Abstract
Two distinct classes of repetitive sequences, interspersed mobile elements and satellite DNAs, shape eukaryotic genomes and drive their evolution. Short arrays of tandem repeats can also be present within nonautonomous miniature inverted repeat transposable elements (MITEs). In the clam Donax trunculus, we characterized a composite, high copy number MITE, named DTC84. It is composed of a central region built of up to five core repeats linked to a microsatellite segment at one array end and flanked by sequences holding short inverted repeats. The modular composition and the conserved putative target site duplication sequence AA at the element termini are equivalent to the composition of several elements found in the cupped oyster Crassostrea virginica and in some insects. A unique feature of D. trunculus element is ordered array of core repeat variants, distinctive by diagnostic changes. Position of variants in the array is fixed, regardless of alterations in the core repeat copy number. Each repeat harbors a palindrome near the junction with the following unit, being a potential hotspot responsible for array length variations. As a consequence, variations in number of tandem repeats and variations in flanking sequences make every sequenced element unique. Core repeats may be thus considered as individual units within the MITE, with flanking sequences representing a “cassette” for internal repeats. Our results demonstrate that onset and spread of tandem repeats can be more intimately linked to processes of transposition than previously thought and suggest that genomes are shaped by interplays within a complex network of repetitive sequences.
Keywords: mobile element, MITE, satellite DNA, tandem repeats, sequence rearrangements, evolution
Introduction
Eukaryotic genomes host two ubiquitous classes of highly abundant repetitive sequences, satellite DNAs (satDNAs) and transposable elements (TEs) (López-Flores and Garrido-Ramos 2012). TEs are sequence segments able to move to new genomic locations and form interspersed repeats if replicated in this process (Finnegan 1989; Kazazian 2004; Jurka et al. 2007). Large number of diverse TEs exists in genomes, grouped into two basic classes based on mechanisms of transposition. Class I elements transpose by RNA-mediated mechanisms, while DNA-mediated processes spread class II elements. Each of them includes autonomous and nonautonomous copies, the former being able to code for all products needed for their own transposition while the later depend on enzymes produced by the first. Passive transposition of a whole palette of sequences is possible due to the ability of mechanisms involved in transposition to recognize DNA secondary structures, such as inverted repeats (Craig 1995; Izsvák et al. 1999; Coates et al. 2011).
satDNAs are tandemly repeated noncoding sequences located in heterochromatic chromosomal compartments (Plohl et al. 2008). Characteristic low sequence variability of satDNAs is considered to be a consequence of a phenomenon called concerted evolution, in which mutations are homogenized among repeats of a family in a genome and fixed among individuals in a population (Dover 1986). Many satDNAs differing in length, sequence, copy number, and origin can coexist in a genome, but processes and possible constraints limiting their onset and persistence are understood only fragmentarily (Meštrović et al. 2006).
Despite differences in structure, organization, mechanisms of spread, and sequence dynamics, growing number of reports indicate traits that link TEs and satDNAs. Internal tandem repeats found in some TEs provoke a hypothesis that their expansion may represent a source of some satDNAs (Noma and Ohtsubo 2000; Gaffney et al. 2003; Macas et al. 2009). In addition, satDNA repeats were found as single units or short arrays interspersed in euchromatic portions of the genome, probably as parts of yet uncharacterized TEs (Cafasso et al. 2003; Brajković et al. 2012). It was suggested that inverted repeats formed by inversion of satDNA monomers can promote interspersed distribution of such units (Mravinac and Plohl 2010). It must be noted that a direction of transition between organizational forms is difficult to assess because shifts from mobile elements to satDNAs can also be anticipated in the opposite direction (Heikkinen et al. 1995; Macas et al. 2009).
Miniature inverted-repeat transposable elements (MITEs) are one group of nonautonomous DNA transposons. They are small (usually up to 600 bp), lack coding potential and/or RNA pol III promoter site, and are featured by terminal or subterminal inverted repeats, ability to fold into secondary structures, and short target site duplication (TSD) sequences formed in the process of insertion (Feschotte et al. 2002). MITEs are usually present in a high copy number in genomes and are widespread in plants, animals, and fungi (Bureau and Wessler 1992; Wang et al. 2010; Fleetwood et al. 2011). They are considered to be derived from larger autonomous elements (Feschotte and Mouchès 2000) and probably propagate through a cut-and-paste mechanism of transposition combined with a gap repair and/or aberrant DNA replication, triggered by secondary structures (Izsvák et al. 1999; Coates et al. 2011).
Some MITE sequences have tandem repeats in their central part. Arrays of variable number of tandem repeats (usually up to 6) are a common trait of MITE-like elements DINE1 (Yang and Barbash 2008), SGM (Miller et al. 2000), mini-me (Wilder and Hollocher 2001), and PERI (Kuhn and Heslop-Harrison 2011), described in Drosophila, and of MINE-2 in some Lepidoptera (Coates et al. 2011). Tandem repeats of these elements are followed by a short microsatellite array at one end. Both modules are embedded between flanking sequences featured by an inverted repeat and the TSD sequence AA. Described modular structure was found in elements of the pearl family detected in the cupped oyster Crassostrea virginica (Gaffney et al. 2003). In addition, internal tandem repeats (core repeats) of pearl share sequence similarity and unit length with several satDNAs widespread in bivalve mollusks (Plohl and Cornudella 1996; López-Flores et al. 2004; Biscotti et al. 2007; Plohl et al. 2010), thus linking TEs and satDNAs in these organisms.
Standard focus in studying TEs is mostly on characterization of sequence traits that might be responsible for their mobility (López-Flores and Garrido-Ramos 2012). The same also holds for repeat-incorporating MITEs while available information is scarce if we consider sequence dynamics, range, and possible causes of variability of tandem repeats residing within them. Here, we characterize a novel MITE element in the clam D. trunculus, DTC84, with focus on tandem repeats residing within it and suggest pathways and mechanisms involved in their evolution.
Materials and Methods
Construction of Partial Genomic Libraries
Donax trunculus genomic DNA was obtained from commercially supplied adult specimens using adjusted phenol/chloroform extraction protocol (Plohl and Cornudella 1997). Following the strategy described by Biscotti et al. (2007), genomic DNA was partially digested (10 μg of DNA, 37 °C/5 min) with 5 U of AluI restriction endonuclease (Fermentas) in order to reveal mass of degraded fragments in a range between 300 and 3,000 bp. The fragments were ligated into the pUC19/SmaI vector. Transformed Escherichia coli DH5α competent cells (Invitrogen) were grown on 90-mm ampicillin-selective plates. After colony transfer, positively charged membranes (Amersham) were probed with digoxigenin-labeled AluI-digested (complete digestion) D. trunculus genomic DNA. Labeling, hybridization, and signal detection were performed as described in the following section. Hybridization was conducted under 65 °C in 20 mM sodium phosphate buffer (pH 7.2), 20% sodium dodecyl sulfate (SDS), allowing ∼80% sequence similarity.
Southern Hybridization and Dot Blot Quantification
For Southern analysis, genomic DNA (2.5 μg/sample) was digested with 20 U of restriction endonucleases overnight, fragments were separated by electrophoresis on 1% agarose gel, and transferred onto a positively charged nylon membrane (Roche). Polymerase chain reaction (PCR)-amplified fragments of interest were labeled with digoxigenin by random priming using the DIG DNA Labeling and Detection Kit (Roche) and used as a hybridization probe. Membranes were hybridized in 20 mM sodium phosphate (pH 7.2), 20% SDS, at low, moderate, and high stringency conditions (60 °C, 65 °C, and 68 °C, respectively). Stringency washing was conducted in 20 mM sodium phosphate buffer, 1% SDS, at the temperature three degrees lower than the hybridization temperature. To detect the hybridization, signal membranes were incubated with anti-digoxigenin alkaline phosphatase conjugate, and chemiluminescent signals induced by CDP-Star (Roche) were captured on X-ray films (Amersham).
The relative genomic contribution of the DTC84 core repeat sequence was determined by dot blot analysis. Serial dilutions of D. trunculus genomic DNA and core repeat sequences were spotted onto a nylon membrane. Hybridization was performed under high (68 °C) and low stringency conditions (60 °C).
PCR Amplification
To amplify core repeats in DTC84 elements, primer pair DTC84AluSatF: TTGCCTGTGACGTCTACTTGTGC and DTC84AluSatR: AGAGGTCACAGGCAACCATCCA was derived according to the DTC84 clone. Amplification was performed with initial denaturation at 94 °C for 5 min, 35 cycles of 94 °C for 30 s, 57 °C for 30 s, 72 °C for 30 s, and final extension at 72 °C for 7 min.
Primers constructed according to sequence segments that flank core repeats DTC84mobF: AACAAGAGCACCGCTGGGCG and DTC84mobR: CGCACGTTTGAAAAACGGGACGTA were used in order to amplify additional copies of DTC84 elements. Amplification was performed with initial denaturation at 94 °C for 5 min, 35 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 1 min, and final extension at 72 °C for 7 min.
All PCR products were cloned into pGEM-T Easy Vector System (Promega), and recombinant clones containing multimers were sequenced. All cloned fragments were sequenced at Macrogen Inc. (Korea) on ABI3730XL DNA Analyzer. Sequences submitted to GenBank obtained the following accession numbers: KC981676–KC981759.
Sequence Analysis
Obtained multimeric DNA sequences were trimmed to exclude primer binding sites from sequence analysis. Sequence editing and alignments were performed using the Geneious 5.4.3 program (Biomatters Ltd.).
Substructures, repeats, and motifs were searched with appropriate applications within the online tool Oligonucleotids repeats finder, developed by Bazin, Kosarev, and Babenko (http://wwwmgs.bionet.nsc.ru/mgs/programs/oligorep/InpForm.htm, last accessed December 20, 2013). To construct phylogenetic networks, program Network (Fluxus Technology Ltd 1999–2012) was used. CENSOR software tool was used for screening query sequences against a reference collection of the Repbase repetitive DNA collection (Jurka et al. 2005). The distribution of nucleotide diversity along the core repeat sequence using a set of complete repeats was calculated as average number of nucleotide differences per site, using a 10-bp window with a sliding step of 1 bp in DnaSP v4.5 (Rozas et al. 2003).
Results
DTC84 MITE Repetitive DNA Family
Recombinant clones enriched in repetitive fraction of D. trunculus genome were initially selected based on a positive correlation between hybridization signal intensity and genomic abundance of repetitive sequences. Sequenced inserts (in total 117 kb in 67 colonies) revealed, among others, 2,707-bp-long fragment named DTC84Alu. This fragment turned to be of interest because it includes a short array composed of five ∼160-bp-long tandem repeats, followed by a microsatellite segment at one array end. In the next screening of initial set of about 1,000 colonies, repeat-specific hybridization probe revealed four additional genomic fragments as positives. Besides single or few repeats organized in tandem, sequencing and sequence alignment of these fragments with DTC84Alu disclosed two ∼50-bp-long conserved segments that flank repeats (thus called core repeats) and the microsatellite. Schematic presentation of sequenced genomic fragments is shown in supplementary figure S1a, Supplementary Material online.
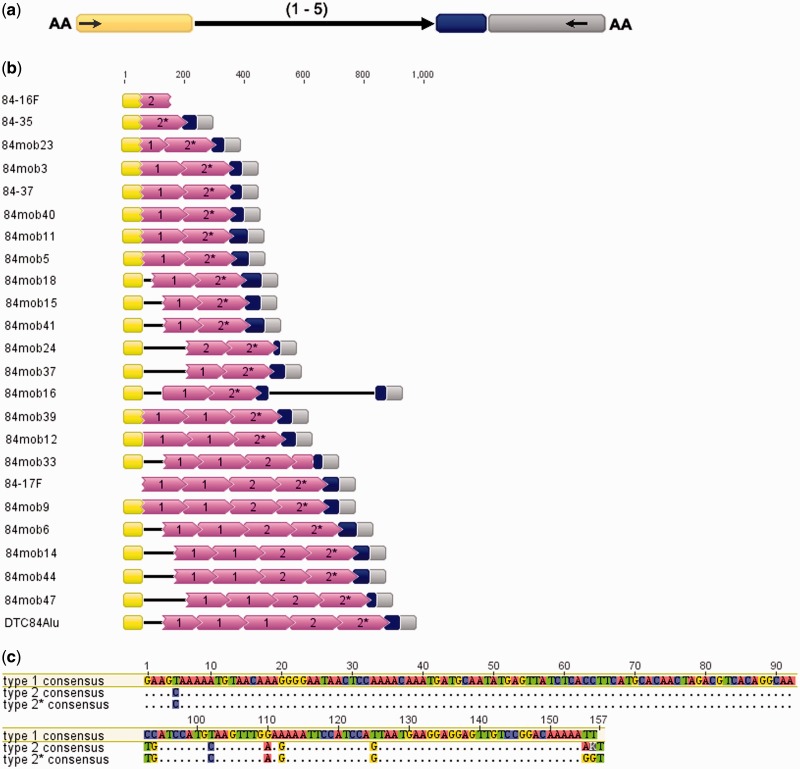
Observed sequence segments have modular composition which includes flanking sequence L + (core repeats)1–5 + (ACGG/ACGA)2–13 microsatellite + flanking sequence R (fig. 1a). In order to make a broader view on sequences between flanking segments, corresponding primers were used in PCR amplification of genomic DNA. In agreement with elements obtained from genomic clones, random selection of 19 amplified fragments revealed conserved sequence and organizational pattern of the new element, named DTC84 after the sequence segment cloned first. All depicted DTC84 elements are shown in figure 1b. Based on the common microsatellite motif ACGG and low sequence similarity (57%) between core repeats of the two elements (supplementary fig. S1b, Supplementary Material online), DTC84 is closest to the CvG MITE-like pearl element from C. virginica (Gaffney et al. 2003).
Fig. 1.—
Modular structure of DTC84 element. (a) Terminal and subterminal inverted repeats of DTC84 and equivalent elements are indicated by arrows within yellow and gray boxes that represent flanking modules. Solid black arrow in the middle of element represents core repeats, while the microsatellite segment is shown as a dark blue box. Conserved putative TSD sequence is given at element ends. (b) DTC84 in cloned genomic fragments (DTC84Alu, 84-16F, 84-17F, 84-35, and 84-37) and in genomic fragments retrieved by PCR amplification of genomic DNA with primers specific for left (yellow boxes) and right (gray boxes) flanking modules. Core repeats are shown as pink boxes, arrowheads indicating the orientation, and microsatellite regions are shown as dark blue boxes. Two types of DTC84 core repeats are indicated, and type 2 core repeat adjacent to the microsatellite is additionally marked by asterisk. Waved line on the left or right end of the core repeat box indicates truncation. Solid black lines represent genomic sequences other than in described modules. (c) Alignment of consensus sequences of different types of core repeats. Dots indicate identity, only differences to the consensus sequence of the first repeat type are shown.
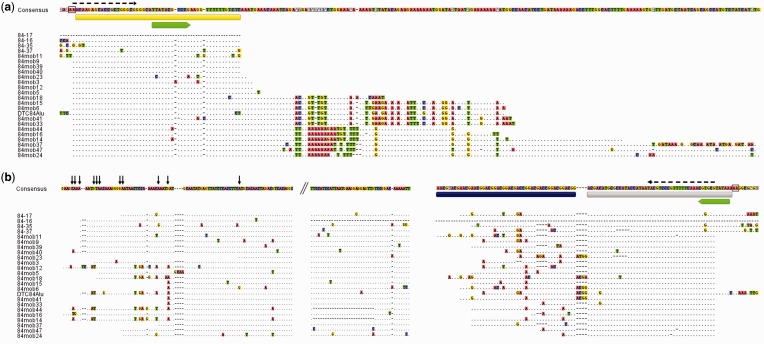
Possibility that mechanisms of transposition are involved in the spread of DTC84 is indicated by observation of AA dinucleotides as putative TSD that defines element ends in the cloned genomic fragments (boxed in fig. 2 consensus sequence). Among other structural features that may be of significance for transposition is 11-bp-long inversely oriented motif positioned at ends of otherwise unrelated sequence modules L and R (figs. 1a and 2, green arrows). This motif is located subterminally in the flanking segment L while it occupies terminal location in R, in agreement with the position of inverted repeats in pearl (Gaffney et al. 2003). As in pearl, complete pol III box A and box B priming sites were not found in DTC84.
Fig. 2.—
Sequence alignment of DTC84 element modules. Consensus sequence is derived according to the majority principle, and only differences to the consensus are shown in each segment. Dots indicate identity and dashes insertions or deletions. (a) Left flanking module (yellow bar) and two groups of accompanying spacer segments. (b) Core repeats, microsatellite region (dark blue bar), and right flanking module (gray bar). For simplicity, only beginning of the first core repeat and ending of the last core repeat in each array is shown, while interruption is indicated by slashes in the consensus sequence. Complete alignment of all core repeats can be seen in supplementary figure S2, Supplementary Material online. Black vertical arrows above the consensus sequence indicate starting positions of truncated repeats. Inverted repeats in flanking modules are indicated by green arrows. AA dinucleotides as putative TSDs are boxed in the consensus sequence. Dashed arrows above the consensus sequence show primer positions.
Additional complexity of DTC84 elements is given by differences in junctions between the flanking module L and core repeats (figs. 1b and 2a). This flanking module can be linked directly to core repeats but it can also accommodate two types of spacer segments, of ∼80 and from 70 to 160 bp, all sharing the first 16 nucleotides (fig. 2a). The common part may represent a residual of the flanking sequence itself, missing when the junction with core repeats is direct. Spacer sequences are not related to other components of DTC84 and do not show any apparent substructure. At the opposite core repeat array end, the microsatellite segment is linked directly to the flanking sequence R (figs. 1b and 2b).
Structured Arrays of DTC84 Core Repeats
Additional set of core repeats was cloned and sequenced after PCR amplification of genomic DNA with core repeat-specific primers. After excision of primer sites, a total of 41 core repeat sequences were extracted from PCR-obtained multimers (up to 5-mer) and compared with 65 core repeats obtained from DTC84 elements described earlier (supplementary fig. S2, Supplementary Material online). It must be noted that genomic environment of PCR-obtained core repeats is not known; they can be incorporated in DTC84 elements but also associated with some other undetermined genomic sequences. All compared core repeats differ mostly in single nucleotide substitutions (supplementary fig. S2, Supplementary Material online) and revealed a consensus length of 156 bp. In addition, sliding window analysis showed that nucleotide differences among core repeat variants are unevenly distributed along the sequence and form regions of reduced variability, approximately in the first half and near the end of the core repeat (supplementary fig. S3, Supplementary Material online).
The first core repeat in DTC84 is regularly 5′ end truncated for 4–67 nt. Only two core repeats out of 24 located at that position are complete (84mob12 and 84mob16). Truncated core repeats start at 11 different nucleotide positions, indicating that they were predominantly formed in independent events (fig. 2b). The distribution of truncation sites is however not quite random and seven core repeats start at the position 8, while the first nucleotide of four core repeats is at the position 42. Curiously, starting nucleotide in these two sets is always A. Although this nucleotide may represent a preferred site of truncation, identity in starting position can also be a consequence of amplification events that followed truncation.
Disregarding the truncated part, all sequenced arrays of DTC84 elements are composed of an integer number of core repeats, all being positioned in the same orientation and forming the same junction with the microsatellite. At its 3′ position, every DTC84 core repeat ends with the 12-nt-long palindrome TTGTCCGGACAA followed by the sequence AAATT, after which starts the next repeat unit. This palindrome is conserved in the majority of core repeats, and mutated variants are rare in this segment (16 out of 106 sequenced core repeats; supplementary fig. S2, Supplementary Material online). Specifically, transition point of the last core repeat into the microsatellite sequence differs in a way that sequence following the palindrome is AAAGGT.
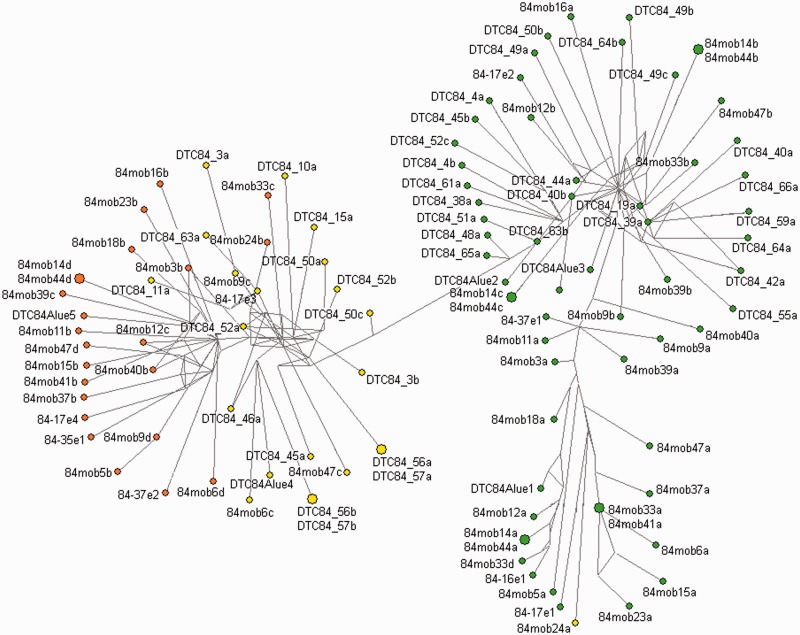
Phylogenetic analysis (fig. 3) combined with visual inspection of aligned core repeats (supplementary fig. S2, Supplementary Material online) revealed clustering and enabled identification of variants according to cluster-specific diagnostic nucleotides. Two types of core repeats are distinctive, based on six most exclusive variable positions. Type 1 variants are recognized by CC-T-G-A-T nucleotides while the same positions are occupied by TG-C-A-G-G nucleotides in variants of the type 2 (fig. 1c and supplementary fig. S2, Supplementary Material online). Although the difference is subtle, type 2 variants can be further subdivided into two groups. As explained in the previous paragraph, those linked directly to the microsatellite (indicated as 2*) differ from others by two additional diagnostic nucleotides located just at the monomer end (fig. 1c and supplementary fig. S2, Supplementary Material online). In the phylogenetic network, these core repeats group mostly in the left node of the type 2 branch (fig. 3).
Fig. 3.—
Grouping of DTC84 core repeats. Type 1 core repeats are indicated by green circles, type 2 are yellow, while type 2* are orange.
Sequenced DTC84 elements are composed of core repeats arranged according to the formula: type 1 (up to 3 consecutive repeats) + type 2 (1–2 repeats) (fig. 1b). It is important to note that no correlation could be derived among core repeat array length and/or composition, truncation point of the first repeat, spacer segments, and number of repeats in the microsatellite array. For example, dimeric core repeats of the arrangement type 1 + type 2 build 11 sequenced DTC84 elements (84mob23, 84mob3, 84-37, 84mob40, 84mob11, 84mob5, 84mob18, 84mob15, 84mob41, 84mob37, and 84mob16; fig. 1b). Despite identical core repeat composition, they differ in all other above-mentioned features (fig. 2). It can be concluded that every studied DTC84 element shows a unique combination of sequences featured in an independent manner.
In core repeat arrays of some DTC84 elements, only type 2 is found (in 84-35, 84mob24, and in the partial segment 84-16; fig. 1b). Following the rule of the first core repeat in array being truncated, in these cases this is the type 2. In other words, core repeats of any type can appear in the truncated form but strictly depending on their position in the array.
Genomic Organization of DTC84 Core Repeats Revealed by Southern Blot Hybridization
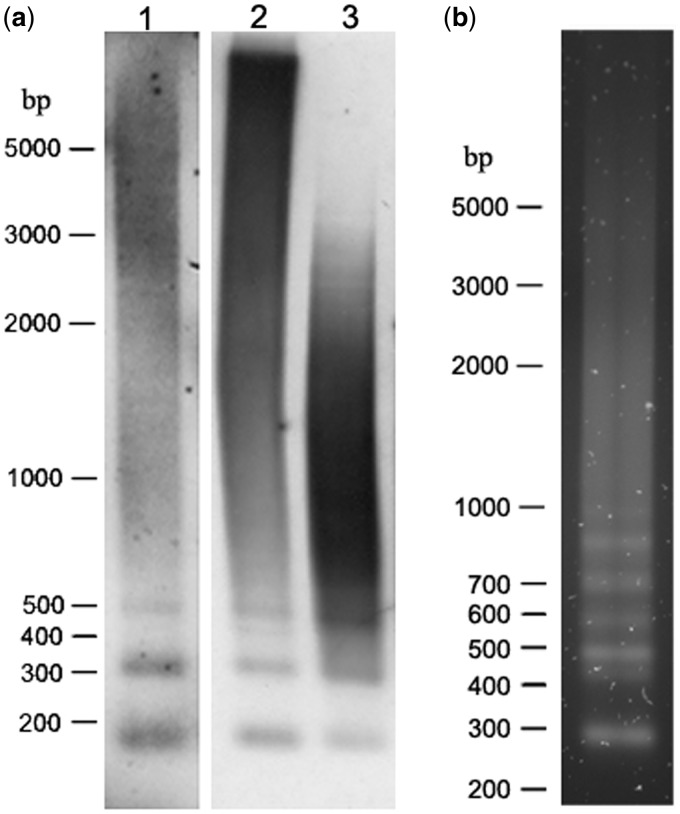
Southern hybridization experiments were performed in efforts to provide a more general view on organizational patterns of core repeats in the D. trunculus genome (fig. 4a). Digestion of genomic DNA with endonuclease MspI, cutting once within the core repeat monomer sequence, and with MboI, which predominantly cuts core repeats of the type 2, revealed short ladders of multimers on Southern blots. Methylation-sensitive endonuclease HpaII (isoschizomer of MspI) revealed slightly less degraded profile (fig. 4a), in agreement with the previous conclusion about methylation of D. trunculus genomic DNA (Petrović et al. 2009). Blurred appearance and hybridization smear of up to about 5 kb could be related to association of core repeats or their segments with other genomic sequences, resulting in a number of hybridizing fragments of different length. In agreement are also short ladders obtained after amplification with PCR primers located in conserved flanking sequences (fig. 4b) and those obtained with primers specific for core repeats (not shown). Obtained results suggest predominant organization of core repeats in short arrays, as observed in the cloned DTC84 representatives.
Fig. 4.—
Southern hybridization of Donax trunculus genomic DNA. (a) Genomic DNA digested with MspI (methylation insensitive, line 1), HpaII (isoshisomer of MspI, methylation sensitive, line 2), and MboI (methylation insensitive, line 3) and hybridized with the core repeat-specific probe. (b) Electrophoretic separation of genomic DNA amplified with PCR primers located in conserved flanking sequences. For the primer position, see figure 2.
According to the dot blot analysis (not shown) performed at high stringency conditions (68 °C), DTC84 core repeats constitute up to 1% of the bivalve genome or 8.9 × 104 copies, considering the genome size reported by Hinegardner (1974). Estimated contribution of these sequences almost double at low stringency conditions (60 °C), indicating that a large number of related sequences should exist in the genome. Genomic abundance estimated to be <1% for homogeneous core repeats constituting DTC84 elements is roughly in agreement with their occurrence in the cloned genomic DNA fragments.
Other Pearl-Related Repetitive Sequences in D. trunculus
Among sequences depicted in initial cloning of D. trunculus genomic DNA, local Blast revealed one fragment, DTC37AluF, which in a stretch of 140 nucleotides shares ∼61% similarity with DTC84 and with the pearl element CvG (Gaffney et al. 2003; supplementary fig. S4a and b, Supplementary Material online). This match stretches over a part of the core repeat and adjacent microsatellite region. The microsatellite array that follows the putative core repeat in DTC37AluF is heterogeneous, built of ACGG motif and its related variants ACTG and ACGA. In addition, DTC37AluF turned to be related to another genomic fragment, DTC41AluF. They share 87% similarity in the cloned core repeat segment, both being truncated at the same nucleotide in the cloning procedure. The microsatellite region differs between them only in length, and it is followed by 94% similar, 50-bp-long R flanking sequence, unrelated to equivalent modules in DTC84 and in CvG. Although not further explored in this work, observed similarity confirms that divergent elements of the DTC84 type exist in the D. trunculus genome, flanked with their distinctive sequences.
In the genomic clone DTC12Alu, we detected 190-bp-long sequence that shows 63% similarity to the CvE element (Gaffney et al. 2003). This fragment incorporates part of the putative core repeat, the microsatellite segment, and adjacent sequence (supplementary fig. S4c, Supplementary Material online). The microsatellite in DTC12Alu is composed of ACCG and ACAG motifs, instead of the ACTG motif found in the original CvE (supplementary fig. S4d, Supplementary Material online).
Discussion
Modular structure of repetitive elements described in this work in the clam D. trunculus corresponds to MITE-like elements of the pearl family, detected in C. virginica, the blood ark Anadara trapezia, the sea urchin Strongylocentrotus purpuratus (Cohen et al. 1985; Gaffney et al. 2003), and in the Mediterranean mussel Mytilus galloprovincialis (Kourtidis et al. 2006). They also share equivalent structure with a group of elements described in Drosophila and some other insects (Locke et al. 1999; Miller et al. 2000; Wilder and Hollocher 2001; Yang and Barbash 2008; Coates et al. 2011; Kuhn and Heslop-Harrison 2011). All these elements are characterized by TSDs and left and right flanking sequences with 10–20-bp-long inverted repeats located at terminal and subterminal positions. In addition, they all have central region composed of a sequence repeated in tandem up to about five times, adjacent at one side to a short tetranucleotide-based microsatellite segment (fig. 1a). Exceptionally, the microsatellite segment is missing in the M. galloprovincialis element (Kourtidis et al. 2006). At the DNA sequence level, in all of them is apparently conserved only the dinucleotide AA as the putative TSD, indicating involvement of a family of related integrases in the process of movement. Based on abundance, TIRs, dinucleotide TSD, and lack or incomplete RNA pol III promoter, these elements are mostly referred as MITE-like TEs. It was recently suggested that they can be alternatively classified as members of Helitron-like TEs, the class exploiting rolling-circle replication in its spread (Yang and Barbash 2008).
Rapid sequence divergence is expected along the whole length of nonautonomous TEs because of the lack of any potential coding function, while secondary structure and architecture of termini are assumed to be major requirements for transposition (Craig 1995; Coates et al. 2011). Comparisons of 12 Drosophila species showed that central region of DINE-1 elements differs among species according to the core repeat sequence and length. The most variable parameter is copy number, varying also among elements of the same type within a species (Yang and Barbash 2008). In C. virginica, core repeats of three elements belonging to the pearl family (CvA, CvE, and CvG) share little or no sequence similarity (Gaffney et al. 2003). However, related sequences can be detected in other species. Sequence similarity between core repeats of CvE and those of the element detected in M. galloprovincialis is relatively high, ∼74–78% (Kourtidis et al. 2006). Sequences related to C. virginica core repeats can be also identified in D. trunculus, CvG being distantly related to DTC84 core repeats and CvE to a fragment of the clone DTC12Alu.
Particularly intriguing is sequential distribution of DTC84 core repeat variants. Based on observed variations in array length and arrangement of core repeats, it is difficult to anticipate evolution of array diversity only as a consequence of accumulation of mutations and subsequent amplification of particular sequence types. Fixed position of the variant 2* and truncation of variants indicate that arrays may be modified by consecutive deletions in the preexisting ancestral array. Excision of core repeats could lead to different truncation of the newly formed first repeat, simply as a result of imprecise mechanisms of sequence turnover. Proneness to alterations by insertions and deletions is additionally evident by finding two types of spacer sequences that separate core repeats and the flanking module L.
The landmark of DTC84 core repeats is a palindrome near the core repeat end, located in the segment with reduced variability compared with the rest of the sequence. Equivalent segment of nonhomologous core repeats of the pearl family CvA also exhibits reduced sequence variability, but, instead of hosting a palindrome, it is characterized by a microsatellite-like motif (different from the microsatellite in continuation of core repeats, Gaffney et al. 2003). CvA-related sequences organized as satDNAs preserve reduced variability in this region and disclose another one, located roughly in the middle of the core repeat (Plohl et al. 2010). Interestingly, two regions of reduced sequence variability were also observed in alignment of core repeats of Drosophila PERI element (Kuhn and Heslop-Harrison 2011).
Putative motifs hidden in conserved sequence segments were observed in many satDNA monomers (e.g., Hall et al. 2003; Meštrović et al. 2006). Their occurrence is usually interpreted as a result of evolution under constraints, most likely due to functional interactions with protein components in chromatin. Experimental evidence is however still missing and the only well-described interaction is between the 17-bp-long CENP-B motif in a subset of human alpha satDNA monomers and the CENP-B protein (Masumoto et al. 1989). This sequence motif is found in apparently unrelated satDNAs of several mammalian species, and its variants were also detected in some invertebrates, including nematodes (Meštrović et al. 2013) and bivalve mollusks (Canapa et al. 2000). The CENP-B protein is likely to be involved in the human centromere assembly (Ohzeki et al. 2002), but its similarity to pogo-like transposases suggests its origin from domesticated mobile elements and a possible role in satDNA sequence rearrangements (Kipling and Warburton 1997; Casola et al. 2008). To the pogo-like subfamily of Tc1/mariner transposons is related one large group of MITEs, abundant in the human and other genomes (Feschotte et al. 2002). These data and the observed putative motifs suggest that transposase-related mechanisms may participate in alterations of tandem repeats of DTC84 and equivalent elements, as well as in formation of satDNAs derived from these sequences.
In addition to sequence motifs, the microsatellite region can also have a role in element structuring and dynamics, or alternatively it can be trailed as a consequence of these processes (Wilder and Hollocher 2001; Coates et al. 2011). A possible significance is supported by existence of the microsatellite repeat motif ACGG in CvG (Gaffney et al. 2003) and in DTC84, despite weak or no relevant sequence similarity between other modules.
The idea about core repeats and satDNA monomers as independent insertion/deletion units is further supported by finding diverse satellite monomers (or their short arrays) on euchromatic genome locations, including in the vicinity of genes (Kuhn et al. 2012; Brajković et al. 2012). Other observations also stress transposition as an intrinsic feature of at least some sequences arranged in tandem. For example, it was proposed that mechanisms of transposition spread human alpha satDNA to new genomic locations (Alkan et al. 2007). In addition, analysis of variability of related satDNAs shared by groups of species led to the model in which bursts of spread are followed by long periods of stasis, a feature pertinent to mobile elements (Meštrović et al. 2006).
It is striking that core repeats of DTC84, pearl-related sequences, and monomers of eight different satDNAs detected in D. trunculus are of similar length, about 160 bp (Plohl and Cornudella 1996, 1997; Petrović and Plohl 2005; Petrović et al. 2009). It was hypothesized that preferred length of satDNA repeats of 140–200 bp and 340 bp is favored by the chromatin structure (Henikoff et al. 2001). Based on observed similarities with core repeats, we can also speculate that the preferred length mirrors specificities of mechanisms involved in initial processes of repeat formation.
Compared with copy number alterations in arrays of tandem repeats, much less can be said about mechanisms responsible for initial tandem duplications of repetitive sequences. In this regard, duplication of entire MITE elements is proposed to be consequence of aberrant DNA replication (Izsvák et al. 1999). Briefly, due to an inverted repeat and/or a palindrome, sequence can be duplicated when DNA polymerase passes through a MITE, followed by excision of the duplicated segment and its reintegration into a new location (fig. 4 in Izsvák et al. 1999). In a case when the duplicated segment is not excised, the result will be formation of a tandem copy. Similarly, another model hypothesizes that internal MITE tandem repeats can be built during DNA replication based on the ability of a sequence to form stem-loop structures (Hikosaka and Kawahara 2004). The later model predicts unidirectional expansion of tandem repeats into long arrays typical for satDNAs. The arrangement of DTC84 core repeats is consistent with consecutive introduction of mutations during stepwise repeat duplication, while observed diversity in composition of variants can be accomplished by subsequent deletions, as explained earlier.
In conclusion, ordered distribution of mutations in the array of a variable number of core repeats in DTC84 is consistent with deletion events in a preformed segment, occurring independently with respect to the flanking modules. In this way, flanking sequences may be considered as a kind of a “cassette” for internal core repeats. One core repeat sequence end is marked by a palindrome, located within a segment of reduced sequence variability. Such segments may have a role in core repeat rearrangements, probably by mechanisms related to transposition. Although limited, similarity of some core repeats and satDNAs characterized previously in D. trunculus and other bivalve mollusks indicates a complex network which links tandem repeats residing inside MITEs and those expanded into arrays of satDNAs.
Supplementary Material
Supplementary figures S1–S4 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Brankica Mravinac, Nevenka Meštrović, and Andrea Luchetti for critical reading and comments on the manuscript. This work was supported by Research Fund of Ministry of Science, Education and Sports of Republic of Croatia, project no. 098-0982913-2756.
Literature Cited
- Alkan C, et al. Organization and evolution of primate centromeric DNA from whole-genome shotgun sequence data. PLoS Comput Biol. 2007;3(9):1807–1818. doi: 10.1371/journal.pcbi.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biscotti MA, et al. Repetitive DNA, molecular cytogenetics and genome organization in the King scallop (Pecten maximus) Gene. 2007;406:91–98. doi: 10.1016/j.gene.2007.06.027. [DOI] [PubMed] [Google Scholar]
- Brajković J, Feliciello I, Bruvo-Mađaric B, Ugarković Đ. Satellite DNA-like elements associated with genes within euchromatin of the beetle Tribolium castaneum. G3. 2012;2:931–941. doi: 10.1534/g3.112.003467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau TE, Wessler SR. Tourist: a large family of small inverted repeat elements frequently associated with maize genes. Plant Cell. 1992;4:1283–1294. doi: 10.1105/tpc.4.10.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafasso D, Cozzolino S, De Luca P, Chinali G. An unusual satellite DNA from Zamia paucijuga (Cycadales) characterised by two different organisations of the repetitive unit in the plant genome. Gene. 2003;311:71–79. doi: 10.1016/s0378-1119(03)00555-9. [DOI] [PubMed] [Google Scholar]
- Canapa A, Barucca M, Cerioni PN, Olmo E. A satellite DNA containing CENP-B box-like motifs is present in the antarctic scallop Adamussium colbecki. Gene. 2000;247:175–180. doi: 10.1016/s0378-1119(00)00101-3. [DOI] [PubMed] [Google Scholar]
- Casola C, Hucks D, Feschotte C. Convergent domestication of pogo-like transposases into centromere-binding proteins in fission yeast and mammals. Mol Biol Evol. 2008;25:29–41. doi: 10.1093/molbev/msm221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates BS, Kroemer JA, Sumerford DV, Hellmich RLL. A novel class of miniature inverted repeat transposable elements (MITEs) that contain hitchhiking (GTCY)n microsatellites. Insect Mol Biol. 2011;20:15–27. doi: 10.1111/j.1365-2583.2010.01046.x. [DOI] [PubMed] [Google Scholar]
- Cohen JB, Hoffman-Liebermann B, Kedes L. Structure and unusual characteristics of a new family of transposable elements in the sea urchin Strongylocentrotus purpuratus. Mol Cell Biol. 1985;5:2804–2813. doi: 10.1128/mcb.5.10.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig NL. Unity in transposition reactions. Science. 1995;270:253–254. doi: 10.1126/science.270.5234.253. [DOI] [PubMed] [Google Scholar]
- Dover GA. Molecular drive in multigene families: How biological novelties arise, spread and are assimilated. Trends Genet. 1986;2:159–165. [Google Scholar]
- Feschotte C, Mouchès C. Evidence that a family of miniature inverted-repeat transposable elements (MITEs) from the Arabidopsis thaliana genome has arisen from a pogo-like DNA transposon. Mol Biol Evol. 2000;17:730–737. doi: 10.1093/oxfordjournals.molbev.a026351. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Zhang X, Wessler S. Miniature inverted-repeat transposable elements (MITEs) and their relationship with established DNA transposons. In: Craig N, editor. Mobile DNA II. Washington (DC): ASM Press; 2002. pp. 1147–1158. [Google Scholar]
- Finnegan DJ. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989;5:103–107. doi: 10.1016/0168-9525(89)90039-5. [DOI] [PubMed] [Google Scholar]
- Fleetwood DJ, et al. Abundant degenerate miniature inverted-repeat transposable elements in genomes of epichloid fungal endophytes of grasses. Genome Biol Evol. 2011;3:1253–1264. doi: 10.1093/gbe/evr098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney PM, Pierce JC, Mackinley AG, Titchen DA, Glenn WK. Pearl, a novel family of putative transposable elements in bivalve mollusks. J Mol Evol. 2003;56:308–316. doi: 10.1007/s00239-002-2402-5. [DOI] [PubMed] [Google Scholar]
- Hall SE, Kettler G, Preuss D. Centromere satellites from Arabidopsis populations: maintenance of conserved and variable domains. Genome Res. 2003;13:195–205. doi: 10.1101/gr.593403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkinen E, Launonen V, Muller E, Bachmann L. The pvB370 BamHI satellite DNA family of the Drosophila virilis group and its evolutionary relation to mobile dispersed genetic pDv elements. J Mol Evol. 1995;41:604–614. doi: 10.1007/BF00175819. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- Hikosaka A, Kawahara A. Lineage-specific tandem repeats riding on a transposable element of MITE in Xenopus evolution: a new mechanism for creating simple sequence repeats. J Mol Evol. 2004;59:738–746. doi: 10.1007/s00239-004-2664-1. [DOI] [PubMed] [Google Scholar]
- Hinegardner R. Cellular DNA content of the Mollusca. Comp Biochem Physiol A Comp Physiol. 1974;47:447–460. doi: 10.1016/0300-9629(74)90008-5. [DOI] [PubMed] [Google Scholar]
- Izsvák Z, et al. Short inverted-repeat transposable elements in teleost fish and implications for a mechanism of their amplification. J Mol Evol. 1999;48:13–21. doi: 10.1007/pl00006440. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Kohany O, Jurka MV. Repetitive sequences in complex genomes: structure and evolution. Annu Rev Genomics Hum Genet. 2007;8:241–259. doi: 10.1146/annurev.genom.8.080706.092416. [DOI] [PubMed] [Google Scholar]
- Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kazazian HH. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Kipling D, Warburton PE. Centromeres, CENP-B and Tigger too. Trends Genet. 1997;13:141–145. doi: 10.1016/s0168-9525(97)01098-6. [DOI] [PubMed] [Google Scholar]
- Kourtidis A, Drosopoulou E, Pantzartzi CN, Chintiroglou CC, Scouras ZG. Three new satellite sequences and a mobile element found inside HSP70 introns of the Mediterranean mussel (Mytilus galloprovincialis) Genome. 2006;49:1451–1458. doi: 10.1139/g06-111. [DOI] [PubMed] [Google Scholar]
- Kuhn GCS, Heslop-Harrison JS. Characterization and genomic organization of PERI, a repetitive DNA in the Drosophila buzzatii cluster related to DINE-1 transposable elements and highly abundant in the sex chromosomes. Cytogenet Genome Res. 2011;132:79–88. doi: 10.1159/000320921. [DOI] [PubMed] [Google Scholar]
- Kuhn GCS, Küttler H, Moreira-Filho O, Heslop-Harrison JS. The 1.688 repetitive DNA of Drosophila: concerted evolution at different genomic scales and association with genes. Mol Biol Evol. 2012;29:7–11. doi: 10.1093/molbev/msr173. [DOI] [PubMed] [Google Scholar]
- Locke J, Howard LT, Aippersbach N, Podemski L, Hodgetts RB. The characterization of DINE-1, a short, interspersed repetitive element present on chromosome and in the centric heterochromatin of Drosophila melanogaster. Chromosoma. 1999;108:356–366. doi: 10.1007/s004120050387. [DOI] [PubMed] [Google Scholar]
- López-Flores I, Garrido-Ramos MA. The repetitive DNA content of eukaryotic genomes. In: Garrido-Ramos MA, editor. Repetitive DNA VII. Basel (Switzerland): Karger Publishers; 2012. pp. 1–28. [DOI] [PubMed] [Google Scholar]
- López-Flores I, et al. The molecular phylogeny of oysters based on a satellite DNA related to transposons. Gene. 2004;339:181–188. doi: 10.1016/j.gene.2004.06.049. [DOI] [PubMed] [Google Scholar]
- Macas J, Koblízková A, Navrátilová A, Neumann P. Hypervariable 3’ UTR region of plant LTR-retrotransposons as a source of novel satellite repeats. Gene. 2009;448:198–206. doi: 10.1016/j.gene.2009.06.014. [DOI] [PubMed] [Google Scholar]
- Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol. 1989;109:1963–1973. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meštrović N, Castagnone-Sereno P, Plohl M. Interplay of selective pressure and stochastic events directs evolution of the MEL172 satellite DNA library in root-knot nematodes. Mol Biol Evol. 2006;23:2316–2325. doi: 10.1093/molbev/msl119. [DOI] [PubMed] [Google Scholar]
- Meštrović N, et al. Conserved DNA motifs, including the CENP-B box-like, are possible promoters of satellite DNA array rearrangements in nematodes. PLoS One. 2013;8:e67328. doi: 10.1371/journal.pone.0067328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WJ, Nagel A, Bachmann J, Bachmann L. Evolutionary dynamics of the SGM transposon family in the Drosophila obscura species group. Mol Biol Evol. 2000;17:1597–1609. doi: 10.1093/oxfordjournals.molbev.a026259. [DOI] [PubMed] [Google Scholar]
- Mravinac B, Plohl M. Parallelism in evolution of highly repetitive DNAs in sibling species. Mol Biol Evol. 2010;27:1857–1867. doi: 10.1093/molbev/msq068. [DOI] [PubMed] [Google Scholar]
- Noma K, Ohtsubo E. Tnat1 and Tnat2 from Arabidopsis thaliana: novel transposable elements with tandem repeat sequences. DNA Res. 2000;7:1–7. doi: 10.1093/dnares/7.1.1. [DOI] [PubMed] [Google Scholar]
- Ohzeki J, Nakano M, Okada T, Masumoto H. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol. 2002;159:765–775. doi: 10.1083/jcb.200207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrović V, Plohl M. Sequence divergence and conservation in organizationally distinct subfamilies of Donax trunculus satellite DNA. Gene. 2005;362:37–43. doi: 10.1016/j.gene.2005.06.044. [DOI] [PubMed] [Google Scholar]
- Petrović V, et al. A GC-rich satellite DNA and karyology of the bivalve mollusk Donax trunculus: a dominance of GC-rich heterochromatin. Cytogenet Genome Res. 2009;124:63–71. doi: 10.1159/000200089. [DOI] [PubMed] [Google Scholar]
- Plohl M, Cornudella L. Characterization of a complex satellite DNA in the mollusc Donax trunculus: analysis of sequence variations and divergence. Gene. 1996;169:157–164. doi: 10.1016/0378-1119(95)00734-2. [DOI] [PubMed] [Google Scholar]
- Plohl M, Cornudella L. Characterization of interrelated sequence motifs in four satellite DNAs and their distribution in the genome of the mollusc Donax trunculus. J Mol Evol. 1997;44:189–198. doi: 10.1007/pl00006135. [DOI] [PubMed] [Google Scholar]
- Plohl M, Luchetti A, Mestrović N, Mantovani B. Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene. 2008;409:72–82. doi: 10.1016/j.gene.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Plohl M, et al. Long-term conservation vs high sequence divergence: the case of an extraordinarily old satellite DNA in bivalve mollusks. Heredity. 2010;104:543–551. doi: 10.1038/hdy.2009.141. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Wang S, Zhang L, Meyer E, Matz MV. Characterization of a group of MITEs with unusual features from two coral genomes. PLoS One. 2010;5:e10700. doi: 10.1371/journal.pone.0010700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder J, Hollocher H. Mobile elements and the genesis of microsatellites in dipterans. Mol Biol Evol. 2001;18:384–392. doi: 10.1093/oxfordjournals.molbev.a003814. [DOI] [PubMed] [Google Scholar]
- Yang H-P, Barbash DA. Abundant and species-specific DINE-1 transposable elements in 12 Drosophila genomes. Genome Biol. 2008;9:R39. doi: 10.1186/gb-2008-9-2-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.