Abstract
Macrophages play a role in innate immune responses to various foreign antigens. Many products from primary tumors influence the activation and transmigration of macrophages. Here, we investigated a migration of macrophages stimulated with cancer cell culture-conditioned medium (CM). Macrophage activation by treatment with CM of B16F10 cells were judged by the increase in protein levels of inducible nitric oxide synthase (iNOS) and cyclooxygenase 2 (COX2). The location where macrophages were at 4 h-incubation with control medium or CM was different from where they were at 5 h-incubation in culture dish. Percentage of superimposed macrophages at every 1 h interval was gradually increased by CM treatment as compared to control. Total coverage of migrated track expressed in coordinates was smaller and total distance of migration was shorter in CM-treated macrophages than that in control. Rac1 activity in CM-treated macrophages was also decreased as compared to that in control. When macrophages were treated with CM in the presence of dexamethasone (Dex), an increase in COX2 protein levels, and a decrease in Rac1 activity and total coverage of migration were reversed. In the meanwhile, biphasic changes were detected by Dex treatment in section distance of migration at each time interval, which was more decreased at early time and then increased at later time. Taken together, data demonstrate that macrophage motility could be reduced in accordance with activation in response to cancer cell products. It suggests that macrophage motility could be a novel marker to monitor cancer-associated inflammatory diseases and the efficacy of anti-inflammatory agents.
Keywords: Macrophage, Cell motility, Culture conditioned medium, Dexamethasone
INTRODUCTION
Cell migration plays a role in many physiological and pathological processes (Singer and Kupfer, 1986). Cell migration is a multistep cycle including extension of a protrusion, formation of stable attachments near the leading edge of the protrusion, translocation of the cell body forward, release of adhesions and retraction at the cell rear (Lauffenburger and Horwitz, 1996). Macrophages also transmigrate through the endothelial wall, basal membranes, and connective tissues to reach infection or inflammation sites (Cougoule et al., 2010; Aflaki et al., 2011).
Macrophages are resident phagocytic cells in lymphoid and nonlymphoid tissues (Gordon, 2002). Macrophages are critical effectors to be involved in steady-state tissue homeostasis, via the clearance of apoptotic cells, the production of growth factors, and the regulation of inflammation and innate immune response (Geissmann et al., 2010). Macrophages are the first line of host defense against microorganisms (Medzhitov, 2008), which is accomplished by phagocytosis and the production of inflammatory cytokines (Gordon, 2002). In addition, inflammatory chemoattractants induced by distant primary tumors influence the attraction of macrophages in secondary sites before metastasis (Hiratsuka et al., 2006). The presence of macrophages within tumors is a sign of a poor prognosis as they enhance angiogenesis and metastases (Balkwill et al., 2005; Condeelis and Pollard, 2006; Mantovani et al., 2008). However, the migration of activated macrophages has not yet been elucidated.
Many cellular factors such as Arp2/3 complex, profiling and FAK, are involved in cell migration (Borisy and Svitkina, 2000; Webb et al., 2002; Luedde, 2010). Cell migration is resulted from the integration and temporal coordination of many different processes in spatially distinct locations of cells (Webb et al., 2002). Wide variety of proteins is involved in forming a complex structure accompanying the re-organization of actin cytoskeleton during cell migration (Chen et al., 2000; Yamazaki et al., 2005). Cytoskeletal reorganization is dependent on small GTPases, including Rac1, Cdc42, and Rap1 (Takai et al., 1995; Takai et al., 2001; Bailey et al., 2009), which is also important in macrophage migration (Aflaki et al., 2011). However, little has been reported about the regulation of Rac1 in activated macrophages.
In the present work, we examined the mobility of macrophages stimulated with cancer cell culture-conditioned medium. In addition, we would like to establish a system testing macrophage motility for the application to screen drug candidates by using dexamethasone with a strong anti-inflammatory efficacy. We found that CM treatment reduced macrophage motility as judged by the percentage of superimposed cells, the coverage of migrated track in coordinate, and total distance of migration. Our data also showed that CM-induced changes in motility were reversed by the treatment with dexamethasone. It suggests that macrophage motility could be used as a novel marker for diagnosis or for the development of drug candidates to cancer-associated inflammatory disease.
MATERIALS AND METHODS
Reagents
Dexamethasone was purchased from Sigma chemical company (St. Louis, MO, USA). Antibody which is reactive with COX2 was obtained from Cayman Chemical Co. (Ann Arbor, MI, USA). Antibody to iNOS was obtained from Millipore Corporation. Except where indicated, all other materials are obtained from the Sigma chemical company (St. Louis, MO, USA).
Cell cultures
B16F10 mouse melanoma cells (H-2b) were obtained from the Korea Institute of Radiological and Medical Science (KIRMS) cell bank (Seoul, Korea). IC-21 mouse macrophage cells (H-2b) were obtained from Korea Research Institute of Bioscience and Biotechnology (Daejeon, Korea). B16F10 and IC- 21 cells were maintained and cultured in Dulbecco’s modified Eagle’s medium (DMEM) and RPMI 1640 medium (GIBCO, Grand Island, NY, USA), respectively, supplemented with 5% heat-inactivated fetal bovine serum (GIBCO, Grand Island, NY, USA), 2 mM L-glutamin, 100 units/ml penicillin and 100 units/ml streptomycin (GIBCO, Grand Island, NY, USA). Cells were incubated in an incubator at 37℃ in an atmosphere of 5% CO2 in air.
Analysis of macrophage migration
IC-21 macrophage culture dishes were focused under live cell imaging light microscope. Cells were treated with cultureconditioned medium (CM) of B16F10 cells. Video image of live macrophages were automatically taken for 12-15 h. Changes in image of each macrophage was analyzed with tracking program, ImageJ plugin MTrackJ (Version 1.5.0).
Measurement of active GTP-bound Rac1
The level of active GTP-bound Rac1 was determined using the GST-pulldown assay (Bos et al., 2001). In brief, cells were harvested and lysed. Then, Rac1-GTP was concentrated by the incubation of 500 μg of clarified cell lysates with GSTPBD of PAK in the presence of glutathione immobilized beads (Santa Cruz Biotechnology, CA) at 4℃ for 1 h with rotation. Unbound proteins were removed by centrifugation. Following washings with 1×PBS, the samples were eluted in 3×SDS sample buffer and run on SDS-PAGE. Rac1-GTP was detected by immunoblot analyses using the Rac1-specific antibodies.
Immunoblot analysis
Cells were lysed in ice-cold lysis buffer containing 0.5% Nonidet P-40 (vol/vol) in 20 mM Tris-HCl (pH 8.3); 150 mM NaCl; protease inhibitors (2 μg/ml aprotinin, pepstatin, and chymostatin; 1 μg/ml leupeptin and pepstatin; 1 mM phenyl-methyl sulfonyl fluoride (PMSF); and 1 mM Na4VO3. Lysates were incubated for 30 minutes on ice before centrifugation at 14,000 rpm for 5 minutes at 4℃. Proteins in the supernatant were denatured by boiling for 5 minutes in sodium dodecyl sulfate (SDS) sample buffer. Proteins were separated by 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membranes. Following transfer, equal loading of protein was verified by Ponceau staining. The membranes were blocked with 5% skim milk in Tris-buffered saline with Tween 20 (TBST) (10 mM Tris-HCl, pH 7.6; 150 mM NaCl; 0.5% Tween 20) and incubated with the indicated antibodies, monoclonal Myc (1:1,000), tubulin (1:5,000), and polyclonal mouse BAFF (1:1,000). Bound antibodies were visualized with HRP-conjugated secondary antibodies with the use of enhanced chemiluminescence (ECL) (Pierce, Rockford, IL).
Statistical analyses
Experimental differences were tested for statistical signifi-cance using ANOVA and Students’ t-test. p value of <0.05 or 0.01 was considered to be significant.
RESULTS
Treatment with cancer cell culture-conditioned medium (CM) decreased macrophage motility
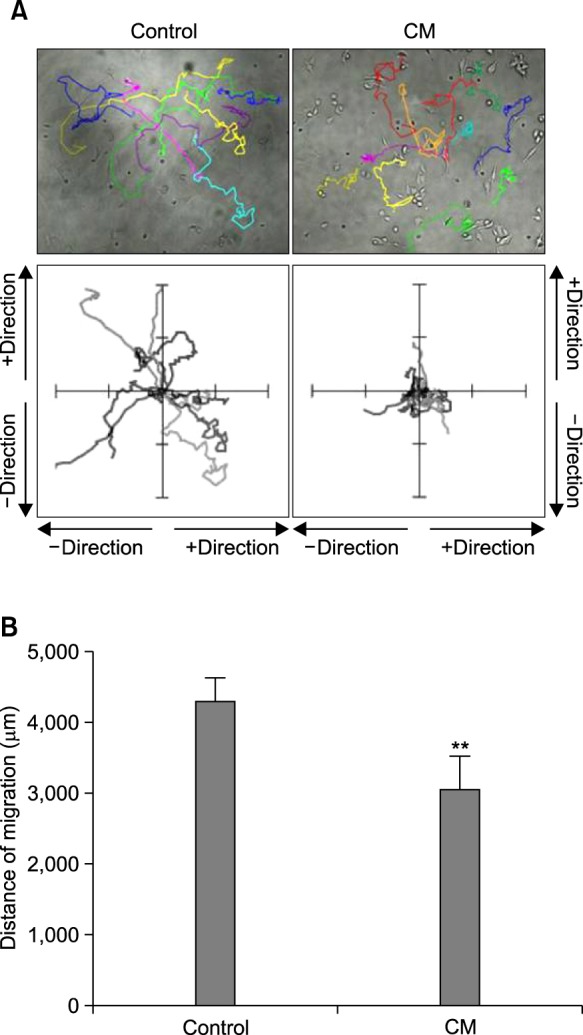
To study the effect of cancer cell culture-conditioned medium (CM) on macrophages, we used IC-21 macrophages. When macrophages were activated by the treatment with CM of B16F10 cells, protein levels of inducible nitric oxide synthase (iNOS) and cyclooxygenase 2 (COX2) were increased (Fig. 1A). We compared the location of macrophages between 4 h and 5 h after incubation. As shown in Fig. 1B, the location at 4 h-incubation was different from that at 5 h-incubation. We measured the number of superimposed cells at every 1 h interval during total incubation for 15 h. As the incubation time passed, percentage of superimposed macrophages was gradually increased by CM treatment as compared to control (Fig. 1C). As we also observed macrophages with various type of movement during incubation (Fig. 2A, top), each migrated track of macrophages was expressed in coordinates. Total coverage of migrated tack was smaller in a group of CMtreated macrophages than that in control (Fig. 2A, bottom). In addition, total distance of migration was significantly shorter in a group of CM-treated macrophages than that in control (Fig. 2B). Data demonstrate that macrophage motility could be reduced in activated macrophages.
Fig. 1. Macrophage motility was increased by the treatment with B16F10 melanoma cell culture-conditioned medium (CM). (A) IC- 21 cells were treated with CM. Each protein of iNOS and COX2 was detected by immunoblot analysis. (B) and (C) CM-treated macrophages were photographed for each time. Cells were shaped with different color, yellow for 4 h-incubation and green for 5 h-incubation. Then, two colors were overlapped to compare the changes in location by movement (B). Percentage of superimposed cells was analyzed at each time interval. Data were the representative of three experiments. Data in the bar graph represent the means ± SED. *p<0.05; **p<0.01, significantly different from control group.
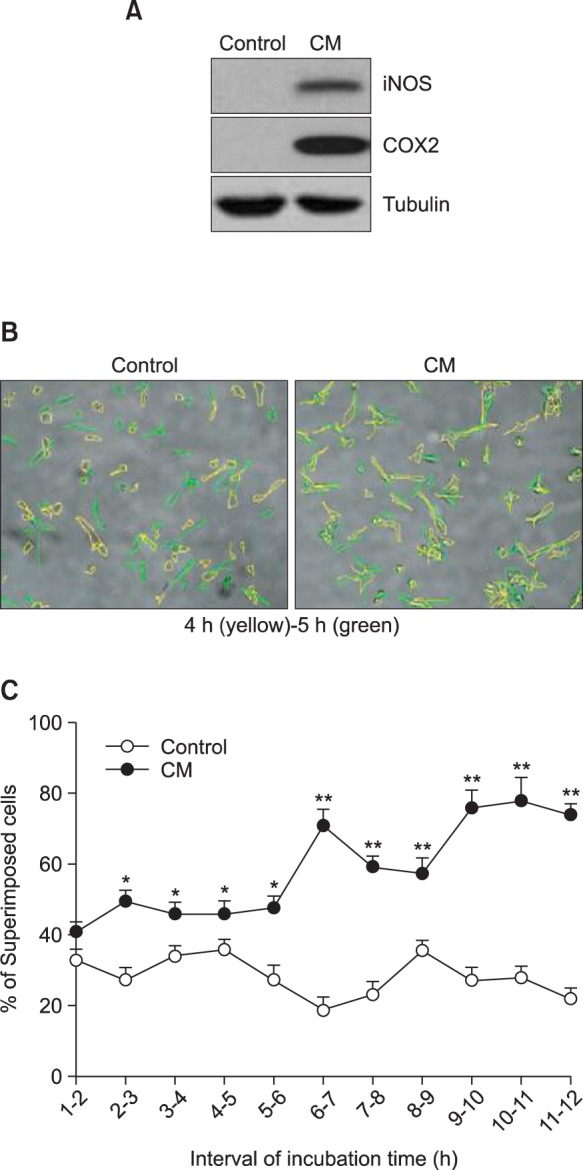
Fig. 2. Total area and distance of macrophage movement were reduced by CM treatment. (A) and (B) IC-21 cells were treated with CM. Macrophage motility was measured and the changes in image of each macrophage were analyzed with tracking program, ImageJ plugin MTrackJ (Version 1.5.0) by the method described in materials and methods. Movement track was presented with colored line (A, top) or in coordinates for each cell (A, bottom). Total distance of macrophages was calculated for 15 h-migration. Data were the representative of three experiments. Data in the bar graph represent the means ± SED. **p<0.01, significantly different from control group (B).
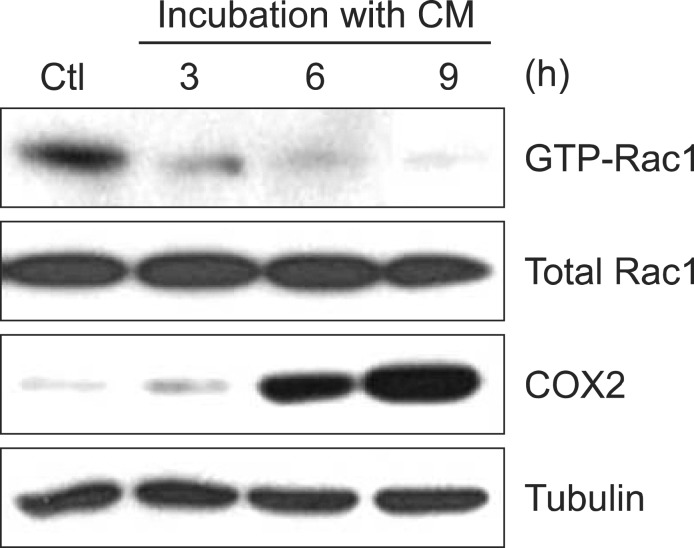
Previous reports showed that various proteins are involved in the course of cell migration and adhesion and form a complex structure accompanying the re-organization of actin cytoskeleton (Chen et al., 2000; Yamazaki et al., 2005). Given that Rac1 plays a role in regulation of actin dynamics (Takai et al., 1995; Takai et al., 2001; Kometani et al., 2004; Yamazaki et al., 2005), we measured Rac1 activity in CM-treated macrophages. As shown in Fig. 3, Rac1 activity in CM-treated macrophages was decreased as compared to that in control macrophages. It suggests that macrophage motility could be regulated byvity in the activated macrophages.
Fig. 3. GTP-bound Rac1 activity was inhibited by CM treatment. IC-21 cells were treated with CM for the indicated times (3, 6, and 9 h). Rac1 activity was measured by GST-Pulldown assay as described in materials and methods. COX2 proteins were detected by immunoblot analysis.
Macrophage motility decreased by CM was reversed by the treatment with dexamethasone
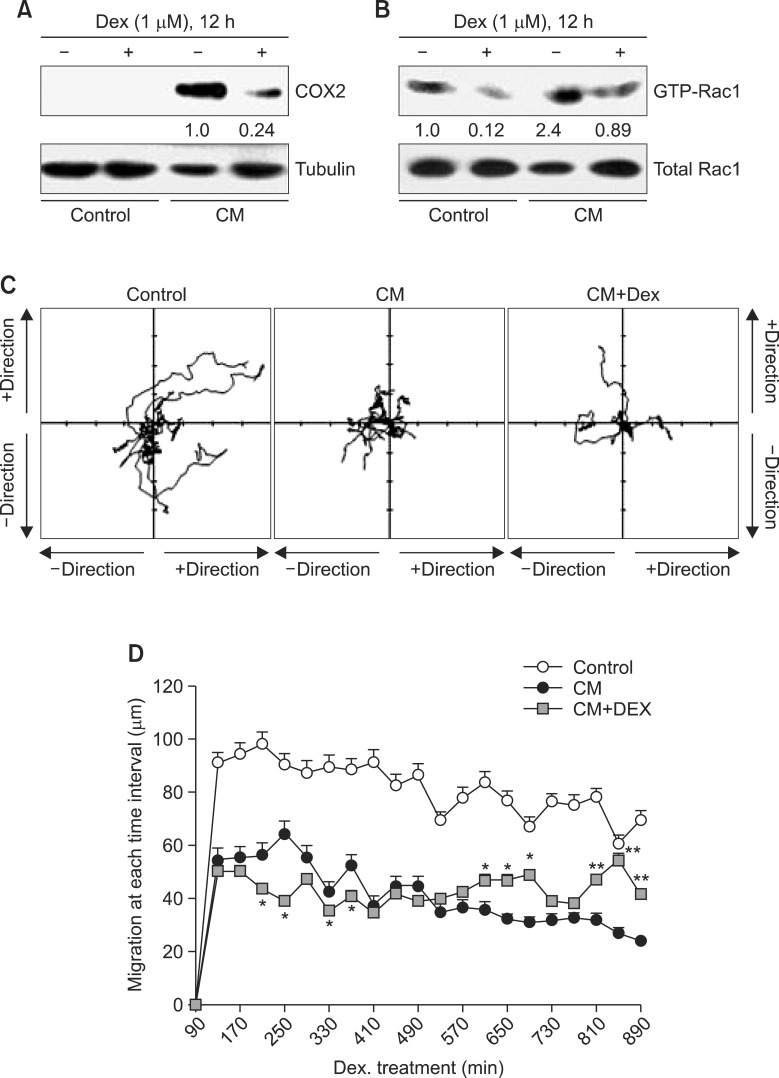
To examine whether macrophage motility is regulated by inflammatory responses, the effect of dexamethasone (Dex), a famous anti-inflammatory agent, was measured in CMtreated IC-21 cells. When IC-21 cells were treated with CM in the presence of Dex, COX2 inflammatory protein levels were significantly decreased as compared to that in CM-treated control group (Fig. 4A). In addition, Rac1 activity was a little increased in Dex-treated group as compared to that in CMtreated control (Fig. 4B). Total shape of coverage on macrophage migration was comparable to the group of CM-treated macrophages (Fig. 4C). In the meanwhile, biphasic changes were detected in section distance of Dex-treated macrophage migration at each time interval (Fig. 4D). Section distance was decreased for about 7 h and then increased at later time after the treatment with Dex. Data demonstrate that a reduction of macrophage motility could be associated with inflammatory activation in response to cancer cell products. It suggests that macrophage motility could be a novel marker to monitor efficacy of anti-inflammatory agents.
Fig. 4. CM-induced decrease in macrophage motility was reversed by the treatment with dexamethasone. (A) and (B) IC-21 cells were treated with CM in the presence or absence of dexamethasone (Dex). COX2 proteins were detected by immunoblot analysis (A). Rac1 activity was measured by GST-Pulldown assay as described in materials and methods (B). Band intensity to control protein was normalized with NIH image analysis software (version 1.62). Fold increase to control group was represented under each band (A and B). Macrophage motility was measured and the changes in image of each macrophage was analyzed with tracking program, ImageJ plugin MTrackJ (Version 1.5.0) by the method described in materials and methods. Movement track was presented in coordinates for each cell (C). Moved distance of macrophages was calculated at every 40 min interval for 15 h. Data were the representative of four experiments. Data in the bar graph represent the means ± SED. *p<0.05; **p<0.01, significantly different from CM-treated group (D).
DISCUSSION
Cell migration plays a role in maintaining physiological homeostasis (Singer and Kupfer, 1986). Transmigration of monocytes and macrophages into inflammation or infection site is critical in the progression of diseases (Cougoule et al., 2010; Aflaki et al., 2011). Cell migration is dependent on many cellular factors such as small Rho GTPase family (RhoA, Cdc42, Rac) involved in signal transduction and the continuous organization of the actin cytoskeleton (Takai et al., 1995; Borisy and Svitkina, 2000; Takai et al., 2001; Webb et al., 2002; Bailey et al., 2009; Luedde, 2010; Aflaki et al., 2011). A little has been answered about the molecular and cellular mechanisms to regulate macrophage motility. Several signaling molecules were reported to be involved in macrophage migration. Matrix metalloproteinase 9 is linked to the recruitment of macrophages in an aortic aneurysm model (Gong et al., 2008). The Src-family protein tyrosine kinases, Hck, Fgr, and Lyn, regulate macrophage migration and degranulation (Baruzzi et al., 2008; Cougoule et al., 2010). In addition, migration of resting macrophages was induced by Rac1 activation (Aflaki et al., 2011). Our data showed that macrophage migration was inhibited in the state of stimulation with CM (Fig. 1,2). Our data also showed that migration in activated macrophages was associated with a decrease in Rac1 activity (Fig. 3). It suggests that Rac1 might regulate migration of macrophages that is resting or activated state.
The presence of macrophages is associated with a poor prognosis in tumors (Balkwill et al., 2005; Condeelis and Pollard, 2006; Mantovani et al., 2008). Therefore, it is becoming a challenge to specifically control tissue infiltration of macrophages (Cougoule et al., 2010). Furthermore, new anti-inflammatory and antitumor-based drugs could be developed by targeting macrophage migration-related molecules (Luster et al., 2005; Mackay, 2008). Our results also showed that Dex reversed CM-induced changes in macrophage migration that was represented by total coverage in coordinates, total distance and Rac1 activity (Fig. 4). However, we still have no information about the reason why Rac1 activity was increased in CM-untreated group. Our data support a previous report that macrophage migration could be a target for drug development. In addition, it suggests that innate immune responses by tumor-producing products could be accompanied with a decrease in macrophage motility for a long time interaction between tumor cells and macrophages in tumor microenvironment.
Overall, although it has not been cleared all of possible signal transduction on CM-activated macrophage migration, this work suggests that CM could negatively regulate macrophage migration for a long time interaction with tumor cells. Data also suggest that CM-induced macrophages may crosstalk to inhibit immune responses in tumor microenvironment through the inhibition of migration. It may provide additional information to develop a new candidate for the regulation of macrophage migration activated by tumor cell-producing products. In addition, data suggest that our experimental system to test macrophage motility could be useful for screening drug candidates.
Acknowledgments
This work was supported by Grants from Mid-career Researcher Program (#2012-R1A2A2A01005449), and Nuclear R&D Program (#2013M2B2A9A03051296 and 2010-0018545) through National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (MEST).
References
- 1.Aflaki E., Balenga N. A., Luschnig-Schratl P., Wolinski H., Povoden S., Chandak P. G., Bogner-Strauss J. G., Eder S., Konya V., Kohlwein S. D., Heinemann A., Kratky D. Impaired Rho GTPase activation abrogates cell polarization and migration in macrophages with defective lipolysis. Cell. Mol. Life Sci. (2011);68:3933–3947. doi: 10.1007/s00018-011-0688-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey C. L., Kelly P., Casey P. J. Activation of Rap1 promotes prostate cancer metastasis. Cancer Res. (2009);69:4962–4968. doi: 10.1158/0008-5472.CAN-08-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balkwill F., Charles K. A., Mantovani A. Smoldering and polarized inflammation in the initiation and promotion of malignant disease. Cancer Cell. (2005);7:211–217. doi: 10.1016/j.ccr.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Baruzzi A, Caveggion E., Berton G. Regulation of phagocyte migration and recruitment by Src-family kinases. Cell. Mol. Life Sci. (2008);65:2175–2190. doi: 10.1007/s00018-008-8005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borisy G. G., Svitkina T. M. Actin machinery: pushing the envelope. Curr. Opin. Cell Biol. (2000);12:104–112. doi: 10.1016/S0955-0674(99)00063-0. [DOI] [PubMed] [Google Scholar]
- 6.Bos J. L., de Rooij J., Reedquist K. A. Rap1 signalling: adhering to new models. Nat. Rev. Mol. Cell Biol. (2001);2:369–377. doi: 10.1038/35073073. [DOI] [PubMed] [Google Scholar]
- 7.Chen H., Bernstein B. W., Bamburg J. R. Regulating actin-filament dynamics in vivo. Trends Biochem. Sci. (2000);25:19–23. doi: 10.1016/S0968-0004(99)01511-X. [DOI] [PubMed] [Google Scholar]
- 8.Condeelis J., Pollard J. W. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. (2006);124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Cougoule C., Le Cabec V., Poincloux R., Al Saati T., Mege J. L., Tabouret G., Lowell C. A., Laviolette-Malirat N., Maridonneau-Parini I. Three-dimensional migration of macrophages requires Hck for podosome organization and extracellular matrix proteolysis. Blood. (2010);115:1444–1452. doi: 10.1182/blood-2009-04-218735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geissmann F., Manz M. G., Jung S., Sieweke M. H., Merad M., Ley K. Development of monocytes, macrophages, and dendritic cells. Science. (2010);327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong Y., Hart E., Shch A., Hoover-Plow J. Inflammatory macrophage migration requires MMP-9 activation by plasminogen in mice. J. Clin. Invest. (2008);118:3012–3024. doi: 10.1172/JCI32750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon S. Pattern recognition receptors: doubling up for the innate immune response. Cell. (2002);111:927–930. doi: 10.1016/S0092-8674(02)01201-1. [DOI] [PubMed] [Google Scholar]
- 13.Hiratsuka S., Watanabe A., Aburatani H., Maru Y. Tumour-mediated upregulation of chemoattractants and recruitment of myeloid cells predetermines lung metastasis. Nat. Cell Biol. . (2006);8:1369–1375. doi: 10.1038/ncb1507. [DOI] [PubMed] [Google Scholar]
- 14.Kometani K., Ishida D., Hattori M., Minato N. Rap1 and SPA-1 in hematologic malignancy. Trends Mol. Med. (2004);10:401–408. doi: 10.1016/j.molmed.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Lauffenburger D. A., Horwitz A. F. Cell migration: a physically integrated molecular process. Cell . (1996);84:359–369. doi: 10.1016/S0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 16.Luedde T. MicroRNA-151 and its hosting gene FAK (focal adhesion kinase) regulate tumor cell migration and spreading of hepatocellular carcinoma. Hepatology . (2010);52:1164–1166. doi: 10.1002/hep.23854. [DOI] [PubMed] [Google Scholar]
- 17.Luster A. D., Alon R., von Andrian U. H. Immune cell migration in inflammation: present and future therapeutic targets. Nat. Immunol. (2005);6:1182–1190. doi: 10.1038/ni1275. [DOI] [PubMed] [Google Scholar]
- 18.Mackay C. R. Moving targets: cell migration inhibitors as new anti-inflammatory therapies. Nat. Immunol. (2008);9:988–998. doi: 10.1038/ni.f.210. [DOI] [PubMed] [Google Scholar]
- 19.Mantovani A., Allavena P., Sica A., Balkwill F. Cancerrelated inflammation. Nature. (2008);454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 20.Medzhitov R. Origin and physiological roles of inflammation. Nature. (2008);454:428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- 21.Singer S. J., Kupfer A. The directed migration of eukaryotic cells. Annu. Rev. Cell Biol. (1986);2:337–365. doi: 10.1146/annurev.cb.02.110186.002005. [DOI] [PubMed] [Google Scholar]
- 22.Takai Y., Sasaki T., Matozaki T. Small GTP-binding proteins. Physiol. Rev. (2001);81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 23.Takai Y., Sasaki T., Tanaka K., Nakanishi H. Rho as a regulator of the cytoskeleton. Trends Biochem. Sci. (1995);20:227–231. doi: 10.1016/S0968-0004(00)89022-2. [DOI] [PubMed] [Google Scholar]
- 24.Webb D. J., Parsons J. T., Horwitz A. F. Adhesion assembly, disassembly and turnover in migrating cells -- over and over and over again. Nat. Cell Biol. (2002);4:E97–100. doi: 10.1038/ncb0402-e97. [DOI] [PubMed] [Google Scholar]
- 25.Yamazaki D., Kurisu S., Takenawa T. Regulation of cancer cell motility through actin reorganization. Cancer Sci. (2005);96:379–386. doi: 10.1111/j.1349-7006.2005.00062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]