et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell. 2011;42:719–730. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LM, Aiken CT, Kaltenbach LS, et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J Cell Biol. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay U, Kaushik S, Varticovski L, Cuervo AM. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008;28:5747–5763. doi: 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador N, Aguado C, Horst M, Knecht E. Import of a cytosolic protein into lysosomes by chaperone-mediated autophagy depends on its folding state. J Biol Chem. 2000;275:27447–27456. doi: 10.1074/jbc.M001394200. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay U, Sridhar S, Kaushik S, Kiffin R, Cuervo AM. Identification of regulators of chaperone-mediated autophagy. Mol Cell. 2010;39:535–547. doi: 10.1016/j.molcel.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali AB, Nin DS, Tam J, Khan M. Role of chaperone mediated autophagy (CMA) in the degradation of misfolded N-CoR protein in non-small cell lung cancer (NSCLC) cells. PLoS One. 2011;6:e25268. doi: 10.1371/journal.pone.0025268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Dice JF, Knecht E. A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J Biol Chem. 1997;272:5606–5615. doi: 10.1074/jbc.272.9.5606. [DOI] [PubMed] [Google Scholar]
- Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci USA. 2006;103:5805–5810. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Massey AC, Mizushima N, Cuervo AM. Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol Biol Cell. 2008;19:2179–2192. doi: 10.1091/mbc.E07-11-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Massey AC, Cuervo AM. Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J. 2006;25:3921–3933. doi: 10.1038/sj.emboj.7601283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiffin R, Christian C, Knecht E, Cuervo AM. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Dice JF. Unique properties of lamp2a compared to other lamp2 isoforms. J Cell Sci. 2000;113 Pt 24:4441–4450. doi: 10.1242/jcs.113.24.4441. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Dice JF. Regulation of lamp2a levels in the lysosomal membrane. Traffic. 2000;1:570–583. doi: 10.1034/j.1600-0854.2000.010707.x. [DOI] [PubMed] [Google Scholar]
- Anguiano J, Garner TP, Mahalingam M, Das BC, Gavathiotis E, Cuervo AM. Chemical modulation of chaperone-mediated autophagy by retinoic acid derivatives. Nat Chem Biol. 2013;9:374–382. doi: 10.1038/nchembio.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu R, Kaushik S, Cannizzo E, et al. Microautophagy of cytosolic proteins by late endosomes. Develop Cell. 2011;20:131–139. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM. Methods to monitor chaperone-mediated autophagy. Meth Enzymol. 2009;452:297–324. doi: 10.1016/S0076-6879(08)03619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Hu W, Lim B, Dice JF. IkappaB is a substrate for a selective pathway of lysosomal proteolysis. Mol Biol Cell. 1998;9:1995–2010. doi: 10.1091/mbc.9.8.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon M, Kiffin R, Koga H, et al. Chaperone-mediated autophagy is required for tumor growth. Sci Transl Med. 2011;3:109ra117. doi: 10.1126/scitranslmed.3003182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Palmer A, Rivett AJ, Knecht E. Degradation of proteasomes by lysosomes in rat liver. Eur J Biochem. 1995;227:792–800. doi: 10.1111/j.1432-1033.1995.tb20203.x. [DOI] [PubMed] [Google Scholar]
- Metrailler S, Schorderet DF, Cottet S. Early apoptosis of rod photoreceptors in Rpe65(−/−) mice is associated with the upregulated expression of lysosomal-mediated autophagic genes. Exp Eye Res. 2012;96:70–81. doi: 10.1016/j.exer.2011.12.019. [DOI] [PubMed] [Google Scholar]
- Dohi E, Tanaka S, Seki T, et al. Hypoxic stress activates chaperone-mediated autophagy and modulates neuronal cell survival. Neurochem Int. 2012;60:431–442. doi: 10.1016/j.neuint.2012.01.020. [DOI] [PubMed] [Google Scholar]
- Ferreira JV, Fofo H, Bejarano E, et al. STUB1/CHIP is required for HIF1A degradation by chaperone-mediated autophagy. Autophagy. 2013;9:1349–1366. doi: 10.4161/auto.25190. [DOI] [PubMed] [Google Scholar]
- Franch HA, Sooparb S, Du J, Brown NS. A mechanism regulating proteolysis of specific proteins during renal tubular cell growth. J Biol Chem. 2001;276:19126–19131. doi: 10.1074/jbc.M101777200. [DOI] [PubMed] [Google Scholar]
- Sooparb S, Price SR, Shaoguang J, Franch HA. Suppression of chaperone-mediated autophagy in the renal cortex during acute diabetes mellitus. Kidney Int. 2004;65:2135–2144. doi: 10.1111/j.1523-1755.2004.00639.x. [DOI] [PubMed] [Google Scholar]
- Yang Q, She H, Gearing M, et al. Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science. 2009;323:124–127. doi: 10.1126/science.1166088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Li P, Lin Y, et al. Lamp-2a facilitates MHC class II presentation of cytoplasmic antigens. Immunity. 2005;22:571–581. doi: 10.1016/j.immuni.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Finn PF, Mesires NT, Vine M, Dice JF. Effects of small molecules on chaperone-mediated autophagy. Autophagy. 2005;1:141–145. doi: 10.4161/auto.1.3.2000. [DOI] [PubMed] [Google Scholar]
- Arias E, Cuervo AM. Chaperone-mediated autophagy in protein quality control. Curr Opin Cell Biol. 2011;23:184–189. doi: 10.1016/j.ceb.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga H, Cuervo AM. Chaperone-mediated autophagy dysfunction in the pathogenesis of neurodegeneration. Neurobiol Dis. 2011;43:29–37. doi: 10.1016/j.nbd.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orenstein SJ, Cuervo AM. Chaperone-mediated autophagy: molecular mechanisms and physiological relevance. Semin Cell Dev Biol. 2010;21:719–726. doi: 10.1016/j.semcdb.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI, Driessen AJ, Hegde RS, Langer T. The life of proteins: the good, the mostly good and the ugly. Nat Struct Mol Biol. 2011;18:1–4. doi: 10.1038/nsmb0111-1. [DOI] [PubMed] [Google Scholar]
- Wong E, Cuervo AM. Neuronal autophagy gone awry: many fixings for the autophagic wrong-doing. Nat Neurosci. 2010;13:805–811. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Kaushik S, et al. Dopamine-modified alpha-synuclein blocks chaperone-mediated autophagy. J Clin Invest. 2008;118:777–788. doi: 10.1172/JCI32806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orenstein SJ, Kuo SH, Tasset I, et al. Interplay of LRRK2 with chaperone-mediated autophagy. Nat Neurosci. 2013;16:394–406. doi: 10.1038/nn.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Martinez-Vicente M, Kruger U, et al. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum Mol Genet. 2009;18:4153–4170. doi: 10.1093/hmg/ddp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak SK, McCormack AL, Manning-Bog AB, Cuervo AM, Di Monte DA. Lysosomal degradation of alpha-synuclein in vivo. J Biol Chem. 2010;285:13621–13629. doi: 10.1074/jbc.M109.074617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogiatzi T, Xilouri M, Vekrellis K, Stefanis L. Wild type alpha-synuclein is degraded by chaperone-mediated autophagy and macroautophagy in neuronal cells. J Biol Chem. 2008;283:23542–23556. doi: 10.1074/jbc.M801992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabuta T, Furuta A, Aoki S, Furuta K, Wada K. Aberrant interaction between Parkinson's disease-associated mutant UCH-L1 and the lysosomal receptor for chaperone-mediated autophagy. J Biol Chem. 2008;283:23731–23738. doi: 10.1074/jbc.M801918200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oueslati A, Fournier M, Lashuel HA. Role of post-translational modifications in modulating the structure, function and toxicity of alpha-synuclein: implications for Parkinson's disease pathogenesis and therapies. Prog Brain Res. 2010;183:115–145. doi: 10.1016/S0079-6123(10)83007-9. [DOI] [PubMed] [Google Scholar]
- Alvarez-Erviti L, Rodriguez-Oroz MC, Cooper JM, et al. Chaperone-mediated autophagy markers in Parkinson disease brains. Arch Neurol. 2010;67:1464–1472. doi: 10.1001/archneurol.2010.198. [DOI] [PubMed] [Google Scholar]
- Malkus KA, Ischiropoulos H. Regional deficiencies in chaperone-mediated autophagy underlie alpha-synuclein aggregation and neurodegeneration. Neurobiol Dis. 2012;46:732–744. doi: 10.1016/j.nbd.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Erviti L, Seow Y, Schapira AH, Rodriguez-Oroz MC, Obeso JA, Cooper JM. Influence of microRNA deregulation on chaperone-mediated autophagy and alpha-synuclein pathology in Parkinson's disease. Cell Death Dis. 2013;4:e545. doi: 10.1038/cddis.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang S, Chen D, Zhang A, Qin X, Yan B. Genetic analysis of the LAMP-2 gene promoter in patients with sporadic Parkinson's disease. Neurosci Lett. 2012;526:63–67. doi: 10.1016/j.neulet.2012.07.044. [DOI] [PubMed] [Google Scholar]
- Xilouri M, Brekk OR, Landeck N, et al. Boosting chaperone-mediated autophagy in vivo mitigates alpha-synuclein-induced neurodegeneration. Brain. 2013;136:2130–2146. doi: 10.1093/brain/awt131. [DOI] [PubMed] [Google Scholar]
- Liu H, Wang P, Song W, Sun X. Degradation of regulator of calcineurin 1 (RCAN1) is mediated by both chaperone-mediated autophagy and ubiquitin proteasome pathways. FASEB J. 2009;23:3383–3392. doi: 10.1096/fj.09-134296. [DOI] [PubMed] [Google Scholar]
- Saha T. LAMP2A overexpression in breast tumors promotes cancer cell survival via chaperone-mediated autophagy. Autophagy. 2012;8:1643–1656. doi: 10.4161/auto.21654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakifahmetoglu-Norberg H, Kim M, Xia HG, et al. Chaperone-mediated autophagy degrades mutant p53. Genes Dev. 2013;27:1718–1730. doi: 10.1101/gad.220897.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch T, Younsi A, Disanza A, et al. Eps8 is recruited to lysosomes and subjected to chaperone-mediated autophagy in cancer cells. Exp Cell Res. 2010;316:1914–1924. doi: 10.1016/j.yexcr.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiffin R, Kaushik S, Zeng M, et al. Altered dynamics of the lysosomal receptor for chaperone-mediated autophagy with age. J Cell Sci. 2007;120:782–791. doi: 10.1242/jcs.001073. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro JA, Kaushik S, Koga H, et al. Inhibitory effect of dietary lipids on chaperone-mediated autophagy. Proc Natl Acad Sci USA. 2012;109:E705–E714. doi: 10.1073/pnas.1113036109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci USA. 2006;103:5905–5910. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008;14:959–965. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E, Cuervo AM. Integration of clearance mechanisms: the proteasome and autophagy. Cold Spring Harb Perspect Biol. 2010;2:a006734. doi: 10.1101/cshperspect.a006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C, Cuervo AM. Selective autophagy: talking with the UPS. Cell Biochem Biophys. 2013;67:3–13. doi: 10.1007/s12013-013-9623-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Massey A, Mizushima N, Cuervo AM. Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol Biol Cell. 2008;19:2179–2192. doi: 10.1091/mbc.E07-11-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga H, Martinez-Vicente M, Macian F, Verkhusha VV, Cuervo AM. A photoconvertible fluorescent reporter to track chaperone-mediated autophagy. Nat Commun. 2011;2:386. doi: 10.1038/ncomms1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey AC, Follenzi A, Kiffin R, Zhang C, Cuervo AM. Early cellular changes after blockage of chaperone-mediated autophagy. Autophagy. 2008;4:442–456. doi: 10.4161/auto.5654. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Palmer A, Rivett AJ, Knecht E. Degradation of proteasomes by lysosomes in rat liver. Eur J Biochem. 1995;227:792–800. doi: 10.1111/j.1432-1033.1995.tb20203.x. [DOI] [PubMed] [Google Scholar]
- Koga H, Martinez-Vicente M, Arias E, Kaushik S, Sulzer D, Cuervo AM. Constitutive upregulation of chaperone-mediated autophagy in Huntington's disease. J Neurosci. 2011;31:18492–18505. doi: 10.1523/JNEUROSCI.3219-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Duden R, Rubinsztein D. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Wong E, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington's Disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp MS, Patel CN, Bersuker K, et al. Indirect inhibition of 26S proteasome activity in a cellular model of Huntington's disease. J Cell Biol. 2012;196:573–587. doi: 10.1083/jcb.201110093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ES, Tan JM, Soong WE, et al. Autophagy-mediated clearance of aggresomes is not a universal phenomenon. Hum Mol Genet. 2008;17:2570–2582. doi: 10.1093/hmg/ddn157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E, Bejarano E, Rakshit M, et al. Molecular determinants of selective clearance of protein inclusions by autophagy. Nat Commun. 2012;3:1240. doi: 10.1038/ncomms2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- Tan JM, Wong ES, Kirkpatrick DS, et al. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum Mol Genet. 2008;17:431–439. doi: 10.1093/hmg/ddm320. [DOI] [PubMed] [Google Scholar]
- Bauer PO, Goswami A, Wong HK, et al. Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein. Nat Biotechnol. 2010;28:256–263. doi: 10.1038/nbt.1608. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Muela N, Koga H, Garcia-Ledo L, et al. Balance between autophagic pathways preserves retinal homeostasis. Aging cell. 2013;12:478–488. doi: 10.1111/acel.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]