Abstract
Spatial organization of chromatin plays an important role at multiple levels of genome regulation. On a global scale its function is evident in processes like metaphase and chromosome segregation. On a detailed level, long range interactions between regulatory elements and promoters are essential for proper gene regulation. Microscopic techniques like FISH can detect chromatin contacts, although the resolution is generally low making detection of enhancer-promoter interaction difficult. The 3C methodology allows for high-resolution analysis of chromatin interactions. 3C is now widely used and has revealed that long-range looping interactions between genomic elements is widespread. However, studying chromatin interactions in large genomic regions by 3C is very labor intensive. This limitation is overcome by the 5C technology. 5C is an adaptation of 3C, in which the concurrent use of thousands of primers permits the simultaneous detection of millions of chromatin contacts. The design of the 5C primers is critical, since this will determine which and how many chromatin interactions will be examined in the assay. Starting material for 5C is a 3C template. To make a 3C template, chromatin interactions in living cells are crosslinked using formaldehyde. Next, chromatin is digested and subsequently ligated under conditions favoring ligation events between crosslinked fragments. This yields a genome-wide 3C library of ligation products representing all chromatin interactions in vivo. 5C then employs multiplex ligation mediated amplification to detect, in a single assay, up to millions of unique ligation products present in the 3C library. The resulting 5C library can be analyzed by microarray analysis or deep sequencing. The observed abundance of a 5C product is a measure of the interaction frequency between the two corresponding chromatin fragments. The power of the 5C technique described in this chapter is the high-throughput, high-resolution and quantitative way in which the spatial organization of chromatin can be examined.
Keywords: Chromosome conformation capture, chromatin looping, chromatin structure, long-range gene regulation, high-throughput
1. Introduction
Tremendous efforts are underway to annotate all genes and other functional elements within the human genome as well as the genomes of several model organisms such as C. elegans and D. melanogaster [e.g 1]. These large-scale efforts will ultimately result in linear maps of genes and elements within the genome. Although these maps will provide crucial information about gene content and regulatory potential encoded within genomes, these maps will not directly reveal which regulatory elements regulate each gene. Identification of functional relationships between regulatory elements and target genes is complicated because elements such as enhancers, repressors and insulators can be located at large genomic distances from their cognate target genes, and in some cases can even be located on other chromosomes (2, 3). During the last couple of years it has been demonstrated that distant regulatory elements regulate genes through direct physical associations with target genes resulting in the formation of chromatin loops [e.g. 4, 5]. Thus, the spatial organization of chromosomes plays a critical role in bringing together functionally related genomic elements. This further implies that mapping spatial organization of genomes will be a powerful approach to determine which (distant) regulatory elements regulate any given gene.
The spatial organization of chromosomes likely plays roles in many other nuclear processes as well. Most notably, the formation of condensed metaphase chromosomes involves organization of chromosomes in topologically reproducible compact shapes, which is essential for faithful chromosome segregation. During interphase chromosomes are less condensed and appear diffuse and unorganized. However, recent studies have revealed that the interphase nucleus also displays significant spatial organization (6, 7). For instance, the three-dimensional arrangement of chromosomes and positioning of genes with respect to each other and to sub-nuclear structures such as the nuclear envelope is correlated with gene activity. However, currently it is unknown whether the three-dimensional organization of the nucleus directly affects gene expression, or whether it is a downstream consequence of gene expression. Comprehensive studies of the spatial organization of chromosomes during interphase and metaphase may provide new insights into these long-standing questions.
The spatial organization of chromosomes can be studied using microscopic methods to visualize the locations of genes inside individual cells. Although modern imaging technologies allow detailed analysis of positioning of specific loci inside living cells, the resolution of light microscopy is in many cases not sufficiently high to detect looping interactions between promoters and enhancers. To overcome these issues we developed the chromosome conformation capture (3C) technology that allows high-resolution analysis of chromatin looping events and chromosome conformation in general (8).
3C is used to measure physical interaction frequencies between small chromatin fragments in vivo (8-10). The method uses formaldehyde crosslinking to capture chromatin interactions in living cells (Fig 1). Chromatin is then digested and subsequently ligated under very dilute conditions to form intra-molecular ligation products of crosslinked restiction fragments specifically. After ligation, crosslinks are reversed and the DNA is purified. The resulting 3C template is a library of genomic ligation products reflecting all chromatin interactions that occur throughout the entire genome. Individual ligation products are detected through semi-quantitative PCR using pairs of primers that recognize specific combinations of ligated restriction fragments. The abundance of a particular ligation product in the 3C library and hence the yield of PCR product is in proportion to the nuclear interaction frequency between the two corresponding restriction fragments.
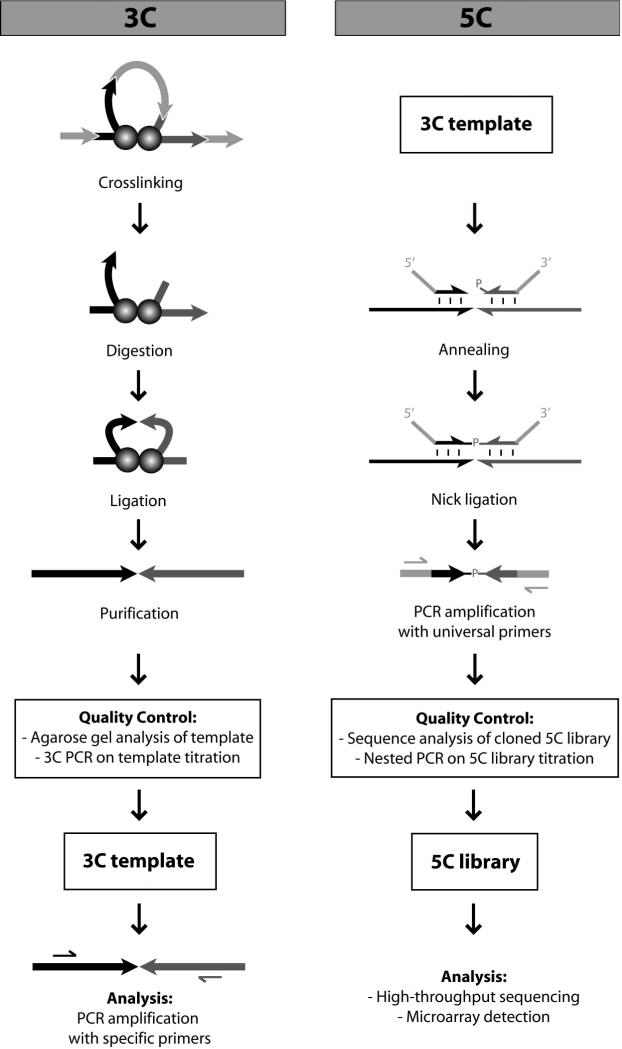
Figure 1.
Overview of the 3C (left) and 5C (right) methods. In 3C, chromatin contacts are crosslinked with formaldehyde. The DNA is digested with a restriction enzyme and is subsequently ligated under conditions favoring intramolecular ligation. After purification, the 3C template is run on gel and titrated in a 3C PCR experiment for quality control. Conventional 3C analysis is done in a one-to-one fashion by performing semi-quantitative PCR using specific primers for individual restriction fragments. The 5C method involves multiplex ligation-mediated amplification of 5C primers detecting 3C ligation products. The quality of a 5C library is checked by nested PCR and by sequencing of individual cloned products. DNA interaction frequencies are examined in a many-to-many fashion by either microarray analysis or deep sequencing of the 5C library.
The 3C method has been proven to be a powerful tool to detect long-range interactions that are involved in gene regulation, both in cis and in trans (3, 11-14). The limitation of this technique is that interactions are analyzed in a “one-to-one” manner. Analysis of large numbers of interactions is time-consuming and labor intensive. Hence, 3C is most appropriate to study interactions between candidate loci located in a relatively small genomic region of up to a few hundred kilobases.
Recently, we have adapted 3C to allow for high-throughput and comprehensive analyses of interaction networks between large numbers of genomic elements (15, 16). The resulting “3C-carbon copy” or “5C” technique combines 3C with highly multiplexed ligation mediated amplification (LMA) and thereby permits millions of interactions to be tested simultaneously in a “many-to-many” fashion. In the 5C method, the relative abundance of the ligation products is detected by forward and reverse 5C primers that are designed to anneal directly up or downstream, respectively, of the newly formed restriction site in a 3C ligation product (Fig 1). After annealing to the 3C template, the primers are ligated by Taq DNA ligase, which specifically ligates nicked DNA. The ligated primer pairs form copies of the unique ligation junctions that characterize 3C ligation products present in the original 3C library, hence the name “3C carbon copy” or 5C. LMA allows for very high levels of multiplexing, since thousands of forward and reverse primers can be combined to detect millions of unique chromatin interactions in a single assay. Using common tails on the 5C primers, all 5C ligation products can be simultaneously amplified with universal primers. The resulting product is a 5C library, that can be subsequently analyzed by either deep-sequencing or microarray analysis.
Under ideal conditions the abundance of a 5C product in the 5C library directly reflects the frequency with which the two corresponding chromatin segments interact in the nucleus. However, the efficiency of formation of 5C products can be biased due to differences in 5C primer annealing efficiency and PCR amplification of 5C ligation products. These biases are minimized by careful design of 5C primers so that they are all of equal length and all have identical melting temperatures. Any remaining technical biases can be corrected for by using a so-called control 5C library. A control 5C library is generated by performing 5C with a special control 3C library as template. The control 3C library is comprised of randomly ligated fragments of the region of interest. As a result, every possible ligation product will be equally represented and any differences in abundance of 5C products in the 5C control library generated with the control 3C library will be due to annealing and amplification differences between 5C primers. Any biases in 5C library composition due to primer differences are removed by dividing the signal for each ligation product in the 5C library by the signal of the corresponding product in the control 5C library. This ratio is a quantitative measure for the interaction frequency of the two corresponding DNA fragments in the nucleus. These quantitative results make the 5C technique extremely powerful.
The 5C method can be used for different types of large-scale studies. The type of study will determine the design of a 5C experiment, since the combination of forward and reverse 5C primers defines the interactions that can be measured in the assay. For example, 5C can be used to determine a profile of chromatin interactions between one or a few fragments of interest and all other fragments within a large genomic domain. This approach can be used to discover the elements involved in regulation of one or a few specific genes. In this case, reverse primers are designed for the fragments containing the transcription start sites of the genes and forward primers are designed for all other fragments within the genomic domain of interest. Other studies can be focused on the identification of the global chromatin conformation of a specific region by determining dense networks of interaction frequencies between every pair of restriction fragments in that region. For this type of analysis, forward and reverse 5C primers are designed in an alternating manner for consecutive restriction fragments within the region of interest. Both types of data generated by 5C will give invaluable information about the spatial organization of chromatin and will provide new insights into the elements and mechanisms involved in long range gene regulation.
2. Materials
2.1 Generation of a 3C template
Deionized autoclaved water for use in all solutions.
7 × 107 - 1 × 108 mammalian cells grown under appropriate conditions.
Cell culture medium.
37% (v/v) formaldehyde (Mallinckrodt, cat. no. 5016-02) (see Note 1). (This product is flammable, can cause skin burns, is an eye irritant and is toxic by inhalation. Therefore, formaldehyde should be handled with protective gear in a chemical fume hood.)
2.5 M glycine (Fisher, cat. no. BP381-1). Store at room temperature.
Lysisbuffer: 10 mM Tris-HCl pH 8.0, 10 mM sodium chloride, 0.2% (v/v) Igepal CA-630 (MP Biomedicals, cat. no. 198596). Store at 4°C.
Protease inhibitor cocktail (Sigma, cat. no. P8340).
Dounce homogenizer (pestle A; VWR, cat. no. KT885300-0002)
10× restriction buffer (NEB).
Restriction enzyme (NEB) (see Note 2).
1% (w/v) SDS (Fisher, cat. no. BP166-500). Store at room temperature.
10% (w/v) SDS. Store at room temperature.
10% (v/v) Triton X-100 (VWR, cat. no. VW3929-2). Store at room temperature.
10× T4 ligation buffer: 500 mM Tris-HCl pH 7.5, 100 mM magnesium chloride, 100 mM DTT. Store at −20°C (see Note 3).
10 mg/ml BSA (NEB, cat. no. B9001S).
100 mM ATP (Sigma, cat. no. A9187). Store at −20°C.
T4 DNA ligase (300 cohesive end units/μl) (Invitrogen, cat. no. 15224-025).
10 mg/ml proteinase K (Invitrogen, cat. no. 25530-031). Dissolve in 1× TE buffer pH 8.0, aliquot and store at −20°C.
Saturated phenol pH 8.0 (Fisher, cat. no. BP-1750-400). (This product is a toxic and corrosive material. Wear protective gear and handle in a chemical fume hood.) Store at 4°C.
Phenol pH 8.0:chloroform (1:1). (This product is a toxic and corrosive material. Wear protective gear and handle in a chemical fume hood.) Store at 4°C.
1× Tris-EDTA (TE) pH 8.0: 10 mM Tris-HCl pH 8.0, 1 mM EDTA pH 8.0. Store at room temperature.
3M sodium acetate pH 5.2. Store at room temperature.
100% ethanol. Store at −20°C.
70% ethanol. Store at room temperature.
10 mg/ml of DNase-free, RNase A (Sigma, cat. no. R6513). Dissolve in water, aliquot and store at −20°C.
Agarose
10× Tris-borate-EDTA (TBE) buffer: 0.89 M Tris base, 0.89 M boric acid, 0.02 M EDTA pH 8.0.
10 mg/ml ethidium bromide. (This product should be regarded as mutagenic to man and could be carcinogenic. Therefore, ethidium bromide should be handled with protective gear.)
4× DNA loading buffer: 10% (w/v) ficoll, 0.17% (w/v) xylene cyanol (see Note 4).
2.2 Generation of a control 3C template
LB medium pH 7.0: 0.01% (w/v) bacto-tryptone, 0.005% (w/v) bacto-yeast extract, 0.01% sodium chloride.
Antibiotic.
Large-construct DNA purification kit (Qiagen, cat. no. 12462).
Chloroform (Mallinckrodt 4440-04). (This product is a toxic and corrosive material. Wear protective gear and handle in a chemical fume hood.)
2.3 Quality control of 3C and control templates
10× PCR buffer: 600 mM Tris-H2SO4 pH 8.9, 180 mM ammonium sulfate. Store at −20°C.
50 mM magnesium sulfate (Sigma, cat. no. M2773). Store at −20°C.
dNTP mix (25 mM each) (Invitrogen, cat. no. 10297-018).
80 μM 3C template titration primers (see Note 5). Dissolve in 1× TE buffer pH 8.0 and store at −20°C.
5 U/μl Taq DNA polymerase (NEB, cat. no. M0267L).
2.5 Preparation of a 5C primer pool
10 U/μl T4 polynucleotide kinase (PNK) (NEB, cat. no. M0201S).
10× T4 polynucleotide kinase reaction buffer (PNK buffer) (NEB, cat. no. M0201S): 700 mM Tris-HCl pH 7.6, 100 mM magnesium chloride, 50 mM DTT.
10 mM ATP (Sigma, cat. no. A9187). Store at −20°C.
2.6 Preparation of a 5C library
1 μg/μl Salmon sperm DNA (SSD) (Invitrogen, cat. no. 15632-011). Dilute in 1× TE pH 8.0, aliquot and store at −20°C (see Note 6).
10× 5C annealing buffer (NEBuffer 4, NEB, cat. no. B7004S): 200 mM Tris-acetate pH 7.9, 500 mM potassium acetate, 100 mM magnesium acetate, 10 mM DTT.
10× Taq DNA ligase buffer (NEB, cat. no. B0208S): 200 mM Tris-HCl pH 7.6, 250 mM potassium acetate, 100 mM magnesium acetate, 100 mM DTT, 10 mM NAD+, 1% (v/v) Triton X-100.
40 U/μl Taq DNA ligase (NEB, cat. no. M0208S).
10× PCR buffer II (Applied Biosystems, cat. no. N8080243).
25 mM magnesium chloride (Applied Biosystems, cat. no. N8080243).
AmpliTaq Gold DNA polymerase (Applied Biosystems, cat. no. N8080243).
80 μM T3 (5’ TATTAACCCTCACTAAAGGGA 3’) and T7 (5’ TAATACGACTCACTATAGCC 3’) PCR primers (see Note 7).
MinElute PCR purification kit (Qiagen, cat. no. 28004).
2.7 Quality control of a 5C library: nested PCR
80 μM nested PCR primers (see Note 8).
2.8 Quality control of a 5C library: cloning and sequence analysis
TOPO TA Cloning Kit (Invitrogen, cat. no. K4500-01).
40 mg/ml Xgal in dimethylformamide (US Biological, cat. no. X1000-05).
QIAprep Spin Miniprep kit (Qiagen, cat. no. 27106).
−21 M13 sequencing primer (5’ TGTAAAACGACGGCCAGT 3’).
Sequencing reagents
3. Methods
3.1 Generation of a 3C template
Grow between 7 × 107 and 1 × 108 cells under the preferred conditions in the appropriate medium.
Aspirate the medium and add 22.5 ml of fresh medium to the cells (see Note 9).
To crosslink chromatin, add 625 μl of 37% formaldehyde to obtain a 1% final concentration. Mix gently and incubate at room temperature for 10 minutes. Gently rock the plates every 2 minutes.
Stop the reaction by adding 1.25 ml of 2.5 M glycine. Mix gently and incubate at room temperature for 5 minutes, followed by incubation on ice for at least 15 minutes to stop crosslinking completely (see Note 10).
Scrape the cells from the plates with a cell scraper and transfer the cells to a 250 ml conical tube.
Centrifuge the crosslinked cells at 450g for 10 minutes. Discard the supernatant (see Note 11).
Mix 2 ml of ice-cold lysisbuffer with 200 μl of protease inhibitor cocktail and add it to the cell pellet. Resuspend well and let the suspension sit on ice for at least 15 minutes to let the cells swell (see Note 12).
Dounce homogenize the cells by stroking 10 times, followed by incubation on ice for 1 minute and subsequently stroking for another 10 times.
Transfer the suspension to 2 1.7 ml centrifuge tubes, spin at 2,000g at room temperature for 5 minutes.
Discard the supernatant and wash each pellet with 1 ml cold 1× restriction buffer. Centrifuge at 2,000g for 5 minutes at room temperature.
Repeat the wash step in step 10.
Discard the supernatant, resuspend each pellet in 500 μl of 1× restriction buffer and pool the suspensions (see Note 13). Divide the supsension into 50 μl aliquots in 1.7 ml centrifuge tubes (~22 tubes) (see Note 14).
Add 312 μl of 1× restriction buffer per tube.
To remove the proteins that are not directly crosslinked to the DNA, add 38 μl of 1% SDS per tube. Mix well while avoiding air bubbles (see Note 15) and incubate at 65°C for 10 minutes (see Note 16).
Quench the SDS by adding 44 μl of 10% Triton X-100 to each tube. Mix well but avoid air bubbles.
Add 400 U of restriction enzyme per tube, mix well and digest the DNA overnight at the manufacturer recommended temperature (see Note 17).
Inactivate the restriction enzyme by adding 86 μl of 10% SDS and incubate at 65°C for 30 minutes (see Note 16 and Note 18).
Meanwhile, prepare a ligation master mix containing N × (745 μl 10% Triton X-100, 745 μl 10× ligation buffer, 80 μl 10 mg/ml BSA, 80 μl 100 mM ATP and 5960 μl water) for N tubes. Aliquot 7.61 ml ligation master mix in 15 ml conical tubes and put on ice.
Transfer the digestion product from Step 17 into each 15 ml conical tube. To ligate the DNA fragments, add 10 μl of T4 DNA ligase per tube. Mix by inverting the tubes several times, spin down shortly and incubate at 16°C for 2 hours.
Add 50 μl of 10 mg/ml proteinase K per tube. Mix by inverting the tubes multiple times, spin down shortly and reverse the crosslinking by incubating at 65°C overnight.
Add 50 μl of 10 mg/ml proteinase K per tube and incubate at 65°C for an additional 2 hours (see Note 19).
Pool two ligation mixtures into 1 clean 50 ml conical tube (~11 tubes). Add 20 ml of phenol pH 8.0 to each tube, vortex for 2 minutes and spin the tubes at 2,500g for 10 minutes at room temperature.
Transfer the aqueous upper phase to a clean 50 ml conical tube. Add 20 ml of phenol pH8.0:chloroform per tube, vortex each tube for 1 minute and spin the tubes at 2,500g for 10 minutes at room temperature (see Note 20).
Pool the aqueous phases into 4 50 ml conical tubes (see Note 21). Adjust the volume to 50 ml per tube with 1× TE pH 8.0 (see Note 22) and transfer the DNA solutions to 4 250-ml screw-cap centrifuge tubes that are suitable for high-speed spinning.
Add 5 ml of 3M sodium acetate pH 5.2 per tube, mix and add 125 ml 100% ice-cold ethanol per tube. Mix gently and precipitate the DNA by incubating at −80°C for at least 1 hour (see Note 23).
Pellet the DNA by spinning at 10,000g for 20 minutes at 4 °C.
Discard the supernatant. Dissolve each pellet in 500 μl of 1× TE buffer pH 8.0 and pool the DNA solutions.
Wash each tube with an additional 500 μl of 1× TE buffer pH 8.0 and pool it with the DNA solution from the previous step. Mix and aliquot 500 μl into 8 fresh 1.7 ml centrifuge tubes.
Add 500 μl of phenol pH8.0:chloroform to each tube, vortex for 1 minute and spin at 18,000g for 5 minutes at room temperature.
Transfer 450 μl of each upper aqueous phase to a fresh 1.7 ml centrifuge tube and repeat step 29.
Transfer 400 μl of each upper aqueous phase to fresh 1.7 centrifuge tubes (see Note 21). Add 40 μl of 3M sodium acetate pH 5.2 and vortex briefly. Add 1 ml of 100% ethanol, mix gently and precipitate the DNA by placing the tubes at −80°C for at least 30 minutes.
Pellet the DNA by spinning at 18,000g for 20 minutes at 4°C.
Aspirate the supernatant and wash the pellets with 1 ml of 70% ethanol. Make sure the pellet is resuspended well to allow the salt in the pellet to dissolve into the ethanol. Spin at 18,000g for 15 minutes at 4°C.
Repeat step 33 at least 5 times or until the volume of the pellet does not decrease anymore (see Note 24).
Remove the last traces of 70% ethanol and air dry the pellets briefly.
Dissolve all pellets in a total volume of 1 ml of 1× TE buffer pH 8.0.
To degrade RNA, add 1 μl of 10 mg/ml of (DNase-free) RNase A and incubate at 37°C for 15 minutes.
Prepare a 0.8% agarose/0.5× TBE gel containing 0.5 μg/ml ethidium bromide.
Load 0.1, 0.2 and 0.4 μl of the 3C template and 150 ng of a molecular weight standard (Fig 2A).
After running the gel, estimate the 3C template concentration by comparing the intensities to the molecular weight standard (see Note 25).
Aliquot the 3C template and store it up to at least 2 years at −20°C.
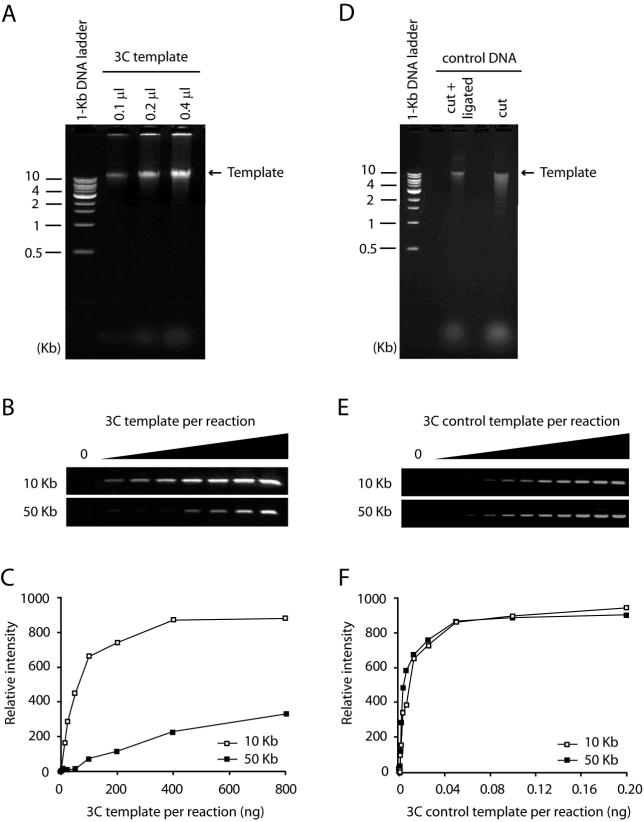
Figure 2.
Quality control of 3C and control templates. (A) Increasing amounts of 3C template were resolved on a 0.8% agarose gel. Typically a 3C template runs as a rather tight band larger than 10 Kb. A DNA smear indicates poor ligation efficiency. (B) Agarose gel analysis and (C) quantification of a 3C template titration in a 3C PCR. Increasing amounts of 3C template were analyzed with two 3C primer pairs. One primer pair interrogates an interaction between fragments that are close (10 Kb) to each other in the linear genome. The other primer pair examines the interaction between two more distant (50 Kb) fragments. A titration curve should demonstrate a linear increase at the beginning of the curve and should reach a plateau with increasing amounts of 3C template. The curve of the PCR product representing the interaction between two adjacent restriction fragments should be above the curve of the PCR product testing the interaction between two, more distant fragments. (D) Digested and religated BAC DNA were resolved on a 0.8% agarose gel. Digested control BAC DNA usually runs as a smear on the gel. Religated control DNA should run as a band above 10 Kb and the smear should be mostly gone. (E) Agarose gel analysis and (F) quantification of a control 3C template titration in a 3C PCR. Increasing amounts of control 3C template were analyzed with the same 3C primer pairs as for the 3C template. The titration curves of both PCR products should look more similar compared to the curves of a 3C template, indicating that all interactions in the control 3C template are represented equally.
3.2 Generation of a control 3C template
Select one or more BAC clones that cover the genomic region of interest with the least possible overlap between the clones and leaving the lowest number of gaps (see Note 26).
Purify the BAC DNA from 500 ml overnight cultures using the large-construct DNA purification kit.
If more than one BAC clone is used, mix the different clones in equimolar amounts (see Note 27).
Estimate the DNA concentration by running 1 μl of BAC DNA and a molecular weight standard of known concentration on a 0.8% agarose/0.5× TBE gel containing 0.5 μg/ml ethidium bromide. The concentration of the BAC DNA should be between 50 and 100 ng/μl.
Prepare the following reaction: 20 μg BAC DNA, 160 μl 10× restriction buffer, 800 U restriction enzyme and add water till 1.6 ml. Mix well and digest the BAC DNA overnight at the manufacturer recommended temperature (see Note 28)
Split the samples in 4× 400 μl in fresh 1.7 ml centrifuge tubes.
Add 500 μl of phenol pH8.0:chloroform, vortex for 30 seconds and spin at 18,000g for 5 minutes at room temperature.
Transfer the aqueous phases to fresh 1.7 ml centrifuge tubes. Add 40 μl of 3M sodium acetate pH 5.2 and vortex briefly. Add 1 ml of 100% ethanol, mix gently and precipitate the DNA by placing the tubes at ×20°C for at least 15 minutes.
Pellet the DNA by centrifuging at 18,000g for 20 minutes at 4°C.
Wash the pellets with 1 ml of 70% ethanol and spin at 18,000g for 15 minutes at 4°C.
Resuspend each pellet in 161 μl water and dissolve the BAC DNA by incubating at 37 °C for 15 minutes.
Take 4 μl of each tube and keep that separate for future gel analysis.
Prepare the following ligation reactions: 157 μl digested BAC DNA, 20 μl T4 ligation buffer, 2 μl 10 mg/ml BSA, 2 μl 100 mM ATP and 19 μl T4 DNA ligase. Incubate at 16°C overnight.
Inactivate the T4 DNA ligase by incubating the reactions at 65°C for 15 minutes.
Add 200 μl phenol pH8.0:chloroform to each tube, vortex for 30 seconds and spin at 18,000g for 5 minutes at room temperature.
Transfer the aqueous phases to fresh 1.7 ml centrifuge tubes and repeat the phenol pH8.0:chloroform extraction once.
Transfer the aqueous phases to fresh 1.7 ml centrifuge tubes. Add 200 μl chloroform, vortex for 30 seconds and spin at 18,000g for 5 minutes at room temperature.
Pool two aqueous phases in one fresh 1.7 ml centrifuge tube rendering two tubes, each containing ~ 350 μl.
Add 35 μl of 3M sodium acetate pH 5.2 and vortex briefly. Add 875 μl of 100% ethanol, mix gently and precipitate the DNA by placing the tubes at −20°C for at least 15 minutes.
Pellet the DNA by centrifuging at 18,000g for 20 minutes at 4°C.
Wash the pellet with 1 ml of 70% ethanol and spin at 18,000g for 15 minutes at 4°C.
Aspirate the supernatant and air dry the pellet briefly.
Resuspend both pellets in a total volume of 200 μl of 1× TE buffer pH 8.0. Dissolve the DNA by incubating at 37°C for 15 minutes.
Prepare a 0.8% agarose/0.5× TBE gel containing 0.5 μg/ml ethidium bromide.
Load 1 μl of the control 3C template, 4 μl of digested BAC DNA from step 12 and a molecular weight standard of known concentration (Fig 2D).
After running the gel, estimate the control 3C template concentration by comparing the intensities to the molecular weight standard (see Note 29).
Aliquot the control 3C template and store it up to at least 2 years at -20°C.
3.3 Quality control of 3C and control templates
Prepare 8-12 twofold serial dilutions of the 3C template starting with 250 ng/μl and ending with a “no template” control. Do the same for the control 3C template starting with a dilution of around 5 ng/μl . The minimum volume of each dilution is 8 μl. Every dilution is used for two separate PCR reactions each containing a specific pair of PCR primers designed to detect a 3C interaction between fragments that are either nearby or far apart on the linear genome (see Note 5).
PCR reactions are set up as follows: 4 μl 3C template dilution, 2.5 μl 10× PCR buffer, 2 μl 50mM magnesium sulfate, 0.2 μl 25 mM dNTP mix, 0.125 μl 80 μM 3C primer 1, 0.125 μl 80 μM 3C primer 2, 0.2 μl 5 U/μl Taq DNA polymerase and 15.85 μl water.
Amplify the DNA products using the following PCR parameters: 1 cycle 5 minutes at 95 °C; 35 cycles 30 seconds at 95°C followed by 30 seconds at 65°C followed by 30 seconds at 72°C; 1 cycle 30 seconds at 95°C followed by 30 seconds at 65°C followed by 8 minutes at 72°C.
Add 8 μl of 4× DNA loading buffer to each PCR reaction and mix by pipetting. Analyze 14 μl of each sample on a 1.5% agarose/0.5× TBE gel containing 0.5 μg/ml ethidium bromide.
Quantify the PCR products using a gel documentation system and plot the quantity of PCR product versus the amount of 3C template used as input material (Fig 2C-D and Fig 2E-F) (see Note 30).
3.4 5C primer design
Two types of 5C primers should be designed: forward 5C primers that anneal directly upstream of the restriction site of the 3C ligation product and reverse 5C primers that anneal exactly downstream of it (Fig 3). Usually, the 5’ half of the restriction site is included at the 3’ end of the forward primer and the 3’ half of the restriction site is incorporated at the 5’ end of the reverse primer (see Note 31 and Note 32).
Only one primer (either a forward or a reverse) is designed per restriction fragment. 5C can only detect a 3C ligation product recognized by a combination of a forward primer and a reverse primer. Therefore, one should carefully decide for which fragments forward primers should be designed and for which fragments reverse primers.
Typically, the sequence specific part of the primers is around 40 nucleotides long. The annealing temperature should be adjusted to ~72°C. Primers with a too low annealing temperature should be discarded. If the annealing temperature of a primer is too high, specific nucleotides should be removed from the 5’ end of forward primers and from the 3’ end of reverse primers till the annealing temperature is ~72°C. The removed nucleotides should be replaced by random nucleotides, so that the total length of each primer remains exactly the same.
It is recommended to exclude primers that anneal to repetitive sequences in the genome, since they are likely to generate excessively large amounts of ligation products (see Note 33).
To ensure the ligation of 5C products, all reverse primers should be modified with a phosphate group at the 5’ end of the primer. This can be done either during synthesis of the individual primers or by phosphorylating a pool of reverse primers with PNK (see below for protocol).
All 5C ligation products are simultaneously amplified with two universal PCR primers. Therefore, common tails should be added to both the 5’ end of 5C forward primers and the 3’ end of 5C reverse primers (Fig 3). Typically, the T7 sequence is used for the forward primer tails and the complementary T3 promoter sequence is applied to the reverse primer tails.
Order the primers as 50 μM stock solutions in 1× TE pH 8.0.
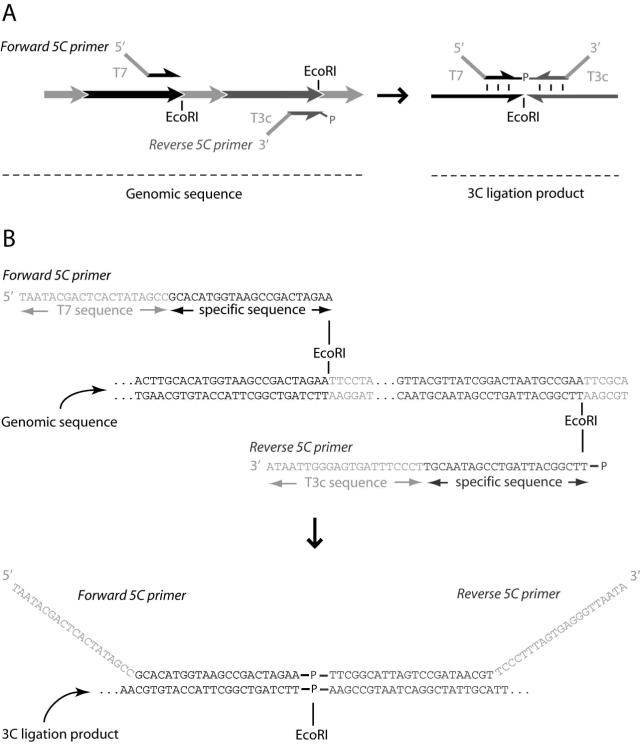
Figure 3.
5C primer design. (A) Diagram and (B) example of 5C primer design. Both forward and reverse 5C primers are designed on the 5’ end of a restriction site in the genomic sequence and include 3 bases of the restriction site. Note, the forward and reverse 5C primers are designed on opposite strands in the genomic sequence (A left, B top), but anneal to the same strand in the 3C template (A right, B bottom). Universal tails are added to the specific sequence of the 5C primers. The T7 sequence is added to the 5’ end of forward 5C primers. The complement of the T3 sequence (T3c) is added to the 3’ end of reverse 5C primers. Only reverse 5C primers are phosphorylated at the 5’ end.
3.5 Preparation of a 5C primer pool
Pool all forward primers in equimolar amounts (see Note 34).
Make a separate pool of all reverse primers.
- If the reverse primers were synthesized without a 5’ phosphate group, perform the following phosphorylation reaction:
- - Add 10 μl PNK buffer, 10 μl 10 mM ATP and 10 μl l PNK to 70 μl reverse primer pool.
- - Incubate at 37°C for 30 minutes.
- - Inactivate PNK by incubating the sample at 65°C for 10 minutes.
Add the forward primers to the reverse primers in a way that all individual primers are present at equimolar amounts.
Aliquot the 5C primer pool and store it at −20°C.
3.6 Preparation of a 5C and control library
Prepare 12 reactions each containing an amount of 3C template that corresponds to ~100,000 genome copies (see Note 35 and Note 36). These 12 reactions include 10 5C reactions for making a 5C library, 1 “no primer” control and 1 “no ligase” control.
Adjust the total DNA quantity in each reaction to 1.5 μg with 1 μg/μl SSD.
Add 1 μl of the appropriate 5C primer pool dilution to each 5C annealing reaction except the “no primer” control (see Note 37).
Add 2 μl of 5C annealing buffer and adjust the final volume to 20 μl with water.
Denature the 3C template and primers by incubating the samples at 95°C for 5 minutes.
Anneal the primers to the 3C template by incubating the samples at 55°C for 16 hours.
Add 20 μl 1× Taq ligase buffer to the “no ligase” control.
Add 20 μl 1× Taq ligase buffer containing 10 units of Taq DNA ligase to all other samples.
Mix by pipetting and continue the incubation at 55°C for 1 hour.
Inactivate the reactions by incubating the samples at 65°C for 10 minutes.
Split each reaction into 4× 6 μl and set up the following PCR reactions: 6 μl 5C ligation product, 2.5 μl 10× PCR buffer II, 1.8 μl 25 mM magnesium chloride, 0.2 μl 25 mM dNTP mix, 0.5 μl 80 μM T7, 0.5 μl 80 μM T3, 0.225 μl AmpliTaq Gold DNA polymerase and 13.275 μl water. Include a water control for the PCR (see Note 38).
Amplify the DNA products using the following PCR parameters: 1 cycle 9 minutes at 95 °C; 24 cycles 30 seconds at 95°C followed by 30 seconds at 65°C followed by 30 seconds at 72°C; 1 cycle 30 seconds at 95°C followed by 30 seconds at 65°C followed by 8 minutes at 72°C.
Pool the PCRs from the samples for the 5C library.
Pool the PCRs from the “no ligase” control and make a separate pool of the PCRs from the “no primer” control.
Run aliquots from the 5C library, the “no ligase” and “no primer” controls and the water control on a 2% agarose/0.5× TBE gel containing 0.5 μg/ml ethidium bromide.
If the control lanes are empty and a clean single band is observed for the 5C library, continue with purifying the 5C library using the MinElute PCR Purification Kit.
Analyze serial dilutions of the purified 5C library and a molecular weight standard of known concentration on a 2% agarose/0.5× TBE gel containing 0.5 μg/ml ethidium bromide.
After running the gel, estimate the 5C library concentration by comparing the intensity to the molecular weight standard.
Aliquot the 5C library and store it at ×20°C.
3.7 Quality control of a 5C library: nested PCR
Prepare 8-12 twofold serial dilutions of the 5C library starting with 2-10 ng/μl and ending with a water (no template) control. Do the same for the control 5C library. The minimum volume of each dilution is 12 μl. Every dilution is used for two separate PCR reactions each containing a specific pair of PCR primers. Each pair of PCR primers is designed to detect a 5C ligation product representing an interaction between fragments that are either nearby or far apart on the linear genome (see Note 8).
Set up the following PCR reactions for each dilution: 6 μl 5C library dilution, 2.5 μl 10× PCR buffer, 2 μl 50mM magnesium sulfate, 0.2 μl 25 mM dNTP mix, 0.125 μl 80 μM 5C nested primer 1, 0.125 μl 80 μM 5C nested primer 2, 0.2 μl 5 U/μl Taq DNA polymerase and 13.85 μl water.
Amplify the DNA products using the following PCR parameters: 1 cycle 5 minutes at 95 °C; 20 cycles 30 seconds at 95°C followed by 30 seconds at 56°C followed by 30 seconds at 72°C; 1 cycle 30 seconds at 95°C followed by 30 seconds at 56°C followed by 8 minutes at 72°C.
Add 8 μl of 4× DNA loading buffer to each PCR reaction and mix by pipetting. Analyze 14 μl of each sample on a 2% agarose/0.5× TBE gel containing 0.5 μg/ml ethidium bromide.
Quantify the PCR products using a gel documentation system and plot the relative quantity of PCR product versus the amount of 5C library used as input material (Fig 4) (see Note 39).
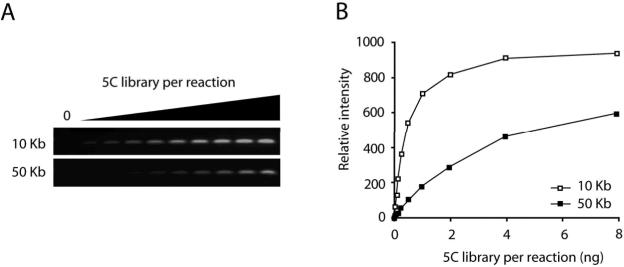
Figure 4.
Quality control of a 5C library by nested PCR. (A) Agarose gel analysis and (B) quantification of a 5C library titration in a nested PCR. Increasing amounts of 5C library were analyzed with two nested primer pairs. One primer pair interrogates an interaction between fragments that are close (10 Kb) to each other in the linear genome. The other primer pair examines the interaction between two more distant (50 Kb) fragments. A titration curve should demonstrate a linear increase at the beginning of the curve and should reach a plateau with increasing amounts of 5C library. The curve of the PCR product testing the interaction between two adjacent restriction fragments should be above the curve of the PCR product testing the interaction between two, more distant fragments. Note, when using a control 5C template, these two titration curves should be more similar.
3.8 Quality control of a 5C library: cloning and sequence analysis
To ensure that none of the 5C primers is misbehaving in the assay, take an aliquot of the purified 5C library and clone the PCR products using the TOPO TA cloning kit.
Inoculate 100 colonies and isolate the plasmids containing the 5C ligation products using a Qiaprep Spin Miniprep kit.
Sequence the inserts using the \m=-\21 M13 sequencing primer and analyze the sequences (see Note 40).
The 5C library is ready for deep sequencing or microarray analysis, if the results from the nested PCR show that the 5C library is a reliable copy of the 3C template and when the sequence analysis of the 5C library does not identify problematic primers. We recommend acquiring specific instructions regarding sample preparations for the particular microarray platform or sequencing platform that will be used.
3.9 Normalization and analysis of the results
Both the number of sequence hits and the intensity on the microarray are a measure for the amount of 5C ligation product in the 5C library. The abundance of specific 5C ligation products in the 5C library is proportionate to the frequency with which the two corresponding restriction fragments interact inside the nucleus. Interaction frequencies are calculated by dividing the amount of a specific 5C ligation product in the 5C library by the amount of the same 5C product in the control 5C library. This ratio is a direct measure for the interaction incidence, and is normalized for any differences in primer efficiency and 5C ligation product amplification efficiency. However, the ratio remains an arbitrary unit, meaning that only the frequencies obtained within a single 5C experiment can be directly compared. To be able to compare interaction frequencies from different 5C experiments, one has to use a common internal control to normalize the different datasets. This internal control can be a set of interactions of which the frequencies are expected to be the same in the different 5C experiments. Including a set of interactions between fragments located within a conserved gene desert region on human chromosome 16 have been used successfully to normalize 5C experiments (15) (see Note 41).
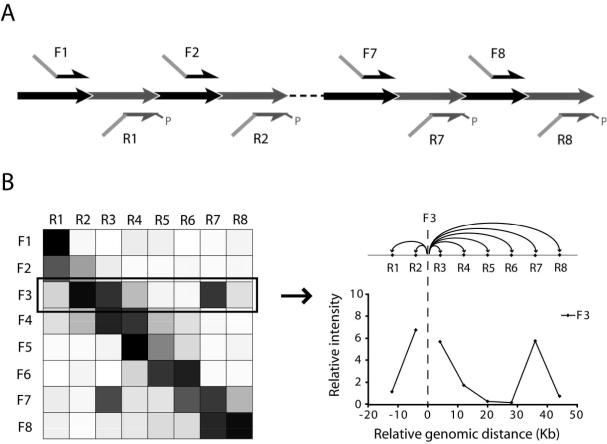
Due to the many-to-many set up, the 5C data is typically presented in a matrix (Fig 5). The color of each cell represents the measured amount of 5C ligation product formed by the corresponding forward and reverse 5C primers on the x- and y-axes. The resulting 5C heatmap can be regarded as a collection of 3C graphs. Consequently, the analysis of data from a 5C experiment is similar to analysis of 3C data (17). 3C graphs of individual 5C primers can be obtained by plotting the values of its corresponding column or row. Only background interactions are detected in the absence of a looping interaction. The interaction frequencies of background interactions are inversely correlated to the genomic distance between the restriction fragments. In the presence of a specific long range interacton, a local peak on top of the background interactions will be detected.
Figure 5.
Expected results from a 5C experiment using an alternating scheme. (A) Diagram of a 5C experiment using alternating forward and reverse 5C primers for consecutive restriction fragments. (B) Expected results are shown in a heatmap, where every box represents an interaction between two fragments corresponding to the primers indicated to the left and the top. The gray level of each box is a measure of interaction frequency as determined by 5C. White corresponds to no interaction, whereas black corresponds to a high interaction frequency. Interactions between neighboring fragments are usually strong and are represented by a diagonal across the heatmap. High interaction frequencies that are off the diagonal represent long range interactions. Data from individual column or row can be translated into a 3C graph for that particular primer.
Footnotes
4. Notes
Formaldehyde older than 6 months – 1 year will result in less efficient crosslinking and should not be used.
There are several criteria to take into consideration when selecting the restriction enzyme. Most importantly, the enzyme should cut efficiently under the conditions of the 3C protocol. The use of BamHI is not recommended, since it has proven to be less efficient under the specific 3C conditions. One can choose enzymes that recognize a 6-mer palindromic sequence and cut every ~4 kb (e.g., EcoRI, HindIII, BglII) or choose enzymes that cut more frequently (e.g., MseI). The choice of enzyme will also depend on the desired resolution and the distribution of the restriction sites within the region of interest.
Avoid multiple freeze-thaw cycles of buffers containing DTT. It is best to store these solutions in aliquots.
The use of bromophenol blue is not recommended, since this dye will run at the same position as the PCR products and will therefore interfere with DNA quantification.
For quality control, 3C templates should be titrated in a 3C PCR experiment. Typically, two head-to-head 3C ligation products are measured by semi-quantitative PCR and agarose gel quantification. One tested interaction should be between two adjacent or nearby restriction fragments (10 kb) and one should be between two, more distant fragments (50-80 kb). The 3C primers have to be designed unidirectionally along the linear genome and 50-150 bp upstream of the 3’ end of the predicted restriction fragment (8). Generally, the 3C primers are 28-32 bp long, have a GC content of approximately 50% and should preferably be unique as determined by BLAST.
Since the 5C technology is very sensitive to contaminations, we recommend to divide all reagents mentioned in section 2.6 into single use aliquots.
The PCR primers should be adjusted if other tail sequences were used in the design of the 5C primers. The primers should also be modified dependent on the detection method used in the assay. For microarray analysis, we recommend using a Cy3 labeled reverse primer to generate labeled antisense 5C products. For sequence analysis, the 5C libraries should be amplified with 5’-phosphorylated primers to allow ligation of linkers used for subsequent sequencing. We advise to consult your microarray or sequencing facility to decide on the proper modification for either analysis method.
For quality control, 5C libraries should be titrated in a nested PCR experiment. Typically, two 5C ligation products are measured by semi-quantitative PCR and agarose gel quantification. One 5C ligation product should correspond to an interaction between two adjacent or nearby restriction fragments (~10 kb) and one to an interaction between two more distant fragments (50-80 kb). The forward nested primer should be designed starting at the first specific nucleotide at the 3’ end of the common tail. The same holds true for the reverse nested primer on the reverse strand of the 5C ligation product. Generally, the 5C nested primers are 19-22 bp long and preferentially have a GC content of approximately 50%.
Cells grown in suspension should be pelleted gently by centrifuging at 300g for 10 minutes. Next, the cells should be resuspended in 45 ml fresh culture medium and the amounts of formaldehyde and glycine in the following 2 steps should be doubled.
It is essential to crosslink the cells for exactly 10 minutes. Shorter incubation times will result in lower detection signals of chromatin interactions, whereas longer incubation times will cause too many crosslinks resulting reduced digestion efficiency.
The experiment can be paused at this point by incubating the cell pellet on dry ice for 20 minutes and storing the pellet at −80°C. Pellets can be stored at −80°C for at least one 1 year.
If fewer cells were used for crosslinking, the volumes in this 3C template protocol should be adjusted accordingly.
The cell lysate should not be viscous. Viscous lysates are caused by insufficient crosslinking due to using formaldehyde that is too old (see Note 1).
Try to avoid sedimentation of the suspension to prevent uneven distribution of the chromatin.
Air bubbles will make it more difficult at a later stage to quench the SDS with Triton.
Longer incubation at 65°C will cause reversal of the crosslinking of the chromatin interactions and should be avoided.
The mixture should appear slightly granular.
The solution should be clear at this stage.
The second proteinase K digestion step increases the 3C template yield after phenol/chloroform extraction.
Both the phenol and the aqueous phase can appear cloudy. The DNA accumulates close to the interface during the first extraction. Take off as much material as possible without transferring any interface material.
The supernatant and interface should both be clear at this stage. If not, perform another phenol pH8.0:chloroform extraction.
Dilution of the solution helps to reduce the amount of salt precipitating in the next step.
The tubes can also be left at −80°C overnight and the protocol can be continued the next day.
PCR reactions with 3C templates are inhibited by residual salt in the template. Thorough desalting is required. Desalting columns are not recommended, since they often “size fractionate” the DNA and can change the nature of the samples.
Typically, the 3C template concentration is around 200 to 250 ng/μl. The 3C template should run as a fairly tight band of more than 10 kb. A DNA smear indicates poor ligation efficiency and material trapped in the wells indicates incomplete DNA digestion. Very little RNA should be present.
It is recommended to use a control 3C template of randomly ligated fragments to create a control 5C library. This is used to correct for differences in annealing efficiencies of the 5C primers and slight amplification biases of the different 5C ligation products in a normal 5C library. For small genomes (e.g., yeast), a control 3C template can be generated by digesting and randomly ligating whole genomic DNA (10). However, the complexity of the ligation mixture resulting from a larger genome (e.g., human) becomes too high to reliably detect individual ligation products. Therefore, a control template should be made by using equimolar amounts of a set of minimally overlapping BAC clones that covers the genomic region of interest.
To determine accurate relative concentrations, perform a real-time quantitative PCR with primers recognizing a common BAC vector region. Agarose gel detection is generally not sufficiently accurate for this purpose.
The volume of the added restriction enzyme should not exceed 10% of the total volume, because the glycerol in the enzyme storage buffer will inhibit enzymatic activity.
Typically, the control 3C template concentration is around 100 ng/μl. Digested BAC DNA appears as a smear of DNA fragments with the larger bands migrating around 10 kb. Ligated BAC DNA should appear as a tight smear migrating just above 10 kb.
A titration curve should demonstrate a linear increase at the beginning of the curve and should reach a plateau with increasing amounts of 3C template. The curve of the PCR product testing the interaction between two adjacent restriction fragments should be above the curve of the PCR product testing the interaction between two, more distant fragments. This validates that the relative amounts of 3C ligation products in the 3C template are consistent with the actual interaction frequencies of the chromatin fragments in vivo. In contrast, both curves will be much more similar for a control 3C template. If excess salt is still present in the 3C template sample, the curve will show an irregular linear phase and /or appear bi-phasic and it is recommended to reprecipitate the 3C template and to extensively wash the DNA pellet with 70% ethanol. The 3C template should be titrated again after reprecipitation.
It is recommended to first read the complete protocol before starting with the 5C primer design.
Importantly, forward and reverse primers will anneal to the same strand of the 3C ligation product formed by a head-to-head ligation of two restriction fragments. Thus, the forward and reverse primers are designed on the 3’ ends of restriction fragments so that they will anneal to different strands on the regular genomic sequence. This approach will prevent detection of partial digestion products and of ligation products resulting from self-circularization of restriction fragments. The latter occurs very frequently in the generation of the 3C library.
5C is much less sensitive to fluctuations in primer efficiency than regular PCR because all amplicons are equal in size and are amplified using a single universal PCR primer pair. In addition, they are designed with equal annealing temperatures. However, it is recommended to make a 3C control template to correct for any differences in annealing efficiencies of the 5C primers and slight amplification biases of the different 5C ligation products in a normal 5C library (see 3.2).
The total primer concentration will remain 50 μM. To calculate the individual primer concentration, divide the total primer concentration by the number of primers in the pool.
For the human genome the amount corresponding to ~100,000 genome copies is ~400 ng. The amount of control 3C template used in the 5C reaction should be determined by a titration experiment (see Note 36).
We recommend performing a (control) 3C template titration experiment before preparing the (control) 5C library. Prepare serial dilutions of the 3C template. Include a “no ligase” control with the highest 3C template concentration as well as a “no template” control and perform the 5C reactions according to the protocol. A titration curve should demonstrate a linear increase at the beginning of the curve and should reach a plateau with increasing amounts of 3C template. The amount of 3C template that will be used to make the 5C library should be in the linear range of the curve.
Typically, 1 fmol of each individual 5C primer is added to the reaction. We recommend performing a primer titration experiment before preparing the 5C library. Prepare serial dilutions of the 5C primer pool. Include a “no ligase” control with the highest primer concentration as well as a “no primer” control at the end and perform the 5C reactions according to the protocol. A titration curve should demonstrate a linear increase at the beginning of the curve and should reach a plateau with increasing amounts of 5C primer pool. The optimal primer concentration is the concentration that corresponds to the point where the curve just reached the plateau. A peak in the curve could be an indication that the amplification primers are quenched in the PCR reaction. This can be resolved by decreasing the number of PCR cycles or by increasing the amount of T7 and T3 primers.
To avoid any artifacts of primers annealing to each other and subsequently being ligated by residual activity of Taq ligase, it is essential to setup the PCR reactions immediately after step 10 and not to store the ligation reactions at 4°C.
A titration curve should demonstrate a linear increase at the beginning of the curve and should reach a plateau with increasing amounts of 5C library. The curve of the PCR product testing the interaction between two adjacent restriction fragments should be above the curve of the PCR product testing the interaction between two, more distant fragments. This confirms that the 5C ligation products in the 5C library form an accurate copy of the 3C template and hence the interaction frequencies in vivo. In contrast, both curves should be more similar for a control 5C library.
Pay attention to individual 5C primer sequences that are overrepresented in the cloned inserts. This could indicate that this primer is somehow misbehaving in the assay. 5C ligation products corresponding to interactions of nearby fragments are expected to be slightly overrepresented in a 5C library. However, in a control 5C library every 5C ligation product should be equally present. If problematic primers are identified by sequence analysis, a new 5C primer pool should be made without the troublesome primers.
Including an internal control has consequences for the experimental setup. First of all, BAC clones covering the region of the internal control are added in equimolar amounts to the BAC clones of the region of interest prior to making a control 3C template. Second, 5C primers are designed for the region of the internal control. Typically, an alternating format is used for the design of these 5C primers.
5. References
- 1.ENCODE-consortium Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dekker J. Gene regulation in the third dimension. Science. 2008;319:1793–1794. doi: 10.1126/science.1152850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and Interaction between Hypersensitive Sites in the Active beta-globin Locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 5.Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- 6.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 7.Sexton T, Schober H, Fraser P, Gasser SM. Gene regulation through nuclear organization. Nat. Struct. Mol. Biol. 2007;14:1049–1055. doi: 10.1038/nsmb1324. [DOI] [PubMed] [Google Scholar]
- 8.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing Chromosome Conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 9.Splinter E, Grosveld F, de Laat W. 3C technology: analyzing the spatial organization of genomic loci in vivo. Methods Enzymol. 2004;375:493–507. doi: 10.1016/s0076-6879(03)75030-7. [DOI] [PubMed] [Google Scholar]
- 10.Miele A, Gheldof N, Tabuchi TM, Dostie J, Dekker J. Mapping chromatin interactions by Chromosome Conformation Capture (3C) In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. Supplement 74. John Wiley & Sons; Hoboken, N.J.: 2006. pp. 21.11.1–21.11-20. [DOI] [PubMed] [Google Scholar]
- 11.Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 12.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 13.Bacher CP, Guggiari M, Brors B, et al. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol. 2006;8:293–299. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- 14.Xu N, Tsai CL, Lee JT. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311:1149–1152. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- 15.Dostie J, Richmond TA, Arnaout RA, et al. Chromosome Conformation Capture Carbon Copy (5C): A Massively Parallel Solution for Mapping Interactions between Genomic Elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dostie J, Dekker J. Mapping networks of physical interactions between genomic elements using 5C technology. Nat. Protoc. 2007;2:988–1002. doi: 10.1038/nprot.2007.116. [DOI] [PubMed] [Google Scholar]
- 17.Dekker J. The 3 C's of Chromosome Conformation Capture: Controls, Controls, Controls. Nat Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]