Abstract
Autophagy is a primarily degradative pathway that takes place in all eukaryotic cells. It is used for recycling cytoplasm to generate macromolecular building blocks and energy under stress conditions, to remove superfluous and damaged organelles to adapt to changing nutrient conditions and to maintain cellular homeostasis. In addition, autophagy plays a critical role in cytoprotection by preventing the accumulation of toxic proteins and through its action in various aspects of immunity including the elimination of invasive microbes and its participation in antigen presentation. The most prevalent form of autophagy is macroautophagy, and during this process, the cell forms a double-membrane sequestering compartment termed the phagophore, which matures into an autophagosome. Following delivery to the vacuole or lysosome, the cargo is degraded and the resulting macromolecules are released back into the cytosol for reuse. The past two decades have resulted in a tremendous increase with regard to the molecular studies of autophagy being carried out in yeast and other eukaryotes. Part of the surge in interest in this topic is due to the connection of autophagy with a wide range of human pathophysiologies including cancer, myopathies, diabetes and neurodegenerative disease. However, there are still many aspects of autophagy that remain unclear, including the process of phagophore formation, the regulatory mechanisms that control its induction and the function of most of the autophagy-related proteins. In this review, we focus on macroautophagy, briefly describing the discovery of this process in mammalian cells, discussing the current views concerning the donor membrane that forms the phagophore, and characterizing the autophagy machinery including the available structural information.
Keywords: autophagosome, autophagy, degradation, lysosome, phagophore, stress, vacuole
Introduction
The definition of autophagy
Autophagy is a cellular process in which cytoplasmic contents are degraded within the lysosome/vacuole, and the resulting macromolecular constituents are recycled1. Macroautophagy is one type of autophagic process in which the substrates are sequestered within cytosolic double-membrane vesicles termed autophagosomes. The substrates of macroautophagy include superfluous and damaged organelles, cytosolic proteins and invasive microbes. Following degradation, the breakdown products are released back into the cytosol in order to recycle the macromolecular constituents and to generate energy to maintain cell viability under unfavorable conditions, and to protect the cell during various conditions of stress2,3. Autophagy is highly conserved from yeast to mammals, both morphologically and with regard to the protein constituents that make up the core autophagy machinery.
Terms and structures
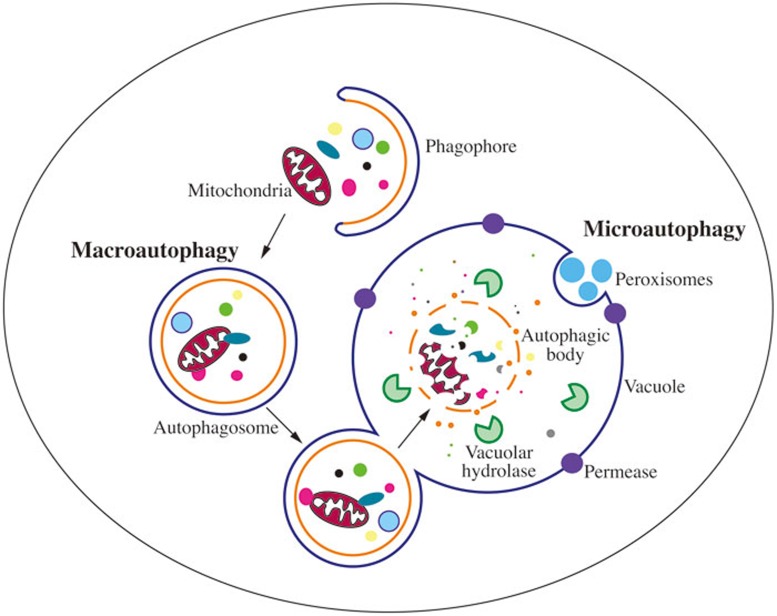
The most readily identifiable feature of macroautophagy is the sequestration of cargo within cytosolic double-membrane vesicles, and many researchers still consider the analysis of macroautophagy by electron microscopy to be the “gold standard” for verifying macroautophagy activity. Below, we present some of the most common terms describing the principle structures of macroautophagy (Figure 1).
Figure 1.
Schematic depiction of the two main types of yeast autophagy. In macroautophagy, random cytoplasm and dysfunctional organelles are sequestered by the expanding phagophore, leading to the formation of the autophagosome. The autophagosome subsequently fuses with the vacuole membrane, releasing the autophagic body into the vacuole lumen. Eventually, the sequestered cargos are degraded or processed by vacuolar hydrolases. In microautophagy, cargos are directly taken up by the invagination of the vacuole membrane, followed by scission, and subsequent lysis, exposing the cargo to vacuolar hydrolases for degradation.
Autophagic body: The inner-membrane-bound structure released into the vacuole lumen after the outer membrane of the autophagosome fuses with vacuolar membrane4. Note that autophagic bodies only appear in yeast and plants due to the large size of the vacuole, and are not found in mammalian lysosomes.
Autophagosome: The completed double-membrane-bound compartment, the product of phagophore expansion and closure, which sequesters cytoplasmic cargos during macroautophagy5,6.
PAS: The phagophore assembly site; a peri-vacuolar location or compartment where the nucleation of the phagophore initiates. Most components of the macroautophagy core machinery locate at least transiently at the PAS in yeast; however, a mammalian equivalent of the PAS has not been identified7,8,9.
Phagophore: The double-membrane structure that functions in the initial sequestering of cargo, and thus the active compartment of macroautophagy. The phagophore further elongates/expands and ultimately closes, generating a completed autophagosome10.
The history of discovery
The term “autophagy” was coined by Christian de Duve at the CIBA Foundation Symposium on Lysosomes in 1963. This was based on his discovery of lysosomes in 1955, which won him the Nobel Prize in Physiology or Medicine in 197411. Autophagy was named morphologically by the observations from electron microscopy of rat hepatic cell lysosomes, where single membrane vesicles, containing cytoplasm or organelles such as mitochondria and endoplasmic reticulum (ER) were observed12,13,14,15. Based on Thomas Ashford's and Keith Porter's early findings in 1962 that revealed the presence of sequestered organelles in rat hepatocytes following their exposure to glucagon12, de Duve and his colleagues confirmed that this hormone can induce autophagy16. Subsequently, other researchers further examined the hormonal and enzymatic regulation of autophagy. For instance, Per Seglen and Paul Gordon discovered the inhibitory role of 3-methyladenine17. As de Duve mentioned in 1966 that autophagy could be selective14, the specific sequestration of organelles including ER18, mitochondria19 and peroxisomes20 by autophagy was demonstrated in the following decade. Although autophagy was initially revealed in mammalian systems, the molecular understanding of it was largely expanded and facilitated through genetic studies in yeast. Genetic screens of macroautophagy-defective mutants in Saccharomyces cerevisiae were carried out by the Ohsumi and Thumm laboratories21,22. Shortly thereafter, the first autophagy-specific gene, APG1 (now ATG123), was identified, and the corresponding gene product, Apg1/Atg1, was characterized as a Ser/Thr protein kinase24. Multiple laboratories working on autophagy primarily in S. cerevisiae, Pichia pastoris and Hansenula polymorpha subsequently discovered more than thirty autophagy-related (ATG) genes. The molecular basis of autophagy was first connected to human diseases by Beth Levine's laboratory through the identification of BECN1/VPS30/ATG6 as a tumor suppressor gene25. Subsequently, a series of studies uncovered the connections between autophagy and pathophysiological conditions, such as pathogen infection26,27,28,29 and neurodegeneration30, and its dual role in cell growth and death31,32.
Nonselective and selective autophagy
There are three primary types of autophagy: microautophagy, macroautophagy and a mechanistically unrelated process, chaperone-mediated autophagy that only occurs in mammalian cells. Both micro and macroautophagy can be selective or nonselective and these processes have been best characterized in yeast33 (Table 1). As noted above, the most distinguishing feature of macroautophagy is the formation of the double-membrane bound phagophore and autophagosome (Figure 1). In contrast, during microautophagy the cargos are sequestered by direct invagination or protusion/septation of the yeast vacuole membrane34. Nonselective autophagy is used for the turnover of bulk cytoplasm under starvation conditions, whereas selective autophagy specifically targets damaged or superfluous organelles, including mitochondria and peroxisomes, as well as invasive microbes; each process involves a core set of machinery, as well as specific components, and accordingly is identified with a unique name — mitophagy for selective mitochondria degradation by autophagy, pexophagy for peroxisomes, xenophagy for microbes, etc.35,36. In this review, we will mainly focus on macroautophagy, since the molecular or physiological studies of nonselective microautophagy are limited, and selective microautophagy shares most of the same machinery with macroautophagy (hereafter autophagy).
Table 1. The main types of autophagy in yeast.
| Name | Cargo | Characteristics | ||
|---|---|---|---|---|
| Macroautophagy | ||||
| Nonselective | Random cytoplasm | Autophagosome formation | ||
| Selective | Cvt pathway | Vacuolar hydrolases | Autophagosome or Cvt vesicle formation; Atg19 and Atg34 are receptors, Atg11 is a scaffold | |
| Mitophagy | Damaged or superfluous mitochondria | Autophagosome formation; Atg32 is a receptor, Atg11 is a scaffold | ||
| Pexophagy | Superfluous peroxisomes | Autophagosome formation; Atg30 and Atg36 are receptors, Atg11 and Atg17 are scaffolds | ||
| Microautophagy | ||||
| Nonselective | Random cytoplasm, vacuole membrane | Vacuole invagination | ||
| Selective | Mitochondria | Vacuole invagination or protrusion | ||
| Peroxisomes | Vacuole invagination or protrusion | |||
| Nuclear membrane | Vacuole invagination, requires Nvj1 and Vac8 | |||
Identification of the PAS and omegasome, and the origin of the phagophore membrane
Identification of the PAS
Atg8 was the first Atg protein characterized to mark the phagophore and autophagosome37,38. Immunofluorescence microscopy revealed that in rich conditions, Atg8 is primarily dispersed in the cytoplasm as small puncta, while it forms larger puncta in the vicinity of the vacuole when yeast cells are switched to starvation conditions; the nature of the smaller puncta is not known, whereas the larger puncta correspond to nascent autophagosomes. Subsequent studies with green fluorescent protein (GFP)-tagged chimeras demonstrated that most of the Atg proteins reside at this location, at least transiently8. Accordingly, this site is proposed to be where the phagophore assembles, and is thus termed the phagophore assembly site, or PAS8,9. The precise nature of the PAS with regard to its protein and membrane composition, or the reason for its perivacuolar localization, is not known, but assembly of the Atg proteins at the PAS occurs in a hierarchical manner39. The PAS forms constitutively, and in vegetative conditions a key component that marks this site is Atg11; upon autophagy induction, the structure transitions into an autophagy-specific PAS and the function of Atg11 is replaced by a ternary complex of Atg17-Atg31-Atg29 that assembles at the PAS along with Atg1 and Atg1340,41. Atg9, which shuttles between the PAS and peripheral sites, plays an important role in directing membrane to the PAS that is needed for autophagosome formation, and it localizes to the PAS at a similar time as the Atg1 kinase complex. Subsequently, the PtdIns3K and Atg12–Atg5-Atg16 complexes are recruited to the PAS; the latter acts in part as an E3 ligase to facilitate the formation of Atg8–PE, one of the last proteins that are recruited to the PAS. In mammalian cells, the colocalization of ATG proteins is observed at multiple sites42,43,44, instead of a single PAS as in yeast.
Origin of the phagophore
Until recently, the dogma was that the phagophore and autophagosome formed de novo, meaning that the sequestering membrane does not form in “one step” from a pre-existing organelle already containing its cargo as occurs throughout the secretory pathway45,46. Regardless of the specific mechanism, autophagosome formation is thought to occur by the expansion of the phagophore, that is, a sheet of membrane that would correspond to the size of a complete autophagosome does not separate from the endomembrane system and simply curl up to form an autophagosome. Rather, after nucleation the phagophore grows through the addition of membrane from one or more donor sources1,47; however, the origin of the phagophore membrane is still under debate. Various lines of data suggest a role for almost every membrane compartment in contributing to formation of the autophagosome, including the ER, mitochondria, the Golgi apparatus and the plasma membrane48,49,50,51,52. As discussed above, there is no absolute equivalent of the yeast PAS in mammalian cells. Instead, there is evidence that there may be at least two separate mechanisms for generating autophagosomes. One mechanism would involve the delivery of membrane from various organelles, as is thought to occur in yeast, and the second utilizes an omega-shaped membrane structure or cradle, derived from phosphatidylinositol-3-phosphate (PtdIns3P)-enriched ER subdomains, termed an omegasome53.
Core molecular machinery of autophagosome formation
The transition of autophagy studies from morphology to molecular machinery relied on the identification of the ATG genes23. As mentioned above, genetic screens for autophagy-defective mutants in yeast have led to the identification of over 30 ATG genes21,22,23,54, many of which have known orthologs in higher eukaryotes.
Among these ATG genes, one subgroup, consisting of approximately 18 genes (Table 2), is shared among the various types of autophagy including nonselective macroautophagy, the cytoplasm-to-vacuole-targeting (Cvt) pathway (a biosynthetic autophagy-like pathway), mitophagy and pexophagy. More specifically, the corresponding gene products of this subgroup are required for autophagosome formation, and thus are termed the core autophagy machinery. The core Atg proteins can be divided into different functional subgroups: (A) the Atg1/ULK complex (Atg1, Atg11, Atg13, Atg17, Atg29 and Atg31) is the initial complex that regulates the induction of autophagosome formation; (B) Atg9 and its cycling system (Atg2, Atg9 and Atg18) play a role in membrane delivery to the expanding phagophore after the assembly of the Atg1 complex at the PAS; (C) the PtdIns 3-kinase (PtdIns3K) complex (Vps34, Vps15, Vps30/Atg6, and Atg14) acts at the stage of vesicle nucleation, and is involved in the recruitment of PtdIns3P-binding proteins to the PAS; (D) two ubiquitin-like (Ubl) conjugation systems: the Atg12 (Atg5, Atg7, Atg10, Atg12 and Atg16) and Atg8 (Atg3, Atg4, Atg7 and Atg8) conjugation systems play roles in vesicle expansion39,55,56,57.
Table 2. Atg/ATG proteins in the core machinery of autophagosome formation.
| Yeast | Mammals | Characteristics and functions | |
|---|---|---|---|
| Atg1/ULK complex |
Atg1 | ULK1/2 | Ser/Thr protein kinase; phosphorylated by M/TORC1; recruitment of Atg proteins to the PAS |
| Atg13 | ATG13 | Regulatory subunit through phosphorylation by M/TORC1 and/or PKA, linker between Atg1 and Atg17 | |
| Atg17 | RB1CC1/FIP200 (functional homolog) | Scaffold protein, ternary complex with Atg29 and Atg31. Phosphorylated by ULK1; scaffold for ULK1/2 and ATG13 | |
| Atg29 | Ternary complex with Atg17 and Atg31 | ||
| Atg31 | Ternary complex with Atg17 and Atg29 | ||
| Atg11 | Scaffold protein in selective autophagy for PAS organization | ||
| |
C12orf44/Atg101 |
Component of the complex with ATG13 and RB1CC1 |
|
| Atg9 and its cycling system |
Atg2 | ATG2 | Interacts with Atg18 |
| Atg9 | ATG9A/B | Transmembrane protein, directs membrane to the phagophore | |
| Atg18 |
WIPI1/2 |
PtdIns3P-binding protein |
|
| PtdIns3K complex |
Vps34 | PIK3C3/VPS34 | PtdIns 3-kinase |
| Vps15 | PIK3R4/VPS15 | Ser/Thr protein kinase | |
| Vps30/Atg6 | BECN1 | Component of PtdIns3K complex I and II | |
| Atg14 |
ATG14 |
Component of PtdIns3K complex I |
|
| Atg8 Ubl conjugation system |
Atg8 | LC3A/B/C, GABARAP, GABARAPL1/2 | Ubl, conjugated to PE |
| Atg7 | ATG7 | E1-like enzyme | |
| Atg3 | ATG3 | E2-like enzyme | |
| Atg4 |
ATG4A/B/C/D |
Deconjugating enzyme, cysteine proteinase |
|
| Atg12 Ubl conjugation system | Atg12 | ATG12 | Ubl |
| Atg7 | ATG7 | E1-like enzyme | |
| Atg10 | ATG10 | E2-like enzyme | |
| Atg16 | ATG16L1 | Interacts with Atg5 and Atg12 | |
| Atg5 | ATG5 | Conjugated by Atg12 |
Characterization and function of the Atg proteins
The autophagy-related proteins have been described in great detail in numerous reviews. Here, we highlight some of the key information.
Yeast Atg1 kinase complex
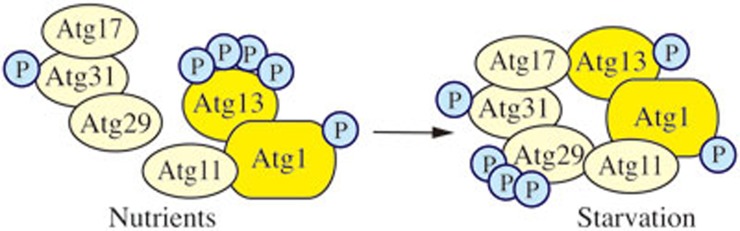
The Atg1 kinase complex regulates the magnitude of autophagy58,59,60,61,62. This complex also plays a role in mediating the retrieval of Atg9 from the PAS63. The Atg1 kinase complex is extensively regulated, receiving input from several signaling pathways. In yeast, much of the regulation centers around nutrient signaling and involves kinases that sense the nutritional status of the cell including the target of rapamycin (TOR), protein kinase A (PKA), Gcn2 and Snf1. The yeast Atg1 kinase complex includes Atg1, a Ser/Thr protein kinase, Atg13, a regulatory subunit, and the Atg17-Atg31-Atg29 complex, which may function in part as a scaffold (Figure 2). Atg1 is the only kinase of the core autophagy machinery; however, despite the fact that it was the first Atg protein to be identified, the key physiological substrate of yeast Atg1 is not known. Atg1 kinase activity is required for autophagosome formation and the Cvt pathway, although there are conflicting data concerning the changes in kinase activity when yeast cells shift from vegetative growth to autophagy-inducing conditions24,64,65. Atg1 recruitment to the PAS may be regulated by PKA-dependent phosphorylation66. TOR also phosphorylates Atg1, and autophosphorylation is important in regulating Atg1 kinase activity67,68.
Figure 2.
Yeast Atg1 kinase complex. The yeast Atg1 kinase complex includes Atg1, a Ser/Thr protein kinase, Atg13, a regulatory subunit required for Atg1 kinase activity, and the Atg17-Atg31-Atg29 complex, which may function in part as a scaffold. Atg31 bridges Atg17 and Atg29, Atg17 binds Atg13, and Atg29 binds Atg11.
Atg13 is required for Atg1 kinase activity. Atg13 is hyperphosphorylated under nutrient-rich conditions, and its phosphorylation is regulated by TOR complex 1 (TORC1)69 and/or PKA70. Atg13 is rapidly, but only partially, dephosphorylated upon autophagy induction71. These data, coupled with affinity isolation studies, led to a model suggesting that the interaction between Atg1 and Atg13 is controlled by Atg13 phosphorylation — dephosphorylation would lead to the formation of an Atg1-Atg13 complex, and subsequent activation of Atg1 kinase activity61. Recent data, however, suggest that Atg1 and Atg13 interact in a constitutive manner72, which is similar to the situation in other organisms.
Atg17-Atg31-Atg29 exists as a stable complex under both vegetative and starvation conditions, suggesting that formation of the complex is not involved in autophagy induction. Atg31 bridges Atg17 and Atg29, Atg17 binds Atg13, and Atg29 binds Atg11; the latter is a scaffold protein that may also be considered part of the Atg1 kinase complex because it binds Atg1 and is involved in the transition of the PAS that occurs during the switch from vegetative to starvation conditions40,41,58,73,74,75. Both Atg29 and Atg31 are phosphoproteins. The phosphorylation of Atg29 appears to be involved in regulation and is proposed to alter the conformation of an inhibitory C-terminal peptide to allow the protein to become active41.
Mammalian ULK1/2 Complex
The mammalian ULK1/2 complex includes ULK1/2 (mammalian homologs of Atg1), ATG13 (a homolog of yeast Atg13), RB1CC1/FIP200 (a putative Atg17 homolog) and C12orf44/ATG101 (the latter component is not conserved in S. cerevisiae)76,77,78,79,80. ULK1/2 interact with ATG13, which directly binds RB1CC1 and mediates the latter's interaction with the ULKs76,81. In mammalian cells, ULK1 kinase can be activated by both AMP-activated protein kinase (AMPK)-dependent (glucose starvation) and -independent (amino acid starvation) processes82. The ULK1/2-ATG13-RB1CC1 interaction is nutrient independent, forming a complex even in nutrient-rich conditions80. In this situation, MTORC1 phosphorylates and inhibits ULK1/2 and ATG13, disrupting the interaction between ULK1 and AMPK82. In contrast, under autophagy-inducing conditions, MTOR is released from the complex, resulting in activation of ULK1/2, which phosphorylates, and presumably activates, ATG13 and RB1CC1. AMBRA1 and BECN1, components of the autophagy-promoting PtdIns3K complexes, are also phosphorylated by ULK1; these modifications result in localization of the lipid kinase complex to the ER and its activation83,84.
Yeast Atg9
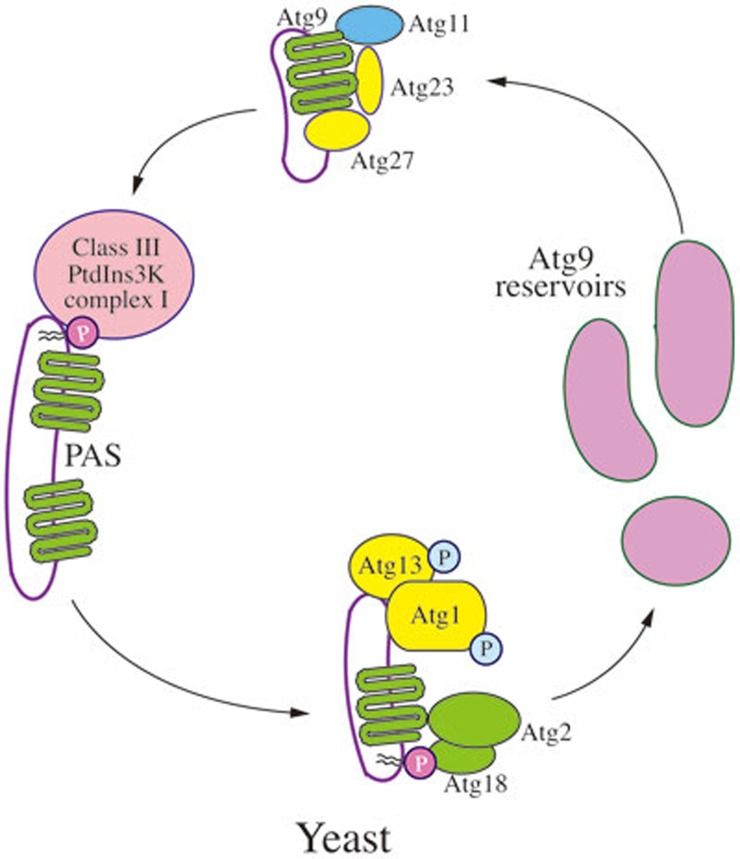
In yeast cells, Atg9 is a transmembrane protein that transits between the PAS and peripheral sites (Figure 3); the latter have been termed Atg9 reservoirs or tubulovesicular clusters, and are proposed to represent sites from which membrane is delivered to the forming phagophore49,63,85, although the exact role of Atg9 in this process is not clear. Atg9 is proposed to consist of six transmembrane domains, with both the amino and carboxyl termini exposed in the cytosol. Atg9 self-interacts and may exist in a complex85,86.
Figure 3.
Trafficking of yeast Atg9. In yeast cells, Atg9 is a transmembrane protein that transits between the PAS and peripheral sites (Atg9 reservoirs). Atg9 localization to the PAS from the peripheral sites depends on Atg11, Atg23 and Atg27. Return to the peripheral sites requires Atg1-Atg13, Atg2 and Atg18; PAS recruitment of the latter involves binding to PtdIns3P, the product of the class III PtdIns3K complex.
Atg9 localization to the PAS from the peripheral sites (referred to as anterograde transport) depends on Atg11, Atg23 and Atg2763,87,88,89. Return to the peripheral sites (retrograde movement) requires Atg1-Atg13, Atg2 and Atg18; PAS recruitment of the latter involves binding to PtdIns3P and thus necessitates the function of Atg14 and the class III PtdIns3K complex63. Atg27 is a type I integral transmembrane protein, while the other components involved in Atg9 movement are soluble and/or peripherally associated with membranes.
Mammalian ATG9
The mammalian Atg9 homolog ATG9A localizes to the trans-Golgi network and late endosomes in nutrient-rich conditions44; another Atg9 ortholog, ATG9B, displays a similar localization, but is expressed only in the placenta and pituitary gland, whereas ATG9A is expressed ubiquitously43. ATG9 appears to have a conserved role in coordinating membrane transport from donor sources to the phagophore and displays a cycling pattern similar to that seen in yeast44,90. ATG9 and ATG16L1 also appear to traffic at least in part on vesicles derived from the plasma membrane, which are delivered to the recycling endosome; the two proteins are initially present in separate vesicular populations that subsequently fuse as part of the process of autophagosome formation91. ATG9 movement is dependent on the activity of the ULK1 and PIK3C3/VPS34 kinases, as well as WIPI2 (a homolog of yeast Atg18).
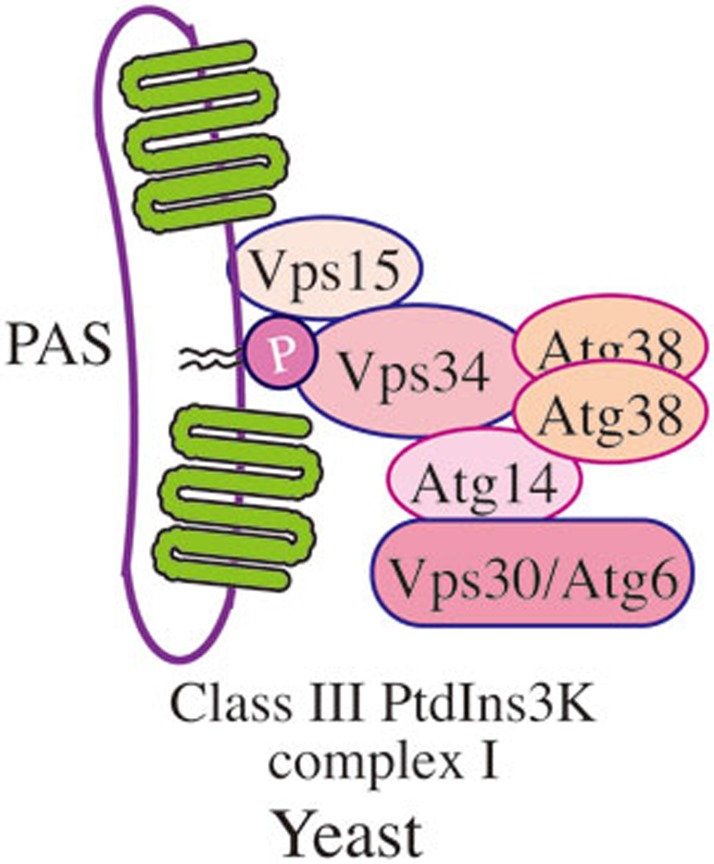
Yeast PtdIns3K complexes I and II
In yeast, Vps15 (a putative regulatory protein kinase that is required for Vps34 membrane association), Vps30/Atg6, Vps34 (the PtdIns3K), Atg14 and Atg38, form the autophagy-specific class III PtdIns3K complex I (Figure 4), which localizes at the PAS92,93,94. One key role of the PtdIns3K complex, the generation of PtdIns3P, is presumably to recruit PtdIns3P-binding proteins such as Atg18 to the PAS. Vps15, Vps34 and Vps30 are present in a second complex that includes Vps38 instead of Atg14. Thus, the latter protein appears to determine the specificity of the PtdIns3K complex I for macroautophagy.
Figure 4.
Yeast PtdIns3K complex I. In yeast, Vps15 (a putative regulatory protein kinase that is required for Vps34 membrane association), Vps30/Atg6, Vps34 (the PtdIns3K), Atg14 and Atg38 form the autophagy-specific class III PtdIns3K complex, which localizes at the PAS. Vps15, Vps34 and Vps30 are present in the second class III PtdIns3K complex that includes Vps38 instead of Atg14.
Mammalian class III PtdIns3K
Mammalian cells have both a class I phosphoinositide 3-kinase and a class III PtdIns3K; the class III enzyme complexes consist of homologs of Vps34 (PIK3C3), Vps15 (PIK3R4) and Vps30 (BECN1)95. These core components are part of at least three different complexes96,97. The ATG14 complex contains ATG14 and is regulated by the binding of BECN1 to AMBRA1 and BCL2, which stimulate and inhibit the complex, respectively. This complex functions similarly to the yeast PtdIns3K complex I. The UVRAG complex replaces ATG14 and AMBRA1 with UVRAG and its positive regulator SH3GLB1, and participates in both endocytosis and macroautophagy. In the third complex, SH3GLB1 is replaced with KIAA0226/Rubicon, which inhibits UVRAG; this complex acts to negatively regulate macroautophagy.
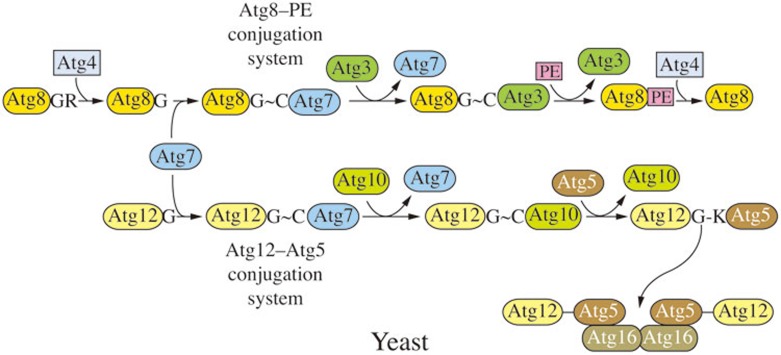
Yeast Ubl conjugation systems
There are two Ubl protein conjugation systems that participate in macroautophagy48. These include two Ubl proteins, Atg8 and Atg12, which are used to generate the conjugation products Atg8–PE and Atg12–Atg5, respectively (Figure 5). The Ubl conjugation systems participate in phagophore expansion, although their functions are not fully understood. The Atg12–Atg5 conjugate, along with a third component, Atg16, may act in part as an E3 ligase to facilitate the conjugation of Atg8 to PE, and the amount of Atg8 can regulate the size of autophagosomes98; however, Atg8 also functions in cargo binding during selective autophagy99.
Figure 5.
Yeast Ubl conjugation systems. There are two Ubl protein conjugations systems, involving Atg8 and Atg12, which are used to generate the conjugation products Atg8–PE and Atg12–Atg5, respectively. Atg8 is initially processed by Atg4, a cysteine protease, and during autophagy, Atg8 is released from Atg8–PE by a second Atg4-dependent cleavage. The Atg12–Atg5 conjugate, along with Atg16, facilitates the conjugation of Atg8 to PE. Both of the Ubl protein conjugation systems share a single activating enzyme, Atg7. The conjugating enzymes are Atg3 for Atg8, and Atg10 for Atg12.
Yeast Atg8 is initially synthesized with a C-terminal arginine residue that is removed by Atg4, a cysteine protease38,100. During autophagy, Atg8 is released from Atg8–PE by a second Atg4-dependent cleavage referred to as deconjugation101, whereas there is no similar cleavage that separates Atg12 from Atg5. Both of the Ubl protein conjugation systems share a single E1-like activating enzyme, Atg7100,102,103,104. The E2-like conjugating enzymes are Atg3 for Atg8, and Atg10 for Atg12102,105.
Mammalian Ubl protein conjugation systems
The Ubl protein conjugation systems are highly conserved from yeast to mammals106,107,108,109. One of the primary differences is the existence of multiple homologs of yeast Atg8, which are divided into two subfamilies, LC3 and GABARAP. LC3 functions at the stage of phagophore elongation, whereas GABARAP proteins act at a later stage of maturation110. There are four mammalian ATG4 homologs, with ATG4B carrying out the primary role in macroautophagy111. As in yeast, this enzyme cleaves after the C-terminal glycine of proLC3 to form cytosolic LC3-I, followed by its subsequent conjugation to PE to generate the membrane-associated LC3-II form112. The GABARAP proteins undergo a similar type of posttranslational modification.
Ligand-receptor-scaffold system
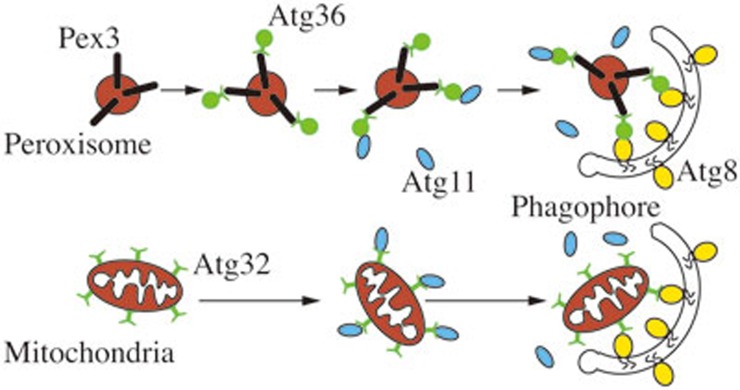
In addition to the core machinery of autophagosome formation, other proteins play significant roles in autophagy, in particular during selective types of macroautophagy. Thus, in S. cerevisiae, Atg19 and its paralog, Atg34, are receptors for the biosynthetic delivery of resident vacuolar hydrolases113,114, Atg32 is a receptor for mitophagy115,116 and Atg36 functions in a similar manner during pexophagy117 (Figure 6). In the case of Atg19, Atg34 and Atg36, the receptor binds a ligand on the cargo, which corresponds to the propeptide of prApe1 for the Cvt pathway, and Pex3 during pexophagy; Atg32 is a mitochondrial membrane protein and does not appear to bind a separate ligand. Each of the receptors binds the scaffold Atg11 and subsequently interacts with Atg8 to link the cargo with the autophagic machinery118.
Figure 6.
Schematic depiction of the mechanism of selective autophagy. Using mitophagy and pexophagy as examples, the model of ligand-receptor-scaffold complex is shown, with Pex3 functioning as a ligand, Atg36 and Atg32 as receptors and Atg11 as the scaffold protein; in the case of mitophagy there does not appear to be a separate ligand on the cargo since Atg32 is an integral membrane protein of the mitochondrial outer membrane.
Selective autophagy: Cvt pathway, pexophagy and mitophagy
In selective autophagy, the overall morphology is largely the same as during nonselective autophagy with one primary distinguishing feature; in selective autophagy, the membrane-bound vesicle targets specific cargos, instead of random cytoplasm. In addition to the core machinery, a cargo ligand (a molecular entity on the surface of the cargo that is recognized by a receptor), receptor and scaffold are involved; the latter act to specifically bind the cargo and provide a bridge between the cargo and the core machinery118,119. Accordingly, during selective autophagy the phagophore is closely apposed to the cargo, and the completed autophagosome excludes bulk cytoplasm. As noted above, there are different types of selective autophagy according to the cargo types. Here we will focus on the Cvt pathway, mitophagy and pexophagy as examples (Table 1).
Cvt pathway
With regard to the details of cargo recognition, the Cvt pathway was the first well-studied type of selective autophagy in yeast; it is also the primary example of a biosynthetic rather than degradative use of autophagy119,120,121,122. The double-membrane sequestering vesicle is termed a Cvt vesicle rather than an autophagosome; as noted above, this vesicle contains selective cargos of the Cvt pathway, but excludes bulk cytoplasm, in accordance with the biosynthetic and constitutive nature of the Cvt pathway compared to starvation-induced autophagy. The Cvt pathway delivers precursor forms of hydrolases, such as Ape1 (aminopeptidase I), Ape4 and Ams1 (α-mannosidase), from the cytoplasm to the vacuole, where they carry out their functions. The formation of the Cvt vesicle at the PAS is driven by the formation of a cargo complex, composed primarily of oligomerized precursor (prApe1) along with a few other resident vacuolar hydrolases123,124,125. Atg19 acts as a cargo receptor that binds the ligand (the prApe1 propeptide) and is thus recruited to the complex113. Subsequently, the interaction between Atg19 and Atg11, a key scaffold that functions in most or all types of selective autophagy in yeast, mediates the delivery of the cargo complex to the PAS99,126. Atg19 also interacts with Atg8, which might play a role in tethering the Cvt complex to the phagophore127 and in directing the selective sequestration of the cargo during vesicle formation99.
Mitophagy
Mitophagy, the selective degradation of mitochondria by autophagy, occurs through both macro- and microautophagic processes. When yeast cells are shifted from a nonfermentable carbon source such as glycerol to a fermentable/preferred carbon source such as glucose, a part of the mitochondria population that is now superfluous is subjected to degradation through mitophagy, and this is particularly enhanced when cells are simultaneously starved for nitrogen. The selective recognition of the cargo is mediated through the mitochondria outer-membrane protein Atg32, which functions as a receptor; this protein is not required for the Cvt pathway or nonselective autophagy128. Similar to the situation with Atg19, Atg32 interacts with Atg11 to recruit mitochondria to the PAS, and also binds to Atg8 to complete vesicle formation116,128 (Figure 6). Atg33 is another protein that is specifically involved in mitophagy115,129. Mitophagy can also be induced by growth in a nonfermentable carbon source when yeast cells grow past the logarithmic phase. Though post-logarithmic-induced mitophagy does not completely share the same mechanism with starvation-induced mitophagy, they are both controlled by two mitogen-activated protein kinases (MAPKs), Slt2 and Hog1129,130. Slt2 affects the recruitment of mitochondria to the PAS, and is also required in pexophagy, whereas Hog1 is mitophagy-specific130. In addition, the absence of the mitochondrial outer membrane protein Uth1 impairs the induction of autophagy by rapamycin treatment or starvation131, suggesting its potential role as a ligand118. Ptc6/Aup1, a yeast mitochondrial protein phosphatase homolog, is required for post-logarithmic phase mitophagy132, and the function of Ptc6 in mitophagy may be based on its regulation of Rtg3-dependent transcription133.
Specific degradation of obsolete mitochondria is also observed in mammalian cells, although Atg32 is not conserved in higher eukaryotes134,135,136. Instead, FUNDC1 and BNIP3, BNIP3L/NIX and SQSTM1/p62 function as receptors in mammalian mitophagy, depending on the inducing conditions, during hypoxia, erythrocyte maturation or damage-induced mitophagy, respectively136,137,138. Moreover, mutations of PARK2/Parkin and PINK1 are linked to Parkinson136,139,140 and Gaucher diseases141. An elegant model for PINK1 stabilization on, and PINK1-dependent recruitment of PARK2 to, the mitochondrial outer membrane, that relies on the E3 ubiquitin ligase activity of PARK2 to induce mitophagy, has been proposed in mammalian cells136.
Pexophagy
Similar to mitophagy, pexophagy happens when the medium of fungi is switched from oleic acid or methanol, where peroxisomes (the only organelle in yeast that can carry out β-oxidation) proliferate, to glucose or nitrogen-starvation medium142. Atg36 and PpAtg30 are receptors for pexophagy in S. cerevisiae and P. pastoris, respectively143,144. In both yeast and mammalian cells Pex3 and Pex14 function as cargo ligands145,146. As with mitophagy, pexophagy is regulated by the Slt2 pathway130.
Structural studies of the Atg proteins
The tremendous amount of molecular genetic data on the yeast Atg proteins has provided considerable information regarding protein-protein interactions; however, detailed structural information at the atomic level has lagged behind. Structural data for the Atg proteins have started to provide direct evidence for protein interactions and are also generating insights about protein function. An increasing amount of crystallography data for the Atg proteins has been reported in recent years. While these data have contributed to the elucidation of autophagy, structural information for many of the Atg proteins is still unavailable. Here, we review the current data for those proteins with available structural information.
Atg1 kinase complex
Atg1, together with the regulatory subunit Atg13, forms the core machinery of the Atg1 kinase complex. Structural data of the entire putative holo-complex (i.e., containing at least Atg1-Atg11-Atg13-Atg17-Atg31-Atg29, Figure 2) are still unavailable. Crystallization of the Atg13 N-terminal domain (NTD) shows a HORMA (Hop1, Rev1 and Mad2) fold147. This domain is important for Atg14 recruitment but not for Atg13 localization or its interaction with Atg1.
In addition to the core machinery, a stable ternary complex composed of Atg17-Atg31-Atg29 (written in the order that reflects the known protein interactions) is also important for the induction of nonselective autophagy. Structural studies41,75,148 reveal that Atg17 is a coiled-coil protein that is made up of four helices. The Atg17 monomer exhibits a crescent shape, which suggests the possibility that it binds curved membranes. The N terminus of Atg17 is involved in binding Atg13. The Atg31-Atg29 proteins are located toward the ends of the Atg17 crescent, where Atg31 serves as a connector bridging Atg17 and Atg29. Atg31 binds to Atg17 through its C-terminal α-helix and interacts with Atg29 via its N-terminal β sandwich. No direct interaction is detected between Atg17 and Atg29. The position of Atg31-Atg29 indicates that this subcomplex may function in regulating Atg17 membrane binding. The ternary complex needs dimerization to be functional, and this is essential for the initiation of autophagy. The dimerization is only mediated by Atg17 helix α4, which is located in the C-terminal region; thus, it does not involve Atg31-Atg29 and may be independent of ternary complex formation. Atg17 dimerization stabilizes the two crescents where they oppose each other, making a “wave” or “S” shape. This two-crescent conformation suggests that the dimer may be involved in both vesicle binding and tethering. Previous studies suggest that the C-terminal domain of Atg1 can also bind vesicles and contains the binding site for Atg13149. The relationship between this domain and Atg17-Atg31-Atg29 has not been determined. Both Atg29 and Atg31 are phosphoproteins, and phosphorylation of Atg29 appears to alter the conformation of the protein to alleviate inhibition by the extreme C terminus as discussed above41. The structure of the C-terminal EAT (“early autophagy targeting/tethering”) domain of Atg1 has not been solved, but based on its predicted helical structure and the observation that it can bind highly curved vesicles, it may sense membrane curvature, dimerize and tether liposomes75.
Atg9 complex
Atg9 was the first identified transmembrane protein in the Atg protein family, but detailed structural information is not available. Similarly, no structural information has been published for the Atg11, Atg23 and Atg27 proteins, which are involved in Atg9 localization to the PAS63,87,88,89 (Figure 3). Atg18, together with Atg2, helps Atg9 cycle from the PAS to peripheral sites63. Although the structure of Atg18 has not been solved, a paralog, Hsv2, which shares high sequence similarity with Atg18, has been studied in different yeast strains150,151,152. Both proteins belong to the “PROPPINs” family, which are defined by β propeller structures and the existence of phosphoinositide binding sites152,153. Hsv2 displays the typical propeller fold that consists of seven β blades. In addition, Hsv2 (and Atg18 along with a second paralog, Atg21) harbor an FRRG motif, which is required for phosphoinositide binding. Detailed analysis indicates that Hsv2 possesses two PtdIns3P binding sites, located in blade 5 and blade 6. Mutations in the corresponding sites in Atg18 result in loss of Atg18 localization. Moreover, the blade 6 β3-β4 loop of Hsv2 proves to be important for protein docking to the membrane. When several hydrophobic residues in this region, or the corresponding residues in Atg18, are mutated, the binding of the protein to liposomes is eliminated152. These results suggest a potential Atg18-docking model: the disk-like Atg18 touches the membrane with its edge; two binding sites interact with PtdIns3P on the membrane surface and the hydrophobic loop penetrates the membrane to stabilize the interaction.
The Atg18 β1-β2 loop and β2-β3 loop in blade 2 are important for PAS targeting151. Blade 2 is located at the opposite position to blade 5 and blade 6, and thus is not involved in membrane docking. Additional studies indicate that the loops in blade 2 are important for Atg18-Atg2 interaction151,154, allowing a refinement of the model for Atg18 membrane binding. While Atg18 binds to the membrane through blades 5 and 6, blade 2, which is located on the opposite side of the protein, interacts with Atg2. The structural data further suggest that the Atg18-Atg2 interaction is independent of membrane docking. Detailed information about the Atg18-Atg2 interaction awaits progress on the crystallization of Atg2, which may also provide insight into their interdependent localization to the PAS.
Two Ubl protein conjugation systems
By far, the most structural information for the Atg proteins concerns those involved in the two Ubl protein conjugation systems (Figure 5). Atg8 is the first identified Ubl Atg protein, and there are several mammalian homologs, comprising the LC3 and GABARAP subfamilies. The crystal structure reveals that the major part of LC3 resembles a ubiquitin core, which consists of a five-stranded β sheet and two α-helices155. Besides the ubiquitin fold, LC3 also contains two N-terminal α-helices and an extended C-terminal tail. As noted above, Atg4 is a cysteine protease that is responsible for removal of the C-terminal residues that follow Gly of Atg8/LC3. The structural analysis of human ATG4B, the homolog of yeast Atg4 that appears to be the most relevant isoform for autophagy, shows that the catalytic residue Cys74 is located in the center of the unique papain-like domain156,157. The catalytic site is masked by a regulatory loop of ATG4B when it is not interacting with its substrate. When LC3 binds ATG4B through its ubiquitin core, both proteins undergo large conformational changes. Trp142 of ATG4B is spatially replaced by Phe119 of LC3, which generates an open conformation of the ATG4B regulatory loop, resulting in exposure of its catalytic site. The opening of the active site is so limited that only the small Gly120 of LC3 can gain access to Cys74, which then attacks the covalent bond between Gly120 and the neighboring residue. At this moment, the open regulatory loop, together with Trp142 of ATG4B, serves as a clamp to stabilize the conformation of LC3-ATG4B157. After the C-terminal amino acid residues are removed, processed LC3 with an exposed Gly120 is released and ATG4B goes back to its auto-inhibited conformation.
LC3/Atg8 is then sent to a non-canonical E1-like enzyme, ATG7/Atg7. The Atg7 monomer is composed of three parts: NTD, adenylation domain (AD) and C-terminal Atg7-specific domain (ECTD)158. The NTD of Atg7 is highly basic, and Atg3, the cognate E2 enzyme, recognizes this positively charged domain. The AD is the key functional domain in Atg7. This domain possesses a long loop region containing the catalytic residue Cys507. ATP, which is important for activation of Atg8, also binds to this domain. The C-terminal tail of the ECTD shows no secondary structure and can interact with Atg8. Biochemical data further show that this interaction is important for recognition of Atg8 by Atg7158,159. Based on the structural and biochemical data, Atg8 is first recognized by the ECTD C-terminal tail of Atg7. Next, the long loop in the Atg7 AD undergoes a large conformational change, which allows Atg8 binding to the AD through its ubiquitin core. At the same time, ATP is also bound to the Atg7 AD. The C-terminal Gly of Atg8 then becomes adenylated. Subsequently, the catalytic Cys507 of Atg7 forms a thioester bond with Atg8. The crystal structure data reveal that Atg7 functions as a homodimer, in which the monomers interact with each other through their AD158. Considering that the NTD of Atg7 is responsible for Atg3 binding, this suggests that Atg8 is activated by one Atg7 and then transferred to Atg3 that is bound to another Atg7160. This trans-transfer model displays higher efficiency than a cis-model and may explain why Atg7 forms a homodimer.
The crystal structure of Atg3 shows a hammer-like conformation with an N-terminal head moiety and a C-terminal handle moiety161. The head moiety shares some similarities to canonical E2 enzymes, while the handle moiety, which is suggested to bind Atg7, is unique to Atg3. Cys234 in Atg3 forms the thioester bond with Atg8, generating the Atg3-Atg8 intermediate. The function of an E3 enzyme in the Atg8 Ubl conjugation system is not fully elucidated. One hypothesis is that the Atg12–Atg5-Atg16 complex, which is the product of the Atg12 Ubl conjugation system in autophagy, acts as the E3 enzyme162. Biochemical studies indicate that the unique catalytic site of Atg3 involves a Thr residue, instead of the more typical Arg, for the transfer of Atg8 to PE163. Furthermore, interaction with the Atg12–Atg5 conjugate results in a change in the orientation of the catalytic cysteine, such that it moves closer to the Thr residue, facilitating the conjugation activity of Atg3.
ATG12 was first crystallized from plant164. The topology of Arabidopsis thaliana ATG12 is similar to other ubiquitin-fold proteins, and ATG12 shares a high degree of structural similarity with LC3/Atg8 except for a unique hydrophobic patch. Similar to Atg8, Atg12 is activated by Atg7. Next, it is sent to another E2 enzyme, Atg10, forming an Atg12–Atg10 intermediate. It is unclear whether Atg3 will compete with Atg10 for interaction with Atg7. Previous studies show that Atg10 bound to Atg7 could be easily replaced by Atg3, but not the reverse, indicating that Atg3 has a higher affinity for binding Atg7159. Atg10 possesses a conserved E2 core that is comprised of four β sheets and two α-helices. However, Atg10 contains two additional accessory α-helices and three additional β sheets, which are important for its normal function165,166. Atg12 is finally conjugated to Lys149 of Atg5, and both structural and biochemical evidence indicate that Atg10 directly interacts with Atg5 through its accessory β5 and β6 sheets.
Atg5 has two Ubl domains, an NTD UblA and C-terminal domain UblB, with a helix-rich domain connecting them, and an extreme N-terminal α-helix165,167. Analysis suggests that Atg5 β7 located in UblB is responsible for interacting with Atg10165. After Atg12 is conjugated to Atg5, no major conformational change is detected168. Atg12–Atg5 prefers to interact with Atg16, forming an Atg12–Atg5-Atg16 complex. Atg16 possesses a coiled-coil C-terminal region, which is responsible for Atg16 homodimer formation169. The NTD of Atg16 is comprised of an α-helix and a loop moiety, and binds Atg5. Structural data reveal that the helix binds to the groove formed by UblA, UblB and the N-terminal α-helix of Atg5, while the loop moiety mainly binds to UblA167.
As mentioned above, previous studies indicate that the Atg12–Atg5-Atg16 complex functions as an E3 ligase in Atg8 lipidation. The structural data of the ATG12–ATG5-ATG16L1N (containing the N-terminal part of ATG16L1) complex reveal that ATG16L1N binds to ATG5 at the opposite side to where ATG12 binds; no interaction is detected between ATG16L1N and ATG12170. Human ATG12 can interact with ATG3 through its Phe108 residue162. Phe154 in plant ATG12 is equivalent to the human ATG12 Phe108, and the former is masked in the ATG12–ATG5 complex168. This observation raises a possible explanation for the function of Atg12–Atg5-Atg16: The complex has two states; in the absence of Atg3–Atg8, the complex is in the closed state where Atg5 buries the Atg3-binding surface of Atg12. When Atg3–Atg8 forms, a conformational change is triggered that exposes the Atg3-binding surface in Atg12. Atg12 then interacts with Atg3 and brings the Atg3–Atg8 intermediate to the membrane, where the Atg12–Atg5-Atg16 complex facilitates Atg8–PE conjugation. Further studies are needed to support and/or refine this model.
The PtdIns3K Complexes
The PtdIns3K complex plays a critical role in autopha-gy through the generation of PtdIns3P, which allows the recruitment of several of the Atg proteins to the PAS. Vps34, the lipid kinase, is the only PtdIns3K in yeast. As described above, Vps34 together with Vps15, Vps30/Atg6, Atg14 and Atg38, form the class III PtdIns3K complex I, which specifically functions in autophagy171.
The structure of Vps34 contains a C2 domain at the N terminus, a helical domain in the middle and a C-terminal catalytic domain. The C2 domain does not affect the catalytic activity of the kinase and is thought to be involved in protein-protein interactions. The crystal structure of Vps34 lacking the C2 domain has been determined172. Vps34 has a highly ordered phosphatidylinositol-binding loop located in its catalytic domain, while the corresponding activation loop that is critical in substrate recognition in phosphoinositide 3-kinases is largely disordered. Vps34 also contains a catalytic loop in this domain. The conserved DRH motif is found in the loop and the His residue in the motif could be the catalytic base. The C-terminal tail that is important for kinase activity is helical. Structural information also reveals that the loop between the last helices (kα11 and kα12) is flexible, which enables the helix to cover/uncover the reaction center. Thus, the final helices help to regulate the activity of Vps34 by covering the catalytic site and preventing it from touching the membrane172.
Vps30/Atg6 forms a complex with Atg14, which, along with Atg38, links it to Vps34. The structures of the separate domains of BECN1, the mammalian ortholog of Vps30, have been solved173,174,175. BECN1 contains three domains, a BH3 domain located at the N terminus, a coiled-coil domain in the middle and a C-terminal evolutionarily conserved domain (ECD). The crystal structure information for the BECN1 ECD indicates that it consists of four α-helices, six β-strands and six loops. One significant feature of the BECN1 ECD concerns loop 4; the main ECD displays an ellipsoid conformation except for the protrusion of loop 4. At the tip of the protrusion, three aromatic amino acids form an “aromatic finger,” which is important for membrane association173. The coiled-coil domain is involved in forming a metastable homodimer that plays a critical role in the interactions of BECN1 with some of its many binding partners175.
Vps15 plays a regulatory role in the complex and its kinase activity is important for Vps34 function. Vps15 contains an N-terminal kinase domain, HEAT repeats in the middle, and a C-terminal WD domain. The structure of the WD domain reveals that it is a seven-bladed propeller176, but the function behind this structure needs to be elucidated with further analysis.
Future directions
Our knowledge of autophagy has expanded significantly during recent years. However, many challenges remain with regard to understanding the detailed mechanism of this process. Perhaps the most complex step of autophagy is that of autophagosome biogenesis. We still do not understand the details of the nucleation process, including the origin of the phagophore membrane or the function of the PAS. For example, how are lipids directed away from their normal destination and into the autophagy pathway? Similarly, there are many fundamental questions concerning phagophore and autophagosome maturation. For example, how is the curvature and size of the phagophore regulated, and what controls the timing of fusion of the completed autophagosome with the lysosome or vacuole? Although the SNARE STX17 was recently shown to bind only to completed autophagosomes177, it is not known what controls the interaction of this protein with the sequestering compartment.
Another challenge in the field relates to the structural studies. Although the structures of some of the Atg/ATG proteins are solved, the crystal structures for many are still unavailable. In addition, x-ray crystallography provides only a static image of a protein and cannot offer any information about disordered regions that may play critical roles in function41. Knowing detailed structural information for the autophagy-related proteins will not only help us better understand the process of autophagy, but can also contribute to rational drug design178. The studies of autophagy regulation are also promising in terms of their potential health applications, and continued studies in this field will provide insight into human diseases and enable the discovery of new therapeutics aimed at their treatment179.
References
- Klionsky DJ, Baehrecke EH, Brumell JH, et al. A comprehensive glossary of autophagy-related molecules and processes (2nd edition) Autophagy. 2011;7:1273–1294. doi: 10.4161/auto.7.11.17661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Osumi M, Ohsumi Y. Analysis of the membrane structures involved in autophagy in yeast by freeze-replica method. Cell Struct Funct. 1995;20:465–471. doi: 10.1247/csf.20.465. [DOI] [PubMed] [Google Scholar]
- Fimia GM, Stoykova A, Romagnoli A, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- Dunn WA., Jr Studies on the mechanisms of autophagy: maturation of the autophagic vacuole. J Cell Biol. 1990;110:1935–1945. doi: 10.1083/jcb.110.6.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Atg9 trafficking in autophagy-related pathways. Autophagy. 2007;3:271–274. doi: 10.4161/auto.3912. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, et al. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Huang WP, Stromhaug PE, Klionsky DJ. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J Biol Chem. 2002;277:763–773. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seglen PO, Gordon PB, Holen I. Non-selective autophagy. Semin Cell Biol. 1990;1:441–448. [PubMed] [Google Scholar]
- de Duve C, Pressman BC, Gianetto R, Wattiaux R, Appelmans F. Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem J. 1955;60:604–617. doi: 10.1042/bj0600604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol. 1962;12:198–202. doi: 10.1083/jcb.12.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SL., Jr Cellular differentiation in the kidneys of newborn mice studied with the electron microscope. J Biophys Biochem Cytol. 1957;3:349–362. doi: 10.1083/jcb.3.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol. 1966;28:435–492. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- Novikoff AB. The proximal tubule cell in experimental hydronephrosis. J Biophys Biochem Cytol. 1959;6:136–138. doi: 10.1083/jcb.6.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deter RL, Baudhuin P, de Duve C. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J Cell Biol. 1967;35:C11–C16. doi: 10.1083/jcb.35.2.c11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci USA. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolender RP, Weibel ER. A morphometric study of the removal of phenobarbital-induced membranes from hepatocytes after cessation of threatment. J Cell Biol. 1973;56:746–761. doi: 10.1083/jcb.56.3.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulaton J, Lockshin RA. Ultrastructural study of the normal degeneration of the intersegmental muscles of Anthereae polyphemus and Manduca sexta (Insecta, Lepidoptera) with particular reference of cellular autophagy. J Morphol. 1977;154:39–57. doi: 10.1002/jmor.1051540104. [DOI] [PubMed] [Google Scholar]
- Veenhuis M, Douma A, Harder W, Osumi M. Degradation and turnover of peroxisomes in the yeast Hansenula polymorpha induced by selective inactivation of peroxisomal enzymes. Arch Microbiol. 1983;134:193–203. doi: 10.1007/BF00407757. [DOI] [PubMed] [Google Scholar]
- Thumm M, Egner R, Koch B, et al. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Cregg JM, Dunn WA, Jr, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Rikihisa Y. Glycogen autophagosomes in polymorphonuclear leukocytes induced by rickettsiae. Anat Rec. 1984;208:319–327. doi: 10.1002/ar.1092080302. [DOI] [PubMed] [Google Scholar]
- Liang XH, Kleeman LK, Jiang HH, et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A, Alexander D, Tallóczy Z, et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Tallóczy Z, Virgin HW IV, Levine B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy. 2006;2:24–29. doi: 10.4161/auto.2176. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, DiFiglia M, Heintz N, et al. Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy. 2005;1:11–22. doi: 10.4161/auto.1.1.1513. [DOI] [PubMed] [Google Scholar]
- Boya P, Gonzalez-Polo RA, Casares N, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Alva A, Su H, et al. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz JB, Schwarz H, Mayer A. Determination of four sequential stages during microautophagy in vitro. J Biol Chem. 2004;279:9987–9996. doi: 10.1074/jbc.M307905200. [DOI] [PubMed] [Google Scholar]
- Deffieu M, Bhatia-Kissova I, Salin B, et al. Glutathione participates in the regulation of mitophagy in yeast. J Biol Chem. 2009;284:14828–14837. doi: 10.1074/jbc.M109.005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn WA, Jr, Cregg JM, Kiel JAKW, et al. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- Huang W-P, Scott SV, Kim J, Klionsky DJ. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J Biol Chem. 2000;275:5845–5851. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- Kirisako T, Baba M, Ishihara N, et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- Cheong H, Klionsky DJ. Dual role of Atg1 in regulation of autophagy-specific PAS assembly in Saccharomyces cerevisiae. Autophagy. 2008;4:724–726. doi: 10.4161/auto.6375. [DOI] [PubMed] [Google Scholar]
- Mao K, Chew LH, Inoue-Aono Y, et al. Atg29 phosphorylation regulates coordination of the Atg17-Atg31-Atg29 complex with the Atg11 scaffold during autophagy initiation. Proc Natl Acad Sci USA. 2013;110:E2875–E2884. doi: 10.1073/pnas.1300064110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Carson AR, Caniggia I, et al. Endothelial nitric-oxide synthase antisense (NOS3AS) gene encodes an autophagy-related protein (APG9-like2) highly expressed in trophoblast. J Biol Chem. 2005;280:18283–18290. doi: 10.1074/jbc.M413957200. [DOI] [PubMed] [Google Scholar]
- Young ARJ, Chan EYW, Hu XW, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- Noda T, Suzuki K, Ohsumi Y. Yeast autophagosomes: de novo formation of a membrane structure. Trends Cell Biol. 2002;12:231–235. doi: 10.1016/s0962-8924(02)02278-x. [DOI] [PubMed] [Google Scholar]
- Kovács AL, Palfia Z, Rez G, Vellai T, Kovács J. Sequestration revisited: integrating traditional electron microscopy, de novo assembly and new results. Autophagy. 2007;3:655–662. doi: 10.4161/auto.4590. [DOI] [PubMed] [Google Scholar]
- Gordon PB, Seglen PO. Prelysosomal convergence of autophagic and endocytic pathways. Biochem Biophys Res Commun. 1988;151:40–47. doi: 10.1016/0006-291x(88)90556-6. [DOI] [PubMed] [Google Scholar]
- Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. EMBO Rep. 2008;9:859–864. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari M, Griffith J, Rieter E, et al. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190:1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart A, Griffith J, Reggiori F. Exit from the Golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2270–2284. doi: 10.1091/mbc.E09-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen W-L, Shintani T, Nair U, et al. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J Cell Biol. 2010;188:101–114. doi: 10.1083/jcb.200904075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R, Jr, Chen PH, Chou CC, Patel J, Jin SV. KCS1 deletion in Saccharomyces cerevisiae leads to a defect in translocation of autophagic proteins and reduces autophagosome formation. Autophagy. 2012;8:1300–1311. doi: 10.4161/auto.20681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Cheong H, Yorimitsu T, Reggiori F, et al. Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Kamada Y, Baba M, et al. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell. 2005;16:2544–2553. doi: 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Kawamata T, Suzuki K, Ohsumi Y. Cis1/Atg31 is required for autophagosome formation in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2007;356:405–410. doi: 10.1016/j.bbrc.2007.02.150. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Kamada Y, Suzuki K, et al. Characterization of a novel autophagy-specific gene, ATG29. Biochem Biophys Res Commun. 2005;338:1884–1889. doi: 10.1016/j.bbrc.2005.10.163. [DOI] [PubMed] [Google Scholar]
- Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- Abeliovich H, Zhang C, Dunn WA, Jr, Shokat KM, Klionsky DJ. Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol Biol Cell. 2003;14:477–490. doi: 10.1091/mbc.E02-07-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair U, Klionsky DJ. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J Biol Chem. 2005;280:41785–41788. doi: 10.1074/jbc.R500016200. [DOI] [PubMed] [Google Scholar]
- Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci USA. 2009;106:17049–17054. doi: 10.1073/pnas.0903316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh YY, Wrasman K, Herman PK. Autophosphorylation within the Atg1 activation loop is required for both kinase activity and the induction of autophagy in Saccharomyces cerevisiae. Genetics. 2010;185:871–882. doi: 10.1534/genetics.110.116566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijanska M, Dohnal I, Reiter W, et al. Activation of Atg1 kinase in autophagy by regulated phosphorylation. Autophagy. 2010;6:1168–1178. doi: 10.4161/auto.6.8.13849. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Yoshino K, Kondo C, et al. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30:1049–1058. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya YV, Stephan JS, Deminoff SJ, Herman PK. An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 2005;102:13933–13938. doi: 10.1073/pnas.0501046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Nice DC III, Nau JJ, et al. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem. 2000;275:25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- Kraft C, Kijanska M, Kalie E, et al. Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J. 2012;31:3691–3703. doi: 10.1038/emboj.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Nair U, Yasumura-Yorimitsu K, Klionsky DJ. A multiple ATG gene knockout strain for yeast two-hybrid analysis. Autophagy. 2009;5:699–705. doi: 10.4161/auto.5.5.8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Noda NN, Fujioka Y, et al. Characterization of the Atg17-Atg29-Atg31 complex specifically required for starvation-induced autophagy in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2009;389:612–615. doi: 10.1016/j.bbrc.2009.09.034. [DOI] [PubMed] [Google Scholar]
- Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012;151:1501–1512. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Sasaki T, Iemura S, et al. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 2009;5:973–979. doi: 10.4161/auto.5.7.9296. [DOI] [PubMed] [Google Scholar]
- Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy. 2009;5:649–662. doi: 10.4161/auto.5.5.8249. [DOI] [PubMed] [Google Scholar]
- Hara T, Takamura A, Kishi C, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro SH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bartolomeo S, Corazzari M, Nazio F, et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol. 2010;191:155–168. doi: 10.1083/jcb.201002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RC, Tian Y, Yuan H, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Shintani T, Nair U, Klionsky DJ. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 2005;1:101–109. doi: 10.4161/auto.1.2.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Baba M, Cao Y, Klionsky DJ. Self-interaction is critical for Atg9 transport and function at the phagophore assembly site during autophagy. Mol Biol Cell. 2008;19:5506–5516. doi: 10.1091/mbc.E08-05-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Song H, Yorimitsu T, et al. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J Cell Biol. 2006;175:925–935. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legakis JE, Yen W-L, Klionsky DJ. A cycling protein complex required for selective autophagy. Autophagy. 2007;3:422–432. doi: 10.4161/auto.4129. [DOI] [PubMed] [Google Scholar]
- Yen WL, Legakis JE, Nair U, Klionsky DJ. Atg27 is required for autophagy-dependent cycling of Atg9. Mol Biol Cell. 2007;18:581–593. doi: 10.1091/mbc.E06-07-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi A, Razi M, Dooley HC, et al. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell. 2012;23:1860–1873. doi: 10.1091/mbc.E11-09-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell. 2013;154:1285–1299. doi: 10.1016/j.cell.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara K, Sekito T, Ohsumi Y. Assortment of phosphatidy-linositol 3-kinase complexes–Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:1527–1539. doi: 10.1091/mbc.E05-09-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki Y, Ku WC, Akioka M, et al. Atg38 is required for autophagy-specific phosphatidylinositol 3-kinase complex integrity. J Cell Biol. 2013;203:299–313. doi: 10.1083/jcb.201304123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parzych KR, Klionsky DJ.An overview of autophagy: Morphology, mechanism, and regulation Antioxid Redox Signal 2013 Aug 2. doi: 10.1089/ars.2013.5371 [DOI] [PMC free article] [PubMed]
- Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Huang W-P, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Dalton VM, Eggerton KP, Scott SV, Klionsky DJ. Apg7p/Cvt2p is required for the cytoplasm-to-vacuole targeting, macroautophagy, and peroxisome degradation pathways. Mol Biol Cell. 1999;10:1337–1351. doi: 10.1091/mbc.10.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Ichimura Y, Okada H, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, et al. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Tanida I, Mizushima N, Kiyooka M, et al. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol Biol Cell. 1999;10:1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Mizushima N, Ogawa Y, et al. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 1999;18:5234–5241. doi: 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem. 1998;273:33889–33892. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- Tanida I, Tanida-Miyake E, Ueno T, Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem. 2001;276:1701–1706. doi: 10.1074/jbc.C000752200. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. Mouse Apg10 as an Apg12-conjugating enzyme: analysis by the conjugation-mediated yeast two-hybrid method. FEBS Lett. 2002;532:450–454. doi: 10.1016/s0014-5793(02)03739-0. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Kuma A, Kobayashi Y, et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- Weidberg H, Shvets E, Shpilka T, et al. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Hou Y, Wang J, et al. Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J Biol Chem. 2011;286:7327–7338. doi: 10.1074/jbc.M110.199059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Yamamoto A, et al. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Noda NN, Kumeta H, et al. Selective transport of α-mannosidase by autophagic pathways: structural basis for cargo recognition by Atg19 and Atg34. J Biol Chem. 2010;285:30026–30033. doi: 10.1074/jbc.M110.143545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Klionsky DJ. Mitophagy in yeast occurs through a selective mechanism. J Biol Chem. 2008;283:32386–32393. doi: 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Motley AM, Nuttall JM, Hettema EH. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. 2012;31:2852–2868. doi: 10.1038/emboj.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijaljica D, Nazarko TY, Brumell JH, et al. Receptor protein complexes are in control of autophagy. Autophagy. 2012;8:1701–1705. doi: 10.4161/auto.21332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch-Day MA, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS Lett. 2010;584:1359–1366. doi: 10.1016/j.febslet.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Cueva R, Yaver DS. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda MN, Scott SV, Hefner-Gravink A, Caffarelli AD, Klionsky DJ. Identification of a cytoplasm to vacuole targeting determinant in aminopeptidase I. J Cell Biol. 1996;132:999–1010. doi: 10.1083/jcb.132.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Hefner-Gravink A, Morano KA, et al. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc Natl Acad Sci USA. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Baba M, Ohsumi Y, Klionsky DJ. Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J Cell Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Scott SV, Oda MN, Klionsky DJ. Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1997;137:609–618. doi: 10.1083/jcb.137.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kamada Y, Stromhaug PE, et al. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol. 2001;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J, Klionsky DJ. Quantitative regulation of vesicle formation in yeast nonspecific autophagy. Autophagy. 2008;4:955–957. doi: 10.4161/auto.6791. [DOI] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Baba M, et al. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol Biol Cell. 2009;20:4730–4738. doi: 10.1091/mbc.E09-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K, Wang K, Zhao M, Xu T, Klionsky DJ. Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae. J Cell Biol. 2011;193:755–767. doi: 10.1083/jcb.201102092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissova I, Deffieu M, Manon S, Camougrand N. Uth1p is involved in the autophagic degradation of mitochondria. J Biol Chem. 2004;279:39068–39074. doi: 10.1074/jbc.M406960200. [DOI] [PubMed] [Google Scholar]
- Tal R, Winter G, Ecker N, Klionsky DJ, Abeliovich H. Aup1p, a yeast mitochondrial protein phosphatase homolog, is required for efficient stationary phase mitophagy and cell survival. J Biol Chem. 2007;282:5617–5624. doi: 10.1074/jbc.M605940200. [DOI] [PubMed] [Google Scholar]
- Journo D, Mor A, Abeliovich H. Aup1-mediated regulation of Rtg3 during mitophagy. J Biol Chem. 2009;284:35885–35895. doi: 10.1074/jbc.M109.048140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen M, Ferguson DJ, Simon AK. Mitochondrial clearance by autophagy in developing erythrocytes: clearly important, but just how much so. Cell Cycle. 2010;9:1901–1906. doi: 10.4161/cc.9.10.11603. [DOI] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak I, Kirkin V, McEwan DG, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak I, Dikic I. Autophagy receptors in developmental clearance of mitochondria. Autophagy. 2011;7:301–303. doi: 10.4161/auto.7.3.14509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- Osellame LD, Duchen MR. Defective quality control mechanisms and accumulation of damaged mitochondria link Gaucher and Parkinson diseases. Autophagy. 2013;9:1633–1635. doi: 10.4161/auto.25878. [DOI] [PubMed] [Google Scholar]
- Hutchins MU, Veenhuis M, Klionsky DJ. Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J Cell Sci. 1999;112:4079–4087. doi: 10.1242/jcs.112.22.4079. [DOI] [PubMed] [Google Scholar]
- Farré JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell. 2008;14:365–376. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarko TY, Farre JC, Subramani S. Peroxisome size provides insights into the function of autophagy-related proteins. Mol Biol Cell. 2009;20:3828–3839. doi: 10.1091/mbc.E09-03-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Kuge S, Fujiki Y. The peroxin Pex14p is involved in LC3-dependent degradation of mammalian peroxisomes. Exp Cell Res. 2008;314:3531–3541. doi: 10.1016/j.yexcr.2008.09.015. [DOI] [PubMed] [Google Scholar]
- Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci USA. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao CC, Ragusa MJ, Stanley RE, Hurley JH. A HORMA domain in Atg13 mediates PI 3-kinase recruitment in autophagy. Proc Natl Acad Sci USA. 2013;110:5486–5491. doi: 10.1073/pnas.1220306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew LH, Setiaputra D, Klionsky DJ, Yip CK. Structural characterization of the Saccharomyces cerevisiae autophagy regulatory complex Atg17-Atg31-Atg29. Autophagy. 2013;9:1467–1474. doi: 10.4161/auto.25687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh YY, Shah KH, Herman PK. An Atg13 protein-mediated self-association of the Atg1 protein kinase is important for the induction of autophagy. J Biol Chem. 2011;286:28931–28939. doi: 10.1074/jbc.M111.250324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krick R, Busse RA, Scacioc A, et al. Structural and functional characterization of the two phosphoinositide binding sites of PROPPINs, a β-propeller protein family. Proc Natl Acad Sci USA. 2012;109:E2042–E2049. doi: 10.1073/pnas.1205128109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Kobayashi T, Yamamoto H, et al. Structure-based analyses reveal distinct binding sites for Atg2 and phosphoinositides in Atg18. J Biol Chem. 2012;287:31681–31690. doi: 10.1074/jbc.M112.397570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskaran S, Ragusa MJ, Boura E, Hurley JH. Two-site recognition of phosphatidylinositol 3-phosphate by PROPPINs in autophagy. Mol Cell. 2012;47:339–348. doi: 10.1016/j.molcel.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove SK, Piper RC, McEwen RK, et al. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J. 2004;23:1922–1933. doi: 10.1038/sj.emboj.7600203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieter E, Vinke F, Bakula D, et al. Atg18 function in autophagy is regulated by specific sites within its b-propeller. J Cell Sci. 2013;126:593–604. doi: 10.1242/jcs.115725. [DOI] [PubMed] [Google Scholar]
- Sugawara K, Suzuki NN, Fujioka Y, et al. The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8. Genes Cells. 2004;9:611–618. doi: 10.1111/j.1356-9597.2004.00750.x. [DOI] [PubMed] [Google Scholar]
- Sugawara K, Suzuki NN, Fujioka Y, et al. Structural basis for the specificity and catalysis of human Atg4B responsible for mammalian autophagy. J Biol Chem. 2005;280:40058–40065. doi: 10.1074/jbc.M509158200. [DOI] [PubMed] [Google Scholar]
- Satoo K, Noda NN, Kumeta H, et al. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009;28:1341–1350. doi: 10.1038/emboj.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda NN, Satoo K, Fujioka Y, et al. Structural basis of Atg8 activation by a homodimeric E1, Atg7. Mol Cell. 2011;44:462–475. doi: 10.1016/j.molcel.2011.08.035. [DOI] [PubMed] [Google Scholar]
- Hong SB, Kim BW, Lee KE, et al. Insights into noncanonical E1 enzyme activation from the structure of autophagic E1 Atg7 with Atg8. Nat Struct Mol Biol. 2011;18:1323–1330. doi: 10.1038/nsmb.2165. [DOI] [PubMed] [Google Scholar]
- Taherbhoy AM, Tait SW, Kaiser SE, et al. Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway. Mol Cell. 2011;44:451–461. doi: 10.1016/j.molcel.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Suzuki NN, Hanada T, et al. The crystal structure of Atg3, an autophagy-related ubiquitin carrier protein (E2) enzyme that mediates Atg8 lipidation. J Biol Chem. 2007;282:8036–8043. doi: 10.1074/jbc.M611473200. [DOI] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, et al. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoh-Nakatogawa M, Matoba K, Asai E, et al. Atg12-Atg5 conjugate enhances E2 activity of Atg3 by rearranging its catalytic site. Nat Struct Mol Biol. 2013;20:433–439. doi: 10.1038/nsmb.2527. [DOI] [PubMed] [Google Scholar]
- Suzuki NN, Yoshimoto K, Fujioka Y, Ohsumi Y, Inagaki F. The crystal structure of plant ATG12 and its biological implication in autophagy. Autophagy. 2005;1:119–126. doi: 10.4161/auto.1.2.1859. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Noda NN, Yamamoto H, et al. Structural insights into Atg10-mediated formation of the autophagy-essential Atg12-Atg5 conjugate. Structure. 2012;20:1244–1254. doi: 10.1016/j.str.2012.04.018. [DOI] [PubMed] [Google Scholar]
- Kaiser SE, Mao K, Taherbhoy AM, et al. Noncanonical E2 recruitment by the autophagy E1 revealed by Atg7-Atg3 and Atg7-Atg10 structures. Nat Struct Mol Biol. 2012;19:1242–1249. doi: 10.1038/nsmb.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita M, Suzuki NN, Obara K, et al. Structure of Atg5·Atg16, a complex essential for autophagy. J Biol Chem. 2007;282:6763–6772. doi: 10.1074/jbc.M609876200. [DOI] [PubMed] [Google Scholar]
- Noda NN, Fujioka Y, Hanada T, Ohsumi Y, Inagaki F. Structure of the Atg12-Atg5 conjugate reveals a platform for stimulating Atg8-PE conjugation. EMBO Rep. 2013;14:206–211. doi: 10.1038/embor.2012.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y, Noda NN, Nakatogawa H, Ohsumi Y, Inagaki F. Dimeric coiled-coil structure of Saccharomyces cerevisiae Atg16 and its functional significance in autophagy. J Biol Chem. 2010;285:1508–1515. doi: 10.1074/jbc.M109.053520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otomo C, Metlagel Z, Takaesu G, Otomo T. Structure of the human ATG12∼ATG5 conjugate required for LC3 lipidation in autophagy. Nat Struct Mol Biol. 2013;20:59–66. doi: 10.1038/nsmb.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Klionsky DJ. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics. 2013;194:341–361. doi: 10.1534/genetics.112.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Tavshanjian B, Oleksy A, et al. Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science. 2010;327:1638–1642. doi: 10.1126/science.1184429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Choi W, Hu W, et al. Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell Res. 2012;22:473–489. doi: 10.1038/cr.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J Biol Chem. 2007;282:13123–13132. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- Li X, He L, Che KH, et al. Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat Commun. 2012;3:662. doi: 10.1038/ncomms1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heenan EJ, Vanhooke JL, Temple BR, et al. Structure and function of Vps15 in the endosomal G protein signaling pathway. Biochemistry. 2009;48:6390–6401. doi: 10.1021/bi900621w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Miller S, Oleksy A, Perisic O, Williams RL. Finding a fitting shoe for Cinderella: searching for an autophagy inhibitor. Autophagy. 2010;6:805–807. doi: 10.4161/auto.6.6.12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012;11:709–730. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]