Introduction
Riboswitches regulate gene expression in cis in response to changing metabolite concentrations in cells. Since flavin mononucleotide1,2 and thiamine pyrophosphate1,3 sensing riboswitches were discovered in 2002, more than 20 classes of metabolite-responsive riboswitches have been identified, primarily controlling essential metabolic or virulence genes in bacteria. Riboswitches are typically found in the mRNA 5′ untranslated region where they control expression usually by regulating premature transcription termination or translation initiation. Some riboswitches additionally control mRNA turnover4 or Rho-dependent termination.5 Regardless of their regulatory mechanism, the modular riboswitch structure is composed of an aptamer domain (ligand binding) and an effector domain (regulation). Ligand binding to the aptamer domain stabilizes an alternate structure and induces an effector domain switch to turn gene expression on or off. Riboswitches offer an efficient and rapid means to respond to changing metabolic conditions in a cell.
Purine Riboswitches
Purine riboswitches refer to a very broad classification, encompassing guanine, adenine, 2′-deoxyguanosine, cyclic-di-GMP, and PreQ16,7 riboswitches. Representative ligand-bound structures from each group reveal conserved interactions with the purine moiety (reviewed in Batey6). Guanine- and adenine-responsive riboswitches are particularly well studied since their discovery8,9 and crystal structure determination10,11 approximately 10 years ago. They share a structurally conserved three-way junction connecting helices P1–P3 (Fig. 1). They exhibit high sequence conservation in the hairpin loops (L2 and L3) that form tertiary contacts and the junction nucleotides (J1/2, J2/3, and J3/1) forming the purine-binding pocket. The guanine and adenine riboswitches can be interconverted by a single base substitution between C and U at position 74 (numbering according to Stoddard et al.13) that forms a Watson–Crick base pair with the purine ligand, demonstrating that the groups are mechanistically equivalent.
Fig. 1.
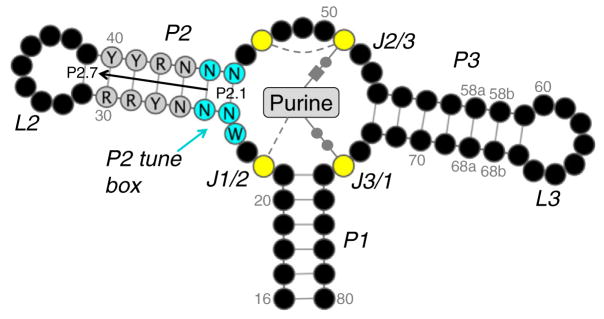
General schematic of the guanine/adenine riboswitch aptamer domain. Nucleotides are shown as black circles, excluding the P2 tune box (cyan), P2 core (gray), and purine-binding pocket (yellow). Bases are numbered according to a scheme in Stoddard et al. The numbering scheme for P2 is labeled (P2.1–P2.7). A purine is represented in the binding pocket with conserved bonds shown. Base pairs with the ligand are drawn using the Leontis–Westhof notation while single hydrogen bonds are shown as gray broken lines. The figure was generated using Varna.12
Same Ligand-Binding Pocket, Different Response Dynamics
Metabolic processes need to be finely tuned for survival and structured RNAs are incredibly amenable to fine-tuning through compensatory mutations and base isostericity.14 This idea is elegantly illustrated by work on RNA thermometers. By varying the length and strength of base pairing, helix melting and regulation are tuned to an optimal temperature response, with virulence, cold- or heat-shock responses with zipper-like structures even offering a graded response.15,16 Similarly, riboswitches are readily adaptable and their affinities can be efficiently tuned to optimize gene expression. Work on the guanine,17 SAM-I riboswitches,18 TPP, and c-di-GMP19 have clearly demonstrated that a single class of riboswitch can have a broad range of kinetic parameters. For instance, Bacillus subtilis contains at least 11 SAM-I riboswitches and their affinity for S-adenosyl-L-methionine (SAM) generally correlates with the regulated genes’ role in import or biosynthesis, allowing B. subtilis to preferentially use exogenous supplies, when available.18 Likewise, variability between organisms was demonstrated for the guanine,17 TPP,19 and c-di-GMP19 riboswitches. Invariably, the ligand-binding sites are highly conserved, indicating that regions outside the binding pocket are responsible for the differences. Clearly, riboswitches are highly adaptable elements, but given the subtle changes and difficulties predicting non-canonical base pair contributions, the molecular basis for activity tuning is poorly understood.
Finding the Tuning Element in Purine Riboswitches
In this issue, Stoddard et al.13 delve into the tuning mechanism in the well-studied purine riboswitches. They cleverly hypothesized that since the purine-binding site is highly conserved, perhaps the adjacent regions bear sequence elements responsible for fine-tuning the ligand-binding kinetics. Using statistical coupling analysis and a set of high-quality structure-guided sequence alignments, they identified co-evolving nucleotides at positions 24 (A or U) and 25, adjacent to the binding pocket. The co-variations could be further subdivided based on combinations of position 24 with the first two base pairs in P2 (P2.1 and P2.2). Can these sequences tune the behavior of purine riboswitches? To probe further, they engineered a subset of these naturally occurring sequences, as well as their non-natural A24/U24 inversions, into a common platform, or “chassis”, to assess binding kinetics, in vivo responses, and structural effects. In all, they demonstrated that alterations as simple interchanging of A24 or U24 with different combinations of P2.1/P2.2 resulted in a gradient of kinetic parameters with natural variants typically tuned for more efficient responses than their non-natural counterparts. The range of parameters achieved by the variations spanned expected values for riboswitches that would function by kinetic to thermodynamic mechanisms, according to previous approximations.20 This tunable gradient manifested in vivo as a near-complete loss of responsiveness despite modest kinetic changes. To investigate possible structural causes, they determined X-ray crystal structures for one natural and two non-natural variants. The purine-binding site was essentially unchanged, but co-variation at positions 24 and 25 could be partially explained by local adjustments. U24 is positioned similarly to A24 but does not stack equivalently between G72 and A73. This change naturally co-occurs with imperfect pairing and a kink at the base of P2, suggesting that one necessarily accommodates the other. This also explains the loss of in vivo control observed in non-natural Position 24–P2.1/P2.2 combinations. Overall, what makes this paper stand out is its systematic experimental validation of these newly identified fine-tuning sequence elements. These data provide strong evidence that the subtle co-variation at the P2 tune box has minimal effect on the structure but can impart a range of effects on binding kinetics in the guanine/ adenine riboswitches.
Outlook
Since the discovery of riboswitches over 10 years ago, the mechanisms of direct ligand recognition have been intensely studied; however, the complexity of riboswitch tertiary structures makes understanding regulatory tuning an arduous task. The work by Stoddard et al. provides direct experimental evidence for evolutionarily important tuning hotspots within riboswitches, where subtle co-variations are sufficient to impart a spectrum of responses. Obviously, this P2 tune box is not the only functional means for tuning in the purine riboswitches and it will be interesting to note whether the other identified co-varying regions are also involved in tuning. Given the power and simplicity of accumulating simple changes in non-coding RNA, it seems likely that many riboswitches will possess similar tuning hotspots where mutations rapidly and efficiently adjust gene regulation.
Acknowledgments
The work is supported by a National Institutes of Health operating grant (GM-086766) to A.K. J.C.G. is supported by a Postdoctoral Fellowship from the Canadian Institutes of Health Research.
References
- 1.Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, et al. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 2.Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci USA. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 4.Caron MP, Bastet L, Lussier A, Simoneau-Roy M, Masse E, Lafontaine DA. Dual-acting riboswitch control of translation initiation and mRNA decay. Proc Natl Acad Sci USA. 2012;109:E3444–E3453. doi: 10.1073/pnas.1214024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hollands K, Proshkin S, Sklyarova S, Epshtein V, Mironov A, Nudler E, Groisman EA. Riboswitch control of Rho-dependent transcription termination. Proc Natl Acad Sci USA. 2012;109:5376–5381. doi: 10.1073/pnas.1112211109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batey RT. Structure and mechanism of purine-binding riboswitches. Q Rev Biophys. 2012;45:345–381. doi: 10.1017/S0033583512000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim JN, Breaker RR. Purine sensing by riboswitches. Biol Cell. 2008;100:1–11. doi: 10.1042/BC20070088. [DOI] [PubMed] [Google Scholar]
- 8.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 9.Mandal M, Breaker RR. Adenine ribos-witches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 10.Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 11.Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, et al. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darty K, Denise A, Ponty Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoddard CD, Widmann J, Trausch JJ, Marcano-Velazques JG, Knight R, Batey RT. Nucleotides adjacent to the ligand-binding pocket are linked to activity tuning in the purine riboswitch. J Mol Biol. 2013;425:1596–1611. doi: 10.1016/j.jmb.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cruz JA, Westhof E. The dynamic landscapes of RNA architecture. Cell. 2009;136:604–609. doi: 10.1016/j.cell.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Kortmann J, Narberhaus F. Bacterial RNA thermometers: molecular zippers and switches. Nat Rev Microbiol. 2012;10:255–265. doi: 10.1038/nrmicro2730. [DOI] [PubMed] [Google Scholar]
- 16.Chowdhury S, Maris C, Allain FH, Narberhaus F. Molecular basis for temperature sensing by an RNA thermometer. EMBO J. 2006;25:2487–2497. doi: 10.1038/sj.emboj.7601128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mulhbacher J, Lafontaine DA. Ligand recognition determinants of guanine riboswitches. Nucleic Acids Res. 2007;35:5568–5580. doi: 10.1093/nar/gkm572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomsic J, McDaniel BA, Grundy FJ, Henkin TM. Natural variability in S-adenosylmethio-nine (SAM)-dependent riboswitches: S-box elements in Bacillus subtilis exhibit differential sensitivity to SAM in vivo and in vitro. J Bacteriol. 2008;190:823–833. doi: 10.1128/JB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baird NJ, Kulshina N, Ferre-D’Amare AR. Riboswitch function: flipping the switch or tuning the dimmer? RNA Biol. 2010;7:328–332. doi: 10.4161/rna.7.3.11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]


