Abstract
The analysis of high-throughput data sets, such as microarray data, often requires that individual variables (genes, for example) be grouped into clusters of variables with highly correlated values across all samples. Gene shaving is an established method for generating such clusters, but is overly sensitive to the input data: changing just one sample can determine whether or not an entire cluster is found. This paper describes a clustering method based on the bootstrap aggregation of gene shaving clusters, which overcomes this and other problems, and applies the new method to a large gene expression microarray dataset from brain tumour samples.
Keywords: bootstrap aggregation, clustering, gene shaving, glioblastoma
1 Introduction
The analysis of high-throughput data sets, such as microarray data, often requires that individual variables (genes, for example) be grouped into small clusters such that all the variables in any given cluster have highly correlated values across all samples. Unlike hierarchical clustering in which all variables are recursively aggregated, no matter how different, into ever-larger clusters to form a tree-like dendrogram, we are interested only in clusters containing variables with very similar measurements in all samples. Such clusters are sometimes called meta-genes or meta-variables, which we use to denote the average measurement for each sample of all the variables in the cluster. Figure 1 shows an example of a gene cluster. One use of these clusters is to infer the relatedness of individual variables from their membership in a common cluster. A second use is to suggest possible functions for individual variables of interest, based on the functions of other variables in the cluster, and to suggest additional related variables that might also be of interest. A third use is to calculate a cluster meta-variable for each sample by averaging the individual variables in the cluster. The cluster meta-variables might yield more robust measurements and tests of sample characteristics than the individual variables. Converting the individual variables into meta-variables also reduces the number of variables, and makes searching high dimensional spaces for interaction effects more tractable.
Figure 1.
The first cluster. The samples (columns) are sorted so that the signed mean gene increases from left to right. The top two rows show colour encoded representations of the batch the sample came from and the chip type used. Neither shows any obvious evidence of batch effects (see online version for colours)
1.1 Gene shaving
Gene shaving (Hastie et al., 2000) is an established method for generating such clusters. Gene shaving is so named because it was first developed in the context of analysing gene expression microarrays, which measure tens of thousands of probe sets for each sample. Each probe set measures the amount of messenger RNA in a sample for a specific gene, but any particular microarray design may contain 0, 1, 2, or more probe sets for any specific gene. Despite its name, gene shaving can be applied to any high-throughput data set containing continuous data.
The gene shaving method begins by computing the first principal component of the data and ranking all variables (genes) by their correlation with that principal component. It then shaves off, or removes, a small number of the variables that are the least correlated with that principal component, recomputes the principal component of the reduced data set, and ranks the remaining variables according to the new principal component. The shaving process is continued until only two variables remain. We call the set of variables remaining at each step in this process a shaving.
The cluster itself is chosen from the set of shavings such that it maximises the gap statistic between the shavings based on the original data and the shavings based on randomly permuted data. Multiple, independently permuted copies of the data are used to estimate the gap statistic.
Unlike other clustering methods, gene shaving allows both positively and negatively correlated variables to belong to the same cluster. (This is not shown in any of the examples in this paper.) When computing the cluster's meta-variable the sign of each variable's correlation must also be considered, so we use the terms signed meta-variable or signed meta-gene.
Once the cluster is chosen, the data set is orthogonalised with respect to the cluster's meta-variable. Additional clusters can then be generated by repeating the entire process until a fixed number of clusters have been generated, or the ‘optimal’ cluster would contain all remaining variables. The idea behind orthogonalisation is that it removes the effect of the generated cluster from the data, but the variables stay in the data set so that, potentially at least, individual variables may belong to multiple clusters.
We developed a fast implementation of gene shaving called GeneClust (Do et al., 2003), consisting of a user-friendly Java based user interface, a fast C implementation of gene shaving, and an R library interface to the C function. We have used GeneClust to analyse numerous high-throughput data sets.
1.2 Limitations of gene shaving
In our experience, gene shaving is overly sensitive to the input data. For example, we applied gene shaving to a large data set containing hundreds of glioma samples. In an early attempt to estimate the sensitivity of gene shaving to the data, we performed leave-one-out cross-validation and noticed that in many cases entire clusters would appear in some data sets but not others, even if we generated hundreds of clusters from each data set. In this example, a known cluster containing genes of interest did not appear in the clusters generated from the entire data set, but did appear as one of the first clusters generated in many of the data sets from which one sample had been left out.
We believe this occurs because the first principal component that is selected is indicative of a general trend across a large number of genes, but the shaving and cluster selection process narrows this down to a much smaller, very specific set of genes. The orthogonalisation of the data set after the cluster is selected not only removes the effect of the selected cluster's meta-variable, but also greatly reduces the influence of similar variables, even though these variables have not been assigned to any cluster, such that they will never be selected as a cluster. Thus, we believe that when the data is changed slightly, the first principal component is also changed only slightly, but the shaving process narrows down to a similar but ultimately different cluster. The orthogonalisation of the data set then allows only whichever cluster was chosen first to be detected, with all other similar clusters orthogonalised away. Since the initial principal component determined for the first cluster may reflect a major trend over most of the variables in the data set, we view this exclusion of all related variables as a major problem.
Although the stated reason for orthogonalising the data set, instead of removing the variables concerned from further consideration, is to allow individual variables to appear in more than one cluster, we have rarely observed this in practice. Thus, orthogonalisation has a significant detrimental impact for little benefit.
Another limitation of gene shaving is that individual variables either belong to a cluster or they do not. It would be helpful to have some measure of how sensitive a variable's membership in a specific cluster is to the data. In many cases, the gap curve for determining the size of a cluster shows two distinct peaks, the relative heights of which will determine whether the generated cluster is small or large. We believe these two peaks correspond to a group of core variables and another group of very similar but slightly distinct variables. Standard gene shaving will make this decision without considering the distribution of the input data, and if the smaller cluster is chosen will not output the additional variables in any cluster due to orthogonalisation.
1.3 Outline
The following section describes a clustering method based on the bootstrap aggregation (bagging) (Breiman, 1996) of gene shaving clusters, which overcomes the limitations of gene shaving described above.
Section 3 describes the application of the new method to a large meta-data set of gene expression microarray data from brain tumour samples.
Section 4 concludes the paper and discusses topics for further research.
2 Bagged gene shaving
The basic idea of bagged gene shaving is very simple:
generate multiple bootstrap resamples of the data set
run gene shaving independently on each resample
cluster variables that frequently co-occur in the output of the different resamples.
The specifics of the first step will depend on the data set concerned, so we will defer any discussion of resampling to discussion of our brain cancer example in Section 3.
The second step is also straight-forward, although we note that even without orthogonalisation of the data set after each cluster is generated, individual variables can sometimes occur in one cluster and sometimes in another. In our brain cancer example, we chose to eliminate the variables in a cluster from the data set instead of orthogonalising against the cluster meta-variable. In the resulting output, variables frequently occur in different clusters.
The final step is the most challenging, in large part because of the fact that variables frequently occur in different clusters. The first challenge, however, is that there is not necessarily any correspondence between similarly numbered clusters generated from different resampled data sets. To determine the degree to which two variables, i and j, co-occur we therefore compute a variable adjacency matrix, Ai,j. Matrix elements are non-negative real values, with 0 indicating the two variables never occur in the same cluster. Larger values mean the two variables co-occur more frequently and/or do so in better clusters, and are thus more adjacent.
To reduce the influence of poor clusters, we incorporate two measures of cluster quality into the adjacency matrix. The first is the percent variance, , explained by a cluster . The total variance, , of the cluster can be decomposed into the variance within the samples, , and the variance between the samples, . The percent variance explained by the cluster is then . The second quality measure is the gap statistic, , which measures the difference between the percent variance explained by generated cluster, , and the percent variance explained by similarly sized clusters obtained from randomly permuted data.
Thus, the adjacency matrix is defined by
| (1) |
That is, the adjacency between two variables is the sum of over all clusters that contain both variables.
Having obtained the adjacency matrix, we must now extract clusters from it. This raises the question of what we mean by a cluster, especially when some variables associate to a greater or lesser degree with a wide range of other variables. We define a cluster to be a set of variables that co-occur with similarly frequency and which show similar patterns of co-occurrence with other variables. Variables will belong to at most one cluster. (This restriction could potentially be lifted. See Section 4 for further discussion.)
To extract a cluster, we find the two variables with the largest entry in the adjacency matrix. (In the case of a tie, an arbitrary pair of variables is chosen.) We then expand the cluster by considering additional variables whose adjacency to any current member of the cluster exceeds a fixed fraction of the new variable's highest adjacency. That is, we exclude variables that are weakly adjacent to the current cluster, but which are much more strongly adjacent to other variables. We also require the new variable to be adjacent to all existing members. The cluster is expanded in this way until no other suitable variables are available.
The potential cluster may contain variables that are strongly adjacent to all other members of the potential cluster, but which show a different pattern of adjacency strengths. This occurs when there are multiple distinct but strongly adjacent groups of variables. Although there is adjacency between all variables concerned, there is noticably greater adjacency between the variables belonging to each subgroup. We chose to separate these into distinct clusters.
We use the Pearson correlation in the adjacency strengths of the variables in the potential cluster to determine whether such subgroups exist and if so which variables belong to the same subgroup as the first variable in the cluster. If the p value of the correlation is close to 0, the variable's pattern of adjacency strengths is highly correlated with that of the first variable and we include it in the first subgroup, whereas if it is close to one we exclude it. Figure 2 shows the distribution of the p values obtained in the analysis of the glioma dataset described below. The large bars at 0 and one indicate that the great majority of p values are close to these limits. The exact p value at which we chose to include or exclude a variable is a parameter of the algorithm. We used 0.05, but the p value distribution suggests that the algorithm is not particularly sensitive to it. We also did not perform this subgroup test for very small clusters (less than 8 variables) because of poor statistical power. Sometimes the subgroup test produces a cluster containing just the first variable. Such clusters are discarded in a subsequent step.
Figure 2.
Histogram of the p values obtained by the subgroup test when applied to the glioma dataset
The remaining variables are output as a cluster, eliminated from further consideration, and the process repeated until all variables have been used or a sufficiently large number of clusters have been generated. The above method generates cohesive clusters, but the order in which they are generated depends only on the maximum adjacency and not at all on cluster size. We define the quality of a cluster , , to be the mean adjacency within the variables belonging to the cluster multiplied by the number of variables in the cluster:
| (2) |
The generated clusters are sorted in order of decreasing .
Adjacencies between clusters are summarised by computing a cluster adjacency matrix, . Each entry in the cluster adjacency matrix is the mean of the adjacencies of the variables in the two clusters concerned:
| (3) |
Our earlier implementation of gene shaving, GeneClust, could extract a few tens of clusters from data sets containing up to about 100 samples in a reasonable time, but had a computational time complexity of O(N3) in the number of samples. Performing gene shaving on a large number of bootstrap resamples of our much larger data set was computationally infeasible.
To enable all of the computations required to be completed in a reasonable time, we have developed a much more efficient command line based implementation. This implementation uses multiple threads to take advantage of the multiple processors available on current workstations and cluster compute nodes. It is also highly optimised to make better use of cache and has improved locality of memory references. Although still O(N3), it is fast enough on current quad-processor machines to use on data sets containing about 1000 samples.
3 Application to glioma dataset
We applied bagged gene shaving to a large collection of gene expression microarray data obtained from infiltrating gliomas (WHO grade II-IV).
3.1 Dataset
The data consists of Affymetrix gene expression microarray results from a total of 739 infiltrating gliomas (WHO grade II-IV). The samples came from 10 different institutions and were measured on three different platforms (U133A, U133Plus2, and HT_U133A). Since the samples from some institutions were collected under different conditions, including different platforms, for the purpose of detecting and mitigating batch effects (Baggerly et al., 2008), we divided the samples into 15 distinct batches, which are summarised in Table 1. We split the data into a training set of 442 samples and a validation set of 297 samples, such that a similar proportion of training and test cases occur in each batch and in each disease grade.
Table 1.
Batch statistics
| Batch | Training samples | Test samples | Platform |
|---|---|---|---|
| AO-LBL | 18 | 14 | HT_U133A |
| AO-MDA | 11 | 13 | U133A |
| Belgium | 12 | 9 | U133Plus2 |
| Collins | 39 | 22 | U133A |
| Duke | 21 | 10 | U133A |
| EORTC | 40 | 25 | U133Plus2 |
| HF | 68 | 37 | U133Plus2 |
| MDA | 31 | 20 | U133A |
| MDA-new | 25 | 13 | U133Plus2 |
| TCGA | 60 | 50 | HT_U133A |
| UCLA | 51 | 31 | U133A |
| UCLA3 | 22 | 20 | U133A |
| UCLA4 | 6 | 3 | U133Plus2 |
| UCLA5 | 21 | 16 | U133A |
| UCSF | 17 | 14 | U133A |
3.2 Data preprocessing and bootstrap resample generation
Due to advances in annotation of the genome and transcriptome, the original Affymetrix probe set definitions are inaccurate (Dai et al., 2005). Consequently, all of the original CEL files were quantified using updated probe set definitions based on version 11 of the Entrez Gene definition. Using these new probe set definitions, there are 11911 probe sets common to the three different platforms used. Unlike the original Affymetrix probe set definitions, each gene is associated with at most one probe set. We will therefore use the term ‘gene’ to include the meaning ‘probe set’ in the remainder of this paper.
To reduce batch effects, the quantified data was combined as follows. The gene measurements for each sample were ranked from 1 to 11911. For the training data, the ranks for each gene were then ranked across the samples in its batch, and scaled to the range 0 to 1 by dividing by the number of samples in the batch. For the test data, each test sample was added individually to the training data for its batch, and the gene ranks were ranked across samples and scaled as described for the training data, and the scaled ranks for the test sample were recorded. (The scaled ranks for the training data obtained when the test sample was included were discarded.)
For resample generation, the final ranking across samples was weighted according to a draw from the Bayesian bootstrap distribution (Rubin, 1981) instead of uniformly. We used Bayesian bootstrap resampling because it was easy to combine with our batch effect mitigation strategy and because it has better theoretical properties for small sample sizes, such as for some of the batches.
3.3 Bagged gene shaving
Bagged gene shaving was performed on the training data as described above using our optimised gene shaving software. We used MD Anderson's large computational cluster to perform gene shaving on multiple resamples at once. The gene shaving for each resample was executed on a single four-processor compute node and required approximately 10 h of elapsed time. We used a total of 256 resamples.
For each resample, we generated a predetermined number of 600 clusters, removing the genes in each discovered cluster instead of orthogonalising against the cluster mean gene. On average, the generated clusters contained a total of 10740 genes per resample. We then combined the cluster information to generate the raw gene adjacency matrix, from which we extracted a predetermined number of 1000 clusters. Of these, 193 contained only a single gene, leaving 807 clusters, which were sorted into decreasing order of Qk.
3.4 Comparison to unbagged shaving
To compare the cluster quality obtained by the two methods, we clustered the 297 test samples using the clusters found by each method when applied to the training data, and calculated the percent variance explained, , for each cluster. Figure 3 shows a histogram comparing the results. The results for the bagged gene clusters are significantly higher than those for unbagged gene shaving (a mean of 59 vs. 38).
Figure 3.
Comparison of the percent variance explained, , of the bagged vs. unbagged clusters when applied to the test data (see online version for colours)
3.5 Results
For reference, Figure 1 shows the first cluster. The samples (columns) are sorted so that the signed mean gene increases from left to right. The top two rows show colour encoded representations of the batch the sample came from and the chip type used. Neither shows any obvious evidence of batch effects. The uniformity of the cluster is due in part to the preprocessing, which reduces the data for all genes into the range from 0 to 1 (which is then centred by gene shaving so that the final range is –0.5 to 0.5).
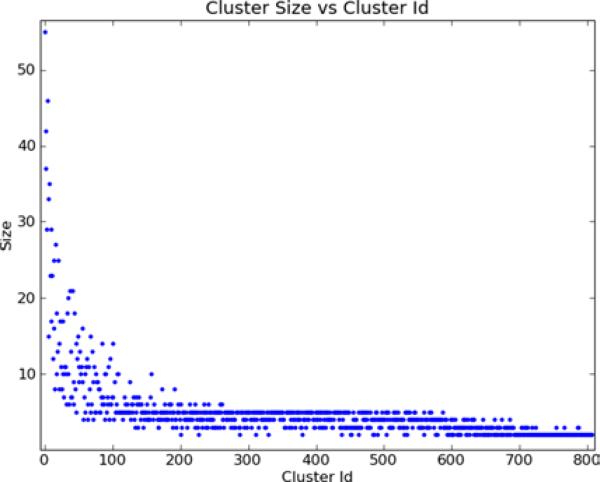
Cluster 1 is the largest cluster and among the most strongly detected clusters. Figure 4 shows the number of genes in each cluster. Since the clusters are sorted in order of decreasing Qk, the larger and more frequently generated clusters occur earlier, as expected. To summarise, there are 161 clusters containing 2 genes, 155 containing 3, 173 containing 4, 194 containing 5, and 124 containing 6 or more genes. The 807 clusters contain in total 3937 genes.
Figure 4.
The number of genes in each cluster (see online version for colours)
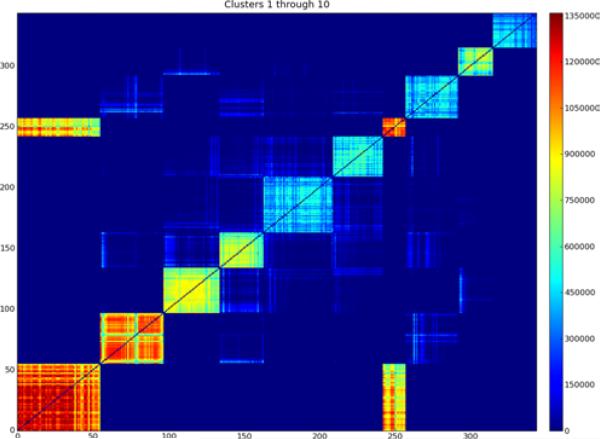
Figure 5 shows the gene adjacency matrix for the first ten clusters. The clusters are clearly discernable as the highly adjacent squares of genes along the diagonal, with cluster 1 on the bottom left and cluster 10 on the upper right. The adjacency scores show that clusters 1, 2, and 7 are either detected the most often, or belong to better clusters as reflected by the clusters’ gap and/or percent variance explained statistics. Clusters 3, 4, and 9 belong to somewhat less well scoring clusters, while clusters 5, 6, 8, and 10 score lower again.
Figure 5.
Adjacency matrix for genes in clusters 1 to 10. Each entry is the sum of the quality scores for all bootstrap clusters in which both genes occur. Although there is noticeable adjacency between many of the clusters, the adjacency within the clusters is much stronger (see online version for colours)
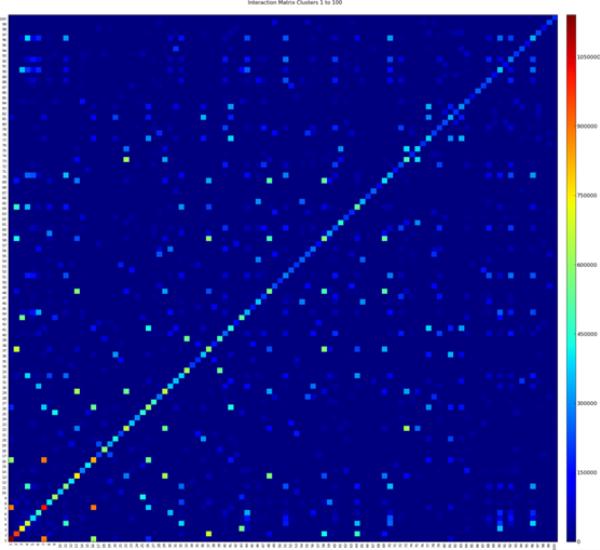
In addition to the strong adjacency between genes within each cluster, there is also noticeable adjacency between genes from different clusters. For example, there is significant adjacency between the genes in cluster 1 and those in cluster 7. The inter-cluster adjacencies indicate that in some resamples, genes belong to a different cluster than the one to which they usually belong. Another possibility is that in some resamples the genes from both clusters are combined into a single, large cluster. Whether clusters 1 and 7 should remain separate or be combined into a single large cluster is a matter of degree. It is clear that the other clusters in Figure 5 are distinct, even though there is a little adjacency between them. We believe that the separation of clusters 1 and 7 is also appropriate, since the adjacency within each is significantly higher and more consistent than the adjacency between them. The cluster adjacency matrix records the average of the gene adjacency either within a cluster, or between any two clusters. Figure 6 shows the cluster adjacency matrix for the first 100 clusters. Cluster 1 is on the bottom left and cluster 100 on the top right. Adjacency between clusters 1 and 7 is clearly visible (on a suitably magnified view), as is adjacency between clusters 7 and 16. The cluster adjacency matrix can be easily queried to determine the clusters that are most adjacent to any cluster of interest. Figure 7 shows the adjacency between the genes in the 10 clusters that have the highest average adjacency with cluster 1. Note that in this example, the clusters are sorted by their average adjacency with cluster 1, not by any measure of their own size or strength. From bottom left to top right, the clusters concerned are numbers 1, 7, 16, 26, 134, 81, 464, 201, 20, and 36. The fifth through eighth clusters in this example are relatively small and hard to discern.
Figure 6.
Matrix of the average adjacency between the first 100 clusters (see online version for colours)
Figure 7.
Adjacency matrix for the 10 clusters with the highest average adjacency with cluster 1 (see online version for colours)
To evaluate the correctness of the clusters, especially those with relatively low adjacency scores that are detected towards the end of the process, we consider separately false positives, in which a gene is assigned to a cluster to which it does not actually belong, and false negatives, in which a gene is not assigned to a cluster to which it does belong. Answering this question is hard, because a cluster is (in general) an artificial construct, invented for our convenience, that means the variables in the cluster are highly correlated. It is not the case that variables in different clusters are uncorrelated. Indeed, the adjacency between clusters 1 and 7 in Figure 5 is indicative of a reasonable degree of correlation between the variables concerned.
When applied to random noise, many other clustering methods, such as hierarchical clustering, will happily produce many alleged clusters, but gene shaving is very unlikely to generate more than one or two such clusters and any that it does produce will have a very small gap statistic, , since that already includes a comparison to random permutations of the data. The bagged version of gene shaving is likely to spread any such weak, random adjacency across the adjacency matrix.
If the original Affymetrix probe set definitions are used, a useful consistency check is available, since many genes are measured by multiple probe sets. In earlier analyses based on these probe sets, we detected many clusters containing multiple probe sets for the same gene, and detected many clusters consisting solely of different probe sets for a single gene. Unfortunately, the revised probe set definitions used in this example contain at most one probe set for each gene, so such a check is not possible.
Nevertheless, there are several clusters containing only very similar genes. Cluster 207, for example, contains only the genes HOXD9, HOXD10, HOXD11, and HOXD13. Cluster 208 contains only the genes HIST1H2BF, HIST1H2BG, HIST1H2AE, and HIST1H3D. Cluster 685 contains only the genes FAM128A and FAM128B. Cluster 695 contains only the genes MT2A and MT4. These results suggest that many hundreds of bagged clusters are potentially indicative of true relationships.
In addition to clusters containing only very similar genes (or multiple probe sets for the same gene if using the original Affymetrix probe sets), we found two other major types of cluster. The first type consists of multiple genes related primarily by function. For example, cluster 1 is associated with the cell cycle, and contains many genes associated with nuclear replication, including the centromere proteins CENPA and CENPN, the kinesins KIF2C, KIF4A, KIF14, KIF20A, and KIF23, and the cell cycle genes CCNA2, CCNB1, and CCNB2.
The second type consists of multiple genes related by genomic location and not by function. For example, cluster 24 contains 17 genes all located on chromosome 10, while cluster 50 contains 15 genes all from cytogenetic band 19p13. These suggest the expression levels of the genes concerned are directly determined by a genome level alteration, such as a copy number change or epigentic modifications to a large region of DNA. Chromosome 10, for example, is well known to be partially or completely lost in many glioblastomas (Fujisawa et al., 2000). (We do not regard clusters containing only very similar genes from the same genomic location, such as clusters 207 and 208 described above, as indicative of genome related expression levels.)
In addition to cluster 24, there are several additional clusters containing only genes from chromosome 10. Figure 8 shows the gene adjacency matrix for these clusters. The first cluster (number 24) is much stronger than the others. It is adjacent to the genes in the fifth cluster shown, although these adjacencies are much weaker than those within cluster 24. The remaining clusters are all fairly small, many containing only two genes, and adjacent among themselves. There is little adjacency between these clusters and the first and fifth clusters.
Figure 8.
Adjacency matrix of clusters containing genes only from chromosome 10 (see online version for colours)
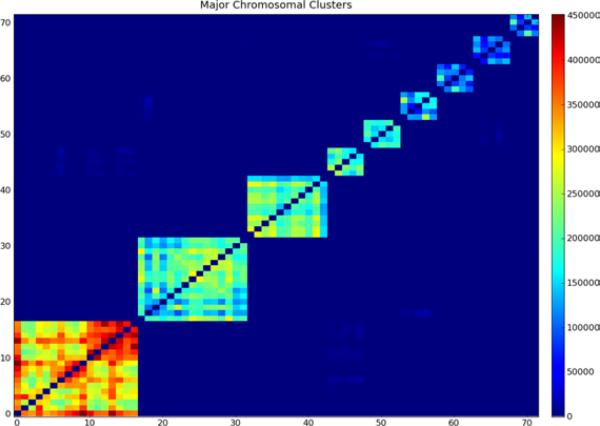
In this data set, we found clusters containing only genes from a single chromosome for 9 different chromosomes. Figure 9 shows the gene adjacency between the first cluster specifically associated with each of these chromosomes. Starting at the bottom left, these clusters are associated with chromosomes 10, 19, Y, 7, 17, 22, 14, 9, and 20. There is strong adjacency within each cluster, but virtually none between the clusters.
Figure 9.
Adjacency matrix for primary clusters associated with a specific chromosome (see online version for colours)
We compared the clusters from bagged gene shaving to the clusters obtained by applying gene shaving with orthogonalisation to the original data. Bagged cluster 1 contains mostly genes from unbagged cluster 2, with ten additional genes not present in any of the unbagged clusters. These additional genes are highly aligned with the functional aspects of the genes in bagged cluster 1. They include kinesian genes KIF14 and KIF23, cell cycle control genes CDC2, CDC6, and CDC20, a gene associated with the initiation of DNA replication (GINS1), a gene thought to be involved in the inhibition of spliceosome assembly during mitosis (MELK), a gene thought to be cell cycle regulator (DLGAP5), as well as two other genes (KIAA0101, TRIP13) whose function in DNA replication or cell cycle control is not known.
Unbagged cluster 2 contains mostly genes (46) from bagged cluster 1, but it also contains all of bagged clusters 7 (15 genes), 16 (8 genes) and 26 (8 genes). The genes in bagged cluster 7 are also associated with cell division and includes the centromere genes CENPE and CENPM, kinesin KIF11, and kinetochore related gene ZWINT. Bagged cluster 16 includes cell division cycle genes CDCA3, CDC25C, and CDC45L, centromere gene CENPF. Bagged cluster 26 contains one kinesian gene (KIF15), a kinetochore component (SPC25), a component of the minichromosome maintenance complex (MCM10), a gene that regulates centriole duplication (PLK4), and genes associated with DNA repair (NEIL3), neuronal proliferation (RACGAP1), and chromatin condensation (NCAPH). All of these clusters are clearly related with nuclear replication. The question remains whether their further division into 4 subclusters is warranted.
In contrast, bagged cluster 2 contains 42 genes, of which 6 are present in unbagged cluster 12 and 1 is from unbagged cluster 1. The 6 genes in unbagged cluster 12 are LAPTM5 (Lysosomal-associated multispanning membrane protein), TYROBP (transmembrane signalling polypeptide), RNASE6 (Ribonuclease), PTPRC (protein tyrosine phosphatase), ITGB2 (integrin chain component), and HCLS1 (substrate of antigen receptor-coupled tyrosine kinase).
The remaining 35 genes, listed in Table 2 are not output in any unbagged cluster. Figure 10 shows a heatmap of bagged cluster 2, including these 35 genes. The high percent variance explained by the cluster, the high adjacency between the genes in the cluster (Figure 5), and the common theme of known immune system related function among many of the genes concerned, strongly suggests that it is a valid cluster. We speculate they are not discovered by unbagged gene shaving because the genes in this cluster are somewhat correlated with those in the first unbagged cluster. Indeed one gene, ARPC1B, from bagged cluster 2 occurs in unbagged cluster 1. Orthogonalisation against cluster 1 makes it hard to detect that these genes are clustered, and further orthogonalisation against those few genes found in cluster 12 makes it impossible.
Table 2.
Missing genes
| ALOX5 | Involved in synthesis of leukotrienes, important mediators in inflammatory response |
| ARHGDIB | Rho GDP dissociation inhibitor (GDI) beta |
| C1orf38 | Chromosome 1 open reading frame 38 |
| C1QA | Complement component 1, q subcomponent, A chain |
| C1QB | Complement component 1, q subcomponent, B chain |
| CD14 | Antigen |
| CD300A | Antigen-like family member A |
| CLEC7A | Membrane receptor with an immunoreceptor tryosine-based activation motif |
| CTSS | Cathepsin S. Key protease responsible for removing invariant chain from MHC class II molecules |
| CYBA | Cytochrome b-245, alpha polypeptide |
| FCER1G | High affinity immunoglobulin epsilon receptor subunit gamma precursor |
| FPR1 | Formyl peptide receptor 1 – involved in neutrophil activation |
| GMFG | Glia maturation factor, gamma |
| GPR65 | G protein-coupled receptor 65. Possible role in activation-induced cell death or T-cell differentiation |
| HLA-DMA | Major histcompatibility complex, class II, DM alpha, anchored in membrane |
| HLA-DRB1 | Major histcompatibility complex, class II, DR beta 1, anchored in membrane, central role in immune system |
| LAIR1 | Leukocyte-associated immunoglobin-like receptor |
| LCP2 | Lymphocyte cytosolic protein 2 |
| LHFPL2 | Transmembrane protein encoding genes |
| LYN | Yamaguchi sarcoma viral related oncogene homolog |
| MS4A4A | Membrane spanning 4A gene |
| MS4A6A | Membrane spanning 4A gene |
| MSR1 | Macrophage scavenger receptor 1 |
| MYO1F | Myosin 1F |
| NCF4 | Neutrophil cytosolic factor 4 |
| NPC2 | Niemann-Pick disease, type C2 |
| PYCARD | Involved in caspase-controlled inflammation and apoptosis pathways |
| RAC2 | Plasma membrane-associated small GTPase |
| RNASE2 | Ribonuclease, RNase A family, 2. Eosinophil-derived neurotoxin |
| SERPINA1 | Serpin peptidase inhibitor, clade A, member 1 |
| SLA | SRC-like-adaptor - negatively regulates T-cell receptor signalling |
| SLC7A7 | Solute carrier family 7, member 7 |
| TLR1 | Toll-like receptor |
| TLR2 | Toll-like receptor 2 |
| VAMP8 | Vesicle-associated membrane protein 8 |
Figure 10.
The second cluster, including the 35 genes not found by unbagged gene shaving. The samples (columns) are sorted so that the signed mean gene increases from left to right. The top two rows show colour encoded representations of the batch the sample came from and the chip type used. Neither shows any obvious evidence of batch effects (see online version for colours)
4 Conclusions
In this paper we described several issues that arise when applying the gene shaving clustering method to large high-throughput data sets. These include the sensitivity of the generated clusters to the input data, in particular the fact that large clusters are simply not found by gene shaving, as well as the lack of information concerning the sensitivity of membership in those clusters that are found to the input data. We described the bagged gene shaving method for overcoming these issues by using bootstrap aggregation of gene shaving results from multiple bootstrap resamplings of the original data, and briefly described a high-performance implementation of gene shaving that makes such computations feasible.
We applied bagged gene shaving to a large, multi-institutional data set of infiltrating glioma samples and showed that bagged gene shaving finds large gene clusters not found by unbagged gene shaving. We also showed that genes that unbagged gene shaving simply lumps together into a single cluster have a definite and pronounced substructure that is resolved into distinct clusters by bagged gene shaving.
The gene clusters found by bagged gene shaving include small clusters of similar genes, large clusters of functionally related genes, and clusters of genes related only by genomic location.
The generated gene clusters can be used in many ways not described in this paper to gain insight into the biology of infiltrating gliomas. The signed meta-gene score computed for each cluster may be more reliable than using individual variables for detecting significant correlations between high-throughput data measurements and sample covariates of interest, such as patient survival. The smaller number of meta-genes also makes searching for high-dimensional interaction effects more tractable.
Methods such as Bayesian network analysis (Pearl, 1988), for example, can be used to obtain a more global view of the significant interactions between variables, but do poorly when applied to data sets containing many highly-correlated variables, since the choice of one specific variable from a set of highly correlated variables will be driven principally by noise. Reducing several highly-correlated variables to a single meta-variable not only eliminates the correlation related problems, but also enables data sets containing more variables to be analysed.
When generating clusters from the variable adjacency matrix, we required that variables belong to at most one cluster. The final clusters we generated also contained only a fraction of the variables in the entire data set, even though the gene shaving of each individual bootstrap resample assigned nearly all variables to clusters. The current clusters could be expanded to include all variables that are more weakly adjacent to the existing variables in the cluster than the current, core members. We are not convinced by the benefits of such an approach. It would destroy the distinction between clusters 1, 7, 16, and 26, which were lumped together by the unbagged method. The current definition produces tight, consistent clusters, and the information about genes that co-occur in multiple clusters is available from the cluster adjacency matrix. Since variables that do not co-occur strongly enough to frequently join the same cluster are much more likely to be false positives, we believe that they should not be included.
Further tools are required to simplify understanding of the hundreds of generated clusters and their interactions. We have fed the genes in individual clusters into various gene ontology and pathway analysis tools but have not found them particularly enlightening compared to quickly scanning the biological roles of the genes concerned.
We would like to develop a database of clusters found across multiple diseases (at least cancers) so that we can more easily identify gene groupings that are common to many diseases and those that are specific to a particular disease. For example, finding a cluster of genes, such as cluster 1, related to proliferation in a cancer data set is not interesting, it is expected. More interesting would be to find genes in that cluster that do not belong to proliferation clusters from any other diseases.
Acknowledgement
This work was supported in part by the NIH/NCI SPORE in Brain Cancer (PP-3A) 1P50CA127001 01A1. Additional support was received from a Neurooncology Grant from the Center for Targeted Therapy at the University of Texas MD Anderson Cancer Center.
Biography
Bradley M. Broom received his PhD in Computer Science in 1988 from the University of Queensland, Australia. He is an Associate Professor in the Department of Bioinformatics and Computational Biology at the University of Texas MD Anderson Cancer Center in Houston, Texas. His research interests include the development of algorithms for robust machine learning from datasets containing many variables but few datapoints.
Erik P. Sulman received his MD, PhD in Cancer Genetics in 2003 from the Temple University in Philadelphia, Pennsylvania. He is currently an Assistant Professor in the Department of Radiation Oncology at the University of Texas MD Anderson Cancer in Houston, Texas. His current research interests include the development of predictive models of survival and treatment response for patients with malignant brain tumours.
Kim-Anh Do received her PhD in statistics from Stanford University in 2000. She is a Professor in the Department of Biostatistics at the University of Texas MD Anderson Cancer Center, and a recipient of the Faculty Scholar Award at MD Anderson in 2003. She is a Fellow of the American Statistical Association and the Royal Statistical Society. She has served as a primary statistician or co-investigator on many National Institutes of Health (NIH) funded grants including the Early Detection Research Network (EDRN) grant, the Prostate SPORE (as Director of the Biostatistics Core), the Breast SPORE, and the Brain SPORE at MD Anderson. Her current research interests are in statistical methodology with focus on the development of clustering and analytical methods for genomic and proteomic expressions.
Mary E. Edgerton received her PhD in Biophysics in 1979 from the University of East Anglia, UK. She received her MD form the Medical College of Pennsylvania in 1994, and complete residency and fellowship in Pathology in 200. She is an Associate Professor in the Department of Pathology at the University of Texas MD Anderson Cancer Center and Adjunct Professor in the School of Health Information Sciences at University of Texas Health Science Center at Houston. Currently, she researches pathway discovery using high throughput molecular data generated from human tissue collections.
Kenneth D. Aldape received his MD in 1991 from the University of California, San Francisco. He is a Professor in the Department of Pathology, University of Texas MD Anderson Cancer Center. Currently, he is the Director of the Brain Tumor Tissue Bank at MD Anderson. His research interests include molecular genetics of brain tumours and prognostic predictive markers in cancer.
Footnotes
Reference to this paper should be made as follows: Broom, B.M., Sulman, E.P., Do, K-A., Edgerton, M.E. and Aldape, K.D. (2010) ‘Bagged gene shaving for the robust clustering of high-throughput data’, Int. J. Bioinformatics Research and Applications, Vol. 6, No. 4, pp.326–343.
Contributor Information
Bradley M. Broom, Department of Bioinformatics and Computational Biology, UT MD Anderson Cancer Center, Houston, Texas 77030, USA.
Erik P. Sulman, Department of Radiation Oncology, UT MD Anderson Cancer Center, Houston, Texas 77030, USA epsulman@mdanderson.org
Kim-Anh Do, Department of Biostatistics, UT MD Anderson Cancer Center, Houston, Texas 77030, USA kimdo@mdanderson.org.
Mary E. Edgerton, Department of Pathology, UT MD Anderson Cancer Center, Houston, Texas 77030, USA medgerton@mdanderson.org
Kenneth D. Aldape, Department of Pathology, UT MD Anderson Cancer Center, Houston, Texas 77030, USA kaldape@mdanderson.org
References
- Baggerly KA, Coombes KR, Neeley ES. Run batch effects potentially compromise the usefulness of genomic signatures for ovarian cancer. J. Clin. Oncol. 2008;26(7):1186–1187. doi: 10.1200/JCO.2007.15.1951. [DOI] [PubMed] [Google Scholar]
- Breiman L. Bagging predictors. Machine Learning. 1996;24(2):123–140. [Google Scholar]
- Dai M, Pinglang W, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, Watson SJ, Meng F. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Research. 2005;33(20) doi: 10.1093/nar/gni179. doi:10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do KA, Broom BM, Wen S. Geneclust. In: Parmigiani G, Garrett ES, Irizarry RA, Zeger SL, editors. The Analysis of Gene Expression Data: Methods and Software. Springer; New York, NY: 2003. pp. 342–361. [Google Scholar]
- Fujisawa H, Reis RM, Nakamura M, Colella S, Yonekawa Y, Kleihaus P, Ohgaki H. Loss of heterozygosity on chromosome 10 is more extensive in primary (De Novo) than in secondary glioblastomas. Lab Invest. 2000;80(1):65–72. doi: 10.1038/labinvest.3780009. [DOI] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Eisen MB, Alizadeh A, Levy R, Staudt L, Chan WC, Botstein D, Brown P. Gene shaving’ as a method for identifying distinct sets of genes with similar expression patterns. Genome Biology. 2000;1(2) doi: 10.1186/gb-2000-1-2-research0003. http://genomebiology.com/2000/1/2/research/0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl J. Probabilistic Reasoning in Intelligent Systems. Morgan Kauffman; San Francisco: 1988. [Google Scholar]
- Rubin DB. The Bayesian bootstrap. The Annals of Statistics. 1981;9(1):130–134. [Google Scholar]