Abstract
It is well known that horizontal gene transfer (HGT) is a major force in the evolution of prokaryotes. During the adaptation of a bacterial population to a new ecological niche, and particularly for intracellular bacteria, selective pressures are shifted and ecological niches reduced, resulting in a lower rate of genetic connectivity. HGT and positive selection are therefore two important evolutionary forces in microbial pathogens that drive adaptation to new hosts. In this study, we use genomic distance analyses, phylogenomic networks, tree topology comparisons, and Bayesian inference methods to investigate to what extent HGT has occurred during the evolution of the genus Rickettsia, the effect of the use of different genomic regions in estimating reticulate evolution and HGT events, and the link of these to host range. We show that ecological specialization restricts recombination occurrence in Rickettsia, but other evolutionary processes and genome architecture are also important for the occurrence of HGT. We found that recombination, genomic rearrangements, and genome conservation all show evidence of network-like evolution at whole-genome scale. We show that reticulation occurred mainly, but not only, during the early Rickettsia radiation, and that core proteome genes of every major functional category have experienced reticulated evolution and possibly HGT. Overall, the evolution of Rickettsia bacteria has been tree-like, with evidence of HGT and reticulated evolution for around 10–25% of the core Rickettsia genome. We present evidence of extensive recombination/incomplete lineage sorting (ILS) during the radiation of the genus, probably linked with the emergence of intracellularity in a wide range of hosts.
Keywords: Rickettsia, horizontal gene transfer, phylogenomic networks, reticulate evolution, bacterial speciation
Introduction
Genetic connectivity is the main speciation criterion in eukaryotes, and under the biological species concept, speciation occurs when two populations lose genetic connectivity and become reproductively isolated (Coyne and Orr 2004). However, the concepts and theories of speciation developed for plants and animals cannot be applied directly to bacteria, because of major differences in their mode of inheritance from typical multicellular eukaryotes (Gevers et al. 2005). Even if bacteria reproduce only clonally, they can exchange DNA horizontally through different processes: transformation, conjugation, transduction, and gene transfer agents (Smith et al. 1993; Medini et al. 2005; Ochman et al. 2005; Norman et al. 2009). Regardless of the process, homologous recombination is perhaps the most important mechanism for integrating donor DNA into a recipient genome in horizontal gene transfer (HGT) (Vulic et al. 1997; Majewski and Cohan 1998; Majewski et al. 2000). Homologous recombination requires incoming DNA to be highly similar to the recipient DNA, and so HGT is usually more common between similar bacteria (Zawadzki et al. 1995; Fraser et al. 2005; Thomas and Nielson 2005; Raymond et al. 2010; Didelot et al. 2011). Other genetic isolation mechanisms have been recently described in bacteria (Carrolo et al. 2009; Budroni et al. 2011; Corander et al. 2012), which consist on clade-associated recombination modification systems that generate differential barriers to DNA exchange that corresponds to population structure and, interestingly, are themselves subject to horizontal transfer.
In addition to HGT, specialization to different ecological niches (ecological speciation) has also been proposed as a major diversification process in bacteria (Cohan 2001, 2006). Even with strictly clonal reproduction, for which traditional ideas of speciation based on reproductive isolation do not apply, specialization to distinct niches can cause the emergence of independently evolving genetic clusters (Barraclough et al. 2012). An extreme case of ecological specialization is the transition to an intracellular lifestyle, and many bacterial groups have taken this route through the establishment of mutualistic, pathogenic, or commensal relationships within eukaryotic hosts. Importantly, populations become fragmented, resulting in isolated allopatric populations with effective population size dramatically reduced because it is constrained by the number of hosts, the number of infected cells, and the cellular space available for growth. In allopatric populations, there is also less opportunity for innovation, as HGT occurs less frequently than in free-living, sympatric bacterial populations (Bordenstein and Reznikoff 2005). Finally, the metabolite-rich intracellular environment relaxes the selective constraints on metabolic genes, which leads to the accumulation of mutations, gradual loss of function, and gene loss (Moran et al. 2009).
Comparisons of closely related species and strains are needed to test the role of HGT in bacterial diversification. Many studies have inferred HGT by comparing more distantly related taxa (Dagan et al. 2008; Kloesges et al. 2011; Schliep et al. 2011), confirming the importance of HGT for acquisition of new functions, but this does not necessarily demonstrates that HGT played a major role in the initial ecological divergence or emergence of barriers to recombination leading to divergence from a common ancestral population. The genus Rickettsia provides an excellent model to study the adaptive and diversifying evolutionary processes that occur during extreme host adaptation and specialization. Some species are associated with a single tick genus (Rickettsia helvetica seems to be associated with the tick genus Ixodes [Burgdorfer et al. 1979; Beati et al. 1993; Fournier et al. 2002]), whereas others, like R. rickettsii, are found in several tick genera (Merhej and Raoult 2011). Between these two extremes, there are certain species that are associated with several species within a single genus, such as R. africae and R. slovaca with various Amblyomma spp. and Dermacentor spp., respectively (Parola and Raoult 2001). Genomic surveys of mobile DNA elements in obligate intracellular bacteria suggest that there is a correlation between ecological specialization and HGT. Strict pathogenic or parasitic bacteria that host-switch (less specialized) harbor mobile DNA in their genomes, whereas stable mutualistic (more specialized) genera such as Buchnera, Blochmannia, and Wigglesworthia lack these elements (Bordenstein and Reznikoff 2005).
Reticulated evolution refers to the lack of independence between different evolutionary lineages and results when two or more independent evolutionary lineages are combined at some level of biological organization: chromosomes, genomes, or species. Given the complexity of interactions of Rickettsia species with very different hosts that can eventually result in niche partition and allopatric populations and speciation (breaking of genetic connectivity) and the overall genome-reductive processes, a key question in their evolution is to estimate to what extent do the above-mentioned mechanisms act in concert on components of the genome, or whether they act separately on different species, genes, or genomic regions producing mosaic histories nontractable as evolutionary lineages. In other words, are bacterial “species” real evolutionary units or a conglomerate of different gene networks resulting from HGT? Two main approaches have been used to detect and quantify HGT and reticulated evolution: phylogenomics to point out incongruent evolutionary histories of genes (Gogarten et al. 1992; Gogarten 1995) and parametric analyses to find genomic regions displaying sequence patterns that could be interpreted as footprint signs of long-term evolution in another mutational (and therefore, genomic) context (Médigue et al. 1991). The aim of this study is to quantify through both phylogenomic and parametric analyses: 1) the extent of reticulate evolution (including potential HGT detectable as recombination events) between host specialist and more generalist Rickettsia; and 2) the differences in number and type of genes/gene families potentially involved in reticulate evolution.
Materials and Methods
Genome Sequences and Alignments
A whole-genome alignment was obtained for 24 Rickettsia species and strains using MAUVE version 2.3.1 using the progressiveMauve algorithm (Darling et al. 2010). The genomes were retrieved from the GenBank database and are listed in supplementary table S1, Supplementary Material online.
Pre-tree Analyses: δ Plots
The C software DeltaStats (Holland et al. 2002) was used to analyze different distance measures derived from whole-genome alignments. This quartet-based distance measurement assesses the “tree-likeness” of distance data before phylogeny estimation and allows the identification of non-tree-like data arising from processes such as lateral gene transfer (Holland et al. 2002). Individual taxa can be ranked by reference to the tree-likeness of the quartets to which they belong. A value of 0 indicates perfectly tree-like data sets, and progressively higher values indicate more reticulation. For genetic distances, the genome alignments were corrected for multiple changes according to the GTR+I+G model (more generalized distance corrections with distributions of rates across sites or proportions of invariant sites were also tried and produced similar values). Genomic conservation and genomic rearrangements distances were calculated using MAUVE. All data were analyzed with the R statistical software package (http://cran.r-project.org, last accessed December 2, 2013) using the nonparametric Wilcoxon rank-sum test (an alternative to the Student’s t-test for small samples; Hollander and Wolfe 1973). All tests were two-tailed, and P values <0.05 were considered significant.
Phylogenomic Network Analyses
We constructed a split decomposition network to check for the signal of recombination between genomes. This method allows the visualization of the ancestral relationships between elements by showing conflicting signal for whole-genome data (genome conservation, genomic content, and genetic distance data) and for core proteome data (concatenated sequences). Networks were determined using the NeighborNet algorithm as implemented in SplitsTree v. 4.8 (Huson and Bryant 2006). The unrooted networks were validated statistically using the Phi test, which calculates the pairwise homoplasy index (PHI) as the mean of the refined incompatibility scores obtained for nearby nucleotide sites along the sequences (Bruen et al. 2006). Rooted phylogenetic networks for the core proteome were calculated using the Galled Network and level-k combinatorial algorithms as implemented in Dendroscope v. 3.2.1 (Huson and Scornavacca 2012).
Orthologous Genes Identification
Orthologous groups of genes from the different Rickettsia genomes were identified using OrthoMCL (Li et al. 2003), with a BlastP E value cutoff of 1 × 10−5 and a default MCL inflation parameter of 1.5. Assignment of protein functions was performed by searching against the RickBase (Altschul et al. 1997), GenBank, and Pfam databases using BLASTP (Blanc et al. 2007b; Punta et al. 2012).
Multiple Coalescence Analysis
To identify which genes and species produce discordant topologies, and therefore possibly evolve in a non-tree-like manner, we explored the genomic tree space of the core proteome for the 606 core gene maximum likelihood (ML) trees. Genes and species with different topologies were detected based on multiple co-inertia analysis (MCOA) as implemented in the R script Phylo-MCOA (de Vienne et al. 2012). This method efficiently captures and compares the similarities in the phylogenetic topologies produced by individual gene trees with no need of a reference species tree. This analysis identifies which genes and species produce discordant topologies and therefore evolve in a different way compared with the majority of trees (outlier genes and species). In order to better explore the discordance in the proteome’s evolution, different thresholds for considering that a species or a gene is a complete outlier were used. The default software value is set to 50% (i.e., species detected as outlier for more than 50% of the genes are seen as complete outliers and genes detected as outliers for more than 50% of the species are considered as complete outliers). We used 5%, 10%, 20%, 30%, and 50% values. Individual gene sequences were aligned using the MUSCLE algorithm (Edgar 2004). ML phylogenetic analyses were conducted using the Phyml package (Guindon and Gascuel 2003), with 1,000 bootstrap replicates, and the GTR+I+G substitution model. Nodal distances between species were used.
Phylogenetic Estimation of Recombination
We used ClonalFrame v. 1.2 (Didelot and Falush 2007) to estimate parameters in the evolutionary process that led to the observed pattern of nucleotide variation. It uses a Bayesian approach which jointly reconstructs the clonal relationships between the taxa in a sample and the location of recombination events that have disrupted the clonal signal. Four independent runs were performed, each consisting of 10,000 Markov chain Monte Carlo (MCMC) iterations, and the first half was discarded as burn-in. Convergence of tree topologies and mixing of the MCMC were found to be satisfactory by manual comparison of the runs. Because recombination can cause genome rearrangements, orthologous regions of one genome may be reordered or inverted relative to another. Mauve identifies conserved segments that appear to be internally free from genome rearrangements (locally collinear blocks, LCBs). The resulting set of LCBs was concatenated by striping out variable regions from the alignment to leave only core alignment blocks longer than 500 nt.
Phylogenetic Concordance Analyses
Bayesian concordance analysis (BCA; Ané et al. 2007) as implemented in the software BUCKy v 1.4.0 (Larget et al. 2010) was used to reconstruct the distribution of phylogenetic trees along the proteome and estimate the proportion of loci for which a given clade is true. In a first step, we carried out an analysis using MrBayes for each individual gene with the GTR+I+G model, employing MCMCMC with one cold and three heated chains, with default parameters for the prior distribution. We used the default values for tuning parameters, running each chain for 1 million generations or until the average standard deviation of split frequencies reached or was inferior to 0.001. We discarded the first 100,000 as burn-in and subsampled every 100th tree. The final number of trees per gene varied, as different loci reached stability at different generation numbers. BCA assumes that all sites within a given locus evolved under the same underlying tree topology. A nonparametric approach is used to model discordance between loci, with no single process assumed to be responsible for incongruence. A prior parameter is used to draw a random number of clusters and then randomly assign loci to clusters. The parameter α measures the a priori level of discordance expected among gene trees. An α = 0 assumes that all loci share the same tree in a single cluster, whereas an infinite α corresponds to assuming complete independence among locus trees. We carried out preliminary analyses with different α values (0.1, 0.5, the default value 1 and 100), with two independent runs each for 1 million generations. The number of estimated clusters and their prior probabilities were the same for all α values, so we used the default for the analyses, which corresponds to a 50% prior probability of loci sharing the same topology. We ran two independent sets of MCMCMC, each run used one cold chain and four heated chains, set burn-in for 100,000 cycles, and retained an additional 900,000 cycles for analysis. BUCKy estimated the posterior probabilities for sets of loci of sharing the same topology and inferred concordance factors (CFs) for clades. The genome-wide CF of a clade is the proportion of loci in the genome that truly contains the clade, as opposed to the statistical support provided by bootstrap values or posterior probabilities. A concordance tree was built from clades with the largest CFs to represent the dominant evolutionary history of Rickettsia.
Results
Pre-tree Analyses: δ Plots
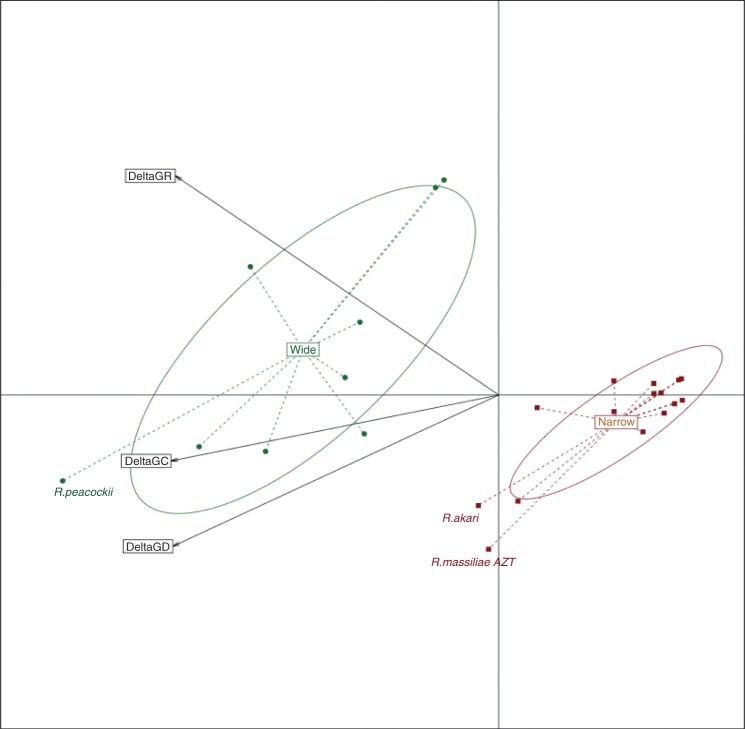
There was a significant effect of ecological niche amplitude on reticulation levels, with wide host range species having significantly higher δ values for all three genomic distance measures than species with narrower niches (Wilcoxon rank-sum test, P ≤ 0.0072; table 1). Principal component analysis (PCA) detected a between-category structure involving two distinct niche-associated groups, represented by locally “extreme” δ values strongly partitioned between ecological categories (wide and narrow ecological niches; fig. 1). This pattern is compatible with a reduction of genetic connectivity (and HGT) expected for fragmented and more specialized populations. There are, however, two species with narrow host ranges that have δ values closer to those of wide species: R. akari and R. massiliae AZT (table 1 and fig. 1). Even if HGT and recombination are not the sole causes of reticulated evolution, these high values imply that even specialized bacteria are still capable of significant genetic exchange.
Table 1.
Host Ranges and Measures of Reticulate Evolution (δ Values) for Rickettsia Species Calculated with δ Plots
| Species/Host Range | δ GC | δ GD | δ GR |
|---|---|---|---|
| R. akari/narrow | 0.082795479 | 0.378926211 | 0.033333333 |
| R. australis/narrow | 0.079534821 | 0.251495143 | 0.03333333 |
| R. Canadensis CA/narrow | 0.076828036 | 0.258589468 | 0.004166667 |
| R. Canadensis McK/narrow | 0.068624935 | 0.229864215 | 0.004166667 |
| R. conorii/narrow | 0.112250287 | 0.200588468 | 0.004166667 |
| R. heilongjiangensis/narrow | 0.121680988 | 0.196572867 | 0.004166667 |
| R. massiliae AZT/narrow | 0.138092916 | 0.177975839 | 0.004166667 |
| R. massiliae MTU/narrow | 0.119862616 | 0.358807859 | 0.004166667 |
| R. montanensis/narrow | 0.151071384 | 0.243803947 | 0.033333333 |
| R. parkeri/narrow | 0.152924254 | 0.292575292 | 0.004166667 |
| R. philipii/narrow | 0.15223778 | 0.200265796 | 0.004166667 |
| R. prowazekii MAD/narrow | 0.074950853 | 0.240241396 | 0.004166667 |
| R. prowazekii Rp22/narrow | 0.074607341 | 0.230598852 | 0.004166667 |
| R. slovaca 13Bnarrow | 0.106402799 | 0.19013078 | 0.004166667 |
| R. slovaca DCnarrow | 0.104094587 | 0.19013078 | 0.004166667 |
| Average | 0.107730605 | 0.242704461 | 0.012121212 |
| R. africae/wide | 0.201992224 | 0.29655889 | 0.101165072 |
| R. bellii OSU/wide | 0.113363604 | 0.234665676 | 0.162406932 |
| R. bellii RML/wide | 0.113974112 | 0.242176629 | 0.162406932 |
| R. felis/wide | 0.116700877 | 0.339836324 | 0.137385287 |
| R. japonica/wide | 0.251233383 | 0.329896265 | 0.089570374 |
| R. peacockii/wide | 0.316822239 | 0.401119108 | 0.135014642 |
| R. rhipicephali/wide | 0.231303473 | 0.383681072 | 0.119843847 |
| R. rickettsii/wide | 0.188528525 | 0.316222358 | 0.180385488 |
| R. typhi/wide | 0.131857171 | 0.3710849 | 0.089570374 |
| Average | 0.185086179 | 0.323915691 | 0.126917752 |
| Test significance | * | * | ** |
Note.—Values in italic were not used to calculate GR mean values nor for statistical test but were kept in the PCA plot. All tests were two-tailed and P values <0.05 were considered significant.
*P < 0.01, **P < 0.001.
Fig. 1.—
Relationship between δ values for whole-genome genetic distances (GD), genomic conservation (synteny, GC), and genomic rearrangements (GR). Rickettsia species were assigned to two categories depending on their host range: wide (with different insect vectors and vertebrate hosts; dark green circles) and narrow (with a single genus insect vector and few vertebrate host; dark red squares). The 24 taxa are projected on the first two PCA axes, which represent 68% and 16% of the total inertia.
Phylogenomic Analyses: Split Networks
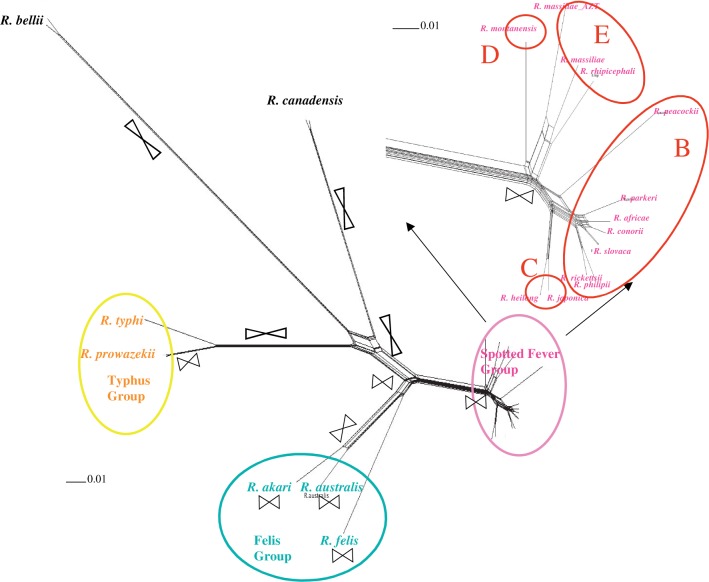
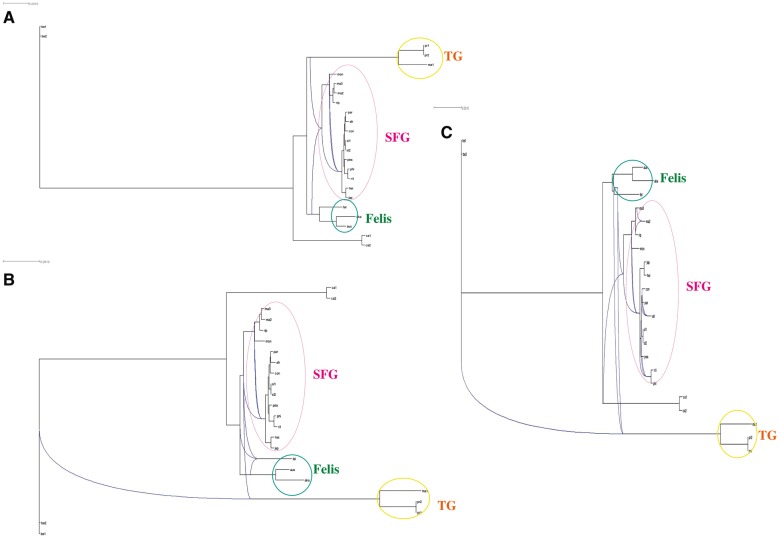
We constructed a split decomposition network to check for the signal of recombination events between the genomes. This method allows the visualization of the ancestral relationships between elements and does show some conflicting phylogenetic signals as indicated by the presence of cycles (splits) in the network (fig. 2). These cycles are highly supported statistically (Phi = 0) and thus are suggestive of the presence of recombination. Reticulation is located far from the tips of the branches, suggesting that recombination occurred mainly before or during species radiation. This is not the case, however, for the spotted fever group (SFG), for which reticulation is much closer to the very short branch tips. It is also worth noting that some reticulation is seen along the branch leading to R. typhi and R. prowazekii. On the other hand, neither genomic rearrangements nor genomic uncorrected distance data resolved the main Rickettsia groups (supplementary figs. S1 and S2, Supplementary Material online). The genome content network (fig. 2) contains 93 splits out of 919 possible edges, indicating that the horizontal phylogenetic signal is confined to ca. 10% of the species’ genomes. The genomic rearrangements network reveals larger horizontal signal, with 46 splits of 282 possible (16% of the genome; supplementary fig. S1, Supplementary Material online). Finally, for the genetic distance network, horizontal phylogenetic signal is present in 17.5% of the species’ DNA (supplementary fig. S2, Supplementary Material online).
Fig. 2.—
Split phylogenomic network obtained for Rickettsia genome content data. Main groups are color-coded in pink (spotted fever group), yellow (typhus group), and blue (Felis group). The main recombination events are mapped as intercrossing symbols on the phylogeny. The SFG detail shows clades corresponding to the cohesion plot for core proteome. Details on recombination events are provided in supplementary material, Supplementary Material online.
Phylogenomic Analyses: Core Proteome
A total number of 1,402 clusters of orthologous protein-coding genes (COGs) were identified. The core genome consists of 606 chromosomal genes, which represent 43.2% of the total. This core set represents a large proportion (606/755) of the total number of ORFs in the smallest genome (R. typhii Wilmington) and roughly half (606/1,204) in the largest genome (R. bellii RML 369-C). The core genome size of Rickettsia we estimated is quite similar to previous reports (Blanc et al. 2007b; Wu et al. 2009), with a length of 678,829 bp, of which 209,785 (31%) were variable. Functional classification of the COGs shows that they belong to all primary functions (supplementary fig. S3, Supplementary Material online), with the majority coding for components of information-processing systems involved in translation, ribosomal structure, and metabolism.
Phylogenomic Analyses: MCOA
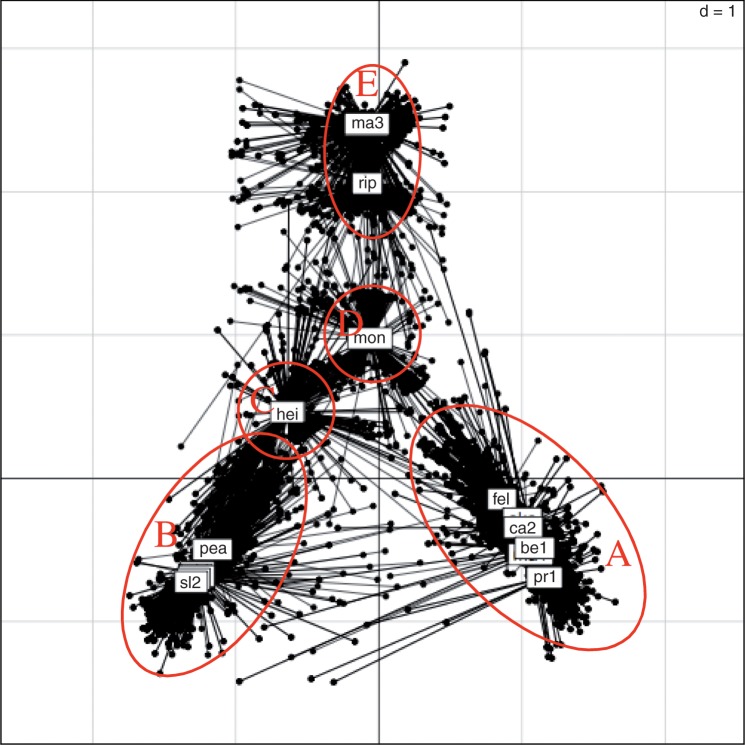
The plot of the reference position of each species, as well as their position in each of the 606 gene trees using the two first axes of the MCOA analysis (cohesion plot), is shown in figure 3. The set of reference positions (square labels) of the species in the cohesion plot is called consensus typology. Interestingly, the typology clusters in the cohesion plot correspond to those of the phylogenomic networks: cluster A corresponds to the TG + Felis group + R. belli + R. canadensis clades in the network (fig. 1); cluster B corresponds to seven species of the R. parkeri–R. philipii–R. peacockii network group; the other two groups roughly correspond to network clusters, but they appear more distantly connected in the cohesion plot (fig. 2). Cluster C groups the oriental species R. heilongjiangensis and R. japonica, while cluster E groups R. rhipicephali and R. massiliae. R. montanensis (cluster D) appears in between typology clusters C and E but closer to C. The spotted fever group clusters (B, C, D, and E) are all grouped in highly reticulated part of the network, and thus their relative positions is different in the cohesion plot.
Fig. 3.—
Cohesion plot for every gene tree, showing the consensus typology estimated by Phylo-MCOA. For each species, lines represent the distance between its reference position (squares) and its position in each individual tree. Long lines represent species which position in a given tree is not concordant with its reference position.
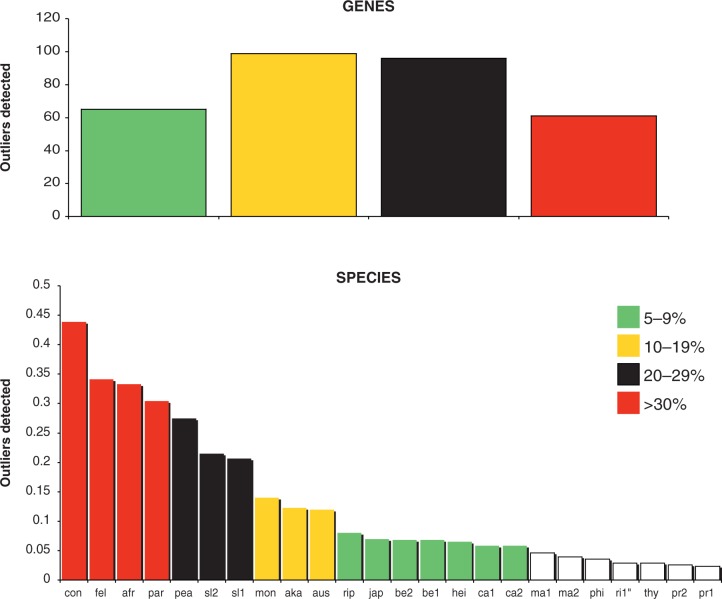
The MCOA scores for genes and species are shown in figure 4. The analysis detected outlier genes in all major functional categories at different thresholds, with the majority (n = 195) having different histories in 10–30% of the trees (fig. 4). This represents more than a quarter (32.18%) of the total, which is higher than the previous estimation (28%) of recombination on the Rickettsia core proteome (Wu et al. 2009). Furthermore, 65 genes have conflicting histories in 5–9% and 61 in 30% or more of the trees. The total number of outlier genes above the 5% threshold represents more than half of the core proteome (n = 321; 53%). On the other hand, 169 genes had the same position in all trees (27.88%). The analyses detected six outlier species in more than 20% of the trees: R. conorii had different phylogenetic position in 44%, followed by R. felis (34%), R. africae (33%), R. parkeri (30%), R. peacockii (27%), R. slovaca D-CWPP (22%), and R. slovaca 13-B (21%). R. montanensis, R. akari, and R. australis had incongruent topologies in more than 10% of the trees, and a further seven species had different histories for 5–9% of the trees (fig. 4).
Fig. 4.—
Bar plots for the Rickettsia tree comparison, showing the score calculated for each gene (top) and each species (bottom). Red bars represent significant outliers according to the threshold chosen (0.2). Nodal distances were used for the Phylo-MCOA analysis.
Phylogenomic Analyses: Rooted Networks
The rooted galled-phylogenetic network (fig. 5A) displays a set of putative recombination events (blue lines) that may explain the differences between the 586 individual gene trees, calculated for clusters present in at least 20% of them. Again, the recombination seems to have occurred during the radiation of Rickettsia and happened more frequently within the SFG and between TG, Felis group, and SFG. When the tree-reconciliation threshold is lower, considering clusters present in at least 15% of the trees, recombination is observed between R. bellii and TG, as well as more recombination between R. felis, R. australis, R. akari, and the SFG (fig. 5B). For an even smaller fraction of genes (threshold 10%), more recombination is observed, particularly more recent events between R. africae with R. conorii, R. parkerii, and R. slovaca; between the two strains of R. massiliae and R. rhipicephali; and between R. rickettsii–R. philipi and the rest of SFG (fig. 5C). Threshold values more than 20% resulted in no reticulation events, indicating that 80% of the genes have consistent histories. Finally, the rooted consensus recombination network (supplementary fig. S4, Supplementary Material online) for all the gene trees also shows a pattern of recombination during the radiation of the group (blue lines) and between same-species strains, except for the non-SFG ones. The recombination network has 69 splits out of 614 edges, which correspond to 11.24% of horizontal signal in the trees.
Fig. 5.—
Galled rooted hybridization networks displaying putative recombination events that reconcile the differences between the 606 individual core gene trees. Decreasing thresholds for network construction reveal increasing numbers of recombination events. Networks display clusters present in at least (A) 20%, (B) 15%, and (C) 10% of the gene trees.
Phylogenomic Analyses: Bayesian Estimation of Recombination
The ratio of recombination to mutation rate ρ/θ was estimated by ClonalFrame to be between 0.025 and 0.037 (mean = 0.034), and the ratio of rates at which sites are altered through recombination and mutation (r/m) was estimated between 0.325 and 0.514 (mean = 0.355). This value falls toward the bottom end of r/m values estimated for several archaea and bacterial groups (Vos and Didelot 2009). We found the mean tract length of recombined fragments to be 77 bp, with a range between 66 and 145 bp. The Bayesian tree computed by Clonal Frame and the full view of recombination events across the genomes is shown in supplementary figure S5, Supplementary Material online. The reconstruction of recombination events on the genome content network (fig. 2) shows that recombination took place mainly during the radiation of the spotted fever group, as no events were detected near the branches. However, the Felis group which includes R. akari, R. felis, and R. australis shows evidence of recombination both during the radiation and more recent events (supplementary fig. S5, Supplementary Material online). Recombination was found to be extensive and ongoing between strains of R. bellii, R. prowazekii, and R. Canadensis, but it is absent between R. massiliae and R. slovaca strains.
Phylogenomic Analyses: Bayesian Phylogenomic Concordance
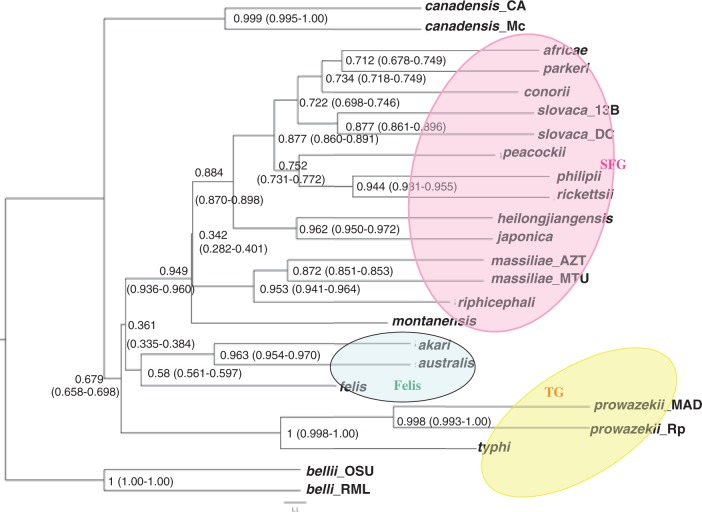
The combined samples for the 606 loci resulted in 1,042,519 single-gene Bayesian topologies and 39,908 different splits. The mean number of clusters (groups of loci sharing the same topology) at the 95% probability region estimated by the BCA was 175 (SD across runs = 1.201; range 163–192). The concordance analysis revealed that only the typhus group clade (R. typhi–R. prowazekii) and the R. bellii clade were supported by 100% of the genes (fig. 6). The lowest CF values are those at the base of the SFG clade, with only 36.1% of the core genes supporting the basal node (fig. 6). Furthermore, even if the relationship between R. montanensis and the rest of the SFG has strong genome support, the base of the rest of the SFG is only represented in 34.2% of the core genes. Similarly, the rest of the internal SFG nodes range from 34% to 84% genome support and only the terminal relationships (species relationships) have higher CF values. This is in agreement with the recombination events detected and consistent with a rapid radiation scenario. Most strain relations are supported by more than 90% of the genes, except for the R. massiliae clade, supported by 87.2% of core genes.
Fig. 6.—
Primary concordance tree for the Rickettsia core genome data set. Numbers are posterior mean CFs and their 95% credibility intervals, obtained with α = 1. Numbers refer to sample-wide CFs and their associated 95% confidence interval (in parentheses). Short branches (branch length given in coalescent units), most affected by ILS, have lowest CFs.
Discussion
Comparisons of closely related bacterial species are needed to test the role of HGT (recombination) in bacterial evolution and eventually speciation (Wiedenbeck and Cohan 2011). In general, studies of diversification in bacteria have not yet attempted to identify and compare the genomes of the most recent products of a radiation. In this study, we used 24 complete published genomes to provide the first global insight into the processes of HGT and diversification in the genus Rickettsia.
Ecology and Reticulated Evolution
We found a consistent pattern of δ values correlation with host range, which implies that population fragmentation and isolation, together with specialization, substantially reduce the genetic connectivity of intracellular bacteria to exchange genes horizontally. Even so, some specialized Rickkettsia species do show reticulation levels more similar to those of generalist ones (R. akari and R. massiliae, both SFG members; fig. 1). The link between ecological niche and reticulation/recombination is not straightforward though, because recombination was detected at deep nodes within the SFG. Furthermore, even if R. felis has multiple genomic rearrangements and was found to recombine extensively (discussed later), it was always found to have intermediate δ-plot values. The δ plots for different genomic regions clearly illustrate that reticulated evolution varies across the genome. R. peacockii had the highest values in the genetic distance and genome conservation δ-plot analyses (table 1 and fig. 1). This is consistent with the presence of 42 copies of the ISRpe1 transposon on the R. peacockii chromosome, which might be associated with a lack of synteny with the genome of closely related Rickettsia species, and numerous deletions via recombination between transposon copies (Felsheim et al. 2009).
Recombination/HGT
A previous phylogenomic analysis for 11 species estimated that recombination occurred frequently on core proteome genes during Rickettsia evolution (Wu et al. 2009). Here, we show that homologous recombination is detectable across the genome, and that, despite being less frequent than mutation (0.035 recombination events per mutation event), it occurred mainly during the radiation of the group and is still happening for some species. No significant positional bias for recombination was detected along the chromosome, and ρ did not correlate positively with the degree of sequence conservation; this suggests that recombination acts similarly on most of the genome. Furthermore, the SFG outlier species detected for core proteome (R. conorii, R. africae, R. parkeri, R. peacockii, and R. slovaca) showed no evidence of recent recombination and are all grouped in the SFG clade, for which recombination was inferred at its basal nodes, suggesting recombination early in their divergence (fig. 4 and supplementary fig. S4, Supplementary Material online).
Similarly, our genomic concordance analysis revealed a primary phylogenetic history across the Rickettsia genome, recovering all the major clades previously identified, but also documented striking phylogenetic discordance on a genome-wide scale. In the case of the taxa where there is an underlying tree-like history (the species tree), the primary phylogenetic history is expected to match the species history, whereas the frequencies of the minor trees should reflect the contributions of incomplete lineage sorting and HGT/recombination. The most striking discordance was found at the base of the SFG, including the node connecting the SFG and the Felis group (supported by 34% and 36% of genes, respectively), and all of which have very short branches (i.e., short estimated coalescent units). The other nodes with CF values <0.5 had also short branches and were ancestral. Short branches, most affected by ILS, have lowest CFs. When ILS is the only process causing discordance, the CF of minor clades conflicting with the “main” topology is completely determined by the coalescent units (Ané 2011). The genome content network is consistent with this, as more splits and very short branches are seen in the central parts of the network, including the SFG cluster (fig. 2). Phylogenetic analyses using 16S and other nuclear genes indicate that the genus Rickettsia evolved approximately 150 Ma, and that a rapid radiation occurred about 50 Ma which led to most of the species of arthropod-related rickettsies (Weinert et al. 2009). The fact that around 10–17.5% of the genome contains horizontal phylogenetic signal and that around 25% of the core genes have different evolutionary histories supports the importance of HGT in the radiation and diversification of Rickettsia bacteria.
Which Genes Are Involved?
It has been proposed that almost all genes are prone to HGT, with only a few that are resistant to it in the laboratory (McInerney and Pisani 2007; Sorek et al. 2007) and probably none that has not been affected by it over the course of evolutionary time (Bapteste et al. 2009). Our core genome analyses reveal that, during Rickettsia evolution, genes belonging to all functional categories show reticulated evolution and possible HGT. In contrast to the assertion that certain (adaptive) traits are more likely to be associated with HGT than others, such as the acquisition of antibiotic resistance or defensive traits in pathogens (Wiedenbeck and Cohan 2011), our results reveal only 14 genes involved in cell wall and membrane biogenesis and only two involved in defense mechanisms (supplementary fig. S3, Supplementary Material online). However, only one species for which recent recombination was detected (R. felis) was retrieved as an outlier by the MCOA analyses for the core proteome. Similarly, the species with higher number of recombination events (R. bellii, R. typhii, R. prowazekii, and R. canadensis) had intermediate or low proportions of outlier genes detected. The incongruence between the core proteome and the whole core genome estimations could result from many factors. In terms of ecology, only one generalist species (R. felis which has a broad range of insect hosts, including fleas, ticks, and lice) was detected as outlier (table 1) and with recent recombination events, whereas the specialist species R. australis and R. akari also show evidence of moderate to low recent recombination events but were not detected as outliers. It is possible that the lack of recombination observed in the SFG results from their specialized life style and narrow host ranges, and that the reticulated evolution observed (including the detection of four outlier species within the group) results from ancient recombination, as shown in our results. Another possibility is that ancestral polymorphisms (i.e., incomplete lineage sorting), which is common after rapid radiations, results in reticulated evolution. If the latter is the case, the extent and evolutionary significance of recombination is hard to evaluate, because of the difficulty in distinguishing hybridization from incomplete lineage sorting.
“Mobilome” and Genome Architecture
The presence of mobile genetic elements (transposable elements, plasmids, and conjugation elements) might also play an important role in the distribution and frequency of recombination. For example, the two plasmids present in R. felis may have been acquired by conjugation and provided evidence for gene transfers between the chromosome and the pRF plasmid (Ogata et al. 2005). While all rickettsial genomes have chromosome-encoded patatin-like phospholipase (pat 1) and the gene organization around pat 1 is similar between different rickettsiae, R. felis possesses an additional paralog pat 2 in the pRF plasmid. Interestingly, a phylogenetic analysis for patatin-like phospholipase genes indicates a close relationship between pat 1 and pat 2 of R. felis that together form an outgroup to pat 1 sequences of other Rickettsia spp., suggesting gene replacement of the chromosomally encoded pat 1 by the plasmid-encoded pat 2 in the lineage leading to R. felis (Blanc et al. 2005). This could also be the case for R. massiliae, for which no recombination events or reticulated evolution by MCOA analyses was detected. It had, however, very high δ values (fig. 1), in particular for genomic rearrangements. Indeed, the genome of R. massiliae shows extensive colinearity with the other SFG genomes except for the tra region. A phylogenetic study gave evidence for horizontal acquisition of the tra cluster by R. massiliae from a species related to R. bellii (Blanc et al. 2007a). Another interesting species is R. conorii, for which 44% of its core proteins had different evolutionary histories, but again no recombination was detected for the core genome. The R. conorii genome contains a large number of orphans, of which 80% are short gene fragments or fusions of short segments from neighboring, deteriorated genes (Amiri et al. 2003). The resulting short repeated sequences located in close proximity to each other are known to play an important role in mediating recombination events in Rickettsia and other species and can result in fusions and partial losses of genic sequences (Andersson and Andersson 1999; Ogata et al. 2000; Ogata et al. 2001; Amiri et al. 2003).
Intraspecific Sampling and Accuracy of Recombination Estimates
In bacteria, each recombination event affects a contiguous region of sequence but leaves the remainder of the circular chromosome unchanged. Due to the rapid evolution of bacterial populations and genomes, the footprint left by a recombination event can quickly be “eroded” by mutation and drift, making very difficult to detect ancient recombination events (discussed earlier). This is of particular concern for the Bayesian inference method implemented in ClonalFrame, which estimates the subset of the genome that has not undergone recombination (i.e., clonal lineage) between closely related bacterial strains. Furthermore, as ClonalFrame does not look for potential sources of descent for each stretch of transferred DNA, it tends to underestimate the number of recombination events that have taken place even between closely related strains (Didelot and Falush 2007). Despite the small number of intraspecific strains analyzed and the underestimation of recombination this entails, the amount of recombination events detected between strains is clearly higher than between species (red bars in supplementary fig. S5, Supplementary Material online). This is also observed for the estimations obtained with BCA (fig. 6) and hybridization network (supplementary fig. S4, Supplementary Material online).
Conclusion
Overall, our results reveal that 1) ecological specialization seems to restrict recombination, both within and between species, but genomic architecture and content of mobile elements are also capital in HGT and recombination potential; 2) different genomic regions contain different levels of HGT and reticulated evolution; 3) genetic distances, genomic rearrangements, and genome conservation (synteny) all show evidence of HGT and network-like evolution at whole and core genome scale; 4) core proteome genes of every major functional categories have experienced reticulated evolution and HGT; 5) recombination events occurred at the origin of the SFG radiation and is still occurring within some strains and has recently occurred between TG and Felis group species; finally 6) we show that HGT events, even if relatively common, still leave a major tree-like history for Rickettsia evolutionary history, but HGT seems to have played an important role during the radiation of the genus.
Supplementary Material
Supplementary figures S1–S5 and table S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The computational work was done using Blades from Aix-Marseille University specific funding program (FIR). The research was funded by a post doctoral Infectiopole Sud grant.
Literature Cited
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiri H, Davids W, Andersson SGE. Birth and death of orphan genes in Rickettsia. Mol Biol Evol. 2003;20:1575–1587. doi: 10.1093/molbev/msg175. [DOI] [PubMed] [Google Scholar]
- Andersson JO, Andersson SGE. Genome degradation is an ongoing process in Rickettsia. Mol Biol Evol. 1999;16:1178–1191. doi: 10.1093/oxfordjournals.molbev.a026208. [DOI] [PubMed] [Google Scholar]
- Ané C. Detecting phylogenetic breakpoints and discordance from genome-wide alignments for species tree reconstruction. Genome Biol Evol. 2011;3:246–258. doi: 10.1093/gbe/evr013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ané C, Larget B, Baum DA, Smith SD, Rokas A. Bayesian estimation of concordance among gene trees. Mol Biol Evol. 2007;24:412–426. doi: 10.1093/molbev/msl170. [DOI] [PubMed] [Google Scholar]
- Bapteste E, et al. Prokaryotic evolution and the tree of life are two different things. Biol Direct. 2009;4:34. doi: 10.1186/1745-6150-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraclough TG, Balbi KJ, Ellis RJ. Evolving concepts of bacterial species. Evol Biol. 2012;39:148–157. [Google Scholar]
- Beati L, Peter O, Burgdorfer W, Aeschlimann A, Raoult D. Confirmation that Rickettsia helvetica sp. nov. is a distinct species of the spotted fever group of rickettsiae. Int J Syst Bacteriol. 1993;43:521–526. doi: 10.1099/00207713-43-3-521. [DOI] [PubMed] [Google Scholar]
- Blanc G, Renesto P, Raoult D. Phylogenic analysis of rickettsial patatin-like protein with conserved phospholipase A2 active sites. Ann N Y Acad Sci. 2005;1063:83–86. doi: 10.1196/annals.1355.012. [DOI] [PubMed] [Google Scholar]
- Blanc G, et al. Lateral gene transfer between obligate intracellular bacteria: evidence from the Rickettsia massiliae genome. Genome Res. 2007a;17:1657–1664. doi: 10.1101/gr.6742107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, et al. Reductive genome evolution from the mother of Rickettsia. PLoS Genet. 2007b;3:e14. doi: 10.1371/journal.pgen.0030014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein SR, Reznikoff WS. Mobile DNA in obligate intracellular bacteria. Nat Rev Microbiol. 2005;3:688–699. doi: 10.1038/nrmicro1233. [DOI] [PubMed] [Google Scholar]
- Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budroni S, et al. Neisseria meningitidis is structured in clades associated with restriction modification systems that modulate homologous recombination. Proc Natl Acad Sci U S A. 2011;108:4494–4499. doi: 10.1073/pnas.1019751108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgdorfer W, Aeschlimann A, Peter O, Hayes SF, Philip RN. Ixodes ricinus: vector of a hitherto undescribed spotted fever group agent in Switzerland. Acta Trop. 1979;36:357–367. [PubMed] [Google Scholar]
- Carrolo M, Pinto FR, Melo-Cristino J, Ramirez M. Pherotypes are driving genetic differentiation within Streptococcus pneumoniae. BMC Microbiol. 2009;9:191. doi: 10.1186/1471-2180-9-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohan FM. Bacterial species and speciation. Syst Biol. 2001;50:513–524. doi: 10.1080/10635150118398. [DOI] [PubMed] [Google Scholar]
- Cohan FM. Towards a conceptual and operational union of bacterial systematics, ecology, and evolution. Philos Trans R Soc Lond B Biol Sci. 2006;361:1985–1996. doi: 10.1098/rstb.2006.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corander J, Connor TR, O’Dwyer CA, Kroll JS, Hanage WP. Population structure in the Neisseria, and the biological significance of fuzzy species. J R Soc Interface. 2012;9:1208–1215. doi: 10.1098/rsif.2011.0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sunderland (MA): Sinauer Associates; 2004. [Google Scholar]
- Dagan T, Artzy-Randrup Y, Martin W. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc Natl Acad Sci U S A. 2008;105:10039–10044. doi: 10.1073/pnas.0800679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vienne DM, Ollier S, Aguileta G. Phylo-MCOA: a fast and efficient method to detect outlier genes and species in phylogenomics using multiple co-inertia analysis. Mol Biol Evol. 2012;29:1587–1598. doi: 10.1093/molbev/msr317. [DOI] [PubMed] [Google Scholar]
- Didelot X, Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics. 2007;175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, et al. Recombination and population structure in Salmonella enterica. PLoS Genet. 2011;7:e1002191. doi: 10.1371/journal.pgen.1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsheim RF, Kurtti TJ, Munderloh UG. Genome sequence of the endosymbiont Rickettsia peacockii and comparison with virulent Rickettsia rickettsii: identification of virulence factors. PLoS One. 2009;4:e8361. doi: 10.1371/journal.pone.0008361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier PE, Fujita H, Takada N, Raoult D. Genetic identification of rickettsiae isolated from ticks in Japan. J Clin Microbiol. 2002;40:2176–2181. doi: 10.1128/JCM.40.6.2176-2181.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C, Hanage WP, Spratt BG. Neutral microepidemic evolution of bacterial pathogens. Proc Natl Acad Sci U S A. 2005;102:1968–1973. doi: 10.1073/pnas.0406993102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, et al. Re-evaluating prokaryotic species. Nat Rev Microbiol. 2005;3:733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- Gogarten JP. The early evolution of cellular life. Trends Ecol Evol. 1995;10:147–151. doi: 10.1016/s0169-5347(00)89024-2. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Starke T, Kibak H, Fishman J, Taiz L. Evolution and isoforms of V-ATPase subunits. J Exp Biol. 1992;172:137–147. doi: 10.1242/jeb.172.1.137. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Holland BR, Huber KT, Dress A, Moulton V. δ Plots: a tool for analyzing phylogenetic distance data. Mol Biol Evol. 2002;19:2051–2059. doi: 10.1093/oxfordjournals.molbev.a004030. [DOI] [PubMed] [Google Scholar]
- Hollander M, Wolfe DA. Nonparametric statistical methods. New York: John Wiley and Sons; 1973. [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks to evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Huson DH, Scornavacca C. Dendroscope 3: an interactive viewer for rooted phylogenetic trees and networks. Syst Biol. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- Kloesges T, Popa O, Martin W, Dagan T. Networks of gene sharing among 329 proteobacterial genomes reveal differences in lateral gene transfer frequency at different phylogenetic depths. Mol Biol Evol. 2011;28:1057–1074. doi: 10.1093/molbev/msq297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larget BR, Kotha SK, Dewey CN, Ané C. BUCKy: gene tree/species tree reconciliation with Bayesian concordance analysis. Bioinformatics. 2010;26:2910–2911. doi: 10.1093/bioinformatics/btq539. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J, Cohan FM. The effect of mismatch repair and heteroduplex formation on sexual isolation in Bacillus. Genetics. 1998;48:13–18. doi: 10.1093/genetics/148.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J, Zawadzki P, Pickerill P, Cohan FM, Dowson CG. Barriers to genetic exchange between bacterial species: Streptococcus pneumoniae transformation. J Bacteriol. 2000;182:1016–1023. doi: 10.1128/jb.182.4.1016-1023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney JO, Pisani D. Genetics—paradigm for life. Science. 2007;318:1390–1391. doi: 10.1126/science.1151657. [DOI] [PubMed] [Google Scholar]
- Médigue C, Rouxel T, Vigier P, Hénaut A, Danchin A. Evidence of horizontal gene transfer in Escherichia coli speciation. J Mol Biol. 1991;222:851–856. doi: 10.1016/0022-2836(91)90575-q. [DOI] [PubMed] [Google Scholar]
- Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. The microbial pan-genome. Curr Opin Genet Dev. 2005;15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Merhej V, Raoult D. Rickettsial evolution in the light of comparative genomics. Biol Rev. 2011;86:379–405. doi: 10.1111/j.1469-185X.2010.00151.x. [DOI] [PubMed] [Google Scholar]
- Moran NA, McLaughlin HJ, Sorek R. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science. 2009;323:379–382. doi: 10.1126/science.1167140. [DOI] [PubMed] [Google Scholar]
- Norman A, Hansen LH, Sorensen SJ. Conjugative plasmids: vessels of the communal gene pool. Philos Trans R Soc Lond B Biol Sci. 2009;364:2275–2289. doi: 10.1098/rstb.2009.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Lerat E, Daubin V. Examining bacterial species under the specter of gene transfer and exchange. Proc Natl Acad Sci U S A. 2005;102:6595–6599. doi: 10.1073/pnas.0502035102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, et al. Selfish DNA in protein coding genes. Science. 2000;290:347–350. doi: 10.1126/science.290.5490.347. [DOI] [PubMed] [Google Scholar]
- Ogata H, et al. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science. 2001;293:2093–2098. doi: 10.1126/science.1061471. [DOI] [PubMed] [Google Scholar]
- Ogata H, et al. The genome sequence of Rickettsia felis identifies the first putative conjugative plasmid in an obligate intracellular parasite. PLoS Biol. 2005;3: e248. doi: 10.1371/journal.pbio.0030248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parola P, Raoult D. Ticks and tickborne bacterial diseases in humans: an emerging infectious threat. Clin Infect Dis. 2001;32:897–928. doi: 10.1086/319347. [DOI] [PubMed] [Google Scholar]
- Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond B, Wyres KL, Sheppard SK, Ellis RJ, Bonsall MB. Environmental factors determining the epidemiology and population genetic structure of the Bacillus cereus group in the field. PLoS Pathog. 2010;6:e1000905. doi: 10.1371/journal.ppat.1000905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliep K, Lopez P, Lapointe FJ, Bapteste E. Harvesting evolutionary signals in a forest of prokaryotic gene trees. Mol Biol Evol. 2011;28:1393–1405. doi: 10.1093/molbev/msq323. [DOI] [PubMed] [Google Scholar]
- Smith JM, Smith NH, O'Rourke M, Spratt BG. How clonal are bacteria? Proc Natl Acad Sci U S A. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, et al. Genome-wide experimental determination of barriers to horizontal gene transfer. Science. 2007;318:1449–1452. doi: 10.1126/science.1147112. [DOI] [PubMed] [Google Scholar]
- Thomas CM, Nielson K. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- Vos M, Didelot X. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 2009;3:199–208. doi: 10.1038/ismej.2008.93. [DOI] [PubMed] [Google Scholar]
- Vulic M, Dionisio F, Taddei F, Radman M. Molecular keys to speciation: DNA polymorphism and the control of genetic exchange in enterobacteria. Proc Natl Acad Sci U S A. 1997;94:9763–9767. doi: 10.1073/pnas.94.18.9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert LA, Werren JH, Aebi A, Stone GN, Jiggins FM. Evolution and diversity of Rickettsia bacteria. BMC Biol. 2009;7:6. doi: 10.1186/1741-7007-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenbeck J, Cohan FM. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol Rev. 2011;35:957–976. doi: 10.1111/j.1574-6976.2011.00292.x. [DOI] [PubMed] [Google Scholar]
- Wu J, Yu T, Bao Q, Zhao F. Evidence of extensive homologous recombination in the core genome of rickettsia. Comp Funct Genomics. 2009:510271. doi: 10.1155/2009/510270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawadzki P, Roberts MS, Cohan FM. The log-linear relationship between sexual isolation and sequence divergence in Bacillus transformation is robust. Genetics. 1995;140:917–932. doi: 10.1093/genetics/140.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.