Abstract
Recent studies of transcription have revealed a level of complexity not previously appreciated even a few years ago, both in the intricate use of post-initiation control and the mass production of rapidly degraded transcripts. Dissection of these pathways requires strategies for precisely following transcripts as they are being produced. Here we present an approach (native elongating transcript sequencing, NET-seq), based on deep sequencing of 3′ ends of nascent transcripts associated with RNA polymerase, to monitor transcription at nucleotide resolution. Application of NET-seq in Saccharomyces cerevisiae reveals that although promoters are generally capable of divergent transcription, the Rpd3S deacetylation complex enforces strong directionality to most promoters by suppressing antisense transcript initiation. Our studies also reveal pervasive polymerase pausing and backtracking throughout the body of transcripts. Average pause density shows prominent peaks at each of the first four nucleosomes, with the peak location occurring in good agreement with in vitro biophysical measurements. Thus, nucleosome-induced pausing represents a major barrier to transcriptional elongation in vivo.
Introduction
Accumulating evidence now reveals that transcription elongation is not a straightforward read-out of the downstream DNA sequence. Co-transcriptional processing events dictate the covalent nature and fate of RNA transcripts1. Indeed many transcripts are targeted co-transcriptionally for rapid degradation and hence are effectively invisible to approaches that monitor mature messenger RNAs2, 3, 4. In addition to these processing events, the strong propensity of RNA polymerase (RNAP) to pause creates barriers to elongation and provides an opportunity for regulation and coordination of co-transcriptional events5, 6. In vitro, RNAP pausing is found to be ubiquitous7. Biophysical approaches have provided a structural and energetic understanding of RNAP pausing which results from both intrinsic properties of the polymerase itself as well as interactions with its DNA template, including the presence of bound proteins (for example, histones)8, 9, 10, 11, 12. In the cell, elongation factors probably alter the energetic landscape of transcription, but the extent and mechanism of RNAP pausing in eukaryotic cells remain largely unknown. Bridging the divide between in vivo and in vitro transcriptional views requires approaches that visualize transcription with comparable precision afforded by in vitro transcriptional assays. More generally, the ability to monitor quantitatively nascent transcripts would provide broad insights into the roles and regulation of transcription initiation, elongation and termination in gene expression.
Historically, two strategies have been used to provide snapshots of transcriptional activity in vivo. In the first approach, RNAP is crosslinked to DNA and RNAP-bound DNA elements are identified by microarrays or deep sequencing13, 14. Although providing a global view of RNAP binding sites, these measurements are of limited spatial and temporal resolution and do not reveal the identity of the transcribed strand or even whether RNAP molecules are engaged in transcription. In the second approach, transcription is halted in vivo and then reinitiated in isolated nuclei under conditions that allow labelling of nascent chains, thereby enabling them to be distinguished from bulk RNA15, 16. Such ‘nuclear run-on’ strategies reveal actively transcribed DNA regions but require extensive manipulations that limit resolution and depend on the efficient re-initiation of transcription under non-physiological conditions.
To monitor the transcriptional states of unperturbed cells, we sought to determine the precise in vivo position of all active RNAP complexes. Here we present an approach (native elongating transcript sequencing, NET-seq) that accomplishes this goal by exploiting the extraordinary stability of the DNA–RNA–RNAP ternary complex17 to capture nascent transcripts directly from live cells without crosslinking. The identity and abundance of the 3′ end of purified transcripts are revealed by deep sequencing18, thus providing a quantitative measure of RNAP density with single nucleotide precision. Using NET-seq, we expose rapidly degraded transcription products, locate the position of RNAP pauses, and identify factors and chromatin structure that regulate these transcription events.
Quantifying transcription at nucleotide resolution
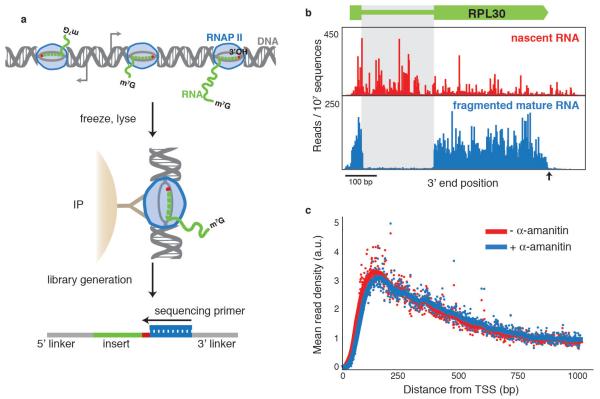
We focused on the transcription by RNAPII of protein-coding genes in the budding yeast, Saccharomyces cerevisiae, although the NET-seq approach should be readily adaptable to other systems. To facilitate purification, we worked with a strain that endogenously expressed a functional variant of RNAPII with a 3×-Flag epitope attached to its third subunit (Rpb3). Log-phase cultures were collected by filtration and flash frozen in liquid nitrogen (Fig. 1a). After cryogenic lysis, RNAPII was efficiently immunoprecipitated (Supplementary Fig. 1a). We prepared the co-purified RNA for deep sequencing using a protocol that allows efficient RNA capture while minimizing bias19, and sequenced 40 bases from the 3′ end. The alignment of these sequences to the yeast genome identified the final nucleotide that was incorporated by RNAPII, and the number of sequencing reads at each position along the genome indicated the density of transcriptionally active RNA polymerases at that site (Fig. 1b, alignment statistics displayed in Supplementary Table 1). A metagene analysis of RNAPII distribution across transcription units shows higher RNAPII density for the first 700 base pairs (bp) from the 5′ end (Fig. 1c), consistent with lower resolution observations seen using a global run-on approach16.
Figure 1.
a, Schematic diagram of NET-seq protocol. A yeast culture is flash frozen and cryogenically lysed. Nascent RNA is co-purified via an immunoprecipitation (IP) of the RNAPII elongation complex. Conversion of RNA into DNA results in a DNA library with the RNA as an insert between DNA sequencing linkers. The sequencing primer is positioned such that the 3′ end of the insert is sequenced. m7G refers to the 7-methylguanosine cap structure at the 5′ end of nascent transcripts.b, The 3′ end of each sequence is mapped to the yeast genome and the number of reads at each nucleotide is plotted at the RPL30 locus for nascent RNA and lightly fragmented mature RNA. Note that for the nascent transcripts, the introns (grey box) and regions after the polyadenylation site (black arrow) are readily detected. c, Metagene analysis for well-expressed genes (n = 471, >1.5 reads per bp in both conditions) of the mean read density (arbitrary units, a.u.) in the presence and absence of transcription inhibitor, α-amanitin. TSS, transcription start site.
Several observations indicate that we are detecting nascent transcription. First, we robustly capture transcripts from introns and regions after polyadenylation sites; areas that are present in nascent transcripts but absent from mature messenger RNAs (Fig. 1b). Second, we verified that transcripts do not associate with RNAPII after cell lysis (Supplementary Table 2). Third, we saw negligible degradation of RNA under the immunoprecipitation conditions. Nevertheless, our library generation protocols prevent detection of co-purified degradation products by requiring that input RNAs have 3′ hydroxyl termini, as hydrolysis and degradation products primarily have terminal phosphates20. Finally, we saw that transcription did not proceed during processing of lysates as addition of the transcription inhibitor α-amanitin to the lysis buffer did not change the RNAPII density (Fig. 1c).
In addition to nascent transcripts, the RNAPII immunoprecipitation captures splicing intermediates (that is, the 5′ exon and the excised lariat). Their 3′ hydroxyl termini allow them to appear in our data at the 3′ ends of exons and introns (Supplementary Fig. 5). These observations indicate the widespread existence of co-transcriptional splicing in yeast and establish NET-seq as a powerful tool for studying such events.
Direct observation of transcription of unstable RNA
NET-seq monitors transcripts regardless of their stability, making it ideally suited to the analysis of unstable transcripts. Recent studies have revealed a class of cryptic unstable transcripts (CUTs) that are short (less than ~700 nucleotides), upstream and antisense to an annotated gene and rapidly degraded by the exosome2, 3, 4, 15, 21. Divergent transcription, yielding the production of antisense CUTs and mature messenger RNAs from the sense direction, is seen at many promoters in both yeast and metazoans. The observation of widespread divergent transcription was surprising and it remains unclear how antisense transcripts initiate and what biological function they may have. It is likely that the nucleosome-free region associated with promoters facilitates antisense transcription. Additionally, it has been suggested that antisense and sense transcription levels are co-dependent15, 21, as transcription in the sense direction could promote upstream antisense transcription (and vice versa) by creating negatively supercoiled DNA and recruiting factors that set permissive histone marks22. Critical evaluation of these hypotheses has been limited by the difficulty in quantitatively monitoring the levels of unstable antisense transcripts.
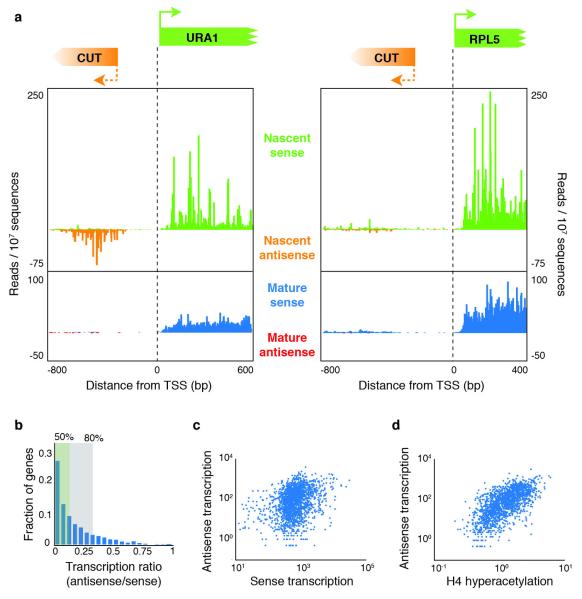
As NET-seq directly monitors the production of transcripts, we were able to quantify the relative amounts of nascent sense and antisense transcripts (Fig. 2a, b). We focused our analysis on promoters between genes encoded on the same strand (tandem genes), because in those instances, antisense transcripts can be clearly differentiated from the stable upstream transcript. To quantify divergent transcription, we integrated the transcript levels for the first 500 bp of transcribed DNA in each direction. Although we clearly observed divergent promoters, the large majority of promoters had much less antisense transcription than sense transcription; for more than half of the promoters, sense transcription was at least eight times higher than antisense transcription, and for 80% of the promoters the sense-to-antisense transcription ratio exceeded threefold (Fig. 2b). Notably, a comparison between the levels of sense and antisense transcription showed only modest correlation (Spearman correlation coefficient, rs = 0.34) (Fig. 2c).
Figure 2.
a, Nascent and mature transcripts initiating from URA1 and RPL5 promoters in the sense and antisense directions. Note that there are cryptic unstable transcripts (CUTs) in the antisense direction for URA1 but not RPL5. b, A histogram of the transcription ratio (antisense/sense transcription levels) for 1,875 genes. The green and grey boxes indicate the subset of genes with a ratio of less than 1:8 and less than 1:3, respectively. c, Antisense transcription levels are plotted versus sense transcription for each tandem gene (Spearman correlation coefficient, rs = 0.34). d, The level of antisense transcription for each promoter is plotted versus the local enrichment for H4 hyperacetylation using available data24 (rs = 0.65).
The above analysis establishes that antisense transcription is not an obligatory consequence of having an active promoter. What then dictates whether a promoter is directional? Transcription initiation is known to occur in nucleosome-free regions; however, we failed to see a correlation when we compared antisense transcription levels with published data23 reporting on promoter nucleosome-free-region size and promoter average nucleosome occupancy (Supplementary Fig. 2a, b). We also investigated whether histone modifications associated with active promoters correlated with antisense transcription, as it was observed that H3 acetylation peaks in regions of antisense transcription in human fibroblasts15. Notably, we found a strong positive correlation (rs = 0.65) between antisense transcription levels and earlier measurements of the levels of H4 (and to a lesser extent H3) acetylation enrichment24 (Fig. 2d and Supplementary Fig. 2c, d).
Rpd3S promotes promoter directionality
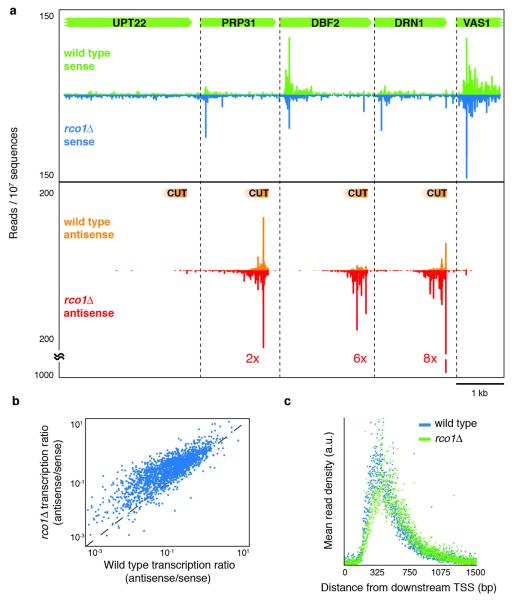
The strong correlation between antisense transcription and H4 acetylation indicates that H4 acetylation may have a causative role in facilitating antisense transcription. To test this, we examined the effect on antisense transcription of loss of RCO1, a required and dedicated subunit of the Rpd3 small (Rpd3S) H4 deacetylation complex25, 26. We focused on Rpd3S, as earlier studies had shown that it contributes to deacetylation of H4 in the 3′ region of transcripts and the large majority of antisense transcripts overlap the 3′ ends of upstream genes. Previous global studies of Rpd3S monitored accumulation of mature stable RNAs and so would not detect the effects of Rco1 on transient RNA species25, 26. Our analysis revealed a pervasive increase (average fourfold) in unstable antisense transcription (Fig. 3a, b). This effect was the dominant transcriptional phenotype that we observed and was specific to antisense transcription: we found no systematic increase in RNAPII density at the beginning of sense transcripts (Supplementary Fig. 3). Importantly, antisense transcripts seen in the rco1δ strain have the same transcription start sites and the same lengths as the wild-type transcripts, indicating that Rco1 is acting at the initiation stage of antisense transcription and does not affect termination (Fig. 3c). Additionally, we observed that deletion of EAF3, another subunit of Rpd3S, mimicked the increases seen in the rco1δ data (rs = 0.88, Supplementary Fig. 4). Thus, the primary function of the Rpd3S histone deacetylase complex seems to be to enforce promoter directionality.
Figure 3.
a, Examples of cryptic unstable transcripts (CUTs, orange data) upstream and antisense of DBF2,DRN1 and VAS1 promoters. The fold increase of CUT transcription in the rco1δ strain is marked. b, The transcription ratio (antisense/sense) in the rco1δ strain is plotted against the transcription ratio in the wild-type strain for each gene. c, A metagene analysis of well-expressed antisense transcription (n = 171, >1 read per bp).
This raises the question of how Rpd3S is recruited to positions designated for suppression of antisense transcription. The Rco1 and Eaf3 components of the Rpd3S complex bind H3 lysine 36 methylation marks made by Set2 and that binding activates the deacetylase activity of Rpd3S (refs 25-27). However, a distinct RNAPII-associated methyltransferase, Set1, has also been implicated in Rpd3S-dependent repression28. Moreover, even in the absence of methylation, RNAPII is capable of recruiting Rpd3S to gene bodies during transcription29.
To investigate how Rpd3S is localized to suppress antisense transcription, we analysed nascent transcripts in cells lacking Set1 or Set2. SET1 deletion caused a weak increase in antisense transcription in a manner that correlated only modestly with the rco1δ and eaf3δ data (rs = 0.36 and rs = 0.38 respectively; Supplementary Fig. 5). In contrast, deletion of SET2 led to a pronounced increased in antisense transcription that was highly correlated with the rco1δ and eaf3δ data (rs = 0.88 and rs = 0.89 respectively; Supplementary Fig. 5). These data together with earlier work on the Set2/Rpd3S pathway indicate that the major mechanism for Rpd3S action on antisense transcription involves Set2 recruitment to elongating RNAPII via Ser2 phosphorylation on its carboxy-terminal domain30. This in turn, through the Set2 methylation activity, allows recruitment of Rpd3S to the 3′ ends of genes, suppressing antisense transcription from downstream nucleosome-free regions. Future challenges will be to explain how histone acetylation in the body of antisense transcripts can affect transcription initiation, and to determine other mechanisms that localize Rpd3S, particularly for the handful of antisense transcripts that do not overlap the 3′ ends of genes.
Pausing occurs throughout transcription elongation
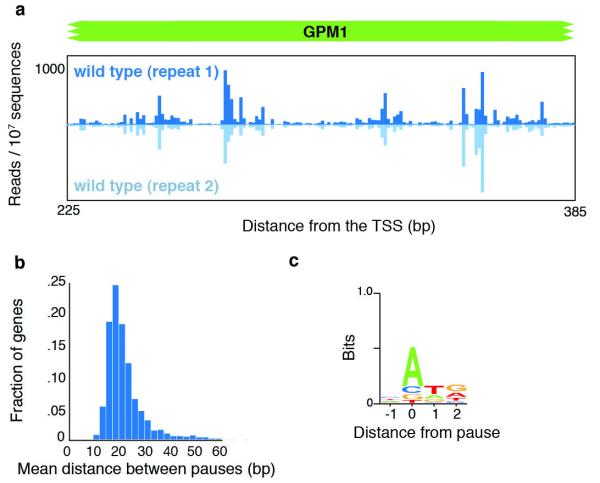
The ability of NET-seq to map the density of nascent transcripts enables in-depth investigation of the extent and sources of RNAP pausing in vivo. Our data revealed strong and highly reproducible spikes in the density of 3′ ends of nascent transcripts along a given gene indicative of RNAPII pause sites (for example, GPM1; Fig. 4a). We developed an algorithm to identify RNAPII pause positions that finds points where the read density is at least three standard deviations above the mean in a local 200-bp window. We found that pauses occur frequently throughout the body of RNA messages and are evenly distributed after the first ~700 bp (Fig. 4b and Supplementary Fig. 6). The high density of pauses was not an artefact of library generation and sequencing biases, as we detected tenfold fewer spikes in data from messenger RNA lightly fragmented by alkaline hydrolysis (Supplementary Fig. 7). Notably, 70% of the more than 2 × 105 pause sites that we identified had an A at the 3′ end of the transcript. Additionally, there was a preference for the pause to be followed immediately by a T and then G (Fig. 4c). None of these biases was seen in the control sample of fragmented mRNA (Supplementary Fig. 7a).
Figure 4.
a, NET-seq data at the GPM1 gene for biological replicates. b, A histogram of the mean distance between pauses for each well-expressed gene (n = 1,006, >2 reads per bp). c, The consensus sequence of the DNA coding strand surrounding pause sites found from all genes.
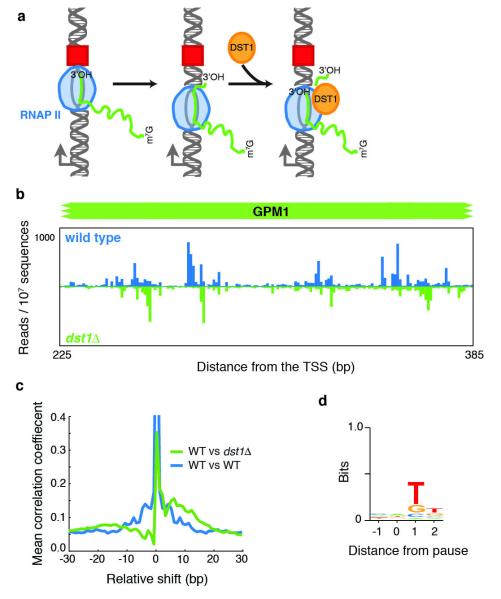
Largely from in vitro studies, one mechanism of RNAP pausing has been shown to involve backtracking: after encountering a blockage, RNAP reverses direction and moves upstream31. In the backtracked state, the 3′ end of the RNA transcript is no longer aligned with the active site and RNAP must either return to the initial pause site or cleave the transcript. The latter option is aided by the presence of the elongation factor TFIIS (Dst1 in yeast) that enhances RNAP’s intrinsic RNA cleavage activity (Fig. 5a)32, 33. Although the role of TFIIS is well established in vitro, its mechanism in vivo has been less explored34, 35, 36.
Figure 5.
a, A schematic describing an existing model for how RNAPII pauses at an obstacle (red square), backtracks and is induced to cleave its transcript through binding to Dst1 (refs 32, 33). b, A comparison of NET-seq data for wild-type and dst1δ strains at the GPM1 gene. c, Mean cross-correlation between the dst1δ and wild-type data of well transcribed genes (n = 770, >2 reads per bp) (green line) was calculated by determining the Pearson’s correlation coefficient at each gene between fixed dst1δ data and shifted wild-type data followed by averaging over all genes. This analysis is compared to the mean autocorrelation of the wild-type data for well transcribed genes (blue line). d, The consensus sequence for all pauses observed in the dst1δ strain.
To investigate the role that backtracking has in pausing in vivo, we deleted DST1 and repeated the NET-seq assay. Notably, we saw a large-scale downstream shift in the position of the pauses, an average of 5-18 bp (Fig. 5b, c). This shift was observed for ~75% of the pauses (Supplementary Fig. 8) and was accompanied by a global change in the sequences surrounding pause sites; the preference for A at the pause was lost and instead there was a strong preference for T immediately downstream of the pause (Fig. 5d). These observations confirm that the observed spikes in NET-seq data result from RNAPII pausing, and indicate that pausing followed by backtracking—which previously had been observed at promoter-proximal pauses35—is prevalent throughout the body of transcripts. Additionally, our studies indicate that Dst1-stimulated RNA cleavage has a strong sequence bias and that a slow step follows cleavage before transcription resumes.
RNAPII pause density peaks before the nucleosome dyad
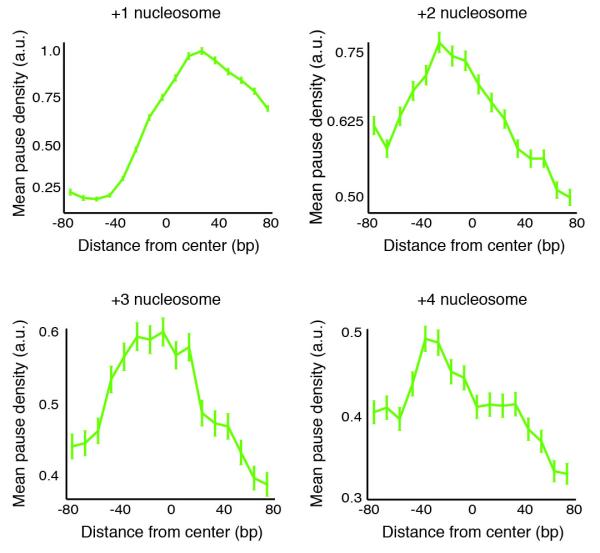
The pauses observed in the dst1δ strain reveal positions where RNAPII began to backtrack and, therefore, represent the primary point of transcriptional blockage. By analysing these pause positions, we can evaluate what induced RNAPII to backtrack. In vitro, nucleosomes induce RNAPII backtracking and TFIIS aids the progression of RNAPII through them10, 12. In vivo, it is unknown whether nucleosomes interfere with transcription, as chromatin remodelling factors could greatly diminish the nucleosome barrier or remove nucleosomes before RNAPII arrival37, 38. Global high-resolution measurements of steady-state nucleosome occupancy revealed that the first few nucleosomes after the transcription start site are phased and well positioned23, 39. Thus, by correlating the relative density of RNAPII pauses with nucleosome positions, we can evaluate whether nucleosomes promote RNAPII pausing in vivo.
We compared the pause positions in the dst1δ strain to the centre positions of nucleosomes using previously published data23. Notably, we saw marked peaks of mean pause density at each of the first four nucleosomes (Fig. 6). The precise position of the point of maximal RNAPII pausing at the +1 nucleosome is obscured because it is located just after the transcription start site where many nascent transcripts are too short for unique alignment to the genome. For the +2, +3 and +4 nucleosomes, however, the pause density peaks just before the nucleosome dyad axis (Fig. 6). As would be expected from RNAPII backtracking, the excess pause density before the nucleosome dyad in the wild-type strain is spread out over the upstream region (Supplementary Fig. 9).
Figure 6.
Plot of mean pause densities in dst1δ data relative to the first four nucleosomes after the transcription start site using available nucleosome positioning data23. Error bars represent one standard deviation.
Our finding that the peak in pause density occurs just before the nucleosome dyad is particularly remarkable as it is in excellent agreement with earlier biophysical measurements. Specifically, optical trapping studies that physically unwrapped the DNA of a nucleosome off the histone core observed that the dyad is the point where the strongest DNA-histone contacts are found40. Moreover, high-resolution optical trapping experiments that followed RNAPII transcribing through a nucleosome found that the RNAPII pause density peaked before the nucleosome dyad10. Taken together, the above observations provide strong evidence that nucleosomes do indeed present a barrier to elongating polymerases in vivo and that this barrier leads to polymerase pausing and backtracking.
Perspective
One of the major surprises in the transcription field in recent years has been the widespread observation of divergent transcription, revealing that the majority of promoters engage in canonical transcription in the sense direction along with the production of unstable transcripts in the antisense direction2, 3, 4, 15, 21. NET-seq provides an ideal tool to look at this phenomenon and uncovers several fundamental properties of divergent transcription. First, most promoters show a strong directionality favouring the sense transcript. Second, suppression of antisense transcripts is enforced by two distinct mechanisms: Rpd3S-mediated deacetylation that prevents antisense initiation, and an independent mechanism, previously characterized to involve the Nrd1-Nab3-Sen1 complex41, that terminates antisense transcripts and shuttles them to the exosome for degradation. Interestingly, sense transcription may also use this termination mechanism, as our data showed an enrichment for transcripts at the 5′ end of genes that mirrors what we observed for antisense transcripts and complements observations that Nrd1 localizes to the 5′ end of genes42. Third, our observations indicate independence between the initiation of the sense and antisense transcripts. Specifically, we found only modest correlation between sense and antisense transcription levels. Moreover, even among the set of antisense transcripts that increased when RCO1 is deleted, no increase in sense transcription levels was seen. These findings argue against models in which antisense transcription serves to promote sense transcription (for example, by unwinding DNA supercoils or by removing nucleosomes).
The potential for RNAP to pause has been apparent for decades, motivating interest in the mechanisms and regulatory roles of pausing in the process of transcription7, 9, 11, 12, 15. NET-seq provides the first in-depth view of pausing in a eukaryotic cell, revealing that transcription is punctuated by pauses throughout the body of all RNA messages. Taking into account both the abundance and magnitude of the pauses, we conclude that RNAPII spends comparable time in a paused state and moving forwards (Supplementary Fig. 10). We establish that nucleosomes induce pausing in vivo, and may be the major source of pausing considering that the increase in pause density at nucleosomes is comparable to the increase in nucleosome occupancy23. Our observation that pausing peaks at the nucleosome dyad reveal a striking similarity between our measurements and optical trap measurements, indicating that the physical forces observed in purified in vitro systems are at play in the cell. NET-seq’s ability to follow the physical basis of transcription in vivo, allowing direct comparison with high-resolution in vitro measurements, may prove to be the most transformative aspect of this approach.
METHODS
Strain construction
All experiments were conducted using derivatives of yeast strain BY4741. Epitope-tagged Rpb3 (C-terminal 3×-Flag) was expressed from its endogenous locus. Deletion strains were made by standard PCR-based methods.
Extract and total RNA preparation
Yeast strains were grown in YEPD at 30 °C with shaking from an initial optical density (OD) of 0.1 to mid-log phase with an OD of 0.6–0.8. Two litres of yeast culture were harvested in turn by filtration onto 0.45-μm-pore-size nitrocellulose filters (Whatman). The culture was scrapped off the filter with a spatula pre-chilled by liquid nitrogen and flash frozen by plunging into liquid nitrogen. Frozen cells were pulverized for six cycles, each of 3 min at 15 Hz, on a Retsch MM301 mixer mill. Sample chambers were pre-chilled in liquid nitrogen and re-chilled between each pulverization cycle.
One gram of ground cells (~ 1 l at 0.7 OD) was added to 5 ml of ice-cold lysis buffer (20 mM HEPES, pH 7.4, 110 mM KOAc, 0.5% Triton X-100, 0.1% Tween 20, 10 mM MnCl2, 50 U ml−1 SUPERase•In (Ambion)) supplemented with protease inhibitor cocktail (1× Complete, EDTA-free, Roche). The experiment using α-amanitin included 10 μg ml−1 α-amanitin (Sigma Aldrich) in the lysis buffer. After re-suspending the lysate by pipetting, 660 units of DNase I (Promega, RQ1 RNase-Free DNase) was added and incubated for 20 min on ice. The lysate was then clarified by centrifugation at 4 °C at 20,000g for 10 min. The supernatant is reserved for immunoprecipitation.
Two-hundred microlitres of clarified lysate is reserved for total RNA purification which was done by the hot acid phenol method. Typical yields were 20 μg.
Native affinity purifications of RNAPII
0.5 ml of Anti-Flag M2 Affinity Gel (Sigma Aldrich) was washed twice with lysis buffer. The clarified lysate was added to the washed gel, incubated at 4 °C and nutated for 2.5 h. The immunoprecipitation was washed 4 × 10 ml with wash buffer (20 mM HEPES, pH 7.4, 110 mM KOAc, 0.5% Triton X-100, 0.1% Tween 20, 50 U ml−1 SUPERase•In (Ambion), 1 mM EDTA). Bound proteins were eluted twice with 150 μl elution buffer (20 mM HEPES, pH 7.4, 110 mM KOAc, 0.5% Triton X-100, 0.1% Tween 20) with 2 mg ml−1 3×-Flag peptide (Sigma Aldrich). RNA from the combined eluates was purified using the miRNeasy kit (Qiagen, 217004). A typical yield from approximately one litre of log-phase yeast culture was 3 μg.
mRNA purification and fragmentation
Polyadenylated mRNA was purified from 50 μg total RNA using magnetic oligo-dT DynaBeads (Invitrogen). Purified RNA was eluted in 20 μl 10 mM Tris, pH 7.0. The purified mRNA was mixed with an equal volume of 2× alkaline fragmentation solution (2 mM EDTA, 10 mM Na2CO3, 90 mM NaHCO3, pH ≈ 9.3) and incubated for 5 min at 95 °C. These conditions yielded lightly fragmented RNA of size distribution similar to that of the nascent RNA. The fragmentation reaction was stopped by the addition 0.56 ml of ice-cold precipitation solution (final 300 mM NaOAc pH 5.5, plus GlycoBlue (Ambion) as a co-precipitant) and RNA was purified by a standard isopropanol precipitation, as follows: after adding 650 μl of isopropanol, samples were placed at −30°C for at least 30 min. Precipitated RNA was pelleted by centrifugation at 4°C at 20,000g for 30 min. The pellet was air dried after a quick wash with 80% ethanol and then re-suspended in 10 mM Tris pH 7.0.
A total of 6.4 μg of fragmented mRNA was dephosphorylated in a 50 μl reaction with 1× T4 polynucleotide kinase buffer without ATP, 0.5 U SUPERase•In (Ambion) and 22.5 units T4 polynucleotide kinase (NEB). The dephosphorylation reaction was incubated at 37°C for 1 h followed by 10 min at 75°C for enzyme heat inactivation. RNA was precipitated with GlycoBlue by standard methods (see above).
RNA linker ligation, fragmentation and size selection
An RNA linker that was 5′ adenylated and 3′-end blocked with a dideoxy-C base (5′-CTGTAGGCACCATCAAT, Integrated DNA Technologies) was ligated onto the 3′ end of the immunoprecipitated RNA, the fragmented mRNA and a synthetic 28-base RNA oligonucleotide (oNTI199, 5′-AUGUACACGGAGUCGACCCGCAACGCGA) similarly to what has been described43. Specifically, 3 μg of each RNA sample was broken into three reactions and diluted to 10 μl with 10 mM Tris, pH 7.0. After a brief denaturation the reactions were brought to 20 μl with a buffer that gave final concentrations of 12% PEG8000, 50 ng μl−1 linker, 1× T4 Rnl2, truncated reaction buffer and 2 units μl−1 of T4 Rnl2, truncated (NEB). The reaction was incubated at 37°C for 3 h. Ligation conditions were systematically optimized to maximize ligation efficiency to ~90% to ensure that the majority of the input RNA was ligated.
Fragmentation of the ligated samples allowed for the final DNA library to contain inserts of a narrow range to reduce any length biases of downstream enzymatic reactions. EDTA was added to all reactions for a final concentration of 17 mM. 20 μl of 2× alkaline fragmentation solution (2 mM EDTA, 10 mM Na2CO3, 90 mM NaHCO3, pH ≈ 9.3) was added to each reaction and incubated at 95°C for 30 min. The reactions were stopped by the addition of 0.56 ml of ice-cold precipitation solution (final 300 mM NaOAc pH 5.5, plus GlycoBlue (Ambion) as a co-precipitant), followed by a standard isopropanol precipitation (see above).
The ligated and fragmented samples were size-selected by gel electrophoresis. The purified reactions along with the oNTI199 RNA oligonucleotide was mixed with 2× Novex TBE-Urea sample prep buffer (Invitrogen) and briefly denatured, then loaded on a Novex denaturing 15% polyacrylamide TBE-urea gel (Invitrogen) and run according to the manufacturer’s instructions. The gel was stained with SYBR Gold (Invitrogen) and the 35–85-nucleotide region was excised. The gel was physically disrupted and either allowed to soak overnight in gel elution buffer (300 mM NaOAc pH 5.5, 1 mM EDTA, 0.1 U μl−1 SUPERase·In) or incubated in 200 μl of water treated with diethylpyrocarbonate (DEPC) for 10 min at 70°C. The gel debris was removed from the water or buffer using a Spin-X column (Corning) and RNA was precipitated with GlycoBlue as a co-precipitant using standard methods.
cDNA synthesis
cDNA synthesis was performed as described with a few modifications19. The primer used for reverse transcription was oLSC003 (5′-pTCGTATGCCGTCTTCTGCTTG•-AATGATACGGCGACCACCGATCCGACGATCATTGATGGTGCCTACAG) where the initial ‘p’ indicates 5′ phosphorylation and ‘•’ indicates the following spacer added for increased flexibility, 18 carbon spacer molecule-CACTCA-18 carbon spacer molecule. Efficient circularization of the RT product was performed as described19 with CircLigase (Epicentre) according to the manufacturer’s directions. Any ligation bias at this step is averaged out as the random fragmentation leaves a range of 5′ ends for each 3′ end. The PCR was performed directly on the circularized product as described19, resulting in DNA with Illumina cluster generation sequences on each end and a sequencing primer binding site positioned so that sequencing would start at the 3′ end. DNA was purified from a PCR reaction that had not reached saturation and was quantified using the Agilent BioAnalyser High Sensitivity DNA assay. DNA was then sequenced on the Illumina Genome Analyser 2 according to the manufacturer’s instructions, using 4-6 pM template for cluster generation and sequencing primer oLSC006 (5′-TCCGACGATCATTGATGGTGCCTACAG).
Data analysis
Data analysis was performed using scripts written in Python 2.6 that are available upon request.
Sequencing analysis
Image data obtained by the Illumina Genome Analyser 2 was analysed using the GAPipeline to extract raw sequences. Matrix and phasing parameters were estimated from a φX control lane.
Sequence alignment
Raw sequences 40 bases long were composed of the cDNA of the fragmented RNA sequence. For RNA fragments smaller than 40 bases, the sequence is followed by part of the 5′ Illumina linker sequence which was removed in silico. Alignments to the yeast genome were performed by the alignment program, Bowtie 0.12.044 (http://bowtie-bio.sourceforge.net/). Bowtie settings were chosen so that three mismatches were allowed and alignments were required to be unique. The shortest sequenced fragments were approximately 18 nucleotides due to the RNA size selection step after ligation and random fragmentation. Eighteen-base-pair sequences would occur by chance every 6.9 × 1010 bp, which is sufficiently rare for 18-bp sequences to be generally uniquely aligned to the 1.2 × 107 bp yeast genome. Alignments were first performed against tRNA and rRNA sequences to remove them. The remaining sequences were aligned against a recent version of the yeast genome downloaded from the Saccharomyces Genome Database (SGD, http://www.yeastgenome.org/) on 11 October 2009. Statistics on sequence alignments are reported in Supplementary Table 1.
Quantifying antisense and sense transcription levels
At tandem promoters sense transcription was determined using available annotated transcription start sites3. To allow for the error involved in these transcription start site measurements, we calculated the sum of the read density in 500-nucleotide windows for the first 700 bases after the transcription start site and chose the highest sum. The antisense transcription was determined by starting 100 bases upstream of the transcription start site and the read density sum in 500-nucleotide-wide windows was calculated for the subsequent 1,000 bases. The highest sum was used for downstream analysis.
Metagene analysis
Each gene included in the analysis is normalized by the mean number of reads in a 400-bp window beginning 100 bases downstream from the transcription start site. A mean read density (MRD) is then calculated for each position over all genes as described below.
where are the reads for the jth gene at the ith position after the transcription start site.
Extracting pause positions
Pauses were identified in previously annotated transcription units3 of well-expressed genes. Pauses were defined as having reads higher than three standard deviations above the mean of the surrounding 200 nucleotides which do not contain pauses. Pauses were required to have at least four reads regardless of the gene’s sequencing coverage. Sequence consensus was calculated by WebLogo 3 (http://weblogo.threeplusone.com/)45.
Comparing pause densities to nucleosome positions
Nucleosome positions23 were assigned as +1, +2, +3 etc according to their position relative to transcription start sites. The mean pause density (MPD) relative to a particular nucleosome was determined by the number of pauses observed at that position (Np) divided by the total number of opportunities it could be observed there (No):
where k is the nucleosome number, g(y) is the binary function indicating whether a pause occurs at y, and are the centre nucleosome positions. For the +2 and +3 nucleosomes, the number of pause opportunities was uniform at every position and was simply the number of genes included in the analysis. The +1 nucleosome analysis required that the number of pause opportunities at each position represent the number of genes where that position occurs after the transcription start site. The error of the pause density was calculated via the standard deviation of the binomial distribution
The densities were then binned by averaging across windows ten nucleotides wide. The error for each bin was calculated by computing the sum of the variances of the binned measurements and calculating the square root.
Supplementary Material
Acknowledgements
We thank C. Guthrie, N. Krogan, S. Luo, G. Schroth, J. Steitz and K. Yamamoto for advice and discussions; D. Breslow, P. Fordyce, A. Frost, J. Huff, M. Kampmann and M. Pufall for critical comments on the manuscript; C. Chu and N. Ingolia for help with sequencing and analysis; and S. Rouskin for help developing the ligation protocol. This research was supported by the Damon Runyon Cancer Research Foundation (DRG-1997-08 to L.S.C.) and by the Howard Hughes Medical Institute (to J.S.W.)
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Contributions
L.S.C. and J.S.W. designed the experiments; L.S.C. performed the experiments and analysed the data; and L.S.C. and J.S.W. interpreted the results and wrote the manuscript.
References
- 1.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Preker P, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 3.Xu Z, et al. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neil H, et al. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 5.Rougvie AE, Lis JT. The RNA polymeraseII molecule at the end of the uninduced hsp70 gene of D.melanogaster is transcriptionallyengaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- 6.Proshkin S, Rahmouni AR, Mironov A, Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328:504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kassavetis GA, Chamberlin MJ. Pausing and termination of transcription within the early region of bacteriophage T7 DNA in vitro. J. Biol. Chem. 1981;256:2777–2786. [PubMed] [Google Scholar]
- 8.Shaevitz JW, Abbondanzieri EA, Landick R, Block SM. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature. 2003;426:684–687. doi: 10.1038/nature02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herbert KM, et al. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 2006;125:1083–1094. doi: 10.1016/j.cell.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal fluctuations govern the transcription dynamics ofRNA polymerase II. Science. 2009;325:626–628. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kireeva ML, Kashlev M. Mechanism of sequence-specific pausing of bacterial RNA polymerase. Proc. Natl Acad. Sci. USA. 2009;106:8900–8905. doi: 10.1073/pnas.0900407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kireeva ML, et al. Nature of the nucleosomal barrier to RNA polymerase II. Mol. Cell. 2005;18:97–108. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 13.Kim TH, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lefrançois P, et al. Efficient yeast ChIP-Seq using multiplex short-read DNA sequencing. BMC Genomics. 2009;10:37. doi: 10.1186/1471-2164-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodríguez-Gil A, et al. The distribution of active RNA polymerase II along the transcribed region is gene-specific and controlled by elongation factors. Nucl. Acids Res. 2010;38:4651–4664. doi: 10.1093/nar/gkq215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai H, Luse DS. Transcription initiation by RNA polymerase II in vitro. Properties of preinitiation, initiation, and elongation complexes. J. Biol. Chem. 1987;262:298–304. [PubMed] [Google Scholar]
- 18.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Markham R, Smith JD. The structure of ribonucleic acids. I. Cyclic nucleotides produced by ribonuclease and by alkaline hydrolysis. Biochem.J. 1952;52:552–557. doi: 10.1042/bj0520552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seila AC, et al. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seila AC, Core LJ, Lis JT, Sharp PA. Divergent transcription: a new feature of active promoters. Cell Cycle. 2009;8:2557–2564. doi: 10.4161/cc.8.16.9305. [DOI] [PubMed] [Google Scholar]
- 23.Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 2010;20:90–100. doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pokholok DK, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 25.Carrozza MJ, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 26.Keogh MC, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 27.Li B, et al. Histone H3 lysine 36 dimethylation (H3K36me2) is sufficient to recruit the Rpd3s histone deacetylase complex and to repress spurious transcription. J. Biol. Chem. 2009;284:7970–7976. doi: 10.1074/jbc.M808220200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinskaya M, Gourvennec S, Morillon A. H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J. 2009;28:1697–1707. doi: 10.1038/emboj.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Govind CK, et al. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol. Cell. 2010;39:234–246. doi: 10.1016/j.molcel.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krogan NJ, et al. Methylation of histone H3by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 32.Izban MG, Luse DS. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J. Biol. Chem. 1992;267:13647–13655. [PubMed] [Google Scholar]
- 33.Reines D, Conaway RC, Conaway JW. Mechanism and regulation of transcriptional elongation by RNA polymerase II. Curr. Opin. Cell Biol. 1999;11:342–346. doi: 10.1016/S0955-0674(99)80047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kulish D, Struhl K. TFIIS enhances transcriptional elongation through an artificial arrest site in vivo. Mol. Cell. Biol. 2001;21:4162–4168. doi: 10.1128/MCB.21.13.4162-4168.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nechaev S, et al. Global analysis of short RNAs reveals widespread promoter proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sigurdsson S, Dirac-Svejstrup AB, Svejstrup JQ. Evidence that transcript cleavage is essential for RNA polymerase II transcription and cell viability. Mol. Cell. 2010;38:202–210. doi: 10.1016/j.molcel.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li B, Carey M, Workman J. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 38.Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaplan N, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall MA, et al. High-resolution dynamicmapping of histone-DNA interactions in a nucleosome. Nature Struct. Mol. Biol. 2009;16:124–129. doi: 10.1038/nsmb.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arigo JT, Eyler DE, Carroll KL, Corden JL. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell. 2006;23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 42.Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. The Nrd1-Nab3- Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nature Struct. Mol. Biol. 2008;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Unrau PJ, Bartel DP. RNA-catalysed nucleotide synthesis. Nature. 1998;395:260–263. doi: 10.1038/26193. [DOI] [PubMed] [Google Scholar]
- 44.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.