Abstract
An enzyme of unknown function within the amidohydrolase superfamily was discovered to catalyze the hydrolysis of the universal second messenger, cyclic-3’, 5’-adenosine monophosphate (cAMP). The enzyme, which we have named CadD, is encoded by the human pathogenic bacterium Leptospira interrogans. Although CadD is annotated as an adenosine deaminase, the protein specifically deaminates cAMP to cyclic-3’, 5’-inosine monophosphate (cIMP) with a kcat/Km of 2.7 ± 0.4 × 105 M−1 s−1 and has no activity on adenosine, adenine, or 5’-adenosine monophosphate (AMP). This is the first identification of a deaminase specific for cAMP. Expression of CadD in Escherichia coli mimics the loss of adenylate cyclase in that it blocks growth on carbon sources that require the cAMP-CRP transcriptional activator complex for expression of the cognate genes. The cIMP reaction product cannot replace cAMP as the ligand for CRP binding to DNA in vitro and cIMP is a very poor competitor of cAMP activation of CRP for DNA binding. Transcriptional analyses indicate that CadD expression represses expression of several cAMP-CRP dependent genes. CadD adds a new activity to the cAMP metabolic network and may be a useful tool in intracellular study of cAMP-dependent processes.
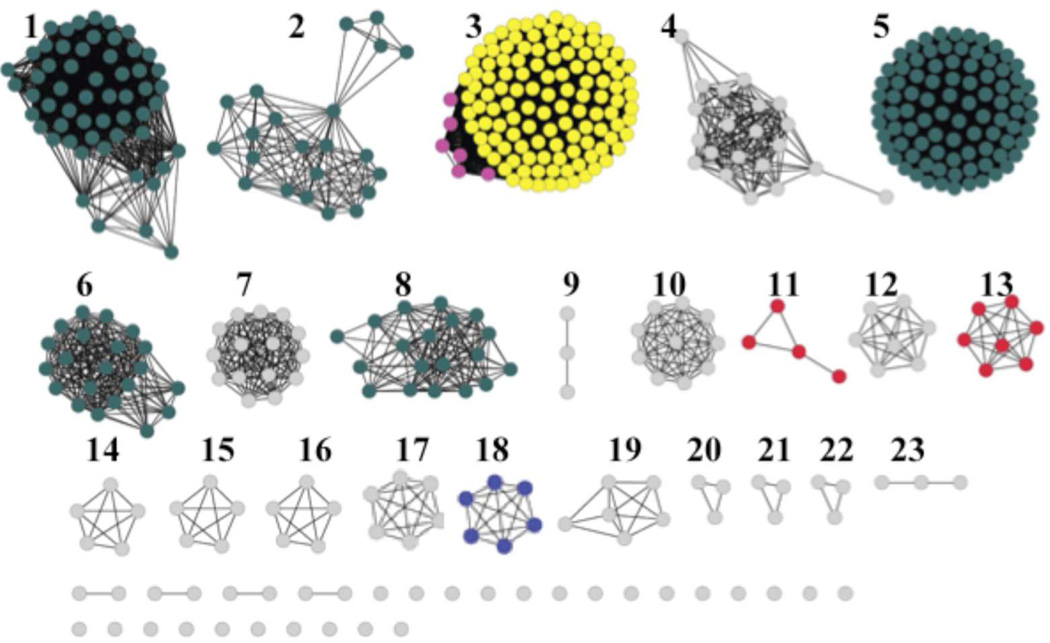
The amidohydrolase (AHS) enzyme superfamily is a well-characterized group of enzymes first identified by Holm and Sander based on the similarities in the three-dimensional structures of phosphotriesterase, adenosine deaminase and urease 1. The proteins of the AHS family fold into a distorted (β/α)8-barrel with conserved metal binding residues at the C-terminal ends of β-strands 1, 4, 5, 6 and 8. These enzymes have either mononuclear or binuclear metal centers which activate a hydrolytic water for nucleophilic attack and are involved in the metabolism of amino acids, sugars, nucleic acids, and organophosphate esters 2. NCBI has divided the AHS into 24 clusters of orthologous groups (COGs) and annotated all bacterial proteins in COG1816 as enzymes that catalyze the deamination of adenosine. The structure and mechanism of action of adenosine deaminases from several organisms have been studied and the sequence similarity network3, for COG1816 is given in Figure 1. Enzymes experimentally verified as adenosine deaminases are found in Group 5, which contains the Escherichia coli K-12 adenosine deaminase (locus tag: b1623). Additional adenosine deaminases are found in Groups 1, 2, 6, and 8. The essential residues for adenosine deaminase activity 2 include an HxHxD motif following β-strand 1, an aspartate following β-strand 3, a glycine following β-strand 4, an HxxE motif following β-strand 5, a histidine following β-strand 6, and a pair of aspartates following β-strand 8 2. Additional functions found in COG1816 include adenine deaminase and cytokinin deaminase in Group 3. Despite an overall amino acid sequence identity of ~30% to E. coli adenosine deaminase, the proteins of Group 18 cannot deaminate adenosine. We have identified a protein of Group 18 of COG1816 that specifically deaminates the signaling molecule cAMP to cyclic-3’, 5’-inosine monophosphate (cIMP) and lacks activity on adenosine. We have named this enzyme CadD for cyclic adenylate deaminase.
Figure 1.
Sequence similarity network created using Cytoscape (www.cytoscape.org) of COG1816 from the amidohydrolase superfamily. Each node in the network represents a single sequence, and each edge (depicted as lines) represents the pairwise connection between two sequences at a BLAST E-value of better than 1 × 10−70. Lengths of edges are not significant, except for tightly clustered groups, which are more closely related than sequences with only a few connections. Groups 1, 2, 5, 6, and 8 contain the adenosine deaminase enzymes including the E. coli adenosine deaminase in Group 5. In Group 3, the sequences depicted in yellow are adenine deaminases whereas the pink sequences represent cytokinin deaminase enzymes. Group 18 (blue) contains CadD, the enzyme studied in this investigation. All proteins of Group 18 are from Leptospira strains.
The universal signaling molecule cAMP is found in all three domains of life where it plays key roles as a secondary messenger that transmits signals to regulate physiological processes such as sugar and lipid metabolism, cardiac function, cell growth and differentiation 4:Antoni, 2012 #11, 5. Action of cAMP is mediated by binding to a variety of cAMP receptor proteins. These receptor proteins are divided into four main classes: cyclic nucleotide-dependent kinases, cyclic nucleotide-gated ion channels, guanine nucleotide exchange factors and bacterial cAMP-dependent transcription factors (Crp) 4. In the case of Crp proteins cAMP binding results in a large well-understood allosteric transition that allows binding to specific DNA sequences 6–8. The deamination product of cAMP, cIMP, has been reported as a compound released into the growth medium by cultures of Corynebacterium murisepticum and Microbacterium sp. 205 9, 10 and a component of mammalian tissue culture cells 11, although in each of these systems the means of cIMP production are unknown. Deamination of cAMP by a broad specificity fungal adenosine deaminase 12 and in toad bladder extracts 13 has been reported. This work describes the discovery and in vivo function of a cAMP deaminase from the human pathogen, Leptospira interrogans 14, a bacterium transmitted by zoonotic contact that can cause potentially fatal diseases such as renal failure and pulmonary hemorrhage 15.
RESULTS AND DISCUSSION
In vitro specificity of CadD
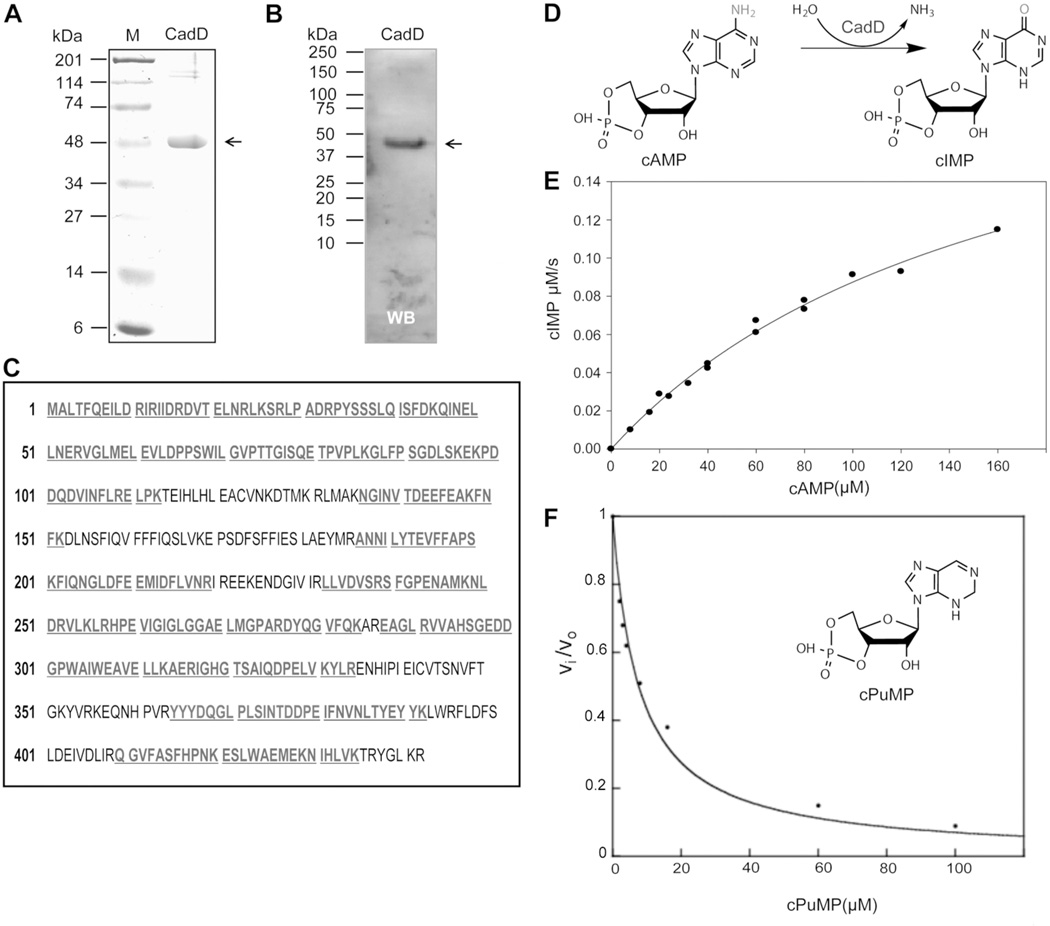
CadD was readily expressed at high levels in E. coli and purified to homogeneity. The protein was screened for deaminase activity against a library of compounds (Methods) and showed activity only with cAMP. CadD deaminated cAMP to cIMP (Figure 2) with values of kcat of 46 ± 4 s−1, Km of 170 ± 20 µM and a kcat/Km of 2.7 ± 0.4 × 105 M−1 s−1. The rate constants for deamination of adenosine, adenine and AMP were <0.01 s−1. Purified CadD contained 0.4 equivalents of Zn2+ and 0.2 equivalents each of Cu2+ and Fe2+. Purine riboside-3’, 5’-cyclic monophosphate (cPuMP) was found to be an inhibitor of CadD activity with an inhibition constant of 1.3 ± 0.5 µM (Figure 2).
Figure 2. CadD purification, identification and enzymatic properties.
Panels A and B. SDS-polyacrylamide gel analyses of purified CadD protein with detection by either Coomassie Blue staining or by western blotting with an anti-hexahistidine antibody. Panel C. Mass spectral identification of the recombinant CadD protein. The peptide fragments matching the database sequence are given in bold and underlined type. Panel D. The CadD reaction. Panel E. CadD deamination of cAMP. The CadD concentration was 5 nM. Panel E. Inhibition of CadD by cPuMP (inset). Inhibition by cPuMP of CadD activity in 25 mM potassium HEPES (pH 7.5). The solid line represents the fit of the data to equation 2.
In vivo specificity of CadD
High levels of CadD activity were readily obtained upon expression in E. coli and thus the protein was not noticeably toxic to growth as expected for an enzyme of narrow specificity. This lack of toxicity allowed us to test if the activity and specificity observed for CadD in vitro extended to an in vivo situation. The cAMP receptor protein (CRP) complex (cAMP-CRP complex) is a global regulator that generally acts as an transcriptional activator in modulating expression of genes involved in various metabolic pathways in E. coli and other bacteria 16–18. DNA binding by CRP requires that the protein first bind cAMP 19, 20. When DNA-bound the cAMP-CRP complex interacts with the α subunit of RNA polymerase to allow transcription to proceed20. The classical role of the cAMP-CRP complex is in determining the “pecking order’ of carbon source utilization where glucose has the highest status 17. For example when E. coli is grown in a mixture of glucose and lactose, the lactose utilization genes (e.g., lacZ encoding β-galactosidase) are not induced and lactose is not catabolized until the glucose has been exhausted. This is because expression of the lactose operon requires the cAMP-CRP complex to activate transcription even when the lactose repressor is inactive and glucose grown cells have very low cAMP levels 17. Inactivation of the genes encoding either the DNA binding protein, CRP, or the cAMP synthetic enzyme, adenylate cyclase (crp and cyaA, respectively), results in strains unable to grow with lactose, other sugars such as maltose and arabinose or low status carbon sources such as fatty acids as sole carbon and energy source 16. The in vitro activity of CadD predicted that its expression in a wild type E. coli strain would block growth on lactose, maltose and arabinose and thereby mimic a cyaA (or crp) mutant strain 16. Corollaries to this prediction are that intracellular cAMP levels should decline upon CadD expression and that cIMP should be unable to replace cAMP as the ligand required for CRP binding to DNA.
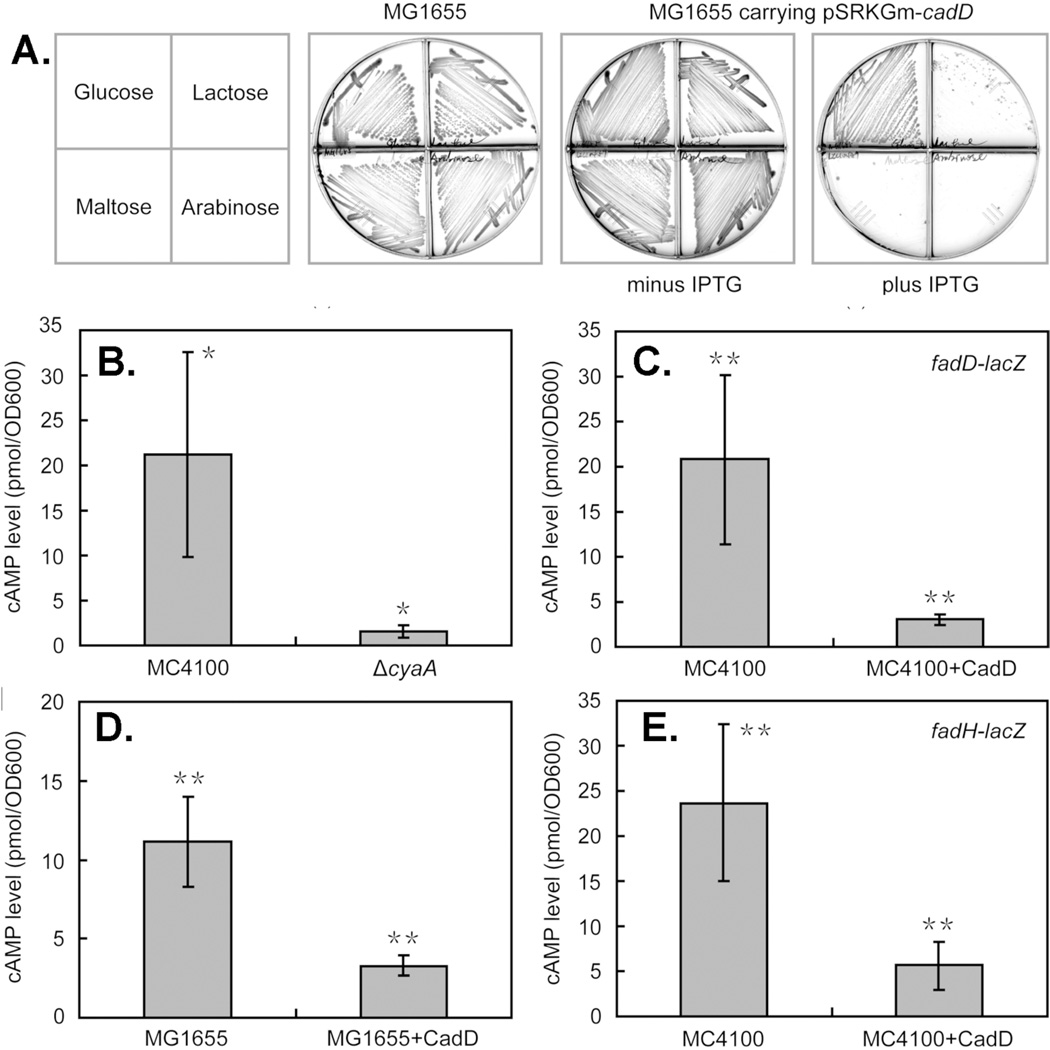
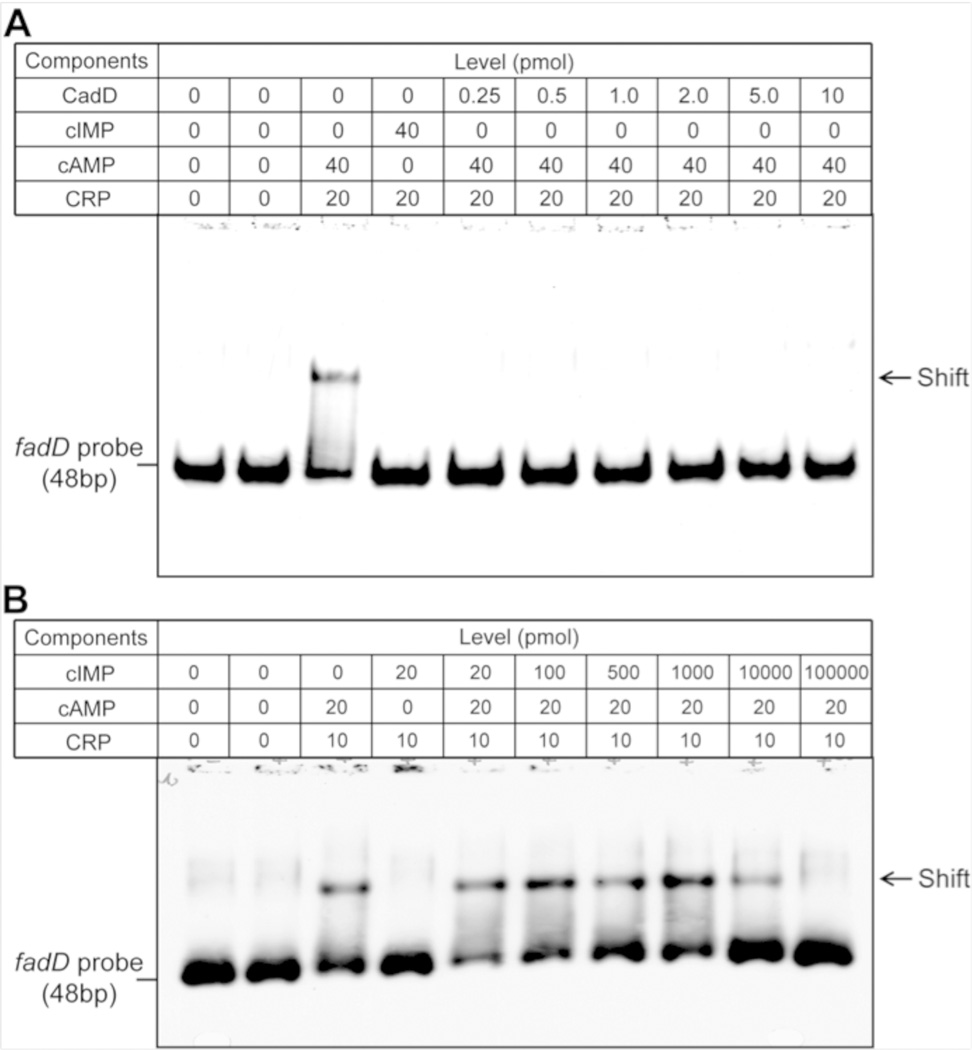
All three predictions have been satisfied. Induction of CadD expression in the wild type strain MG1655 blocked growth on lactose, maltose and arabinose but not on glucose whereas cultures lacking inducer grew well on all four sugars (Figure 3). Moreover, CadD expression resulted in severe decreases in intracellular cAMP levels as expected from the carbon source data (Figure 3). The third prediction was buttressed by an early report that cIMP is a poor replacement for cAMP in allowing CRP to bind DNA 21. However, the DNA used was a synthetic copolymer that bound the cAMP-CRP complex poorly and thereby resulted in an insensitive assay of poor precision. Other early work was based on an indirect in vitro system in which cIMP was reported to be unable to replace cAMP 22. However, no details or data were given. To obtain quantitative data using purified CRP and a DNA containing a known cAMPCRP complex binding site, we performed electromobility shift assays of the binding reaction and found that cIMP addition failed to allow CRP to the bind DNA even at twice the concentration that sufficed for cAMP (Figure 4). Moreover, addition of purified CadD to the reactions containing both cAMP and CRP blocked formation of the ternary complex with DNA. A final question was whether or not cIMP could effectively compete with the cAMP-CRP complex for DNA binding. Competition was seen only when cIMP was in 500-fold excess over cAMP and a 5000-fold excess of cIMP was needed to abolish DNA binding (Figure 4).
Figure 3. CadD expression mimics adenylate cyclase deficiency.
Panel A. Growth of E. coli wild type strain MG1655 on various carbon sources in the presence or absence of CadD expression. Glucose does not require cAMP for its utilization whereas cAMP is required for expression of the genes encoding the proteins required to utilize the other sugars. IPTG (0.5 mM) was added for induction of CadD expression. The plates are sectored by plastic walls to prevent cross-feeding. Panels B-E. The cytosolic levels of cAMP in the presence or absence of CadD expression were determined in various strains. For comparison panel B shows the effects of deleting cyaA, the gene encoding adenylate cyclase. * denotes p<0.05 whereas ** denotes p<0.01. Two E. coli K-12 backgrounds were used. Strain MC4100 was used in much of the early work on cAMP control, but is unable to grow on lactose or arabinose due to deletion of the lacZYA and araBAD operons, respectively. Hence, the wild type strain MG1655 was used to assay response to these sugars. Strain MC4100 was used for the LacZ fusion studies because its lacZYA deletion eliminates background β-galactosidase activity. The genomes of both strains have been fully sequenced and their cyaA and crp genes and flanking sequences are identical 34.
Figure 4. Effects of cAMP, cIMP and CadD on DNA binding by CRP protein.
Panels A and B are electrophoretic mobility assays done using 0.2 pmol of a dioxigenin-labeled fadD probe and CRP at the levels given in a total volume of 20 µL. In panel A the effects of cIMP and cadD were assayed whereas panel B tested the ability of cIMP to displace cAMP from CRP and thereby prevent DNA binding.
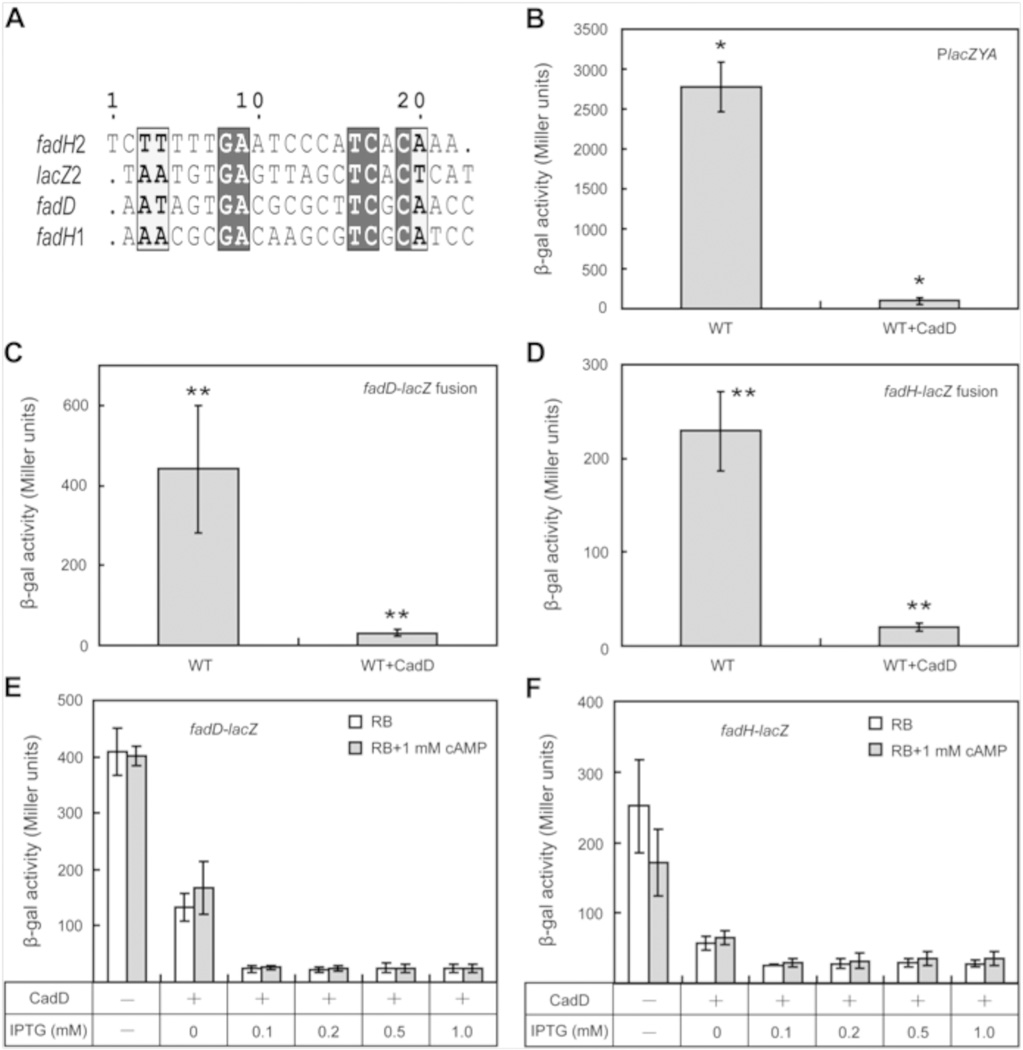
To bridge between the growth phenotypes and DNA binding assays we assayed the effects on CadD expression on transcription from three CRP-cAMP dependent promoters, those of lacZYA and two fatty acid β-oxidation pathway genes, fadD and fadH. Expression of the β-oxidation genes was measured by use of transcriptional fusions to lacZ constructed in the E. coli chromosome. Hence, expression of all three genes could be assayed by β-galactosidase activity. Transcription from all three promoters was strongly inhibited by expression of CadD (Figure 5). We attempted to reverse the inhibition by addition of exogenous cAMP which enters E. coli by an unknown and inefficient mechanism. However, this was largely unsuccessful, although a small increase in fadD transcription was seen when CadD was expressed at basal levels suggesting that at induced levels of CadD expression, cAMP deamination was more rapid than cAMP entry (Figure 5). Note that the growth rate and yield of E. coli expressing CadD on glucose as sole carbon source was essentially the same as the strain lacking the amidohydrolase, a finding consistent with the substrate specificity seen in vitro.
Figure 5. Effects of CadD expression on transcription of cAMP-CRP dependent genes.
Panel A. Alignment of the CRP binding sites. Panels B-D are the effects of CadD expression on transcription of lacZ and two fatty acid β-oxidation genes fabD and fabH. Panels E and F are the effect of CadD induction and exogenous cAMP addition on transcription of the fad genes. Expression of CadD and LacZ was induced by IPTG (0.5 mM) whereas the β-oxidation genes were induced by 5 mM oleic acid. * denotes p< 0.001 whereas ** denotes p< 0.0001. Strain MG1655 was assayed for lacZ whereas the fad strains were the chromosomal integrants described previously.
Binding of cAMP and cIMP to L. interrogans Cyclic Nucleotide Binding Proteins
Two of the four cloned putative L. interrogans cyclic nucleotide binding proteins produced soluble proteins that allowed further studies. As indicated by shifts in the denaturation temperature of the protein encoded by LIC10791 (UniProt:Q72U71) in the presence of cAMP, cAMP is capable of binding to and stabilizing the protein, but cIMP does not affect protein stability. The melting temperature of protein Q72U71 in the absence of ligand was 52.9 ± 0.4°C. Addition of cIMP gave at most only a slight shift in the melting temperature with a Tm of 53.0 ± 0.6°C whereas in the presence of cAMP the denaturation temperature of protein Q72U71 was shifted to 62.9 ± 0.7° C. In contrast, the protein encoded by LIC10237 (UniProt:Q72VQ9) did not show a significant shift in denaturation temperature in the presence of either cAMP or cIMP. When protein Q72U71 was analyzed by isothermal titration calorimetry, an association equilibrium constant of 6.1 × 105 M was observed for cAMP (Figure S2). In contrast when titrated with cIMP no binding isotherm was observed. Titration of protein Q72VQ9 with either cAMP or cIMP produced no binding isotherms indicating that the protein bound neither ligand.
Implications of cIMP
Conversion of cAMP to cIMP essentially inactivates the cyclic nucleotide as an E. coli signaling molecule and cIMP competes extremely weakly with cAMP for CRP binding (Figures 4 and 5). Indeed, given the numerous contacts between the ligand and protein in the crystal structures of the E. coli cAMP-CRP complex 6, 8, it is surprising that CRP binds cIMP so weakly since both salt bridges and eight out of ten hydrogen bonds of the cAMPCRP complex should be retained in the cIMP-CRP complex 7. It seems likely that the intersubunit hydrogen bonds formed between the adenosine amino group and the hydroxyl groups of Thr127 and Serl28 play a role in facilitating the other ligand-CRP interactions. The lack of DNA binding of the cIMP-CRP is readily explained by the fact that the cAMP amino group hydrogen bonds to Thr127 and Serl28 which are involved in a coil-to-helix transition that converts CRP into its DNA binding state 8. Indeed, although CRP binds cGMP binds more strongly than cIMP, the guanine base cannot make the key contacts with Thr-127 and Ser-128 and hence, like the cIMP-CRP complex, the cGMP-CRP complex remains in a conformation that precludes DNA-binding 8.
The conversion of cAMP to cIMP suggests the possibility that L. interrogans uses cIMP as a signaling molecule. As a preliminary test of this possibility the genes encoding four CRP homologues were expressed, two of which gave soluble proteins amenable to further analysis. One of these proteins bound cAMP, but not cIMP, whereas the other protein bound neither nucleotide. Note that the L. interrogans genome encodes 18 putative adenylate cyclases, the most of any known bacterial pathogen 23. This plethora of putative cAMP producing enzymes suggests an important function for cAMP in this bacterium. In a microarray study that assayed transcriptional regulation of genes upon shift from low osmolarity to a higher osmolarity increased transcription of locus LIC12327, which encodes a putative adenylate cyclase was observed 24. Such an osmotic shift has been associated with triggering of L. interrogans pathogenicity 24. Note that CadD may not be the only hydrolytic enzyme that L. interrogans has to manipulate cAMP levels. Locus LIC11606 encodes a protein that has all the conserved residues typical of bacterial phosphodiesterases 25, 26. However, we were unable to detect phosphodiesterase activity with cAMP, cGMP, cIMP or cCMP as substrates by NMR spectroscopy.
METHODS
Materials
All chemicals were purchased from Sigma-Aldrich except purine riboside-3’, 5’-cyclic monophosphate (cPuMP, Figure 2) which was purchased from Axxora. The PCR primers used are given in Table 1S.
Cloning of LIC10459 from Leptospira interrogans
The gene having locus tag LIC10459 encoding Uniprot protein Q72V44 was cloned from Leptospira interrogans serovar Copenhageni str. Fiocruz L1-130 genomic DNA 27 from the ATCC. Restriction sites for NdeI (overlapping the initiation codon) and HindIII (downstream of the termination codon) were inserted into the forward and reverse primers, respectively. The PCR product was purified with a PCR clean-up system (Qiagen), digested with NdeI and HindIII and ligated into vector pET30a(+) which had been previously digested with NdeI and HindIII. The LIC10459 termination codon was removed from the 3’-end to produce a C-terminal hexahistidine-tagged version of Uniprot protein Q72V44 that we have named CadD. The fidelity of gene amplification was confirmed by sequencing. The coding sequence of the pET30a construct was ligated into pSRKGm, a moderate copy number broad-host-range expression plasmid 28 in which it was expressed from an intact lacZYA promoter under stringent IPTG inducible control. The plasmid used to express and purify CRP and the fadD and fadH transcriptional reporter strains were described previously 29.
For expression and purification of CadD the CadD-pET30a(+)construct was transformed into BL21(DE3) competent cells. A single colony was used to inoculate a 5 mL overnight culture of LB medium containing 50 µg/mL kanamycin. Each overnight culture was used to inoculate 1 L of LB medium containing 50 µg/mL kanamycin and the cultures were grown at 37 °C and when an OD600 of 0.6 was reached were supplemented with 1.0 mM ZnCl2 and 50 µM isopropyl D-thiogalactopyranoside. At the time of induction, the temperature was lowered to 20 °C and shaken for 18 h prior to harvesting of the cells by centrifugation. The cells were resuspended in 20 mM potassium (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer (pH 7.5) containing 20 mM imidazole, 120 mM ammonium sulfate, 0.1 mg/mL phenylmethylsulfonyl fluoride and 0.5 mg/mL bovine pancreatic deoxyribonuclease I and then lysed by sonication. The soluble protein fraction resulting from centrifugation to remove cellular debris was loaded onto a HisTrap column (GE Healthcare) and eluted with a gradient of imidazole.
CadD Activity Screen
CadD (1 µM) was screened for deaminase activity in 20 mM potassium HEPES (pH 7.5) against a library of compounds by UV-Vis spectrometry. The compounds screened included adenosine, 2'-deoxyadenosine, 3'-deoxyadenosine, 2', 5'- deoxyadenosine, 5'-deoxyadenosine, 3', 5'-cyclic AMP, ADP, ATP, N6-methyl-2'- deoxyadenosine, zeatin riboside, 7-deazaadenine, 8-oxoadenosine, S-adenosyl-homocysteine, Sadenosyl- methionine, adenine, N-butyladenine, 6-methoxypurine, isoguanosine, benzyladenine, 2-chloroadenosine, 1-(6-amino-9H-purin-9-yl)-1-deoxy-N-ethyl-β-D-ribofuranonamide, 1-(4- nitrophenyl)-3-phenyl-1H-pyrazol-5-ol, N,N-dimethyladenine, 2-chloroadenine, N6- methyladenine, 2-dimethylaminoadenine, N6-ethenoadenine, 6-chloropurine, 6-methylpurine, 8- oxoadenine, 2,6-diaminopurine, 6-methylthiopurine, 6-mercaptopurine, cytosine, O6- methylguanine, 7-methylguanine, 6-deoxyaminofutalosine, puromycin, pterin, 5- methyldeoxycytidine, cytidine, 2'-deoxycytidine, 2,4-diaminopyrimidine, 4,6- diaminopyrimidine, toxopyrimidine, 4-aminopyrimidine, 2'-deoxyguanosine, 5'- methylthioadenosine, 5'-carboxylic acid adenosine, guanine, isoguanine, acetyl adenine, 2'- deoxy-3', 5'-diphosphoadenosine, 2'-deoxy-5'-AMP, 2'-AMP, 3'-AMP, 2', 5'- diphosphoadenosine, 3', 5'-diphosphoadenosine, acetyl-AMP, and cyclic-2', 3'-AMP. The substrate concentration was 80 µM in each case. Enzymatic activity was monitored through changes in absorbance between 240–300 nm on a SPECTRAmax Plus spectrophotometer (Molecular Devices).
Production of cIMP from cAMP by CadD
CadD was incubated with cyclic-3, 5’-AMP until the reaction was complete as confirmed by changes in the UV absorbance. Standards of cIMP and cAMP were loaded onto a C18 column in 20 mM ammonium acetate (pH 6.0) containing 5% buffer B. Compounds were eluted with a gradient of buffer B (20 mM ammonium acetate, pH 6.0, in 80% acetonitrile). After the standards had been run, the product of the CadD reaction with cAMP and was run followed by a run with cIMP alone. Samples of peak fractions detected by UV absorbance were dried and analyzed by ESI mass spectrometry. The cAMP peak fractions had an observed m/z of 328.022 for the M-H ion (theoretical m/z of 328.045 for C10H11N5O6P). The cIMP peak fractions had an observed m/z of 329.053 for the MH ion (theoretical m/z of 329.029 for C10H10N4O7P).
Expression of Putative L. interrogans Cyclic-Nucleotide Binding Proteins
The E. coli and M. tuberculosis CRPs, locus tags b3357 and Rv3676, respectively, were used to search the genome of Leptospira interrogans for cAMP-binding proteins. The search revealed 10 proteins having more than 20% sequence identity to the characterized CRPs. The LIC10791, LIC10268, LIC11484, and LIC10237 loci were chosen for cloning because the encoded proteins had the highest sequence similarities 25–30% identical residues) with the known CRP proteins.
The L. interrogans genes with locus tags LIC10459, LIC10791, LIC10268, LIC11484, encoding putative cyclic nucleotide binding proteins were PCR amplified from L. interrogans genomic DNA as described above. The PCR products were purified, digested with NcoI and HindIII, and ligated into vector pET-30a(+) digested with the same restriction enzymes. The proteins encoded by these constructs were obtained as described above for CadD. Only two of the constructs (those of LIC10237 and LIC10791) gave soluble proteins, C-terminal hexahistidine tagged versions of UniProt proteins Q72U71 and Q72VQ9, respectively.
Measurement of Kinetic Constants
Assays were conducted with substrate concentrations of 3–200 µM. Deamination of cAMP was monitored by following the decrease in absorbance at 269 nm. Difference extinction coefficients were calculated by subtracting the extinction coefficient of the product from the extinction coefficient of the substrate (Δε269 = 3430 M−1 cm−1). Assays were performed in 25 mM potassium HEPES (pH 7.5) at 30 °C. Inhibition of cAMP deaminase activity by cPuMP was determined by addition of 25 nM CadD to 180 #x000B5;M cAMP and concentrations of cPuMP ranging from 4 to 152 µM.
Fluorescence Monitoring of Thermal Denaturation
Proteins Q72U71 and Q72VQ9 were tested for binding of cAMP and cIMP by monitoring fluorescence in the presence of Sypro Orange (Invitrogen) 30 which is supplied as a 5000X concentrated solution in DMSO. In a 50 µL assay 10 µM protein, 200 µM ligand, 25 mM potassium HEPES (pH 7.5), 10X Sypro Orange, and 10% DMSO were combined and relative fluorescence units were monitored. Ten duplicate reactions were run simultaneously.
Isothermal Titration Calorimetry
Proteins Q72U71 (36 µM) and Q72VQ9 (50 µM) were titrated with cAMP (0.25 mM) or cIMP (1 mM) to determine the association equilibrium constants.
Metal Analysis
The metal content of the proteins was determined by ICP-MS 31. Protein samples for ICP-MS were digested with HNO3 by refluxing for ~45 minutes to prevent protein precipitation during the measurement. The protein concentration was adjusted to ~1.0 µM with 1% (v/v) HNO3.
Data Analysis
Steady state kinetic data were analyzed using Softmax Pro, version 5.4. Kinetic parameters were determined by fitting the data to equation 1 using the nonlinear least-squares fitting program in SigmaPlot 9.0. In this equation A is the substrate concentration, Km is the Michaelis constant, v is the velocity of the reaction and kcat is the turnover number. The inhibition constant for cPuMP was determined by fitting the plot of residual enzyme activity against inhibitor concentration to equation 2. In this equation, vi is the residual enzyme activity in the presence of cPuMP, vo is the enzyme activity in the absence of inhibitor, and I is the inhibitor concentration.
| (1) |
| (2) |
Electrophoretic Mobility Shift Assays
Binding of cAMP-CRP to the fadD promoter CRP binding site was measured essentially as previously reported 29 with minor modifications. The digoxigenin-labeled fadD probe (0.2 pmol) was incubated with CRP protein purified as previously described in the presence or absence of cyclic nucleotide for 20 min at room temperature and then analyzed by electrophoresis on native 7.5% polyacrylamide gels PAGE. To test the effects of CadD on binding of fadD promoter cAMP-CRP complex various amounts of enzyme was added to the gel shift incubations.
Bacterial Media and Growth Conditions
The medium for carbon source utilization was M9 salts 32 containing 0.4% carbohydrate. The medium for cAMP determinations and gene expression assays by β-galactosidase activity (assayed as previously described) was RB medium (10% tryptone and 1% yeast extract) because growth on amino acids results in high levels of cAMP 17.
Determination of cAMP levels
The cAMP Direct Enzyme Immunoassay kit (Sigma) was used to assay the cAMP levels in mid-log phase bacterial cultures in RB medium. The bacterial cultures were adjusted to an absorbance at 600 nm of 1.0 and 1 mL of cell suspension was washed three times (once in the same medium, and the other twice in minimal medium M9) by centrifugation and resuspension. The air-dried bacterial pellets were dissolved in 300 µL of 0.1 M HCl to block endogenous phosphodiesterase activity and then placed in a boiling water bath for 20 min for release of cAMP 33. The boiled supernatants were centrifuged (12,000×g, 20 min) and assayed. A series of dilutions of a cAMP standard to give 200, 50, 12.5, 3.12 or 0.78 pmol/mL) were used to obtain a standard curve. The assay protocol recommended by the manufacturer for the bacterial and cAMP standards was followed and absorbance of each microtiter plate well was read at 405 nm. The net absorbance bound for each well was calculated by subtracting the nonspecific binding of wells lacking cAMP from the experimental samples. A standard curve was plotted and the cAMP levels of the bacterial samples were determined by interpolation.
Supplementary Material
Acknowledgments
This research was supported by a grant from the Robert A Welch foundation (A-840) and by a cooperative agreement from the National Institute of General Medical Sciences (U54GM093342). We thank R. Kumar for valuable discussions.
Footnotes
Supporting Information
Oligonucleotides used in this work and isothermal titration calorimetry data. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Holm L, Sander C. An evolutionary treasure: unification of a broad set of amidohydrolases related to urease. Proteins. 1997;28:72–82. [PubMed] [Google Scholar]
- 2.Seibert CM, Raushel FM. Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry. 2005;44:6383–6391. doi: 10.1021/bi047326v. [DOI] [PubMed] [Google Scholar]
- 3.Atkinson HJ, Morris JH, Ferrin TE, Babbitt PC. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS One. 2009;4:e4345. doi: 10.1371/journal.pone.0004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sassone-Corsi P. The cyclic AMP pathway. Cold Spring Harb Perspect Biol. 2012;4:a011148. doi: 10.1101/cshperspect.a011148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu J, Bao Q, Wu J, Wang H, Li D, Xi Y, Wang S, Yu S, Qu J. CSCDB: the cAMP and cGMP signaling components database. Genomics. 2008;92:60–64. doi: 10.1016/j.ygeno.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 6.McKay DB, Weber IT, Steitz TA. Structure of catabolite gene activator protein at 2.9-A resolution. Incorporation of amino acid sequence and interactions with cyclic AMP. J Biol Chem. 1982;257:9518–9524. [PubMed] [Google Scholar]
- 7.Weber IT, Steitz TA. Structure of a complex of catabolite gene activator protein and cyclic AMP refined at 2.5 A resolution. J Mol Biol. 1987;198:311–326. doi: 10.1016/0022-2836(87)90315-9. [DOI] [PubMed] [Google Scholar]
- 8.Popovych N, Tzeng SR, Tonelli M, Ebright RH, Kalodimos CG. Structural basis for cAMP-mediated allosteric control of the catabolite activator protein. Proc Natl Acad Sci U S A. 2009;106:6927–6932. doi: 10.1073/pnas.0900595106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newton RPK, EE Overton A. Mass spectrometric identification of cyclic nucleotides released by the bacterium Corynebacterium murisepticum into the culture medium. Rapid Commun Mass Spectrom. 1998;12:729–735. [Google Scholar]
- 10.Ishiyama J. Isolation of inosine 3': 5'-monophosphate from bacterial culture medium. J Cyclic Nucleotide Res. 1976;2:21–24. [PubMed] [Google Scholar]
- 11.Newton RP, Kingston EE, Hakeem NA, Salih SG, Beynon JH, Moyse CD. Extraction, purification, identification and metabolism of 3',5'-cyclic UMP: 3',5'-cyclic IMP and 3',5'-cyclic dTMP from rat tissues. Biochem J. 1986;236:431–439. doi: 10.1042/bj2360431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolfenden R, Sharpless TK, Allan R. Substrate binding by adenosine deaminase. Specificity, pH dependence, and competition by mercurials. J Biol Chem. 1967;242:977–983. [PubMed] [Google Scholar]
- 13.Ferguson DR, Price RH. Studies on the metabolism of cyclic nucleotides by the toad bladder: the metabolic products of cAMP. FEBS Lett. 1973;34:204–206. doi: 10.1016/0014-5793(73)80794-x. [DOI] [PubMed] [Google Scholar]
- 14.Ko AI, Goarant C, Picardeau M. Leptospira : the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat Rev Microbiol. 2009;7:736–747. doi: 10.1038/nrmicro2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levett PN. Leptospirosis. Clin Microbiol Rev. 2001;14:296–326. doi: 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brickman E, Soll L, Beckwith J. Genetic characterization of mutations which affect catabolite-sensitive operons in Escherichia coli including deletions of the gene for adenyl cyclase. J Bacteriol. 1973;116:582–587. doi: 10.1128/jb.116.2.582-587.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorke B, Stulke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 18.Gosset G, Zhang Z, Nayyar S, Cuevas WA, Saier MH., Jr Transcriptome analysis of Crp-dependent catabolite control of gene expression in Escherichia coli . J Bacteriol. 2004;186:3516–3524. doi: 10.1128/JB.186.11.3516-3524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP) J Mol Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 20.Lawson CL, Swigon D, Murakami KS, Darst SA, Berman HM, Ebright RH. Catabolite activator protein: DNA binding and transcription activation. Curr Opin Struct Biol. 2004;14:10–20. doi: 10.1016/j.sbi.2004.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krakow JS. Cyclic adenosine monophosphate receptor: effect of cyclic AMP analogues on DNA binding and proteolytic inactivation. Biochim Biophys Acta. 1975;383:345–350. doi: 10.1016/0005-2787(75)90303-2. [DOI] [PubMed] [Google Scholar]
- 22.Anderson WB, Perlman RL, Pastan I. Effect of adenosine 3',5'-monophosphate analogues on the activity of the cyclic adenosine 3',5'-monophosphate receptor in Escherichia coli. J Biol Chem. 1972;247:2717–2722. [PubMed] [Google Scholar]
- 23.Galperin MY. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 2005;5:35. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsunaga J, Medeiros MA, Sanchez Y, Werneid KF, Ko AI. Osmotic regulation of expression of two extracellular matrix-binding proteins and a haemolysin of Leptospira interrogans: differential effects on LigA and Sph2 extracellular release. Microbiology. 2007;153:3390–3398. doi: 10.1099/mic.0.2007/007948-0. [DOI] [PubMed] [Google Scholar]
- 25.Kim YG, Jeong JH, Ha NC, Kim KJ. Structural and functional analysis of the Lmo2642 cyclic nucleotide phosphodiesterase from Listeria monocytogenes. Proteins. 2011;79:1205–1214. doi: 10.1002/prot.22954. [DOI] [PubMed] [Google Scholar]
- 26.Shenoy AR, Sreenath N, Podobnik M, Kovacevic M, Visweswariah SS. The Rv0805 gene from Mycobacterium tuberculosis encodes a 3',5'-cyclic nucleotide phosphodiesterase: biochemical and mutational analysis. Biochemistry. 2005;44:15695–15704. doi: 10.1021/bi0512391. [DOI] [PubMed] [Google Scholar]
- 27.Nascimento AL, Ko AI, Martins EA, Monteiro-Vitorello CB, Ho PL, Haake DA, Verjovski-Almeida S, Hartskeerl RA, Marques MV, Oliveira MC, Menck CF, Leite LC, Carrer H, Coutinho LL, Degrave WM, Dellagostin OA, El-Dorry H, Ferro ES, Ferro MI, Furlan LR, Gamberini M, Giglioti EA, Goes-Neto A, Goldman GH, Goldman MH, Harakava R, Jeronimo SM, Junqueirade-Azevedo IL, Kimura ET, Kuramae EE, Lemos EG, Lemos MV, Marino CL, Nunes LR, de Oliveira RC, Pereira GG, Reis MS, Schriefer A, Siqueira WJ, Sommer P, Tsai SM, Simpson AJ, Ferro JA, Camargo LE, Kitajima JP, Setubal JC, Van Sluys MA. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J Bacteriol. 2004;186:2164–2172. doi: 10.1128/JB.186.7.2164-2172.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan SR, Gaines J, Roop RM, 2nd, Farrand SK. Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl Environ Microbiol. 2008;74:5053–5062. doi: 10.1128/AEM.01098-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng Y, Cronan JE. Crosstalk of Escherichia coli FadR with global regulators in expression of fatty acid transport genes. PLoS One. 2012;7:e46275. doi: 10.1371/journal.pone.0046275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo MC, Aulabaugh A, Jin G, Cowling R, Bard J, Malamas M, Ellestad G. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal Biochem. 2004;332:153–159. doi: 10.1016/j.ab.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 31.Hall RS, Xiang DF, Xu C, Raushel FM. N-Acetyl-D-glucosamine-6-phosphate deacetylase: substrate activation via a single divalent metal ion. Biochemistry. 2007;46:7942–7952. doi: 10.1021/bi700543x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller J. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 33.Haggerty DM, Schleif RF. Kinetics of the onset of catabolite repression in Escherichia coli as determined by lac messenger ribonucleic acid initiations and intracellular cyclic adenosine 3',5'-monophosphate levels. J Bacteriol. 1975;123:946–953. doi: 10.1128/jb.123.3.946-953.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferenci T, Zhou Z, Betteridge T, Ren Y, Liu Y, Feng L, Reeves PR, Wang L. Genomic sequencing reveals regulatory mutations and recombinational events in the widely used MC4100 lineage of Escherichia coli K-12. J Bacteriol. 2009;191:4025–4029. doi: 10.1128/JB.00118-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.