Key Points
Zinc-finger nucleases simultaneously and permanently inactivate HIV coreceptors ccr5 and cxcr4 resulting in HIV-resistant CD4+ T cells.
These HIV-resistant cells may be used to achieve a functional cure for HIV in humans.
Abstract
HIV-1 entry into CD4+ T cells requires binding of the virus to CD4 followed by engagement of either the C-C chemokine receptor 5 (CCR5) or C-X-C chemokine receptor 4 (CXCR4) coreceptor. Pharmacologic blockade or genetic inactivation of either coreceptor protects cells from infection by viruses that exclusively use the targeted coreceptor. We have used zinc-finger nucleases to drive the simultaneous genetic modification of both ccr5 and cxcr4 in primary human CD4+ T cells. These gene-modified cells proliferated normally and were resistant to both CCR5- and CXCR4-using HIV-1 in vitro. When introduced into a humanized mouse model of HIV-1 infection, these coreceptor negative cells engraft and traffic normally, and are protected from infection with CCR5- and CXCR4-using HIV-1 strains. These data suggest that simultaneous disruption of the HIV coreceptors may provide a useful approach for the long-term, drug-free treatment of established HIV-1 infections.
Introduction
Potent antiretroviral therapy (ART) suppresses circulating levels of HIV to below the limit of detection, dramatically delaying progression to AIDS.1,2 However, the side effects, cost, daily adherence requirement, and immune dysfunction that persist despite ART underscore the need for strategies to suppress HIV infection and reconstitute immune function without ART.1-5 One approach involves rendering CD4+ T cells resistant to HIV by permanent inactivation of the viral entry receptors. To enter a cell, HIV must bind its primary receptor CD4, and 1 of 2 coreceptors, C-C chemokine receptor 5 (CCR5) or C-X-C chemokine receptor 4 (CXCR4) (×4).6-13 The majority of transmitted viruses use R5 to enter cells14-16 while X4-using viruses are seen in half of late-stage infections17,18 and are associated with a more rapid disease progression.19,20 Individuals lacking functional R5 due to the naturally occurring ccr5Δ32 mutation are highly resistant to HIV infection,21-23 and, recently, an HIV-positive male who received a bone marrow transplant from a ccr5Δ32 homozygous donor appears to be cured of HIV.24,25 As allogeneic stem cell transplants prior to this case have not had any effect on HIV infection,26,27 the response seen in this patient highlights the importance of the coreceptors in maintaining HIV infection, and supports the idea that permanent coreceptor inactivation in infected individuals may provide drug-free control of HIV.
In this study, we have used zinc-finger nucleases (ZFNs) to simultaneously inactivate the ccr5 and cxcr4 genes. ZFNs are chimeric proteins that function as a pair and act in a sequence-specific manner to introduce a double-stranded break at a predetermined site in the genome.28 As these double-stranded breaks are often repaired by error-prone nonhomologous end joining,29 ZFN cleavage introduces mutations that produce truncated or nonfunctional proteins. The use of ZFNs to target the coreceptors is particularly attractive, as ZFNs need only be transiently expressed, and generate stable, heritable mutations in target cells. We and others have shown that permanent inactivation of ccr5 or cxcr4 alone can be achieved in CD4+ T cells and protects from infection with viruses that exploit the targeted coreceptor.30-33 Additionally, a recent phase 1 study examining the safety of R5-ZFN–modified CD4+ T cells in HIV-infected individuals showed that not only is this approach safe and well tolerated, but that these gene-modified cells are long-lived and capable of trafficking to the rectal mucosa, a major site of HIV replication (C.H.J., University of Pennsylvania, written communication, May 23, 2013). To make this approach more broadly applicable to patients with both R5 and X4-HIV, we sought to determine the feasibility of simultaneous modification of both HIV coreceptors in CD4+ T cells by ZFNs and its effect on HIV infection.
Methods
Cell culture and ZFN treatment
SupT1 T cells expressing multiple copies of CCR5 introduced by lentiviral transduction (SupT1-R5), and primary human CD4+ T cells, were maintained in RPMI (Invitrogen) supplemented with 10% fetal bovine serum. CD4+ T cells were grown in the presence of human interleukin-2 (IL-2) (100 IU/mL). We used R5 and X4 zinc-finger proteins (ZFPs) from our previous studies.31,32 However, the ZFN nuclease domains were the ELD/KKR variant which were modified to function as obligate heterodimers with enhanced cleavage activity.34 To deliver ZFNs, we simultaneously transduced cells with 2 Ad5/F35 vectors encoding either the R5- or X4-ZFNs or an Ad5/F35 green fluorescent protein (GFP)-expressing control vector.31,32 This is a chimeric adenoviral vector based on the AdEasy vector system (Promega) with an E1/E3-deleted backbone and a chimeric fiber gene composed of a serotype 5 fiber tail domain, and serotype 35 shaft and knob domains.35 CD4+ T cells were activated 18 to 24 hours prior to vector transduction with anti-CD3/anti-CD28–coated magnetic beads.32 We determined vector multiplicity of infections (MOIs) using the 293T cell line and measured mutation frequencies by surveyor nuclease (Cel1), T7 endonuclease I assay (T7E1), or deep sequencing.36-38
Virus production and infection
Pseudoviruses mediate 1 round of infection as the viral glycoprotein gene is only supplied in trans. We generated HIV pseudoviruses encoding a GFP reporter, bearing either the HIV glycoprotein (Env) or the vesicular stomatitis virus glycoprotein (VSV-G) by cotransfection of the viral glycoprotein and the pNL4.3-Δenv-vpr+-eGFP-HIV backbone39 into 293T cells using calcium phosphate. We harvested supernatants after 72 hours and concentrated virus by ultracentrifugation.40 The HIV pseudoviruses were made using the R5 and X4-HIV envs JRFL and TYBE,41 respectively. For SupT1-R5 pseudovirus infections, we infected 1 × 105 cells in a 96-well v-bottom plate with 500 ng of HIV-1 p24 or 5 to 10 ng of HIV-VSV-G p24. Samples were spun at 1200g for 90 minutes at 25°C then transferred to 37°C. Infection was measured at 96 hours by flow cytometric analysis of GFP-positive cells.
Replication competent HIV-1 was made in primary CD4+ T cells40 and we infected SupT1-R5 cells and primary cells with 50 to 100 ng of p24 per million cells 5 to 7 days following ZFN treatment. The replication competent HIV-1 strains used were the R5-virus BaL and the X4-viruses BK132 and HxB2. All CD4+ T cell infections were performed using cells from 3 independent donors. For all infections where cells were challenged simultaneously with R5 and X4-HIV, the viruses were mixed in a 1:1 ratio normalized by HIV-1 p24.
Flow cytometry
Cells were stained at room temperature in fluorescence-activated cell sorter (FACS) buffer (phosphate-buffered saline−/−, 2% fetal bovine serum, 2mM EDTA). SupT1-R5 cells and CD4+ T cells were stained with Live/Dead Aqua and QD655-CD3 (clone S4.1) (Invitrogen), fluorescein isothiocyanate–CD45 (clone H130) (Biolegend), Alexa Fluor 700–CD4 (clone RPA-T4), Pacific Blue–CD8 (clone RPA-T8), allophycocyanin–CCR5 (clone 2D7), and phycoerythrin–CXCR4 (clone 12G5) (BD Biosciences). Samples were run on an LSR II (BD Biosciences) and analyzed using FlowJo 10.0.4 (TreeStar).
HIV-1–humanized mouse challenge
Twelve- to 14-week-old NOD.Cg-PrkdcscidIl2rgtm1Wjl/Szj (NSG) immunodeficient mice received either unmodified CD4 T cells, R5-ZFN–treated CD4 T cells, or R5/X4-ZFN–treated CD4 T cells. Each group contained 18 animals and each animal received 1.8 × 107 cells. Mice were randomly assigned to control for cage, sex, and birth-cohort effects. The animals were maintained in a defined flora facility at the University of Pennsylvania with approval of the Institutional Animal Care and Use Committee. We measured peripheral blood CD4 counts 21 days postinfusion to assess engraftment by staining for CD45+/CD3+/CD4+/CD8− cells. Three days later, half the animals in each group received 5 × 104 CD4+ T cells infected with the R5-virus BaL, and 5.0 × 104 CD4+ T cells infected with the X4-virus Bk132. Control animals received 1 × 105 uninfected CD4+ T cells. We performed retro-orbital bleeds to assess CD4 counts and measure gene disruption. Mice were killed following the development of xenogeneic-graft-versus-host disease. Cardiac punctures and splenectomies were performed on all mice following euthanasia. We passed spleens through a 70-μm strainer to obtain a single-cell suspension for measuring gene modification and CD4 count. ZFN-induced mutation frequencies were determined by performing deep sequencing at the R5 and X4-ZFN target sites.36
Off-target site analysis
We previously used systematic evolution of ligands by exponential enrichment (SELEX) to determine the in vitro binding preference of each ZFP.31,32 Using the information on CCR5- and CXCR4-binding preferences, we generated a position-weight matrix which we aligned to the human genome to look for sites with similarity to each of the 4 possible combinations of CCR5-CXCR4 cross heterodimers (R5-left with X4-right; R5-left with X4-left; R5-right with X4-right; R5-right with X4-left). We allowed up to a 4-bp mismatch compared with the SELEX consensus sequence, and considered sites with a 5-bp or 6-bp spacer between each ZFP pair. We performed deep sequencing at these sites to identify off-target activity.36 Off-target activity was defined as an insertion/deletion (indel) involving the target region (20 bp) centered at the nuclease-binding sites at a frequency of >0.1%, and with a calculated ratio of cleavage (Z − M)/M of >2, where Z represents the percentage of indels in ZFN-treated samples, and M represents the percentage of indels in mock-treated samples.
Results
Simultaneous ZFN disruption of ccr5 and cxcr4 in a T-cell line protects from R5 and X4-HIV infection
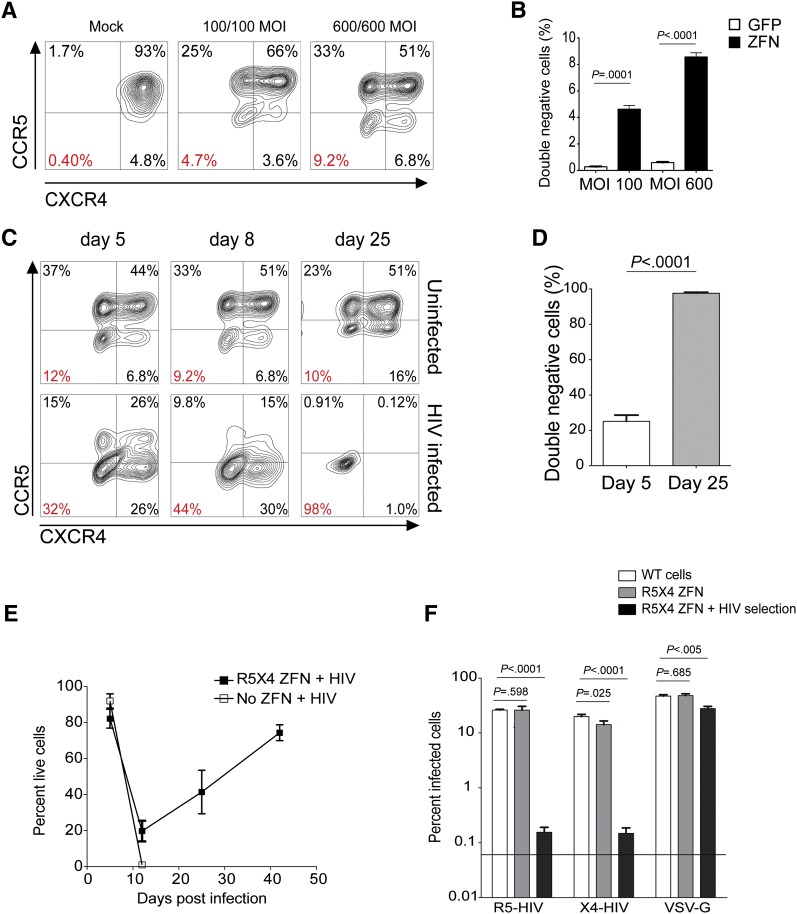
To determine whether 2 ZFN pairs targeting the HIV-coreceptors ccr5 (R5) and cxcr4 (×4) could abolish expression of both coreceptors, we cotransduced the human SupT1-R5 T-cell line with increasing amounts of the Ad5/F35 vectors encoding the R5-ZFN and X4-ZFN. Coadministration of both (R5/X4) ZFNs caused a dose-dependent reduction in cell-surface expression of both coreceptors with 9% of cells no longer expressing either coreceptor at the highest MOI, while delivery of the same vector encoding GFP had no effect on surface R5 or X4 levels (Figure 1A-B). Next, we assessed the ability of ccr5 and cxcr4 codisruption to protect cells from infection with R5- and X4-using HIV-1 strains. We challenged untreated or R5/X4-ZFN–treated SupT1-R5 cells simultaneously with the R5- and X4-HIV strains BaL and BK132, respectively, and monitored surface coreceptor expression for 42 days. The proportion of double (R5/X4)–negative cells increased over the course of the infection such that by 25 days postinfection, 96% to 99% of the R5/X4-ZFN–treated cells challenged with HIV no longer expressed either coreceptor (Figure 1C-D). In contrast, the proportion of double-negative cells in the uninfected R5/X4-ZFN–treated group did not change significantly over this time (Figure 1C top panels). We also monitored cell viability and while untreated cells all died by 10 days postinfection, a portion of the R5/X4-ZFN–treated cells survived the infection with cell numbers increasing exponentially over time (Figure 1E). The coreceptor-negative cells that survived HIV infection were rechallenged with pseudovirus bearing either HIV Env or VSV-G to confirm that the mechanism of resistance was due to inhibition of HIV Env-specific entry. These cells were highly resistant to either R5 or X4-HIV pseudovirus rechallenge, displaying 170-fold and 92-fold decreases in reinfection with R5- and X4-HIV, respectively (Figure 1F). Of note, low levels of residual infection (∼0.1%) were observed in the R5/X4-ZFN group prechallenged with HIV. However, the majority of this residual infection occurred in the 1% to 4% of cells still expressing CCR5 and CXCR4 (data not shown). Additionally, these coreceptor-negative cells were readily infected by HIV expressing VSV-G HIV, a virus that is not dependent upon R5 or X4 for infection (Figure 1F). Taken together, these data suggest that simultaneous delivery of 2 ZFN pairs is an efficient and viable strategy to disrupt ccr5 and cxcr4, resulting in cells resistant to infection with both R5- and X4-using HIV.
Figure 1.
Simultaneous ZFN modification of ccr5 and cxcr4 protects SupT1-R5 T cells from infection with viruses that use either CCR5 or CXCR4. (A) Surface expression of CCR5 (R5) and CXCR4 (×4) on SupT1-R5 cells following delivery of increasing MOIs of the Ad R5- and X4-ZFNs. Percentage of cells lacking both coreceptors is labeled in red. (B) Proportion of cells lacking both surface R5 and X4 (double-negative cells) following simultaneous treatment with the R5 (MOI 600) and X4 (MOI 600) ZFNs as measured by FACS. (C) Dual ZFN-treated cells were challenged with a mix of R5- and X4-using HIV and surface expression of R5 and X4 was measured over time in infected and uninfected R5/X4-ZFN–treated cells. (D) The proportion of double-negative cells following R5/X4-ZFN treatment and subsequent HIV infection was measured by FACS 5 and 25 days postinfection. (E) Cell viability after infection of mock (no ZFN) or R5/X4-ZFN–treated supT1-R5 cells with a mix of R5- and X4-HIV. Viability was measured by FACS following treatment with a viability dye. (F) Dual ZFN-treated supT1-R5 cells previously challenged with HIV and no longer expressing either coreceptor (▪) as shown in panel C, were rechallenged with either R5-HIV, X4-HIV, or VSV-G-HIV pseudoviruses expressing GFP. HIV pseudovirus rechallenge of previously HIV-selected double-negative cells resulted in 170-fold and 92-fold decreases in infection by R5 and X4-HIV, respectively, whereas VSV-G pseudovirus infection was decreased only ∼1.7-fold. All graphs represent the mean (±SEM) of 4 independent ZFN treatments and 4 independent infection experiments; P values calculated using the Student t test.
ZFNs achieve simultaneous disruption of ccr5 and cxcr4 in human CD4+ T cells
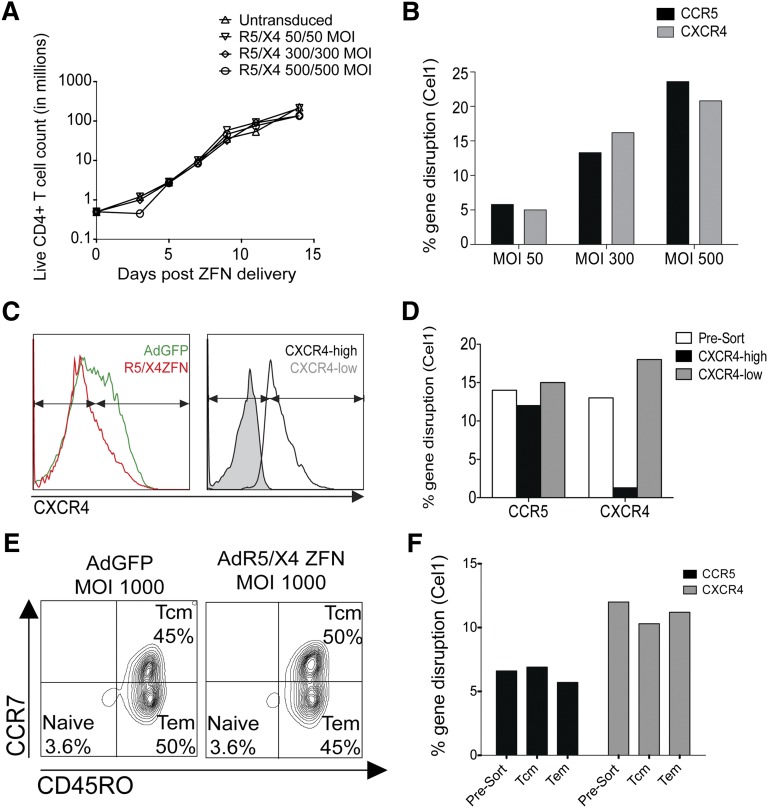
To determine the feasibility of this approach in primary cells, CD4+ T cells from healthy human donors were simultaneously transduced with the adenoviral vectors encoding the R5 and X4-ZFNs. Transducing cells with MOIs of up to 500 of each ZFN resulted in modification of ∼20% of all R5 and X4 genes as measured by the Cel1 assay, without significant impact on cell growth compared with untransduced controls (Figure 2A-B). As surface expression of CCR5 is undetectable on most primary CD4+ T cells, we could not determine the proportion of cells lacking both coreceptors by flow cytometry. To exclude the possibility that coadministration of both ZFNs in primary cells results in R5 and X4 gene modification in mutually exclusive cells, we delivered both ZFNs to CD4+ T cells and sorted them by FACS based on surface CXCR4 expression into CXCR4-high and CXCR4-low populations (Figure 2C). We then measured levels of R5 and X4 gene modification in both of these populations. Whereas cxcr4 gene modification was greatly reduced in the X4-high population following sorting, levels of ccr5 modification were similar in the X4-high and X4-low populations (Figure 2D), suggesting that adenoviral dual ZFN treatment achieved dual modification of ccr5 and cxcr4 in primary CD4+ T cells.
Figure 2.
ZFNs simultaneously disrupt ccr5 and cxcr4 in primary human CD4+ T cells. (A) Dual ZFN treatment does not result in a significant growth defect as determined by live primary CD4+ T-cell count following simultaneous delivery of increasing amounts of the R5- and X4-ZFNs. Data are from 1 of 3 independent experiments. (B) ccr5 and cxcr4 gene modification increases in a dose-dependent manner following R5/X4-ZFN treatment, as measured by the Cel1 assay 7 to 10 days post-ZFN treatment. Data are from 1 of 3 independent experiments. (C) Primary CD4+ T cells were sorted by CXCR4 expression into CXCR4-high and CXCR4-low populations (left) following simultaneous R5/X4-ZFN treatment. Successful sorting was confirmed by repeat FACS (right panel). Arrows indicate gating strategy. (D) ccr5 and cxcr4 gene disruption was measured by cel1 following R5/X4-ZFN treatment and subsequent sorting by FACS into CXCR4-high and -low populations. ccr5 disruption is similar in the X4-high and X4-low populations, suggesting ccr5 and cxcr4 disruption are not occurring in mutually exclusive cells. Data are from 1 of 3 independent experiments. (E) CD4+ T cells treated with equal MOIs of the AdGFP or the R5- and X4-ZFNs were stained with the memory markers CCR7 and CD45RO Tcm [CD45RO+ CCR7+] and Tem [CD45RO+CCR7−]). (F) ccr5 and cxcr4 gene modification in Tcm and Tem subsets was similar suggesting that long-lived Tcm cells can be efficiently rendered HIV-resistant. Data shown are from 1 of 3 independent experiments. Tcm, central memory T cell; Tem, effector memory T cell.
Depletion of central memory CD4+ T cells is a hallmark of HIV pathogenesis and progression to AIDS.42 As such, protection of this long-lived subset is important for the success of any HIV gene therapy approach. We therefore determined whether central memory CD4+ T cells could be modified by the R5- and X4-ZFNs. We first transduced CD4+ T cells with both ZFNs, expanded them in vitro for 10 days, and then sorted the cells by FACS into central and effector memory populations based on surface expression of the memory markers CCR7 and CD45RO43 (Figure 2E). CD4+ T-cell activation using CD3/CD28-coated beads results in differentiation of ∼97% of cells into a memory phenotype, with roughly half of all cells being either central or effector memory cells, and the proportion of effector and central memory subtypes is not affected by treatment with the adenoviral vector or the ZFN (data not shown). We detected both ccr5 and cxcr4 modification (Cel1) in the central memory (CCR7+, CD45RO+) and effector memory (CCR7−, CD45RO+) populations at similar levels (Figure 2E), suggesting that we can effectively target the coreceptors in these critically important T-cell subsets.
Survival advantage of dual ZFN-treated CD4+ T cells in vitro
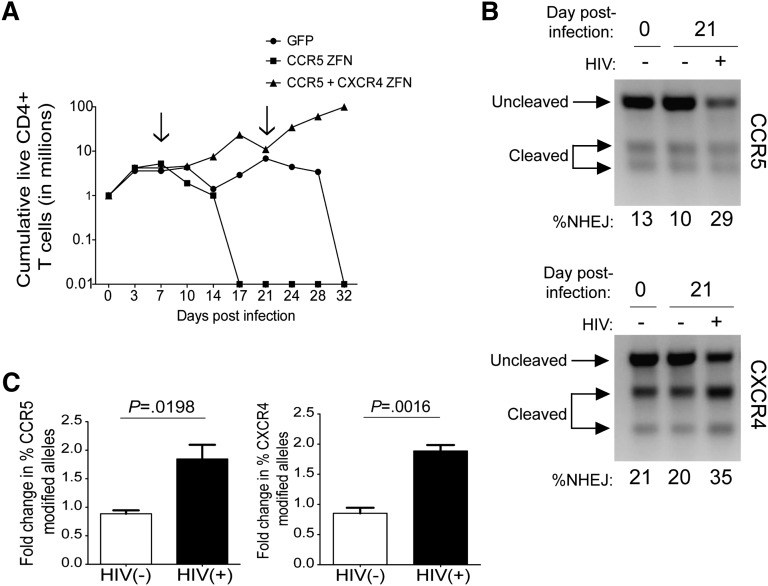
To determine whether the level of gene modification seen in primary cells rendered them resistant to both forms of HIV-1, we treated CD4+ T cells with both ZFNs, the R5-ZFN alone, or with a GFP control. We then challenged all 3 groups with a mix of R5- and X4-using HIV-1 and monitored cell growth and viability. By 32 days postinfection, there were no detectable live cells in the groups that received either no ZFN or the R5-ZFN alone, whereas cells from the R5/X4 ZFN group continued to expand (Figure 3A). We measured levels of ccr5 and cxcr4 gene modification 3 weeks into the infection in the R5/X4-ZFN–treated group and observed increases in both ccr5 (1.8-fold) and cxcr4 (1.9-fold) gene modification (Figure 3B-C) when compared with the start of the infection. The preservation of cell growth and increases in gene modification seen only in the R5/X4-ZFNgroup in the presence of HIV suggest that ZFN modification of both coreceptors provides a significant survival advantage in the presence of R5- and X4-using HIV.
Figure 3.
Primary CD4+ T cells treated with the R5 and X4-ZFNs and challenged with HIV have a survival advantage in vitro. (A) Primary CD4+ T cells treated with GFP, the R5-ZFN, or 2 vectors encoding the R5 and X4-ZFNs were challenged with a mix of 2o HIV strains: the R5-using virus BaL and the X4-using virus HxB2. Dual ZFN treatment conferred a significant survival advantage in vitro compared with both the GFP and R5-ZFN control. Live cells were counted every 3 to 4 days for 32 days. Arrows indicate the time points where cells were reactivated with anti-CD3/CD28 magnetic beads. Data are from 1 of 3 independent experiments. (B) T7E1 analysis of ccr5 (top panel) and cxcr4 (bottom panel) gene modification reveals that HIV challenge enriches for ccr5 and cxcr4 disrupted cells. Analysis was performed 0 and 21 days postinfection. Data are from 1 of 3 independent experiments. (C) The proportion of modified ccr5 and cxcr4 alleles in cells treated with both ZFNs increases approximately twofold in the presence or absence of HIV, as measured by the T7E1 assay. Data are shown as the mean ± SEM of 3 independent experiments. P values calculated using the Student t test.
Analysis of off-target cleavage following dual ZFN administration
Simultaneous administration of 2 ZFN pairs creates the theoretical possibility of forming 4 different ZFN cross-heterodimers made up of the left and right halves of the 2 unique ZFN pairs. These ZFN cross-heterodimers may subsequently bind to unintended target sites, resulting in more off-target gene modification compared with delivery of a single ZFN pair. As the target sequence contributes to ZFN binding and thus specificity,44 we previously determined the in vitro DNA binding preference of each CCR5 and CXCR4 ZFP using SELEX.31,32 We constructed a position-weight matrix for each 12-bp ZFP binding site, and by comparing these sequences against the human genome to identify potential off-target binding sites, identified sites that were in fact independently cleaved by the R5- and X4-ZFNs.31,32 In this study, we took a similar approach to identify off-target sites that could result from the binding and cleavage of a ZFN heterodimer consisting of one-half R5-ZFN and one-half X4-ZFN. We performed deep sequencing at these predicted sites in primary CD4+ T cells following treatment with both the R5- and X4-ZFNs compared with otherwise identical untransduced controls (supplemental Table 1, available on the Blood Web site). Using genomic DNA from samples where on-target modification of ccr5 and cxcr4 were 28% and 20%, respectively, we analyzed 4000 to 20 000 reads per predicted off-target site but failed to detect significant levels of modification at any of the 40 predicted off-target sites (supplemental Table 1). These results suggests that cross-heterodimerization of the R5- and X4-ZFN pairs following delivery to primary CD4+ T cells does not result in detectable off-target activity at the 40 sites examined. Moreover, the use of recently described orthogonal heterodimeric nuclease domains that further restrict cleavage34 could further address this concern.
Dual coreceptor disruption protects CD4+ T cells following HIV infection in vivo
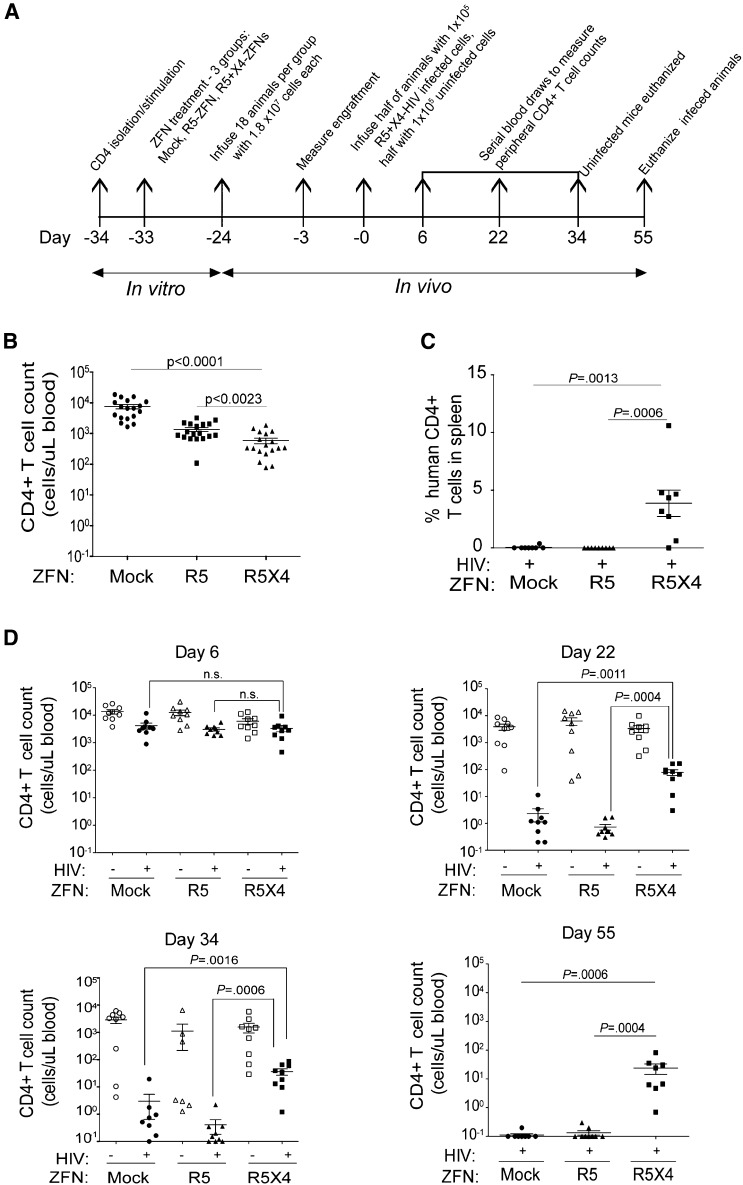
NOD.Cg-PrkdcscidIl2rgtm1Wjl/Szj (NSG) immunodeficient mice are a useful model for studying HIV-1 in vivo as their immune systems can be reconstituted using human cells that are susceptible to HIV infection.45 To assess whether our approach protects against infection with both R5- and X4-HIV in vivo, we treated primary CD4+ T cells with no ZFN, the R5-ZFN alone, or with the R5- and X4-ZFNs simultaneously. Three groups of 18 mice each were infused with 1.8 × 107 cells per animal from each treatment group (Figure 4A). We observed CD4+ T-cell engraftment across all groups 21 days postinfusion (Figure 4B).We then infected half the animals in each group by simultaneously infusing 5 × 104 unmodified (no ZFN) CD4+ T cells infected with the R5-virus BaL and 5 × 104 unmodified CD4+ T cells infected with the X4-virus BK132. Of note, while the BK132 HIV-1 swarm primarily uses CXCR4 to enter cells, it can also achieve very low levels of infection using CCR546 as evidenced by the outgrowth of R5-HIV when this virus was used to challenge X4-ZFN–treated CD4 T cells.32 Prior to infusion, the frequency of HIV p24+ cells was 58% and 44% in the R5- and X4-HIV–infected cells, respectively. Control animals in each group received an infusion of 1 × 105 uninfected, unmodified CD4+ T cells. Animals that received R5-ZFN– or R5/X4-ZFN–treated cells had 5.5- and 12.8-fold lower CD4 counts, respectively, than animals that received untransduced cells when measured 3 days prior to infection (Figure 4B). However, this difference in CD4 counts was no longer present at the time of the first peripheral blood sampling 6 days postinfection (Figure 4D). This slight engraftment difference was not seen previously31,32 and may be donor specific. Although CCR5 ablation alone was previously shown to protect against challenge with CCR5-using HIV,31 animals that received both ZFNs were better able to maintain their CD4+ T-cell counts following challenge with R5- and X4-HIV (Figure 4D). Specifically, CD4 counts in animals from the infected R5/X4-ZFN group were 35- and100-fold higher than those in animals from the mock and R5-ZFN–infected groups, respectively, by 22 days postinfection, and these differences increased to an average 200-fold difference 55 days postinfection (Figure 4D). Additionally, we measured the proportion of human CD4+ T cells in the spleens of infected animals at time of sacrifice. In animals that received dual ZFN-treated cells, up to 10% of all cells populating the spleen were human CD4+ T cells whereas we could not detect any human CD4+ T cells in spleens from animals that received either no ZFN or the R5-ZFN alone (Figure 4C).
Figure 4.
Dual R5- and X4-ZFN treatment confers protection in vivo. (A) Experimental timeline of the in vivo study is shown. (B) All 3 treatment groups were successfully engrafted as measured 21 days postinfusion and 3 days prior to infection with a slight decrease in CD4+ T-cell counts in ZFN groups compared with the GFP control. (C) Dual ZFN treatment preserves CD4+ T cells in the spleen compared with controls. Spleens were obtained from infected animals from all animals at the time of euthanasia. Human CD4+ T cells were defined as CD45+CD3+CD4+CD8− T cells by FACS. (D) Dual ZFN treatment confers long-term protection against HIV as measured by preservation of peripheral blood CD4+ T-cell counts. All uninfected animals and 1 infected animal in the R5/X4-ZFN–treated group were killed 34 days postinfection due xenogeneic graft-versus-host disease. Infected animals were followed for 55 days after infection. All statistical analyses were performed using a Mann-Whitney U test. Error bars indicate the mean ± SEM.
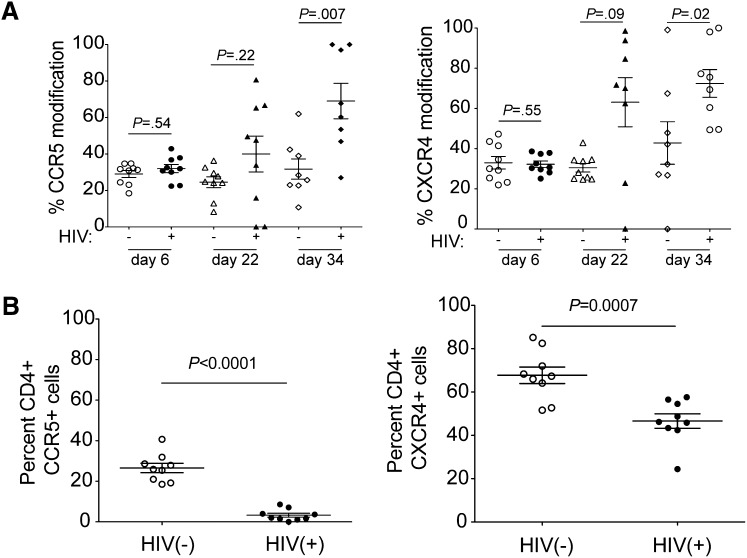
To determine the mechanism of CD4 count maintenance observed in the R5/X4-ZFN group, we performed deep sequencing at the ZFN cut sites in CD4+ T cells from animals in the R5/X4-ZFN group to measure changes in the proportion of ZFN-induced indels. There was no change in ccr5 or cxcr4 gene modification in uninfected animals that received both ZFNs. However, on average, 69% of all ccr5 genes and 73% of all cxcr4 genes were mutated in CD4+ T cells from the infected R5/X4-ZFN animals by 34 days postinfection (Figure 5A). This represents a 2.2-fold increase in ccr5-modification and a 2.3-fold increase in cxcr4 modification compared with 6 days postinfection. Additionally, we observed significantly lower surface expression of both coreceptors on cells from the infected R5/X4-ZFN–treated mice 34 days postinfection (Figure 5B). These data provide strong evidence that simultaneous disruption of ccr5 and cxcr4 using ZFNs is capable of generating a pool of cells that are resistant to the major forms of HIV in vivo, and these cells can engraft, traffic normally to the spleen, and have a significant survival advantage in the presence of R5- and X4-using HIV.
Figure 5.
Cells lacking both ccr5 and cxcr4 following R5/X4-ZFN treatment have a survival advantage in vivo in the presence of HIV. (A) The frequency of human ccr5- and cxcr4-modified genes significantly increases in the R5/X4-ZFN treatment group over the course of the in vivo infection. ccr5 and cxcr4 disruption are stable over time in the absence of HIV infection suggesting there is no significant adverse effect of dual ZFN treatment. We performed Illumina deep sequencing of the R5 and X4-ZFN target sites and identified ZFN-induced mutations at these sites in 8 of 9 uninfected and 8 of 9 infected animals. We were unable to obtain sequence information from 1 infected animal due to limiting quantities of CD4+ T cells, and killed 1 uninfected animal following the development of GVHD, and this animal was thus excluded. (B) We stained whole blood from infected and uninfected animals in the R5/X4-ZFN treatment group 34 days postinfection with human antibodies to identify human CD4+ T cells and determine surface levels of R5 and X4. Both CCR5 and CXCR4 expression were significantly reduced on dual ZFN-treated cells in the presence of HIV challenge suggesting that coreceptor negative cells have an in vivo survival advantage due to ZFN treatment.
Discussion
Genetic approaches to control HIV infection have so far involved the removal of host genes required by the virus, or the introduction of antiviral genes that interfere with virus replication.47,48 Although both are attractive options, the latter approach could be hampered by the potential immunogenicity of foreign transgenes and the diversity of the HIV viral quasispecies in chronically infected individuals, which may allow for the outgrowth of viral variants resistant to these antiviral genes. Removal of essential host factors, such as the coreceptors, may present a greater challenge for the virus to overcome as suggested by the natural HIV resistance in ccr5Δ32 homozygotes and the functional cure achieved following the transplantation of ccr5Δ32 stem cells.24,25 Genetic editing through the use of coreceptor-specific ZFNs clearly renders a fraction of cells coreceptor-negative, as HIV-1 challenge of ZFN-treated cells results in preferential survival of edited cells lacking the targeted coreceptor.31,32 Treatment with R5-specific ZFNs results in permanent modification of 30% and 50% of ccr5 alleles in human hematopoietic stem cells (HSCs) and T cells, respectively,31,36 with a biallelic modification frequency of up to half of that.36 The efficiency of biallelic disruption becomes even more important when attempting to disrupt both ccr5 and cxcr4, necessitating the inactivation of 4 genes in a given cell to render it resistant to infection by virtually all HIV-1 strains.
Although inactivation of the viral coreceptors appears to be a logical approach to control HIV infection, what is less obvious is the amount of coreceptor ablation required to observe a therapeutic effect. Data from ccr5Δ32 heterozygotes suggest that even partial coreceptor ablation may be of clinical benefit as these individuals display delayed disease progression following HIV infection.49 As such, the current ZFN technology may provide a therapeutic benefit despite not completely eliminating coreceptor expression. In support of this, in a recent phase 1 study of HIV-infected patients who received autologous R5-ZFN–modified CD4+ T cells, a number of these patients exhibited viral load decreases compared with pretreatment levels following a 12-week interruption of ART (C.H.J., University of Pennsylvania, written communication, May 23, 2013). Of note, the viral load in 1 of these patients reached undetectable levels prior to the reinstatement of ART and further analysis revealed this patient to be a ccr5Δ32 heterozygote. Despite the modest number of patients enrolled in this study, it is tempting to speculate that even the relatively low levels of coreceptor ablation achieved by ZFN modification may be of therapeutic benefit, and that this benefit will only increase as the expression of either coreceptor is further reduced.
A potential limitation associated with any approach that targets CCR5 alone is the presence of virus strains that use CXCR4. Clinical failure associated with the use of the CCR5 antagonist maraviroc is either the result of an outgrowth of preexisting virus strains that use CXCR4 and continue to replicate despite CCR5 blockade,50 or mutations that arise in the viral glycoprotein allowing it to use the drug-bound form of CCR5,51 a resistance pathway not available to the virus when ccr5 is disrupted by ZFN technology. The fact that X4 HIV-1 strains can be present at low levels in chronically infected individuals provides a powerful rationale for using a strategy that targets both ccr5 and cxcr4.52 Although R5 ZFNs have been used to ablate ccr5 in HSCs to provide a self-renewing population of CCR5-negative cells, CXCR4 signaling plays a critical role in the bone marrow retention of HSCs.53,54 As a result, loss of CXCR4 expression on HSCs may result in their unwanted egress into the peripheral blood. Although less is known about the effects of the specific loss of CXCR4 expression on CD4+ T cells, a recent study of T-cell–specific cxcr4 knockout mice showed that these animals are born in normal numbers and display humoral and cellular responses indistinguishable from those of wild-type mice,55 suggesting that loss of CXCR4 expression on CD4+ T cells may be immunologically tolerated.
Our work and that of others provides compelling evidence for the ability of ZFN-mediated coreceptor ablation to protect CD4+ T cells and provide virologic control of HIV infections. However, a number of questions remain regarding the clinical efficacy of this approach and the ease with which it can be implemented. Although the original transplant using ccr5Δ32 cells resulted in a cure, other factors may have contributed to this outcome including reductions in reservoir size by the conditioning chemotherapy and graft-versus-host disease experienced by the patient. In fact, a recent study demonstrated that allogeneic stem cell transplant with a reduced-intensity chemotherapy regimen followed by multi-year ART, decreased the peripheral blood HIV reservoir size.56 Additionally, due to the efficacy of ART, future studies must determine whether ZFN-treated cells can sufficiently control viral replication, restore normal immune function, and decrease the viral reservoir, as these features of HIV infection must be impacted if we hope to achieve a cure,4,57 functional or otherwise, in HIV-infected individuals.
Supplementary Material
Acknowledgments
We thank Beatrice Hahn and Nicholas Parrish for providing HIV-1 virus stocks, and James Hoxie for generously providing the SupT1-R5 cell line. We also thank the University of Pennsylvania’s Center for Aids Research (CFAR) supported Human Immunology core for human CD4+ T cells, and the Stem Cell and Xenograft core. Additionally, we thank Xiaolong Fong for generously providing the Ad5/F35 vector, Frederick Bushman, Jason Wojcechowskyj, Nicholas Parrish, and Zahra Parker for helpful comments and technical advice. We also thank David Shivak, Kenneth Kim, and Jenny Jiacheng Yan at Sangamo Bioscienes Inc. for help in analyzing on- and off-target gene modification in ZFN-treated samples.
This work was supported by National Institutes of Health (NIH) grant U19 AI082628.
Footnotes
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: C.A.D., C.B.W., G.A.D.-D., J.L.R., P.D.G., C.H.J., M.C.H., and R.W.D. conceived and designed experiments; C.A.D., J.W., J.D., and A.J.S. carried out experiments; C.A.D., C.B.W., J.W., and R.W.D. analyzed and interpreted data; J.W., G.A.D.-D., J.L.R., P.D.G., C.H.J., and M.C.H. contributed reagents and analytical tools; and C.A.D., C.B.W., and R.W.D. wrote the manuscript.
Conflict-of-interest disclosure: J.W., J.D., P.D.G., and M.C.H. are employed by Sangamo Biosciences. The remaining authors declare no competing financial interests.
Correspondence: Robert W. Doms, Children’s Hospital of Philadelphia, 3401 Civic Center Blvd, 5th Floor Main Building, Philadelphia PA 19104; e-mail: domsr@e-mail.chop.edu.
References
- 1.Detels R, Muñoz A, McFarlane G, et al. Multicenter AIDS Cohort Study Investigators. Effectiveness of potent antiretroviral therapy on time to AIDS and death in men with known HIV infection duration. JAMA. 1998;280(17):1497–1503. doi: 10.1001/jama.280.17.1497. [DOI] [PubMed] [Google Scholar]
- 2.Palella FJ, Jr, Delaney KM, Moorman AC, et al. HIV Outpatient Study Investigators. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N Engl J Med. 1998;338(13):853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 3.Durand CM, Blankson JN, Siliciano RF. Developing strategies for HIV-1 eradication. Trends Immunol. 2012;33(11):554–562. doi: 10.1016/j.it.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siliciano JD, Siliciano RF. HIV-1 eradication strategies: design and assessment. Curr Opin HIV AIDS. 2013;8(4):318–325. doi: 10.1097/COH.0b013e328361eaca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deeks SG. HIV infection, inflammation, immunosenescence, and aging. Annu Rev Med. 2011;62:141–155. doi: 10.1146/annurev-med-042909-093756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maddon PJ, Dalgleish AG, McDougal JS, Clapham PR, Weiss RA, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47(3):333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 7.McDougal JS, Maddon PJ, Dalgleish AG, et al. The T4 glycoprotein is a cell-surface receptor for the AIDS virus. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 2):703–711. doi: 10.1101/sqb.1986.051.01.083. [DOI] [PubMed] [Google Scholar]
- 8.Alkhatib G, Combadiere C, Broder CC, et al. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272(5270):1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 9.Choe H, Farzan M, Sun Y, et al. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85(7):1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 10.Deng H, Liu R, Ellmeier W, et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381(6584):661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 11.Doranz BJ, Rucker J, Yi Y, et al. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85(7):1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 12.Dragic T, Litwin V, Allaway GP, et al. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381(6584):667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 13.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272(5263):872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 14.Schuitemaker H, Koot M, Kootstra NA, et al. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol. 1992;66(3):1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu T, Mo H, Wang N, et al. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261(5125):1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
- 16.Keele BF, Giorgi EE, Salazar-Gonzalez JF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A. 2008;105(21):7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann C. The epidemiology of HIV coreceptor tropism. Eur J Med Res. 2007;12(9):385–390. [PubMed] [Google Scholar]
- 18.Hunt PW, Harrigan PR, Huang W, et al. Prevalence of CXCR4 tropism among antiretroviral-treated HIV-1-infected patients with detectable viremia. J Infect Dis. 2006;194(7):926–930. doi: 10.1086/507312. [DOI] [PubMed] [Google Scholar]
- 19.Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR. Change in coreceptor use correlates with disease progression in HIV-1—infected individuals. J Exp Med. 1997;185(4):621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koot M, Keet IP, Vos AH, et al. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med. 1993;118(9):681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 21.Huang Y, Paxton WA, Wolinsky SM, et al. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2(11):1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 22.Liu R, Paxton WA, Choe S, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86(3):367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 23.Samson M, Libert F, Doranz BJ, et al. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382(6593):722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 24.Allers K, Hütter G, Hofmann J, et al. Evidence for the cure of HIV infection by CCR5Δ32/Δ32 stem cell transplantation. Blood. 2011;117(10):2791–2799. doi: 10.1182/blood-2010-09-309591. [DOI] [PubMed] [Google Scholar]
- 25.Hütter G, Nowak D, Mossner M, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360(7):692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 26.Hütter G, Zaia JA. Allogeneic haematopoietic stem cell transplantation in patients with human immunodeficiency virus: the experiences of more than 25 years. Clin Exp Immunol. 2011;163(3):284–295. doi: 10.1111/j.1365-2249.2010.04312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoxie JA, June CH. Novel cell and gene therapies for HIV. Cold Spring Harb Perspect Med. 2012;2(10) [DOI] [PMC free article] [PubMed]
- 28.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 29.Liang F, Han M, Romanienko PJ, Jasin M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc Natl Acad Sci U S A. 1998;95(9):5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holt N, Wang J, Kim K, et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28(8):839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez EE, Wang J, Miller JC, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26(7):808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilen CB, Wang J, Tilton JC, et al. Engineering HIV-resistant human CD4+ T cells with CXCR4-specific zinc-finger nucleases. PLoS Pathog. 2011;7(4):e1002020. doi: 10.1371/journal.ppat.1002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan J, Wang J, Crain K, et al. Zinc-finger nuclease editing of human cxcr4 promotes HIV-1 CD4(+) T cell resistance and enrichment. Mol Ther. 2012;20(4):849–859. doi: 10.1038/mt.2011.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doyon Y, Vo TD, Mendel MC, et al. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods. 2011;8(1):74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 35.Nilsson M, Ljungberg J, Richter J, et al. Development of an adenoviral vector system with adenovirus serotype 35 tropism; efficient transient gene transfer into primary malignant hematopoietic cells. J Gene Med. 2004;6(6):631–641. doi: 10.1002/jgm.543. [DOI] [PubMed] [Google Scholar]
- 36.Li L, Krymskaya L, Wang J, et al. Genomic editing of the HIV-1 coreceptor CCR5 in adult hematopoietic stem and progenitor cells using zinc finger nucleases. Mol Ther. 2013;21(6):1259–1269. doi: 10.1038/mt.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- 38.Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30(5):460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H, Zhou Y, Alcock C, et al. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J Virol. 2004;78(4):1718–1729. doi: 10.1128/JVI.78.4.1718-1729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parrish NF, Wilen CB, Banks LB, et al. Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin α4β7. PLoS Pathog. 2012;8(5):e1002686. doi: 10.1371/journal.ppat.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yi Y, Chen W, Frank I, et al. An unusual syncytia-inducing human immunodeficiency virus type 1 primary isolate from the central nervous system that is restricted to CXCR4, replicates efficiently in macrophages, and induces neuronal apoptosis. J Neurovirol. 2003;9(4):432–441. doi: 10.1080/13550280390218706. [DOI] [PubMed] [Google Scholar]
- 42.Brenchley JM, Silvestri G, Douek DC. Nonprogressive and progressive primate immunodeficiency lentivirus infections. Immunity. 2010;32(6):737–742. doi: 10.1016/j.immuni.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfaff JM, Wilen CB, Harrison JE, et al. HIV-1 resistance to CCR5 antagonists associated with highly efficient use of CCR5 and altered tropism on primary CD4+ T cells. J Virol. 2010;84(13):6505–6514. doi: 10.1128/JVI.00374-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pauwels K, Podevin N, Breyer D, Carroll D, Herman P. Engineering nucleases for gene targeting: safety and regulatory considerations. New Biotechnol. 2013 doi: 10.1016/j.nbt.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 45.Van Duyne R, Pedati C, Guendel I, et al. The utilization of humanized mouse models for the study of human retroviral infections. Retrovirology. 2009;6:76. doi: 10.1186/1742-4690-6-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Broder CC, Collman RG. Chemokine receptors and HIV. J Leukoc Biol. 1997;62(1):20–29. doi: 10.1002/jlb.62.1.20. [DOI] [PubMed] [Google Scholar]
- 47.Rossi JJ, June CH, Kohn DB. Genetic therapies against HIV. Nat Biotechnol. 2007;25(12):1444–1454. doi: 10.1038/nbt1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kiem H-P, Jerome KR, Deeks SG, McCune JM. Hematopoietic-stem-cell-based gene therapy for HIV disease. Cell Stem Cell. 2012;10(2):137–147. doi: 10.1016/j.stem.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dean M, Carrington M, Winkler C, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273(5283):1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 50.Fätkenheuer G, Nelson M, Lazzarin A, et al. MOTIVATE 1 and MOTIVATE 2 Study Teams. Subgroup analyses of maraviroc in previously treated R5 HIV-1 infection. N Engl J Med. 2008;359(14):1442–1455. doi: 10.1056/NEJMoa0803154. [DOI] [PubMed] [Google Scholar]
- 51.Moore JP, Kuritzkes DR. A pièce de resistance: how HIV-1 escapes small molecule CCR5 inhibitors. Curr Opin HIV AIDS. 2009;4(2):118–124. doi: 10.1097/COH.0b013e3283223d46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burnett JC, Zaia JA, Rossi JJ. Creating genetic resistance to HIV. Curr Opin Immunol. 2012;24(5):625–632. doi: 10.1016/j.coi.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aiuti A, Webb IJ, Bleul C, Springer T, Gutierrez-Ramos JC. The chemokine SDF-1 is a chemoattractant for human CD34+ hematopoietic progenitor cells and provides a new mechanism to explain the mobilization of CD34+ progenitors to peripheral blood. J Exp Med. 1997;185(1):111–120. doi: 10.1084/jem.185.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peled A, Petit I, Kollet O, et al. Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science. 1999;283(5403):845–848. doi: 10.1126/science.283.5403.845. [DOI] [PubMed] [Google Scholar]
- 55.Chung S-H, Seki K, Choi B-I, et al. CXC chemokine receptor 4 expressed in T cells plays an important role in the development of collagen-induced arthritis. Arthritis Res Ther. 2010;12(5):R188. doi: 10.1186/ar3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Henrich TJ, Hu Z, Li JZ, et al. Long-term reduction in peripheral blood HIV type 1 reservoirs following reduced-intensity conditioning allogeneic stem cell transplantation. J Infect Dis. 2013;207(11):1694–1702. doi: 10.1093/infdis/jit086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Margolis DM, Hazuda DJ. Combined approaches for HIV cure. Curr Opin HIV AIDS. 2013;8(3):230–235. doi: 10.1097/COH.0b013e32835ef089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.