Abstract
Nested genes are fairly common within the mammalian nervous system, yet few studies have examined whether the guest and host genes might be coordinately regulated. TIMPs inhibit extracellular matrix proteolysis mediated by metzincin proteases. TIMP-2 is the only TIMP not nested within a synapsin gene. It does, however, serve as a host for DDC8, a testis-specific gene whose expression is up-regulated during spermatogenesis. Here, we demonstrate that DDC8 is not testis-specific. Furthermore, DDC8 expression in non-neural and neural tissues mimics that of TIMP-2, including its up-regulation in response to traumatic brain injury, suggesting a potential regulatory relationship. The most striking observation is that the TIMP-2 knockout mouse brain contains TIMP-2 mRNA encoding exons 2-5, which are down-stream of DDC8, but not exon 1 which contains the signal sequence and cysteine residue required for MMP inhibition, indicating a functional knockout. That TIMP-2 transcripts in wild-type brain contain DDC8 sequence suggests alternative splicing between the two genes.
Keywords: alternative splicing, DDC8, gene nesting, knockout, TIMP-2
INTRODUCTION
Recent bioinformatics studies indicate that alternative polyadenylation and splicing have played a major role in genome evolution by increasing transcriptome diversity. Alternative polyadenylation modifies the 3’ UTR, thus influencing transcript tissue distribution (5) or developmental regulation (14). In contrast, alternative splicing, which affects the transcript coding region, enables exons to be joined in various combinations to produce distinct mature transcripts. A study of human expressed sequence tags (ESTs) showed that ~60% of the investigated genes were subject to alternative splicing (45) and the majority (74%) of these changes modified the protein product. Interestingly, genes expressed within the nervous system were particularly prone to alternative splicing.
Although alternative splicing usually occurs between exons of a single gene, splicing between a “nested” gene, a gene that is contained within another gene, and its host gene further increases protein diversity. Gene nesting was first identified in Drosophila (27) and subsequently demonstrated in humans (39). Most nested genes (~60%) are found completely within an intron of and in the opposite orientation to the host gene. Many nested genes are intronless or have their entire coding region contained within one exon. The nested gene is usually expressed at higher levels than the host gene (7); thus, when expressed, it negatively influences its host via antisense-mediated inhibition (19, 54, 58). Only in one case have the host (neurofibromin 1) and nested (oligodendrocyte myelin glycoprotein) genes been reported to have similar functions (i.e., growth suppression) (24). Approximately 60% of nested genes are conserved in mouse and human with a significant proportion expressed in a tissue specific manner (60).
A well-conserved guest/host relationship is that of the tissue inhibitor of metalloproteinases (TIMPs) within the synapsin gene family (i.e., TIMP-1/synapsin 1, TIMP-3/synapsin 3 and TIMP-4/synapsin 2) which is maintained in human, mouse and Drosophila (15, 17, 49). TIMPs are small (20-30 kDa) secreted molecules that are primarily recognized for their inhibition of matrix metalloproteinase (MMP) proteolytic activity (4). In addition to MMP inhibition, TIMPs play a role in MMP activation. In particular, the activation of proMMP-2 by MT1-MMP requires TIMP-2 (6). In a number of systems it has been demonstrated that TIMPs also possess MMP-independent functions (2, 43, 51, 52). These functions are thought to reside in the TIMP carboxy terminus (21).
While TIMP-2 is the only TIMP not nested within a synapsin gene, it does serve as a host for a gene, DDC8. DDC8 (differential display clone 8) was isolated in a screen to identify genes differentially expressed during spermatogenesis (8, 29). The DDC8 cDNA is 1965 bp that encodes a protein with a calculated molecular weight of 62 kDa. The predicted hydrophilic protein shows similarities to structural and cytoskeletal proteins, including trichohyalin (37), non-muscle caldesmon (26), myosin heavy chain C and spectrin. DDC8 was reported to be testis-specific, yet no northern blot or RNase protection data was presented to substantiate its tissue specificity. Like DDC8, TIMP-2 is expressed in testis (23) where it is thought to play a role in germ cell migration through the seminiferous epithelium (41, 42, 55).
Studies were undertaken to determine whether a relationship exists between TIMP-2 and DDC8. Here, we demonstrate that DDC8 is not testis-specific and its expression mimics that of TIMP-2 in non-neural and neural tissues. Furthermore, TIMP-2 knockout mice (57) possess TIMP-2 mRNA. Given that this mRNA contains DDC8 sequence, it suggests alternative splicing between DDC8 and TIMP-2. The presence of TIMP-2 transcripts containing DDC8 in wild-type mice indicates that the DDC8/TIMP-2 splicing is not due to secondary effects as a consequence of the altered genomic structure in knockout mice. The functional significance of the DDC8/TIMP-2 relationship warrants further investigation.
Methods
Animal Care and Treatment
Mice bearing a targeted disruption of the TIMP-2 gene have been described elsewhere (57). With the exception of primary cultured embryonic fibroblasts, TIMP-2-/- mice and wild-type littermates were obtained from heterozygous matings. Genotyping was performed as previously described (35). Animals were euthanized by decapitation in the absence of anesthesia, tissue quickly removed and frozen on dry ice. These methods conform to NIH guidelines for the humane euthanasia of vertebrate animals in accordance with an approved University of Vermont Institutional Animal Care and Use Committee protocol. PCR experiments are representative of at least three animals and in situ hybridization are representative of two animals in two independent hybridization experiments.
Western blot analysis
SDS-PAGE and western blot analysis was performed as previously described (51). Primary TIMP-2 antibodies were used to epitopes in loop 1 (rabbit polyclonal, 3,000X; Triple Point Biologics; Forest Grove, OR), loop 6 (sheep polyclonal, 1,000X; Biogenesis; Kingston, NH), and carboxy-terminus (rabbit polyclonal, 1,500X; Chemicon, Temecula, CA). Horseradish peroxidase-conjugated secondary antibodies (3,000X) were obtained from Jackson ImmunoResearch (West Grove, PA). Antibody specificity was demonstrated by incubating antibodies with a 400-fold molar excess of recombinant rat TIMP-2 protein purified from transfected HEK293T cells. Antibodies were incubated with protein for 30 min at 4 °C with shaking prior to addition to blots.
Northern Blot Analysis
Northern blot analysis with 25 μg total RNA was performed exactly as previously described (32). For TIMP-2, a full-length rat cDNA probe was used. A 417 bp DDC8 exon 3 PCR amplicon (see Fig. 2C) was cloned into the TA vector (Invitrogen; Carlsbad, CA) and sequenced prior to use as a 32P-dCTP labeled probe. To confirm RNA integrity in samples lacking DDC8 hybridization and equal loading of RNA per lane, the blot was hybridized with the ubiquitously expressed, non-developmentally regulated gene cyclophilin (13, 38). Molecular sizes were determined relative to RNA molecular weight standards (Invitrogen). Blots were exposed to film (BioMax MR; Kodak, Rochester, NY) at -80 °C with an intensifying screen for 2 days.
Fig. 2. TIMP-2 mRNA contains DDC8 sequence.
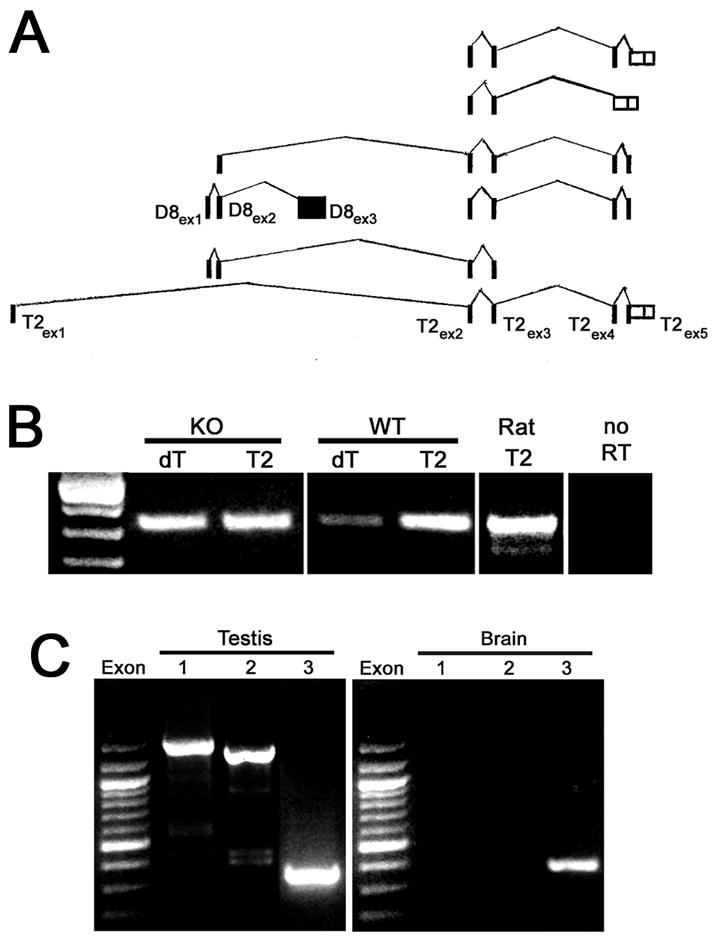
A) EST database analysis indicating potential alternative splicing between DDC8 and its host gene TIMP-2. B) First strand cDNA was primed either with oligo (dT) or with an oligonucleotide specific to TIMP-2 exon 2 (T2). The cDNAs were then amplified with primers within DDC8 exon 3. The oligo (dT) cDNA demonstrates that DDC8 is expressed in adult TIMP-2-/- (KO) and wild-type (WT) brain. Amplification of the TIMP-2 primed cDNA reveals the presence of TIMP-2 transcripts containing DDC8 sequence in adult TIMP-2-/- and wild-type mice, and rat brain. TIMP-2 primed cDNA in the absence of reverse transcriptase (no RT) fails to amplify a DDC8 exon 3 product, suggesting the products did not arise from contaminating genomic DNA. C) PCR with oligo (dT) primed adult cDNA amplifies the three DDC8 exons in testis, but only exon 3 in brain indicating that DDC8 mRNA expressed in brain differs from that expressed in testis.
Reverse transcription Polymerase Chain Reaction (rtPCR)
Total cellular RNA was prepared using a modification of the procedure by (11) (RNA STAT-60; Tel-Test B, Friendswood, TX). Contaminating genomic DNA was digested in 40 mM Tris-HCl (pH 7.9), 10 mM NaCl, 6 mM MgCl2, 10 mM CaCl2, 10 mM DTT, 40U RNasin ribonuclease inhibitor (Promega; Madison, WI) and 2U RQ1 RNase-free DNase (Promega) for 30 min at 37 °C. Following digestion, samples were extracted with phenol:chloroform, chloroform and precipitated. Total RNA (1-2 μg) was reversed transcribed using either oligo (dT) or a gene specific primer to the 5’ end of TIMP-2 exon 2 (5’-CTCCTTGTCGCTCACTGCTTTGGC-3’; 365-388 bp; X62622) according to manufacturer’s instructions (SuperScript II first-strand synthesis system; Invitrogen). Double stranded cDNA was amplified from 1 μl of transcription reaction using HotStar Taq Master Mix (Qiagen; Valenica, CA). Amplification was preceded by an initial denaturation of 94 °C for 5 min. The cycling parameters used were as follows: DDC8 (94 °C/45 sec, 54 °C/45 sec, 72 °C/45 sec extension for exon 3 and 1min 45 sec extension for exon 1 or 2; 35 cycles); TIMP-2 (49 °C annealing for 30 cycles); cyclophilin (49 °C annealing for 28 cycles) followed by a final extension of 72 °C for 5 min. The primers used are shown in Table 1. Ten μls of amplified product was electrophoresed on 1.6% agarose gels and visualized with ethidium bromide.
Table 1.
rt-PCR gene-specific primers
| Specificty | Sequence | Positiona | Amplicon size |
|---|---|---|---|
| DDC8 exon 1 | 5’-GACCACCTTAGGACAAGATT-3’ | 47-66 | |
| 5’-CAGGGATGTATTTATCATGG3’ | 1902-1921 | 1874 bp | |
| DDC8 exon 2 | 5’-GTAATGTGTCCTCATTGGC-3’ | 239-257 | |
| 5’-CAGGGATGTATTTATCATGG-3’ | 1902-1921 | 1682 bp | |
| DDC8 exon 3 | 5’-AGAAACAGGCAGACTCTGTA-3’ | 1504-1523 | |
| 5’-CAGGGATGTATTTATCATGG-3’ | 1902-1921 | 417 bp | |
| TIMP-2 exon 1 | 5’-ATGGGCGCCGCGGCC-3’ | 227-241 | |
| 5’-CTACGTCTGCATTGCAAAAC-3’ | 337-356 | 129 bp | |
| TIMP-2 exon 2 | 5’-GTAGTGATCAGGGCCAA-3’ | 353-369 | |
| 5’-CTTTATCTGCTTGATCTCA-3’ | 439-457 | 104 bp | |
| TIMP-2 exon 3 | 5’-ATGTTCAAAGGACCTGAC-3’ | 458-475 | |
| 5’-GCAATTAGATATTCCTTCTT-3’ | 545-564 | 106 bp | |
| TIMP-2 exon 4 | 5’-AGGCGGAAGGAGATGG-3’ | 570-585 | |
| 5’-TGCACTCGCAGCCCAT-3’ | 674-689 | 119 bp | |
| TIMP-2 exon 5 | 5’-CATGCTACATCTCCTCC-3’ | 714-730 | |
| 5’-GGGTCCTCGATGTCAAG-3’ | 869-885 | 171 bp | |
| TIMP-2 | 5’-GTAGTGATCAGGGCCA-3’ | 353-369 | |
| 5’-CCTTCIGCCTTYCCTGC-3’ | 563-579 | 210 bp | |
| Cyclophilin | 5’-AGACATGGTCAACCCCACCGTG-3’ | 39-60 | |
| 5’-TTAGAGTTGTCCACAGTCGGAG-3’ | 516-537 | 498 bp |
Reverse zymography
Primary cultured fibroblasts were prepared from embryonic day 14 (E14) TIMP-2-/- and wild-type mice. The skin was placed dermis side down on 60 mm plates in DMEM supplemented with 10% FCS for 48 hrs to allow fibroblasts to migrate out of the tissue. The tissue was removed and cells cultured for 7 days until 75-85% confluent. Cells were washed with PBS three times and cultured for an additional 12 hr in OPTIMEM serum-free medium (Invitrogen). Conditioned medium was removed, centrifuged to remove cells, and processed for reverse zymography as previously described (51). Recombinant human TIMP-2 (Chemicon) was used as a positive control.
Stab Injury
The cortical stab injury was performed as previously described (33). Young adult (postnatal day 45) Sprague-Dawley rats were anesthetized by intraperitoneal injection of chloral hydrate (420 mg/kg weight). A 1 mm2 craniostomy was performed above the right cerebral hemisphere (4.5 mm caudal of Bregma, 3.5 mm lateral of the midline). A 27 gauge needle was briefly dipped into the fluorescent chromagen Fast Blue (1%), inserted 4 mm through the cortex, into the underlying thalamus, held in place for 1 min, and then withdrawn. Fast Blue is used to identify the injury site when sectioning the brain. The craniostomy hole was sealed with dental wax and the incision closed with surgical staples. Following surgery, the animals were placed on thermal pads, with recovery from anesthesia generally occurring within 3 hrs. The animals displayed no obvious discomfort or neurologic abnormalities as a result of the surgery. Animals were euthanized at 2, 4, 7, and 14 days post surgery.
In situ hybridization
In situ hybridization was performed exactly as previously described (32). Near-adjacent (12 μm) sections were hybridized with antisense and sense 33P-UTP labeled cRNA probes for DDC8 (417 bp, 1504-1921 of Y09878) and TIMP-2 (232 bp, 350-582 of X62622). Following hybridization, the slides were initially exposed to autoradiographic film (BioMax MR) at 4 °C for either 3 days (TIMP-2) or 11 days (DDC8). Autoradiograms were scanned using an Epson Expression 800 scanner with transparency adapter and imported into Adobe Photoshop (Adobe Systems Inc., San Jose, CA). For higher resolution, the slides were dipped in NTB emulsion (Kodak), developed after 6 days (TIMP-2) or 22 days (DDC8), examined with a Nikon E800 microscope (Micro Video Instruments; Avon, MA), and images captured with a Spot RT digital camera (Diagnostic Instruments; Sterling Heights, MI).
Identification of DDC8 homologs
Proteins sharing ancestors with DDC8 were identified using: 1) BLASTp with DDC8 human protein (gi 89043091) and its paralog (gi 20521976) as the query, 2) TBLASTn searches of vertebrate genomes, and 3) TBLASTn searches of vertebrate EST databases (3).
Sequence Alignment and Phylogenetic Analysis
Protein sequences were aligned using progressive alignment T-Coffee with default parameter settings (47). Phylogenetic trees were constructed using a Bayesian (MrBayes 3.1; (28), maximum parsimony (PROTPARS; (20), and neighbor joining algorithm (PROTDIST/NEIGHBOR; (20) methods. Confidence in each clade was determined by three parameters: 1) bootstrap support under maximum parsimony on 100 samples, 2) bootstrap support using the neighbor joining algorithm on 100 samples, and 3) the posterior probability obtained from MCMC simulation using Mr. Bayes program. The SEQBOOT and CONSENSE programs of the PHYLIP package (20) was used for the generation of bootstrapped data sets and consensus tree reconstructions, respectively.
Gene Structure
Gene structure was inferred using Aceview (http://www.ncbi.nlm.nih.gov/AceView/) and Mapviewer (http://www.ncbi.nlm.nih.gov/mapview/) on the genomes of: human (build 36.1), mouse (build 36.1), rat (build 3.1), cow (build 2.1), chicken (build 1.1) and chimpanzee (build 1.1).
Results
Adult TIMP-2-/- mice possesses TIMP-2 mRNA and protein, but lack MMP inhibitory activity
Soloway and colleagues developed mice carrying a targeted mutation in the TIMP-2 gene by deletion of a 4.4 kb genomic fragment containing the first coding exon and additional 5’ sequences (Fig. 1A) (57). No TIMP-2 mRNA was detected by northern blot analysis of mouse lung, a tissue which abundantly expresses TIMP-2, indicating a null mutation. The only phenotype observed by these investigators was the impairment of proMMP-2 activation. However, we have identified behavioral (31, 35) and histological (35, 51) alterations in the TIMP-2-/- mice.
Fig. 1. TIMP-2-/- mice possess TIMP-2 mRNA.
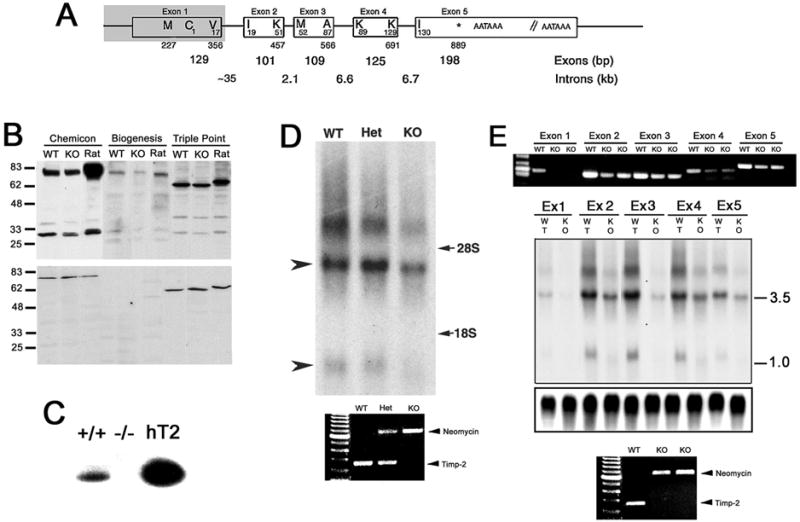
A) Genomic structure demonstrating mouse TIMP-2 intron-exon junctions (9). Amino acid resides are indicated within the boxes and the corresponding nucleotide sequence are indicated below (GenBank Accession mRNA: X62622, GeneID: 21858). The shaded area represents the genomic sequence deleted in TIMP-2-/- mice (57). B) Western blot analysis of adult mouse brain (20 μg crude homogenate) from wild-type (WT) and littermate TIMP-2-/- (KO) mice, and adult rat brain using antibodies to three distinct TIMP-2 epitopes. In addition to identifying TIMP-2 (28 kDa protein), additional proteins of 78 kDa (Chemicon and Biogenesis) and 62 kDa (Triple Point) are identified (upper blot). TIMP-2 expression is reduced, but not abolished in TIMP-2-/- mice. Pre-incubation of antibodies with recombinant TIMP-2 protein abrogates (28 kDa TIMP-2 protein) or significantly diminishes (78 and 62 kDa proteins) immunoreactivity (lower blot), thus confirming antibody specificity. C) Reverse zymography of conditioned media from E14 fibroblasts demonstrates that wild-type (+/+), but not knockout (-/-) cells possess MMP-inhibitory activity, indicating a functional knockout. Human recombinant TIMP-2 (hT2) was used as a positive control. D) Northern blot of adult brain (25 μg total RNA) from wild-type, heterozygous (Het) and TIMP-2-/- littermates hybridized with a full-length TIMP-2 probe. The 1.0 kb transcript is not readily detectable (lower arrowhead), while the 3.5 kb transcript (upper arrowhead) is reduced, but not absent in TIMP-2-/- mice. An additional ~7.0 kb transcript of unknown origin is also detected in wild-type brain, but is almost absent in TIMP-2-/- brain. The positions of 28S and 18S rRNA are indicated on the right. PCR of genomic DNA from these animals demonstrating their genotype is shown below the blot. E) PCR with primers specific to each TIMP-2 exon using adult brain from wild-type and 2 TIMP-2-/- mice (one littermate and one from another litter). No exon 1 product is obtained; however, products for the remaining 4 exons are obtained from TIMP-2-/- mice. The expression of these exons is reduced approximately 50% relative to wild-type mice. Northern blot analysis of adult brain (25 μg total RNA) hybridized with exon specific probes detects the 3.5 kb transcript in TIMP-2-/- mice. The different expression level of the 5 exons in wild-type brain suggests TIMP-2 variants with alternative exon usage (see Fig 2A). Cyclophilin was used to verify equal loading of RNA per lane (lower portion blot). PCR of genomic DNA from these animals demonstrating their genotype is shown below the blot.
During the analysis of the TIMP-2-/- mice, immunohistochemical staining was observed within the brain and spinal cord. Western blot analysis using antibodies to three distinct epitopes of TIMP-2 confirmed the histological observation (Fig. 1B). The molecular weight of TIMP-2 is 21.5 kDa, while the molecular weight of TIMP-1 is 28.5 kDa. The three TIMP-2 antibodies used in the study recognized a protein of 28 kDa in adult wild-type and TIMP-2-/- mouse brain, as well as adult rat brain. Therefore, there was significant concern that the antibodies were actually reacting with TIMP-1, thus explaining the immunoreactivity in TIMP-2-/- mice. However, these antibodies failed to identify recombinant human TIMP-1 protein, but showed significant reactivity with recombinant human TIMP-2 protein, confirming antibody specificity towards TIMP-2 (data not shown). In addition to the 28 kDa TIMP-2 protein, a 78 kDa protein was identified with two of the antibodies and a 62 kDa protein with the third. Immunoreactivity of the 28 kDa protein was completely blocked while the 78 and 62 kDa proteins was dramatically reduced by pre-incubation with recombinant TIMP-2 protein, further confirming antibody specificity.
Reverse and gelatin zymography were performed to verify that mice were indeed TIMP-2-deficient (Fig. 1C). In reverse zymography, samples are electrophoresed on a non-denaturing SDS-PAGE containing gelatin and conditioned medium from baby hamster kidney cells, which express MMPs. The MMPs degrade the gelatin in all regions of the gel except where there is TIMP activity. MMP-inhibitory activity was detected in embryonic day 14 (E14) wild-type, but not TIMP-2-/- fibroblasts. In addition, the 28 kDa protein possess MMP-inhibitory activity in wild-type, but not TIMP-2-/-, brain (not shown). The reduction in proMMP-2 activation in TIMP-2-/- mice was corroborated (57) using fluorescently caged gelatin as a substrate (40). Taken together, these data indicate that although TIMP-2 protein is present in TIMP-2-/- mice, the mice are phenotypically deficient in both MMP inhibition and proMMP-2 activation.
Northern blot analysis was performed to confirm the absence of TIMP-2 mRNA in the adult TIMP-2-/- brain. Unexpectedly, this analysis revealed the presence of TIMP-2 mRNA (Fig. 1D). In contrast to wild-type and heterozygous littermates in which both the 3.5 and 1.0 kb TIMP-2 transcripts were detected, only the 3.5 kb transcript was readily apparent in TIMP-2-/- brain. Expression of this transcript was reduced by 50% relative to wild-type littermates. Only a slight size reduction in the 3.5 kb transcript was detected since just 356 bp of coding sequence was deleted. A previously unreported, additional TIMP-2 transcript of ~7.0 kb was detected in all three genotypes. PCR amplification using two different primer sets substantiated the genotypes of the animals. Because the hybridization probe used was to full length TIMP-2, rtPCR was performed with primers specific to each of the five coding TIMP-2 exons to determine which exons were expressed in TIMP-2-/- mice (Fig. 1E). The absence of an exon 1 product from TIMP-2-/- brain verified the Soloway targeting strategy. However, amplification products for the remaining four exons were obtained. PCR amplification of reverse transcription reactions lacking enzyme failed to produce a product for any of the exons (data not shown). Also, amplification of first strand cDNA with primers that flanked exon junctions only generated a single product. Taken together, it indicates that the amplicons did not originate from genomic DNA in the RNA preparation. Sequencing authenticated each amplicon obtained from TIMP-2-/- mice as TIMP-2. These amplicons were then used as northern hybridization probes to quantitate the relative expression of each exon. In wild-type mice, exons 2, 3, and 4 were expressed at greater levels than exon 5. Interestingly, exon 1 was expressed at the lowest levels. In TIMP-2-/- mice, exons 2 and 4 were expressed at greater levels, with lower expression of exon 5 and the least expression of exon 3. This differential expression is not likely due to different hybridization efficiencies since each probe was approximately the same length (see Figs. 1A, 1E PCR). Rather, it suggests alternative exon usage.
The nested gene DDC8 is contained within TIMP-2 mRNA transcripts
Since the gene DDC8 is nested within the first intron of TIMP-2, we sought to determine whether its promoter could be responsible for TIMP-2 mRNA production in TIMP-2-/- mice. In silico analysis of the EST database revealed several potential splice junctions between DDC8 and TIMP-2 (Fig. 2A) as well as DDC8 paralogs (discussed below). DDC8 is encoded by three exons with the entire coding region contained within exon 3. Although no EST identified splicing between DDC8 exon 3 and TIMP-2, we nonetheless proceeded to determine whether a relationship between DDC8 and TIMP-2 existed. Semi-quantitative rtPCR with oligo (dT) primed cDNA from adult brain and primers within exon 3 demonstrated that DDC8 was not testis-specific, as previously reported (8) (Fig. 2B). While DDC8 was detected in both genotypes, expression was greater in TIMP-2-/- than wild-type brain. Sequencing authenticated the amplicons as DDC8. Surprisingly, when cDNA was primed with an oligonucleotide specific to the 5’ most end of TIMP-2 exon 2 and PCR amplification performed with DDC8 exon 3 primers, a product was obtained that was authenticated as DDC8. Due to concerns that this relationship resulted from secondary effects as a consequence of altered genomic structure in knockout mice, wild-type mice were likewise analyzed. TIMP-2 mRNA similarly contained DDC8 sequence in wild-type mice. DDC8 sequence was also detected within rat TIMP-2 mRNA. That the product was more abundant in rat than mouse is correlated with the more abundant expression of the 28 kDa and 78 kDa proteins in rat brain (Fig. 1B). Next, the expression of DDC8 exons 1 and 2, which contain the 5’ UTR, was determined. Using oligo (dT) primed cDNA, exons 1 and 2 were detected in adult wild-type testis, but not brain (Fig. 1C). Similar results were obtained with TIMP-2 primed cDNA (data not shown). These data suggest that DDC8 is expressed in brain and that TIMP-2 mRNA transcripts contain DDC8 sequence, but that the DDC8 5’ UTR in brain differs from that in testis.
The spatial expression of DDC8 mRNA mimics that of TIMP-2 mRNA
To determine the spatial distribution of DDC8 mRNA in adult wild-type mice, northern blot analysis was performed (Fig. 3A). Using the 417 bp amplicon that recognized DDC8 in testis and brain (Fig. 2C) as a probe, a discrete 2.0 kb mRNA transcript and a diffuse ~4.4 kb transcript was detected only in testis. Given that mRNA represents a small (~1%) fraction of total RNA (i.e., mRNA, rRNA, tRNA, and hnRNA), the lack of signal in other tissues is likely due to insensitivity of detection. Therefore, semi-quantitative PCR was performed to determine DDC8 tissue distribution (Fig. 3B). As expected from the northern blot, DDC8 was most abundantly expressed in testis. However, DDC8 expression was also detected in brain, lung, and kidney. This expression pattern is similar to that reported by microarray analysis (56), http://expression.gnf.org). With the exception of the increased expression in kidney, DDC8 expression was similar to that of TIMP-2 reported here and by others (48, 59). DDC8 expression in brain was further investigated by examining specific brain regions (Fig. 3C). DDC8 expression was greatest in the brainstem, cerebral cortex, and thalamus with lesser expression in cerebellum and hippocampus. Similar to its expression in non-neural tissues, the expression of DDC8 in the various brain regions mimicked that of TIMP-2. Notable exceptions include the hippocampus and cerebellum in which DDC8 expression is greater than TIMP-2. Taken together, these data further suggest a relationship between DDC8 and TIMP-2.
Fig. 3. DDC8 expression is not testis-specific.
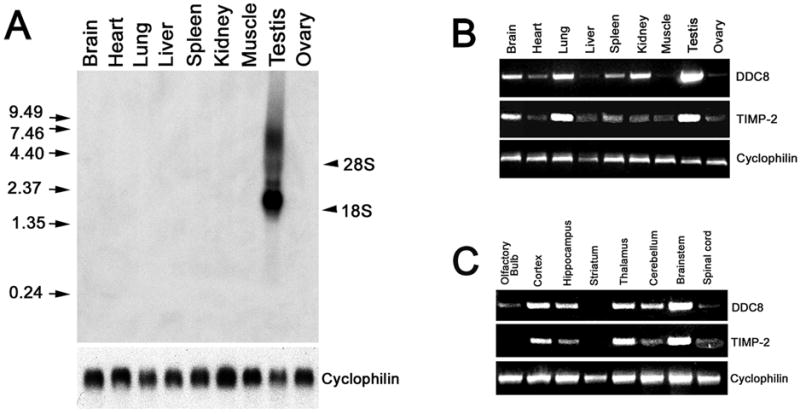
A) Northern blot analysis of adult wild-type tissues (25 μg total RNA) detects a DDC8 mRNA transcript of 2.0 kb, as well as a diffuse band of ~4.4 kb, only in testis. RNA integrity and equal loading of RNA per lane is demonstrated using the ubiquitously expressed gene cyclophilin (below blot). The positions of 28S and 18S rRNA are indicated on the right. RNA molecular weight standards are indicated on the left. B) Semi-quantitative PCR of oligo (dT) primed cDNA (from 2 μg total RNA) demonstrates that DDC8 is not testis-specific. DDC8 is also abundantly expressed in brain, lung, and kidney. Like DDC8, TIMP-2 expression is abundant in testis, lung, and brain. It should be noted that DDC8 requires 35 cycles while TIMP-2 only 30 cycles to be within the linear amplification range. Cyclophilin (28 cycles of amplification) was used to verify equivalent reverse transcription between samples. C) Semi-quantitative PCR of oligo (dT) primed cDNA (from 1.25 μg total RNA) reveals that DDC8 expression is similar to that of TIMP-2 within the brain, with greatest expression in the brainstem, thalamus, and cerebral cortex. DDC8 expression is greater than TIMP-2 in the cerebellum and hippocampus. Cyclophilin was used to verify equivalent reverse transcription between samples.
The spatial distribution of DDC8 in brain throughout embryonic (Fig. 4A) and postnatal (Fig. 4B) murine development was examined by in situ hybridization. DDC8 expression was detected at E14, the first time point examined (Fig. 4A). Within non-neural tissues, DDC8 expression was enriched in the kidney (Figs. 4Aa,c) and small intestine (Figs. 4Aa-c). Within the nervous system, expression was particularly prominent in the most superficial layers of the posterior cerebral cortex at E14 (Fig. 4Aa) and the retina at E18 (Fig. 4Ac). Postnatally, a diffuse hybridization signal was detected throughout the brain parenchyma (Figs. 4Ba-i). As observed embryonically, DDC8 expression was enriched in the posterior cerebral cortex at postnatal day 0 (P0) (Fig. 4Ba), but expression declined dramatically by P7 (Fig. 4Bb). Cerebellar expression was most prominent during the first two postnatal weeks, which coincides with the period of granule cell migration and synaptogenesis (Figs. 4Ba-c). DDC8 expression throughout the brain appeared maximal at P21 (Fig. 4Bd) and then gradually declined to reach adult levels by P60 (Fig. 4Be-i). As revealed by northern blot analysis and rtPCR, DDC8 expression in testis was far greater than that in brain (Figs. 4Bj, k). Because DDC8 is also expressed in rat (Figs. 1B, 2B), its expression was examined (Fig. 4C) and compared to that of TIMP-2 (Fig. 4D). At E14, DDC8 expression was detected within the forebrain and hindbrain (Fig. 4Ca) in a pattern similar to that previously observed for TIMP-2 (18). DDC8 expression in the rat cerebral cortex at E18 mimicked that observed at E14 and P0 in the mouse (Fig. 4Cb). At P14, DDC8 expression was enriched in the olfactory bulb, striatum, hippocampus, and cerebellar Purkinje cells (Fig. 4Cc). TIMP-2 expression was enriched in the posterior cerebral cortex in the E16 rat (Fig. 4Da) similar to that observed for DDC8 in the mouse and rat. In contrast to DDC8, TIMP-2 expression was significantly greater in the brain (Figs. 4Db,c) and reduced in testis (Figs. 4Dd,e).
Fig. 4. DDC8 expression in the brain is similar to that of TIMP-2.
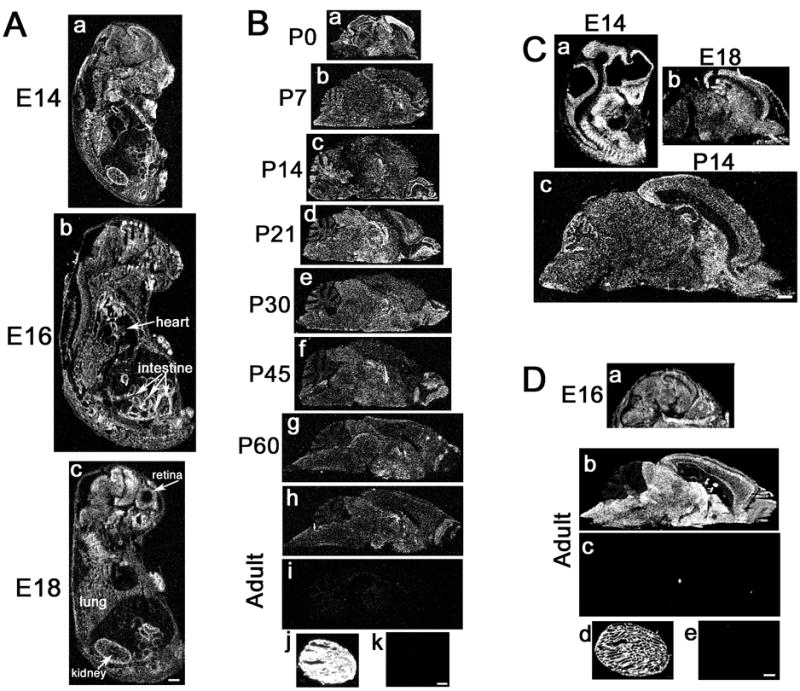
In situ hybridization at the indicated embryonic (A) and postnatal (B) time points of mouse development reveals the spatial distribution of DDC8 expression. DDC8 is expressed throughout brain development, with maximal expression at P21. DDC8 (Bj) is more abundantly expressed than TIMP-2 (Dd) in the adult testis. DDC8 expression (C) mimics that of TIMP-2 (D) in the rat. Sense controls for DDC8 (Bi, Bk) and TIMP-2 (Dc, De) demonstrate hybridization specificity in the adult brain and testis, respectively. Hybridized slides were apposed to autoradiographic film for either 3 days (TIMP-2) or 11 days (DDC8). Scale bars = 1 mm.
Closer examination of emulsion-coated sections revealed that the spatial expression of DDC8 was comparable, but not identical, to that of TIMP-2 (Fig. 5). Both genes showed similar distributions in the retina, including abundant expression in the outer plexiform layer with lesser expression in the ganglion cell layer (Fig. 5Aa,b), confirming previous reports of TIMP-2 immunoreactivity in the interphotoreceptor matrix and Müller glia (1, 22). In contrast, expression in the hippocampus and cerebral cortex were remarkably different. DDC8 expression was detected throughout the hippocampal formation, including all CA subfields and the dentate gyrus (Fig. 5Ac), while a low level of TIMP-2 expression was restricted to CA1 (Fig. 5Ad). Conversely, DDC8 expression was diffusely expressed throughout all cortical layers (Fig. 5Ae), while TIMP-2 expression was enriched in layer III and the white mater (Fig. 5Af). Expression in testis also differed. DDC8 was primarily expressed within the seminiferous tubules within spermatids (Fig. 5Ag, i), while TIMP-2 was expressed in Sertoli and Leydig cells (Fig. 5Ah), corroborating the proposed expression of DDC8 (8) and previous report of TIMP-2 expression in testis (23).
Fig. 5. DDC8 expression is up-regulated in response to intracranial injury.
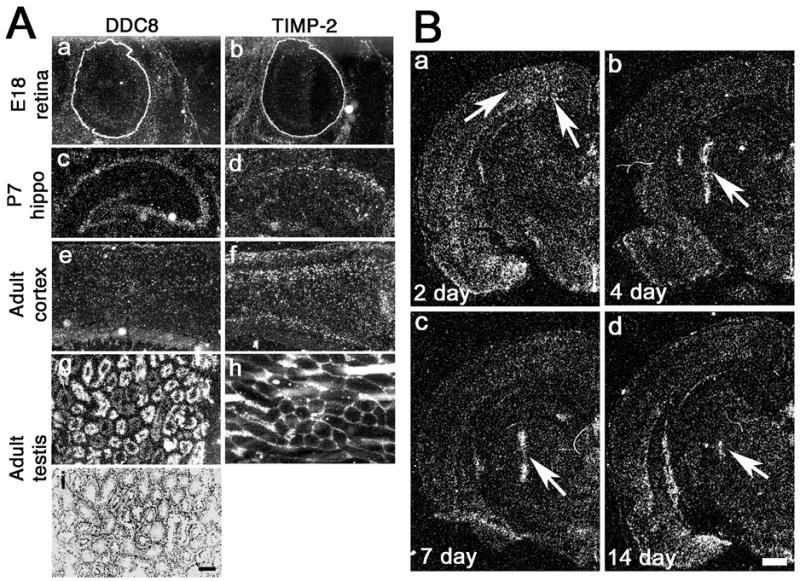
A) High magnification emulsion-coated sections show that DDC8 expression is similar to TIMP-2 in the mouse retina, but differs in the hippocampus (hippo), cerebral cortex, and testis. Cresyl violet stained section (i) reveals testis morphology. Hybridized slides were exposed to autoradiographic emulsion for either 6 days (TIMP-2) or 22 days (DDC8). B) Coronal sections show that DDC8 expression is up-regulated (arrows) after a penetrating stab injury to the rat brain. Scale bars = 250 μm (A), 1 mm (B).
Despite these differences in expression patterns, the spatial distribution of DDC8 and TIMP-2 were sufficiently similar to suggest that their expression might be coordinately regulated. Previously, we demonstrated that TIMP-2 expression is increased in response to a penetrating injury to the rat cerebral cortex (30). Therefore, we sought to determine whether DDC8’s expression would be similarly up-regulated in response to injury (Fig. 5B). At 2 days after injury, TIMP-2 expression is moderately increased in the penumbra of the injury. TIMP-2 expression peaks at 4 days post-injury and declines only slightly at 7 days post-injury. At both time points expression is largely restricted to the region immediately adjacent to the needle tract. Two weeks after injury, only a weak hybridization signal for TIMP-2 remains at the injury site. Similar to TIMP-2, DDC8 expression was moderately up-regulated 2 days after injury (Fig. 5Ba), but was markedly increased at 4 days (Fig. 5Bb) and 7 days (Fig. 5Bc) post-injury. At 14 days post-injury, only limited DDC8 expression was detectable (Fig. 5Bd). The use of radioactive in situ hybridization precludes the determination of whether both genes are co-expressed. While TIMP-2 is primarily expressed by microglial cells at the injury, the cellular source(s) of DDC8 is currently unresolved. Combined in situ hybridization / immunohistochemistry was inconclusive as to whether DDC8 was expressed in infiltrated inflammatory cells as well as GFAP-positive astrocytes (data not shown).
DDC8 homologs are represented broadly among vertebrates
In silico experiments were performed to determine whether particular sequence variations might be responsible for differences in DDC8 expression patterns observed in mouse and rat (i.e., expression in mouse cerebellar granule cells, but rat Purkinje cells). This analysis revealed that in addition to mouse and rat, the DDC8 protein was present in primates (e.g., human and macaque) (Fig. 6). Sequence similarity between mouse and rat (80% identity, 86% similarity) was much greater than that between mouse and human (42% identity, 56% similarity), suggesting evolutionary divergence. However, NCBI Aceview and Mapviewer data confirmed the presence of DDC8 nesting within three of the four TIMP-2 genes (e.g., human, mouse, rat). We were unable to confirm DDC8 nesting within Macaca fascicularis TIMP-2 because the genome sequence is incomplete.
Fig. 6. DDC8 orthologs exist in multiple species.
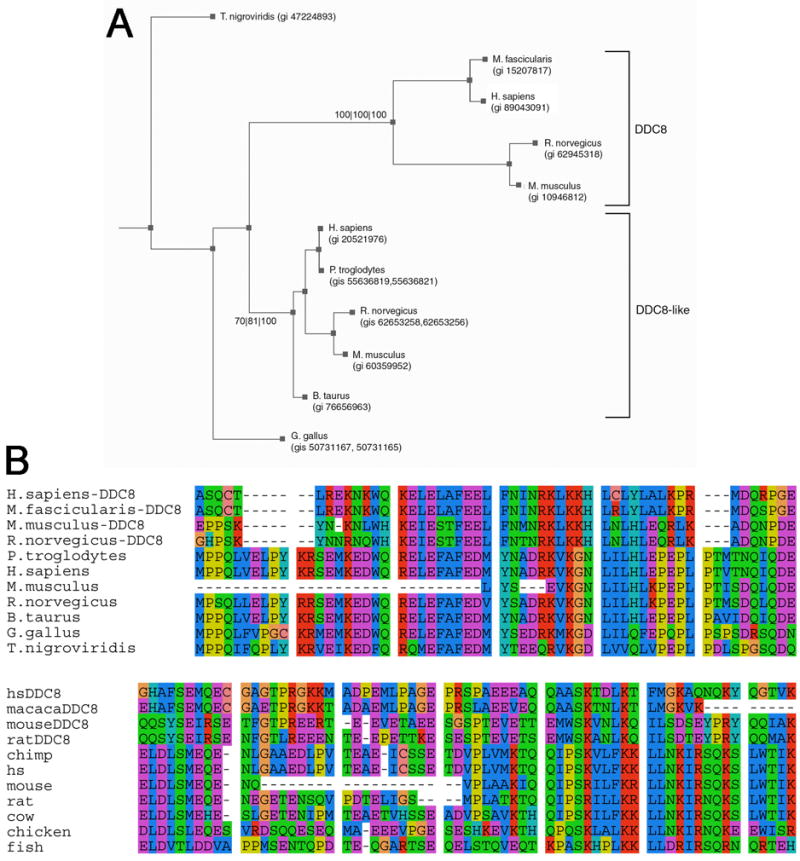
A) BLASTp, and TBLASTn searches of vertebrate genome and EST databases identified two families of DDC8 genes. One clade, termed DDC8, is supported at 100/100/100 (each value represents the statistical confidence from each of three methods used – see Experimental Design). Three of these genes (e.g., human, mouse, and rat) are nested within the TIMP-2 gene. In addition, a previously unreported clade (here termed DDC8-like), which is supported at a confidence of 70/81/100, is present in more species than the DDC8 clade. Some species have two GI numbers indicated because the sequence similarity extended through more than more gene. B) The most conserved region of the protein sequence alignment for the DDC8 and DDC8-like proteins is shown. This region of mouse DDC8 shows 86% similarity to rat, 60% similarity to macaque, and 57% similarity to human DDC8, but does not correspond to any known functional domain.
The database analysis revealed the presence of a DDC8 ortholog in fish, birds, and mammals (Fig. 6A). Based on the magnitude of the e-values (smaller than 4e-7) and on visual inspection of alignments (Fig. 6B), we conclude that these sequences share ancestry with DDC8 and are, therefore, referred to as DDC8-like. Humans and rodents each have two representatives from the DDC8 family, suggesting that a gene duplication preceded the divergence of primates and rodents. The DDC8 clade may have arisen either from a gene duplication following the divergence of fish from terrestrial vertebrates or, alternatively, from a gene duplication prior to the divergence of fish and terrestrial vertebrates, followed by loss of DDC8 orthologs from fish and birds. The DDC8 gene is located on chromosome 17, 11, and 10, while the DDC8-like gene is located on chromosome 11, 9, and 8 in human, mouse, and rat, respectively. These orthologs share 17%, 27%, and 14% similarity, respectively. The true structure of the DDC8-like gene is not clear since we were unable to determine whether this is a single gene or two shorter genes close together in the genome.
As previously reported (8) and confirmed here, DDC8, and to a lesser degree DDC8-like proteins, share similarity to a number of cytoskeletal proteins. However, no functional domains were identifiable either in the DDC8 or DDC8-like proteins. Therefore, the exact function of these proteins is not known at the present time.
Database analysis also revealed a relationship between the DDC8-like gene and the gene Josd3. The DDC8-like gene is in close proximity to the Josd3 (Josephin containing domain 3) gene in human, mouse, rat, cow, chimpanzee and chicken. However, the significance of this gene arrangement is unknown.
Discussion
Two novel observations are described in the present report. First, TIMP-2-/- mice possess mRNA encoding TIMP-2 exons 2-5 not deleted in the targeting strategy (57). Second, TIMP-2 mRNA in both wild-type and knockout brain contains sequence for DDC8, a gene which is nested within the first intron of TIMP-2. Taken together, these data suggest that DDC8 may serve as the source of TIMP-2 mRNA.
TIMP-2-/- mice are biologically functional knockouts despite the presence of TIMP-2 mRNA
Soloway and colleagues devised an ingenious targeting strategy that would ensure the creation of a biologically functional knockout (57). Targeted disruption of TIMP-2 by removal of the entire gene was not possible due to its large size (~50 kb). Thus, they removed exon 1 which contains the signal peptide and the terminal cysteine residue of the mature protein required for interaction with MMPs. Hence, even if additional promoter elements were present either up-stream of the region deleted or down-stream within the ~35 kb intron 1, the product would not be secreted and would lack MMP-inhibitory activity. Data presented here and elsewhere demonstrates that TIMP-2-/- mice lack a secreted TIMP-2 immunoreactive protein product, lack TIMP-2-mediated MMP inhibitory activity, and are deficient in proMMP-2 activation, and; thus, are functional TIMP-2 knockouts even though TIMP-2 mRNA and protein products seem to be present.
One curious observation was the presence of the 3.5 kb, but not detectable expression of the 1.0 kb, TIMP-2 transcript in TIMP-2-/- brain. A previous report (25) and numerous EST sequences substantiate the generation of the two transcripts via alternative polyadenylation. If only one TIMP-2 promoter was present, then its deletion should disrupt both transcripts. It is also intriguing that the 3.5 kb transcript persists in the TIMP-2-/- brain because its expression, unlike that of the 1.0 kb transcript, is up-regulated during brain development (18). These data suggest the presence of additional regulatory elements. Furthermore, northern hybridization with exon-specific probes showed that the expression of the 3.5 kb transcript in the TIMP-2-/- brain was reduced to a greater degree with some exons (i.e., exons 3 and 5) than others (i.e., exons 2 and 4), suggesting alternative splicing in TIMP-2. This may explain how the single 3.5 kb transcript could drive the expression of both the 28 kDa and 78 kDa proteins.
Two “TIMP-2” protein products are detected in brain. The 28 kDa protein possess MMP-inhibitory activity in wild-type, but not TIMP-2-/-, brain, confirming this protein as TIMP-2. The identity of the 78 kDa protein is currently unknown, but two possibilities have been ruled out. Although TIMPs are known to form SDS stable dimers (12), several observations suggest this is not the basis of the larger protein. First, the molecular weight of the 78 kDa protein is inconsistent with TIMP-2 dimers or trimers. Second, the 78 kDa protein does not dissociate into smaller proteins after heating in the presence of 20 mM EDTA or 50 mM DTT, suggesting a single protein. Finally, the fact that 78 kDa protein is developmentally up-regulated while the 28kDa TIMP-2 is constitutively expressed (unpublished observation) is inconsistent with multiplexing of the 28 kDa protein. It is interesting to note that the developmental expression of the 78 kDa protein mimics that of the 3.5 kb mRNA transcript, while the 28 kDa TIMP-2 is constitutively expressed similar to the 1.0 kb transcript. Another possibility that was ruled out was that the 78 kDa protein represented the “large inhibitor of metalloproteinase” (LIMP) (10, 12). LIMP is a 76 kDa stable complex of proMMP-2 bound to TIMP-2 and, thus, would be immunoreactive with TIMP-2 antibodies. LIMP is capable of inhibiting MMP-1, -2, and –3. By reverse zymography, the 78 kDa protein lacks detectable MMP-inhibitory activity, suggesting it is not LIMP.
DDC8 may serve as the source of TIMP-2 mRNA and protein in TIMP-2-/- brain
The most parsimonious explanation for the presence of TIMP-2 mRNA in the TIMP-2-/- brain is that it arises from the DDC8 promoter. The presence of TIMP-2 mRNA with DDC8 sequence in wild-type brain indicates that this is not an epiphenomenon of knockout genetic manipulation. Nothing is known about DDC8, other than that predicted from the cDNA sequence. Nonetheless, several of its features are intriguing and relevant to the current study. Here, we demonstrate northern hybridization for DDC8, which was not previously reported (8). In addition to the 2.0 kb mRNA, which is consistent with the 1965 bp cDNA, an additional diffuse transcript of ~4.4 kb was detected. In addition, we detected a previously unreported 7.0 kb transcript that hybridized with TIMP-2 in wild-type brain. This transcript is larger than expected by joining the 3.5 kb TIMP-2 and 2.0 kb DDC8 messages, yet smaller than joining with the 4.4 kb DDC8 transcript. It is highly unlikely that the 7.0 kb transcript is the source of the 28 and 78 kDa proteins because it was almost completely abrogated in TIMP-2-/- mice. The 5’ UTR of DDC8 is encoded by exons 1 and 2. These exons were detected in testis, but not brain, suggesting that DDC8 uses an alternative 5’ UTR. The use of a common promoter, but different 5’ UTR can be explained by the presence of a potential 5’ splice site 20 bp into DDC8 exon 3. DDC8 also contains a very short 3’ UTR (55 bp) containing two putative overlapping non-canonical polyadenylation sequences (i.e., GATAAA and AATACA). Although these sequences only differ from the canonical consensus sequence AAUAAA (50) at one residue, non-canonical sequences are rare among mRNAs (<1.5%), likely due to their lower efficiency of polyadenylation (11%) and cleavage (30%) (53). Our ability to detect DDC8 by rtPCR of oligo (dT) primed cDNA and northern hybridization demonstrates appropriate polyadenylation and cleavage.
Unlike most nested genes, DDC8 is in the same orientation and is less abundantly expressed than its host, with the exception of testis; thus, it is not capable of antisense-mediated inhibition. Rather, the spatial expression and temporal regulation of DDC8 mimics that of TIMP-2, suggesting a positive relationship. This hypothesis is substantiated by the similar expression of DDC8 and TIMP-2 in response to a penetrating injury to the cerebral cortex. Splicing of DDC8 (calculated molecular weight of 62 kDa) with the remaining exons of TIMP-2 (19 kDa due to the loss of only 17 amino acid residues in exon 1) would result in a protein mass of 81 kDa, similar to that observed in our study. Like the majority of nested genes, the DDC8 coding region is restricted to one exon and, hence, is not subject to splice variants. However, DDC8 contains five potential translation initiation sites. The reported predicted molecular mass was based on the largest open reading frame and the initiation of translation at the third ATG codon (8). Yet, the sequence surrounding this codon is unfavorable for translation initiation (36). Because DDC8 lacks a signal sequence, DDC8 and the DDC8/TIMP-2 proteins would be localized intracellularly. This is substantiated by the observation that TIMP-2 is more abundantly expressed intracellularly than on the cell surface (34). Previously, we demonstrated that TIMP-2 induces cell cycle arrest (51). Although DDC8 contains multiple nuclear localization sequences, we have never observed TIMP-2 expression within the nucleus. DDC8 has homology to cytoskeletal proteins and TIMP-2 induces neurite outgrowth (51). Experiments are required to determine the translation start site and the function of DDC8 in brain to determine whether it may contribute to TIMP-2’s activities.
Bioinformatics analysis reveals a relationship between DDC8 and two divergent genes
Database analysis identified a well-defined set of DDC8-like orthologs, suggesting a gene duplication occurred during evolution of this protein family. An inferred phylogeny of these sequences exhibited two clear clades, both with strong statistical support. Although it is apparent that a gene duplication preceded divergence of primates and rodents, the precise placement of the duplication is not clear. Furthermore, given the restricted region of similarity between the orthologs (125 of 533 amino acid residues), low sequence similarity (~20%), and the lack of identifiable conserved domains within this region, the significance of the DDC8 orthologs is currently unknown.
In addition to identifying the DDC8 orthologs, the database analysis identified a conserved relationship between DDC8-like and josd3 genes. With the exception of the presence of a Josephin domain, no data exists on the function of the josd3 protein. The Josephin domain is a conserved module named after the Machado–Joseph disease, also known as spinocerebellar ataxia type 3, one of several hereditary autosomal dominant neurodegenerative disorders caused by expansion of a polyglutamine repeat in the affected gene product. The Josephin domain, which is present in at least 30 predicted proteins, binds ubiquitin, consistent with its deubiqutinating activity (44). Ataxin-3/Josephin and sertolin, a marker of cell-cell interaction during spermatogenesis (46), share homology with thrombospondin, an extracellular matrix protein, known to be involved in cell adhesion, proliferation, and migration. It is interesting to note that TIMP-2-/- mice display a loss of balance and motor coordination (35) similar to spinocerebellar ataxia (16). However, this is likely purely coincidental because josd3 and the DDC8-like genes are not located on the same chromosome as TIMP-2. Similar to the nesting of TIMPs within synapsins and DDC8 within TIMP-2, the close proximity of DDC8-like and josd3 genes may be happenstance, reflecting evolutionary multiplication of genome sections, or it may represent a specific organizational or regulatory relationship which is yet to be identified.
Several questions remain unresolved in this study (e.g., what 5’UTR is used by DDC8 in the brain, what is the correct DDC8 translation start site, does alternative TIMP-2 exon usage exist, does the 78 kDa protein represent a splice between DDC8 and TIMP-2, and can DDC8 regulate TIMP-2 expression?). Future studies need to be undertaken to answer these questions. The most critical goal is to identify the element(s) responsible for the generation of TIMP-2 mRNA transcripts in the knockout mouse and determine the phenotypic effect of their deletion.
Supplementary Material
Acknowledgments
We thank Dr. Paul D. Soloway for kindly providing TIMP-2-/- mice, Dr. Leonor Perez-Martinez for performing the fibroblast cultures and reverse zymography, Benjamin Bakondi for the initial DDC8/TIMP-2 PCR, and Garrett D. Langlois for assistance with cryostat sectioning. We also thank Kim Pruitt from NCBI for help in determining the gene structure of the DDC8-like proteins, Dr. Jeff Bond for assistance with the bioinformatics analysis, and Dr. Greg Gilmartin for critical review of the manuscript. This work was supported by NS045225 co-funded by NINDS and NCRR (DMJ). Genotyping was performed using equipment provided at the University of Vermont Neuroscience Center of Biomedical Research Excellence Molecular Core facility (NIH NCRR P20 RR16435). Sequence analysis was performed in the Vermont Cancer Center DNA Analysis Facility and was supported, in part, by grant P30 CA22435 from the NCI. The phylogenetic and sequence analyses were performed at the Bioinformatics core of the Vermont Genetics Network supported by grant P20 RR16462 from the INBRE Program of the NCRR.
References
- 1.Ahuja S, Ahuja P, Caffe AR, Ekstrom P, Abrahamson M, van Veen T. rd1 mouse retina shows imbalance in cellular distribution and levels of TIMP-1/MMP-9, TIMP-2/MMP-2 and sulfated glycosaminoglycans. Ophthalmic Res. 2006;38:125–136. doi: 10.1159/000090533. [DOI] [PubMed] [Google Scholar]
- 2.Akahane T, Akahane M, Shah A, Connor CM, Thorgeirsson UP. TIMP-1 inhibits microvascular endothelial cell migration by MMP-dependent and MMP-independent mechanisms. Exp Cell Res. 2004;310:158–167. doi: 10.1016/j.yexcr.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker AH, Edwards DR, Murphy G. Metalloproteinase inhibitors: biological actions and therapeutic opportunities. J Cell Sci. 2002;115:3719–3727. doi: 10.1242/jcs.00063. [DOI] [PubMed] [Google Scholar]
- 5.Beaudoing E, Gautheret D. Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. Genome Res. 2001;11:1520–1526. doi: 10.1101/gr.190501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler GS, Butler MJ, Atkinson SJ, Will H, Tamura T, van Westrum SS, Crabbe T, Clements J, d’Ortho M-P, Murphy G. The TIMP2 membrane type 1 metalloproteinase “receptor” regulates the concentration and efficient activation of progelatinase A. JBiolChem. 1998;273:871–880. doi: 10.1074/jbc.273.2.871. [DOI] [PubMed] [Google Scholar]
- 7.Castillo-Davis CI, Mekhedov SL, Hartl DL, Koonin EV, Kondrashov FA. Selection for short introns in highly expressed genes. Nat Genet. 2002;31:415–418. doi: 10.1038/ng940. [DOI] [PubMed] [Google Scholar]
- 8.Catalano RD, Vlad M, Kennedy RC. Differential display to identify and isolate novel genes expressed during spermatogenesis. Mol Hum Reprod. 1997;3:215–221. doi: 10.1093/molehr/3.3.215. [DOI] [PubMed] [Google Scholar]
- 9.Caterina JJ, Yamada S, Caterina NCM, Longenecker G, Holmbäck K, Shi J, Yermovsky AE, Engler JA, Birkedal-Hansen H. Inactivating mutation of the mouse Tissue Inhibitor of Metalloproteinases-2 (Timp-2) gene alters proMMP-2 activation. J Biol Chem. 2000;275:26416–26422. doi: 10.1074/jbc.M001271200. [DOI] [PubMed] [Google Scholar]
- 10.Cawston TE, Curry VA, Clark IM, Hazleman BL. Identification of a new metalloproteinase inhibitor that forms tight-binding complexes with collagenase. Biochem J. 1990;269:183–187. doi: 10.1042/bj2690183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5298. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 12.Curry VA, Clark IM, Bigg H, Cawston TE. Large inhibitor of metalloproteinases (LIMP) contains tissue inhibitor of metalloproteinases (TIMP)-2 bound to 72,000-M(r) progelatinase. Biochem J. 1992;285:143–147. doi: 10.1042/bj2850143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danielson P, Forss-Peter S, Brow MA, Calavetta L, Douglass J, Milner RJ, Sutcliffe JG. p1B15: A cDNA clone of the rat mRNA encoding cyclophilin. DNA. 1988;7:261–267. doi: 10.1089/dna.1988.7.261. [DOI] [PubMed] [Google Scholar]
- 14.de Moor CH, Meijer H, Lissenden S. Mechanisms of translational control by the 3’ UTR in development and differentiation. Semin Cell Dev Biol. 2005;16:49–58. doi: 10.1016/j.semcdb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Derry JM, Barnard PJ. Physical linkage of the A-raf-1, properdin, synapsin I, and TIMP genes on the human and mouse X chromosomes. Genomics. 1992;12:632–638. doi: 10.1016/0888-7543(92)90286-2. [DOI] [PubMed] [Google Scholar]
- 16.Duenas AM, Goold R, Giunti P. Molecular pathogenesis of spinocerebellar ataxias. Brain. 2006;129:1357–1370. doi: 10.1093/brain/awl081. [DOI] [PubMed] [Google Scholar]
- 17.Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, Ainscough R, Almeida JP, Babbage A, Bagguley C, Bailey J, Barlow K, Bates KN, Beasley O, Bird CP, Blakey S, Bridgeman AM, Buck D, Burgess J, Burrill WD, O’Brien KP, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- 18.Fager N, Jaworski DM. Differential spatial distribution and temporal regulation of tissue inhibitor of metalloproteinase mRNA expression during rat central nervous system development. Mech Dev. 2000;98:105–109. doi: 10.1016/s0925-4773(00)00437-8. [DOI] [PubMed] [Google Scholar]
- 19.Farrell CM, Lukens LN. Naturally occurring antisense transcripts are present in chick embryo chondrocytes simultaneously with the down-regulation of the a1 (I) collagen gene. J Biol Chem. 1995;270:3400–3408. doi: 10.1074/jbc.270.7.3400. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein J. Phylogeny Inference Package, version 3.6. Department of Genome Sciences; University of Washington, Seattle: 2005. Distributed by author. [Google Scholar]
- 21.Fernandez CA, Butterfield C, Jackson G, Moses MA. Structural and functional uncoupling of the enzymatic and angiogenic inhibitory activities of tissue inhibitor of metalloproteinase-2 (TIMP-2): loop 6 is a novel angiogenesis inhibitor. J Biol Chem. 2003;278:40989–40995. doi: 10.1074/jbc.M306176200. [DOI] [PubMed] [Google Scholar]
- 22.Gariano RF, Hu D, Helms J. Expression of angiogenesis-related genes during retinal development. Gene Expr Patterns. 2006;6:187–192. doi: 10.1016/j.modgep.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Grima J, Calcagno K, Cheng CY. Purification, cDNA cloning, and developmental changes in the steady-state mRNA level of rat testicular tissue inhibitor of metalloproteases-2 (TIMP-2) J Androl. 1996;17:263–275. [PubMed] [Google Scholar]
- 24.Habib AA, Gulcher JR, Hognason T, Zheng L, Stefansson K. The OMgp gene, a second growth suppressor within the NF1 gene. Oncogene. 1998;16:1525–1531. doi: 10.1038/sj.onc.1201683. [DOI] [PubMed] [Google Scholar]
- 25.Hammani K, Blakis A, Morsette D, Bowcock AM, Schmutte C, Henriet P, DeClerck YA. Structure and characterization of the human tissue inhibitor of metalloproteinase-2 gene. J Biol Chem. 1996;271:25498–25505. doi: 10.1074/jbc.271.41.25498. [DOI] [PubMed] [Google Scholar]
- 26.Hayashi K, Fujio Y, Kato I, Sobue K. Structural and functional relationships between h- and l-caldesmons. J Biol Chem. 1991;266:355–361. [PubMed] [Google Scholar]
- 27.Henikoff S, Keene MA, Fechtel K, Fristrom JW. Gene within a gene: nested Drosophila genes encode unrelated proteins on opposite DNA strands. Cell. 1986;44:33–42. doi: 10.1016/0092-8674(86)90482-4. [DOI] [PubMed] [Google Scholar]
- 28.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatiocs. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 29.Iguchi N, Tobias JW, Hecht NB. Expression profiling reveals meiotic male germ cell mRNAs that are translationally up- and down-regulated. Proc Natl Acad Sci USA. 2006;103:7712–7717. doi: 10.1073/pnas.0510999103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaworski DM. Differential regulation of tissue inhibitor of metalloproteinases mRNA expression in response to intracranial injury. Glia. 2000;30:199–208. doi: 10.1002/(sici)1098-1136(200004)30:2<199::aid-glia9>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 31.Jaworski DM, Boone J, Caterina J, Soloway P, Falls WA. Prepulse inhibition and fear-potentiated startle are altered in tissue inhibitor of metalloproteinase-2 (TIMP-2) knockout mice. Brain Res. 2005;1051:81–89. doi: 10.1016/j.brainres.2005.05.057. [DOI] [PubMed] [Google Scholar]
- 32.Jaworski DM, Kelly GM, Hockfield S. BEHAB, a new member of the proteoglycan tandem repeat family of hyaluronan-binding proteins that is restricted to brain. J Cell Biol. 1994;125:495–509. doi: 10.1083/jcb.125.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaworski DM, Kelly GM, Hockfield S. Intracranial injury acutely induces the expression of the secreted isoform of the CNS-specific hyaluronan-binding protein BEHAB/brevican. Exp Neurol. 1999;157:327–337. doi: 10.1006/exnr.1999.7062. [DOI] [PubMed] [Google Scholar]
- 34.Jaworski DM, Pérez-Martínez L. Tissue inhibitor of metalloproteinase-2 (TIMP-2) expression is regulated by multiple neural differentiation signals. J Neurochem. 2006;98:234–247. doi: 10.1111/j.1471-4159.2006.03855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaworski DM, Soloway P, Caterina J, Falls WA. Tissue inhibitor of metalloproteinase-2 (TIMP-2)-deficient mice display motor deficits. J Neurobiol. 2006;66:82–94. doi: 10.1002/neu.20205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984;12:857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee SC, Kim IG, Marekov LN, O’Keefe EJ, Parry DA, Steinert PM. The structure of human trichohyalin. J Biol Chem. 1993;268:12164–12176. [PubMed] [Google Scholar]
- 38.Lenoir D, Battenberg E, Kiel M, Bloom FE, Milner RJ. The brain-specific gene 1B236 is expressed postnatally in the developing rat brain. JNeurosci. 1986;6:522–530. doi: 10.1523/JNEUROSCI.06-02-00522.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levinson B, Kenwrick S, Lakich D, Hammonds GJ, Gitschier J. A transcribed gene in an intron of the human factor VIII gene. Genomics. 1990;7:1–11. doi: 10.1016/0888-7543(90)90512-s. [DOI] [PubMed] [Google Scholar]
- 40.Lluri G, Langlois GD, McClellan B, Soloway PD, Jaworski DM. Tissue inhibitor of metalloproteinase-2 (TIMP-2) regulates neuromuscular junction development via a β1 integrin-mediated mechanism. J Neurobiol. 2006 doi: 10.1002/neu.20315. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Longin J, Guillaumot P, Chauvin MA, Morera AM, Le Magueresse-Battistoni B. MT1-MMP in rat testicular development and the control of Sertoli cell proMMP-2 activation. J Cell Sci. 2001;114:2125–2134. doi: 10.1242/jcs.114.11.2125. [DOI] [PubMed] [Google Scholar]
- 42.Longin J, Le Magueresse-Battistoni B. Evidence that MMP-2 and TIMP-2 are at play in the FSH-induced changes in Sertoli cells. Mol Cell Endocrinol. 2002;189:25–35. doi: 10.1016/s0303-7207(01)00756-0. [DOI] [PubMed] [Google Scholar]
- 43.Lovelock JD, Baker AH, Gao F, Dong JF, Bergeron AL, McPheat W, Sivasubramanian N, Mann DL. Heterogeneous effects of tissue inhibitors of matrix metalloproteinases on cardiac fibroblasts. Am J Physiol Heart Circ Physiol. 2005;288:H461–468. doi: 10.1152/ajpheart.00402.2004. [DOI] [PubMed] [Google Scholar]
- 44.Mao Y, Senic-Matuglia F, Di Fiore PP, Polo S, Hodsdon ME, De Camilli P. Deubiquitinating function of ataxin-3: insights from the solution structure of the Josephin domain. Proc Natl Acad Sci USA. 2005;102:12700–12705. doi: 10.1073/pnas.0506344102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Modrek B, Resch A, Grasso C, Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mruk DD, Cheng CY. Sertolin is a novel gene marker of cell-cell interactions in the rat testis. J Biol Chem. 1999;274:27056–27068. doi: 10.1074/jbc.274.38.27056. [DOI] [PubMed] [Google Scholar]
- 47.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 48.Nuttall RK, Sampieri CL, Pennington CJ, Gill SE, Schultz GA, Edwards DR. Expression analysis of the entire MMP and TIMP gene families during mouse tissue development. FEBS Lett. 2004;563:129–134. doi: 10.1016/S0014-5793(04)00281-9. [DOI] [PubMed] [Google Scholar]
- 49.Pohar N, Godenschwege TA, Buchner E. Invertebrate tissue inhibitor of metalloproteinase: structure and nested gene organization within the synapsin locus is conserved from Drosophila to human. Genomics. 1999;57:293–296. doi: 10.1006/geno.1999.5776. [DOI] [PubMed] [Google Scholar]
- 50.Proudfoot NJ, Brownlee GG. 3’ non-coding region sequences in eukaryotic messenger RNA. Nature. 1976;263:211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- 51.Pérez-Martínez L, Jaworski DM. Tissue inhibitor of metalloproteinase-2 promotes neuronal differentiation by acting as an anti-mitogenic signal. J Neurosci. 2005;25:4917–4929. doi: 10.1523/JNEUROSCI.5066-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seo DW, Li H, Guedez L, Wingfield PT, Diaz T, Salloum R, Wei BY, Stetler-Stevenson WG. TIMP-2 mediated inhibition of angiogenesis: an MMP-independent mechanism. Cell. 2003;114:171–180. doi: 10.1016/s0092-8674(03)00551-8. [DOI] [PubMed] [Google Scholar]
- 53.Sheets MD, Ogg SC, Wickens MP. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 1990;18:5799–5805. doi: 10.1093/nar/18.19.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silverman TA, Noguchi M, Safer B. Role of sequences within the first intron in the regulation of expression of eukaryotic initiation factor 2a. J Biol Chem. 1992;267:9738–9742. [PubMed] [Google Scholar]
- 55.Siu MK, Cheng CY. Interactions of proteases, protease inhibitors, and the β1 integrin/laminin γ3 protein complex in the regulation of ectoplasmic specialization dynamics in the rat testis. Biol Reprod. 2004;70:945–964. doi: 10.1095/biolreprod.103.023606. [DOI] [PubMed] [Google Scholar]
- 56.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, Patapoutian A, Hampton GM, Schultz PG, Hogenesch JB. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Z, Juttermann R, Soloway PD. TIMP-2 is required for efficient activation of proMMP-2 in vivo. J Biol Chem. 2000;275:26411–26415. doi: 10.1074/jbc.M001270200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wutz A, Smrzka OW, Schweifer N, Schellander K, Wagner EF, Barlow DP. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature. 1997;389:745–749. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]
- 59.Young DA, Phillips BW, Lundy C, Nuttall RK, Hogan A, Schultz GA, Leco KJ, Clark IM, Edwards DR. Identification of an initiator-like element essential for the expression of the tissue inhibitor of metalloproteinases-4 (Timp-4) gene. Biochem J. 2002;364:89–99. doi: 10.1042/bj3640089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu P, Ma D, Xu M. Nested genes in the human genome. Genomics. 2005;86:414–422. doi: 10.1016/j.ygeno.2005.06.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


