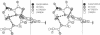Abstract
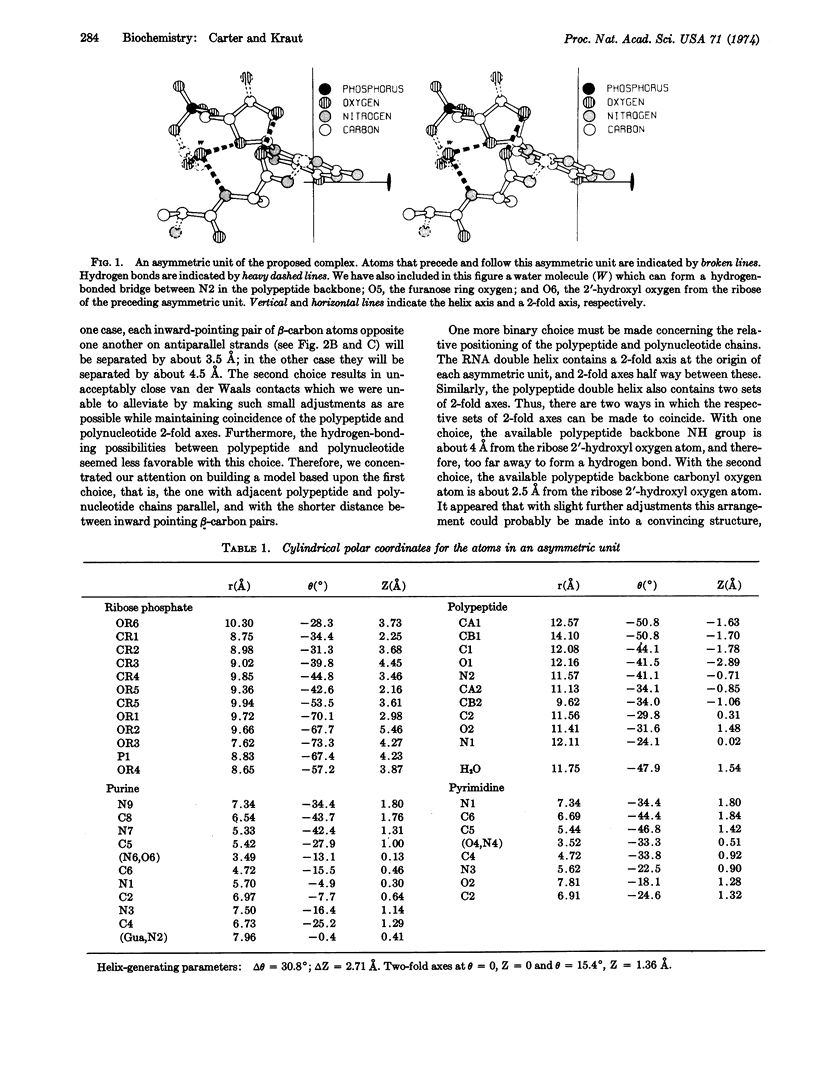
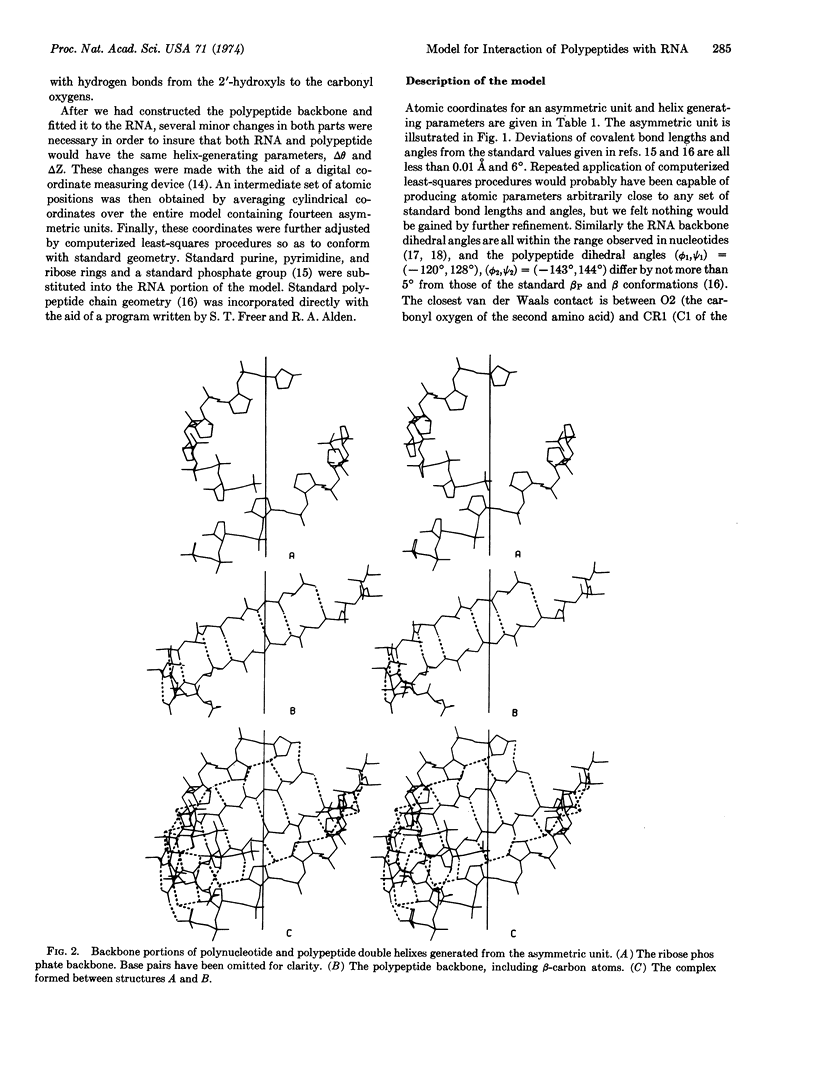
Pairs of antiparallel β polypeptide-chain segments in known protein structures are usually observed to form right-handed double helixes with helix parameters in the same range as those of nucleic acids. We have constructed a model containing only standard bond lengths, bond angles, and dihedral angles in which such a polypeptide double helix fits precisely into the minor groove of an RNA double helix with identical helix parameters. The geometry of the RNA portion is essentially a hybrid between those of the A and A′ forms. Hydrogen bonds can be made between the ribose 2′-hydroxyls and polypeptide carbonyl oxygens. Since such precise complementarity between the stable conformations of RNA and polypeptides is unlikely to be merely coincidental, we propose that it played a fundamental role in the initiation of precellular evolution. Specificially, we propose that the two double-helical structures are mutually catalytic for assembly of one another from activated precursors in the prebiotic soup, and moreover that they provide some degree of genetic coding.
Keywords: precellular evolution, L-amino acids in proteins, double-helical conformations, twisted-β-sheets
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alden R. A., Birktoft J. J., Kraut J., Robertus J. D., Wright C. S. Atomic coordinates for subtilisin BPN' (or Novo). Biochem Biophys Res Commun. 1971 Oct 15;45(2):337–344. doi: 10.1016/0006-291x(71)90823-0. [DOI] [PubMed] [Google Scholar]
- Arnott S., Hukins D. W. Conservation of conformation in mono and poly-nucleotides. Nature. 1969 Nov 29;224(5222):886–888. doi: 10.1038/224886a0. [DOI] [PubMed] [Google Scholar]
- Arnott S., Hukins D. W., Dover S. D. Optimised parameters for RNA double-helices. Biochem Biophys Res Commun. 1972 Sep 26;48(6):1392–1399. doi: 10.1016/0006-291x(72)90867-4. [DOI] [PubMed] [Google Scholar]
- Blake C. C., Koenig D. F., Mair G. A., North A. C., Phillips D. C., Sarma V. R. Structure of hen egg-white lysozyme. A three-dimensional Fourier synthesis at 2 Angstrom resolution. Nature. 1965 May 22;206(4986):757–761. doi: 10.1038/206757a0. [DOI] [PubMed] [Google Scholar]
- Chothia C. Conformation of twisted beta-pleated sheets in proteins. J Mol Biol. 1973 Apr 5;75(2):295–302. doi: 10.1016/0022-2836(73)90022-3. [DOI] [PubMed] [Google Scholar]
- Day R. O., Seeman N. C., Rosenberg J. M., Rich A. A crystalline fragment of the double helix: the structure of the dinucleoside phosphate guanylyl-3',5'-cytidine. Proc Natl Acad Sci U S A. 1973 Mar;70(3):849–853. doi: 10.1073/pnas.70.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Quigley G. J., Suddath F. L., McPherson A., Sneden D., Kim J. J., Weinzierl J., Rich A. Three-dimensional structure of yeast phenylalanine transfer RNA: folding of the polynucleotide chain. Science. 1973 Jan 19;179(4070):285–288. doi: 10.1126/science.179.4070.285. [DOI] [PubMed] [Google Scholar]
- Mathews F. S., Argos P., Levine M. The structure of cytochrome b 5 at 2.0 Angstrom resolution. Cold Spring Harb Symp Quant Biol. 1972;36:387–395. doi: 10.1101/sqb.1972.036.01.050. [DOI] [PubMed] [Google Scholar]
- Quiocho F. A., Lipscomb W. N. Carboxypeptidase A: a protein and an enzyme. Adv Protein Chem. 1971;25:1–78. doi: 10.1016/s0065-3233(08)60278-8. [DOI] [PubMed] [Google Scholar]
- Ramachandran G. N., Sasisekharan V. Conformation of polypeptides and proteins. Adv Protein Chem. 1968;23:283–438. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- Rohrer D. C., Sundaralingam M. Stereochemistry of nucleic acids and their constituents. XII. The crystal and molecular structure of alpha-D-2'-deoxyadenosine monohydrate. J Am Chem Soc. 1970 Aug 12;92(16):4956–4962. doi: 10.1021/ja00719a032. [DOI] [PubMed] [Google Scholar]
- Rosenberg J. M., Seeman N. C., Kim J. J., Suddath F. L., Nicholas H. B., Rich A. Double helix at atomic resolution. Nature. 1973 May 18;243(5403):150–154. doi: 10.1038/243150a0. [DOI] [PubMed] [Google Scholar]
- Rossmann M. G., Adams M. J., Buehner M., Ford G. C., Hackert M. L., Lentz P. J., Jr, McPherson A., Jr, Schevitz R. W., Smiley I. E. Structural constraints of possible mechanisms of lactate dehydrogenase as shown by high resolution studies of the apoenzyme and a variety of enzyme complexes. Cold Spring Harb Symp Quant Biol. 1972;36:179–191. doi: 10.1101/sqb.1972.036.01.025. [DOI] [PubMed] [Google Scholar]
- Salemme F. R., Fehr D. G. A device for the rapid measurement of molecular model co-ordinates for x-ray crystallography. J Mol Biol. 1972 Oct 14;70(3):697–700. doi: 10.1016/0022-2836(72)90568-2. [DOI] [PubMed] [Google Scholar]
- Saunders M. A., Rohlfing D. L. Polyamino acids: preparation from reported proportions of "prebiotic" and extraterrestrial amino acids. Science. 1972 Apr 14;176(4031):172–173. doi: 10.1126/science.176.4031.172. [DOI] [PubMed] [Google Scholar]
- Sulston J., Lohrmann R., Orgel L. E., Miles H. T. Nonenzymatic synthesis of oligoadenylates on a polyuridylic acid template. Proc Natl Acad Sci U S A. 1968 Mar;59(3):726–733. doi: 10.1073/pnas.59.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff H. W., Tsernoglou D., Hanson A. W., Knox J. R., Lee B., Richards F. M. The three-dimensional structure of ribonuclease-S. Interpretation of an electron density map at a nominal resolution of 2 A. J Biol Chem. 1970 Jan 25;245(2):305–328. [PubMed] [Google Scholar]