Abstract
High-throughput technologies are widely used, for example to assay genetic variants, gene and protein expression, and epigenetic modifications. One often overlooked complication with such studies is batch effects, which occur because measurements are affected by laboratory conditions, reagent lots and personnel differences. This becomes a major problem when batch effects are correlated with an outcome of interest and lead to incorrect conclusions. Using both published studies and our own analyses, we argue that batch effects (as well as other technical and biological artefacts) are widespread and critical to address. We review experimental and computational approaches for doing so.
Many technologies used in biology — including high-throughput ones such as microarrays, bead chips, mass spectrometers and second-generation sequencing — depend on a complicated set of reagents and hardware, along with highly trained personnel, to produce accurate measurements. When these conditions vary during the course of an experiment, many of the quantities being measured will be simultaneously affected by both biological and non-biological factors. Here we focus on batch effects, a common and powerful source of variation in high-throughput experiments.
Batch effects are sub-groups of measurements that have qualitatively different behaviour across conditions and are unrelated to the biological or scientific variables in a study. For example, batch effects may occur if a subset of experiments was run on Monday and another set on Tuesday, if two technicians were responsible for different subsets of the experiments or if two different lots of reagents, chips or instruments were used. These effects are not exclusive to high-throughput biology and genomics research1, and batch effects also affect low-dimensional molecular measurements, such as northern blots and quantitative PCR. Although batch effects are difficult or impossible to detect in low-dimensional assays, high-throughput technologies provide enough data to detect and even remove them. However, if not properly dealt with, these effects can have a particularly strong and pervasive impact. Specific examples have been documented in published studies2,3 in which the biological variables were extremely correlated with technical variables, which subsequently led to serious concerns about the validity of the biological conclusions4,5.
Normalization is a data analysis technique that adjusts global properties of measurements for individual samples so that they can be more appropriately compared. Including a normalization step is now standard in data analysis of gene expression experiments6. But normalization does not remove batch effects, which affect specific subsets of genes and may affect different genes in different ways. In some cases, these normalization procedures may even exacerbate technical artefacts in high-throughput measurements, as batch and other technical effects violate the assumptions of normalization methods. Although specific normalization methods have been developed for microarray studies that take into account study design7 or otherwise correct for the batch problem8, they are still not widely used.
As described here, in surveying a large body of published data involving high-throughput studies and a number of technology platforms, we have found evidence of batch effects. In many cases we have found that these can lead to erroneous biological conclusions, supporting the conclusions of previous publications4,5. Here we analyse the extent of the problem, review the critical downstream consequences of batch effects and describe experimental and computational solutions to reduce their impact on high-throughput data.
An illustration of batch effects
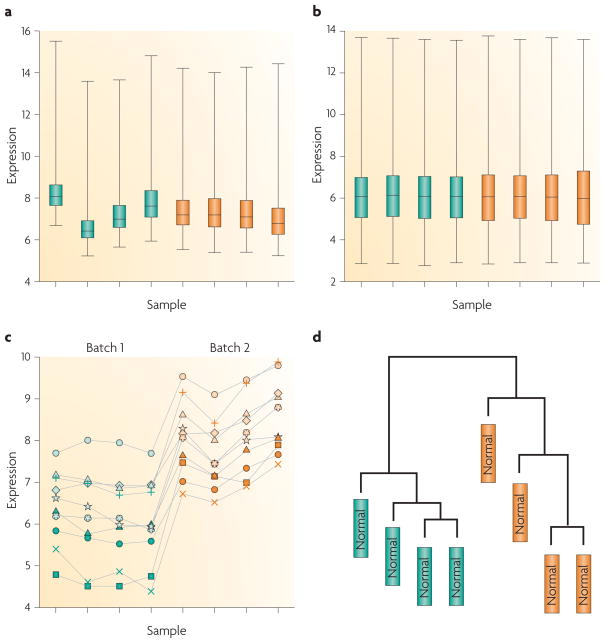
To introduce the batch effect problem we used data from a previously published bladder cancer study9 (FIG. 1). In this study, microarray expression profiling was used to examine the gene expression patterns in superficial transitional cell carcinoma (sTCC) with and without surrounding carcinoma in situ (CIS). Hierarchical cluster analysis separated the sTCC samples according to the presence or absence of CIS. However, the presence or absence of CIS was strongly confounded with processing date, as reported in REF. 10. In high-throughput studies it is typical for global properties of the raw data distribution to vary strongly across arrays, as they do for this data set (FIG. 1a). After normalization, these global differences are greatly reduced (FIG. 1b), and the sTCC study properly normalized the data. However, it is typical to observe substantial batch effects on subsets of specific genes that are not addressed by normalization (FIG. 1c). In gene expression studies, the greatest source of differential expression is nearly always across batches rather than across biological groups, which can lead to confusing or incorrect biological conclusions owing to the influence of technical artefacts. For example, the control samples in the sTCC study clustered perfectly by the processing date (FIG. 1d), and the processing date was confounded with the presence/absence status.
Figure 1. Demonstration of normalization and surviving batch effects.
For a published bladder cancer microarray data set obtained using an Affymetrix platform9, we obtained the raw data for only the normal samples. Here, green and orange represent two different processing dates, a | Box plot of raw gene expression data (log base 2). b | Box plot of data processed with RMA, a widely used preprocessing algorithm for Affymetrix data27. RMA applies quantile normalization — a technique that forces the distribution of the raw signal intensities from the microarray data to be the same in all samples28. c | Example often genes that are susceptible to batch effects even after normalization. Hundreds of genes show similar behaviour but, for clarity, are not shown. d | Clustering of samples after normalization. Note that the samples perfectly cluster by processing date.
Group and date are only surrogates
The processing of samples using protocols that differ among laboratories has been linked to batch effects. In such cases, the samples that have been processed using the same protocol are known as processing groups. For example, multiple laboratory comparisons of microarray experiments have shown strong laboratory-specific effects11. In addition, in nearly every gene expression study, large variations are associated with the processing date12, and in microarray studies focusing on copy number variation, large effects are associated with DNA preparation groups13. The processing group and date are therefore commonly used to account for batch effects. However, in a typical experiment these are probably only surrogates for other sources of variation, such as ozone levels, laboratory temperatures and reagent quality12,14. Unfortunately, many possible sources of batch effects are not recorded, and data analysts are left with just processing group and date as surrogates.
One way to quantify the affect of non-biological variables is to examine the principal components of the data. Principal components are estimates of the most common patterns that exist across features. For example, if most genes in a microarray study are differentially expressed with respect to cancer status, the first principal component will be highly correlated with cancer status. Principal components capture both biological and technical variability and, in some cases, principal components can be estimated after the biological variables have been accounted for15. In this case, the principal components primarily quantify the effects of artefacts on the high-throughput data. Principal components can be compared to known variables, such as processing group or time. If the principal components do not correlate with these known variables, there may be an alternative, unmeasured source of batch effects in the data.
Examination of public data
In addition to the example described above involving the sTCC study, we examined the extent of batch effects for eight other published or publicly available data sets (TABLE 1) using the following approach. First, we identified a surrogate for batch effects (such as date or processing group) for each data set. We then used simple linear models to measure the level of confounding between this surrogate and the study outcome (for example, case or control) when available. Note that the more confounding there is, the more likely it is that batch variability can be confused with biological variability. We then summarized the susceptibility to batch effects for each high-throughput feature. We did this by quantifying the association between observed values and the surrogates using analysis of variance models. If the association p-value was below 0.01, we declared the feature as susceptible to batch effects. Next, we identified the principal components that were most correlated with the surrogate and with the outcome, again using analysis of variance models. Finally, we identified associations between the feature measurements (for example, copy number and gene expression levels) and the outcome of interest for each study. The outcomes of this analysis are described below.
Table 1.
Batch effects seen for a range of high-throughput technologies
| Study description* | Known variable used as a surrogate
|
Principal components used as a surrogate
|
Association with outcome Significant features (%)‡‡ | Refs | ||||
|---|---|---|---|---|---|---|---|---|
| Surrogate‡ | Confounding (%)§ | Susceptible features(%%)|| | Principal components rank of surrogate (correlation)¶ | Principal components rank of outcome (correlation)# | Susceptible features (%)** | |||
| Data set 1: gene expression microarray, Affymetrix(Np = 22,283) | Date | 29.7 | 50.5 | 1 (0.570) | 1 (0.649) | 91.6 | 71.9 | 9 |
|
| ||||||||
| Data set 2: gene expression, Affymetrix (Np = 4167) | Date | 77.6 | 73.7 | 1 (0.922) | 1 (0.668) | 98.5 | 62.2 | 2 |
|
| ||||||||
| Data set 3: mass spectrometry (Np = 15,154) | Processing group | 100 | 51.7 | 2 (0.344) | 2 (0.344) | 99.7 | 51.7 | 3 |
|
| ||||||||
| Data set 4: copy number variation, Affymetrix (Np = 945,806) | Date | 29.2 | 99.5 | 2 (0.921) | 3 (0.485) | 99.8 | 98.8 | 16 |
|
| ||||||||
| Data set 5: copy number variation, Affymetrix (Np = 945,806) | Date | 12.2 | 83.8 | 1 (0.553) | 1 (0.137) | 99.8 | 74.1 | 17 |
|
| ||||||||
| Data set 6: gene expression, Affymetrix (Np = 22,277) | Processing group | NA | 83.8 | 5 (0.369) | NA | 97.1 | NA | 18 |
|
| ||||||||
| Data set 7: gene expression, Agilent (Np = 17,594) | Date | NA | 62.8 | 2 (0.248) | NA | 96.7 | NA | 18 |
|
| ||||||||
| Data set 8: DNA methylation, Agilent (Np = 27,578) | Processing group | NA | 78.6 | 3 (0.381) | NA | 99.8 | NA | 18 |
|
| ||||||||
| Data set 9: DNA sequencing, Solexa (Np = 2,886) | Date | 24.2 | 32.1 | 2 (0.846) | 2 (0.213) | 72.7 | 16.9 | 1000 Genomes Project |
The first three rows represent studies for which batch effects have been described in the literature4,5,10. Rows four and five are from genome-wide association study data sets. Rows six to eight represent data from The Cancer Genome Atlas (TCGA). Finally, the last row represents second-generation sequencing data from the 1000 Genomes Project. Details for each data set and the analyses used to construct the table are included in Supplementary information S1 (box).
Study description includes the application, platform and number of features (Np).
A known variable was used as a surrogate for batch effect.
Level of confounding between surrogate and biological outcome of interest. We use a generalized R2 statistic for categorical data. The correlation ranges from 0% (no confounding) to 100% (completely confounded).
For each feature of the technology (for example, genes), we computed an F-statistic to test for association by stratifying measurements by the surrogate. p-values were obtained and, because of multiple comparisons, false discovery rates (FDRs) were obtained using the Benjamini-Hochberg procedure. A feature obtaining an FDR below 5% was considered susceptible to batch effects.
Principal components analysis was performed on the feature level data. The principal components were ranked in decreasing order of the variability that they explained. We computed the association (using R2) between the surrogate and the first five principal components. We report the rank of the component with the highest correlation; the correlation is given in parenthesis.
As for ¶but using the biological outcome of interest instead of the surrogate.
As for ¶but using principal components to define batch.
As for ¶but using biological outcome. NA, not available.
We re-examined data sets from three studies for which batch effects have been reported (TABLE 1). The first was the sTCC study9 described above. The second was a microarray data set from a study examining population differences in gene expression2. The conclusion of the original paper, that the expression of 1,097 of 4,197 genes differs between populations of European and Asian descent, was questioned in another publication4 because the populations and processing dates were highly correlated. In fact, more differences were found when comparing data from two processing dates while keeping the population fixed. The third was a mass spectrometry data set that was used to develop a statistical procedure, based on proteomic patterns in serum, to distinguish neoplastic diseases from non-neoplastic diseases within the ovary3. Concerns about the conclusions of this paper were raised in another publication, which showed that outcome was confounded with run date5.
To further illustrate the ubiquity and potential hazards associated with batch effects, we also carried out analyses on representative publicly available data sets that have been established using a range of high-throughput technologies. In addition to the three studies described above, we examined: data from a study of copy number variation in HapMap populations16; a study of copy number variation in a genome-wide association study of bipolar disorder17; gene expression from an ovarian cancer study from The Cancer Genome Atlas (TCGA)18 produced using two platforms (Affymetrix and Agilent); methylation data from the same TCGA ovarian cancer samples produced using Illumina BeadChips; and second-generation sequencing data from a study comparing unrelated HapMap individuals (these data were a subset of the data from the 1000 Genomes Project).
We found batch effects for all of these data sets, and substantial percentages (32.1–99.5%) of measured features showed statistically significant associations with processing date, irrespective of biological phenotype (TABLE 1). This suggests that batch effects influence a large percentage of the measurements from genomic technologies. Next, we computed the first five principal components of the feature data (the principal components were ordered by the amount of variability explained). Ideally these principal components would correlate with the biological variables of interest, as the principal components represent the largest sources of signal in the data. Instead, for all of the studied data sets, the surrogates for batch (date or processing group) were strongly correlated with one of the top principal components (TABLE 1). In general, the correlation with the top principal components was not as high for the biological outcome as it was for the surrogates. This suggests that technical variability was more influential than biological variability across a range of experimental conditions and technologies.
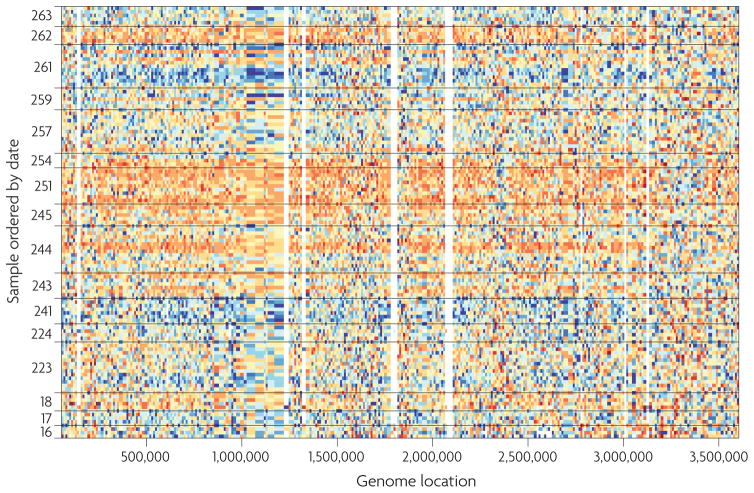
For most of the data sets examined, neither date nor biological factors was perfectly associated with the top principal components, suggesting that other unknown sources of batch variability are present. This implies that accounting for date or processing group might not be sufficient to capture and remove batch effects. For example, we did a further analysis of second-generation sequencing data that were generated by the 1000 Genomes Project (FIG. 2). We found that 32% of the features were associated with date but up to 73% were associated with the second principal component. Note that the principal components cannot be explained by biology because only 17% of the features are associated with the biological outcome.
Figure 2. Batch effects for second-generation sequencing data from the 1000 Genomes Project.
Each row is a different HapMap sample processed in the same facility with the same platform. See Supplementary information SI (box) for a description of the data represented here. The samples are ordered by processing date with horizontal lines dividing the different dates. We show a 3.5 Mb region from chromosome 16. Coverage data from each feature were standardized across samples: blue represents three standard deviations below average and orange represents three standard deviations above average. Various batch effects can be observed, and the largest one occurs between days 243 and 251 (the large orange horizontalstreak).
Downstream consequences
In the most benign cases, batch effects will lead to increased variability and decreased power to detect a real biological signal15. Of more concern are cases in which batch effects are confounded with an outcome of interest and result in misleading biological or clinical conclusions. An example of confounding is when all of the cases are processed on one day and all of the controls are processed on another. We have shown that in a typical high-throughput experiment, one can expect a substantial percentage of features to show statistically significant differences when comparing across batches, even when no real biological differences are present (FIG. 1; TABLE 1). Therefore, if one is not aware of the batch effect, a confounded experiment will lead to incorrect biological conclusions because results due to batch will be impossible to distinguish from real biological effects. As an example, we consider the proteomics study mentioned above3. These published results and further confirmation19 led to the development of a ‘home-brew’ diagnostic assay for ovarian cancer. However, in this study the biological variable of interest (neoplastic disease within the ovary) was extremely correlated with processing day5. Furthermore, batch effects were identified as a major driver of these results. Fortunately, objections raised after the assay was advertised led the US Food and Drug Administration to block use of the assay, pending further validation20.
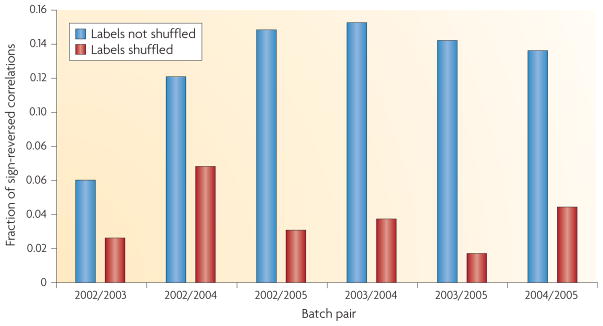
A more subtle consequence of the batch effect relates to correlations between features. These correlations are implicitly or explicitly used in various applications. For example, in systems biology, gene-gene expression correlations are used to analyse or predict pathways. Rank-based classification methods21 are another example of how correlations between features can be used. However, we find that batch effects are strong enough to change not only mean levels of gene expression between batches but also correlations and relative rankings between the expression of pairs of genes. In some cases, the direction of significant positive correlation between genes is completely reversed in different batches. For example, we found an effect of this type in our analysis of the gene expression data set from REF. 2 (TABLE 1). Here, genes that show significant positive correlations in the direction of their gene expression changes in samples from one batch are significantly negatively correlated in samples from a second batch (FIG. 3).
Figure 3. Batch effects also change the correlations between genes.
We normalized every gene in the second gene expression data set2 in TABLE 1 to mean 0, variance 1 within each batch. (The 2006 batch was omitted owing to small sample size.) We identified all significant correlations (p < 0.05) between pairs of genes within each batch using a linear model. We looked at genes that showed a significant correlation in two batches and counted the fraction of times that the correlation changed between the two batches. A large percentage of significant correlations reversed signs across batches, suggesting that the correlation structure between genes changes substantially across batches. To confirm this phenomenon is due to batch, we repeated the process—looking for significant correlations that changed sign across batches—but with the batch labels randomly permuted. With random batches, a much smaller fraction of significant correlations change signs. This suggests that correlation patterns differ by batch, which would affect rank-based prediction methods as well as system biology approaches that rely on between-gene correlation to estimate pathways.
If batch effects go undetected, they can lead to substantial misallocation of resources and lack of reproducibility22. In general, technology that has been developed for the prediction of clinical outcomes using data that show batch effects may produce results that are more variable than expected. Batch effects were shown to have strong adverse effects on predictors built with methods that are naive to these effects10; the result is lower-than-expected classification rates, which might put patients classified with these technologies at risk.
Experimental design solutions
The first step in addressing batch and other technical artefacts is careful study design23. Experiments that run over long periods of time and large-scale experiments that are run across different laboratories are highly likely to be susceptible. But even smaller studies performed within a single laboratory may span several days or include personnel changes.
High-throughput experiments should be designed to distribute batches and other potential sources of experimental variation across biological groups8. For example, in a study comparing a molecular profile in tumour samples versus healthy controls, the tumour and healthy samples should be equally distributed between multiple laboratories and across different processing times22. These steps can help to minimize the probability of confounding between biological and batch effects.
However, even in a perfectly designed study, batch will strongly influence the measured high-throughput data. Information about changes in personnel, reagents, storage and laboratories should be recorded and passed onto data analysts. As it is generally impossible to record all potential sources of batch effects, statistical modelling solutions — as described below — are needed to reduce the impact of batch effects on biological conclusions.
Statistical solutions
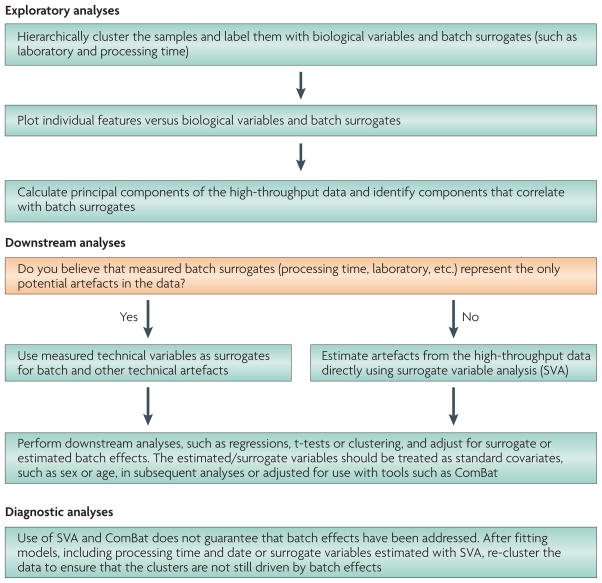
After a high-throughput study has been performed, the statistical approach for dealing with batch effects consists of two key steps. Exploratory analyses must be carried out to identify the existence of batch effects and quantify their effect, as well as the effect of other technical artefacts in the data. Downstream statistical analyses must then be adjusted to account for these unwanted effects (FIG. 4).
Figure 4. Key steps in the statistical analysis of batch effects.
The first step is exploratory data analysis to identify and quantify potential batch effects and other artefacts. The second step is to use known or estimated surrogates of the artefacts to adjust downstream analyses. The final step is to carry out diagnostic analyses.
The first step in the exploratory statistical analysis of batch effects is to identify and quantify batch effects using principal components analysis or visualization techniques, such as hierarchical clustering dendrograms (FIG. 1d) or multidimensional scaling24. Hierarchical clustering of samples25 labelled both with biological groups and known batch surrogates reveals whether the major differences are due to biology or batch (FIG. 1d). It is also useful to plot the levels of individual features (the expression levels of specific genes, probes, proteins, and so on) versus biological variables and batch variables, such as processing group or time (FIG. 1c). Plotting individual features versus biological and known batch variables is crucial, as the bulk distribution of normalized data may seem correct even when batch effects exist (FIG. 1b). A useful way to summarize these feature-level effects is to calculate the principal components of the feature data26. The principal components can also be plotted against known batch variables, such as processing group or time, to determine whether, on average, the high-dimensional feature data are correlated with batch. An example R script is included in the code and data for this article (see ‘Further Information’).
Strong batch effects may exist when: the samples cluster by processing group or time; a large number of features are highly associated with processing group or time; or principal components are associated with batch processing group or time. If strong batch effects exist, they must be accounted for in downstream statistical analyses.
Most downstream statistical analyses performed on high-throughput data rely on linear models, either explicitly or implicitly. However, there are also other solutions, such as those provided for copy number variation microarrays13 that do not use linear models. Because these latter solutions are typically specific to each application, we do not review them here. For analyses using linear models, batch effects can be modelled in one of two ways. If exploratory analyses and prior knowledge suggest that simple surrogates, such as processing time, capture all of the batch effects, these surrogates can be directly incorporated into the models that are used to compare groups. The simplest approach is to include processing group and time as variables in the linear model for association between the high-dimensional features and the outcome variables8,12. See the ComBat website for a discussion of this approach.
In many cases, processing time is a useful surrogate but does not explain all of the technical artefacts and variability that are seen in high-throughput data. When the true sources of batch effects are unknown or cannot be adequately modelled with processing group or date, it may be more appropriate to use methods such as surrogate variable analysis (SVA)15 (see ‘Further Information’). SVA estimates the sources of batch effects directly from the high-throughput data so that downstream significance analyses can be corrected. Variables estimated with SVA can then be incorporated into the linear model that relates the outcome to the high-dimensional feature data, in the same way as processing year or group could be included. An advantage of SVA is that surrogate variables are estimated instead of pre-specified, which means that the important potential batch variables do not have to be known in advance.
These approaches are most effective when batch effects are not highly confounded, or correlated, with the biological variables of interest. To identify potential sources of confounding, biological variables and sample characteristics can be compared to processing group and time. If the biological variables are highly correlated with processing group or time, it is difficult to determine whether observed differences across biological groups are due to biology or artefacts. At a minimum, analyses should report the processing group and time of all samples in a study along with the biological variables of interest so that results can be independently verified.
Conclusion
There has been substantial progress in identifying and accounting for batch effects, but substantial challenges remain. Foremost among these challenges is the need for consistent reporting of the most common potential sources of batch effects, including processing group and date. Experimental designs should also consistently distribute biological groups equally across processing groups and times. Close collaborations between laboratory biologists and data analysts are also needed so that the specific sources of batch effects can be isolated and the dependence on surrogates can be reduced. Targeted experiments may be necessary to determine the precise sources of non-biological signal for each specific technology. Finally, there is a need to incorporate adjustment for batch effects as a standard step in the analysis of high-throughput data analysis along with normalization, exploratory analysis and significance calculation.
Supplementary Material
Acknowledgments
We thank the referees for helpful comments and suggestions. One referee in particular went beyond the call of duty to help us improve clarity. We thank the TCGA and 1000 Genomes Project for making the data public. The GoKinD collection of DNA was genotyped through the Genetic Association Information Network (GAIN) programme with the support of the Foundation for the National Institutes of Health and The National Institute of Diabetes and Digestive and Kidney Diseases. The work of J.T.L., H.C.B., B.L. and R.A.I. is partially funded by US National Institutes of Health grants GM0083084, HG004059 and HG005220.
Glossary
- Confounded
An extraneous variable (for example, processing data) is said to be confounded with the outcome of interest (for example, disease state) when it correlates both with the outcome and with an independent variable of interest (for example, gene expression)
- Feature
The general name given to the measurement unit in high-throughput technologies. Examples of features include probes for the genes represented on microarray mass-to-charge (m/z) ratios for which intensities are measured in mass spectrometry, and loci for which coverage is reported for sequencing technologies
- Hierarchical clustering
A statistical method in which objects (for example, gene expression profiles for different individuals) are grouped into a hierarchy, which is visualized in a dendrogram. Objects close to each other in the hierarchy, measured by tracing the branch heights, are also close by some measure of distance — for example, individuals with similar expression profiles will be close together in terms of branch lengths
- Linear models
Statistical models in which the effect of independent variables and error terms are expressed as additive terms. For example, when modelling the outcomes in a case-control study, the effect of a typical case is added to the typical control level. Variation around these levels is explained by additive error. Linear models motivate many widely used statistical methods, such as t-tests and analysis of variance. Many popular genomics software tools are also based on linear models
- Normalization
Methods used to adjust measurements so that they can be appropriately compared among samples. For example, gene expression levels measured by quantitative PCR are typically normalized to one or more housekeeping genes or ribosomal RNA. In microarray analysis, methods such as quantile normalization manipulate global characteristics of the data
- Principal components
Patterns in high-dimensional data that explain a large percentage of the variation across features. The top principal component is the most ubiquitous pattern in a set of high-dimensional data. Principal components are sometimes called eigengenes when estimated from microarray gene expression data
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
1000 Genomes Project: http://www.1000genomes.org
Code and data for this article: http://rafalab.jhsph.edu/batch
ComBat: http://jlab.byu.edu//ComBat/Abstract.html
Description of surrogate variable analysis:
http://www.biostat.jhsph.edu/~jleek/sva/index.html
The Cancer Genome Atlas: http://www.genome.gov/17516564
Contributor Information
Jeffrey T. Leek, Department of Biostatistics, Johns Hopkins Bloomberg School of Public Health, 615 North Wolfe Street, Baltimore, Maryland 21205-2179, USA
Robert B. Scharpf, Department of Oncology, Johns Hopkins University, Baltimore, Maryland 21205-2013, USA
Héctor Corrada Bravo, Department of Biostatistics, Johns Hopkins Bloomberg School of Public Health, 615 North Wolfe Street, Baltimore, Maryland 21205-2179, USA. Department of Computer Science, University of Maryland, College Park, Maryland 20742, USA.
David Simcha, Biomedical Engineering Department, Johns Hopkins University, 3400 N. Charles St, Baltimore, Maryland 212218, USA.
Benjamin Langmead, Department of Biostatistics, Johns Hopkins Bloomberg School of Public Health, 615 North Wolfe Street, Baltimore, Maryland 21205-2179, USA.
W. Evan Johnson, Department of Statistics, Brigham Young University, Provo, Utah 84602-6575, USA.
Donald Geman, Department of Applied Mathematics and Statistics, Johns Hopkins University, Baltimore, Maryland 21218-2682, USA.
Keith Baggerly, Department of Bioinformatics and Computational Biology, The University of Texas M.D. Anderson Cancer Center, P. O. Box 301402, Houston, Texas 77230, USA.
Rafael A. Irizarry, Department of Biostatistics, Johns Hopkins Bloomberg School of Public Health, 615 North Wolfe Street, Baltimore, Maryland 21205-2179, USA
References
- 1.Youden WJ. Enduring values. Technometrics. 1972;14:1–11. [Google Scholar]
- 2.Spielman RS, et al. Common genetic variants account for differences in gene expression among ethnic groups. Nature Genet. 2007;39:226–231. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petricoin EF, et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359:572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 4.Akey JM, Biswas S, Leek JT, Storey JD. On the design and analysis of gene expression studies in human populations. Nature Genet. 2007;39:807–808. doi: 10.1038/ng0707-807. author reply 808–809. [DOI] [PubMed] [Google Scholar]
- 5.Baggerly KA, Edmonson SR, Morris JS, Coombes KR. High-resolution serum proteomic patterns for ovarian cancer detection. Endocr Relat Cancer. 2004;11:583–584. doi: 10.1677/erc.1.00868. author reply 585–587. [DOI] [PubMed] [Google Scholar]
- 6.Allison DB, Cui XQ, Page CP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nature Rev Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 7.Mecham BH, Nelson PS, Storey JD. Supervised normalization of microarrays. Bioinformatics. 2010;26:1308–1315. doi: 10.1093/bioinformatics/btq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 9.Dyrskjot L, et al. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 2004;64:4040–4048. doi: 10.1158/0008-5472.CAN-03-3620. [DOI] [PubMed] [Google Scholar]
- 10.Zilliox MJ, Irizarry RA. A gene expression bar code for microarray data. Nature Methods. 2007;4:911–913. doi: 10.1038/nmeth1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irizarry RA, et al. Multiple-laboratory comparison of microarray platforms. Nature Methods. 2005;2:345–350. doi: 10.1038/nmeth756. [DOI] [PubMed] [Google Scholar]
- 12.Scherer A. In: Batch Effects and Noise in Micorarray Experiments: Sources and Solutions. Scherer A, editor. John Wiley and Sons; Chichester, UK: 2009. [Google Scholar]
- 13.Scharpf RB, et al. A multilevel model to address batch effects in copy number estimation using SNP arrays. Biostatistics. 2010 Jul 12; doi: 10.1093/biostatistics/kxq043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fare TL, et al. Effects of atmospheric ozone on microarray data quality. Anal Chem. 2003;75:4672–4675. doi: 10.1021/ac034241b. [DOI] [PubMed] [Google Scholar]
- 15.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:e161. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 17.Dick DM, et al. Genomewide linkage analyses of bipolar disorder: a new sample of 250 pedigrees from the National Institute of Mental Health Genetics Initiative. Am J Hum Genet. 2003;73:107–114. doi: 10.1086/376562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conrads TP, et al. High-resolution serum proteomic features for ovarian cancer detection. Endocr Relat Cancer. 2004;11:163–178. doi: 10.1677/erc.0.0110163. [DOI] [PubMed] [Google Scholar]
- 20.Ransohoff DF. Lessons from controversy: ovarian cancer screening and serum proteomics. J Natl Cancer Inst. 2005;97:315–319. doi: 10.1093/jnci/dji054. [DOI] [PubMed] [Google Scholar]
- 21.Liu HC, et al. Cross-generation and cross-laboratory predictions of Affymetrix microarrays by rank-based methods. J Biomed Inform. 2008;41:570–579. doi: 10.1016/j.jbi.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Baggerly KA, Coombes KR, Neeley ES. Run batch effects potentially compromise the usefulness of genomic signatures for ovarian cancer. J Clin Oncol. 2008;26:1186–1187. doi: 10.1200/JCO.2007.15.1951. author reply 1187–1188. [DOI] [PubMed] [Google Scholar]
- 23.Hu J, Coombes KR, Morris JS, Baggerly KA. The importance of experimental design in proteomic mass spectrometry experiments: some cautionary tales. Brief Funct Genomic Proteomic. 2005;3:322–331. doi: 10.1093/bfgp/3.4.322. [DOI] [PubMed] [Google Scholar]
- 24.Cox MAA, Cox TF. In: Handbook of Data Visualization. Chen C-H, Hardle WK, Unwin A, editors. Springer; Berlin: 2008. pp. 315–347. [Google Scholar]
- 25.Sokal RR, Smeath PHA. Principles of Numerical Taxonomy. WH Freeman; San Francisco: 1963. [Google Scholar]
- 26.Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci USA. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 28.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.