Abstract
The year of 2013 marked the 50th anniversary of C de Duve's coining of the term “autophagy” for the degradation process of cytoplasmic constituents in the lysosome/vacuole. This year we regretfully lost this great scientist, who contributed much during the early years of research to the field of autophagy. Soon after the discovery of lysosomes by de Duve, electron microscopy revealed autophagy as a means of delivering intracellular components to the lysosome. For a long time after the discovery of autophagy, studies failed to yield any significant advances at a molecular level in our understanding of this fundamental pathway of degradation. The first breakthrough was made in the early 1990s, as autophagy was discovered in yeast subjected to starvation by microscopic observation. Next, a genetic effort to address the poorly understood problem of autophagy led to the discovery of many autophagy-defective mutants. Subsequent identification of autophagy-related genes in yeast revealed unique sets of molecules involved in membrane dynamics during autophagy. ATG homologs were subsequently found in various organisms, indicating that the fundamental mechanism of autophagy is well conserved among eukaryotes. These findings brought revolutionary changes to research in this field. For instance, the last 10 years have seen remarkable progress in our understanding of autophagy, not only in terms of the molecular mechanisms of autophagy, but also with regard to its broad physiological roles and relevance to health and disease. Now our knowledge of autophagy is dramatically expanding day by day. Here, the historical landmarks underpinning the explosion of autophagy research are described with a particular focus on the contribution of yeast as a model organism.
Keywords: autophagy, yeast, ATG
Introduction
The broadly dynamic nature of life becomes more and more apparent with every advance in scientific understanding. In addition to metabolites, cellular machineries, proteins and organelles themselves are maintained in equilibrium between continuous synthesis and degradation, which is one essential difference between life and man-made machinery. A constant supply of chemicals and energy from the external environment is necessary for the maintenance of this equilibrium state. The most prolific source of such chemicals encountered in animals is the digestion of protein taken as food in the digestive tract, which is topologically held outside of the body. In terms of intracellular proteins, pioneering work by R. Schoenheimer presented an important concept, that of “protein turnover” in the body1. His prescient studies using isotopic labeling of proteins were unconventional at the time, but stimulated much debate and subsequent work in the field of intracellular protein metabolism2. However, people still believed for many years that proteins are stable in vivo and that degradation does not play an important role in protein homeostasis. In the 1970s, trials to estimate individual protein lifetimes by degradation rate within the cell through the injection of purified proteins into cells or by pulse chase experiments indicated that every protein in vivo has a distinct half-life time broadly ranging from a few minutes to more than 100 days3,4. We now know that the proteins constituting our bodies are replaced almost completely every 1-2 months without any apparent change in physical appearance. We still do not completely understand what exactly determines the unique lifetimes of proteins, how and why this phenomenon occurs, or indeed the physiological significance behind such intricate and distinct patterns of protein degradation. However, it is clear that in order to adapt to continuous or abrupt changes in environmental conditions protein turnover must play an essential role.
In nature, starvation, which is simply a shortage in nutrient supply, is the most frequent and serious threat to the maintenance and preservation of life. In order to endure this adversity, the recycling of proteins is a primary line of defense by which cells can reserve amino acids for the synthesis of the minimal complement of proteins essential for survival. At an early stage of evolution, cells must have acquired a certain mechanism of intracellular protein degradation that was refined over the course of evolution; a striking example of this is that all organisms on Earth possess protease without exception. Now it is known that even under normal conditions most amino acids for protein synthesis in our body are derived from the degradation of a cell's own proteins, indicating that recycling is an intrinsic feature of life.
At the same time, intracellular protein degradation could be a dangerous process. Indiscriminate degradation alongside protein synthesis within the same compartment could conceivably cause a futile cycle resulting in the waste of energy. However, there are at least four key strategies that have evolved to avoid this problem: first, protease activity is tightly regulated, and the activation of hydrolytic enzyme occurs only when necessary. Second, protease forms a large caged complex resembling a barrel, confining its degradative activity. Third, targets are selectively modified prior to degradation, allowing a degree of control over what is broken down. Finally, hydrolytic enzymes are sequestered into a membrane compartment, physically segregating this activity within the cell. The ubiquitin/proteasome system is responsible for cases 2 and 3. Case 4 is dependent upon the lysosomal degradation system. Segregation of hydrolases by a membrane inevitably gives rise to another problem, which is how to access the substrates destined for degradation by the hydrolytic enzymes. Therefore, degradation via the lysosomal system, which depends to a large degree upon autophagy, necessarily involves certain membrane-mediated processes.
Discovery of the lysosome and autophagy
The uncovering of the mechanisms of intracellular protein degradation was brought about by the discovery of the lysosome by C de Duve5. During cell fractionation of rat liver homogenate by consecutive centrifugation, he found a latency of acid phosphatase activity. Further analyses revealed a novel organelle enwrapping an acid phosphatase and also various kinds of hydrolytic enzymes with optimal activity at acidic pH. He named this unique organelle, identified purely by biochemical studies, the “lysosome”, referring to its role as an organelle for lytic function6.
Soon afterwards, electron microscopic studies were employed, identifying lysosomes in situ within cells and tissue. A Novikoff and colleagues first observed an isolated lysosome-enriched fraction, and then found similar structures in a thin section of liver tissue, which proved the lysosome as a morphological entity7. The early observation of lysosomes in various cells and tissues revealed that lysosomes are morphologically quite heterogeneous among various cell types.
At that time the process of endocytosis had been studied by electron microscopy (EM), which revealed that cells in which endocytosis is prolific, such as macrophages and leukocytes, contain a lot of lysosomes. In the process of endocytosis, foreign materials from outside the cell are first taken up into a phagosome, which then fuses with the lysosome, yielding a body known as the phagolysosome within which degradation of the target proceeds8. These observations provided an understanding of the route from the outside to the lysosomes.
However, the degradation process of intracellular components was more difficult to analyze. The first important finding was made by S Clark, who observed kidney tubule cells from new born mice, and found, in addition to lysosomes, irregular shaped vacuoles containing amorphous materials and occasionally mitochondria9. A Novikoff et al., also observed similar acid phosphatase-positive membrane structures containing mitochondria, ER, and ribosomes7. These membrane structures could be induced by certain chemical treatments or stress conditions. T P Ashfold and K R Porter next showed that treatment with a hormone, glucagon, results in the increased frequency of lysosomes containing cytoplasmic components10. Such treatments were helpful when describing the morphological evolution of these cytoplasmic component-containing membrane structures over time. Soon afterwards, A U Arstila and B F Trump skillfully showed that a double membrane-bound structure containing a portion of cytoplasm and organelles without hydrolytic enzymes known as the autophagosome is formed at first11. This structure is subsequently observed as a single membrane structure, referred to as the autophagolysosome, showing various stages of organelle degradation by lysosomal enzymes11. Based upon these observations, C de Duve defined this mode of delivery of cytoplasmic materials to the lysosomes for degradation as “autophagy” (from the Greek for self-eating) in 196312, which was at the time contrasted with “heterophagy”, the route from the outside to the lysosomes, now generally called endocytosis.
Since these important findings, autophagy has been reported in various tissues and organs of animals and even plants13. The most critical event in autophagy is the sequestration of a portion of cytoplasm by forming a new compartment, the autophagosome. Autophagosome formation relies on a series of dynamic membrane events: in the cytoplasm a small membrane sac (the phagophore or isolation membrane) first appears, which extends to form a cup-shaped structure that subsequently seals, resulting in a double membrane-bound structure, the autophagosome, enclosing a portion of the cytoplasm. This autophagosome then fuses with the lysosome, and the inner membrane and its contents are degraded by lysosomal hydrolases. Finally, the autophagolysosome becomes a residual body and degradation products such as amino acids are transported back to the cytoplasm and reused as building blocks or a source of energy. The detailed mechanism of membrane biogenesis and the origin of autophagosome are important questions that remain to be answered (see review by Yoshimori in this issue).
Before the autophagy-related genes
Early conceptions of the physiological role of autophagy
As mentioned above, de Duve and others found that glucagon induces autophagy in the liver14. This finding stimulated interest in the physiological role of autophagy. One such subsequent study, by U Pfeifer et al., examined the degradation of long-lived proteins by autophagy, revealing the relationship between autophagy and the circadian rhythm15,16. In this work, the authors found that autophagy is inhibited by feeding and induced by fasting between meals, reinforcing that autophagy is tightly regulated by nutrient conditions. U Pfeifer et al., also showed that by 48 h of starvation 30-40% of liver proteins are degraded16.
Around the same time, G Mortimore's group was working intensively on the regulation of autophagy by nutrients, examining the release of amino acids from proteins in perfused rat liver. First, they found that the protein degradation rate is significantly increased when amino acids in the perfusion solution were decreased. In contrast, increased levels of amino acids in the solution suppressed degradation to one-third of the normal, basal rate. These experiments clearly showed that protein degradation via autophagy is strictly controlled by amino-acid level17. Furthermore, Mortimore asked which amino acids individually or in combination have an inhibitory effect on autophagy, and showed that eight amino acids, Leu, Tyr, Phe, Gln, Pro, His, Trp, and Met, suppress autophagy18. Soon, P O Seglen performed a similar experiment by using hepatocyte culture and while arriving at a similar conclusion, also showed that Leu most strongly inhibits autophagy19. Recently, the effects of amino acids on autophagy have been reexamined in relation to mTORC1 activation, and several fascinating models of action have been proposed20,21. Still, further experiments are necessary to obtain a nuanced understanding of the regulation of autophagy at a molecular level (see review by Guan in this issue).
In contrast to the stimulatory effect of glucagon, several groups reported that insulin suppresses autophagy14, which fits well with the known control functions of these hormones in catabolism and anabolism, respectively. The regulation of autophagy by these hormones is a physiologically important subject, and thus their mechanisms of action have long been of great interest. With the recent development of powerful tools such as mass spectrometry, we can now analyze the dynamic behavior of various metabolites, including amino acids. Further systematic analyses will provide detailed insights into the regulation and the close relationship between autophagy and cellular metabolism.
Since the acidic pH of the lysosome is critical to its function, several reagents which raise the lysosomal pH also inhibit autophagic protein degradation, such as ammonium ions, chloroquine and bafilomycin, which were used to estimate lysosomal degradation. However, these compounds could broadly affect a range of cellular functions, limiting the usefulness of such studies.
In a screen for specific organic compounds inhibiting autophagy, P O Seglen reported the inhibitory activity of 3-methyladenine22, and this compound has been used ever since as an inhibitor of autophagy. Later it was proved to be, like wortmannin, an inhibitor of type III phosphatidylinositol 3-kinase (PI-3K), which is essential for autophagosome formation23. However, 3-methyladenine inhibits not only Class III PI-3K but also Class I kinase, and the concentration required for effective inhibition is very high. Treatment with inhibitors such as leupeptin, a cathepsin inhibitor, induces the formation of many vacuoles containing various cytoplasmic enzymes by blocking degradation in the lysosome, and biochemical analysis revealed that these represent non-selective enwrapping of cytosolic enzymes24.
Involvement of protein phosphorylation in autophagy was also proposed by P O Seglen by using various protein kinase inhibitors25,26, but the targets and kinases involved at various stages of autophagy remained unknown for a long time, which hampered progress in the elucidation of the role of phosphorylation. Now many efforts in searching for specific inhibitors of autophagy are in progress based upon recent advances in biochemical and structural studies of the autophagic machinery.
Different modes of autophagy
The process stated above, involving the autopha-gosomal sequestration of targets to be degraded, is called macroautophagy, which is the major intracellular route to the lysosome. Macroautophagy was initially understood to target cytosolic components and organelles non-selectively. However, ribosomes, mitochondria and other organelles were observed in the autophagolysosome from the early days of autophagy research, which suggests that autophagy is a good means of degrading large supramolecular structures and organelles efficiently in one fell swoop. Necessarily, questions remained as to whether such components are sequestered by chance or selectively delivered to the autophagosome. Selective uptake of proliferated organelles by autophagy following nutrient changes or chemical treatments was shown in early studies tackling this problem. Such works showed that excessive peroxisomes or ER were selectively enwrapped into autophagosomes and degraded27,28. These findings indicated that autophagy plays a role in the control of the quantity of organelles. At present one of the most interesting matters in the field concerns the selective degradation of organelles. Recent progress in the molecular mechanism of selective autophagy is discussed in more detail elsewhere (see review by Okamoto in the same special issue).
The occasional transfer of cytosolic components to the lysosomal compartment by direct invagination of the lysosomal membrane without prior sequestration into an autophagosome has also been observed. This phenomenon is known as microautophagy because a smaller portion of cytoplasm is degraded when compared to macroautophagy29. Detailed studies of microautophagy in mammals remain scarce, but recently its mechanism and significance have attracted much attention. Microautophagy by the vacuolar membrane in several yeast species, such as Pichia pastoris and Hansenula polymorpha, has been intensively studied as a model system of selective autophagy30.
In 1978 J F Dice reported the selective degradation of cytosolic proteins by the lysosome31. A typical substrate for this system, RNase A, was subsequently investigated in further detail. For efficient uptake, the 20-amino acid residues of the N-terminal domain are necessary, and further systematic analysis indicated that the degradation signal resides within the amino-acid sequence of a KFERQ motif which acts to facilitate direct transport of the substrate across the lysosomal membrane. Recently this pathway was named chaperon-mediated autophagy (CMA), the molecular details and physiological roles of which have been intensively studied by A M Cuervo and are presented elsewhere (see review by Cuervo in this issue).
As described above, autophagy is now used as a general term of degradation in the lysosome/vacuole, but hereafter in this review macroautophagy is referred to simply as autophagy.
Difficulties in the study of autophagy
For a long time autophagy was primarily analyzed morphologically by EM through the examination of autophagosomes and autophagolysosomes in ultrathin sections. Lysosome-related membrane structures are quite heterogeneous in shape and content, and therefore precise interpretation of images requires long-term experience. While the phenomenon of autophagy could be observed in cells, for many years there was no good method to quantitatively estimate the extent of autophagy. The release of amino acids, such as Val or Leu, from pre-labeled cellular proteins was frequently monitored as an indicator of autophagy32, but it is hard to estimate how much of this release really reflects autophagic degradation. Moreover, the autophagosome is a transient organelle existing for less than 10 min before fusing with the lysosome, resulting in the appearance of autophagolysosomes at various stages of degradation. Therefore, the biochemical study of autophagy also presents a significant challenge to researchers. Although lysosomal enzymes (especially proteases) have been intensely studied, identification of lysosomal membrane proteins has progressed slowly. V-type ATPase, a proton pump responsible for acidification of the lysosomes, is a typical example, being identified and characterized only in the 1980s33. It is needless to mention that at the early stages of autophagy research neither proteins specifically localizing to the autophagosome nor proteins necessary for autophagy had been identified. Thus, the most crucial problem was a lack of a specific marker of the autophagosome or autophagolysosome for morphological and biochemical studies. A simple and good model system would therefore be essential to elucidate the molecular mechanism of autophagy.
After the ATG genes
Discovery of autophagy in yeast
In the yeast S. cerevisiae, the vacuole is the only visible organelle by phase contrast microscopy, and functions as a storage compartment for amino acids and ions. The vacuole was assumed to be equivalent to the lysosome in mammals, since it is an acidic compartment containing many kinds of hydrolytic enzymes34. When yeast cells are challenged by nitrogen-depleted conditions, sporulation and meiotic cell division are induced. It is clear that this remarkable cell differentiation and remodeling process in the absence of nitrogen supply must require bulk protein degradation of a cell's own proteins. The vacuole is a fairly large compartment (ca 3 μm in diameter) of low refractive index due to its very low protein content. If a vacuolar proteinase-deficient mutant encounters nitrogen-starvation, materials delivered to the vacuole may become observable due to the blocking of normal degradation. Indeed Y Ohsumi found dramatic morphological changes in the vacuoles upon starvation. After 30 min of starvation spherical bodies vigorously moving around appeared in the vacuole, which gradually increased in number and finally filled the vacuole35. EM revealed that these bodies, termed autophagic bodies, were single membrane-bound vesicles averaging 500 nm in diameter and enclosing a portion of cytoplasm, containing ribosomes and occasionally various cytoplasmic structures such as mitochondria36.
The progression of autophagy can therefore be monitored by the accumulation of autophagic bodies in the vacuole under a light microscope. In wild type cells these bodies are immediately disintegrated by vacuolar hydrolases.
Further, EM studies by M Baba et al. revealed a double membrane structure of the same size as autophagic bodies in the cytoplasm, the yeast autophagosome, and also fusion between the outer membrane of the autophagosome and the vacuolar membrane37. This fusion resulted in a single membrane-bound autophagic body, in the vacuolar lumen. This series of membrane dynamics is topologically exactly the same as the process of autophagy in mammals except that the vacuole is much larger than the lysosome. Soon after the same process was shown to be induced under conditions of carbon-, sulfate-, phosphate- and single auxotrophic amino-acid-starvation35. EM analyses of autophagosomes and autophagic bodies revealed several features unique to these membranes. In thin section images, the double-layered autophagosomal membranes are thinner than any other organelle membranes and are tightly associated without any intermembrane space. Also, freeze-fracture images have indicated the limited presence of intramembrane particles in the outer membrane, whereas almost completely none were observed in the inner membrane, showing that the two membranes are differentiated and that the autophagosomal membrane is specialized for the sequestration of a portion of the cytoplasm37.
During morphological studies, T Noda in Ohsumi's lab developed an elegant assay of autophagic activity in yeast38. While a decrease in cytoplasmic proteins by autophagy is hard to quantify, the power of this system was in the measurement of an autophagy-dependent increase in enzyme activity. By constructing a cell harboring an engineered pro-form of vacuolar phosphatase in the cytosol (Pho8Δ60), the maturation of this enzyme can be monitored as it is taken up into autophagosomes, transported into the vacuole by autophagy and subsequently processed to become an active enzyme. This gain of activity forms the reporter of autophagic activity, which for example dramatically increases during autophagy-inducing conditions such as starvation. This important development enabled a simple but reliable estimation of autophagy activity in a quantitative manner.
Genetic analysis of autophagy
Y Ohsumi next adopted a genetic approach in order to dissect the autophagic process. At that time nothing was known about either the physiological roles of autophagy, or indeed the phenotypes of autophagy-defective mutants in yeast. Tsukada's simple approach, using light microscopic selection to obtain mutants that fail to accumulate autophagic bodies under nitrogen-starvation conditions, yielded the first autophagy-defective mutant, apg139. The mutant was indeed defective in starvation-induced protein degradation, and its homozygous diploid was sporulation defective. However, it grew normally in a rich medium and showed no obvious defects in a range of functions including secretion and endocytosis. One obvious phenotype, however, was that apg1 mutant cells died when subjected to long periods of nitrogen starvation39. To obtain further apg mutants this loss of viability phenotype was used as a primary screen, with intial candidates subsequently examined by the microscopic method described above as a secondary screen, resulting in the identification of about one hundred autophagy-defective mutants. Genetic analyses of these apg mutants revealed 15 complementation groups39. These mutants showed quite similar phenotypes to apg1 and were otherwise indistinguishable from each other. EM analyses suggested that these mutants were characterized by defects in autophagosome formation. The strategy of the screen was based upon an assumption that autophagy is not essential for cell growth, and accordingly all of these mutations did not affect vegetative growth in a rich medium. The loss of viability phenotype is caused by the complete loss of starvation-induced autophagy, which indicates that these apg mutants represent null mutations. It should be emphasized that the first screen was highly effective, resulting in the isolation of most of the genes essential for autophagy, greatly facilitating subsequent molecular studies of autophagy40.
Around the same time M Thumm isolated six autophagy-defective aut mutants by screening for colonies defective in cytoplasmic enzyme degradation using antibody staining41. D Klionsky, who originally had been interested in the transport of the vacuolar enzyme α-aminopeptidase I to the vacuole, known as the Cvt pathway, isolated many mutants defective in the transport of this enzyme42. Electron microscopic studies of this pathway revealed that it comprises almost exactly the same membrane dynamics as macroautophagy43, and therefore the Cvt pathway has since been studied as an excellent model of selective autophagy, although it is a constitutive and biosynthetic pathway of vacuolar enzymes rather than a degradative pathway. Dr Klionsky's group successfully isolated a number of cvt mutants defective in this pathway, which turned out to be mostly allelic to apg mutants (see below, and also review by Klionsky in this issue).
Several groups also assessed autophagy in different yeast species, obtaining mutants to which they ascribed their own names, for example naming genes after glucose-mediated selective autophagy44,45, and peroxisome degradation (PAG30, PAZ46, and PDD47). This confusing situation was settled by using a new, unified system of gene nomenclature, with ATG genes being used to name autophagy genes (see review by Klionsky in this issue). Now the number of ATG genes has increased to more than 37, consisting of a range of genes responsible for the core machinery of autophagosome formation as well as those genes required specifically for selective modes of autophagy.
Isolation and characterization of autophagy genes
Cloning of ATG genes was undertaken by screening a genomic library for DNA fragments complementing the phenotype of atg mutants. Ohsumi's group first isolated ATG1, then ATG5, ATG6, ATG13 and so on48. Examination of the amino-acid sequence of Atg1 indicated that this protein is a Ser/Thr protein kinase49. ATG6 was found to be allelic to VPS3050, which was identified at almost the same time, as a factor essential for the vacuolar protein sorting (Vps) pathway. But all other ATG genes turned out to be unknown genes at the time, indicating that most genes specifically involved in autophagy had never been investigated even in yeast. Before the unification of nomenclature, Thumm also cloned AUT genes41, while Klionsky's group obtained several CVT genes42. Owing to the availability of the complete genome sequence made possible by the yeast genome project, the process of the identification of ATG genes was highly accelerated, and most of the ATG genes were cloned within a very short period. However, their predicted primary sequences offered few clues as to the functions of the encoded proteins.
When the cloning of ATG genes neared conclusion, the relationships among Atg proteins were described within a brief period, revealing that the proteins function in several functional units as outlined below.
The Atg12 conjugation system
The first striking finding was the discovery of a ubiquitin-like protein conjugation system by N Mizushima51. Investigation of Atg12 by western blot analyses showed a band of high molecular weight in addition to a band corresponding to the predicted molecular size of Atg12. This high molecular mass band was not observed in atg5, atg7, or atg10 mutants. Further studies disclosed the molecular bases of this phenomenon. Atg12 is a unique ubiquitin-like molecule, 2.5 times larger than ubiquitin, and is synthesized as an active form without the processing of its C-terminus. The C-terminal glycine of Atg12 is first activated by the E1 enzyme Atg7, and is then transferred to an E2 enzyme, Atg10, before finally forming a conjugate with Atg5 via an isopeptide bond with a lysine residue at the middle of Atg551. This Atg12-Atg5 conjugate is essential for autophagy. Atg16 was soon found to act as a protein necessary for the dimerization of two Atg12-Atg5 conjugates through its ability to interact with the Atg12-Atg5 conjugate and form Atg16 homodimers52,53. The precise roles of Atg12-Atg5 conjugate remained unclear, but it is well established that it plays an essential part in the Atg8 system, as described below.
The Atg8 lipidation system
Soon after the identification of the Atg12 conjugation system, another conjugation system was discovered among the Atg proteins. Atg8 is a small hydrophilic protein and its expression is upregulated upon starvation54. The intracellular localization of Atg8 changes upon starvation and it at least partly localizes to the autophagosome and autophagic body membrane54. Atg8 therefore offered great promise as a good marker of the autophagosomal membrane and a key molecule during autophagosome formation. Biochemical studies revealed a tightly membrane-associated form of Atg854, and additional work illustrated the role of a unique ubiqutination-like modification system55. Atg8 is synthesized as a precursor, and is then processed by a novel cysteine protease, Atg4, to become a C-terminally glycine exposed form. Following this, processed Atg8 is activated by the E1 enzyme Atg7, before the Atg8 moiety is transferred to the E2 enzyme Atg3. Atg8 does not show a modified form when analyzed by conventional SDS-PAGE, but mass spectrometry of the tightly membrane-bound modified form of Atg8 proved that Atg8 forms a conjugate with the head group of a membrane phospholipid, phosphatidylethanolamine (PE). The target of this conjugation system was therefore not a protein, but intriguingly a phospholipid. The processing enzyme Atg4 also cleaves Atg8-PE, thereby acting as a deconjugation enzyme56, which is indeed shown to be essential for autophagosome formation55. The amount of Atg8-PE determines the size of autophagosome, suggesting that Atg8-PE is involved in the elongation step of isolation membrane formation57.
Atg7 is a unique E1 enzyme in that it activates two ubiquitin-like molecules, Atg12 and Atg8, and transfers them to different E2 enzymes. Recently Noda et al. reported the structural basis for the action of this enzyme58. As suggested by a common E1 enzyme, these two conjugation systems function in a closely related manner58, and it is evident that the Atg12 system is required for Atg8-PE formation55. In fact, Atg12-Atg5 functions as an E3-like enzyme to enhance the Atg8 lipidation reaction59,60. The structural basis of Atg3 activation by the Atg12-Atg5 conjugate has also recently been elucidated60.
These findings indicate that among Atg gene products eight Atg proteins are involved in these conjugation reactions. The precise role of the lipidated form of Atg8 in autophagosome formation has not yet been elucidated, but it has been observed that Atg8-PE is involved in tethering and hemi-fusion of membrane vesicles in vitro, and mutational analysis demonstrated that these activities are required for autophagosome formation in vivo61.
Atg1 kinase complex
ATG1, the first ATG gene identified, encodes a Ser/Thr kinase essential for autophagy49. Atg13, which binds to Atg1, is highly phosphorylated by TORC1 kinase under growing conditions, and upon starvation or rapamycin treatment is rapidly dephosphorylated62. The affinity of Atg13 for Atg1 increases when this dephosphorylation occurs, and the resulting binding enhances the kinase activity of Atg1. Atg17 was next identified as a binding partner of Atg1-Atg1363, and Atg29 and Atg31 were found to interact with Atg1764. Atg17, Atg29, and Atg31 form a stable 1:1:1 complex regardless of nutrient conditions65,66. Upon starvation, Atg13-Atg1 forms a pentameric complex with Atg17-Atg31-Atg29, which serves as a scaffold for the further recruitment of other Atg proteins. Atg17, Atg29, and Atg31 are specifically required for starvation-induced autophagy, but not for the Cvt pathway. Atg1 plays a central role in the scaffold complex, but its kinase activity is not required for this assembly.
The PI-3K Complex and Atg18-Atg2
Shortly after the identification of the two conjugation systems, Ohsumi's group found a PI-3K complex essential for autophagy50. Yeast has a sole PI-3K, Vps34, which forms two complexes. Complex I is necessary for autophagy, whereas complex II is required for the Vps pathway. Complex I consists of Vps34, Vps15, Vps30/Atg6 and Atg14, while complex II contains Vps38 instead of Atg14. Three subunits are common between the two complexes, and functional specificity is provided by the presence of Atg14 or Vps38 for the respective pathways. Atg14 and Vps38 dictate the localization of the kinase complexes67. Quite recently a novel subunit of Complex I, Atg38, was reported68. PI3P, the product of PI-3K, was found to be essential for autophagosome formation and resides on the autophagosomal membrane, preferentially on the inner membrane of the autophagosome69. PI3P is thought to recruit effector proteins required for autophagosome formation.
Atg18 binds both PI3P and PI3,5P2 and has two distinct functions, playing roles in both autophagy and vacuolar morphology. When bound to PI3,5P2, Atg18 is important for the regulation of vacuolar size, while a portion of Atg18 forms a complex with Atg2 and functions in autophagosome formation in a PI3P-dependent manner70,71. Precise determination of the site of PI3P production by Complex I, and the roles of the Atg2-Atg18 complex are important problems to be answered in ongoing research.
Atg9
Among the Atg proteins, Atg9 is the sole multispanning membrane protein essential for autophagosome formation72, and it has therefore been postulated that this protein plays a key role in the delivery of membrane lipids necessary for autophagosome formation by recycling between a yet unidentified reservoir and the isolation membrane. However, recent work by H Yamamoto et al. reported that the majority of Atg9 exists on small, mobile cytoplasmic membrane vesicles derived from the Golgi apparatus in a process depending upon Atg23 and Atg2773. A small number of these vesicles participate at the early stages of autophagosome formation, generating a nucleation site for membrane biogenesis, as discussed below.
Other modes of autophagy in yeast
The Cvt pathway as a model of selective autophagy
D Klionsky has been working for many years on the cytoplasm-to-vacuole transport pathway, known as the Cvt pathway, which is responsible for the delivery of the vacuolar resident proteins, α-aminopeptidase I and α-mannosidase 174. Although the Cvt pathway is a constitutive and biosynthetic pathway, genes essential for this pathway (the CVT genes) were found to be common with the APG genes. Electron microscopic analyses proved that membrane dynamics of the Cvt pathway are quite similar to those of macroautophagy, which is consistent with its requirement for most of the Atg machinery43,75. The most obvious difference is the size of the double membrane structures of these two pathways: the Cvt vesicles, which functionally correspond to autophagosomes and specifically enwrap the Cvt complex (α-aminopeptidase 1 complex and Ty1 virus-like particles)76 with the exclusion of cytoplasmic proteins, are much smaller (ca 150 nm in diameter) than the autophagosome (ca 500 nm). The specific nature of the Cvt pathway means that it serves as a good model system for studying selective autophagy. In addition to the requirement for the core autophagy machinery, several specific factors for the Cvt pathway were identified. Among these, Atg19 and Atg34 are receptor proteins of the selective cargos α-aminopeptidase 1 and α-mannosidase 1, respectively77. The identification of Atg11, which functions as a scaffold protein bringing the core Atg proteins and the target of selective degradation together, was an important development in the field of selective autophagy78,79.
Under starvation conditions the Cvt complex is preferentially enclosed by autophagosomes, suggesting that even non-selective autophagy has some preference of targets for sequestration into autophagosomes.
Mitophagy and Pexophagy
The selective degradation of peroxisomes in methylotrophic yeasts, such as P. pastoris and H. polymorpha, had also been well studied80. The peroxisomes of these yeasts tremendously proliferate when the microorganisms are cultured in medium containing methanol as the sole carbon source. When these cells are shifted to ethanol or glucose, proliferated peroxisomes are degraded by autophagy. W Dunn showed that P. pastoris utilizes different modes of autophagy, macroautophagy and microautophagy, depending upon the carbon source81. Such selective autophagy of proliferated peroxisomes requires several specific factors for pexophagy80. Recently, studies of the selective degradation of mitochondria (mitophagy) in yeast identified a mitochondrial membrane protein, Atg32, which plays an essential role in this organelle's turnover82,83.
Core machinery of autophagosome formation in yeast
As discussed above, the 18 Atg proteins essential for starvation-induced non-selective autophagy were identified within a short period and divided into six functional units: Atg1 kinase and its regulators, PI-3k complex, the Atg12 conjugation system, the Atg8 lipidation system, Atg9 and the Atg18-Atg2 complex. These Atg proteins function in an extremely tightly concerted manner in the process of autophagosome formation. The next problem that required investigation was how and where these proteins function in vivo. K Suzuki in Ohsumi's lab investigated the localization of Atg proteins by generating fusions of Atg proteins with GFP and visually investigating their intracellular behavior using fluorescence microscopy84. Atg8-GFP was the first Atg protein to be visualized in such a manner, appearing as a perivacuolar dot structure, to which most Atg proteins were subsequently shown to at least partly colocalize. This dot structure is detectable even in certain atg mutants, indicating that it is not an isolation membrane or an autophagsome, and it was therefore named the pre-autophagosomal structure (PAS), referring to its role as an assembly of Atg proteins mediating autophagosome formation84. Mostly only one PAS per cell is observed even under starvation conditions. It is not known how and why the PAS in yeast is associated with the vacuolar membrane. The organization of the PAS was systematically analyzed by quantifying the PAS recruitment of each Atg protein in the absence of every single ATG gene85,86. This analysis showed a hierarchical relationship among the functional units. The discovery of the PAS was an important landmark in the description of the epistatic relationships among the functional units of Atg proteins and substantiation of the molecular machinery in vivo86,87.
Recent studies have revealed that this hierarchy reflects the order in which Atg proteins are recruited to the PAS86,87. Upon starvation, inhibition of TORC1 kinase results in dephosphorylation of Atg13, which binds to Atg1, and forms a scaffold complex, Atg1-Atg13/Atg17-Atg31-Atg29. This complex assembles at a site on the vacuolar membrane, after which a small number of Atg9 vesicles join the PAS via an interaction between Atg17 and Atg9. After this no further Atg9 vesicles join the PAS, indicating that Atg9 vesicles are not the major lipid source for autophagosomal membranes. Next, PI-3K complex I is recruited to the PAS, and the Atg2-Atg18 complex joins the PAS through the binding of Atg18 to PI3P. The Atg12-Atg5-Atg16 complex also localizes to the PAS in a PI3P-dependent manner, although this is independent of Atg2-Atg18 binding. Finally, the Atg12-Atg5-Atg16 complex facilitates the formation of Atg8-PE at the PAS, which is necessary for the expansion of the isolation membrane. All Atg proteins therefore organize to the PAS in a concerted and temporally regulated manner in order to generate the precursor membrane necessary for isolation membrane formation.
Originally the PAS was defined by fluorescence microscopy as a dot structure representing an assembly of Atg proteins84. Since in yeast the Cvt pathway is constitutively active, the PAS is formed in an Atg11-dependent manner under growing conditions, with Atg11 being essential for the Cvt pathway but not for macroautophagy. In the absence of Atg11, the dynamics of starvation-induced PAS assembly become easily detectable. In atg11Δ the PAS is not seen in growing conditions, but is rapidly induced by starvation or rapamycin treatment79. However, the PAS is also observed to disassemble immediately following the addition of the nutrient for which cells were initially starved. Now it is thought that the PAS is in fact not simply a stoichiometric assembly of Atg proteins, but rather a dynamic structure changing in composition over time and at various stages prior to the formation of the isolation membrane. The number of Atg protein molecules associating to the PAS has been shown to vary from a dozen to a few hundred88, while the amount of Atg8 is known to oscillate every 10 min57. Intriguingly, a recent study has suggested that at a very early stage of autophagosome formation Atg9 vesicles are recruited to the PAS, which means that the PAS exists on some sort of precursor membrane before the formation of the isolation membrane. Many Atg proteins in the PAS may change interacting partners throughout this process prior to the emergence of the isolation membrane, and play further roles as the isolation membrane expands. Such spatiotemporal dynamics of Atg proteins may be regulated by phosphorylation or dephosphorylation. So far the PAS in yeast has not been observed by EM, and is too small to distinguish from the isolation membrane or autophagosome by fluorescence microscopy.
Rapid development of autophagy studies
ATG genes in other organisms
During his time in Ohsumi's lab, N Mizushima began a search for the homolog of ATG12 in humans. After identifying a candidate homolog by database searches, he successfully showed that hAtg12 forms a conjugate with hATG589, similar to the conjugation of Atg12 to Atg5 in yeast, indicating the system is well conserved throughout eukaryotes. The counterpart of Atg16, hATG16L, was also found and is characterized by the presence of a long C-terminal extension of WD repeats. Human Atg16L facilitates the binding of two Atg12-Atg5 conjugates by forming a dimer, as observed in yeast90.
T Yoshimori, also in Ohsumi's lab at that time, was investigating LC3, the mammalian homolog of Atg8, and found two forms: LC3-I, a processed form of nascent LC3 by the homolog of yeast Atg4, hATG4, and LC3-II, a lipidated form just like Atg8-PE in yeast91. Yoshimori observed that LC3-II localizes to the autophagosomal membrane, which provided the first good marker of the autophagosomal membrane in mammals. In addition, the amount of LC3-II correlates with the number of autophagosomes, which has been used as an indicator of autophagy. Like yeast Atg8, LC3 localizes to intermediate structures as well as the complete autophagosome, and thereby a portion of LC3 is delivered to the lysosome. Meanwhile, the Atg12-Atg5-Atg16L complex resides only on the convex surface of the isolation membrane and dissociates from the membrane once the autophagosome is completed92,93. GFP-Atg5 was therefore successfully employed as a good marker of the structures of early stage of autophagososome formation.
In addition to Atg proteins of the two conjugation systems, other Atg proteins have been identified in mammals. This process took some time due to the rather low amino-acid sequence similarity between yeast and mammalian homologs. But ultimately almost all counterparts of yeast Atg proteins were found in mammals. These include the ULK1 complex94, the PI-3k complex95,96,97, Atg998, the WIPI-Atg2 complex99, and the two conjugation systems. This preservation of functional units strongly indicates that the fundamental mechanisms of autophagy were acquired at a very early stage of eukaryotic evolution and have since been conserved, a fact that has significantly facilitated further studies of autophagy in mammals.
In contrast to yeast, where a single gene encodes each Atg protein, many Atg proteins in higher eukaryotes, such as Atg1, Atg14, Atg2, Atg18, Atg16 and Atg8 are characterized by paralogy, most likely arising by gene duplication events. This diversification may be a reflection of the increasingly complex nature of autophagy in mammalian cells, possibly due to further regulation or specific roles for each paralog in the diverse modes of autophagy found among various tissue and cell types. While Atg17, Atg29, and Atg31 in yeast have no homologs in mammals, the mammalian proteins FIP200 and Atg101 seem to perform a similar function. Though additional proteins required for autophagy in mammals remain to be identified, it is clear that the basic hierarchical order of events have been conserved100 (see details in review by Klionsky in this issue).
Genetic manipulation of autophagy in mammals
Soon after the discovery of the Atg12 conjugation system in mice, N Mizushima successfully generated a transgenic mouse systemically expressing GFP-LC3, fluorescence microscopy observation of which allowed the easy visualization of the autophagosome. GFP-positive structures correspond to autophagosomes and autophagolysosomes, thus where and how much autophagy occurs in a whole body could be assessed. Using this mouse model, Mizushima showed that during fasting every organ is characterized by differences in the kinetics and extent of autophagy induction93,101.
Mizushima also successfully generated the first mouse knocked out for an Atg gene, which was Atg5102. This mouse was able to produce offspring at a normal rate, but infant mice died within 24 hours of birth, demonstrating that autophagy is essential for survival during the neonatal stage of development in mammals. These GFP-LC3 transgenic and Atg5 knockout mice proved to be so powerful in the detection and investigation of the physiological significance of autophagy in mammals that they were distributed all over the world, contributing much to our understanding of autophagy in mammals through their use in research.
Up to now the disruption of nine Atg genes have been reported in genetically modified mice. Among these knockout mice, the phenotypes of Atg3, Atg7, Atg9 and Atg16L1 knockouts are similar to that of Atg5, while Beclin 1, Ambra1, and FIP200 knockout mice are unable to produce homozygous offspring due to early embryonic lethality, suggesting that these genes have other functions in addition to autophagy103. Meanwhile ULK1 knockout mice suffered slight anemic symptoms.
Identification of ATG genes was subsequently undertaken in a range of organisms, generally by simple database searches or by searching for molecules interacting with known Atg proteins. Disruption of ATG genes was also initiated in model organisms such as Drosophila. Ohsumi's and D Vierstra's groups identified most ATG gene homologs in the higher plant model organism, Arabidopsis thaliana, and showed these genes also participate in autophagy in plants104,105, reinforcing the highly conserved and fundamental role of autophagy in eukaryotic life.
Recently, a forward genetics approach to the identification of autophagy genes in Caenorhabditis elegans revealed that in addition to known ATG gene homologs, several novel genes essential for autophagy are present in this organism106. Some of these novel genes are conserved in higher eukaryotes, suggesting that evolution has seen the appearance of additional autophagy genes among higher eukaryotes (see review by Zhang in this issue).
Important roles of autophagy in higher eukaryotes
As described above, autophagy is an essential mechanism for survival through its role in the recycling of proteins and macromolecules. However, lipids (lipid bodies as well as membrane lipids), nucleic acids, and carbohydrates are also delivered to the lysosome/vacuole in addition to proteins via autophagy. This underscores the body of evidence indicating that autophagy must contribute directly to cell metabolism by supplying cellular building blocks as well as sources of energy.
On the other hand, recent progress in autophagy research in various organisms has revealed another important role of autophagy. To understand the role of autophagy in specific organs or tissues, many tissue-specific knockout mice have been generated. Mizushima generated a neuronal cell-specific knockout mouse in which the basal level of autophagy in the brain was greatly reduced. This caused the accumulation of ubiquitinated proteins and p62, both known to be specific cargoes of autophagy in mammals, and ultimately resulted in neurodegeneration107. Komatsu in Kominami's group demonstrated that the absence of autophagy in liver tissue results in liver hypertrophy and tumor formation108. Mizushima also succeeded in showing that autophagy is induced upon fertilization and is necessary for early embryonic development in mice by using oocytes completely depleted of Atg5109. Since these important works, many studies of knockout mice continue to provide important insights into the many physiological functions of autophagy, suggesting important roles for autophagy beyond the supply of nutrients. Degradation itself or elimination of harmful or excessive materials from the cytoplasm is mediated by autophagy and is crucial for life. In multicellular organisms the process of cell division takes longer when compared to that of unicellular eukaryotes like yeast; this increased cellular life span creates a greater need for stringent intracellular quality control, which is known as clearance. Neuronal cells are an extreme example, as these cells do not divide and are instead retained throughout the life of the organism, strongly emphasizing the need for intracellular housekeeping. One facet of such quality control is the critical role of autophagy in the elimination of misfolded or damaged proteins, as well as aggregate-prone proteins. Every protein in the cell is involved in interactions with other proteins, forming transient complexes in a repeating cycle of association and dissociation in response to metabolic changes. It is highly likely that autophagy is involved in the regulation of such protein dynamics within the cell.
B Levine identified Beclin 1, a mammalian homolog of yeast Atg6110. She proposed, for the first time, that autophagy can be important for tumor suppression, which was later confirmed by her own studies and those of others, which triggered a significant expansion of research into the role of autophagy in tumor growth and survival.
From the early days of autophagy research, it has been clear that excessive or damaged organelles are the target of autophagy. Recently the selective degradation of organelles has been dissected at the molecular level. Two groups, led by Okamoto and Kanki, independently reported that the mitochondrial outer membrane protein Atg32 functions as a receptor for mitochondrial degradation by autophagy (mitophagy)82,111. Atg32 interacts with Atg8 and Atg11. Like Atg32, Atg19, a receptor protein for the Cvt pathway, has a specific motif (named AIM or LIR in mammals) that binds to a hydrophobic pocket in Atg8 and its homologs, which represents an important mode of interaction for selective autophagy.
Meanwhile, mitophagy in mammalian cells has attracted strong interest from researchers, and R Youle's group has intensively studied mitophagy in mammals and shown that PINK1 and Parkin play critical roles in the elimination of damaged mitochondria, and that mitophagy in mammals is dependent upon ubiquitination112 (see review by Okamoto in the same special issue).
Another important and unexpected finding is that autophagy also acts as a defense mechanism against bacterial and viral infection. It is known that Group A Streptococcus enters cells via endocytosis, subsequently escaping to the cytoplasm by secretion of streptolysin O. In 2004 I Nakagawa showed that these bacteria in the cytoplasm are immediately captured in an autophagosome, which fuses with other autophagosomes to form a huge membrane structure in which bacteria are ultimately digested after fusion with a lysosome113. Autophagy is furthermore implicated in innate immunity. Salmonella is also a target of the autophagosome, but it has also been found that Listeria and Shigella are able to escape from degradation by autophagy. Autophagy against intracellular pathogens (called Xenophagy) has consequently burgeoned as an exciting research field promising a range of novel means of combating infectious diseases114.
As briefly mentioned above, selective autophagy, as a means of quality control within the cell, is a major subject in autophagy research at present. So many novel questions remain to be answered, not only regarding their physiological significance, but also the underlying molecular mechanisms allowing targets to be selectively wrapped into autophagosomes.
Autophagy research community
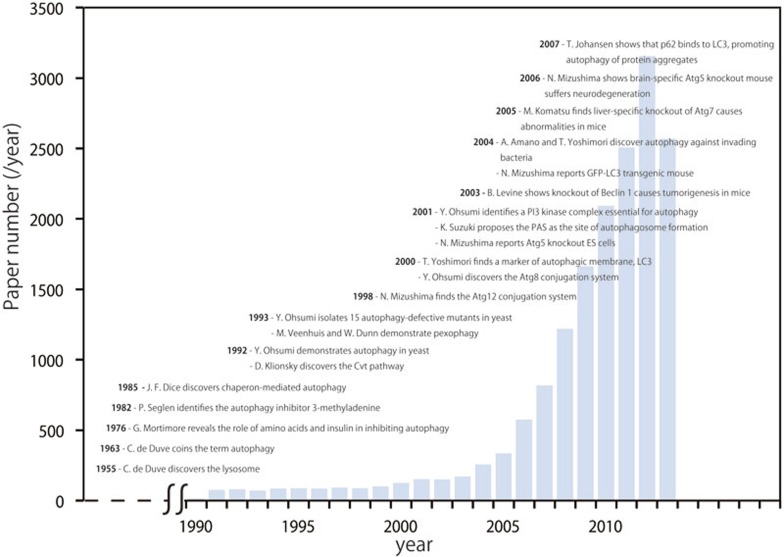
At the conference level, autophagy was first addressed when the first Gordon Research conference on lysosomes was held in 1967, with several talks on autophagy. Following this, P O Seglen organized sessions on autophagy at the International Committee of Proteolysis conferences, which covered non-lysosomal and lysosomal protein turnover; and the 11th meeting at Turku were attended by E Kominami, P Codogno, A Meijer, W Dunn, G Mortimore, and J F Dice. In 1997 Y Ohsumi organized the 1st International Symposium on Autophagy (ISA) at Okazaki, Japan. The ISA, which deals exclusively with autophagy is getting bigger and bigger, and the 6th ISA was held in Okinawa, 2012, with more than 300 participants from all over the world. The Gordon Research Conference on autophagy was arranged by D Klionsky in 2003, and in the following year the first Keystone Symposium on Autophagy was organized by B Levine. Recently, an EMBO conference on autophagy was organized by S Tooze. In addition to the explosion in autophagy work published in journals (Figure 1), the expanding number of international meetings addressing autophagy research is a reflection of the popularity of this exciting field.
Figure 1.
The growth of autophagy research and historical landmarks.
Another point worthy of special mention is the success of Autophagy journal under the tremendous efforts of D Kilonsky, which has now established itself as a high-impact journal.
Conclusion and perspective
As described above, 50 years have passed since studies on autophagy began. Since autophagy in mammalian cells is characterized by such a sophisticated series of membrane phenomena, scientists remained skeptical for many years that autophagy is the major degradative pathway of proteins and organelles. Identification of ATG genes in yeast was a genuine turning point in the modern biology of autophagy. Autophagy has now become an enormously hot field in biology. Over the past two decades we have learned much about the molecular mechanisms of autophagy and its physiological significance. However, we are still at the early stages of autophagy research, and the field continues to grow in both volume and impact. It is now abundantly clear that degradation is a fundamental cellular function, just as essential to the functioning of life as synthesis. Time and further work will reveal the relevance of autophagy to more and more physiological events. There are so many questions that remain to be answered about the unique membrane dynamics that constitute autophagy, and there are still a great deal of mysteries to be uncovered before we truly understand the molecular mechanisms of autophagy. Essential in this undertaking will be the development of various robust systems to work with as well as the further improvement of devices allowing us to visualize the dynamic machinery of autophagy in higher spatiotemporal resolution. More information about the composition and fine localization of lipid molecules within the isolation membrane and autophagosome will also greatly assist research efforts, as will biochemical, structural and biological analyses of Atg and related proteins. Ultimately, with a complete understanding of the mechanisms of autophagy, reconstitution of the process should be an objective for scientists working in the field.
References
- Schoenheimer R.The Dynamic State of Body ConstituentsThe Edward K. Dunham Lectures for the Promotion of the Medical Sciences. Harvard University Press, 1942
- Schoenheimer R, Ratner S, Rittenberg D. The process of continuous deamination and reamination of amino acids in the proteins of normal animals. Science. 1939;89:272–273. doi: 10.1126/science.89.2308.272. [DOI] [PubMed] [Google Scholar]
- Ganschow RE, Schimke RT. Independent genetic control of the catalytic activity and the rate of degradation of catalase in mice. J Biol Chem. 1969;244:4649–4658. [PubMed] [Google Scholar]
- Kalish F, Chovick N, Dice JF. Rapid in vivo degradation of glycoproteins isolated from cytosol. J Biol Chem. 1979;254:4475–4481. [PubMed] [Google Scholar]
- de Duve C, Pressman BC, Gianetto R, Wattiaux R, Appelmans F. Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem J. 1955;60:604–617. doi: 10.1042/bj0600604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Duve C. The lysosome turns fifty. Nat Cell Biol. 2005;7:847–849. doi: 10.1038/ncb0905-847. [DOI] [PubMed] [Google Scholar]
- Novikoff AB, Beaufay H, de Duve C. Electron microscopy of lysosome-rich fractions from rat liver. J Biophys Biochem Cytol. 1956;2:179–184. [PMC free article] [PubMed] [Google Scholar]
- de Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol. 1966;28:435–492. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- Clark SLJ. Cellular differentiation in the kidneys of newborn mice studies with the electron microscope. J Biophys Biochem Cytol. 1957;3:349–362. doi: 10.1083/jcb.3.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol. 1962;12:198–202. doi: 10.1083/jcb.12.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arstila AU, Trump BF. Studies on cellular autophagocytosis. The formation of autophagic vacuoles in the liver after glucagon administration. Am J Pathol. 1968;53:687–733. [PMC free article] [PubMed] [Google Scholar]
- de Duve C, Foundation C.Ciba Foundation Symposium: LysosomeDe Reuck A, Cameron MP, eds. Little, Brown, 1963
- Matile P. Biochemistry and function of vacuoles. Annu Rev Plant Physiol. 1978;29:193–213. [Google Scholar]
- Deter RL, Baudhuin P, de Duve C. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J Cell Biol. 1967;35:C11–C16. doi: 10.1083/jcb.35.2.c11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer U, Strauss P. Autophagic vacuoles in heart muscle and liver. A comparative morphometric study including circadian variations in meal-fed rats. J Mol Cell Cardiol. 1981;13:37–49. doi: 10.1016/0022-2828(81)90227-3. [DOI] [PubMed] [Google Scholar]
- Pfeifer U, Warmuth-Metz M. Inhibition by insulin of cellular autophagy in proximal tubular cells of rat kidney. Am J Physiol. 1983;244:E109–E114. doi: 10.1152/ajpendo.1983.244.2.E109. [DOI] [PubMed] [Google Scholar]
- Mortimore GE, Ward WF. Behavior of the lysosomal system during organ perfusion. An inquiry into the mechanism of hepatic proteolysis. Front Biol. 1976;45:157–184. [PubMed] [Google Scholar]
- Mortimore GE, Hutson NJ, Surmacz CA. Quantitative correlation between proteolysis and macro- and microautophagy in mouse hepatocytes during starvation and refeeding. Proc Natl Acad Sci USA. 1983;80:2179–2183. doi: 10.1073/pnas.80.8.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seglen PO, Gordon PB, Poli A. Amino acid inhibition of the autophagic/lysosomal pathway of protein degradation in isolated rat hepatocytes. Biochim Biophys Acta. 1980;630:103–118. doi: 10.1016/0304-4165(80)90141-5. [DOI] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen JH, Zoncu R, Kim D, Sabatini DM. Defective regulation of autophagy upon leucine deprivation reveals a targetable liability of human melanoma cells in vitro and in vivo. Cancer Cell. 2011;19:613–628. doi: 10.1016/j.ccr.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci USA. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelarova H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- Furuya N, Kanazawa T, Fujimura S, Ueno T, Kominami E, Kadowaki M. Leupeptin-induced appearance of partial fragment of betaine homocysteine methyltransferase during autophagic maturation in rat hepatocytes. J Biochem. 2001;129:313–320. doi: 10.1093/oxfordjournals.jbchem.a002859. [DOI] [PubMed] [Google Scholar]
- Holen I, Gordon PB, Seglen PO. Protein kinase-dependent effects of okadaic acid on hepatocytic autophagy and cytoskeletal integrity. Biochem J. 1992;284:633–636. doi: 10.1042/bj2840633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holen I, Gordon PB, Seglen PO. Inhibition of hepatocytic autophagy by okadaic acid and other protein phosphatase inhibitors. Eur J Biochem. 1993;215:113–122. doi: 10.1111/j.1432-1033.1993.tb18013.x. [DOI] [PubMed] [Google Scholar]
- Masaki R, Yamamoto A, Tashiro Y. Cytochrome P-450 and NADPH-cytochrome P-450 reductase are degraded in the autolysosomes in rat liver. J Cell Biol. 1987;104:1207–1215. doi: 10.1083/jcb.104.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Klei IJ, Harder W, Veenhuis M. Biosynthesis and assembly of alcohol oxidase, a peroxisomal matrix protein in methylotrophic yeasts: a review. Yeast. 1991;7:195–209. doi: 10.1002/yea.320070302. [DOI] [PubMed] [Google Scholar]
- Marzella L, Ahlberg J, Glaumann H. Autophagy, heterophagy, microautophagy and crinophagy as the means for intracellular degradation. Virchows Archiv B Cell Pathology Zell-pathologie. 1981;36:219–234. doi: 10.1007/BF02912068. [DOI] [PubMed] [Google Scholar]
- Sakai Y, Koller A, Rangell LK, Keller GA, Subramani S. Peroxisome degradation by microautophagy in Pichia pastoris: identification of specific steps and morphological intermediates. J Cell Biol. 1998;141:625–636. doi: 10.1083/jcb.141.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dice JF, Walker CD, Byrne B, Cardiel A. General characteristics of protein degradation in diabetes and starvation. Proc Natl Acad Sci USA. 1978;75:2093–2097. doi: 10.1073/pnas.75.5.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimore GE, Woodside KH, Henry JE. Compartmentation of free valine and its relation to protein turnover in perfused rat liver. J Biol Chem. 1972;247:2776–2784. [PubMed] [Google Scholar]
- Kakinuma Y, Ohsumi Y, Anraku Y. Properties of H+-translocating adenosine triphosphatase in vacuolar membranes of Saccharomyces cerevisiae. J Biol Chem. 1981;256:10859–10863. [PubMed] [Google Scholar]
- Jones EW. Vacuolar proteases and proteolytic artifacts in Saccharomyces cerevisiae. Methods Enzymol. 2002;351:127–150. doi: 10.1016/s0076-6879(02)51844-9. [DOI] [PubMed] [Google Scholar]
- Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Takeshige K, Baba N, Ohsumi Y. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J Cell Biol. 1994;124:903–913. doi: 10.1083/jcb.124.6.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Osumi M, Ohsumi Y. Analysis of the membrane structures involved in autophagy in yeast by freeze-replica method. Cell Struct Funct. 1995;20:465–471. doi: 10.1247/csf.20.465. [DOI] [PubMed] [Google Scholar]
- Noda T, Klionsky DJ. The quantitative Pho8Delta60 assay of nonspecific autophagy. Methods Enzymol. 2008;451:33–42. doi: 10.1016/S0076-6879(08)03203-5. [DOI] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- Thumm M, Egner R, Koch B, et al. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Tuttle DL, Shi YJ, Ralph GS, Dunn WAJ. Glucose-induced microautophagy in Pichia pastoris requires the alpha-subunit of phosphofructokinase. J Cell Sci. 1997;110:1935–1945. doi: 10.1242/jcs.110.16.1935. [DOI] [PubMed] [Google Scholar]
- Yuan W, Stromhaug PE, Dunn WAJ. Glucose-induced autophagy of peroxisomes in Pichia pastoris requires a unique E1-like protein. Mol Biol Cell. 1999;10:1353–1366. doi: 10.1091/mbc.10.5.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaiyama H, Oku M, Baba M, et al. Paz2 and 13 other PAZ gene products regulate vacuolar engulfment of peroxisomes during micropexophagy. Genes Cells. 2002;7:75–90. doi: 10.1046/j.1356-9597.2001.00499.x. [DOI] [PubMed] [Google Scholar]
- Titorenko VI, Keizer I, Harder W, Veenhuis M. Isolation and characterization of mutants impaired in the selective degradation of peroxisomes in the yeast Hansenula polymorpha. J Bacteriol. 1995;177:357–363. doi: 10.1128/jb.177.2.357-363.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y. Protein turnover. IUBMB Life. 2006;58:363–369. doi: 10.1080/15216540600758539. [DOI] [PubMed] [Google Scholar]
- Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, et al. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18:3888–3896. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Mizushima N, Ishihara N, Ohsumi Y. Formation of the approximately 350-kDa Apg12-Apg5.Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem. 2002;277:18619–18625. doi: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- Kirisako T, Baba M, Ishihara N, et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Kirisako T, Ichimura Y, Okada H, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Matoba K, Sawada R, et al. Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7. Nat Struct Mol Biol. 2012;19:1250–1256. doi: 10.1038/nsmb.2451. [DOI] [PubMed] [Google Scholar]
- Hanada T, Noda NN, Satomi Y, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- Sakoh-Nakatogawa M, Matoba K, Asai E, et al. Atg12-Atg5 conjugate enhances E2 activity of Atg3 by rearranging its catalytic site. Nat Struct Mol Biol. 2013;20:433–439. doi: 10.1038/nsmb.2527. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Kamada Y, Baba M, Takikawa H, Sasaki M, Ohsumi Y. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell. 2005;16:2544–2553. doi: 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell. 2008;19:2039–2050. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Noda NN, Fujioka Y, Suzuki K, Inagaki F, Ohsumi Y. Characterization of the Atg17-Atg29-Atg31 complex specifically required for starvation-induced autophagy in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2009;389:612–615. doi: 10.1016/j.bbrc.2009.09.034. [DOI] [PubMed] [Google Scholar]
- Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012;151:1501–1512. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara K, Sekito T, Ohsumi Y. Assortment of phosphatidylinositol 3-kinase complexes--Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:1527–1539. doi: 10.1091/mbc.E05-09-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, Ku WC, Akioka M, et al. Atg38 is required for autophagy-specific phosphatidylinositol 3-kinase complex integrity. J Cell Biol. 2013;203:299–313. doi: 10.1083/jcb.201304123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara K, Noda T, Niimi K, Ohsumi Y. Transport of phosphatidylinositol 3-phosphate into the vacuole via autophagic membranes in Saccharomyces cerevisiae. Genes Cells. 2008;13:537–547. doi: 10.1111/j.1365-2443.2008.01188.x. [DOI] [PubMed] [Google Scholar]
- Stromhaug PE, Reggiori F, Guan J, Wang CW, Klionsky DJ. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol Biol Cell. 2004;15:3553–3566. doi: 10.1091/mbc.E04-02-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krick R, Henke S, Tolstrup J, Thumm M. Dissecting the localization and function of Atg18, Atg21 and Ygr223c. Autophagy. 2008;4:896–910. doi: 10.4161/auto.6801. [DOI] [PubMed] [Google Scholar]
- Noda T, Kim J, Huang WP, et al. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol. 2000;148:465–480. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Kakuta S, Watanabe TM, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins MU, Klionsky DJ. Vacuolar localization of oligomeric alpha-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J Biol Chem. 2001;276:20491–20498. doi: 10.1074/jbc.M101150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Baba M, Ohsumi Y, Klionsky DJ. Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J Cell Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Morimoto M, Kondo C, Ohsumi Y. Selective autophagy regulates insertional mutagenesis by the Ty1 retrotransposon in Saccharomyces cerevisiae. Dev Cell. 2011;21:358–365. doi: 10.1016/j.devcel.2011.06.023. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kondo C, Morimoto M, Ohsumi Y. Selective transport of alpha-mannosidase by autophagic pathways: identification of a novel receptor, Atg34p. J Biol Chem. 2010;285:30019–30025. doi: 10.1074/jbc.M110.143511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005;16:1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem. 2004;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn WAJ, Cregg JM, Kiel JA, et al. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- Tuttle DL, Dunn WAJ. Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J Cell Sci. 1995;108:25–35. doi: 10.1242/jcs.108.1.25. [DOI] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Klionsky DJ. Atg32 is a tag for mitochondria degradation in yeast. Autophagy. 2009;5:1201–1202. doi: 10.4161/auto.5.8.9747. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Ohsumi Y. [The pre-autophagosomal structure (PAS), required for the formation of autophagosome membrane] Seikagaku. 2003;75:492–499. [PubMed] [Google Scholar]
- Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Noda T, Ohsumi Y. Interrelationships among Atg proteins during autophagy in Saccharomyces cerevisiae. Yeast. 2004;21:1057–1065. doi: 10.1002/yea.1152. [DOI] [PubMed] [Google Scholar]
- Geng J, Baba M, Nair U, Klionsky DJ. Quantitative analysis of autophagy-related protein stoichiometry by fluorescence microscopy. J Cell Biol. 2008;182:129–140. doi: 10.1083/jcb.200711112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential forautophagy. J Biol Chem. 1998;273:33889–33892. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Kuma A, Kobayashi Y, et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro SH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Li X, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AR, Chan EY, Hu XW, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- Velikkakath AK, Nishimura T, Oita E, Ishihara N, Mizushima N. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol Biol Cell. 2012;23:896–909. doi: 10.1091/mbc.E11-09-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama-Honda I, Itakura E, Fujiwara TK, Mizushima N. Temporal analysis of recruitment of mammalian ATG proteins to the autophagosome formation site. Autophagy. 2013;9:1491–1499. doi: 10.4161/auto.25529. [DOI] [PubMed] [Google Scholar]
- Kuma A, Mizushima N. Chromosomal mapping of the GFP-LC3 transgene in GFP-LC3 mice. Autophagy. 2008;4:61–62. doi: 10.4161/auto.4846. [DOI] [PubMed] [Google Scholar]
- Kuma A, Mizushima N. Physiological role of autophagy as an intracellular recycling system: with an emphasis on nutrient metabolism. Semin Cell Dev Biol. 2010;21:683–690. doi: 10.1016/j.semcdb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem. 2002;277:33105–33114. doi: 10.1074/jbc.M204630200. [DOI] [PubMed] [Google Scholar]
- Hanaoka H, Noda T, Shirano Y, et al. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002;129:1181–1193. doi: 10.1104/pp.011024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez A, Levine B. Autophagy in C. elegans. WormBook. 2009. pp. 1–26. [DOI] [PubMed]
- Hara T, Nakamura K, Matsui M, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, Mizushima N. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–120. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa I, Amano A, Mizushima N, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- Randow F, Munz C. Autophagy in the regulation of pathogen replication and adaptive immunity. Trends Immunol. 2012;33:475–487. doi: 10.1016/j.it.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]