Abstract
Background
Pine wilt disease (PWD) caused by the pinewood nematode Bursaphelenchus xylophilus is one of the most serious forest diseases in the world. The role of B. xylophilus-associated bacteria in PWD and their interaction with the nematode, have recently been under substantial investigation. Several studies report a potential contribution of the bacteria for the PWD development, either as a helper to enhance the pathogenicity of the nematode or as a pathogenic agent expressing interesting traits related to lifestyle host-adaptation.
Results
We investigated the nematode-bacteria interaction under a severe oxidative stress (OS) condition using a pro-oxidant hydrogen peroxide and explored the adhesion ability of these bacteria to the cuticle surface of the nematodes. Our results clearly demonstrated a beneficial effect of the Serratia spp. (isolates LCN-4, LCN-16 and PWN-146) to B. xylophilus under the OS condition. Serratia spp. was found to be extremely OS-resistant, and promote survival of B. xylophilus and down-regulate two B. xylophilus catalase genes (Bxy-ctl-1 and Bxy-ctl-2). In addition, we show that the virulent isolate (Ka4) of B. xylophilus survives better than the avirulent (C14-5) isolate under the OS condition. The bacterial effect was transverse for both B. xylophilus isolates. We could not observe a strong and specific adhesion of these bacteria on the B. xylophilus cuticle surface.
Conclusions
We report, for the first time, that B. xylophilus associated bacteria may assist the nematode opportunistically in the disease, and that a virulent B. xylophilus isolate displayed a higher tolerance towards the OS conditions than an avirulent isolate.
Keywords: Bursaphelenchus xylophilus, Catalase, Oxidative stress, Pine wilt disease
Background
Pine wilt disease (PWD), caused by the migratory plant parasitic nematode Bursaphelenchus xylophilus (the pinewood nematode, PWN), is one of the most serious global forest diseases [1]. B. xylophilus and its vector beetles are listed as worldwide quarantine pests [2,3]. Under laboratory conditions, B. xylophilus has been reported to be sufficient for PWD development [4]. However, because of their ubiquitous existence in the PWD environments, some bacteria have also been thought to be involved in the disease development. For example, some B. xylophilus-associated bacteria are beneficial to B. xylophilus growth and reproduction [5], and others have been suggested or demonstrated to produce interesting bacterial traits that may contribute to B. xylophilus pathogenic potential and, ultimately, to PWD development [6-9].
Plant oxidative burst comprises in the production of reactive oxygen species (ROS) as a result of the interaction between plant cell receptors and pathogen-elicitors immediately after pathogen invasion [10-12]. Being relatively stable and permeable to the cell membrane, hydrogen peroxide (H2O2) is the most predominant ROS in plant oxidative burst [13,14]. In addition, H2O2 leads to the formation of the radical OH, which is extremely reactive and for which there is no scavenging system [15]. H2O2 was found to be transversal in different plant-pathogen systems, being a fundamental diffusible signal in plant resistance to pathogens (i.e. involved in cell-wall reinforcement or induction of defence-related genes in healthy adjacent tissues) [16].
Plant pathogens have evolved different evasion features to protect themselves against plant oxidative stress (OS) [17]. Bacterial defences include production of extracellular polysaccharides (EPS) coating and periplasmic catalases, and cytoplasmic catalase and superoxide dismutases (SOD) to counteract ROS before and after entering bacterial cells [18,19]. Other factors are related to the production of polyesters, poly-(3-hydroxyalkanoate) (PHA) also known as protective molecules [18], or phytotoxins (i.e. coronatine in Pseudomonas syringae) that are able to manipulate or down regulate plant-defences for bacteria successful establishment [20]. In plant- or animal-parasitic nematodes, antioxidant enzymes have been found to be the important weapons against oxidative stress of their plant- or animal-hosts [21]. Molinari [22] detected different antioxidant enzymes in Meloidogyne incognita, M. hapla, Globodera rostochiensis, G. pallida, Heterodera schachtii, H. carotae, and Xiphinema index and their relationship with life stages. Robertson et al. [23] and Jones et al. [24] have studied, the role of host ROS breakdown by peroxiredoxins (PXN) and glutathione peroxidases (GXP) in G. rostochiensis, respectively. Bellafiore et al. [25] reported the presence of several detoxifying enzymes, in particular glutathione S-transferases (GST), in the secretome of M. incognita as means of controlling the global oxidative status and potential nematode virulence.
Pinus thunbergii[26] and P. pinaster[27] are the B. xylophilus-susceptible pine trees found in Japan and Europe (Portugal) to respectively, respond to a strong oxidative burst in the earliest stages of nematode invasion. Most likely, B. xylophilus has developed an efficient antioxidant system to diminish the deleterious effects of oxidative burst in their invasion and colonization [28], as well as other plant parasitic nematodes [29]. Our study aimed to understand the tolerance of the B. xylophilus-associated bacteria under the OS condition and its interaction with the nematode. Also, we explored the bacterial attachment to the nematode cuticle for dissemination purposes.
Results
B. xylophilus and associated Serratia in stress conditions
Firstly, we examined the OS resistance of three B. xylophilus-associated bacteria (Serratia spp. LCN-4, LCN-16 and PWN-146) [8] and a control E. coli strain, OP50. Compared to the control strain, all three Serratia spp. were shown to comparably tolerate different concentrations of H2O2 ranging from 15 to 40 mM, (Figure 1). Moreover, the three isolates were able to survive up to 100 mM H2O2, (data not shown).
Figure 1.
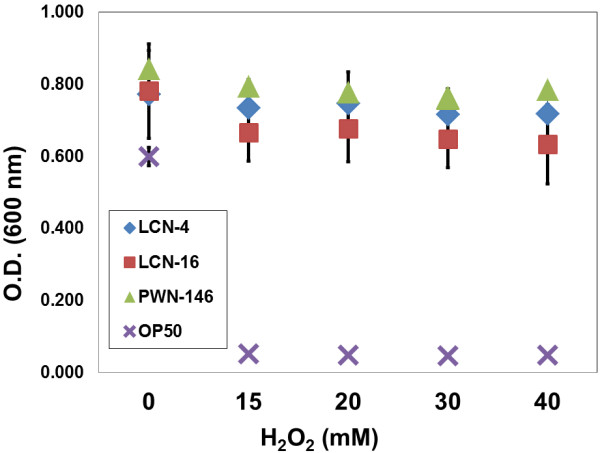
Three Bursaphelenchus xylophilus-associated bacteria (Serratia spp. LCN-4, LCN-16 and PWN-146) have strong resistance against the oxidative stress by H2O2. Average ± S.E. are from 3 biological replications composed of 3 technical replicate. There is no significant difference within the Serratia spp., but between Serratia spp. and E. coli OP50 (p < 0.05). Control E. coli OP50 could not survive under strong oxidative stress conditions.
Next, we examined the OS resistance of the two B. xylophilus isolates with and without bacteria (Figure 2). In the absence of bacteria (surface-sterilized nematode), B. xylophilus isolates Ka4 (virulent) are more resistant to OS than the C14-5 (avirulent) (p < 0.05). At 15 and 20 mM, B. xylophilus Ka4 presented 73% less mortality than B. xylophilus C14-5. The difference of their mortality was 32% and 12% in 30 and 40 mM H2O2. To test the effect of bacteria on B. xylophilus survival under these conditions, we treated B. xylophilus with Serratia spp. (isolates LCN-4, LCN-16 and PWN-146) and E. coli OP50 for 1 h, washed away bacteria by excess and measured their OS resistance. In the presence of Serratia spp., both Ka4 and C14-5 were able to survive at all H2O2 concentrations tested, with mortality rates lower than 10%. Similar to the previous results of Serratia spp. under the OS conditions (Figure 1), there was no significant difference between the OS treatments of three bacterial isolates in association with B. xylophilus (p > 0.05). Serratia spp. PWN-146 was selected for further experiments. In the presence of the E. coli OP50, the mortality of the avirulent C14-5 isolate was higher and similar to that in nematode alone conditions (p > 0.05). For virulent Ka4, association with the control strain lead to similar results at 40 mM H2O2. At 30 mM H2O2, there was a significant difference with Ka4 alone (68%), with control OP50 (38%), although under the same oxidant conditions, survival of E. coli OP50 was significantly reduced (Figure 2). Under the other H2O2 conditions, treatment Ka4 in association with OP50 was almost similar to Ka4 alone. In non-stress conditions, all treatments were statistically equal, indicating that the bacteria used were not harmful to the nematodes.
Figure 2.
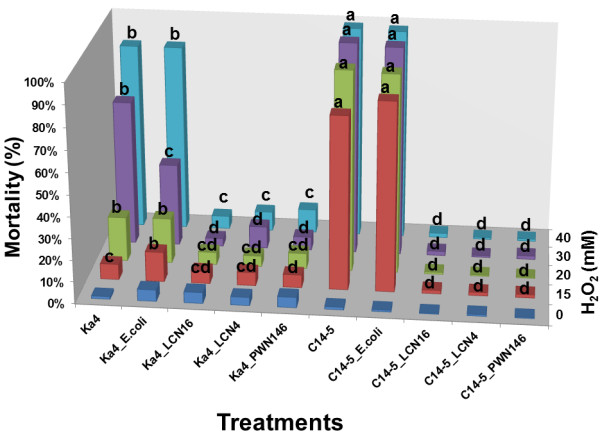
Mortality percentages of Bursaphelenchus xylophilus virulent (Ka4) and avirulent (C14-5), with and without bacteria (Serratia spp. LCN-4, LCN-16 and PWN-146, and E. coli OP50) under oxidative stress conditions. For each H2O2 condition, columns with different letters reflect statistical differences (p < 0.05). In control conditions (0 mM H2O2), no statistical differences were found between all treatments.
Observation of the nematode-bacteria association
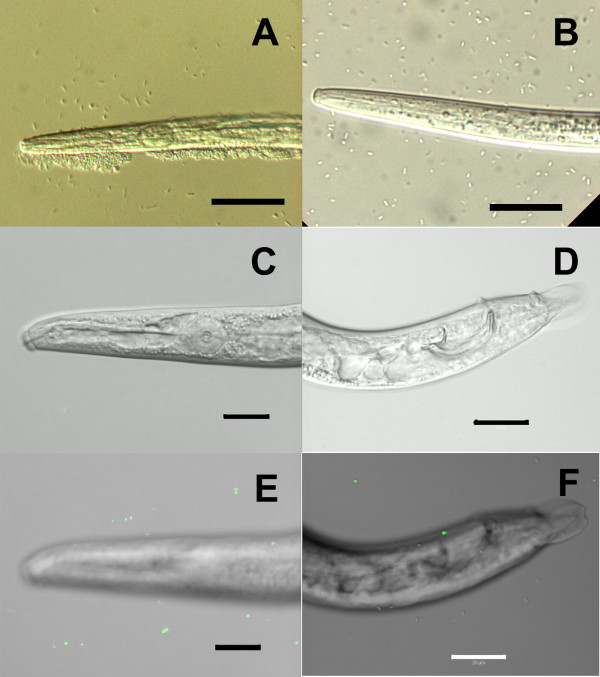
After 1 h contact between B. xylophilus and its associated bacteria, microcolonies were found along the nematode body (Figure 3A). After extensive washing, bacteria were still present in lesser amounts, and scarcely attached to the nematode cuticle (Figure 3B). In order to test if the bacterial adhesion to the nematode became stronger, and if the nematode could uptake bacteria into its body, we performed co-culturing of the nematodes with the GFP-labelled bacteria on the same plate for 24 h. Successful GFP-labelling of B. xylophilus-associated bacteria was only obtained for Serratia spp. LCN-4 and Serratia spp. LCN-16. Serratia spp. PWN-146 were previously found to be multi-drug resistant to the antibiotics available to select for GFP-containing minitransposons [8]. After 24 h contact with Serratia spp. LCN-16, the density of nematode-attached bacteria was sparse (Figure 3C-F), and also no GFP fluorescence signal was detected in the nematode (Figure 3C-F). Taken together, the adhesion of these bacteria to the nematode surface and organs seems to be weak and non-specific.
Figure 3.
Observation of Serratia sp. LCN-16 in association with Bursaphelenchus xylophilus after 1 h and 24 h contact. (A, B) Differential interference contrast (DIC) microscope images of B. xylophilus, treated by 1 h contact of bacteria before (A) and after (B) washing with sterile DW. (C-F) DIC and fluorescence-merged images of B. xylophilus, treated by 24 h contact of bacteria and washed with sterile DW. The images of the head (C) and tail (D) region were captured in a single focal plane . Serial-section images were acquired and stacked, showing surfaces of the head (E) and tail (F) region. Scale bars, (A), (B), 30 μm; (C)-(F), 20 μm.
Relative gene expression of Bxy-ctl-1 and Bxy-ctl-2
Using the C. elegans catalases (Ce-CTL-1, Ce-CTL-2 and Ce-CTL-3) as the search queries, only two catalases were predicted in the B. xylophilus genome, Bxy-CTL-1 (BUX.s00579.159) and Bxy-CTL-2 (BUX.s01109.377) [30]. Both cDNA sequences presented open reading frames (ORF). The longest ORF for Bxy-ctl-1 encodes a 513 aa protein with the molecular weight of ~59kDa. The cDNA to sequence of Bxy-ctl-2 encoded a 512 aa protein with the molecular weight of ~ 55 kDa. Both Bxy-CTL-1 and Bxy-CTL-2 were predicted as non-secretory peroxisomal proteins. However, according to Shinya et al.[31], Bxy-CTL-2 was secreted after pine wood extract stimulation. BlastP search for both catalases retrieved very similar orthologous catalases (62-64% maximum identity and e-value 0.0) from different species of Caenorhabditis and other animal parasitic nematodes, suggesting the catalases are conserved among the phylum Nematoda (Additional file 1: Figure S1 and Additional file 2: Figure S2).
The relative gene expression of catalase genes of B. xylophilus Ka4 and C14-5 with or without Serratia spp. PWN-146 was studied under stress conditions (Figure 4). After 24 h exposure to 15 mM H2O2, the expression levels of Bxy-ctl-1 and Bxy-ctl-2 genes in the B. xylophilus Ka4 and C14-5 were measured (Figure 4A and 4B). While virulent Ka4 catalases (Bxy-ctl-1 and Bxy-ctl-2) were significantly (p < 0.05 and p < 0.01, respectively) up-regulated by nearly 2-2.5-fold compared to the non-stress condition (Figure 4A) The expression of Bxy-ctl-1 in the avirulent C14-5 was unchanged and the expression of Bxy-ctl-2 was slightly reduced (p < 0.05) (Figure 4B). These results seem to support the observations denoted in Figure 2. In the presence of the associated bacteria Serratia spp. PWN-146, the relative expression of Ka4 Bxy-ctl-1 was highly suppressed (p < 0.01), nearly 0.5-fold less than under non-stress conditions. Under the same conditions, Ka4 expression of Bxy-ctl-2 was not affected. The expression levels of both catalases in the avirulent C14-5 showed no significant induction or suppression. In the presence of control strain E. coli OP50, the expression level of Bxy-ctl-1 in the Ka4 was induced four-fold under stress conditions, and Bxy-ctl-2 expression level remained unchanged under non-stress conditions. Similar result was obtained for C14-5, in which E. coli OP50 induced 5 times more Bxy-ctl-1 expression under stress conditions, explaining the results obtained in Figure 2. The expression levels of Bxy-ctl-2 were also induced (p < 0.05), nearly 1.5-fold (Figure 4B).
Figure 4.
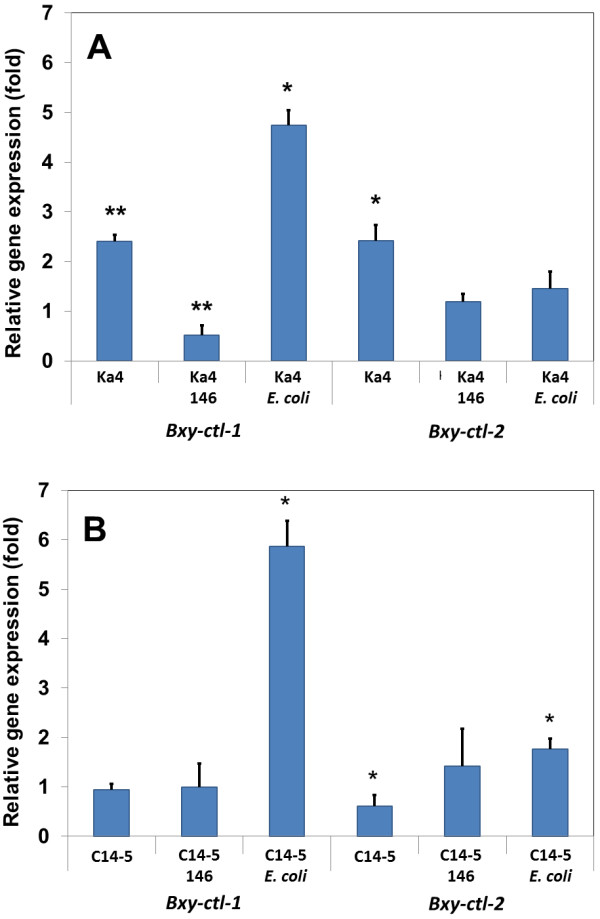
Relative gene expression changes of Bxy-ctl-1 and Bxy-ctl-2 H2O2 treatment for 24 h.Bursaphelenchus xylophilus Ka4 (virulent) and C14-5 (avirulent) with and without bacteria (A and B) (Serratia spp. PWN-146 and E. coli OP50). *p < 0.05; **p < 0.01, compared to a normalized value of 1.00 for control nematode without H2O2.
Discussion
Tolerance to host-mediated OS is an essential characteristic of plant-associated organisms. In this study, we tested if B. xylophilus-associated bacteria could tolerate prolonged oxidative stress conditions with or without the nematode, in an attempt to understand their behaviour in the oxidative burst conditions of the host tree in the early stages of PWD.
Plant-associated bacteria, beneficial or pathogenic, have developed efficient detoxification systems to cope with host-ROS [19,32]. This study demonstrates that Serratia spp. LCN-4 and LCN-16 (S. proteamaculans, 100% identity) and PWN-146 (S. marcescens, 99% identity) associated to B. xylophilus could sustain growth independently, and promote the survival of the nematodes under strong OS conditions. This result indicates, again, a beneficial and a potential helper effect to B. xylophilus. Vicente et al. [8] reported that some B. xylophilus-associated bacteria displayed plant pathogenic traits potentially related with PWD symptoms and B. xylophilus pathogenicity such as high cellulolytic activity, biofilm formation, EPS exudation and siderophores production. In fact, some of these traits are used by environmental bacteria as protectants against OS (i.e. EPS or biofilm). More recently, Chen et al. [9] showed that B. xylophilus-associated bacteria could support the nematode in the degradation of host xenobiotics. Based on our results, we suggest that B. xylophilus-associated Serratia spp. has evolved an elaborate detoxifying system to express several antioxidant enzymes to cope with H2O2-mediated OS.
In this study, we measured the transcript levels of two catalases in B. xylophilus in the presence of H2O2. PWN catalase genes presented a high protein similarity with other nematode catalases, evidencing the conserved nature of this enzyme [21]. Cap’n’collar (Cnc) transcription factors are broadly conserved in eukaryotes except for plant and fungi [33]. C. elegans CnC transcription factor SKN-1 regulates cellular differentiation of the pharynx and intestine during early embryogenesis, and also controls expression of many antioxidative and detoxification enzymes such as CTLs, GPXs and GSTs [34,35]. In C. elegans four pathways (p38 MAPK, Insulin/IGF-1 pathway, WDR-23 ubiquitin pathway, and GSK-3 pathway) are known to control SKN-1 activity and the genomic structures of these pathways are fully conserved in B. xylophilus[30]. Bacterial effect was transversal to virulent and avirulent B. xylophilus. Relative gene expression of catalase genes in B. xylophilus show that without bacteria, the basal expression of the both non-secreted Bxy-ctl-1 and secreted Bxy-ctl-2 genes in the virulent isolate Ka4, were higher than the avirulent C14-5 by 2.5-fold, which explains their differential tolerance level to H2O2. Further investigation on the detoxifying system of B. xylophilus is imperative. When interacting with Serratia spp. PWN-146, both virulent and avirulent B. xylophilus catalase levels decreased to levels comparable to non-stress condition, which is also in agreement with mortality test results (Figure 2).
The correlation between virulence and the ability to cope with oxidative stress has been found in the plant parasitic nematode Melodoigyne incognita[15,29]. Virulent B. xylophilus Ka4 was more tolerant to H2O2 than the avirulent B. xylophilus strain C14-5. Hirao et al. [26] reported that the susceptible P. thunbergii reacts to PWN invasion with a strong oxidative burst, which implies that virulent B. xylophilus must possess an efficient antioxidant system to cope with these conditions. Shinya et al.[36] suggested that potential ROS scavengers GST and GAPDH are localized on the surface coat of B. xylophilus. Li et al. [37] proposed 2-cysteine peroxiredoxin on the nematode cuticle of B. xylophilus, as another antioxidant agent in opposing oxidative burst. Recently, 12 anti-oxidant proteins were identified in the B. xylophilus secretome after plant extract stimuli, namely peroxiredoxin, catalase, glutathione peroxidase, nucleoredoxin-like protein, SOD, and thioredoxin [32]. In this context, it is essential to further investigate the possible relation between virulence of B. xylophilus and its tolerance to oxidative stress, which was shown for the first time in this study.
To explore the bacterial interaction with B. xylophilus, we have studied bacteria attachment to the nematode cuticle, an important characteristic that, to our knowledge, has not been reported before. In our experiments, the associated-bacteria were not found to strongly attach to the cuticle of B. xylophilus. After 24 h contact with a high concentration of GFP-tagged Serratia spp. LCN-16, only a few bacteria could be detected on PWN cuticle (Figure 3). Shinya et al.[36] have shown the presence of few bacteria on the nematode cuticle even after vigorous washing by scanning electron microscopy (SEM). B. xylophilus associated bacteria are reported to be carried on the nematode’s surface, and in average 290 were counted on the cuticle of PWN isolated from diseased trees [7]. If bacteria are not attached to the nematode surface, how can they be transported by B. xylophilus from and into a pine tree? A possible explanation could be that these bacteria are transported within the nematode [38]. However, the possible point of entry in B. xylophilus, the stylet opening, is very small compared with the bacteria size.
Serratia is an environmental ubiquitous Gram-negative bacterium, mostly free-living with an opportunistic lifestyle but also a pathogenic agent to plants, insects and humans [39]. In the plant context, S. proteamaculans is usually identified as an endophytic bacterium living in poplar trees [40], characterized by colonizing in harmony and even expresses PGP (plant growth promoting) traits to promote host health. S. marcescens is also reported as a pathogenic agent of curcubit yellow vine disease [41]. In both cases, these Serratia species are well adapted to the host plant (or tree) conditions, either as endophytes or pathogens, and are able to evade or suppress plant defences [42]. We could not ascertain a strong attachment of associated-Serratia and B. xylophilus. It is not unlike that these bacteria may assist the nematode in an opportunistic or facultative way, and that perhaps these bacteria could be indeed host endophytes. This hypothesis can explain why diverse bacterial communities are associated to B. xylophilus, and why they possess such interesting traits and host-related lifestyle. Moreover, it can help to explain the contrasting results obtained in pathogenicity tests conducted previously [8]. In this scenario, these multi-species consortia that present some in vitro plant-pathogenic traits that could aid the nematode inside the tree and contribute to PWD development as well [3], they could be asymptomatic endophytes that can become pathogenic as soon as the host tree is weakened [42]. Nevertheless, the host-colonizing ability of these bacteria requires further investigation.
Conclusions
This is the first report to show that B. xylophilus-associated Serratia species can assist the nematode survival under prolonged OS conditions, revealing a possible synergism between both organisms. This beneficial effect of bacteria towards nematode resilience to OS has significant influence on PWD development. This disease is presently occurring in a variety of countries/climate zones, and might be influenced by much more various biotic and abiotic factors than previously thought.
Methods
Bursaphelenchus xylophilus isolates and culturing
Two B. xylophilus isolates, virulent Ka4 and avirulent C14-5 [43], were used in this study. Nematodes were cultured in Botrytis cinerea grown on autoclaved barley seeds at 25°C. Prior to the experiments, nematodes were extracted overnight using the Baermann funnel technique at 25°C. Nematodes were washed three times with sterilized distilled water (DW), pelleted in-between by centrifugation at 1,000 rpm during 10 min, surface cleaned with 3% L-lactic acid during 30 s, and finally washed with DW [44]. Mix-staged nematodes were used in all experiments.
Bacteria strains and culturing
Bacterial strains and isolates used in this study are listed in Table 1. All bacteria were grown and maintained in LB plates at 28°C or 37°C (in the case of E. coli strains) for routine use, and in 30% (w/v) glycerol at -80°C for long-term storage. The antibiotics used in this study were: gentamycin (10–30 μg/ml), kanamycin (50 μg/ml) and ampicillin (100 μg/ml).
Table 1.
Bacterial strains and isolates used in this study
| Bacteria used in this study | Genotype or Phenotype | Source or Reference |
|---|---|---|
|
Serratia spp. LCN-4 (100% Max. Identity: S. proteamaculans) |
AmpR; EryR |
Bacterium associated with long lab culturing PWN. [8,45] |
|
Serratia spp. LCN-16 (100% Max. Identity: S. proteamaculans) |
AmpR; EryR |
Bacterium associated with PWN freshly isolated from wilting tree. [8,45] |
|
Serratia spp. PWN-146 (99% Max. Identity: S. marcescens) |
AmpR; EryR; KmR;TetR; RifR |
|
|
Escherichia coli OP50 |
|
WormBase http://www.wormbase.org |
|
mini – TN7 tagging system |
|
|
|
Escherichia coli S17::λpir (deliver) |
pBK-miniTN7-gfp2; GmR; KmR |
[46-48] |
| Escherichia coli SM10::λpir (helper) | pUX-BF13, AmpR | [47] |
R – resistance; Amp – ampicillin; Ery – erytromycin; Km – kanamycin; Tet – tetracyclin; Gm – gentamycin; Rif – rifampycin.
Tagging bacteria with GFP
Bacteria (LCN-16 and PWN-146) were labelled with GFP with the vectors pBK-miniTn7-ΩGm, pBK-miniTn7-gfp2, pUX-BF13 by triparental mating method as described previously [49]. Briefly, 100 μl overnight cultures of bacteria strains and isolates (LB with appropriate antibiotics) were mixed, in 1:1:1 proportion (SM17, SM10, and LCN-16 or PWN-146), pipetted onto a 13-mm cellulose acetate filter membrane and placed on non-selective LB medium. Plates were incubated overnight at 28°C. In the following day, filters were placed into a sterile microcentrifuge tube containing 0.2 ml of 0.9% NaCl and vortexed for cell suspension. Aliquots of 100 μl of each suspension was plated onto LB with selective antibiotic (30 μg/ml gentamycin) and overnight incubated at 28°C.
Bacteria association to nematode
Bacteria isolates (LCN-4, LCN-16 and PWN-146) and strain (OP50) were grown overnight in LB broth at 28°C or 37°C, pelleted at 10,000 rpm for 5 min, washed twice with sterilized DW, and adjusted OD600 for 1.00 (± 107-108 CFU/ml). Two approaches were used to associate bacteria with B. xylophilus. The first approach consisted in the observation of 1 h contact bacterial association with B. xylophilus, before and after washing nematodes for the oxidative stress tests. Firstly, nematodes were surface sterilized and the concentration adjusted to 150 nematodes per 50 μl of sterilized DW. Nematode-bacteria association was performed by 1 h contact between surface cleaned nematodes and 1 ml of bacterial suspension (concentrations were adjusted as described above) and in accordance to Han et al. [50] procedure. Afterwards, bacteria suspension was removed by pelleting the nematodes at low speed rotation (800 × g, 5 min), and then hand-picked with a nematode picker (steel wire) and transferred into a drop of sodium azide (1 M) on the centre of the agar pad [51], covered and sealed with a silicon grease-rimmed coverslip for viewing by Nomarski DIC optics.
The second approach consisted in co-culturing of B. xylophilus Ka4 with GFP-tagged bacteria (LCN-16-GFP; PWN-146-GFP) in 0.1% MEA plate seeded with B. cinerea. Firstly, nematodes were cultured on the 0.1% MEA plate for three-days, and then 500 μl of bacterial suspension (concentrations were adjusted as described above) were added and co-cultured for 24 h at 28°C. Afterwards nematodes were extracted, washed and mounted on the agar pad as described above. GFP-tagged bacteria were observed with a ZEISS Axiovert 200 microscope equipped with a confocal laser-scanning module.
Oxidative stress tolerance tests
To test bacteria tolerance to the oxidative agent, 100 μl of freshly prepared H2O2 and 10 μl of bacteria (concentrations were adjusted as described above) were placed into each well of a 96-well plate and at a total volume of 110 μl per well. Final concentrations of H2O2 were 0, 15, 20, 30 and 40 mM. After 24 h, the plates were read in a multi-spectrophotometer (Viento, Dainippon Sumitomo Pharma, Japan) at OD600. For each B. xylophilus associated bacteria and control E. coli. Three independent biological replicates with three technical replicas per experiment were used for each treatment.
To test nematode and bacteria association in H2O2 oxidative conditions, first, nematodes were surface sterilized and the concentration was adjusted to 150 nematodes per 50 μl of sterilized DW, and performed 1 h nematode-bacteria association as described above. After 1 h contact with bacteria, nematodes were washed and re-suspended in sterilized DW. A 96-well plate was prepared as follows: each well received 50 μl of different H2O2 concentrations (prepared previously in double) and 50 μl of each treatment (nematode-bacteria association, nematode alone and control (DW). Three independent biological replicates with three technical replicas per experiment were used for each treatment. . Mortality of nematodes was scored after 24 h. Nematodes were considered dead, if no movements were observed after mechanical stimulation.
Gene expression analysis of B. xylophilus catalases
Catalase (CTL) was selected as the antioxidant enzyme to infer gene expression differences toward the effect of H2O2 in the nematode-bacteria association. The amino acid sequences of C. elegans catalases (Ce-CTL-1, -2, -3) were obtained from WormBase (http://www.wormbase.org/), and used as templates for a TBLASTN search in the B. xylophilus Ka4 genome. The retrieved best matches were predicted as Bxy-CTL-1 and Bxy-CTL-2 of B. xylophilus. Predictions about general topology, domain/family, and active sites conserved were made using online tools available at Expasy WWW pages (http://www.expasy.org/tools/).
Gene expression of Bxy-ctl-1 and Bxy-ctl-2 were analysed by qRT-PCR using SYBR® green assay. Total RNA was extracted from 24 h-stressed nematodes (treatments: nematodes alone and nematode-bacteria association) in 15 mM H2O2, using CellAmp Direct RNA Prep Kit for RT-PCR (Real time) (Takara Bio Inc., Japan) and following manufacturer’s instructions. The concentration and quality was measured using NanoVue plus spectophotometer (GE Healthcare Life Sciences, USA). Total RNA (adjusted for concentration of 50 ng/μl) was reverse transcribed using Oligo dT primer and PrimeScript RT enzyme from PrimeScript™ RT reagent Kit (Perfect Real Time) (Takara Bio Inc., Japan). Quantitative RT-PCR was performed using CFX96™ Real-Time (Bio-Rad), and SYBR Premix Ex TaqTM II (Tli RnaseH Plus) kit (Takara Bio Inc., Japan). The housekeeping actin gene Bxy-act-1 was used as an internal control gene for calculation of relative expression levels of each antioxidant gene [52]. Primers were designed using Prime 3 software [53] and tested for specificity prior to qPCR. The primers used for Bxy-act-1, Bxy-ctl-1 and Bxy-ctl-2 genes amplification were listed in Additional file 3: Table S1. Two independent biological replicates with two technical replicas per experiment were used for each qPCR test. No template controls (NTC) were prepared for each qPCR run. Thermal cycling conditions were: initial denaturation at 95°C for 30 sec; 39 cycles of denaturation at 95°C for 5 sec, annealing and extension at 60°C for 30°C; followed by the melting curve. A single peak at the melting temperature of the PCR-product confirmed primer specificity.
Relative gene expression of each gene were analysed using ΔΔCT Method [52]. The data were analysed with Ct values in normal and stress conditions and using the following equation: ΔΔCT = (CT,Target ‒ CT,actin)normal ‒ (CT, Target ‒ CT,Actin)stress. The fold change in Bxy-ctl-1 and Bxy-ctl-2 was normalized to Bxy-act-1 and relative to the expression at normal conditions, was calculated for each sample using the equation above.
Statistical analysis
Statistical analysis was performed using SPSS 11.5. Data represent the mean ± standard error (SE). Statistical significance was inferred by one-way ANOVA and post hoc multi-comparison Duncan test.
Competing interests
The authors declare that they have no competing interests.
Author contributions
Conceived and designed the experiments: CSLV, KH. Performed the experiments: CSLV, YI, KH. Analyzed the data: CSLV, YI, KH. Wrote the paper: CSLV, MM, KH. All authors read and approved the final manuscript.
Supplementary Material
Alignment of deduced amino acid sequences from catalase 1 (CTL-1) with the top matches in database. Residues conserved are highlighted in dark grey and marked by an asterisk. Bursaphelenchus xylophilus CTL-1; Caenorhabditis elegans CTL-1 (CAA74393.1); C. remanei CTL-3 (XP_003102502.1); C. briggsae hypothetical protein (XP_002631620.1); Ditylenchus destructor CTL (AFJ15102.1).
Alignment of deduced amino acid sequences from catalase 2 (CTL2) with the top matches in database. Residues conserved are highlighted in dark grey and marked by an asterisk. Bursaphelenchus xylophilus CTL-2; Caenorhabditis elegans CTL-3 (NP741058.1); C. brenneri CTL-2 (EGT40792.1); Haemonchus contortus CTL (AAT28330.1); Ditylenchus destructor CTL (AFJ15102.1).
Primers used in this study.
Contributor Information
Cláudia S L Vicente, Email: cvicente@uevora.pt.
Yoriko Ikuyo, Email: chironokiba@gmail.com.
Manuel Mota, Email: mmota@uevora.pt.
Koichi Hasegawa, Email: koichihasegawa@isc.chubu.ac.jp.
Acknowledgements
B. xylophilus strains Ka4 and C14-5 were provided by FFPRI, Tsukuba Japan. The plasmids pBK-miniTn7-ΩGm, pBK-miniTn7-gfp2, pUX-BF13 were provided by Professor Søren Molin, Danmarks Tekniske Universitet. This work was supported by the Chubu Science and Technology Center fellowship to Cláudia Sofia Leite Vicente; Heiwa Nakajima Foundation, international joint research grant; the European Project REPHRAME - Development of improved methods for detection, control and eradication of pine wood nematode in support of EU Plant Health policy, European Union Seventh Framework Programme FP7-KBBE-2010-4; Portuguese national scientific Portuguese national scientific agency FCT (Fundação para a Ciência e Tecnologia)/project PTDC/BIA-MIC/3768/2012 (FCOMP-01-0124-FEDER-028368); and FEDER Funds through the Operational Programme for Competitiveness Factors - COMPETE and National Funds through FCT - Foundation for Science and Technology under the Strategic Project PEst-C/AGR/UI0115/2011.
References
- Mamiya Y. Pathology of the pine wilt disease caused by Bursaphelenchus xylophilus. Annu Rev Plant Physiol Plant Mol Biol. 1983;13:201–220. doi: 10.1146/annurev.py.21.090183.001221. [DOI] [PubMed] [Google Scholar]
- Mota MM, Vieira P. Pine Wilt Disease: A Worldwide Threat to Forest Ecosystems. Netherlands: Springer; 2008. [Google Scholar]
- Zhao B, Futai K, Sutherland JR, Takeuchi Y. Pine Wilt Disease. Kato Bunmeisha: Springer; 2008. [Google Scholar]
- Zhu LH, Ye J, Negi S, Xu XL, Wang ZL. Pathogenicity of aseptic Bursaphelenchus xylophilus. PLoS One. 2012;13:e38095. doi: 10.1371/journal.pone.0038095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao BG, Liu Y, Lin F. Effects of bacteria associated with pine wood nematode (Bursaphelenchus xylophilus) on development and egg production of the nematode. J Phytopathol. 2007;13:26–30. doi: 10.1111/j.1439-0434.2006.01188.x. [DOI] [Google Scholar]
- Kawazu K, Zhang H, Yamashita H, Kanzaki H. Relationship between the pathogenecity of pine wood nematode, Bursaphelenchus xylophilus, and phenylacetic acid production. Biosci Biotech Biochem. 1996;13:1413–1415. doi: 10.1271/bbb.60.1413. [DOI] [PubMed] [Google Scholar]
- Zhao BGZ, Ang HLW, An SFH, An ZMH. Distribution and pathogenicity of bacteria species carried by Bursaphelenchus xylophilus in China. Nematology. 2003;13:899–906. doi: 10.1163/156854103773040817. [DOI] [Google Scholar]
- Vicente CSL, Nascimento F, Espada M, Barbosa P, Mota M, Glick BR, Oliveira S. Characterization of bacteria associated with pinewood nematode Bursaphelenchus xylophilus. PloS one. 2012;13:e46661. doi: 10.1371/journal.pone.0046661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng XY, Tian XL, Wang YS, Lin RM, Mao ZC, Chen N, Xie BY. Metagenomic analysis of the pinewood nematode microbiome reveals a symbiotic relationship critical for xenobiotics degradation. Scientific reports. 1869;13:3. doi: 10.1038/srep01869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehdy MC. Active oxygen species in plant defense against pathogens. Plant Physiol. 1994;13:467–472. doi: 10.1104/pp.105.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolwell GP, Butt VS, Davies DR, Zimmerlin A. The origin of the oxidative burst in plants. Free radical Res. 1995;13:517–532. doi: 10.3109/10715769509065273. [DOI] [PubMed] [Google Scholar]
- Torres MA, Jones JDG, Dangl JL. Reactive oxygen species signaling in response to pathogens. Plant Physiol. 2006;13:373–378. doi: 10.1104/pp.106.079467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA. ROS in biotic interactions. Physiol plantarum. 2010;13:414–429. doi: 10.1111/j.1399-3054.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- Quan LJ, Zhang B, Shi WS, Li HY. Hydrogen peroxide in plants: a versatile molecule of the reactive oxygen species network. J Integrative Plant Biol. 2008;13:2–18. doi: 10.1111/j.1744-7909.2007.00599.x. [DOI] [PubMed] [Google Scholar]
- Dubreuil G, Deleury E, Magliano M, Jaouannet M, Abad P, Rosso MN. Peroxiredoxins from the plant parasitic root-knot nematode, Meloidogyne incognita, are required for successful development within the host. Int J Parasitol. 2011;13:385–396. doi: 10.1016/j.ijpara.2010.10.008. [DOI] [PubMed] [Google Scholar]
- Lamb C, Dixon R. The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol. 1997;13:251–275. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- Shetty NP, Jørgensen HJL, Jensen JD, Collinge DB, Shetty HS. Roles of reactive oxygen species in interactions between plants and pathogens. Eur J Plant Pathol. 2008;13:267–280. doi: 10.1007/s10658-008-9302-5. [DOI] [Google Scholar]
- Fones H, Preston GM. Reactive oxygen and oxidative stress tolerance in plant pathogenic Pseudomonas. FEMS microbiology letters. 2012;13:1–8. doi: 10.1111/j.1574-6968.2011.02449.x. [DOI] [PubMed] [Google Scholar]
- Guo M, Block A, Bryan CD, Becker DF, Alfano JR. Pseudomonas syringae catalases are collectively required for plant pathogenesis. J Bacteriol. 2012;13:5054–5064. doi: 10.1128/JB.00999-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiga Y, Uppalapati SR, Ishiga T, Elavarthi S, Martin B, Bender CL. Involvement of coronatine-inducible reactive oxygen species in bacterial speck disease of tomato. Plant Signaling and Behavior. 2009;13:237–239. doi: 10.4161/psb.4.3.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkle-Dührsen K, Kampkötter A. Antioxidant enzyme families in parasitic nematodes. Mol Biochem Parasitol. 2001;13:129–142. doi: 10.1016/S0166-6851(01)00252-3. [DOI] [PubMed] [Google Scholar]
- Molinari S. Changes of catalase and SOD activities in the early response of tomato to Meloidogyne attack. Nematol Mediterr. 1998;13:167–172. [Google Scholar]
- Robertson L, Robertson WM, Sobczak M, Helder J, Tetaud E, Ariyanayagam MR, Ferguson MAJ, Fairlamb A, Jones JT. Cloning, expression and functional characterisation of a peroxiredoxin from the potato cyst nematode Globodera rostochiensis. Mol Biochem Parasitol. 2000;13:41–49. doi: 10.1016/S0166-6851(00)00295-4. [DOI] [PubMed] [Google Scholar]
- Jones J, Reavy B, Smant G, Prior A. Glutathione peroxidases of the potato cyst nematode Globodera Rostochiensis. Gene. 2004;13:47–54. doi: 10.1016/j.gene.2003.09.051. [DOI] [PubMed] [Google Scholar]
- Bellafiore S, Shen Z, Rosso MN, Abad P, Shih P, Briggs SP. Direct identification of the Meloidogyne incognita secretome reveals proteins with host cell reprogramming potential. PLoS pathogens. 2008;13:e1000192. doi: 10.1371/journal.ppat.1000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao T, Fukatsu E, Watanabe A. Characterization of resistance to pine wood nematode infection in Pinus thunbergii using suppression subtractive hybridization. BMC plant biology. 2012;13:13. doi: 10.1186/1471-2229-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos CSS, Vascocelos MW. Identification of genes differentially expressed in Pinus pinaster and Pinus pinea after infection with pine wood nematode. Eur J Plant Pathol. 2012;13:407–418. doi: 10.1007/s10658-011-9886-z. [DOI] [Google Scholar]
- Shinya R, Morisaka H, Takeuchi Y, Futai K, Ueda M. Making headway in understanding pine wilt disease: What do we perceive in the postgenomic era? J Biosci Bioeng. 2013;13:1–8. doi: 10.1016/j.jbiosc.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Molinari S. Antioxidant enzymes in (a)virulent populations of root-knot nematodes. Nematology. 2009;13:689–697. doi: 10.1163/156854108X399317. [DOI] [Google Scholar]
- Kikuchi T, Cotton JA, Dalzell JJ, Hasegawa K, Kanzaki N, McVeigh P, Takanashi T, Tsai IJ, Aseffa SA, Cock PJA, Otto TD, Hunt M, Reid AJ, Sanchez-Flores A, Tsuchihara K, Yokoi T, Larsson MC, Miwa J, Maule AG, Sahashi N, Jones JT, Berriman M. Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathog. 2011;13:e1002219. doi: 10.1371/journal.ppat.1002219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya R, Morisaka H, Kikuchi T, Takeuchi Y, Ueda M, Futai K. Secretome analysis of pine wood nematode Bursaphelenchus xylophilus reveals the tangled roots of parasitism and its potential for molecular mimicry. PloS one. 2013;13:e67377. doi: 10.1371/journal.pone.0067377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamet A, Sigaud S, Van de Sype G, Puppo A, Hérouart D. Expression of the bacterial catalase genes during Sinorhizobium meliloti-Medicago sativa symbiosis and their crucial role during the infection process. Mol Plant Microbe In. 2003;13:217–225. doi: 10.1094/MPMI.2003.16.3.217. [DOI] [PubMed] [Google Scholar]
- Sykiotis GP, Bohmann D. Stress-activated Cap’n’collar transcription factors in 43. aging and human disease. Sci Signal. 2010;13:re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman B, Eaton BA, Priess JR. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell. 1992;13:1061–1075. doi: 10.1016/0092-8674(92)90078-Q. [DOI] [PubMed] [Google Scholar]
- Park SK, Tedesco PM, Johnson TE. Oxidative stress and longevity in Caenorhabditis elegans as mediated by SKN-1. Aging Cell. 2009;13:258–269. doi: 10.1111/j.1474-9726.2009.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya R, Morisaka H, Takeuchi Y, Ueda M, Futai K. Comparison of the surface coat proteins of the pine wood nematode appeared during host pine infection and in vitro culture by a proteomic approach. Phytopathol. 2010;13:1289–1297. doi: 10.1094/PHYTO-04-10-0109. [DOI] [PubMed] [Google Scholar]
- Li Z, Liu X, Chu Y, Wang Y, Zhang Q, Zhou X. Cloning and characterization of a 2-Cys peroxiredoxin in the pine wood nematode, Bursaphelenchus xylophilus, a putative genetic factor facilitating the infestation. Int J Biol Scie. 2011;13:823–836. doi: 10.7150/ijbs.7.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XQ, Yuan WM, Tian XJ, Fan B, Fang X, Ye JR, Ding XL. Specific and functional diversity of endophytic bacteria from pine wood nematode Bursaphelenchus xylophilus with different virulence. Int J Biol Sci. 2013;13:34–44. doi: 10.7150/ijbs.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimont F, Grimont PAD. The Genus Serratia. Proc Natl Acad Sci USA. 2006;13:219–244. [Google Scholar]
- Taghavi S, Garafola C, Monchy S, Newman L, Hoffman A, Weyens N, Barac T, Vangronsveld J, Lelie D. Genome survey and characterization of endophytic bacteria exhibiting a beneficial effect on growth and development of poplar trees. App Environ Microbiol. 2009;13:748–757. doi: 10.1128/AEM.02239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Weyant R, Steigerwalt AG, White LA, Melcher U, Bruton BD, Pair SD, Mitchell FL, Fletcher J. Genotyping of Serratia marcescens strains associated with cucurbit yellow vine disease by repetitive elements-based polymerase chain reaction and DNA-DNA hybridization. Phytopathol. 2003;13:1240–1246. doi: 10.1094/PHYTO.2003.93.10.1240. [DOI] [PubMed] [Google Scholar]
- Schulz B, Boyle C. The endophytic continuum. Mycol Res. 2005;13:661–686. doi: 10.1017/S095375620500273X. [DOI] [PubMed] [Google Scholar]
- Aikawa T, Kikuchi T. Estimation of virulence of Bursaphelenchus xylophilus (Nematoda: Aphelenchoididae) based on its reproductive ability. Nematology. 2007;13:371–377. doi: 10.1163/156854107781352007. [DOI] [Google Scholar]
- Takemoto S. Population ecology of Bursaphelenchus xylophilus. Kato Bunmeisha: Springer; 2008. (Pine Wilt Disease). Volume 108. [Google Scholar]
- Vicente CSL, Nascimento F, Espada M, Mota M, Oliveira S. Bacteria associated with the pinewood nematode Bursaphelenchus xylophilus collected in Portugal. A van Leeuw J Microb. 2011;13(100):477–481. doi: 10.1007/s10482-011-9602-1. [DOI] [PubMed] [Google Scholar]
- Kock B, Jensen LE, Nybroe O. A panel of Tn7-based vectors for insertion of the gfp marker gene or for delivery of cloned DNA into Gram-negative bacteria at a neutral chromosomal site. J Microbiol Meth. 2001;13:187–195. doi: 10.1016/S0167-7012(01)00246-9. [DOI] [PubMed] [Google Scholar]
- Højberg O, Schnider U, Winterler HV, Sørensen J, Haas D. Oxygen-sensing reporter strain of Pseudomonas fluorescens for monitoring the distribution of low-oxygen habitats in soil. App Environ Microbiol. 1999;13:4085–4093. doi: 10.1128/aem.65.9.4085-4093.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Lies DP, Fu H, Roberts GP. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of Gram-negative bacteria. Gene. 1991;13:167–168. doi: 10.1016/0378-1119(91)90604-A. [DOI] [PubMed] [Google Scholar]
- Lambertsen L, Sternberg C, Molin S. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ Microbiol. 2004;13:726–732. doi: 10.1111/j.1462-2920.2004.00605.x. [DOI] [PubMed] [Google Scholar]
- Han ZM, Hong YD, Zhao BG. A study on pathogenicity of bacteria carried by pine wood nematodes. J Phytopathol. 2003;13:683–689. doi: 10.1046/j.1439-0434.2003.00790.x. [DOI] [Google Scholar]
- Shaham S. Methods in cell biology. The C. elegans Research Community, WormBook; 2006. (WormBook). doi/10.1895/wormbook.1.7.1, http://www.wormbook.org. [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;13:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletzki H. Primer3 on the www for general users and for biologist programmers. Humana PressKrawetz S, Misener S; 2000. pp. 365–386. (Bioinformatics Methods and Protocols: Methods in Molecular Biology Totowa). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of deduced amino acid sequences from catalase 1 (CTL-1) with the top matches in database. Residues conserved are highlighted in dark grey and marked by an asterisk. Bursaphelenchus xylophilus CTL-1; Caenorhabditis elegans CTL-1 (CAA74393.1); C. remanei CTL-3 (XP_003102502.1); C. briggsae hypothetical protein (XP_002631620.1); Ditylenchus destructor CTL (AFJ15102.1).
Alignment of deduced amino acid sequences from catalase 2 (CTL2) with the top matches in database. Residues conserved are highlighted in dark grey and marked by an asterisk. Bursaphelenchus xylophilus CTL-2; Caenorhabditis elegans CTL-3 (NP741058.1); C. brenneri CTL-2 (EGT40792.1); Haemonchus contortus CTL (AAT28330.1); Ditylenchus destructor CTL (AFJ15102.1).
Primers used in this study.