Abstract
The serpin family comprises a structurally similar, yet functionally diverse, set of proteins. Named originally for their function as serine proteinase inhibitors, many of its members are not inhibitors but rather chaperones, involved in storage, transport, and other roles. Serpins are found in genomes of all kingdoms, with 36 human protein-coding genes and five pseudogenes. The mouse has 60 Serpin functional genes, many of which are orthologous to human SERPIN genes and some of which have expanded into multiple paralogous genes. Serpins are found in tissues throughout the body; whereas most are extracellular, there is a class of intracellular serpins. Serpins appear to have roles in inflammation, immune function, tumorigenesis, blood clotting, dementia, and cancer metastasis. Further characterization of these proteins will likely reveal potential biomarkers and therapeutic targets for disease.
Keywords: Serpins, Serine protease inhibitor, Chaperone, Blood clotting, Thrombolysis, Complement, Cell death, Metastatic cancer
Introduction
Serpins represent the largest and most functionally diverse family of protease inhibitors. The name serpin originates from the first described function of this family, viz., serine proteinase inhibitors. In their native state, serpins exist as monomeric proteins. Most serpin family members inhibit serine proteinases of the chymotrypsin family [1], thereby inhibiting proteolytic cascades. However, some serpins exhibit functions unrelated to inhibition of catalytic activity, such as hormone transport and other mechanisms.
Approximately 1,500 serpin sequences have been identified; they are found in the genomes of all five kingdoms [2]. There are 36 identified human putatively functional protein-coding genes [3]. The serpin superfamily is divided into groups called clades according to their sequence similarity. Clades are classified as A–P, with clades A–I representing human serpins [4].
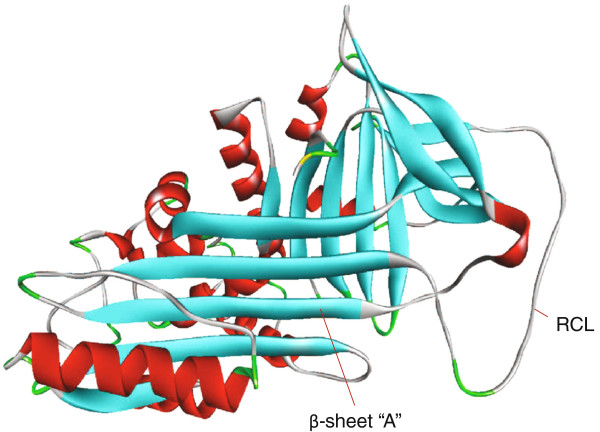
Serpins have well-conserved secondary structures with an exposed reactive center loop (RCL) (Figure 1), which interacts with the protease active site to inhibit protease activity [5]. The ability for serpins to undergo conformational change is crucial for their function, in which serpins act via a suicide substrate inhibitory mechanism [2,4]. Although most serpins selectively inhibit serine proteases, some inhibit cysteine proteases, such as caspases and cathespins; others perform hormone transport and blood pressure regulation [4]. Serpins play important physiological roles in hormone transport, corticosteroid binding, coagulation, and blood pressure regulation.
Figure 1.
Native SERPINA1. Native SERPINA1 with labeled structural elements: β sheet a and reactive center loop (RCL); α helices in red, β sheets in turquoise, turns in green. (Adapted from PDB 1HP7).
Serpin nomenclature
Initially named for tissue location or function (Table 1), a nomenclature committee convened in 1999 with the goal of standardizing serpin gene nomenclature [4]. ‘SERPIN’ was designated as the gene symbol for humans and other species because it is well known and used in the literature and as a keyword [4]. Serpins were not named for activity or function due to the diversity of member structure and tissue distribution. In 2005, proteinase in human gene names was replaced with the term peptidase; however, ‘serpin’ remains the stem because the name was designated prior to this change. The current classification of serpins involves division into clades that are based on phylogenetic relationships (Figure 2). There are 16 clades labeled A–P. Human serpins are represented in the first nine clades (i.e., A–I), with a variety of members being in each clade. Clades are phylogenetically unique and it is important to recognize that no relationships between the clade letters are implied by their order [4]. Some serpins are classified as orphans because they do not group with any other clade. It is likely that they will form clades as new serpins are identified. An example to help illustrate the nomenclature would be α-1-antitrypsin. This was assigned to the first clade, giving it the symbol SERPINA1 with the ‘A’ referencing the clade and the ‘1’ referencing the gene number within the clade [4].
Table 1.
SERPIN aliases and function
| Clade name | Clade | Serpin gene name | Known aliases | Biological function | References |
|---|---|---|---|---|---|
| Alpha 1 proteinase inhibitor antitrypsin |
A |
SERPINA1 |
Alpha 1PI |
Inflammation, complement activation, apoptosis |
17 |
|
SERPINA2 |
|
|
|
||
|
SERPINA3 |
Alpha1 ACT |
Apoptosis, Alzheimer’s disease, prohormone conversion, inflammation and complement activation |
3,16 |
||
|
SERPINA4 |
PI4, KST, KAL |
Kidney function, inflammation, and complement activation |
18 |
||
|
SERPINA5 |
PCI |
Coagulation, inflammation, complement activation, sperm development |
|
||
|
SERPINA6 |
CBG |
Hormone transport |
16 |
||
|
SERPINA7 |
TBG |
Hormone transport |
|
||
|
AGT |
SERPINA8 |
Blood pressure regulation, renal development |
19 |
||
|
SERPINA9 |
Centurin |
B cell development |
3 |
||
|
SERPINA10 |
PZI |
Inhibition of activated factors Z and XI |
3 |
||
|
SERPINA11 |
|
|
|
||
|
SERPINA12 |
Vaspin |
Inhibits kallikrein; unknown role in insulin sensitivity |
20,21 |
||
|
SERPINA13P |
|
|
|
||
|
SERPINB1 |
PI2, LEI, MNEI, EI |
Inflammation, complement activation |
3 |
||
|
SERPINB2 |
PAI2, placental PAI, monocyte ARG serpin, PLANH2 |
Fibrinolysis, elastase inhibitor |
3 |
||
| ov Serpins |
B |
SERPINB3 |
SCCA1, SCC |
Inhibition of cathepsins, tumor promotion |
32 |
|
SERPINB4 |
SCCA2, PI11 |
Inhibition of cathepsins and chymase |
33 |
||
|
SERPINB5 |
Maspin, PI5 |
Tumor cell invasion, angiogenesis |
3 |
||
|
SERPINB6 |
DFNB91, PI6, CAP |
Cathepsin inhibitor |
34 |
||
|
SERPINB7 |
Megsin |
Renal development, mesangial cell proliferation |
35 |
||
|
SERPINB8 |
PI8, CAP2 |
Uncharacterized |
3 |
||
|
SERPINB8P1 |
|
|
|
||
|
SERPINB9 |
PI9, CAP3 |
|
|
||
|
SERPINB10 |
PI10, BOMAPIN |
Hematapoetic and myeloid development |
35 |
||
|
SERPINB11 |
EPIPIN |
Uncharacterized |
36 |
||
|
SERPINB12 |
YUKOPIN |
Trypsin inhibition |
37 |
||
|
SERPINB13 |
PI13, headpin, HUR7, hurpin |
|
|
||
| Antithrombin |
C |
SERPINC1 |
AT3, ATIII, antithrombin3 |
Coagulation, angiogenesis |
38 |
| Heparin cofactor |
D |
SERPIND1 |
HCFII, HCF2, heparin cofactor II, HLS2 |
Coagulation |
40 |
| Nexin/plasminogen activator inhibitor 1 |
E |
SERPINE1 |
PAI, PLANH1 |
Angiogenesis, fibrinolysis |
3 |
|
SERPINE2 |
PI7, GDN, ‘glial-derived nexin 1,’ nexin, PN1, PNI |
Neurotrophic factor |
41 |
||
|
SERPINE3 |
|
|
|
||
| Alpha 2 antiplasmin pigment epithelium derived factor |
F |
SERPINF1 |
PEDF |
Neurotrophic factor, angiogenesis |
16 |
|
SERPINF2 |
Alpha 2AP, PLI, A2AP, AAP, ‘alpha-2-antiplasmin,’ ALPHA‒2‒PI, ‘alpha‒2‒plasmin inhibitor,’ API |
Fibrinolysis |
3 |
||
| C1 inhibitor |
G |
SERPING1 |
C1NH,C1–INH, C1IN, HAE1, HAE2, ‘plasma protease C1 inhibitor’ |
Microbial infection |
42 |
| Heat shock protein |
H |
SERPINH1 |
CBP1, CBP2, collagen, HSP47 |
Chaperone |
43 |
| Neuroserpin |
I |
SERPINI1 |
Neuroserpin, PI12 |
Neurotrophic factor |
46 |
| SERPINI2 | Pancpin, PI14, TSA2004, MEPI | Tumor cell invasion | 47 |
SERPINS are divided into clades and gene name with known aliases and biological function are provided. Alternative name information was determined using HGNC (http://www.genenames.org) and/or MGI (http://www.informatics.jax.org).
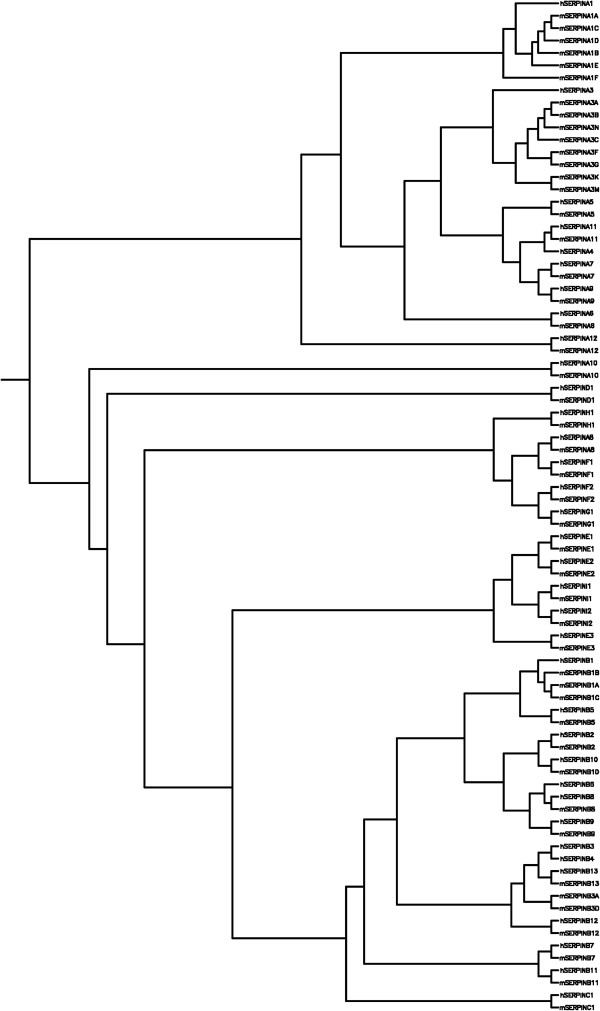
Figure 2.
SERPIN phylogenetic tree. Phylogenetic tree of human and mouse serpin proteins. Protein sequences were aligned using TCOFFEE and analysed using neighbour-joining methods with 10,000 bootstrap replicates in the Phylip package.
Structure function
Serpins have a metastable structure that is required for their function. It consists of a highly conserved secondary structure with three β-sheets (A, B, and C), nine α-helices and a RCL (Figure 1), which serve as bait for target proteases [4,6]. Well-conserved throughout the serpin family, the tertiary structure of scaffold allows for a conformational change critical to protease inhibitor activity [4]. In their native state, serpins exist as monomeric proteins. A serpin molecule consists of a single 330- to 500-amino acid polypeptide chain that has conserved secondary helices and sheets. To inhibit proteolytic activity, the serpin acts as a suicide substrate for the protease [4]. This is accomplished by the RCL of the serpin interacting with the protease's active site [6].
Serpins can exist in several forms, viz., active, latent, cleaved, delta, and polymeric. Each form is defined by the RCL, which is the moiety required for inhibitory activity. The active form (or the native state) has an exposed RCL that allows it to interact with the protease. The RCL forms an exposed extension located above the molecule. Following proteolysis, the amino acid terminus of the RCL inserts into the A β sheet forming a fourth strand. This process is called the ‘stressed (S) to relaxed (R) transition’ [3] used to inhibit proteases, resulting in the cleaved form. The cleaved form is necessary for inhibition of proteases resulting in an irreversible covalent complex with the target protease thus inactivating both the serpin and the target. Some serpins bind cofactors and/or glycosaminoglycans to maximize protease inhibition, which can vastly increase inhibitory potential [7].
The native form of serpins has low thermal stability indicating that it is not the most stable conformation; rather, native serpins are metastable. However, not all serpins undergo this transition. Serpins can transition to the latent form from the active form and back to the active form from the latent form. The latent form does not possess inhibitory activity but it can convert to the active form through denaturation and refolding [4]. Consequently, it can be considered a control mechanism in regulating homeostasis for certain serpins [3]. Alternatively, the latent state caused by a mutation can be pathological [3].
The delta form is an intermediate conformation between latent and native state where the RCL inserts into the A β sheet and one of the helices unwinds and completes hydrogen bonding of the β sheet [3]. Little is known about the function of this conformation; however, it is likely that this favors polymeric or latent conformation transition rather than native. The polymeric form has a loop sheet mechanism whereby the RCL that would be inserted into the same serpin is instead inserted into the A β sheet of another serpin forming a long chain of these molecules [3]. However, this mechanism of polymerization has recently been challenged in favor of that of a domain-swapping model [8]. Serpins are unique in that their native state (active form) is not the most kinetically stable; rather, it is ‘metastable’. By incorporating the RCL into their A β sheet, either by cleavage for inhibition of target protease or spontaneous latency, they become more stable [9]. For an excellent minireview on kinetics of serpins, see Silverman et al. [4].
Evolution
Whereas serpins have highly conserved secondary and tertiary structures upon which they are grouped, they often share little amino acid sequence similarity. They do, however, share a highly conserved core, especially in the shutter domain including Ser56 and Ser53 [10], which is thought to be critical in determining tertiary structure and conformational flexibility.
Due to the numerous, yet distinct, processes regulated by serpins and their widespread functions, serpins offer a unique perspective for protein evolution. Members of the serpin family tend to group phylogenetically by species rather than by function. Therefore, evolution of the serpin family was likely driven by speciation to fill their physiological roles rather than by coevolution with the serine proteases (which group by function) [10]. Numerous serpin genes are also found in clusters on the same chromosomes, reflecting earlier gene-duplication events and potentially indicating a common precursor [11,12]. Interestingly, these genes are functionally divergent, despite their chromosomal proximity [7]. In addition, serpins have distinct patterns of introns and exons. These patterns may contain information regarding phylogenetic signals and be evolutionarily related based on relative intron positioning [13,14].
The distribution of serpins in eukaryotes suggests that they arose early in eukaryotic evolution [1]. Extensive gene clustering indicates that numerous serpins in close proximity on the same chromosome may have arisen as a result of duplications from a common precursor [12]; however, the evolution of these proximal genes gave way to vastly divergent functions.
Intracellular serpins of clade B are ancestral to most extracellular serpins [15,16] and each inhibitory serpin contains a highly conserved hinge region [16] within the RCL. Clade F serpins specifically share ancestry with a sea lamprey serpin. Clade P is specific to plant serpins which form a discrete clade. At the time of divergence between Viridiplantae and fungi/Metazoa groups, there was likely only one serpin gene [16]; however, the ancestral homolog from prokaryote or fungi has not yet been identified [16].
There are eight human serpin pseudogenes listed in Table 2. SERPINA15P has been named in succession for the A clade with the parent gene SERPINA6 according to Ensembl and SERPINE2 is the parent gene for SERPINE4P, again named in sequence of the E clade. There are ten mouse pseudogenes listed (Table 3) which remain uncharacterized.
Table 2.
Human SERPIN genes
| Gene | Entrez gene ID | Chromosomal location | # Exons | # Amino acids | # Alternative transcripts |
|---|---|---|---|---|---|
|
SERPINA1 |
5265 |
14q32.1 |
5 |
418 |
10 |
|
SERPINA2P
|
390502 |
14q32.13 |
NA |
NA |
NA |
|
SERPINA3 |
12 |
14q32.1 |
5 |
423 |
0 |
|
SERPINA4 |
5267 |
14q331–q32.1 |
5 |
427 |
0 |
|
SERPINA5 |
5104 |
14q32.1 |
6 |
406 |
0 |
|
SERPINA6 |
866 |
14q32.1 |
5 |
405 |
0 |
|
SERPINA7 |
6906 |
Xq22.2 |
5 |
415 |
0 |
|
SERPINA7P1 |
100422644 |
Xq22.3 |
NA |
NA |
NA |
|
SERPINA9 |
327657 |
14q32.13 |
6 |
435 |
1 |
|
SERPINA10 |
51156 |
14q32.13 |
5 |
444 |
1 |
|
SERPINA11 |
256394 |
14q32.13 |
5 |
422 |
0 |
|
SERPINA12 |
145264 |
14q32.13 |
6 |
414 |
0 |
|
SERPINA13P
|
388007 |
14q32.13 |
5 |
NA |
NA |
|
SERPINA15P
|
* |
8q23.3 |
NA |
NA |
NA |
|
SERPINB1 |
1992 |
6p25 |
7 |
379 |
0 |
|
SERPINB2 |
5055 |
18q21.3 |
9 |
415 |
1 |
|
SERPINB3 |
6317 |
18q21.3 |
8 |
390 |
0 |
|
SERPINB4 |
6318 |
18q21.3 |
8 |
390 |
0 |
|
SERPINB5 |
5268 |
18q21.3 |
7 |
375 |
0 |
|
SERPINB6 |
5269 |
6p25 |
7 |
376 |
1 |
|
SERPINB7 |
8710 |
18q21.33 |
8 |
380 |
3 |
|
SERPINB8 |
5271 |
18q21.3 |
7 |
242 |
1 |
|
SERPINB8P1
|
11029 |
6p25 |
NA |
NA |
NA |
|
SERPINB9 |
5272 |
6p25 |
7 |
376 |
0 |
|
SERPINB9P
|
221756 |
6p25.2 |
NA |
NA |
NA |
|
SERPINB10 |
5273 |
18q21.3 |
7 |
397 |
0 |
|
SERPINB11 |
89778 |
18q21.33 |
8 |
392 |
0 |
|
SERPINB12 |
89777 |
18q21.33 |
7 |
405 |
0 |
|
SERPINB13 |
5275 |
18q21.3–q22 |
8 |
391 |
0 |
|
SERPINC1 |
462 |
1q23–q25.1 |
7 |
464 |
0 |
|
SERPIND1 |
3053 |
22q11.21 |
4 |
499 |
0 |
|
SERPINE1 |
5054 |
7q21.3–q22 |
9 |
402 |
1 |
|
SERPINE2 |
5070 |
2q33–q35 |
9 |
397 |
2 |
|
SERPINE3 |
647174 |
13q14.3 |
7 |
424 |
0 |
|
SERPINE4P
|
* |
15q12 |
NA |
NA |
NA |
|
SERPINF1 |
5176 |
17p13.3 |
8 |
418 |
0 |
|
SERPINF2 |
5345 |
17p13 |
9 |
491 |
2 |
|
SERPING1 |
710 |
11q12–q13.1 |
8 |
500 |
1 |
|
SERPINH1 |
871 |
11q13.5 |
5 |
418 |
1 |
|
SERPINH1P1
|
158172 |
9p13.3 |
NA |
NA |
NA |
|
SERPINI1 |
5274 |
3q26.1 |
9 |
410 |
1 |
|
SERPINI2 |
5276 |
3q26 |
9 |
405 |
0 |
| AGT | 183 | 1q42.2 | 5 | 485 | 0 |
The human SERPIN family with indicated gene symbol, gene ID, chromosomal location, exon number, alternative transcript number, and number of amino acids.Gene names not italicized are used here simply to underscore that these are pseudogenes for which little or no information is provided. Records are from the National Center for Biotechnology Information (NCBI) gene database.
*Found in Ensembl.
Table 3.
Mouse Serpin genes
| Mouse gene | Entrez gene ID | Chromosomal location | # Exons | # Amino acids | # Alternative transcripts |
|---|---|---|---|---|---|
|
Serpin1a |
20700 |
12; 12 51.0 cM |
7 |
413 |
2 |
|
Serpin1b |
20701 |
12; 12 51.0 cM |
5 |
413 |
1 |
|
Serpin1c |
20702 |
12; 12 51.0 cM |
5 |
413 |
0 |
|
Serpin1d |
20703 |
12; 12 51.0 cM |
5 |
413 |
0 |
|
Serpin1e |
20704 |
12 E; 12 |
5 |
413 |
1 |
|
Serpin1f |
68348 |
12 E; 12 |
6 |
411 |
2 |
|
Serpina2-ps*
|
NA
|
12
|
NA
|
NA
|
NA
|
|
Serpina3d-ps
|
435318 |
12 E; 12
|
NA
|
NA
|
0
|
|
Serpina3e-ps
|
628883
|
12 E; 12
|
NA
|
NA
|
0
|
|
Serpina3a |
74069 |
12 E; 12 |
5 |
422 |
1 |
|
Serpina3b |
271047 |
12 E; 12 |
5 |
420 |
0 |
|
Serpina3c |
16625 |
12 E; 12 |
5 |
417 |
0 |
|
Serpina3f |
238393 |
12 E; 12 |
6 |
445 |
2 |
|
Serpina3g |
20715 |
12 E; 12 |
6 |
440 |
3 |
|
Serpina3h-ps
|
546546
|
12 E; 12
|
5
|
NA
|
1
|
|
Serpina3i |
628900 |
12 E; 12 |
4 |
408 |
1 |
|
Serpina3j |
238393 |
12 E; 12 |
4 |
420 |
1 |
|
Serpina3k |
20714 |
12 E; 12 15.5 cM |
5 |
418 |
0 |
|
Serpina3l-ps
|
628916
|
12 E; 12
|
NA
|
NA
|
0
|
|
Serpina3m |
20717 |
12 E; 12 |
5 |
418 |
0 |
|
Serpina3n |
20716 |
12; 12 F1 |
5 |
418 |
1 |
|
Serpina4-ps
|
321018
|
12 E; 12
|
NA
|
NA
|
NA
|
|
Serpina5 |
268591 |
12 F1 |
5 |
405 |
0 |
|
Serpina6 |
12401 |
12 51.0 cM |
5 |
397 |
1 |
|
Serpina7 |
331535 |
X F1; X |
6 |
426 |
2 |
|
Agt |
11606 |
8 E2; 8 72.81 cM |
5 |
482 |
0 |
|
Serpina9 |
71907 |
12 |
5 |
418 |
1 |
|
Serpina10 |
217847 |
12 |
5 |
448 |
1 |
|
Serpina11 |
380780 |
12 |
5 |
427 |
4 |
|
Serpina12 |
68054 |
12; 12 F1 |
5 |
413 |
0 |
|
Serpina13-ps*
|
NA
|
12
|
NA
|
NA
|
NA
|
|
Serpinb1a |
66222 |
13 A4; 13 12.0 cM |
8 |
379 |
0 |
|
Serpinb1b |
282663 |
13 A3.3, 13 12.6 cM |
7 |
382 |
0 |
|
Serpinb1c |
380839 |
13 A3.3, 12 12.2 cM |
7 |
375 |
1 |
|
Serpinb1-ps
|
282665
|
13 A3.2, 13 13.76 cM
|
NA
|
NA
|
0
|
|
Serpinb2 |
18788 |
1 E2.1; 1 61.1 cM |
11 |
415 |
3 |
|
Serpinb3a |
20248 |
1 E2.1; 1 |
8 |
387 |
0 |
|
Serpinb3b |
383548 |
1 E2.1; 1 |
8 |
387 |
0 |
|
Serpinb3c |
381286 |
1; 1 E1-E2 |
8 |
386 |
0 |
|
Serpinb3d |
394252 |
1E2.1; 1 |
7 |
387 |
0 |
|
Serpinb3-ps1
|
NA
|
1
|
NA
|
NA
|
NA
|
|
Serpinb3-ps2
|
NA
|
1
|
NA
|
NA
|
NA
|
|
Serpinb3-ps3
|
NA
|
1
|
NA
|
NA
|
NA
|
|
Serpinb5 |
20724 |
1 E2.1; 1 |
8 |
375 |
2 |
|
Serpinb6a |
20719 |
1313 A3.3; 13 14.0 cM |
11 |
378 |
16 |
|
Serpinb6b |
20708 |
13 A3.3; 13 13.78 cM |
7 |
377 |
2 |
|
Serpinb6c |
97848 |
13 A3.3; 13 13.99 cM |
7 |
378 |
0 |
|
Serpinb6d |
238568 |
13 A3.3; 13 13.94 cM |
6 |
375 |
0 |
|
Serpinb6e |
435350 |
13 A3.3; 13 13.98 cM |
8 |
429 |
1 |
|
Serpinb7 |
116872 |
1; 1 D |
9 |
380 |
1 |
|
Serpinb8 |
20725 |
1; 1 D |
7 |
374 |
3 |
|
Serpinb8-ps1*
|
NA
|
1
|
NA
|
NA
|
NA
|
|
Serpinb9 |
20723 |
13 A3.3; 13 12.4 cM |
8 |
374 |
0 |
|
Serpinb9b |
20706 |
13 A3.3; 13 13.79 cM |
7 |
377 |
0 |
|
Serpinb9c |
20707 |
13 A3.3; 13 12.82 cM |
8 |
387 |
2 |
|
Serpinb9d |
20726 |
13 A3.3; 13 12.83 cM |
7 |
377 |
0 |
|
Serpinb9e |
20710 |
13 A3.3; 13 12.84 cM |
7 |
377 |
0 |
|
Serpinb9f |
20709 |
13 A3.3; 13 12.86 cM |
7 |
377 |
0 |
|
Serpinb9g |
93806 |
13 A3.3; 13 13.9 cM |
7 |
377 |
0 |
|
Serpinb10 |
241197 |
1 E2. 1; 1 |
8 |
357 |
1 |
|
Serpinb11 |
66957 |
1 E2. 1; 1 |
8 |
388 |
0 |
|
Serpinb12 |
71869 |
1; 1 D |
9 |
423 |
2 |
|
Serpinb13 |
241196 |
1 E2. 1; 1 |
9 |
389 |
1 |
|
Serpinc1 |
11905 |
1H2.1 84.6 cM |
8 |
465 |
0 |
|
Serpind1 |
15160 |
16 A3; 16 9.5 cM |
5 |
478 |
1 |
|
Serpine1 |
18787 |
5 G2; 5 |
9 |
402 |
1 |
|
Serpine2 |
20720 |
1 C4; 1 48.6 cM |
9 |
397 |
1 |
|
Serpine3 |
319433 |
14 D1; 14 |
9 |
401 |
0 |
|
Serpinf1 |
20317 |
11 b5; 11 |
8 |
417 |
7 |
|
Serpinf2 |
18816 |
11 B5; 11 |
11 |
491 |
3 |
|
Serping1 |
12258 |
2 D; 2 |
8 |
504 |
0 |
|
Serpinh1 |
12406 |
7 E2; 7 |
6 |
417 |
1 |
|
Serpini1 |
20713 |
3 E3; 3 |
7 |
410 |
2 |
| Serpini2 | 67931 | 3 E3; 3 | 8 | 405 | 0 |
The mouse serpin family with indicated gene symbol, gene ID, chromosomal location, exon number, alternative transcript number, and number of amino acids. Gene names ending in "-ps" indicate a pseudogene for which little or no information is provided. Records are from the National Center for Biotechnology Information (NCBI) gene database.
*=UCSC genome browser.
Methods
Protein sequences for human serpins were accessed from Uniprot through the HUGO Gene Nomenclature Committee website (http://www.genenames.org). Sequences were retrieved from the National Center for Biotechnology Information (NCBI) gene database (http://www.ncbi.nlm.nih.gov/gene) referenced through the HUGO Gene Nomenclature Committee website (http://www.genenames.org) for humans and MGI website (http://www.informatics.jax.org) for mouse. All sequences were aligned using the most accurate settings of T-Coffee (http://tcoffee.crg.cat/) and phylogenetic trees were constructed using neighbor-joining methods with 1000 replicate bootstrap in PHYLIP 3.69 (http://evolution.genetics.washington.edu/phylip.html) (Figure 2). Expression data were determined using Genecards (http://www.genecards.org) and alternative name information was determined using HGNC (http://www.genenames.org) or MGI (http://www.informatics.jax.org).
Human and mouse serpin isoforms
Clade A
Clade A serpins are classified as antitrypsin-like, extracellular proteins. They are the largest of the eight clades of extracellular serpins. The SERPINA clade has eleven human genes (1, 3–12) and two pseudogenes.
SERPINA1 is an inhibitory serpin formerly known as antitrypsin. It plays a role in the inhibition of neutrophil elastase [3,17].
SERPINA2 was initially classified as a pseudogene; however, recent evidence indicates that it produces an active transcript that encodes a protein located in the endoplasmic reticulum [18]. A study that sequenced SERPINA2 genes across multiple ethnic groups indicated that in addition to active SERPINA2 protein, there is a haplotype characterized by a partial deletion which has patterns suggestive of positive selection for loss-of-function of SERPINA2 protein. They suggest that the partial pseudogenization in humans may indicate an ongoing process of pseudogenization [19].
SERPINA3 is an inhibitory protein formerly known as antichymotrypsin. It inhibits chymotrypsin and cathepsin G [3,16]. This serpin is normally found in blood, liver, kidney, and lung.
SERPINA4 is an inhibitory protein formerly known as kallistatin (PI4), which inhibits kallikrein [20]. It is expressed in blood, liver, kidney, and heart.
SERPINA5, formerly a protein C inhibitor, inhibits active protein C. It is present in blood, kidney and liver.
SERPINA6 was formerly known as corticosteroid-binding globulin. It is a non-inhibitory protein that binds hormones, i.e., cortisol [16].
SERPINA7, formerly thyroxine-binding globulin, is involved in non-inhibitory thyroid hormone transport. It is expressed in blood, kidney, and heart.
SERPINA8 is now referred to as angiotensinogen (AGT), which is a hormone precursor. It has a distinct serpin domain (phylogenetically unrelated to other clade A members in the current analysis) and a distinct, smaller, agt domain. This particular serpin domain appears to be more closely associated with SERPINF and SERPING [21].
SERPINA9 appears to have a role in naïve B cell maintenance. Formerly called centerin, it is expressed in the plasma and liver.
SERPINA10 is an inhibitory protein responsible for inhibition of activated coagulation factors Z and XI [3]. Formerly known as protein Z-dependent proteinase inhibitor, it is expressed in blood and liver.
SERPINA11 is likely a pseudogene and is uncharacterized.
SERPINA12, formerly vaspin, inhibits kallikrein [22] and plays a role in insulin sensitivity [23]. It appears to be expressed in plasma, platelets, liver and heart.
In the mouse (Table 3), Serpina1 has been expanded to include six members, a–f. Serpina3 has been expanded to include nine members, a–c and f–n. The other clade a members are orthologous to human genes. Serpina8, now known as Agt in the mouse, is vital for the development and function of the renin-angiotensin system [24]. It is orthologous to AGT in humans.
Clade B
Clade B consists of intracellular serpins, including ov-serpins, which are ancestral to the extracellular serpins [16]. Members of this subfamily have shorter C and N termini than typical A members and also lack the secretory signal peptide sequence [4]. There are 13 human genes in clade B and one pseudogene. Serpins in clade B are important in inflammation and immune system function as well as mucous production [25]. SERPINB1, B6, B7, and B9 are involved in immune system function with roles in neutrophil and megakaryocyte development [26,27], as well as in the inhibition of the cytotoxic granule protease granzyme B [28]. SERPINB3 and its close homolog B4 are inhibitors that have roles in mucous production [29] and are expressed in epithelial tissues, such as tongue, tonsils, uterus, cervix, and vagina as well as in the upper respiratory tract and thymus [30].
Despite elusive function, SERPINB3 appears to have a role in apoptotic regulation and immunity, which implicates B3 in tumor metastasis and autoimmunity [30]. SERPINB5 has been shown to inhibit metastasis as a tumor suppressor in breast and prostate cancer [30,31]. In addition, multiple serpins in the B clade have been associated with oral squamous cell carcinoma, specifically SERPINB12, SERPINB13, SERPINB4, SERPINB3, SERPINB11, SERPINB7, and SERPINB2 [32]. Less is known about SERPINB10–B13. However, recent evidence points to a role for SERPINB13 in autoimmune diabetes progression and in inflammation [33].
SERPINB1 is an inhibitor of neutrophil elastase. It was formerly called monocyte neutrophil elastase inhibitor and is expressed ubiquitously.
SERPINB2 inhibits PLAU (uPA). It was formerly called plasminogen activator inhibitor 2 (PAI2) and is expressed in blood, kidney, and liver.
SERPINB3 is a cross-class inhibitor of cathepsin L and V [34]. Formerly referred to as squamous cell carcinoma antigen 1, it is expressed in blood, immune cells, kidney, lung, heart, and brain as well as numerous mucosal cells.
SERPINB4 was formerly known as squamous cell carcinoma antigen 2; it was discovered with SERPINB3 [25]. It is a cross-class inhibitor of cathepsin G and chymase [35] and is found in plasma, platelets, kidney, and heart, as well as saliva.
SERPINB5 is a non-inhibitory protein formerly called maspin. It is likely expressed in blood, kidney, liver, lung, as well as saliva.
SERPINB6, formerly called proteinase inhibitor 6 (PI6), is an inhibitor of granule protease, cathepsin G [36]. It is expressed ubiquitously.
SERPINB7 is involved in mesangial cell proliferation [37]. Formerly called megsin, it is expressed in blood and liver.
SERPINB8 is an inhibitory protein. Formerly called proteinase inhibitor 8 (PI8), it is expressed in blood and heart.
SERPINB9 is an inhibitory protein. Formerly called proteinase inhibitor 9 (PI9), it is expressed in blood, liver, lung, and heart.
SERPINB10 is an inhibitory protein involved in hematopoietic and myeloid development [37]. Formerly called bomapin, it expressed in blood and possibly in the brain.
SERPINB11 is a non-inhibitory serpin in human but retains trypsin inhibitory activity in mice [38]. It appears not to exhibit tissue-specific expression; however, it is expressed in HEK cells.
SERPINB12 is a trypsin inhibitor formerly known as yukopin [39]. It is expressed in blood, kidney, liver, heart, and brain.
SERPINB13, formerly known as hurpin, is expressed in blood, kidney, and saliva.
In clade b, mouse Serpinb1 has been expanded to include three members a–c; Serpinb3 as well as Serpinb6 have each expanded to include four members, a–d. In mice, Serpinb4 is not listed; however, it appears that SERPINB3 and SERPINB4 are equally related to Serpinb3a, Serpinb3b, Serpinb3c, and Serpinb3d, despite the initial theory that Serpinb3d is the mouse homolog of human SERPINB3 and Serpinb3c is the mouse homolog of SERPINB4. Serpinb9 has been expanded to seven members and one pseudogene. Interestingly, Serpinb11 is an active proteinase inhibitor, whereas the human ortholog is inactive.
Clade C
Serpin clade C consists of only one serpin member, SERPINC1, more commonly known as antithrombin. SERPINC1 inhibits coagulation factors IX and X [40]. It is expressed in blood, kidney, liver, lung, heart, brain, as well as saliva.
Serpinc1 gene encodes antithrombin and is orthologous to human SERPINC1.
Clade D
Clade D has one serpin member, SERPIND1, which is an extracellular protein also known as heparin cofactor II [41]. It is an inhibitor of thrombin [42] and is expressed in blood, kidney, liver, and heart.
Serpind1 encodes heparin cofactor II and is orthologous to SERPIND1.
Clade E
Clade E has three members, E1, E2, and E3, all of which are extracellular.
SERPINE1, also known as plasminogen activator inhibitor-1 (PAI1), inhibits thrombin. It is expressed in blood, liver, and heart.
SERPINE2 is a glial-derived nexin that is important in recovery of nerve structure and function [43]. It is expressed in blood, liver, kidney, and brain.
Little is known about the function of SERPINE3.
The mouse genes in clade e (Serpine1–3) are orthologous to human SERPINE1–3.
Clade F
There are two members in SERPIN clade F.
SERPINF1 (or pigment epithelium-derived factor (PEDF)) regulates angiogenesis and is an example of a non-inhibitory serpin. It is also thought to be a neurotrophic factor [16], and appears to be expressed in blood, liver, kidney, heart, and possibly lung.
SERPINF2, also known as α-2-antiplasmin, is an inhibitor of fibrinolysis. It is found in blood, kidney, liver, and heart.
Mouse Serpinf1 and f2 genes are orthologous to the human SERPINF1 and SERPINF2 genes, respectively.
Clade G
Clade G consists of one inhibitory serpin.
SERPING1 is a complement I esterase inhibitor [44] formerly called C1 inhibitor. It is expressed in blood, liver, kidney, lung, heart, and brain.
Mouse Serping1 encodes C1 inhibitor and is orthologous to SERPING1.
Clade H
Clade H consists of one member.
SERPINH1, also known as 47-kDa heat shock protein (HSP47), does not act as a proteinase inhibitor, but rather as a chaperone for collagen [45]. It is expressed in blood, liver and heart.
Mouse Serpinh1 encodes HSP47 and is orthologous to SERPINH1. Knockouts of Serpinh1 in mice are lethal [46] and missense mutations are associated with osteogenesis imperfecta [47].
Clade I
Clade I consists of two extracellular proteins. Serpins in clade I include the following.
SERPINI1 is a neuroserpin inhibitor of PLAT (tPA), PLAU (uPA), and plasmin [48]. It is expressed in liver and possibly plasma.
SERPINI2, previously known as pancipin, has an unknown protein target but may be involved in pancreatic dysfunction [49]. It is found in platelets and plasma as well as the heart.
The genes Serpini1 and Serpini2 encode mouse neuroserpin and pancipin, respectively. These are orthologous to SERPINI1 and SERPINI2 in the human.
Clades J–P
Clades j–p represent viral, nematode, horseshoe crab, blood fluke, and plant serpins [16] and will not be described further in this update.
Serpins associated with disease
Serpin polymorphisms have been associated with in many disease states, including blood clotting disorders, emphysema, cirrhosis, and dementia [15,16,50] as well as tumorigenesis and metastasis.
Mutations in SERPINA1 result in a decrease in circulating α-1-antitrypsin which is associated with emphysema and hepatocellular carcinoma [51]. Serpins are implicated in regulation of the cardiovascular system. For example, SERPINA4 depletion is related to renal and cardiovascular injury [52], SERPINA8 variations are integral to the normal function of the renin-angiotensin system and have been found to regulate blood pressure [53], and a SERPINA10 polymorphism was found to increase the risk of venous thromboembolism [54,55]. SERPINA3 deficiency is associated with emphysema [56].
Many SERPINBs are implicated in immune function and dysfunction. In many of these cases, intracellular serpins cause autoimmune antibody production, inflammation, neutropenia, and cancer metastasis [25]. SERPINC1 deficiency has been correlated with autoimmune disease, especially in patients producing antinuclear antibodies, such as those with systemic lupus erythematosus [30]. Interestingly, a SERPINA6 polymorphism has been associated with chronic fatigue syndrome [57], which is thought to be an immune disorder. SERPINA7 deficiency is associated with hyperthyroidism, and high SERPINA12 levels have been associated with insulin resistance [23].
Mutations in SERPINH1, as well as in SERPINF1, are associated with osteogenesis imperfecta [47,58].
Serpins appear to influence protein aggregation. In this respect, SERPINI1 expression has been correlated with dementia [4]. In addition, SERPINA5 accumulation has been identified in plaques in multiple sclerosis [59] and SERPINA3 polymerization may accelerate onset and severity of Alzheimer's disease [30].
Many serpins have been implicated in cancer progression including SERPINBs (on the 18q21 locus) in oral squamous cell carcinoma [25]. Breast and prostate cancer metastases are also closely associated with SERPINB5 [60,61]. In addition, SERPINE1 appears to have a role in tumor progression [62] and metastasis [63]. Further, SERPINI2 may play a possible role in breast and pancreatic cancer metastasis [49]. Adult gliomas have significant associations with SERPINI1 [64], although its role is unknown. In addition, SERPINI1 has also been proposed as one of five biomarkers in hepatocellular carcinoma [65]. Another potential biomarker includes SERPINA9, which has been found to be strongly expressed in B cell lymphomas [66].
Mouse models of human disease
There are numerous mouse models used to study the role of SERPINs in disease. Some examples include knockout of Serpinag3 used in studying T cells in immunology [67], hepatic specific knockout of Serpinc1, which exhibits coagulopathy [68], and Agt knockout to study blood pressure regulation and the renin-angiotensin system where adipocyte-specific knockout of agt caused decreased systolic blood pressure [69]. Serpinb1 knockout mice show neutropenia [70].
Gene variants in SERPINS
A large number of human variants of serpin genes have been found. For example, NCBI's dbSNP database (http://www.ncbi.nlm.nih.gov/snp) has 621 entries for SNPs of SERPINA1 alone (accessed October 2013). In addition, several groups have developed specific databases for individual SERPIN genes. These include databases for SERPINA1[71], SERPINC3[72], and SERPING1[73]. A number of pathologies in humans have been attributed to SERPIN gene variants, and often multiple deleterious mutations are known for each gene. Although a full listing of disease-causing SERPIN mutations is beyond the scope of this review, a sample of their scope is provided here. Mutations in the SERPINA1 gene have been linked with early-onset pulmonary emphysema, neonatal hepatitis, liver cirrhosis, and sometimes panniculitis and vasculitis [74,75]. SERPINA5 mutations have been linked with increased papillary thyroid cancer risk [76], and mutations in SERPINA10 have been linked to pregnancy complications [77]. Predisposition to familial venous thromboembolic disease has been linked to mutations in SERPINC1[78,79]. Finally, SNP variants for the SERPING1 gene have been shown to be associated with hereditary angioedema [80].
Conclusions
Serpins are a large class of diverse proteins, which contribute to numerous physiological and pathological conditions. Identification of serpins in immunological functions, pathology due to polymerization, and cancer metastasis underscores their diverse functions and physiological and pathological importance, and gene mutations often lead to loss-of-function and pathology in affected individuals. However, there is still much to learn about the functions and evolutionary development of serpins. Because of numerous biological functions and pathological states associated with serpins, further characterization of these proteins and mechanistic information will provide insight into potential biomarker identification and therapeutic targets.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CH carried out the sequence alignments and drafted the manuscript. BJ participated in the sequence alignment and analysis. MM reviewed mouse gene/protein data and the nomenclature for accuracy and completeness. MW reviewed human gene/protein data and nomenclature for accuracy and completeness. DT, GS and DWN reviewed and edited the manuscript. VV designed the study and reviewed data and manuscript. All authors read and approved the final manuscript.
Contributor Information
Claire Heit, Email: claire.heit@ucdenver.edu.
Brian C Jackson, Email: Brian.Jackson@ucdenver.edu.
Monica McAndrews, Email: monica.mcandrews@jax.org.
Mathew W Wright, Email: hgnc@genenames.org.
David C Thompson, Email: David.Thompson@ucdenver.edu.
Gary A Silverman, Email: gsilverman@upmc.edu.
Daniel W Nebert, Email: dan.nebert@uc.edu.
Vasilis Vasiliou, Email: vasilis.vasiliou@ucdenver.edu.
Acknowledgements
This work was supported, in part, by the following NIH grants: R24 AA022057, NIEHS P30 ES06096, HG000330, U41HG003345 and also by a Welcome Trust grant no. 099129/Z/12/Z. Fellowship assistance for BCJ (F31 AA020728) is acknowledged. We would like to thank Konstandinos Vasiliou for his assistance.
References
- Wright HT. Introns and higher-order structure in the evolution of serpins. J Mol Evol. 1993;7:136–143. doi: 10.1007/BF00166249. [DOI] [Google Scholar]
- Potempa J, Korzus E, Travis J. The serpin superfamily of proteinase inhibitors: structure, function, and regulation. J Biol Chem. 1994;7:15957–15960. [PubMed] [Google Scholar]
- Law RH, Zhang Q, McGowan S, Buckle AM, Silverman GA, Wong W, Rosado CJ, Langendorf CG, Pike RN, Bird PI, Whisstock JC. An overview of the serpin superfamily. Genome Biol. 2006;7:216. doi: 10.1186/gb-2006-7-5-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PG, Irving JA, Lomas DA, Luke CJ, Moyer RW, Pemberton PA, Remold-O'Donnell E, Salvesen GS, Travis J, Whisstock JC. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins: evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J Biol Chem. 2001;7:33293–33296. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- Schechter I, Berger A. On the size of the active site in proteases: I. Papain. Biochem Biophys Res Commun. 1967;7:157–162. doi: 10.1016/S0006-291X(67)80055-X. [DOI] [PubMed] [Google Scholar]
- Huber R, Carrell RW. Implications of the three-dimensional structure of alpha 1-antitrypsin for structure and function of serpins. Biochemistry-US. 1989;7:8951–8966. doi: 10.1021/bi00449a001. [DOI] [PubMed] [Google Scholar]
- Rein CM, Desai UR, Church FC. Serpin-glycosaminoglycan interactions. Methods Enzymol. 2011;7:105–137. doi: 10.1016/B978-0-12-385950-1.00007-9. [DOI] [PubMed] [Google Scholar]
- Yamasaki M, Sendall TJ, Pearce MC, Whisstock JC, Huntington JA. Molecular basis of alpha1-antitrypsin deficiency revealed by the structure of a domain-swapped trimer. EMBO Rep. 2011;7:1011–1017. doi: 10.1038/embor.2011.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntington JA. Serpin structure, function and dysfunction. J Thromb Haemost. 2011;7(1):26–34. doi: 10.1111/j.1538-7836.2011.04360.x. [DOI] [PubMed] [Google Scholar]
- Krem MM, Di Cera E. Conserved ser residues, the shutter region, and speciation in serpin evolution. J Biol Chem. 2003;7:37810–37814. doi: 10.1074/jbc.M305088200. [DOI] [PubMed] [Google Scholar]
- Billingsley GD, Walter MA, Hammond GL, Cox DW. Physical mapping of four serpin genes: alpha 1-antitrypsin, alpha 1-antichymotrypsin, corticosteroid-binding globulin, and protein C inhibitor, within a 280-kb region on chromosome I4q32.1. Am J Hum Genet. 1993;7:343–353. [PMC free article] [PubMed] [Google Scholar]
- Rollini P, Fournier RE. A 370-kb cosmid contig of the serpin gene cluster on human chromosome 14q32.1: molecular linkage of the genes encoding alpha 1-antichymotrypsin, protein C inhibitor, kallistatin, alpha 1-antitrypsin, and corticosteroid-binding globulin. Genomics. 1997;7:409–415. doi: 10.1006/geno.1997.5077. [DOI] [PubMed] [Google Scholar]
- Long M, de Souza SJ, Gilbert W. Evolution of the intron-exon structure of eukaryotic genes. Curr Opin Genet Dev. 1995;7:774–778. doi: 10.1016/0959-437X(95)80010-3. [DOI] [PubMed] [Google Scholar]
- Logsdon JM Jr, Stoltzfus A, Doolittle WF. Molecular evolution: recent cases of spliceosomal intron gain? Curr Biol. 1998;7:R560–R563. doi: 10.1016/S0960-9822(07)00361-2. [DOI] [PubMed] [Google Scholar]
- Clarke EP, Cates GA, Ball EH, Sanwal BD. A collagen-binding protein in the endoplasmic reticulum of myoblasts exhibits relationship with serine protease inhibitors. J Biol Chem. 1991;7:17230–17235. [PubMed] [Google Scholar]
- Irving JA, Pike RN, Lesk AM, Whisstock JC. Phylogeny of the serpin superfamily: implications of patterns of amino acid conservation for structure and function. Genome Res. 2000;7:1845–1864. doi: 10.1101/gr.GR-1478R. [DOI] [PubMed] [Google Scholar]
- Clemmensen SN, Jacobsen LC, Rorvig S, Askaa B, Christenson K, Iversen M, Jorgensen MH, Larsen MT, van Deurs B, Ostergaard O, Heegaard NH, Cowland JB, Borregaard N. Alpha-1-antitrypsin is produced by human neutrophil granulocytes and their precursors and liberated during granule exocytosis. Eur J Haematol. 2011;7:517–530. doi: 10.1111/j.1600-0609.2011.01601.x. [DOI] [PubMed] [Google Scholar]
- Marques PI, Ferreira Z, Martins M, Figueiredo J, Silva DI, Castro P, Morales-Hojas R, Simoes-Correia J, Seixas S. SERPINA2 is a novel gene with a divergent function from SERPINA1. PLoS One. 2013;7:e66889. doi: 10.1371/journal.pone.0066889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seixas S, Suriano G, Carvalho F, Seruca R, Rocha J, Di Rienzo A. Sequence diversity at the proximal 14q32.1 SERPIN subcluster: evidence for natural selection favoring the pseudogenization of SERPINA2. Mol Biol Evol. 2007;7:587–598. doi: 10.1093/molbev/msl187. [DOI] [PubMed] [Google Scholar]
- Chao J, Schmaier A, Chen LM, Yang Z, Chao L. Kallistatin, a novel human tissue kallikrein inhibitor: levels in body fluids, blood cells, and tissues in health and disease. J Lab Clin Med. 1996;7:612–620. doi: 10.1016/S0022-2143(96)90152-3. [DOI] [PubMed] [Google Scholar]
- Paterson MA, Horvath AJ, Pike RN, Coughlin PB. Molecular characterization of centerin, a germinal centre cell serpin. Biochem J. 2007;7:489–494. doi: 10.1042/BJ20070174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiker JT, Kloting N, Kovacs P, Kuettner EB, Strater N, Schultz S, Kern M, Stumvoll M, Bluher M, Beck-Sickinger AG. Vaspin inhibits kallikrein 7 by serpin mechanism. Cell Mol Life Sci. 2013;7:2569–2583. doi: 10.1007/s00018-013-1258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teshigawara S, Wada J, Hida K, Nakatsuka A, Eguchi J, Murakami K, Kanzaki M, Inoue K, Terami T, Katayama A, Iseda I, Matsushita Y, Miyatake N, McDonald JF, Hotta K, Makino H. Serum vaspin concentrations are closely related to insulin resistance, and rs77060950 at SERPINA12 genetically defines distinct group with higher serum levels in Japanese population. J Clin Endocrinol Metab. 2012;7:E1202–E1207. doi: 10.1210/jc.2011-3297. [DOI] [PubMed] [Google Scholar]
- Hilgers KF, Norwood VF, Gomez RA. Angiotensin's role in renal development. Semin Nephrol. 1997;7:492–501. [PubMed] [Google Scholar]
- Vidalino L, Doria A, Quarta S, Zen M, Gatta A, Pontisso P. SERPINB3, apoptosis and autoimmunity. Autoimmun Rev. 2009;7:108–112. doi: 10.1016/j.autrev.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Tsujimoto M, Tsuruoka N, Ishida N, Kurihara T, Iwasa F, Yamashiro K, Rogi T, Kodama S, Katsuragi N, Adachi M, Katayama T, Nakao M, Yamaichi K, Hashino J, Haruyama M, Miura K, Nakanishi T, Nakazato H, Teramura M, Mizoguchi H, Yamaguchi N. Purification, cDNA cloning, and characterization of a new serpin with megakaryocyte maturation activity. J Biol Chem. 1997;7:15373–15380. doi: 10.1074/jbc.272.24.15373. [DOI] [PubMed] [Google Scholar]
- Miyata T, Inagi R, Nangaku M, Imasawa T, Sato M, Izuhara Y, Suzuki D, Yoshino A, Onogi H, Kimura M, Sugiyama S, Kurokawa K. Overexpression of the serpin megsin induces progressive mesangial cell proliferation and expansion. J Clin Invest. 2002;7:585–593. doi: 10.1172/JCI14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Bird CH, Sutton V, McDonald L, Coughlin PB, De Jong TA, Trapani JA, Bird PI. A cytosolic granzyme B inhibitor related to the viral apoptotic regulator cytokine response modifier A is present in cytotoxic lymphocytes. J Biol Chem. 1996;7:27802–27809. doi: 10.1074/jbc.271.44.27802. [DOI] [PubMed] [Google Scholar]
- Sivaprasad U, Askew DJ, Ericksen MB, Gibson AM, Stier MT, Brandt EB, Bass SA, Daines MO, Chakir J, Stringer KF, Wert SE, Whitsett JA, Le Cras TD, Wills-Karp M, Silverman GA, Khurana Hershey GK. A nonredundant role for mouse serpinb3a in the induction of mucus production in asthma. J Allergy Clin Immunol. 2011;7:254–261. doi: 10.1016/j.jaci.2010.10.009. 261 e251-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto M, Iaccarino L, Ghirardello A, Bassi N, Pontisso P, Punzi L, Shoenfeld Y, Doria A. Serpins, immunity and autoimmunity: old molecules, new functions. Clin Rev Allergy Immunol. 2013;7(2):267–280. doi: 10.1007/s12016-013-8353-3. [DOI] [PubMed] [Google Scholar]
- Zou Z, Anisowicz A, Hendrix MJ, Thor A, Neveu M, Sheng S, Rafidi K, Seftor E, Sager R. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science. 1994;7:526–529. doi: 10.1126/science.8290962. [DOI] [PubMed] [Google Scholar]
- Shiiba M, Nomura H, Shinozuka K, Saito K, Kouzu Y, Kasamatsu A, Sakamoto Y, Murano A, Ono K, Ogawara K, Uzawa K, Tanzawa H. Down-regulated expression of SERPIN genes located on chromosome 18q21 in oral squamous cell carcinomas. Oncol Rep. 2010;7:241–249. doi: 10.3892/or_00000852. [DOI] [PubMed] [Google Scholar]
- Baldzizhar R, Fedorchuk C, Jha M, Rathinam C, Henegariu O, Czyzyk J. Anti-serpin antibody-mediated regulation of proteases in autoimmune diabetes. J Biol Chem. 2013;7:1612–1619. doi: 10.1074/jbc.M112.409664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schick C, Pemberton PA, Shi GP, Kamachi Y, Cataltepe S, Bartuski AJ, Gornstein ER, Bromme D, Chapman HA, Silverman GA. Cross-class inhibition of the cysteine proteinases cathepsins K, L, and S by the serpin squamous cell carcinoma antigen 1: a kinetic analysis. Biochemistry-US. 1998;7:5258–5266. doi: 10.1021/bi972521d. [DOI] [PubMed] [Google Scholar]
- Schick C, Kamachi Y, Bartuski AJ, Cataltepe S, Schechter NM, Pemberton PA, Silverman GA. Squamous cell carcinoma antigen 2 is a novel serpin that inhibits the chymotrypsin-like proteinases cathepsin G and mast cell chymase. J Biol Chem. 1997;7:1849–1855. doi: 10.1074/jbc.272.3.1849. [DOI] [PubMed] [Google Scholar]
- Scott FL, Hirst CE, Sun J, Bird CH, Bottomley SP, Bird PI. The intracellular serpin proteinase inhibitor 6 is expressed in monocytes and granulocytes and is a potent inhibitor of the azurophilic granule protease, cathepsin G. Blood. 1999;7:2089–2097. [PubMed] [Google Scholar]
- Xia Y, Zhang Y, Shi W, Liu S, Chen Y, Liang X, Ye Z. Overexpression of megsin induces mesangial cell proliferation and excretion of type IV collagen in vitro. Cell Immunol. 2011;7:413–417. doi: 10.1016/j.cellimm.2011.08.009. [DOI] [PubMed] [Google Scholar]
- Askew DJ, Cataltepe S, Kumar V, Edwards C, Pace SM, Howarth RN, Pak SC, Askew YS, Bromme D, Luke CJ, Whisstock JC, Silverman GA. SERPINB11 Is a new noninhibitory intracellular serpin: common single nucleotide polymorphisms in the scaffold impair conformational change. J Biol Chem. 2007;7:24948–24960. doi: 10.1074/jbc.M703182200. [DOI] [PubMed] [Google Scholar]
- Askew YS, Pak SC, Luke CJ, Askew DJ, Cataltepe S, Mills DR, Kato H, Lehoczky J, Dewar K, Birren B, Silverman GA. SERPINB12 is a novel member of the human ov-serpin family that is widely expressed and inhibits trypsin-like serine proteinases. J Biol Chem. 2001;7:49320–49330. doi: 10.1074/jbc.M108879200. [DOI] [PubMed] [Google Scholar]
- Huntington JA. Shape-shifting serpins–advantages of a mobile mechanism. Trends Biochem Sci. 2006;7:427–435. doi: 10.1016/j.tibs.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Vicente CP, He L, Pavao MS, Tollefsen DM. Antithrombotic activity of dermatan sulfate in heparin cofactor II-deficient mice. Blood. 2004;7:3965–3970. doi: 10.1182/blood-2004-02-0598. [DOI] [PubMed] [Google Scholar]
- Rau JC, Deans C, Hoffman MR, Thomas DB, Malcom GT, Zieske AW, Strong JP, Koch GG, Church FC. Heparin cofactor II in atherosclerotic lesions from the pathobiological determinants of atherosclerosis in youth (PDAY) study. Exp Mol Pathol. 2009;7:178–183. doi: 10.1016/j.yexmp.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lino MM, Atanasoski S, Kvajo M, Fayard B, Moreno E, Brenner HR, Suter U, Monard D. Mice lacking protease nexin-1 show delayed structural and functional recovery after sciatic nerve crush. J Neurosci. 2007;7:3677–3685. doi: 10.1523/JNEUROSCI.0277-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinrohr L, Harmat V, Dobo J, Lorincz Z, Gal P, Zavodszky P. C1 inhibitor serpin domain structure reveals the likely mechanism of heparin potentiation and conformational disease. J Biol Chem. 2007;7:21100–21109. doi: 10.1074/jbc.M700841200. [DOI] [PubMed] [Google Scholar]
- Widmer C, Gebauer JM, Brunstein E, Rosenbaum S, Zaucke F, Drogemuller C, Leeb T, Baumann U. Molecular basis for the action of the collagen-specific chaperone Hsp47/SERPINH1 and its structure-specific client recognition. Proc Natl Acad Sci U S A. 2012;7:13243–13247. doi: 10.1073/pnas.1208072109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai N, Hosokawa M, Itohara S, Adachi E, Matsushita T, Hosokawa N, Nagata K. Embryonic lethality of molecular chaperone hsp47 knockout mice is associated with defects in collagen biosynthesis. J Cell Biol. 2000;7:1499–1506. doi: 10.1083/jcb.150.6.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen HE, Schwarze U, Pyott SM, Al Swaid A, Al Balwi M, Alrasheed S, Pepin MG, Weis MA, Eyre DR, Byers PH. Homozygosity for a missense mutation in SERPINH1, which encodes the collagen chaperone protein HSP47, results in severe recessive osteogenesis imperfecta. Am J Hum Genet. 2010;7:389–398. doi: 10.1016/j.ajhg.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder T, Cinelli P, Baici A, Pennella A, Krueger SR, Schrimpf SP, Meins M, Sonderegger P. The axonally secreted serine proteinase inhibitor, neuroserpin, inhibits plasminogen activators and plasmin but not thrombin. J Biol Chem. 1998;7:2312–2321. doi: 10.1074/jbc.273.4.2312. [DOI] [PubMed] [Google Scholar]
- Ozaki K, Nagata M, Suzuki M, Fujiwara T, Miyoshi Y, Ishikawa O, Ohigashi H, Imaoka S, Takahashi E, Nakamura Y. Isolation and characterization of a novel human pancreas-specific gene, pancpin, that is down-regulated in pancreatic cancer cells. Genes Chromosomes Cancer. 1998;7:179–185. doi: 10.1002/(SICI)1098-2264(199807)22:3<179::AID-GCC3>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Carrell RW, Lomas DA. Conformational disease. Lancet. 1997;7:134–138. doi: 10.1016/S0140-6736(97)02073-4. [DOI] [PubMed] [Google Scholar]
- Saunders DN, Tindall EA, Shearer RF, Roberson J, Decker A, Wilson JA, Hayes VM. A novel SERPINA1 mutation causing serum alpha(1)-antitrypsin deficiency. PLoS One. 2012;7:e51762. doi: 10.1371/journal.pone.0051762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bledsoe G, Hagiwara M, Shen B, Chao L, Chao J. Depletion of endogenous kallistatin exacerbates renal and cardiovascular oxidative stress, inflammation, and organ remodeling. Am J Physiol Renal Physiol. 2012;7:F1230–F1238. doi: 10.1152/ajprenal.00257.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeunemaitre X, Gimenez-Roqueplo AP, Celerier J, Corvol P. Angiotensinogen variants and human hypertension. Curr Hypertens Rep. 1999;7:31–41. doi: 10.1007/s11906-999-0071-0. [DOI] [PubMed] [Google Scholar]
- Van de Water N, Tan T, Ashton F, O'Grady A, Day T, Browett P, Ockelford P, Harper P. Mutations within the protein Z-dependent protease inhibitor gene are associated with venous thromboembolic disease: a new form of thrombophilia. Br J Haematol. 2004;7:190–194. doi: 10.1111/j.1365-2141.2004.05189.x. [DOI] [PubMed] [Google Scholar]
- Corral J, Gonzalez-Conejero R, Soria JM, Gonzalez-Porras JR, Perez-Ceballos E, Lecumberri R, Roldan V, Souto JC, Minano A, Hernandez-Espinosa D, Alberca I, Fontcuberta J, Vicente V. A nonsense polymorphism in the protein Z-dependent protease inhibitor increases the risk for venous thrombosis. Blood. 2006;7:177–183. doi: 10.1182/blood-2005-08-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooptu B, Hazes B, Chang WS, Dafforn TR, Carrell RW, Read RJ, Lomas DA. Inactive conformation of the serpin alpha(1)-antichymotrypsin indicates two-stage insertion of the reactive loop: implications for inhibitory function and conformational disease. Proc Natl Acad Sci U S A. 2000;7:67–72. doi: 10.1073/pnas.97.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torpy DJ, Bachmann AW, Gartside M, Grice JE, Harris JM, Clifton P, Easteal S, Jackson RV, Whitworth JA. Association between chronic fatigue syndrome and the corticosteroid-binding globulin gene ALA SER224 polymorphism. Endocr Res. 2004;7:417–429. doi: 10.1081/ERC-200035599. [DOI] [PubMed] [Google Scholar]
- Homan EP, Rauch F, Grafe I, Lietman C, Doll JA, Dawson B, Bertin T, Napierala D, Morello R, Gibbs R, White L, Miki R, Cohn DH, Crawford S, Travers R, Glorieux FH, Lee B. Mutations in SERPINF1 cause osteogenesis imperfecta type VI. J Bone Miner Res. 2011;7:2798–2803. doi: 10.1002/jbmr.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MH, Hwang SI, Roy DB, Lundgren DH, Price JV, Ousman SS, Fernald GH, Gerlitz B, Robinson WH, Baranzini SE, Grinnell BW, Raine CS, Sobel RA, Han DK, Steinman L. Proteomic analysis of active multiple sclerosis lesions reveals therapeutic targets. Nature. 2008;7:1076–1081. doi: 10.1038/nature06559. [DOI] [PubMed] [Google Scholar]
- Cao D, Zhang Q, Wu LS, Salaria SN, Winter JW, Hruban RH, Goggins MS, Abbruzzese JL, Maitra A, Ho L. Prognostic significance of maspin in pancreatic ductal adenocarcinoma: tissue microarray analysis of 223 surgically resected cases. Mod Pathol. 2007;7:570–578. doi: 10.1038/modpathol.3800772. [DOI] [PubMed] [Google Scholar]
- Vecchi M, Confalonieri S, Nuciforo P, Vigano MA, Capra M, Bianchi M, Nicosia D, Bianchi F, Galimberti V, Viale G, Palermo G, Riccardi A, Campanini R, Daidone MG, Pierotti MA, Pece S, Di Fiore PP. Breast cancer metastases are molecularly distinct from their primary tumors. Oncogene. 2008;7:2148–2158. doi: 10.1038/sj.onc.1210858. [DOI] [PubMed] [Google Scholar]
- Jing Y, Kovacs K, Kurisetty V, Jiang Z, Tsinoremas N, Merchan JR. Role of plasminogen activator inhibitor-1 in urokinase's paradoxical in vivo tumor suppressing or promoting effects. Mol Cancer Res. 2012;7:1271–1281. doi: 10.1158/1541-7786.MCR-12-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RM, Bernstein D, Higgins SP, Higgins CE, Higgins PJ. SERPINE1 expression discriminates site-specific metastasis in human melanoma. Exp Dermatol. 2012;7:551–554. doi: 10.1111/j.1600-0625.2012.01523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaraman P, Brenner AV, Butler MA, Wang SS, Pfeiffer RM, Ruder AM, Linet MS, Yeager M, Wang Z, Orr N, Fine HA, Kwon D, Thomas G, Rothman N, Inskip PD, Chanock SJ. Common variation in genes related to innate immunity and risk of adult glioma. Cancer Epidemiol Biomarkers Prev. 2009;7:1651–1658. doi: 10.1158/1055-9965.EPI-08-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia HL, Ye QH, Qin LX, Budhu A, Forgues M, Chen Y, Liu YK, Sun HC, Wang L, Lu HZ, Shen F, Tang ZY, Wang XW. Gene expression profiling reveals potential biomarkers of human hepatocellular carcinoma. Clin Cancer Res. 2007;7:1133–1139. doi: 10.1158/1078-0432.CCR-06-1025. [DOI] [PubMed] [Google Scholar]
- Paterson MA, Hosking PS, Coughlin PB. Expression of the serpin centerin defines a germinal center phenotype in B-cell lymphomas. Am J Clin Pathol. 2008;7:117–126. doi: 10.1309/9QKE68QU7B825A3U. [DOI] [PubMed] [Google Scholar]
- Byrne SM, Aucher A, Alyahya S, Elder M, Olson ST, Davis DM, Ashton-Rickardt PG. Cathepsin B controls the persistence of memory CD8+ T lymphocytes. J Immunol. 2012;7:1133–1143. doi: 10.4049/jimmunol.1003406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safdar H, Cheung KL, Salvatori D, Versteeg HH, Laghmani el H, Wagenaar GT, Reitsma PH, van Vlijmen BJ. Acute and severe coagulopathy in adult mice following silencing of hepatic antithrombin and protein C production. Blood. 2013;7:4413–4416. doi: 10.1182/blood-2012-11-465674. [DOI] [PubMed] [Google Scholar]
- Yiannikouris F, Karounos M, Charnigo R, English VL, Rateri DL, Daugherty A, Cassis LA. Adipocyte-specific deficiency of angiotensinogen decreases plasma angiotensinogen concentration and systolic blood pressure in mice. Am J Physiol Regul Integr Comp Physiol. 2012;7:R244–R251. doi: 10.1152/ajpregu.00323.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann M, Pham CT, Benarafa C. SerpinB1 is critical for neutrophil survival through cell-autonomous inhibition of cathepsin G . Blood. 2013;7:3900–3907. doi: 10.1182/blood-2012-09-455022. S3901-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaimidou S, van Baal S, Smith TD, Mitropoulos K, Ljujic M, Radojkovic D, Cotton RG, Patrinos GP. A1ATVar: a relational database of human SERPINA1 gene variants leading to alpha1-antitrypsin deficiency and application of the VariVis software. Hum Mutat. 2009;7:308–313. doi: 10.1002/humu.20857. [DOI] [PubMed] [Google Scholar]
- Lane DA, Olds RJ, Thein SL. Antithrombin III: summary of first database update. Nucleic Acids Res. 1994;7:3556–3559. [PMC free article] [PubMed] [Google Scholar]
- Kalmar L, Hegedus T, Farkas H, Nagy M, Tordai A. HAEdb: a novel interactive, locus-specific mutation database for the C1 inhibitor gene. Hum Mutat. 2005;7:1–5. doi: 10.1002/humu.20112. [DOI] [PubMed] [Google Scholar]
- Sabina J, Tobias W. Augmentation therapy with alpha1-antitrypsin: novel perspectives. Cardiovasc Hematol Disord Drug Targets. 2013;7:90–98. doi: 10.2174/1871529x11313020002. [DOI] [PubMed] [Google Scholar]
- Bornhorst JA, Greene DN, Ashwood ER, Grenache DG. α1-Antitrypsin phenotypes and associated serum protein concentrations in a large clinical population. Chest. 2013;7:1000–1008. doi: 10.1378/chest.12-0564. [DOI] [PubMed] [Google Scholar]
- Brenner AV, Neta G, Sturgis EM, Pfeiffer RM, Hutchinson A, Yeager M, Xu L, Zhou C, Wheeler W, Tucker MA, Chanock SJ, Sigurdson AJ. Common single nucleotide polymorphisms in genes related to immune function and risk of papillary thyroid cancer. PLoS One. 2013;7:e57243. doi: 10.1371/journal.pone.0057243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almawi WY, Al-Shaikh FS, Melemedjian OK, Almawi AW. Protein Z, an anticoagulant protein with expanding role in reproductive biology. Reproduction. 2013;7:R73–R80. doi: 10.1530/REP-13-0072. [DOI] [PubMed] [Google Scholar]
- Maruyama K, Morishita E, Karato M, Kadono T, Sekiya A, Goto Y, Sato T, Nomoto H, Omi W, Tsuzura S, Imai H, Asakura H, Ohtake S, Nakao S. Antithrombin deficiency in three Japanese families: one novel and two reported point mutations in the antithrombin gene. Thromb Res. 2013;7:e118–e123. doi: 10.1016/j.thromres.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Hepner M, Karlaftis V. Antithrombin. Methods Mol Biol. 2013;7:355–364. doi: 10.1007/978-1-62703-339-8_28. [DOI] [PubMed] [Google Scholar]
- Bork K, Davis-Lorton M. Overview of hereditary angioedema caused by C1-inhibitor deficiency: assessment and clinical management. Eur Ann Allergy Clin Immunol. 2013;7:7–16. [PubMed] [Google Scholar]