Abstract
Exposure to environmental xenoestrogens is a major health concern due to the ability of these compounds to perturb estrogen receptor (ER) signaling and act as endocrine disrupting compounds (EDCs). Inappropriate exposure to EDCs during development, even at low doses, can predispose individuals to an increased lifetime risk of disease, including cancer. Recent data indicate that perinatal exposure to EDCs increases cancer risk by (re)programming the epigenome via alterations in DNA and histone methylation. We, and others have begun to dissect the mechanisms by which xenoestrogens disrupt the epigenetic machinery to reprogram the epigenome and induce developmental reprogramming. Our studies revealed that xenoestrogens induce nongenomic ER signaling to activate PI3K/AKT, resulting in AKT phosphorylation and inactivation of the histone methyltransferase EZH2, thus providing a direct link to disruption of the epigenome. Other epigenetic “readers, writers, and erasers” may also be targeted by nongenomic signaling, suggesting this is a central mechanism by which xenoestrogens and other EDCs disrupt the epigenome to induce developmental reprogramming. Elucidating mechanisms of developmental reprogramming of the epigenome is important for understanding how environmental exposures increase cancer risk, and provides a rationale for developing epigenetic interventions that can reverse the effects of environmental exposures to reduce cancer risk.
BACKGROUND
Nongenomic Signaling by Nuclear Hormone Receptors
The nuclear hormone receptor (NHR) superfamily comprises of receptors for thyroid and steroid hormones, retinoids and vitamin D, and also orphan receptors with unknown ligands. For steroids, NHR activity via the genomic pathway involves hormone binding to cytosolic receptors, nuclear translocation and the subsequent binding of the ligand-activated hormone receptors to hormone responsive elements in chromatin to modulate gene transcription. In addition to this classic genomic mechanism, it is now appreciated that when liganded to NHRs including the estrogen, progesterone, androgen, aryl hydrocarbon, and peroxisome proliferator-activated receptors, endogenous and environmental hormones activate nongenomic or membrane-initiated signaling pathways that activate several kinase cascades. This rapid and transient nongenomic signaling occurs in seconds to minutes, compared to genomic signaling, which occurs over hours to days (1-8).
Many of the kinases and pathways involved in nongenomic signaling have been identified. Activation of nongenomic signaling by NHRs primarily initiates at the cell membrane. Estrogen receptors (ERα and ERβ), progesterone receptors (PR-A and PR-B), and the androgen receptor (AR) have been shown to localize to the cell membrane in various cell types (9). The E domain of the NHRs has a conserved palmitoylation motif, which is responsible for membrane localization (9). In the case of the ER, palmitoylation enhances membrane localization, interaction with caveolin-1, and nongenomic activities including activation of MAPK and PI3K signaling (10).
Activation of PI3K/AKT and MAPK signaling are well-characterized examples of nongenomic signaling pathways activated by many NHR. Several studies have shown that PI3K/AKT and MAPK signaling mediates numerous functions evoked by activation of NHRs such as cell growth, motility, differentiation, survival and apoptosis (11, 12). Estrogen receptors have been shown to reside in the lipid rafts (13, 14) where association between ER and PI3K occur to activate PI3K/AKT (15). In the case of PI3K signaling, ERα (but not ERβ) directly interacts with the p85α regulatory subunit of PI3K and this interaction is required to induce non-genomic signaling in breast and endothelial cells (16-18). Complex interactions between AR, p85α, and Src are involved in AR activation of PI3K signaling (4). Membrane-localized AR is found in caveolae, where this NHR interacts with caveolin-1 and c-Src to facilitate activation of c-Src/PI3K/AKT cascade and subsequent activation of eNOS (19). Other studies have also demonstrated that AR and ER associate with Src to activate MAPK signaling leading to cell survival and proliferation (20-22). In addition to PI3K and MAPK, numerous studies have demonstrated that non-genomic signaling of membrane-localized ERs activates other signaling cascades including PKA; PLC/PKC; JAK/STAT; Pak1; casein kinase I-γ2; sphingosine kinase; CaMKIV, tyrosine kinase Src, p21ras, adenylyl cyclase and PKG (23-25).
Several reports from our group and others have demonstrated that in addition to endogenous estrogen (17-β estradiol), low concentrations of xenoestrogens such as bisphenol A (BPA), genistein (GEN), and diethylstilbestrol (DES), can induce nongenomic ER signaling. Acting in pico to nanomolar concentrations, these xenoestrogens are potent activators of nongenomic signaling as exemplified by raising intracellular calcium levels, activating eNOS and signaling cascades such as PI3K/AKT and MAPK in cells and tissues, including the uterus and prostate (5, 26-34). Importantly, these experimental xenoestrogen doses fall within the range of what had been reported in human studies. For example, 0.3 to 5 ng/ml (~1-20 nM) BPA has been detected in adult and fetal human plasma, urine, and breast milk (35). In the case of genistein, serum levels ranged from 2μg/L (7 nM) to 25μg/L (92.5 nM) in Asian women and non-vegetarian women, respectively (36). Xenoestrogen exposure at critical windows of development can have wide-spread deleterious effects. Xenoestrogens such as BPA, GEN and DES that function as EDCs can disrupt normal organogenesis, reduce fecundity, alter sexual behavior and memory, and cause malformations and decreased sperm mobility. Early life exposures to xenoestrogens and other EDCs can increase susceptibility to chronic diseases such as obesity, diabetes mellitus, asthma, and cancer (37).
The role of non-genomic signaling by xenoestrogens in mediating these adverse effects is just becoming known. Importantly, nongenomic signaling provides a mechanism by which NHR activation by xenoestrogens and other EDCs can induce phosphorylation (and perhaps other post-translational modifications) of numerous downstream proteins and alter their activity. For example, recent studies from our group have demonstrated that 17-β estradiol , BPA, GEN and DES evoke rapid non-genomic signaling that regulates the histone methyltransferase (HMT) enhancer of Zeste homolog 2 (EZH2) in breast, uterus and prostate cells and tissues (26, 27). Interestingly, activation of non-genomic signaling by xenoestrogens exhibits tissue-specificity; BPA for example, activates nongenomic PI3K/AKT signaling in the prostate but not in the uterus (26).
Xenoestrogen-induced Developmental Reprogramming Increases Susceptibility to Cancer
Exposure of developing tissues or organs to an adverse stimulus or insult during critical periods of development can permanently reprogram normal physiological responses in a manner which promotes diseases later in life. This process, termed developmental reprogramming, is now known to increase risk in adulthood for many diseases, including cancer. Multiple lines of evidence from human and animal studies have established that epigenetic alterations induced by developmental reprogramming are responsible for the altered patterns of gene expression in adulthood that underlie this increased cancer risk. For example, in the uterus, perinatal EDC exposure reprograms the expression of many estrogen-responsive genes, so that in the adult uterus these genes become hyper-responsive to estrogen, increasing the risk for development of hormone-dependent tumors such as endometrial hyperplasia/carcinoma and uterine leiomyoma (38, 39). Developmental reprogramming of cancer susceptibility by environmental exposures has recently been reviewed (40), and will not be covered in detail here where we focus on the signaling pathways by which EDCs engage the cell's epigenetic machinery to induce developmental reprogramming of the epigenome to increase cancer risk.
Nongenomic Signaling Regulates the Activity of “Readers, Writers and Erasers” of the Epigenome During Developmental Reprogramming
Epigenetic alterations are now appreciated to contribute to the underlying mechanisms by which environmental exposures influence health and disease, including susceptibility to cancer induced by developmental reprogramming. Histone modification, one of the best characterized epigenetic modification, directly impacts DNA accessibility and chromatin structure to regulate gene expression. It is now thought that combinatorial sets of specific histone modifications are written by HMTs (‘writers’), removed by histone demethylases (HMDs) (‘erasers’), and recognized by effector proteins (‘readers’) which are recruited and bind to histone modifications via specific domains, to constitute a ‘histone code’ (41, 42). These “reading, writing, and erasing” activities remodel chromatin to regulate biological processes such as transcription, DNA replication and repair.
The mechanisms that regulate epigenetic “readers, writers and erasers” are only now beginning to be understood. We initially postulated that these epigenetic programmers were targets for non-genomic signaling cascades, which would provide a direct link between environmental exposures and alterations in the epigenome. For instance, IGF-R signaling to PI3K/AKT induces EZH2 phosphorylation at serine 21, which inhibits its HMT activity, decreasing lysine 27 trimethylation of histone H3 (H3K27me3) and consequently, increasing gene expression due to loss of this repressive methyl mark (43). Importantly, we showed that xenoestrogen-induced activation of PI3K/AKT by non-genomic signaling also caused AKT to phosphorylate EZH2 at serine 21, inactivating its HMT activity and reducing global levels of H3K27me3. This discovery provided a direct linkage between xenoestrogen-induced NHR signaling and modulation of the cell's epigenetic machinery.
Other links between cell signaling pathways and epigenetic programmers also exist. Wnt5a has been shown to activate Nemo-like kinase (NLK) through CaMKII and MAPKKK TAK1/TAB2 signaling. Activated NLK can phosphorylate the HMT SETDB1, which leads to the formation of an active co-repressor complex, an increase in the H3K9me3 SETDB1 repressive methyl mark and silencing of target genes such as PPARγ (44). Neurotrophic factors such as BDNF and NGF activate neuronal NOS which nitrosylates GAPDH, enabling it to bind to Siah and translocate to the nucleus. In a ternary complex that comprises of GAPDH-Siah and the HMT SUV39H1, Siah ubiquitinates SUV39H1, resulting in loss of SUV39H1 HMT activity and a decrease in the repressive H3K9me3 methyl mark, thus facilitating activation of target genes (45).
Several other HMTs can also be regulated by phosphorylation. For example, PR-Set7 is phosphorylated by CDK1/cyclin B complex at serine 29 and though this phosphorylation has no impact on its HMT activity, it removes PR-Set7 from mitotic chromosomes and confers protein stability during mitosis (46). SU(VAR)3-9 is phosphorylated by chromosomal kinase JIL-1 at the N-terminus but this PTM does not alter its ability to repress transcription (47). Cyclin-dependent kinase 1 (CDK1) phosphorylates EZH2 at threonine 345 and 487 to target EZH2 for ubiquitination and subsequent proteasomal degradation (48). The latter PTM is also critical for EZH2 interaction with PRC2 components SUZ12 and EED and its HMT activity (43). Carm1 is phosphorylated at serine 217 by an unidentified kinase but this PTM is essential for S-adenosylmethionine binding and HMT activity (49). ENX2 is phosphorylated in vivo but neither the site nor the biological function of this PTM is understood (50).
Mammalian histone demethylases (HDMs) can also be post-translationally modified via phosphorylation (51). One recent example is PHF2, a JmjC demethylase, which converts to an active H3K9me2 demethylase when phosphorylated by PKA. Activated PHF2 then associates with ARID5B to induce demethylation of methylated ARID5B. This PTM also mediates targeting of PHF2-ARID5B complex to promoters where it removes the dimethyl H3K9 mark (52). Another example is PHF8, which is a H4K20me1 demethylase that can function as a cell cycle regulator, partially based on its demethylase activity. When phosphorylated by CDK1, PHF8 dissociates from chromatin in prophase. Concomitantly, increased expression of Pr-Set7 is observed which leads to a surge of H4K20me1 mark that has the ability to interact with the Condensin II complex (53). The H3K36 demethylase Rph1 has been reported to repress transcription of PHR1 gene in a HDM-dependent manner. Rph1 is phosphorylated at serine 652 by a Rad53 kinase dependent manner and this PTM potentially triggers the dissociation of Rph1 from chromatin and modulates transcriptional de-repression of PHR1 gene during DNA damage (54).
Phosphorylation of chromatin effector proteins (epigenetic “readers”) has been described as in the case for Heterochromatin protein 1γ (HP1γ). HP1γ binds to H3K9me3 via its chromodomain, and has been shown to be targeted by PKA for phosphorylation at serine 83. This phosphorylation event is essential for HP1γ's localization in the euchromatin and interaction with Ku70 but functionally impaired its silencing activity (55). Another HP1 homologue, HP1β, is phosphorylated by casein kinase 2 in response to DNA damage. Phosphorylation at its chromodomain leads to rapid release of HP1β from H3K9me3, followed by re-association at later times (56, 57).
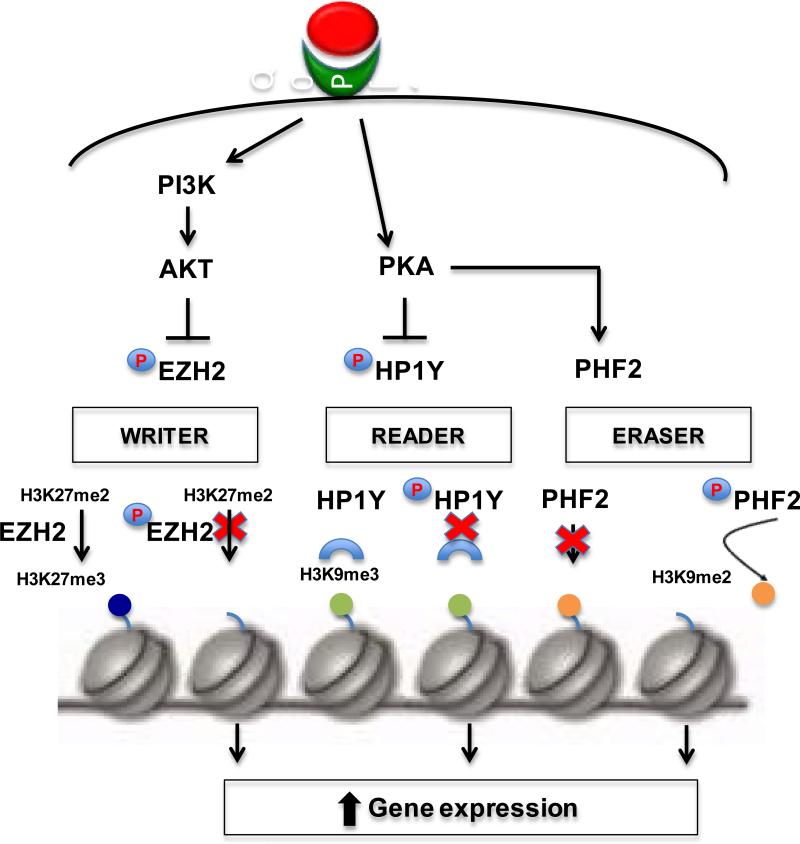
The examples above demonstrate that many epigenetic “readers, writers, and erasers” can be targeted by kinases activated by nongenomic signaling, suggesting that non-genomic signaling may be a central mechanism by which EDCs engage the developing epigenome to induce developmental reprogramming (Figure 1). Since we still know relatively little about how epigenetic programmers are regulated, conceivably, most, if not all ‘readers, writer, and erasers” may be regulated by post-translational modifications such as phosphorylation, making them targets for non-genomic signaling. A summary is provided in Table 1 of key nongenomic signaling pathways and “readers, writers and erasers” known to be, or potentially, regulated by kinases in non-genomic signaling pathways.
Figure 1. Non-genomic signaling pathways that modulate the activity of epigenetic ‘readers, writers, and erasers’.
Endogenous and environmental ligands bind to NHRs to activate nongenomic signaling and kinase cascades. Activated kinases such as AKT and PKA phosphorylate and inhibit the activity of epigenetic “writers” such as the HMT EZH2 and “readers” such as HP1γ respectively. The “eraser” HDM PHF2, also a PKA substrate, becomes activated when phosphorylated. In all three scenarios, the result is increased gene expression, due to loss of, or inability to read, repressive histone methyl marks.
Table 1.
Examples of signaling cascades activated by ligand-activated hormone receptors which impinge on epigenetic regulators, altering gene expression to result in increased tumor susceptibility
| Examples | |
|---|---|
| Ligand ↓ |
17-β estradiol, xenoestrogens (e.g. BPA, DES Genistein) |
| Nuclear Hormone Receptor ↓ |
ER, PR, AR, Ahr, PPARy |
| Non-genomic Signaling ↓ |
Pl3K/AKT, PKA, MAPK, PLC/PKC, PKG, JAK/STAT, Pak1, casein kinase I-γ2, sphingosine kinase, CaMKIV, tyrosine kinase Src, p21ras, adenylyl cyclase etc. |
| Writers, Erasers, Readers ↓ |
EZH2, SETDB1, SUV39H1, PR-Set7, SU(VAR)3−9, Carm1, ENX2, PHF2, PHF8, Rph1, HP1Y, HP1β |
| (re) Programming of the Epigenome ↓ |
Altered H3K27me3, H3K9me2/3, H4K20me1, H3R17me2, H3K36me2/3 |
| Altered gene expression ↓ |
HOXA10, PDE4D4, LTF, FOS, HMGN5, CALB3, GRIA2, GDF10, MMP3 |
| Increased susceptibility to tumorigenesis ↓ |
Uterine and Prostate cancer |
CLINICAL-TRANSLATIONAL ADVANCES
Exposures to EDCs such as xenoestrogens are a major health concern because of the ubiquitous nature of exposures to some of these compounds, and the potential for these exposures, especially when they occur during key developmental windows, to cause life-long changes in the epigenome that increase susceptibility to diseases of adulthood including diabetes, obesity, metabolic syndrome, infertility, and cancer. Importantly, the persistent epigenetic changes induced by developmental reprogramming are present prior to disease onset, presenting an opportunity for developing screens for epigenetic alterations that identify individuals at increased risk, and the development of effective interventions to reduce cancer risk. Just as importantly, the epigenetic alterations induced by developmental reprogramming may be reversible with epigenetic therapy, or life-style interventions such as diet and exercise. In this regard, now is the time for additional effort geared towards identifying tissue- and disease-specific epigenetic signatures induced by developmental reprogramming to determine if these epigenetic “fingerprints” can be employed as reliable biomarkers of environmental exposure and disease risk. The identification of such epigenetic biomarkers will be useful not only for identifying at-risk individuals, but to determine if epigenetic (or other) therapies can reverse the effects of developmental reprogramming to decrease cancer risk.
Acknowledgments
GRANT SUPPORT: This work is supported by grants from the National Institute of Environmental Health Sciences to Cheryl L. Walker (R01ES008263 and RC2ES018789).
Footnotes
DISCLOSURE OF CONFLICTS STATEMENT: The authors have nothing to disclose.
REFERENCES
- 1.Dong B, Cheng W, Li W, Zheng J, Wu D, Matsumura F, et al. FRET analysis of protein tyrosine kinase c-Src activation mediated via aryl hydrocarbon receptor. Biochim Biophys Acta. 2011;1810:427–31. doi: 10.1016/j.bbagen.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faivre EJ, Lange CA. Progesterone receptors upregulate Wnt-1 to induce epidermal growth factor receptor transactivation and c-Src-dependent sustained activation of Erk1/2 mitogen-activated protein kinase in breast cancer cells. Mol Cell Biol. 2007;27:466–80. doi: 10.1128/MCB.01539-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jimenez R, Sanchez M, Zarzuelo MJ, Romero M, Quintela AM, Lopez-Sepulveda R, et al. Endothelium-dependent vasodilator effects of peroxisome proliferator-activated receptor beta agonists via the phosphatidyl-inositol-3 kinase-Akt pathway. J Pharmacol Exp Ther. 2010;332:554–61. doi: 10.1124/jpet.109.159806. [DOI] [PubMed] [Google Scholar]
- 4.Sun M, Yang L, Feldman RI, Sun XM, Bhalla KN, Jove R, et al. Activation of phosphatidylinositol 3-kinase/Akt pathway by androgen through interaction of p85alpha, androgen receptor, and Src. J Biol Chem. 2003;278:42992–3000. doi: 10.1074/jbc.M306295200. [DOI] [PubMed] [Google Scholar]
- 5.Watson CS, Alyea RA, Jeng YJ, Kochukov MY. Nongenomic actions of low concentration estrogens and xenoestrogens on multiple tissues. Mol Cell Endocrinol. 2007;274:1–7. doi: 10.1016/j.mce.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boonyaratanakornkit V, Scott MP, Ribon V, Sherman L, Anderson SM, Maller JL, et al. Progesterone receptor contains a proline-rich motif that directly interacts with SH3 domains and activates c-Src family tyrosine kinases. Mol Cell. 2001;8:269–80. doi: 10.1016/s1097-2765(01)00304-5. [DOI] [PubMed] [Google Scholar]
- 7.Gardner OS, Dewar BJ, Graves LM. Activation of mitogen-activated protein kinases by peroxisome proliferator-activated receptor ligands: an example of nongenomic signaling. Mol Pharmacol. 2005;68:933–41. doi: 10.1124/mol.105.012260. [DOI] [PubMed] [Google Scholar]
- 8.Sciullo EM, Vogel CF, Li W, Matsumura F. Initial and extended inflammatory messages of the nongenomic signaling pathway of the TCDD-activated Ah receptor in U937 macrophages. Arch Biochem Biophys. 2008;480:143–55. doi: 10.1016/j.abb.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 9.Pedram A, Razandi M, Sainson RC, Kim JK, Hughes CC, Levin ER. A conserved mechanism for steroid receptor translocation to the plasma membrane. J Biol Chem. 2007;282:22278–88. doi: 10.1074/jbc.M611877200. [DOI] [PubMed] [Google Scholar]
- 10.Acconcia F, Ascenzi P, Bocedi A, Spisni E, Tomasi V, Trentalance A, et al. Palmitoylation-dependent estrogen receptor alpha membrane localization: regulation by 17beta-estradiol. Mol Biol Cell. 2005;16:231–7. doi: 10.1091/mbc.E04-07-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 12.Zassadowski F, Rochette-Egly C, Chomienne C, Cassinat B. Regulation of the transcriptional activity of nuclear receptors by the MEK/ERK1/2 pathway. Cell Signal. 2012;24:2369–77. doi: 10.1016/j.cellsig.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Gilad LA, Schwartz B. Association of estrogen receptor beta with plasma-membrane caveola components: implication in control of vitamin D receptor. J Mol Endocrinol. 2007;38:603–18. doi: 10.1677/JME-06-0040. [DOI] [PubMed] [Google Scholar]
- 14.Chambliss KL, Yuhanna IS, Mineo C, Liu P, German Z, Sherman TS, et al. Estrogen receptor alpha and endothelial nitric oxide synthase are organized into a functional signaling module in caveolae. Circ Res. 2000;87:E44–52. doi: 10.1161/01.res.87.11.e44. [DOI] [PubMed] [Google Scholar]
- 15.Losel RM, Falkenstein E, Feuring M, Schultz A, Tillmann HC, Rossol-Haseroth K, et al. Nongenomic steroid action: controversies, questions, and answers. Physiol Rev. 2003;83:965–1016. doi: 10.1152/physrev.00003.2003. [DOI] [PubMed] [Google Scholar]
- 16.Simoncini T, Hafezi-Moghadam A, Brazil DP, Ley K, Chin WW, Liao JK. Interaction of oestrogen receptor with the regulatory subunit of phosphatidylinositol-3-OH kinase. Nature. 2000;407:538–41. doi: 10.1038/35035131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castoria G, Migliaccio A, Bilancio A, Di Domenico M, de Falco A, Lombardi M, et al. PI3-kinase in concert with Src promotes the S-phase entry of oestradiol-stimulated MCF-7 cells. Embo J. 2001;20:6050–9. doi: 10.1093/emboj/20.21.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun M, Paciga JE, Feldman RI, Yuan Z, Coppola D, Lu YY, et al. Phosphatidylinositol-3-OH Kinase (PI3K)/AKT2, activated in breast cancer, regulates and is induced by estrogen receptor alpha (ERalpha) via interaction between ERalpha and PI3K. Cancer Res. 2001;61:5985–91. [PubMed] [Google Scholar]
- 19.Yu J, Akishita M, Eto M, Koizumi H, Hashimoto R, Ogawa S, et al. Src kinase-mediates androgen receptor-dependent non-genomic activation of signaling cascade leading to endothelial nitric oxide synthase. Biochem Biophys Res Commun. 2012;424:538–43. doi: 10.1016/j.bbrc.2012.06.151. [DOI] [PubMed] [Google Scholar]
- 20.Migliaccio A, Castoria G, Di Domenico M, de Falco A, Bilancio A, Lombardi M, et al. Steroid-induced androgen receptor-oestradiol receptor beta-Src complex triggers prostate cancer cell proliferation. Embo J. 2000;19:5406–17. doi: 10.1093/emboj/19.20.5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kousteni S, Bellido T, Plotkin LI, O'Brien CA, Bodenner DL, Han L, et al. Nongenotropic, sex-nonspecific signaling through the estrogen or androgen receptors: dissociation from transcriptional activity. Cell. 2001;104:719–30. [PubMed] [Google Scholar]
- 22.Heinlein CA, Chang C. The roles of androgen receptors and androgen-binding proteins in nongenomic androgen actions. Mol Endocrinol. 2002;16:2181–7. doi: 10.1210/me.2002-0070. [DOI] [PubMed] [Google Scholar]
- 23.Acconcia F, Kumar R. Signaling regulation of genomic and nongenomic functions of estrogen receptors. Cancer Lett. 2006;238:1–14. doi: 10.1016/j.canlet.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 24.Keung W, Chan ML, Ho EY, Vanhoutte PM, Man RY. Non-genomic activation of adenylyl cyclase and protein kinase G by 17beta-estradiol in vascular smooth muscle of the rat superior mesenteric artery. Pharmacol Res. 2011;64:509–16. doi: 10.1016/j.phrs.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 25.Li X, Zhang S, Safe S. Activation of kinase pathways in MCF-7 cells by 17beta-estradiol and structurally diverse estrogenic compounds. J Steroid Biochem Mol Biol. 2006;98:122–32. doi: 10.1016/j.jsbmb.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 26.Greathouse KL, Bredfeldt T, Everitt JI, Lin K, Berry T, Kannan K, et al. Environmental estrogens differentially engage the histone methyltransferase EZH2 to increase risk of uterine tumorigenesis. Mol Cancer Res. 2012;10:546–57. doi: 10.1158/1541-7786.MCR-11-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bredfeldt TG, Greathouse KL, Safe SH, Hung MC, Bedford MT, Walker CL. Xenoestrogen-induced regulation of EZH2 and histone methylation via estrogen receptor signaling to PI3K/AKT. Mol Endocrinol. 2010;24:993–1006. doi: 10.1210/me.2009-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouskine A, Nebout M, Brucker-Davis F, Benahmed M, Fenichel P. Low doses of bisphenol A promote human seminoma cell proliferation by activating PKA and PKG via a membrane G-protein-coupled estrogen receptor. Environ Health Perspect. 2009;117:1053–8. doi: 10.1289/ehp.0800367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watson CS, Bulayeva NN, Wozniak AL, Alyea RA. Xenoestrogens are potent activators of nongenomic estrogenic responses. Steroids. 2007;72:124–34. doi: 10.1016/j.steroids.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeng YJ, Kochukov MY, Watson CS. Membrane estrogen receptor-alpha-mediated nongenomic actions of phytoestrogens in GH3/B6/F10 pituitary tumor cells. J Mol Signal. 2009;4:2. doi: 10.1186/1750-2187-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bulayeva NN, Gametchu B, Watson CS. Quantitative measurement of estrogen-induced ERK 1 and 2 activation via multiple membrane-initiated signaling pathways. Steroids. 2004;69:181–92. doi: 10.1016/j.steroids.2003.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu D, Jiang H, Grange RW. Genistein activates the 3',5'-cyclic adenosine monophosphate signaling pathway in vascular endothelial cells and protects endothelial barrier function. Endocrinology. 2005;146:1312–20. doi: 10.1210/en.2004-1221. [DOI] [PubMed] [Google Scholar]
- 33.Liu D, Homan LL, Dillon JS. Genistein acutely stimulates nitric oxide synthesis in vascular endothelial cells by a cyclic adenosine 5'-monophosphate-dependent mechanism. Endocrinology. 2004;145:5532–9. doi: 10.1210/en.2004-0102. [DOI] [PubMed] [Google Scholar]
- 34.Maggiolini M, Vivacqua A, Fasanella G, Recchia AG, Sisci D, Pezzi V, et al. The G protein-coupled receptor GPR30 mediates c-fos up-regulation by 17beta-estradiol and phytoestrogens in breast cancer cells. J Biol Chem. 2004;279:27008–16. doi: 10.1074/jbc.M403588200. [DOI] [PubMed] [Google Scholar]
- 35.Welshons WV, Nagel SC, vom Saal FS. Large effects from small exposures. III. Endocrine mechanisms mediating effects of bisphenol A at levels of human exposure. Endocrinology. 2006;147:S56–69. doi: 10.1210/en.2005-1159. [DOI] [PubMed] [Google Scholar]
- 36.Verkasalo PK, Appleby PN, Davey GK, Key TJ. Soy milk intake and plasma sex hormones: a cross-sectional study in pre- and postmenopausal women (EPIC-Oxford). Nutr Cancer. 2001;40:79–86. doi: 10.1207/S15327914NC402_1. [DOI] [PubMed] [Google Scholar]
- 37.De Coster S, van Larebeke N. Endocrine-disrupting chemicals: associated disorders and mechanisms of action. J Environ Public Health. 2012;2012:713696. doi: 10.1155/2012/713696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greathouse KL, Cook JD, Lin K, Davis BJ, Berry TD, Bredfeldt TG, et al. Identification of uterine leiomyoma genes developmentally reprogrammed by neonatal exposure to diethylstilbestrol. Reprod Sci. 2008;15:765–78. doi: 10.1177/1933719108322440. [DOI] [PubMed] [Google Scholar]
- 39.Cook JD, Davis BJ, Cai SL, Barrett JC, Conti CJ, Walker CL. Interaction between genetic susceptibility and early-life environmental exposure determines tumor-suppressor-gene penetrance. Proc Natl Acad Sci U S A. 2005;102:8644–9. doi: 10.1073/pnas.0503218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walker CL, Ho SM. Developmental reprogramming of cancer susceptibility. Nat Rev Cancer. 2012;12:479–86. doi: 10.1038/nrc3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 42.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 43.Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT, et al. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science. 2005;310:306–10. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- 44.Takada I, Mihara M, Suzawa M, Ohtake F, Kobayashi S, Igarashi M, et al. A histone lysine methyltransferase activated by non-canonical Wnt signalling suppresses PPAR-gamma transactivation. Nat Cell Biol. 2007;9:1273–85. doi: 10.1038/ncb1647. [DOI] [PubMed] [Google Scholar]
- 45.Sen N, Snyder SH. Neurotrophin-mediated degradation of histone methyltransferase by S-nitrosylation cascade regulates neuronal differentiation. Proc Natl Acad Sci U S A. 2011;108:20178–83. doi: 10.1073/pnas.1117820108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu S, Wang W, Kong X, Congdon LM, Yokomori K, Kirschner MW, et al. Dynamic regulation of the PR-Set7 histone methyltransferase is required for normal cell cycle progression. Genes Dev. 2010;24:2531–42. doi: 10.1101/gad.1984210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boeke J, Regnard C, Cai W, Johansen J, Johansen KM, Becker PB, et al. Phosphorylation of SU(VAR)3-9 by the chromosomal kinase JIL-1. PLoS One. 2010;5:e10042. doi: 10.1371/journal.pone.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu SC, Zhang Y. Cyclin-dependent kinase 1 (CDK1)-mediated phosphorylation of enhancer of zeste 2 (Ezh2) regulates its stability. J Biol Chem. 2011;286:28511–9. doi: 10.1074/jbc.M111.240515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feng Q, He B, Jung SY, Song Y, Qin J, Tsai SY, et al. Biochemical control of CARM1 enzymatic activity by phosphorylation. J Biol Chem. 2009;284:36167–74. doi: 10.1074/jbc.M109.065524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogawa M, Hiraoka Y, Aiso S. The Polycomb-group protein ENX-2 interacts with ZAP-70. Immunol Lett. 2003;86:57–61. doi: 10.1016/s0165-2478(02)00293-6. [DOI] [PubMed] [Google Scholar]
- 51.Lan F, Nottke AC, Shi Y. Mechanisms involved in the regulation of histone lysine demethylases. Curr Opin Cell Biol. 2008;20:316–25. doi: 10.1016/j.ceb.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baba A, Ohtake F, Okuno Y, Yokota K, Okada M, Imai Y, et al. PKA-dependent regulation of the histone lysine demethylase complex PHF2-ARID5B. Nat Cell Biol. 2011;13:668–75. doi: 10.1038/ncb2228. [DOI] [PubMed] [Google Scholar]
- 53.Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466:508–12. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang CY, Hsu PH, Chou DF, Pan CY, Wang LC, Huang WC, et al. The histone H3K36 demethylase Rph1/KDM4 regulates the expression of the photoreactivation gene PHR1. Nucleic Acids Res. 2011;39:4151–65. doi: 10.1093/nar/gkr040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lomberk G, Bensi D, Fernandez-Zapico ME, Urrutia R. Evidence for the existence of an HP1-mediated subcode within the histone code. Nat Cell Biol. 2006;8:407–15. doi: 10.1038/ncb1383. [DOI] [PubMed] [Google Scholar]
- 56.Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–6. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- 57.Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, et al. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol. 2009;11:1376–82. doi: 10.1038/ncb1982. [DOI] [PMC free article] [PubMed] [Google Scholar]