SUMMARY
The replicative machinery encounters many impediments, some of which can be overcome by lesion bypass or replication restart pathways, leaving repair for a later time. However, interstrand crosslinks (ICLs), which preclude DNA unwinding, are considered absolute blocks to replication. Current models suggest that fork collisions, either from one or both sides of an ICL, initiate repair processes required for resumption of replication. To test these proposals, we developed a single molecule technique for visualizing encounters of replication forks with ICLs, as they occur in living cells. Surprisingly, the most frequent patterns were consistent with replication traverse of an ICL, without lesion repair. The traverse frequency was strongly reduced by inactivation of the translocase and DNA binding activities of the FANCM/MHF complex. The results indicate that translocase-based mechanisms enable DNA synthesis to continue past ICLs, and that these lesions are not always absolute blocks to replication.
INTRODUCTION
DNA replication is inhibited by a multitude of impediments, some introduced by radiation, others by endogenous and exogenous reactive compounds. Replication forks may be stalled by encounters with DNA damage, necessitating repair prior to the resumption of DNA synthesis (McGlynn and Lloyd, 2002). Alternatively, lesion bypass or replication restart pathways may be engaged (Lehmann and Fuchs, 2006; Sale et al., 2012). These require opening of the helix, which is possible with single strand adducts. However, interstrand crosslinks (ICLs), which covalently join both strands of the duplex, are absolute blocks to DNA unwinding. Consequently, they are considered insurmountable barriers to the replication apparatus. This feature, combined with the requirement for two cycles of repair, underlies the high toxicity of crosslinking agents such as cisplatin and mitomycin C, which are widely used in cancer chemotherapy (Deans and West, 2011).
Cells from patients with Fanconi Anemia (FA) are characterized by genomic instability and hypersensitivity to crosslinking compounds (Kee and D’Andrea, 2010). At this time 16 FA proteins have been identified, with roles in DNA repair, the response to replication stress, and cell signaling (Gari and Constantinou, 2009). Some form the core complex (A, B, C, E, F, G, L), a multifunctional assembly that acts as a ubiquitin ligase that monoubiquitinates FANCD2 and FANCI (Gregory et al., 2003). Individual proteins in the complex participate in survival signaling pathways in hematopoietic cells (Bagby and Alter, 2006). FANCN/PALB2, FANCO/RAD51C, FANCP/SLX4, FANCD1/BRCA2, and FANCJ/BACH1/BRIP, are thought to function downstream of activated FANCD2/I in repair, homologous recombination, and replication fork reconstruction. The most recently recognized Fanconi protein, XPF, is a structure specific endonuclease found in complex with ERCC1, which functions in nucleotide excision and ICL repair (Bogliolo et al., 2013). Cells lacking either XPF or ERCC1 are very sensitive to crosslinking compounds. FANCM, a DNA translocase, associates with the core proteins, but also exists in an additional complex with the MHF1/2 proteins, independent of the FA core components. This protein participates in multiple DNA transactions, including the recovery of stalled replication forks (Collis et al., 2008; Meetei et al., 2005; Schwab et al., 2010; Yan et al., 2010).
Longstanding models of ICL repair envision encounters with a single replication fork as a trigger for repair. This involves cleavage of one of the template strands on both sides of the ICL (unhooking), and gap filling to form a crosslink remnant, which might, or might not, be removed prior to the resumption of replication (Kuraoka et al., 2000; Muniandy et al., 2010; Niedernhofer et al., 2004; Thompson and Hinz, 2009) (Fig. S1). Elegant work from the Walter laboratory, based on replication of a crosslinked plasmid in Xenopus egg extracts, indicates that collisions of replication forks on both sides of an ICL precede repair (Fig. S1) (Raschle et al., 2008). FA proteins participate in unhooking, as well as bypass replication across the crosslink remnant (Knipscheer et al., 2009; Long et al., 2011). In another study, also with Xenopus egg extracts and plasmids, both single and double fork collisions were observed (Le et al., 2011).
Although the plasmid systems describe replication of crosslinked DNA with high resolution, they cannot predict the relative probability of these two models in mammalian genomes, with origins of replication that can be widely separated. We have developed a DNA fiber based approach to visualize encounters of replication forks with genomic ICLs, as they occur in living cells. Remarkably, we found that neither the single fork nor the double fork collision models account for the majority of the replication patterns. Instead we found a new pattern, one we term replication traverse.
RESULTS
Digoxigenin tagged TMP forms a high frequency of ICLs
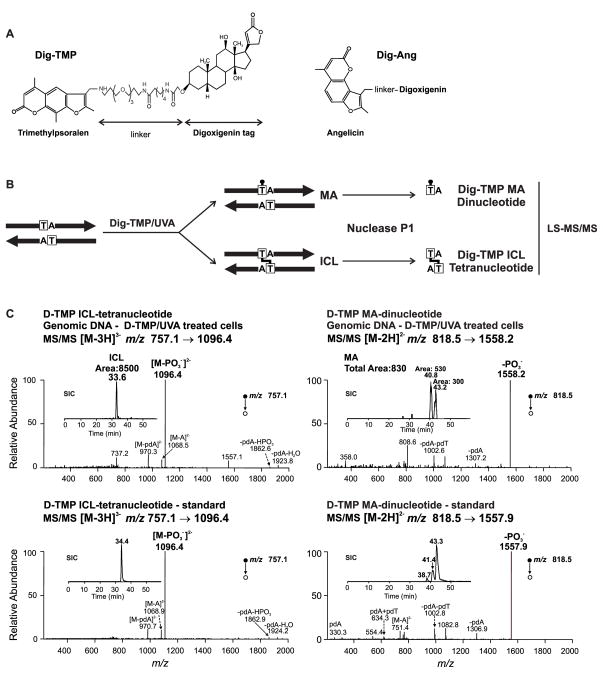
Visualization of replication fork encounters with genomic ICLs requires an agent that yields a high frequency of ICLs relative to monoadducts (MAs) and can be detected by imaging technologies. Photoactive psoralens can be linked to antigens, which enable the detection of psoralen: DNA adducts by immunofluorescence (Thazhathveetil et al., 2007). Furthermore, in contrast to cisplatin and mitomycin C, which generate largely single strand adducts, psoralens can form a high frequency of ICLs relative to MAs (Lai et al., 2008).
We covalently linked digoxigenin (Dig) to angelicin (Ang), which forms only MAs, and to trimethylpsoralen (TMP), which can form ICLs and MAs (Fig. 1A). To determine the relative frequency of ICLs and MAs, DNA was extracted from cells immediately following treatment with Dig-TMP/UVA. The ICL: MA ratio was measured by LC-MS/MS and estimated to be approximately 10:1 (Cao et al., 2008) (Fig. 1B, C Fig S2, S3).
Fig. 1. Cell treatment with Dig-TMP/UVA generates primarily ICLs.
A. Dig-TMP, Dig-Ang. B. Sample preparation prior to analysis by LC-MS/MS. C. Determination of the ICL: MA ratio by LC-MS/MS analysis of genomic DNA (top) from cells treated with Dig-TMP/UVA and in a duplex oligodeoxyribonucleotide calibration standard (lower). Top left: the product-ion spectrum of the ESI-produced [M-3H]3− ion (m/z 757.1) of the tetranucleotide carrying the Dig-TMP ICL. Inset: the selected-ion chromatogram (SIC) for monitoring the m/z 757.1 →1096.4. Top right: the product-ion spectrum of the [M-2H]2− ion (m/z 818.5) of the dinucleotide carrying the Dig-TMP MA. Inset: the SIC for monitoring the m/z 818.5→1558.2 transition. The LC-MS/MS data for the standards are in the lower panels. Note the relative peak areas of the ICL and MAs in the SICs for the genomic DNA sample. See also Figures S2–S3.
Visualization of replication fork encounters with ICLs
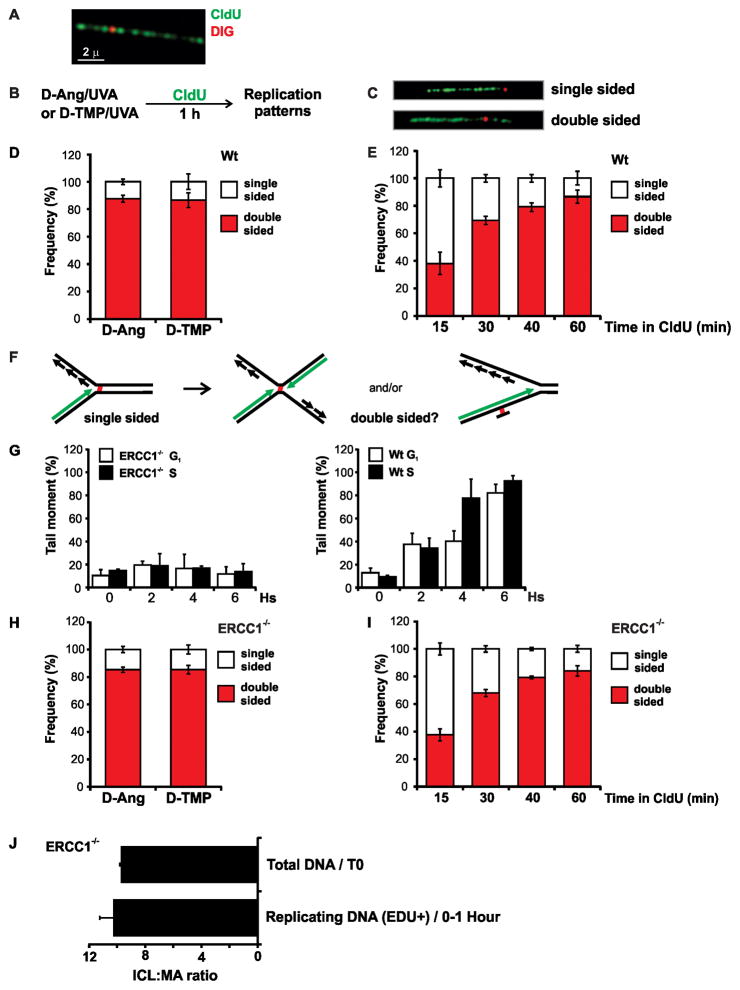
CHO cells were incubated with CldU for 24 hs to label DNA, after which they were treated with UVA in the presence of various concentrations of Dig-TMP. Genomic DNA was spread onto glass slides and the CldU displayed by immunofluorescence (green) and Dig-TMP with immunoquantum dots (red) (Kad et al., 2010) (Fig. 2A). The frequency of Dig signals on the fibers was proportional to the Dig-TMP concentration (not shown). Conditions were chosen for subsequent experiments such that Dig-TMP signals were observed on approximately 20–25% of the fibers.
Fig. 2. Replication patterns in the vicinity of Dig-TMP and Dig-Ang adducts.
A. Immunoquantum dot visualization of Dig-TMP (red) covalently linked to a DNA fiber labeled with CldU (green). B. Experimental design. C. Single and double sided replication patterns. D. Frequency of replication patterns in repair proficient cells. E. Frequency of single and double sided patterns as a function of pulse labeling time in wild type cells. F. Possible explanations for the double sided patterns. Dual fork collision, or partial repair of the ICL, could generate a single strand adduct which could be bypassed by the fork. For simplicity only the leading strand is depicted in green, but both the leading and lagging strands are labeled in the experiment. G. CHO cells were synchronized in G1 or S phase, treated with TMP/UVA and analyzed by an alkaline comet assay to determine the efficiency of unhooking (crosslink release). H. Pattern frequencies in ERCC1−/ − cells. I. Frequency of replication patterns as a function of pulse time in ERCC1−/ − cells. J. ICL: MA ratio in DNA harvested immediately after TMP/UVA treatment, and in replication tracts labeled with EdU for 1 hr. Data are presented as mean ±SD. See also Figure S4A, Table S1.
In our initial experiments we monitored replication in the vicinity of MAs. Repair proficient CHO cells were treated with Dig-Ang/UVA, followed by 1 hr incubation with CldU (Fig. 2B). Two patterns were observed. In one, the replication tract terminated on one side, while in the other the CldU label appeared on both sides, of a Dig-Ang signal (Fig. 2C, S4A). The latter pattern accounted for approximately 90% of events (Fig. 2D).
The experiment was then performed with cells exposed to Dig-TMP/UVA. Unexpectedly, a pattern distribution similar to that with Dig-Ang was obtained (Fig. 2D). This experiment was repeated, varying the duration of the labeling period. The frequency of the double sided patterns was proportional to the length of the incubation with the CldU, less than 40% after 15 minutes, reaching an 85–90% plateau after 60 min (Fig. 2E). There was no further increase on extending the incubation to 2 hr (not shown). These data indicated that single sided events could be converted to double sided over time.
The nearly identical results with angelicin and TMP raised the possibility that unhooking of ICLs to produce a crosslink remnant MA occurred during the CldU labeling (Fig. 2F). To address this concern it was essential to determine the status of the ICLs at the time of the fork encounters. We considered this in three ways. First, we characterized the patterns in ERCC1−/ − cells which cannot unhook crosslinked DNA (Kuraoka et al., 2000) in either G1 or S phase cells, as monitored by alkaline comet assays (Fig. 2G). If ICL unhooking explained the double sided patterns in wild type cells, then a shift to single sided patterns would be expected in ERCC1−/ − cells. In contrast to this prediction, double sided patterns were again dominant in the vicinity of the Dig-TMP adducts (Fig. 2H), reaching a plateau after 60 min (Fig. 2I). The same pattern distribution was recovered with fibers from cells treated with Dig-Ang (Fig. 2H).
LC-MS/MS analysis of TMP adducts on replication tracts
Crosslink release has been ascribed to activities other than ERCC1/XPF (Knipscheer et al., 2009; Smeaton et al., 2008; Smogorzewska et al., 2010). Thus it was desirable to have a more direct assessment of the ICLs embedded in the replication tracts. It has been shown by LC-MS/MS that psoralen monoadducts are not removed in ERCC1−/ − cells (Liu and Wang, 2013). If, during replication, there had been an appreciable unhooking of ICLs (Sengerova et al., 2012; Smeaton et al., 2008), there would be a decline in the ICL: MA ratio in the DNA in the replication tracts, relative to the ratio in DNA harvested from cells immediately after treatment. Accordingly, we treated ERCC1−/ − cells with TMP/UVA and incubated for 1 hr with EdU. Genomic DNA was extracted, sheared, and EdU containing fragments conjugated to biotin by click chemistry and collected by streptavidin/magnetic bead capture (Supplementary Methods). The EdU+ DNA was examined by LC-MS/MS. Comparison of the ICL: MA ratio for the EdU+ DNA to that obtained immediately after exposure to TMP/UVA indicated that they were nearly identical (about 10:1 in both samples) (Fig 2J). Thus, there was no appreciable decline in this ratio on the DNA replicated during the EdU incubation.
Replication in the vicinity of Dig-TMP adducts is on crosslinked template strands
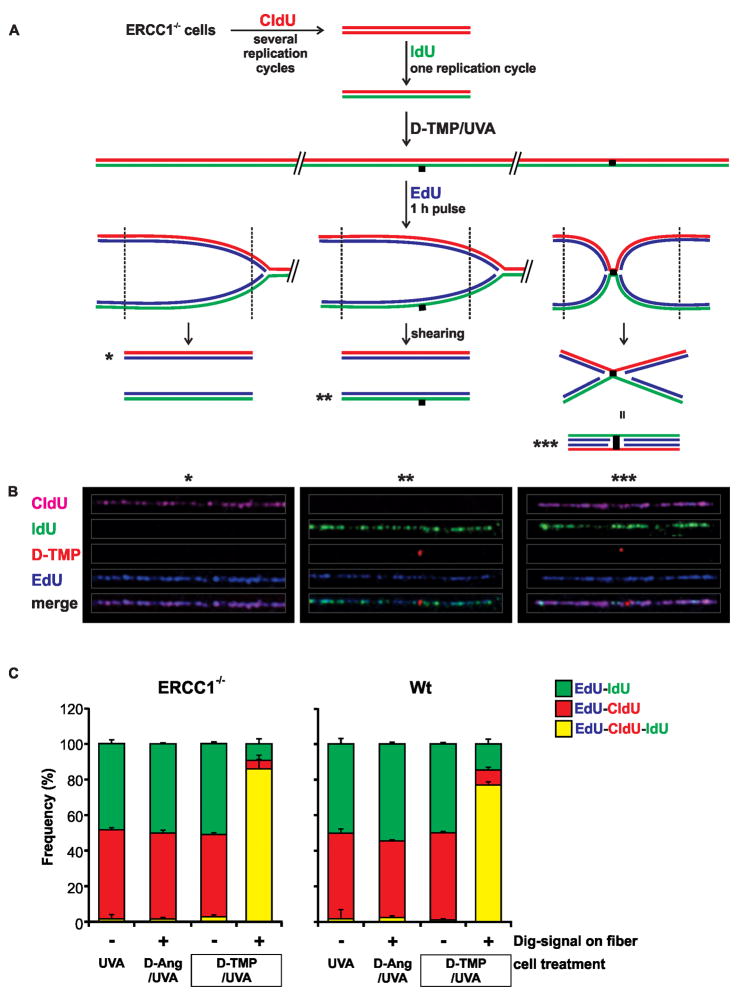
The LC-MS/MS analysis was of the population of TMP adducts associated with replication tracts. In order to address the issue of ICL integrity at the single molecule level, we asked if the template strands were crosslinked during replication in the vicinity of individual Dig-TMP adducts. Our approach was based on a prior inquiry into strand distribution during replication (Meselson and Stahl, 1958). Template strands were differentially labeled by incubation of ERCC1−/ − cells with CldU for several replication cycles, followed by incubation with IdU for a single cycle (Fig 3A). The cells were exposed to Dig-Ang/UVA or Dig-TMP/UVA, followed by 60 min incubation with EdU. They were lysed and, as in the earlier experiment (Hanawalt, 2004), the DNA sheared to release replication products from adjacent unreplicated DNA (Fig 3A). Fibers were spread (Fig S4B) and the relationship of the EdU to the other labels determined. Segregation of EdU tracts with either CldU or IdU would be expected on DNA without crosslinks. That is, EdU tracts: from mock treated cells (UVA only), or; from cells treated with Dig-Ang/UVA, or; on fibers without Dig-TMP, isolated from cells treated with Dig-TMP/UVA (these provided an internal control for the influence of all experimental manipulations) (Fig 3A*, B*), or; on fibers with Dig-TMP spots in which the Dig signal represented a single strand adduct (Fig 3A**, B**). However, parental template strands covalently joined by an intact Dig-TMP ICL could not be split by the EdU marked daughter strands, and these patterns would be marked by all three labels (Fig. 3A***, B***).
Fig. 3. ICL status at the time of fork encounter.
A. Experimental scheme. ERCC1−/ −cells were incubated with CldU (red) for several generations. Then the cells were incubated with IdU (green) for one cycle. The medium was changed and the cells treated with Dig-TMP/UVA, followed by incubation for 1 hr with EdU (blue). Cells were lysed and the DNA sheared prior to spreading on slides and immunostaining. The anticipated patterns are shown. B. Examples of daughter (blue) tracts associated with parental strands. The CldU tracts have been pseudo-colored purple to facilitate visualization of the Dig signal in the merged image. Blue tracts: without Dig-TMP (*); with a Dig-TMP MA signal on a green fiber (**); with a Dig-TMP ICL on a purple/green fiber (***). C. Distribution of EdU (blue) tracts on fibers from ERCC1−/ − cells, or repair proficient wild type cells, treated with Dig-TMP/UVA, Dig-Ang/UVA, or UVA only. The two bars on the right represent fibers without, or with, Dig-TMP signals from cells treated with Dig-TMP/UVA. Data are presented as mean ±SD. See also Figure S4B, Table S2.
In fibers from mock treated cells (UVA only), or from cells with Dig-Ang signals there was almost complete segregation of EdU tracts with either CldU or IdU (less than 2% triple color tracts) (Fig 3C). On fibers without Dig-TMP, isolated from cells treated with Dig-TMP/UVA, there were less than 3% triple color patterns. However, of the fibers with Dig-TMP embedded in EdU tracts, 86% were associated with both CldU and IdU (Fig. 3C), consistent with replication on either side of a Dig-TMP ICL that linked the two parental strands. In wild type cells the frequency of Dig-TMP triple color tracts was 77.5% (Fig 3C), which, although not a statistically significant difference, probably reflected some unhooking during the EdU incubation. These data indicated that the great majority of Dig-TMP adducts embedded in the EdU replication tracts in the ERCC1−/ − cells were intact ICLs. Furthermore, this experiment clearly distinguished replication in the vicinity of adducts formed by Dig-TMP from that in the vicinity of Dig-Ang MAs.
Replication patterns in the vicinity of ICLs
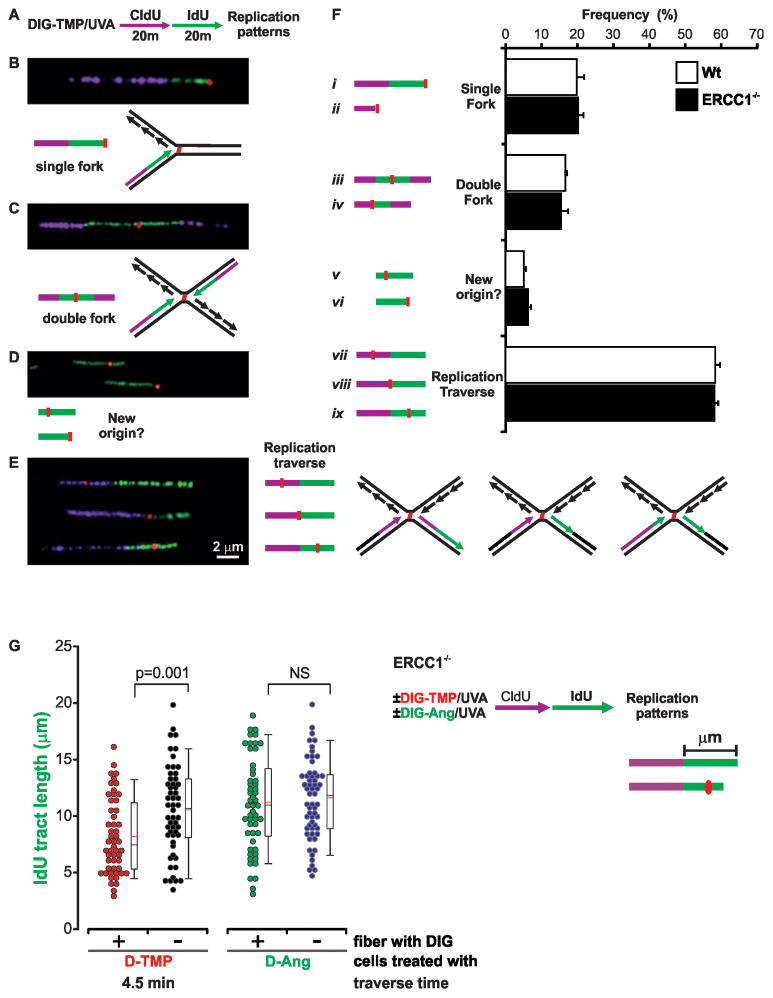
These results argued that the double sided patterns represented replication in the vicinity of intact ICLs. We tested the possibility that these reflected dual fork collisions with a sequential double labeling protocol, which identifies the direction of replication. ERCC1−/ − cells were treated with Dig-TMP/UVA, and then pulsed with CldU for 20 min, followed by 20 min of IdU (Fig. 4A). The simplest prediction of the dual fork model was the ICL embedded in the second label, flanked on both sides by the first label (Fig. 4C).
Fig. 4. Replication traverse of ICLs.
A. Sequential labeling defines the direction of replication in the vicinity of ICLs. Cells were treated with Dig-TMP/UVA and then pulsed for 20 min with CldU (purple), followed by a 20 min pulse of IdU (green). Pseudo-coloring as in Fig 3. B, C, D, E. Images and interpretation of replication patterns in the vicinity of Dig-TMP signals. Another pattern (not shown), in which a Dig-TMP signal appeared in a new origin (purple flanked by green), appeared at a frequency of less than 1 %. F. Quantitation of replication patterns in Wt and ERCC1−/ − cells, mean ±SD. G. Time cost of replication traverse. The lengths of IdU tracts, with or without a Dig-TMP signal, from cells treated with Dig-TMP, were measured and the time difference calculated. The same analysis was performed with IdU tracts from cells treated with Dig-Ang. See also Figure S5, Table S3.
Approximately 20% of the patterns recovered in this experiment were single sided (Fig. 4B, Fi–ii). Somewhat less than 20% were in accord with the dual fork model (Fig. 4C, 4Fiii–iv). There was also a minor pattern in which the Dig-TMP was in an IdU tract unlinked to a CldU tract (Fig. 4D, Fv,vi). However, in the largest group (>50%) the two tracts were contiguous, with the Dig-TMP signal embedded in one or the other (Fig. 4E, Fvii–ix, S5A). Identical results were obtained in repair proficient cells. The patterns in Fig 4E were unexpected and not predicted by either the single or double fork collision models. Instead, they indicated that replication had continued on the side of the ICL distal to the fork encounter.
Time cost to replication in the vicinity of ICLs
As noted above, the conditions of Dig-TMP/UVA treatment yielded about 20–25% of the fibers with Dig-TMP signals. Consequently, many dual label tracts were on DNA fibers without ICLs. These, as also noted, served as an internal control for the influence of experimental manipulation on replication on DNA without psoralen adducts. We measured the lengths of the second color (IdU) tracts with and without an embedded Dig-TMP. The tracts containing Dig-TMP were shorter than those without. The length difference was converted into a time difference (Experimental Procedures). This comparison revealed that the time cost to replication imposed by the ICL was about 4.5 min (Fig 4G). This difference varied according to cell line, about 6 min in DT40 cells and 7 min in MEFs (Fig S5B). Analysis of fibers from cells treated with Dig-Ang/UVA revealed a cost of less than a minute, although the difference did not reach statistical significance. Thus the ICLs could be distinguished from the MAs by the length of time required to generate replication tracts on both sides.
A minor fraction of ICL encounters activate dormant origins of replication
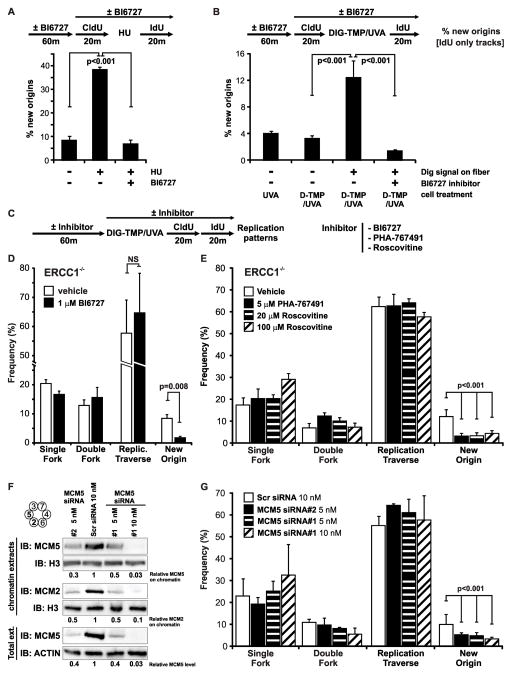
Dormant origins can be activated by replication stress (Blow and Ge, 2009). A possible explanation for the results in Fig 4E is that collision of a single fork with an ICL was followed by the activation of a nearby dormant origin on the distal side (Fig. S6). To test this possibility we asked if dormant origins were activated by Dig-TMP/UVA treatment of the cells, and if inhibitors of this process would influence the replication patterns in Fig 4. Treatment with hydroxyurea (HU) can activate dormant origins, defined as tracts of the second label unconnected to those of the first label. Inhibitors of the Polo-Like-Kinase-1, PLK-1, block this process (Song et al., 2011)(Schwab et al., 2010). We confirmed the activity of the PLK-1 inhibitor BI 6727 against the activation of dormant origins following HU treatment of the cells used in our experiments (Fig. 5A). Cells were pulsed with CldU followed by Dig-TMP/UVA, after which they were pulsed with IdU. The replication patterns on fibers with and without Dig-TMP were examined. The frequency of IdU tracts, unconnected to CldU tracts, was low in fibers from untreated cells and also in fibers, isolated from treated cells, but lacking Dig-TMP signals. However, the frequency was increased in fibers with Dig-TMP signals, and this frequency was reduced to control levels by exposure to BI 6727 (Fig. 5B). These results demonstrated that Dig-TMP/UVA treatment could provoke the de novo activation of origins. It was of interest that the unconnected IdU tracts appeared only on fibers with Dig-TMP signals, indicating that the signal for activation of dormant origins following replication stress was transmitted in cis, as proposed previously (Blow et al., 2011; Yekezare et al., 2013).
Fig. 5. Dormant origin activation does not explain the traverse patterns.
A. The PLK-1 inhibitor BI6727 blocks dormant origin activation induced by HU treatment. After a 20 min CldU pulse, cells were incubated for 12 hours in the presence of 2 mM HU, followed by an IdU pulse. Experiments were done in parallel with or without BI6727 (Methods). B. Dormant origin activation by Dig-TMP/UVA treatment is blocked by BI6727. C, D, E. The frequency of fork traverse patterns is not affected by the inhibitors BI6727, PHA-767491, or roscovitine. Cells were treated with Dig-TMP/UVA, followed by sequential pulses of CldU and IdU, all in the presence of drugs or vehicle. F. Western blot analysis of MCM5 and MCM2 in total cell extracts or in chromatin, following treatment of cells with siRNAs against MCM5. G. The frequency of fork traverse patterns is unaffected by knockdown of MCM5. Data are presented as mean ±SD (p<0.001, chi-square test). See also Figure S6, Table S4.
The double pulse experiment of Fig 4 was repeated in the presence of BI6727. The frequency of the minor patterns in which the Dig-TMP was in the isolated IdU tracts was reduced, but the drug treatment did not diminish the frequency of any of the contiguous double label patterns (Fig. 5C, D). We also performed the experiment in the presence of PHA-767491, an inhibitor of CDC7. This kinase is required for activation of origins of replication, but not for replication fork progression (Montagnoli et al., 2008). There was a decline in isolated second color tracts, but again the levels of contiguous double label patterns were unaffected (Fig 5E). The same results were obtained in the presence of two concentrations of roscovitine, a cyclin dependent kinase inhibitor that blocks dormant origin activation (Schwab et al., 2010) (Fig 5E).
In G1 phase cells the replicative helicase MCM complex is loaded as a double hexamer, encircling the duplex (Evrin et al., 2009; Remus et al., 2009). Many more MCM complexes are loaded (10–20 fold) than are actually used in S phase, and it has been proposed that these “extra” double hexamers would be available to start de novo replication in the event of fork collapse (Blow et al., 2011; Remus et al., 2009). MCM proteins can be reduced, by siRNA treatment of cells, to levels that support unstressed replication, but preclude the activation of dormant origins (Ibarra et al., 2008). We treated cells with different concentrations of two different siRNA against MCM5 that reduced total MCM5 protein to less than 5% of controls (Fig 5F), and the level of chromatin bound MCM5 protein to 3% of controls. Chromatin bound MCM2 was also sharply reduced (Fig 5F). While the reduction in MCM complex proteins suppressed the frequency of isolated second color tracts, there was no effect on the frequency of the other patterns (Fig 5G).
Based on the results of these experiments it seemed unlikely that the replication patterns of Fig 4E could be explained by the activation of dormant origins. Consequently, these patterns, also incompatible with models of single or double fork collisions, were consistent with a fourth, unanticipated, scenario- replication traverse of the ICL.
Replication traverse of ICLs is promoted by the FANCM translocase activity
While the traverse patterns in Fig 4E are in accord with replication restart models (Lehmann and Fuchs, 2006; Lin et al., 2011), there is an obvious difference between the enzymatic requirements for fork passage past a MA, and an ICL, which would block progression of the replicative helicase. Thus, another activity(s) would be required.
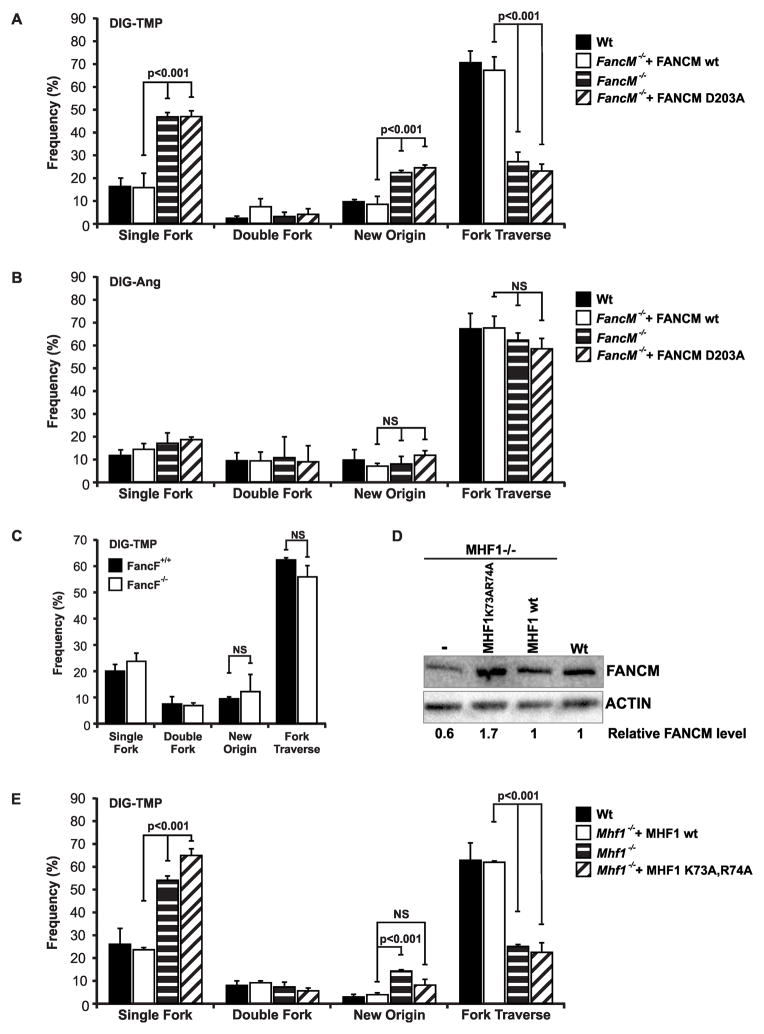
Movement past an ICL in an intact duplex by a bacterial DNA translocase has been shown in vitro (Bastia et al., 2008) raising the possibility that a similar enzyme, able to move along DNA without unwinding, might promote traverse of an ICL in vivo. The FA translocase, FANCM, responds to replication stress (Meetei et al., 2005). Consequently, the double pulse experiment was performed in FANCM−/ − and wild type mouse embryonic fibroblasts (MEFs), and in FANCM−/ − DT40 cells, and in those cells complemented by expression of the wild type translocase (Fig. 6A, Fig. S7A). In the FANCM−/ − cells there was a decline in the frequency of traverse patterns, and an increase in the single sided patterns. In the DT40 cells there was also an increase in the frequency of the isolated,” dormant origin”, IdU tracts, consistent with a recent report (Schwab et al., 2010). There was no change in the frequency of dual fork collisions. The same results were obtained in FANCM−/ − DT40 cells expressing a version of FANCM with a mutation in the ATPase domain (FANCM D203A/-). This variant has no translocase activity, but associates with the FA core complex, which is able to monoubiquitinate FANCD2 (Rosado et al., 2009). In contrast, in cells treated with Dig-Ang, FANCM deficiency had no effect on the replication patterns (Fig. 6B). Thus, the translocase activity of FANCM was specifically required for replication traverse of ICLs, but was not required for the progression of the replication apparatus past angelicin MAs.
Fig. 6. Replication traverse of ICLs, but not MAs, is promoted by FANCM translocase activity.
A. Replication fork traverse of ICLs is reduced in FANCM−/ − cells. DT40 wild type or FANCM−/ −, or FANCM−/ − cells expressing wild type FANCM, or a translocase mutant version (D203A), were treated with Dig-TMP/UVA followed by sequential pulses of CldU and IdU. B. FANCM deficiency does not influence the frequency of replication patterns in cells treated with Dig-Ang. C. FANCF deficiency does not influence the frequency of replication patterns in cells treated with Dig-TMP. (D) Relative FANCM protein levels in MHF1−/ − cells and complemented cells total cell extracts. E. Traverse patterns are reduced in cells deficient in MHF1 or expressing a DNA binding mutant of MHF1(K73A,R74A). Data are presented as mean ±SD (p<0.001, chi-square test). See also Figure S7, Table S5.
As FANCM interacts with FA core proteins (Yan et al., 2010), we asked if the frequency of ICL traverse patterns would be influenced by deficiencies in those proteins. This would eliminate the core complex and the ubiquitination of FANCD2. However, we found that the frequency of the patterns was unaffected in cells lacking FANCF, FANCA, FANCE, or FANCG (Fig. 6C, S7 B–E). Thus, neither the core complex, nor ubiquitinated FANCD2, was required for replication traverse of ICLs.
The FANCM/MHF complex is important for replication traverse of ICLs
Independent of the FA core proteins, FANCM forms a complex with MHF1/2, the interactions with which are important for cellular resistance to crosslinking agents (Ciccia et al., 2007; Wang et al., 2013; Yan et al., 2010). The MHF1/MHF2 complex binds double stranded DNA, and enhances the DNA binding and fork reversal activities of FANCM. The level of FANCM protein was reduced to 60 % of wild type in DT40 knockout cells, but restored in knockout cells complemented with the wild type MHF1 gene (Fig. 6D). Knockout of MHF1 resulted in a replication pattern distribution very similar to that in FANCM −/ − cells, while knockout cells expressing wild type MHF1 gave the wild type distribution (Fig. 6E). Because the decline in FANCM levels could contribute to the shift in replication patterns, we also performed the experiment in knockout cells expressing a mutant version of MHF1, with two alanine substitutions of positively charged amino acid residues (K73A/R74A). The FANCM/MHF1(K73A/R74A) /MHF2 complex does not bind double strand DNA, or forked DNA structures. DT40 MHF1 knockout cells expressing this variant have levels of FANCM and MHF2 that exceed those in wild type cells (Yan et al., 2010) (Fig. 6D). The replication patterns in these cells were the same as in FANCM knockout cells (Fig. 6E). Thus, the DNA binding activity of the FANCM/MHF1/2 complex was required for replication traverse of the ICLs.
DISCUSSION
DNA fiber assays have been used to study replication dynamics in cells responding to DNA damaging agents (Merrick et al., 2004a). However, visualization of both replication tracts and DNA adducts on fibers has received little attention. We have taken advantage of antigen tagged psoralens to follow replication in the vicinity of MAs and ICLs.
In order to accurately interpret results based on this approach it was necessary to clarify the status of the Dig-TMP adducts at the time of fork collisions. This was a pressing concern given our observation that the pulse labeling patterns per se could not distinguish replication on templates containing angelicin or TMP adducts. However, several lines of evidence support the conclusion that the most fork encounters with TMP reaction products were with intact ICLs: 1. the experiments were performed in ERCC1−/ − cells, which are deficient in ICL repair; 2. the LC-MS/MS analysis of TMP adducts on fragments containing the EdU label after 1 hr of replication indicated no difference in ICL: MA ratio relative to DNA harvested immediately after exposure of cells to TMP/UVA; 3. the daughter strand label did not split the substantial majority of the differentially labeled parental template strands in ERCC1−/ − cells when a Dig-TMP adduct was embedded in the replication tract; 4. in contrast, parental strands carrying single strand adducts formed by Dig-Ang were split by the daughter tracts; 5. FANCM deficiency altered the distribution of replication patterns in the vicinity of Dig-TMP, but not Dig-Ang, adducts.
Replication of DNA can be uncoupled from the repair of single strand lesions that block polymerases (Lehmann and Fuchs, 2006; Rupp and Howard-Flanders, 1968; Yeeles and Marians, 2011). Replicative helicases can drive past single strand adducts, opening the helix to permit the restart of synthesis. ICLs, which are absolute blocks to helicases, have always been regarded, intuitively, as absolute blocks to replication. However, our data counter this common belief, and indicate that DNA synthesis can resume past an ICL, leaving behind still crosslinked parental strands. Furthermore, despite three other options, this is the major pathway (Fig. 4). Thus, models in which replication is uncoupled from repair of single strand lesions can be extended to ICLs (Lehmann and Fuchs, 2006). The uncoupling cannot be due solely to the replicative MCM helicase, which encircles the leading parental template strand and would be blocked by the ICL (Fu et al., 2011). Rather, our results indicate that replication traverse is promoted by the translocase and DNA binding activities of FANCM/MHF.
In theory, established pathways could account for the traverse patterns. However, the data in Fig 5 argue against a role for dormant origin activation. Furthermore, some form of template switching, similar to Break Induced Replication, also seems unlikely. In yeast these pathways can take hours (Malkova et al., 2005), not the few minutes associated with the traverse patterns.
Although any explanation for our observations would be purely speculative, we suggest two possible scenarios based on activities of FANCM and features of the MCM replicative helicase complex. The MCM complex has a lock washer, gapped ring structure (Boos et al., 2012). Replication is initiated by melting of the duplex and enclosure by the MCM complex of single strand DNA (Fu et al., 2011; Hashimoto et al., 2012). After recruitment to a fork stalled by an ICL, FANCM/MHF might translocate the MCM complex past the ICL onto the unreplicated duplex, prior to the resumption of unidirectional replication. This scenario is similar to that proposed to explain the movement of the E. coli helicase/translocase DNAB from a single strand region across a duplex oligonucleotide carrying ICLs (Bastia et al., 2008), and consistent with data with other replicative helicases (Jeong et al., 2013).
In another model, FANCM/MHF might serve as a key component of a signal transduction pathway (Blackford et al., 2012; Collis et al., 2008) which would be triggered by the encounter of the replication fork with an ICL. The translocase activity would be required for the signaling function, but FANCM would not actually translocate from one side of the fork to the other. The signaling would eventually activate the replication restart machinery on the distal side of the ICL.
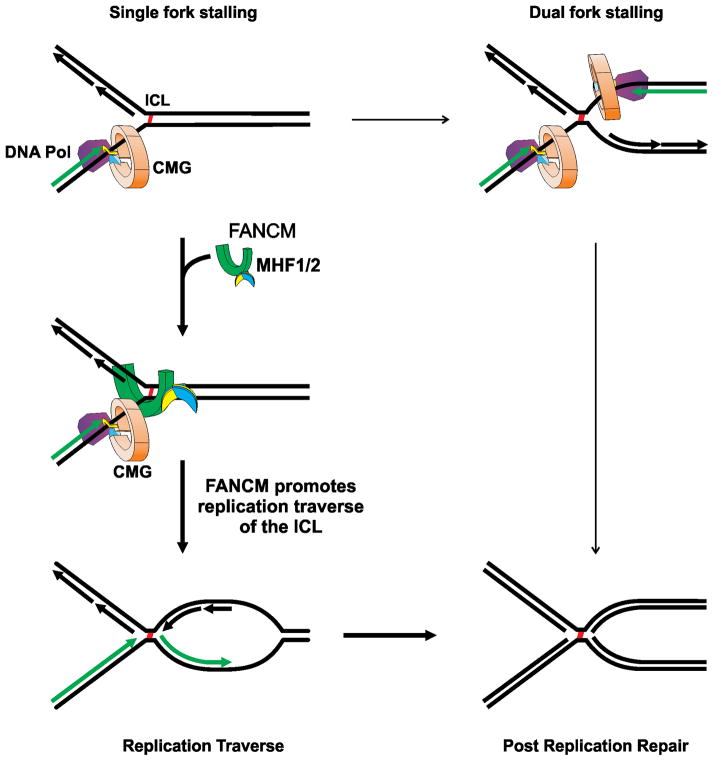
As with the replication restart models for classical single strand adducts (Lehmann and Fuchs, 2006), ICLs would enter postreplication repair pathways sometime after replication traverse. The FA proteins could participate in this process (Knipscheer et al., 2009). Postreplication repair also follows the dual fork collisions described by the Walter group. A central prediction of their model is that bypass synthesis across an ICL remnant involves extension of a leading daughter strand (Raschle et al., 2008) (Fig. S1). Scenarios based on our findings are also consistent with ICL remnant bypass by extension of a daughter strand. In both the double fork and replication traverse models, after Okazaki fragment ligation, the DNA structure around the ICL would be the same. Thus it is possible that the dynamics of post replication repair would be similar, if not identical, in both models (Fig. 7).
Fig. 7. Replication traverse of ICLs.
Following the encounter of a single replication fork, a second fork may collide with the same ICL from the other side (Raschle et al., 2008). More frequently, recruitment of FANCM/MHF1/2 to the stalled fork is followed by replication restart on the distal side of the ICL. After Okazaki fragment ligation the DNA structures formed by either pathway are identical.
ICL traverse patterns were observed in cells of chicken, mouse, hamster, and human origin, some primary, some tumor derived. Thus it would seem that this is not unique to a particular species or a peculiarity of tumorigenesis. Orthologs of FANCM are found in species that appeared long before the emergence of the complete FA pathway in vertebrates (Meetei et al., 2005), while the MHF proteins are found in yeast. The traverse promoting activity of FANCM/MHF was independent of the FA core proteins, implying that this pathway emerged well before the appearance of vertebrate lineages. We suggest that the traverse activity be added to the functions of FANCM/MHF as it responds to replication stress, in support of the cellular imperative to complete S phase. The FANCM/MHF translocase may prove to be a useful target for improved chemotherapy with crosslinking drugs.
EXPERIMENTAL PROCEDURES
Materials
Dig-TMP and Dig-Ang were synthesized as described (Thazhathveetil et al., 2007). HPLC purified oligodeoxyribonucleotides (ODNs) were purchased from Integrated DNA Technologies, Inc. (Coralville, IA). Antibodies were from commercial suppliers: rat anti-BrdU [CldU] (Abcam), mouse anti-BrdU [IdU] (BD), chicken anti digoxigenin (Abcam), rabbit anti-MCM5 and rabbit anti-MCM2 (Abcam), Dylight 649 goat anti-rat, Dylight 488 goat anti-mouse (Jackson ImmunoResearch), Qdot 655 goat anti-chicken, Qdot 565 Streptavidin Conjugate (Molecular Probes). Azide-biotin was from Click Chemistry Tools. BI 6727 was obtained from Chemie Tek (Indianapolis, IN), HU from Sigma, Roscovitine from Cell Signaling Technology, and PHA-767491 from Selleckchem. The siRNA pool oligos (ON-TARGETplus) Mcm5-1i, 5-GGAGGUAGCUGAUGAGGUGTT-3; Mcm5-2i, 5-GGAUCUGGCCAGCUUUGAUTT-3; were purchased from Dharmacon. The transfection reagent was Lipofectamine RNAi MAX (Invitrogen).
Cells
Cells were: Chinese hamster ovary (CHO) wild-type AA8, CHO ERCC1−/ − (Rolig et al., 1997), and CHO FANCG−/ − (Tebbs et al., 2005), FANCM−/ − mouse embryo fibroblasts (MEFs) (Bakker et al., 2009), FANCF−/ − and FANCE−/ − patient derived cells, and derivatives complemented by stable expression of the wild type gene (de Winter et al., 2000), lymphoblastoid FANCA−/ − cells, and cells complemented with the wild type gene, wild type chicken DT40 cells, and FANCM−/ −, and FANCM−/ − DT40 cells complemented with wild type or mutant versions of the gene, and DT40 MHF1−/ − or MHF1−/ − cells expressing wild type or mutant MHF1(K73A/R74A) versions of the gene.
LC-ESI-MS/MS determination of Dig-TMP ICL/MA ratio in genomic DNA
Oligonucleotides containing a Dig-TMP ICL or a MA were synthesized (Lai et al., 2008). CHO AA8 cells were treated with Dig-TMP/ UVA as below. DNA was purified and digested with nuclease P1 to generate mononucleotides (unmodified), dinucleotides (monoadducts), and tetranucleotides (ICLs). The digestion mixtures were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Agilent Technologies, Santa Clara, CA) and an LTQ linear ion-trap mass spectrometer (Thermo Fisher Scientific, San Jose, CA). The mass spectrometer was operated in the negative-ion mode, and the instrument was set up to acquire the tandem mass spectra (MS/MS) for the fragmentation of the [M-3H]3− and [M-2H]2− ions of the Dig-TMP ICL-containing tetranucleotide, as well as the [M-2H]2− and [M-H] − ions of Dig-TMP MA-containing dinucleotide (Cao et al., 2008).
DNA spreading
Cells were incubated for 1 hour with 5 μM Dig-TMP or 20 μM Dig-Ang in the dark before UVA irradiation in a Rayonet chamber at 3 J/cm2. In single pulse experiments labeling was with 20 μM CldU for various times as indicated. In double pulse experiments cells were incubated in 20 μM CldU for 20 min, and then in 100 μM IdU for 20 min. The DT40 cells were treated with 20 μM CldU for 40 min, followed by 250 μM IdU for 20 min. In experiments with inhibitors, cells were pre-treated with compounds for 1 hour which were included during CldU and IdU labeling. Cells were mixed with lysis buffer [0.5% SDS in 200 mM Tris-HCl (pH 7.5), 50 mM EDTA] on a glass slide. After tilting, the slides were air-dried, fixed in 3:1 methanol/acetic acid, incubated in 2.5 M HCl for 60 min, neutralized in 0.4 M Tris-HCl (pH 7.5) for 5 min, washed in PBS, and immunostained (Merrick et al., 2004b). Antibodies and dilutions were: rat anti-BrdU [CldU], 1: 200; Dylight 649 goat anti-rat, 1:100; mouse anti-BrdU [IdU], 1:40 and chicken anti digoxigenin, 1:200; Dylight 488 goat anti-mouse, 1:100 and Qdot 655 goat anti-chicken, 1:2500. Imaging was carried out using a Zeiss Axiovert 200M microscope with the Axio Vision software packages (Zeiss, Germany). The quantum dot signal was imaged with a Qdot 655 filter. The numbers of fields and events for individual experiments are shown in Supplementary Tables.
Time cost of replication traverse
From images collected for dual pulse experiments (Fig 4, 6, S5A), we measured the length (in microns) of the IdU tracts in fibers with (LWD) and without (LWOD) a Dig signal on the IdU tract. The traverse time TT was estimated as: TT = [LWOD- LWD]/RR, and Replication rate (RR) was calculated as RR= LWOD/pulse time.
Assessment of ICL integrity during replication
CHO cells were exposed to 10 μM CldU for 72 hrs, the medium changed and the cells incubated with 10 μM IdU for 20 hrs, then exposed to UVA with or without 5 μM Dig-TMP, or 20 μM Dig-Ang, followed by 60 min incubation with 10 μM EdU. The cells were spun down, lysed in 0.6% SDS, 10 mM EDTA, 10 mM Tris pH 7.4, and the DNA sheared by passage through a 16 gauge needle. NaCl was added to a final concentration of 1 M, the tubes were inverted several times, and incubated at 4°C for 5 hours. The lysate was centrifuged at 17,000g for 30 minutes to remove aggregates (Hirt, 1967) and the DNA in the supernatant was spread onto microscope slides. EdU was biotinylated by click chemistry and detected by a Qdot 565 streptavidin conjugate (Salic and Mitchison, 2008).
Supplementary Material
HIGHLIGHTS.
Single molecule imaging reveals patterns of replication fork encounters with ICLs
Single and double fork collisions, and dormant origin activation, can be detected
Replication traverse of an ICL is the most frequent pattern
FANCM/MHF translocase and DNA binding activities mediate replication traverse of ICLs
Acknowledgments
This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging (AG000688-07). We thank Drs Kurt Andresen, John Wilson, Michael Leffak, and Ranjan Sen for helpful discussions, and Drs KJ Patel, Wojciech Niedzwiedz, Minoru Takata, and Steve West for the generous gift of cell lines. Dedicated to the memory of Johan de Winter, our friend and colleague.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bagby GC, Alter BP. Fanconi anemia. Semin Hematol. 2006;43:147–156. doi: 10.1053/j.seminhematol.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Bakker ST, van de Vrugt HJ, Rooimans MA, Oostra AB, Steltenpool J, Delzenne-Goette E, van der Wal A, van dV, Joenje H, te RH, de Winter JP. Fancm-deficient mice reveal unique features of Fanconi anemia complementation group M. Hum Mol Genet. 2009;18:3484–3495. doi: 10.1093/hmg/ddp297. [DOI] [PubMed] [Google Scholar]
- Bastia D, Zzaman S, Krings G, Saxena M, Peng X, Greenberg MM. Replication termination mechanism as revealed by Tus-mediated polar arrest of a sliding helicase. Proc Natl Acad Sci U S A. 2008;105:12831–12836. doi: 10.1073/pnas.0805898105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackford AN, Schwab RA, Nieminuszczy J, Deans AJ, West SC, Niedzwiedz W. The DNA translocase activity of FANCM protects stalled replication forks. Hum Mol Genet. 2012;21:2005–2016. doi: 10.1093/hmg/dds013. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Ge XQ. A model for DNA replication showing how dormant origins safeguard against replication fork failure. EMBO Rep. 2009;10:406–412. doi: 10.1038/embor.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Ge XQ, Jackson DA. How dormant origins promote complete genome replication. Trends Biochem Sci. 2011;36:405–414. doi: 10.1016/j.tibs.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogliolo M, Schuster B, Stoepker C, Derkunt B, Su Y, Raams A, Trujillo JP, Minguillon J, Ramirez MJ, Pujol R, Casado JA, Banos R, Rio P, Knies K, Zuniga S, Benitez J, Bueren JA, Jaspers NG, Scharer OD, de Winter JP, Schindler D, Surralles J. Mutations in ERCC4, Encoding the DNA-Repair Endonuclease XPF, Cause Fanconi Anemia. Am J Hum Genet. 2013;92:800–806. doi: 10.1016/j.ajhg.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos D, Frigola J, Diffley JF. Activation of the replicative DNA helicase: breaking up is hard to do. Curr Opin Cell Biol. 2012;24:423–430. doi: 10.1016/j.ceb.2012.01.011. [DOI] [PubMed] [Google Scholar]
- Cao H, Hearst JE, Corash L, Wang Y. LC-MS/MS for the detection of DNA interstrand cross-links formed by 8-methoxypsoralen and UVA irradiation in human cells. Anal Chem. 2008;80:2932–2938. doi: 10.1021/ac7023969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, Laghmani eH, Joenje H, McDonald N, de Winter JP, Wang W, West SC. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol Cell. 2007;25:331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Collis SJ, Ciccia A, Deans AJ, Horejsi Z, Martin JS, Maslen SL, Skehel JM, Elledge SJ, West SC, Boulton SJ. FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol Cell. 2008;32:313–324. doi: 10.1016/j.molcel.2008.10.014. [DOI] [PubMed] [Google Scholar]
- de Winter JP, van Der WL, de Groot J, Stone S, Waisfisz Q, Arwert F, Scheper RJ, Kruyt FA, Hoatlin ME, Joenje H. The Fanconi anemia protein FANCF forms a nuclear complex with FANCA, FANCC and FANCG. Hum Mol Genet. 2000;9:2665–2674. doi: 10.1093/hmg/9.18.2665. [DOI] [PubMed] [Google Scholar]
- Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C. A double-hexameric MCM2–7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci U S A. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YV, Yardimci H, Long DT, Ho TV, Guainazzi A, Bermudez VP, Hurwitz J, van OA, Scharer OD, Walter JC. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell. 2011;146:931–941. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K, Constantinou A. The role of the Fanconi anemia network in the response to DNA replication stress. Crit Rev Biochem Mol Biol. 2009;44:292–325. doi: 10.1080/10409230903154150. [DOI] [PubMed] [Google Scholar]
- Gregory RC, Taniguchi T, D’Andrea AD. Regulation of the Fanconi anemia pathway by monoubiquitination. Semin Cancer Biol. 2003;13:77–82. doi: 10.1016/s1044-579x(02)00102-5. [DOI] [PubMed] [Google Scholar]
- Hanawalt PC. Density matters: the semiconservative replication of DNA. Proc Natl Acad Sci U S A. 2004;101:17889–17894. doi: 10.1073/pnas.0407539101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Puddu F, Costanzo V. RAD51- and MRE11-dependent reassembly of uncoupled CMG helicase complex at collapsed replication forks. Nat Struct Mol Biol. 2012;19:17–24. doi: 10.1038/nsmb.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Ibarra A, Schwob E, Mendez J. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc Natl Acad Sci U S A. 2008;105:8956–8961. doi: 10.1073/pnas.0803978105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong YJ, Rajagopal V, Patel SS. Switching from single-stranded to double-stranded DNA limits the unwinding processivity of ring-shaped T7 DNA helicase. Nucleic Acids Res. 2013;41:4219–4229. doi: 10.1093/nar/gkt133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kad NM, Wang H, Kennedy GG, Warshaw DM, Van HB. Collaborative dynamic DNA scanning by nucleotide excision repair proteins investigated by single- molecule imaging of quantum-dot-labeled proteins. Mol Cell. 2010;37:702–713. doi: 10.1016/j.molcel.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, D’Andrea AD. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev. 2010;24:1680–1694. doi: 10.1101/gad.1955310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraoka I, Kobertz WR, Ariza RR, Biggerstaff M, Essigmann JM, Wood RD. Repair of an interstrand DNA crosslink initiated by ERCC1-XPF repair/recombination nuclease. J Biol Chem. 2000;275:26632–26636. doi: 10.1074/jbc.C000337200. [DOI] [PubMed] [Google Scholar]
- Lai C, Cao H, Hearst JE, Corash L, Luo H, Wang Y. Quantitative analysis of DNA interstrand cross-links and monoadducts formed in human cells induced by psoralens and UVA irradiation. Anal Chem. 2008;80:8790–8798. doi: 10.1021/ac801520m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le BC, Hennion M, Arimondo PB, Hyrien O. Replication-fork stalling and processing at a single psoralen interstrand crosslink in Xenopus egg extracts. PLoS ONE. 2011;6:e18554. doi: 10.1371/journal.pone.0018554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann AR, Fuchs RP. Gaps and forks in DNA replication: Rediscovering old models. DNA Repair (Amst) 2006;5:1495–1498. doi: 10.1016/j.dnarep.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Lin JR, Zeman MK, Chen JY, Yee MC, Cimprich KA. SHPRH and HLTF act in a damage-specific manner to coordinate different forms of postreplication repair and prevent mutagenesis. Mol Cell. 2011;42:237–249. doi: 10.1016/j.molcel.2011.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Wang Y. A quantitative mass spectrometry-based approach for assessing the repair of 8-methoxypsoralen-induced DNA interstrand cross-links and monoadducts in Mammalian cells. Anal Chem. 2013;85:6732–6739. doi: 10.1021/ac4012232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long DT, Raschle M, Joukov V, Walter JC. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science. 2011;333:84–87. doi: 10.1126/science.1204258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, Naylor ML, Yamaguchi M, Ira G, Haber JE. RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol Cell Biol. 2005;25:933–944. doi: 10.1128/MCB.25.3.933-944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG. Recombinational repair and restart of damaged replication forks. Nat Rev Mol Cell Biol. 2002;3:859–870. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]
- Meetei AR, Medhurst AL, Ling C, Xue Y, Singh TR, Bier P, Steltenpool J, Stone S, Dokal I, Mathew CG, Hoatlin M, Joenje H, de Winter JP, Wang W. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick CJ, Jackson D, Diffley JF. Visualization of altered replication dynamics after DNA damage in human cells. J Biol Chem. 2004a;279:20067–20075. doi: 10.1074/jbc.M400022200. [DOI] [PubMed] [Google Scholar]
- Merrick CJ, Jackson D, Diffley JF. Visualization of altered replication dynamics after DNA damage in human cells. J Biol Chem. 2004b;279:20067–20075. doi: 10.1074/jbc.M400022200. [DOI] [PubMed] [Google Scholar]
- Meselson M, Stahl FW. THE REPLICATION OF DNA IN ESCHERICHIA COLI. Proc Natl Acad Sci U S A. 1958;44:671–682. doi: 10.1073/pnas.44.7.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnoli A, Valsasina B, Croci V, Menichincheri M, Rainoldi S, Marchesi V, Tibolla M, Tenca P, Brotherton D, Albanese C, Patton V, Alzani R, Ciavolella A, Sola F, Molinari A, Volpi D, Avanzi N, Fiorentini F, Cattoni M, Healy S, Ballinari D, Pesenti E, Isacchi A, Moll J, Bensimon A, Vanotti E, Santocanale C. A Cdc7 kinase inhibitor restricts initiation of DNA replication and has antitumor activity. Nat Chem Biol. 2008;4:357–365. doi: 10.1038/nchembio.90. [DOI] [PubMed] [Google Scholar]
- Muniandy PA, Liu J, Majumdar A, Liu ST, Seidman MM. DNA interstrand crosslink repair in mammalian cells: step by step. Crit Rev Biochem Mol Biol. 2010;45:23–49. doi: 10.3109/10409230903501819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, de Wit J, Jaspers NG, Beverloo HB, Hoeijmakers JH, Kanaar R. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol Cell Biol. 2004;24:5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschle M, Knipsheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, Walter JC. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF. Concerted loading of Mcm2–7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolig RL, Layher SK, Santi B, Adair GM, Gu F, Rainbow AJ, Nairn RS. Survival, mutagenesis, and host cell reactivation in a Chinese hamster ovary cell ERCC1 knock-out mutant. Mutagenesis. 1997;12:277–283. doi: 10.1093/mutage/12.4.277. [DOI] [PubMed] [Google Scholar]
- Rosado IV, Niedzwiedz W, Alpi AF, Patel KJ. The Walker B motif in avian FANCM is required to limit sister chromatid exchanges but is dispensable for DNA crosslink repair. Nucleic Acids Res. 2009;37:4360–4370. doi: 10.1093/nar/gkp365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp WD, Howard-Flanders P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J Mol Biol. 1968;31:291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab RA, Blackford AN, Niedzwiedz W. ATR activation and replication fork restart are defective in FANCM-deficient cells. EMBO J. 2010;29:806–818. doi: 10.1038/emboj.2009.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengerova B, Allerston CK, Abu M, Lee SY, Hartley J, Kiakos K, Schofield CJ, Hartley JA, Gileadi O, McHugh PJ. Characterization of the human SNM1A and SNM1B/Apollo DNA repair exonucleases. J Biol Chem. 2012;287:26254–26267. doi: 10.1074/jbc.M112.367243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeaton MB, Hlavin EM, McGregor MT, Noronha AM, Wilds CJ, Miller PS. Distortion-Dependent Unhooking of Interstrand Cross-Links in Mammalian Cell Extracts. Biochemistry. 2008;47:9920–9930. doi: 10.1021/bi800925e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Desetty R, Saito TT, Schlabach M, Lach FP, Sowa ME, Clark AB, Kunkel TA, Harper JW, Colaiacovo MP, Elledge SJ. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B, Liu XS, Davis K, Liu X. Plk1 phosphorylation of Orc2 promotes DNA replication under conditions of stress. Mol Cell Biol. 2011;31:4844–4856. doi: 10.1128/MCB.06110-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebbs RS, Hinz JM, Yamada NA, Wilson JB, Salazar EP, Thomas CB, Jones IM, Jones NJ, Thompson LH. New insights into the Fanconi anemia pathway from an isogenic FancG hamster CHO mutant. DNA Repair (Amst) 2005;4:11–22. doi: 10.1016/j.dnarep.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Thazhathveetil AK, Liu ST, Indig FE, Seidman MM. Psoralen conjugates for visualization of genomic interstrand cross-links localized by laser photoactivation. Bioconjug Chem. 2007;18:431–437. doi: 10.1021/bc060309t. [DOI] [PubMed] [Google Scholar]
- Thompson LH, Hinz JM. Cellular and molecular consequences of defective Fanconi anemia proteins in replication-coupled DNA repair: Mechanistic insights. Mutat Res. 2009;668:54–72. doi: 10.1016/j.mrfmmm.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Leung JW, Jiang Y, Lowery MG, Do H, Vasquez KM, Chen J, Wang W, Li L. FANCM and FAAP24 Maintain Genome Stability via Cooperative as Well as Unique Functions. Mol Cell. 2013;49:997–1009. doi: 10.1016/j.molcel.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Delannoy M, Ling C, Daee D, Osman F, Muniandy PA, Shen X, Oostra AB, Du H, Steltenpool J, Lin T, Schuster B, Decaillet C, Stasiak A, Stasiak AZ, Stone S, Hoatlin ME, Schindler D, Woodcock CL, Joenje H, Sen R, de Winter JP, Li L, Seidman MM, Whitby MC, Myung K, Constantinou A, Wang W. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol Cell. 2010;37:865–878. doi: 10.1016/j.molcel.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles JT, Marians KJ. The Escherichia coli replisome is inherently DNA damage tolerant. Science. 2011;334:235–238. doi: 10.1126/science.1209111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekezare M, Gomez-Gonzalez B, Diffley JF. Controlling DNA replication origins in response to DNA damage - inhibit globally, activate locally. J Cell Sci. 2013;126:1297–1306. doi: 10.1242/jcs.096701. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.