Abstract
Limiting the precise study of the biochemical impact of whole molecule extracellular matrix (ECM) proteins on stem cell differentiation is the lack of 3D in vitro models that can accommodate many different types of ECM. Here we sought to generate such a system while maintaining consistent mechanical properties and supporting stem cell survival. To this end, we used native chemical ligation to crosslink poly(ethylene glycol) macromonomers under mild conditions while entrapping ECM proteins (termed ECM composites) and stem cells. Sufficiently low concentrations of ECM were used to maintain constant storage moduli and pore size. Viability of stem cells in composites was maintained over multiple weeks. ECM of composites encompassed stem cells and directed the formation of distinct structures dependent on ECM type. Thus, we introduce a powerful approach to study the biochemical impact of multiple ECM proteins (either alone or in combination) on stem cell behavior.
Keywords: stem cells, extracellular matrix, native chemical ligation, differentiation, poly(ethylene glycol) hydrogel
INTRODUCTION
With evolution, the emergence of extracellular matrix proteins (ECM) enabled cells to move from unicellularity to multicellularity and assume new, advanced functionality.1,2 This complex cellular functionality of cells can be lost or altered when cells are removed from tissue and expanded outside the body on 2D planar surfaces. Thus, cell biologists and engineers have joined forces to create 3D scaffolds, exhibiting properties of the native ECM for expanding isolated cells outside the native tissue. Arguably the most prevalent advances have been made with the aid of synthetic hydrogels, which can be tuned to roughly approximate the material properties of many tissues.3–5 To these polymer substrates, polypeptides6 have been added that encode ECM-based signaling cues. Such cues include cell-binding ligands,7,8 immobilized growth factors,9,10 and Ephephirin dual ligands11,12 to stimulate, inhibit or otherwise direct advanced cell functions.
The use of chemically synthesized ECM-based peptides is generally limited to short sequences that lack spatial selectivity necessary to produce all cell-signaling ligands/epitopes of an intact ECM protein. To bypass this limitation, many have used native ECM extracted from tissue to create 3D scaffolds in vitro.13–18 However, with the exception of type I collagen, hyaluronan, fibrin, and laminin/entactin complex, ECM proteins isolated from tissues cannot reassemble reliably into robust 3D scaffolds capable of supporting cellular populations. Extracted ECM proteins that cannot reassemble into 3D scaffolds outside the body have been incorporated into natural or synthetic scaffolds.19,20 For example, type I collagen has been utilized as a supporting scaffold for other ECM proteins purified from tissue21,22 or lyophilized ECM mixtures extracted from decellularized tissues.23 However, in these cases it is challenging to distinguish biochemical effects of the added ECM components from that of the supporting ECM (i.e., type I collagen) scaffold. To overcome the limitations of synthetic and natural ECM-based scaffolds, we have generated ECM composites by entrapping two different families of ECM proteins, fibrillar collagen type I and reticular laminin, within chemoselectively crosslinked poly(ethylene glycol) (PEG) hydrogels. In this way, ECM proteins are secured in 3D space without modification while the biological inert PEG serves to maintain consistent mechanical integrity and transport properties.
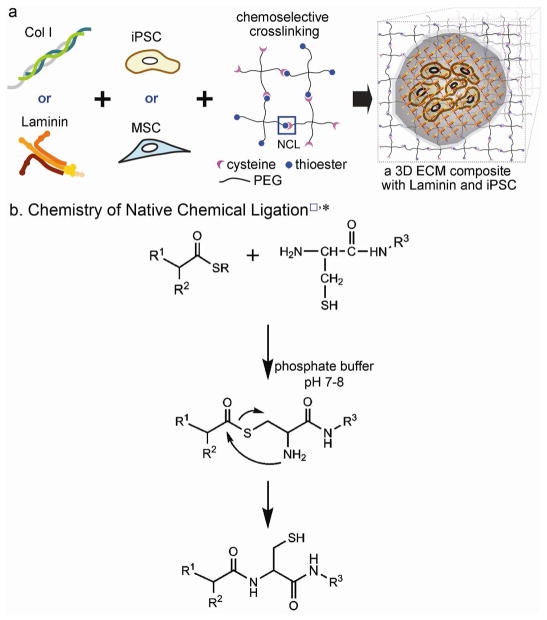
To incorporate whole molecule ECM in 3D hydrogels without altering the presentation of ECM to encapsulated cells, we utilized native chemical ligation (NCL, Figure 1 and S1) to orthogonally crosslink PEG precursors under mild conditions. NCL was originally developed for chemically synthesizing large polypeptides containing more than 300 residues from two or more unprotected synthetic peptide segments.24 The chemistry proceeds via the reaction of a C-terminal thioester and an N-terminal cysteine to form a native peptide bond. It is the formation of this native bond and the fact that the reaction occurs in aqueous solution that makes NCL highly attractive for crosslinking hydrogel precursors intended as scaffolds for biological applications. Further, the reaction is exceptionally chemoselective, and side reactions other than the terminal reactive groups do not occur.
Figure 1.
(a) Schematic representation of producing 3D ECM composites that are composed of whole molecule ECM, stem cells, and PEG crosslinked by NCL. A hypothetical inner view of a 3D Laminin composite encapsulating murine iPSCs is provided to show that the chemoselective crosslinking secured encapsulated stem cells engaged with ECMs in a 3D hydrogel. (b) The scheme of NCL chemistry (*, □ NCL in a) is reproduced from Ref. 30 with permission.
Previously, NCL was utilized to stiffen β-sheet fibrillizing peptide hydrogels,25 to encapsulate insulin secreting MIN6 β cells in PEG hydrogels,26 and to immobilize PEG on titanium surfaces.27 Here, we chemoselectively crosslink PEG precursors without employing ultraviolet (UV) light and toxic radicals such as photoinitators, while incorporating whole molecule ECM to encapsulate multipotent or pluripotent stem cells (see Figure 1). We use this technology to examine the impact of 3D, ECM-based environments on stem cell behavior. The functionality of stem cells is altered with exposure to ECM in 2D culture systems28–31 but until now, it has been difficult to conduct similar studies in 3D scaffolds. Thus this technology holds promise for understanding the biochemical impact of ECM molecules, in 3D context, on the behavior of a variety of cell types including stem cells with the potential to change the way we culture cells in vitro and to predict how cells might behave when delivered to the body for regenerative purposes.
MATERIALS AND METHODS
Materials
Four-armed poly(ethylene glycol) (PEG) with amine end groups (molecular weight 20k: PEG-4A) was purchased from Laysan Bio (cat# 4arm-NH2-20K, Arab, AL). Ethyl-3-mercaptopropionate (EMP) was purchased from TCI America (cat# M1038, Portland, OR). Benzotriazol-1-yl-oxy-tris-(dimethylamino)-phosphonium hexafluorophosphate (BOP) was purchased from Peptide International (cat# KBP-1060-PI, Louisville, KY). The protected cysteine (Boc-Cys(Trt)-OH) was purchased from EMD Millipore (cat# 853005). Diethyl ether, N,N-dimethlyformamide (DMF), acetonitrile, trifluoroacetic acid (TFA), dichloromethane (DCM), methanol, Ellman’s reagent (5,5′-Dithio-bis-(2-nitrobenzoic acid), DTNB, cat# 22582), and Cysteine HCl (cat# 44889) were purchased from Fisher Scientific (Hanover Park, IL). Anhydrous ethyl acetate, succinic anhydride, N,N-diisopropylethylamine (DIEA), 4-dimethylaminopyridine (DMAP), triisopropylsilane (TIS), 1,2-ethanedithiol (EDT), and pyridine were purchased from Sigma-Aldrich (Milwaukee, WI). Rat tail collagen type I and mouse laminin were purchased from BD (cat# 354236 and 354259, respectively, Franklin Lakes, NJ).
Synthesis of 4-armed PEG conjugated with thioester and cysteine groups
Cysteine- and thioester-terminated 4-armed PEG macromonomers were synthesized using previously published protocols.32 Briefly, EMP-SA (0.117 g, 0.5 mmol, details described in Supporting Information) was added to 2 mL DCM containing PEG-4A (1 g, 0.2 mmol amine), BOP (0.111 g, 0.25 mmol), and DIEA (0.174 mL, 1 mmol) in a scintillation vial. This mixture (PEG-thioester) was mixed for 2 h at room temperature (RT), concentrated with a stream of Ar, and dissolved in 25 mL of methanol. This clear solution of PEG-thioester was placed into a cooler with dry ice over an hour, centrifuged at −9°C/1600 g for 20 min, and decanted. Fresh methanol was added and the mixture placed into the cooler with dry ice. This was repeated three more times, where the last step was repeated with diethyl ether. A white precipitation of PEG-thioester was stored at −20°C overnight, then lyophilized for 2 days. This PEG-thioester sample was dialyzed in Milli-Q water (resistivity >18.2 MΩ·cm) over 4 days. A small amount of sample was taken for thin layer chromatography (TLC) to confirm conjugation of EMP-SA to PEG-4A and for nuclear magnetic resonance (NMR) spectroscopy to verify chemical structures of PEG-thioester: 1 H NMR (CDCl3, 400 MHz) δ 4.15 (2H, q, -CH2-CH3), 3.74-3.41 (m, -O-CH2-CH2-O-), 3.12 (2H, t, -S-CH2-), 2.92 (2H, t, -S-CO-CH2-), 2.61 (2H, t, -S-CH2-CH2-), 2.52 (2H, t, -CH2-COOH), 1.26 (3H, t, -CH2CH3). For synthesis of PEG-cysteine (PEG-Cys), Boc-Cys(Trt)-OH (0.07 g, 0.15 mmol) in 4 mL of DCM was added to PEG-4A (0.5 g, 0.1 mmol amine) with BOP (0.067 g, 0.15) and DIEA (0.0265 mL, 0.15 mmol) to a scintillation vial. The subsequent procedures were identical to those for preparation of PEG-thioester. After lyophilization, the side chain protecting group of cysteine was removed by dissolving lyophilized PEG-Cys (protected) in a deprotection cocktail (TFA:TIS:EDT = 28:1:1, volume ratio) for 2 h at RT. The deprotection cocktail was removed by rotary evaporation and washing steps with methanol and diethyl ether were repeated. Lyophilized PEG-Cys (deprotected) was dissolved in 25 mL of 0.01 M NH4HCO3 buffer per 0.2 mmol PEG-NH2, yielding TFA salt-free conjugates. This neutralized PEG-Cys was frozen and lyophilized in the same procedure. After lyophilization, a small amount of sample was taken for thin layer chromatography (TLC) to confirm deprotection of Cys residue in PEG-Cys and MALDI time-of-flight mass spectrometry to verify conjugation of Cys to PEG-4A. PEG-Cys and PEG-thioester were stored at −20°C until experimentation.
Formation of hydrogels and ECM composites
Two different kinds of PEG hydrogels were formed. First, PEG-Cys was dissolved in water and PEG-thioester was dissolved in 70 mM NaHCO3. A 15 μL aliquot of each of the two precursors were mixed (1:1), cast in an UV-sterilized dialysis tube (cat# 69562, Pierce, Rockford, IL), and incubated at 37°C for 15 min. Then, these hydrogels were transferred to a well of a 24-well plate containing 1 mL of buffer (reaction buffer for Ellman’s assay or PBS for oscillating rheometry and thermogravimetric analysis (TGA)), where the hydrogels were in a free-floating condition without exposure to air. To produce an ECM composite, PEG-Cys was dissolved in α-MEM (cat# 12000-022, Invitrogen, Carlsbad, CA) supplemented with 1% non-essential amino acids (cat# 11140-050, Gibco/Invitrogen) and 10% fetal bovine serum (FBS, cat# 30070.03, Thermo Scientific), while PEG-thioester was dissolved in α-MEM supplemented with additional NaHCO3 to adjust the final concentration of NaHCO3 to 35 mM. Either rat tail collagen type I (Col I) or mouse laminin was neutralized and polymerized at 37°C/5% CO2 for 2 h. In each neutralization and mixing step, the pH of the mixtures was verified by pH indicator strips (from pH 5.0 to 10.0 with 0.5 increment, cat# 9588, EMD Millipore, Germany), confirming that the mixture pH was between 7.5 to 8.0. These viscous solutions were mixed and cast with a 200 μL blunt pipette tip onto sterilized dialysis tubes. To remove NCL by-products (for details, see Figure S1.), ECM composites were crosslinked without immersing dialysis tubes on a 48-well plate for 30 min at 37°C/5% CO2, then moved to a well containing 500 μL of fresh cell culture medium on a 48-well plate. This procedure was repeated three times at 30 min intervals, followed by transferring ECM composites to a well on a 24-well plate containing 1 mL of fresh medium. The final concentrations of PEG and ECM were 40 and 2 mg/mL, respectively, for all subsequent studies. For oscillating rheometry and TGA, ECM composites were kept in α-MEM medium at 37°C/5% CO2 for 7 days (medium change every 2 day).
Encapsulation of hMSCs and miPSCs in ECM composites
Mesenchymal stem cells were derived from H1 human embryonic stem cells (hMSCs33). hMSC were cultured using α-MEM medium at 37°C/5% CO2. hMSCs were plated at 4.0×105 cells/175 cm2 and harvested at 60–70% confluence. α-MEM culture medium was changed one day after plating and every 2 days thereafter. hMSCs were harvested (passages from 9 to 10) via application of 0.25% Trypsin in HBSS (cat# 25-054-CI, Mediatech, Manassas, VA) and pelleted using a centrifuge at 300 g for 5 min. Supernatant was removed and hMSCs were resuspended in fresh medium.
Murine induced pluripotent stem cells (miPSCs)34 were cultured using DMEM (cat# 10569-010, Invitrogen, Carlsbad, CA) or GMEM (cat# 51492C, Sigma-Aldrich) supplemented with 1% non-essential amino acids (cat# 11140-050, Gibco/Invitrogen), 1% β-mercaptoethanol (35 μL/50mL PBS), 10% fetal bovine serum (cat# 26140, Gibco/Invitrogen) at 37°C/5% CO2. miPSCs were plated at 2.5×104 cells/10 cm2 with medium containing 106 unit/mL leukemia inhibitory factor (LIF, cat# ESG1107, EMD Millipore, Billerica, MA) and harvested at 60–70% confluence. Culture medium was changed daily. miPSCs were harvested (passages from 19 to 22) via application of 0.05% Trypsin/EDTA in HBSS (cat# 25300-054, Gibco/Invitrogen) and pelleted using a centrifuge at 300 g for 5 min. Supernatant was removed and miPSCs were resuspended in fresh medium.
For encapsulation, 1×105 hMSCs or 3×105 miPSCs were transferred to an Eppendorf tube, cells were centrifuged at 200 g for 5 min, resuspended with polymerized/viscous Col I or Laminin solutions (described in formation of hydrogels and ECM composites above), and incubated for 15 min at 37°C/5% CO2 (the initial viability is shown in Figure S2.) This cell-ECM mixture was then combined with both PEG precursors, followed by casting on dialysis tubes. The cell-ECM mixtures were left on a 48-well plate for NCL and removal of NCL by-products, as described in formation of hydrogels and ECM composites. Culture medium was changed every 2 days and 1 day for hMSCs and miPSCs (without adding LIF), respectively.
Ellman’s assay
After transferring PEG hydrogels (dissolved in Milli-Q water) to a well containing 1 mL reaction buffer (100 mM sodium phosphate dibasic at pH 8.0, containing 1 mM EDTA35) on a 24-well plate, hydrogels were stored at 37°C/5% CO2 for 2 h. The reaction buffer immersing PEG hydrogels was taken to wells containing DTNB on a 96-well plate and a total of 100 μL mixture was agitated on a rocker for 15 min at RT. Absorbance at 412 nm was taken using Infinite M1000 Pro microplate reader (Tecan, Switzerland).
Oscillating rheometry
An ARES-LS2 rheometer (TA Instruments, New Castle, DE) was used to measure the viscoelasticity of PEG hydrogels or acellular ECM composites. The upper plate (parallel plate, 8 mm diameter) was lowered until it was in conformal contact with the top surface of PEG hydrogels or acellular ECM composites. Frequency sweeping measurements were performed from 0.1 to 10 Hz at 1.0% strain, and two (PEG hydrogels) or three (acellular ECM composites) independent samples were tested at each condition. As a control for PEG hydrogels, 100 μL of 5 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP-HCl, cat# c4706, Sigma-Aldrich) in PBS was added for 5 min with agitation at RT. These PEG hydrogels were washed with PBS five times. Negative control samples for acellular ECM composites were mixed with 20 mM acetic acid for Col I composite or PBS for Laminin composites.
Thermogravimetric analysis (TGA) and pore size determination
A Q500 Thermogravimetric Analyzer (TA Instruments, New Castle, DE) was used to measure the equilibrium degree of swelling for each ECM composite to estimate pore size. The center of each ECM composite was cut with 5 mm biopsy punch, then placed into a platinum pan. ECM composite samples were heated at 10°C/min up to 120°C in nitrogen chamber. The mass of ECM composites after swelling was measured before heating in the chamber and dry mass was measured at the end of heating. The swelling ratio was calculated as previously reported (more details in Supporting Information), applying the density of PEG (1.08 g/cm3) and the number-average molecular weight of PEG (22,000 or 11,000 g/mol36–40).
Quantitative and qualitative analysis of encapsulated stem cells in ECM composites
The viability of encapsulated stem cells were tested with Live/Dead® cell viability kit from Invitrogen (cat# L-3224), which was assessed by manual counting of 4 random fields of view (each image contains approximately 200 to 500 cells) per ECM composite. At designated time points (day 1, 7, 14, and 21), cells were incubated with 10 μM Hoechst 33342 (cat# 83218, Anaspec, San Jose, CA) in α-MEM for 45 min at 37°C/5% CO2, followed by adding Live/Dead dyes (2 μM/1 μM in α-MEM) for 5 min. ECM composites were kept in α-MEM (hMSCs) or DMEM (miPSCs) at 37°C/5% CO2 until imaging analysis. Once ECM composites were transferred to a 35 mm imaging dish, wide-field fluorescence microscopy was used to identify live/dead cells from 4 randomly selected regions per ECM composite. Optical sections were taken at 30 μm intervals to a depth of 300 μm for each gel using an IX71 inverted deconvolution microscope (Olympus, Center Valley, PA).
Visualizing cell-ECM interactions
Three weeks after culture initiation, ECM composites were fixed with 4% paraformaldehyde (PFA) for 15 min and then permeablized with 0.1 % Triton-X in 1% BSA in PBS for 1 min on a rocker at RT. Following incubation overnight at 4°C in blocking buffer (5% BSA with 2% goat serum in PBS), Col I and Laminin were probed with rabbit anti-collagen type I (cat# AB755P, EMD Millipore, Billerica, MA) and rat anti-laminin-1 A & B chains (cat# MAB1904, Chemicon, Billerica, MA) antibodies, respectively. The primary antibodies were diluted at a 1:80 dilution in blocking buffer and incubated for 1 h on a rocker at RT. The antibodies against Col I or Laminin were detected by incubating with a secondary antibody, goat anti-rabbit FITC-conjugated (cat# 31583, Thermo Fisher Scientific, Rockford, IL) and goat anti-rat FITC-conjugated (cat# 3010-02, Southern Biotech, Birmingham, AL) at a 1:100 dilution in blocking buffer for 45 min on a rocker at RT, followed by incubation with TRITC-conjugated phalloidin (cat# FAK100, EMD Millipore, Billerica, MA) at a 1:100 dilution in blocking buffer for 45 min on a rocker at RT. ECM composites were stored in 2.5% 1,4-diazabicyclo[2,2,2]octane (DABCO, cat# D27802, Sigma-Aldrich) and 0.005% 4′,6-diamidino-2-phenylindole (DAPI, cat# D9542, Sigma-Aldrich) in 50% glycerol in PBS. Using a custom multiphoton workstation at the University of Wisconsin Laboratory for Optical and Computational Instrumentation (LOCI), the stained samples were imaged using a TE300 inverted microscope (Nikon, Tokyo, Japan) equipped with a Plan Apo VC 20× (NA 0.75; Nikon Instruments, Tokyo, Japan) objective lens and a mode-locked Ti:Sapphire laser (Spectra-Physics® Mai Tai®, Mountain View, CA). The excitation wavelength was tuned to 890 nm, while either a 520/35 or a 580/LP emission filter (Thin Film Imaging, Greenfield, MA) was used to detect FITC and TRITC, respectively. For detecting DAPI signals, the excitation wavelength was set to 780 nm and 457/50 emission filter was used. Images of 512 × 512 pixels were acquired using WiscScan software (LOCI). 3D reconstructions were generated using Imaris 7.6.1 software (Bitplane Inc., South Windsor, CT).
Statistical analysis
Statistical analyses were performed by ANOVA with Tukey’s post hoc test for multiple comparisons or Student’s t-test for independent two samples, where p values < 0.05 were considered significant. At least three independent pilot experiments were performed before the final measurements were recorded.
RESULTS
Modification of PEG-precursors
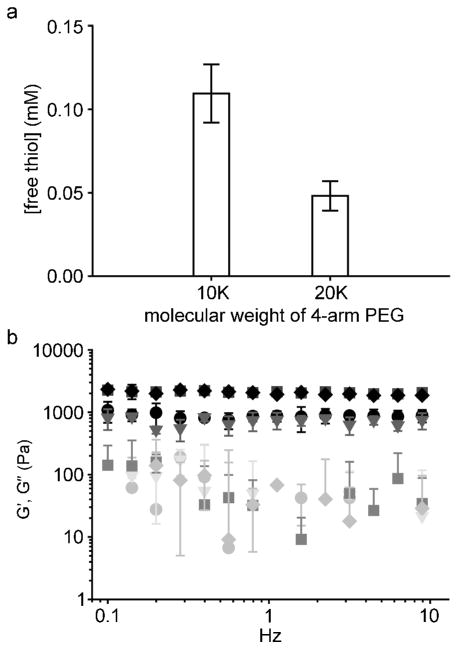
To produce ECM composites by crosslinking PEG-precursors, the protocol described by Hu et al. was initially used with 4-armed PEG-NH2 10,000 Da.32 Rapid polymerization was attained and physical entrapment of ECM was successful. However, when hMSCs were encapsulated into the ECM composites, the viability after 24 h was quite poor (Figure S3. PEG-NCL and Col I composite, % viability less than 80%). Based on previous studies,41–46 we hypothesized that increasing the molecular weight between crosslinks and thereby decreasing the crosslinking density of the PEG hydrogel would increase the average pore size within the hydrogel. The PEG20K hydrogel would therefore harbor decreased stiffness and increased rate of removal of the leaving group of the NCL compared to the PEG10K hydrogel (Figure S1 provides a chemical description of PEG-NCL crosslinking and associated leaving group). These mechanical and chemical changes were predicted to reduce cytotoxic factors and thereby improve viability. Indeed, the average pore size of PEG hydrogels formed using 10,000 Da PEG starting material was 3.9 ± 0.5 nm compared to 13.9 ± 8.5 nm for 20,000 Da PEG starting material (see Table 1). In addition, upon changing the molecular weight of 4-armed PEG-NH2, from 10,000 to 20,000 Da, the concentration of the sulfhydryl containing leaving group decreased by approximately half (Figure 2a). To determine whether the molecular weight change impacted on the efficiency of NCL, oscillating rheometry was conducted on hydrogels with or without exposure to TCEP-HCl. TCEP-HCl disrupts disulfide bonds from PEG-Cys but not the native amide bond between PEG-thioester and PEG-Cys. Indeed, storage and loss moduli did not differ between TCEP-HCl control groups (PEG20K, TCEP (▼, ▽) and PEG10K, TCEP (◆, ◇)) and PEG hydrogel groups (PEG20K (●, ○) and PEG10K (■, □)) at both molecular weights (Figure 2b) indicating the majority of hydrogel polymerization proceeded via NCL. However, there was a significant difference in storage modulus (G′ at 1 Hz) between PEG 10,000 Da and 20,000 Da (Student’s t-test, p < 0.05, Figure 3c) indicating hydrogels synthesized with the larger (20,000 Da) PEG are less stiff than those formed with the 10,000 Da counterpart. Therefore, the increase of molecular weight of PEG with the corresponding decrease in crosslinking density contributed to the lowered stiffness, which improved short and long-term viability of stem cells (described in results below). Therefore, the 20,000 Da 4-armed PEG macromonomers, which exhibited minimal cytotoxicity to cultured hMSCs (Figure S4 in Supporting Information), was used for all subsequent studies.
Table 1.
Estimated pore sizes of ECM composites. TGA was performed after 7 days in culture medium (α-MEM) at 37°C/5% CO2. The final concentration of PEG and whole molecule ECM in each composite was 40 and 2 mg/mL, respectively.
| ECM composite (n=3) | Average pore diameter (nm) |
|---|---|
| Col I | 12.3 ± 4.0 |
| Laminin | 14.6 ± 13.0 |
| no ECM (PEG20k) | 13.9 ± 8.5 |
| no ECM (PEG10k) | 3.9 ± 0.5 |
Figure 2.
Characterization of PEG hydrogels of varied molecular weight. Upon changing the molecular weight of four-armed PEG, the concentration of the leaving group was reduced by approximately half (a), measured by Ellman’s assay; mean±SD, n = 4. Oscillating rheometry showed that the storage modulus of PEG hydrogels was reduced approximately by half with respect to the change of the molecular weight of four-armed PEG from 10,000 to 20,000 Da (b). G′ (filled symbols) and G″ (open symbols) are shown for PEG20K (●, ○), PEG20K, TCEP (▼, ▽), PEG10K (■, □), and PEG10K, TCEP (◆, ◇). The extensive overlap indicates that the storage modulus of PEG before and after TCEP treatment was not significantly altered; mean±SD, n = 2. The final concentration of PEG was 40 mg/mL.
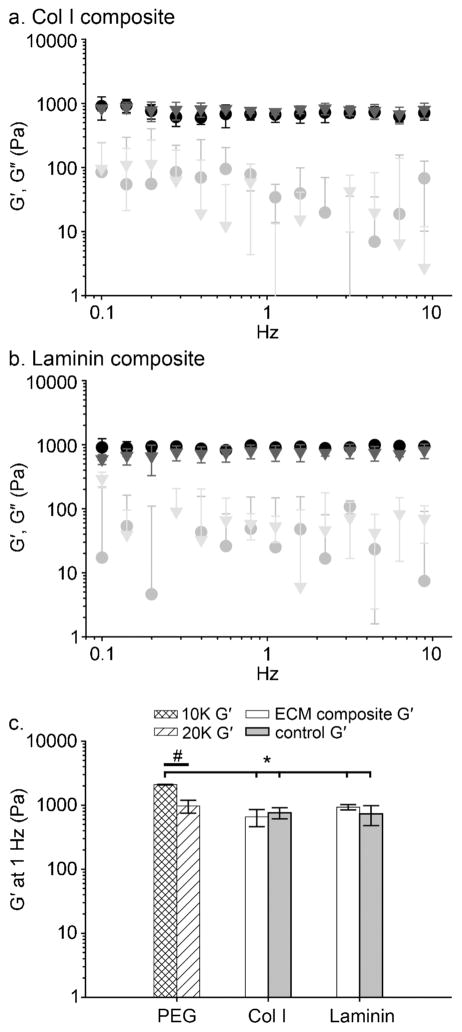
Figure 3.
Oscillating rheometry of ECM composites. The storage modulus of ECM composites was not significantly altered upon incorporating whole molecule ECM or buffer-only negative controls; 20 mM acetic acid for Col I composite or PBS for Laminin composite. G′ (filled symbols) and G″ (open symbols) are shown for Col I composite (●, ○) and its negative control (▼, ▽) (a). A Laminin composite (●, ○) and its negative control (▼, ▽) are shown in (b). In Figure (a) and (b), the extensive overlap indicates that the storage moduli of ECM composites were not significantly altered compared to their respective negative control groups. The final concentration of PEG and whole molecule ECM in each composite was 40 and 2 mg/mL, respectively; mean±SD, n = 3. In (c), the storage modulus of PEG10K, PEG20K (the data from Figure 2b), ECM composites, and their respective control groups (the data from Figure 3a and b) at 1 Hz were plotted. Brackets indicate statistically significant differences between PEG 10,000 Da and ECM composites, ANOVA Tukey’s HSD post hoc test (* p<0.05, mean±SD). A bar indicates statistically significant difference between PEG 10,000 Da and 20,000 Da, Student’s t-test (# p<0.05, mean±SD)
Viscoelasticity of ECM composites
Pore size, stiffness and degradation of the extracellular matrices or scaffolds can impact on stem cell behaviors and lineage progression.5,47–51 In an attempt to discern the impact of the biochemical signals of ECM alone on stem cell behavior, we sought to modulate the ECM composite such that material properties remain consistent between composites. First, the estimated pore size was calculated for hydrogels containing Col I or Laminin up to 2 mg/mL or 4.8% dry weight. No significant differences in pore size were noted with the addition of Col I or Laminin at least up to the maximum concentration used here (2 mg/mL, Table 1). Of note, small pore size was a deliberate design constraint for this application as our intent was to entrap ECM-stem cell aggregates in a PEG-confined space. The PEG-confined spaces containing stem cells and ECM were much larger (approximately 150 – 250 μm from immunofluorescence images). Migration of cells and/or diffusion of ECM through the pores are not preferred as elements of the aggregate (i.e., microenvironment) would be lost. Next, using oscillating rheometry, shear storage and loss moduli of ECM composites and their respective control hydrogels were measured. As shown in Figure 3a and b, the storage moduli of ECM composites and corresponding control groups (20 mM acetic acid for Col I composites or PBS for Laminin composites) were not significantly different. These results indicate that PEG hydrogels (40 mg/mL) maintained consistent storage moduli with up to 2 mg/mL of Col I or Laminin. A majority of loss modulus values (G″, open symbols) were below 100 Pa and some were absent at some frequencies (negative values recorded from the rheometer, Figure 3a and b). These results show that tan δ (= G″/G′) of ECM composites is near or below 0.1 and no crossing of G′ and G″ occurred at any measured frequency, indicating the formation of elastic hydrogels. In comparison to PEG hydrogels (20,000 Da) without whole molecule ECMs, the storage moduli of ECM composites at 1 Hz were not significantly different, as shown in Figure 3c. Collectively, these data indicate that up to 2 mg/mL of Col I or Laminin in chemoselectively polymerized PEG hydrogels can be added without significantly altering the pore size or stiffness.
Viability and morphology of encapsulated hMSCs in 3D ECM composites
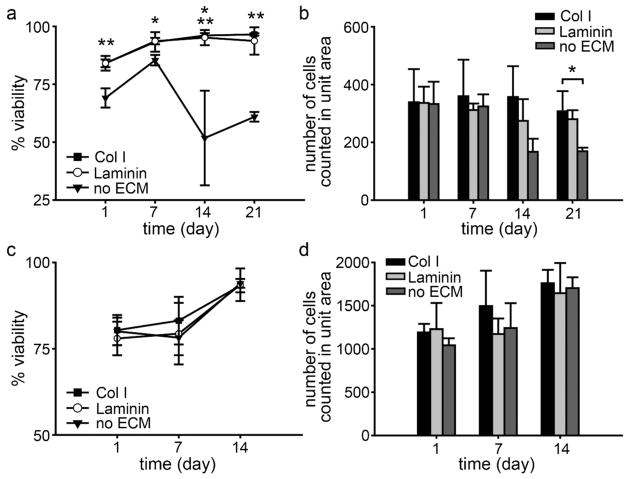
To determine the long-term survival of stem cells encapsulated in 3D ECM composites, the viability of hMSCs was assessed in either Col I or Laminin composites at 1, 7, 14, and 21 days in culture. The viability of encapsulated hMSCs was over 84% in both ECM composites and 69% in no ECM hydrogels at day 1, indicating the NCL polymerization process was relatively mild. From day 7, hMSC viability was significantly higher in ECM composites (containing either Col I or Laminin, 93% or above) compared to that of hMSCs in PEG hydrogels (no ECM control, 85% at day 7 in Figure 4a) indicating that entrapped ECM enhanced long-term viability of hMSC.
Figure 4.
The viability of encapsulated hMSCs (a) and miPSCs (c) in ECM composites. The total number of counted hMSCs (b) and miPSCs (d) from a focal plane in a unit area of 0.143 mm2; mean±SD, n = 3. ** p<0.01 and *p<0.05 with ANOVA Tukey’s post hoc test, compared to no ECM. In (a), at day 14, p<0.01 between no ECM and Col I and p<0.05 between no ECM and Laminin.
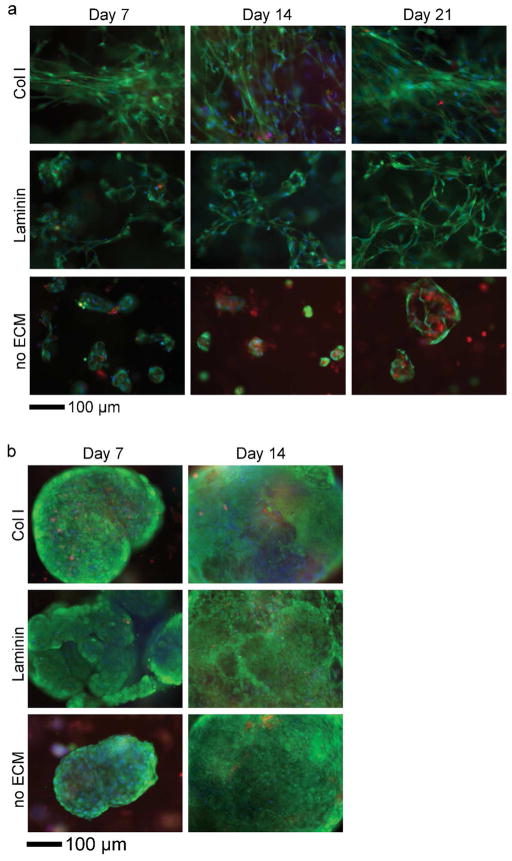
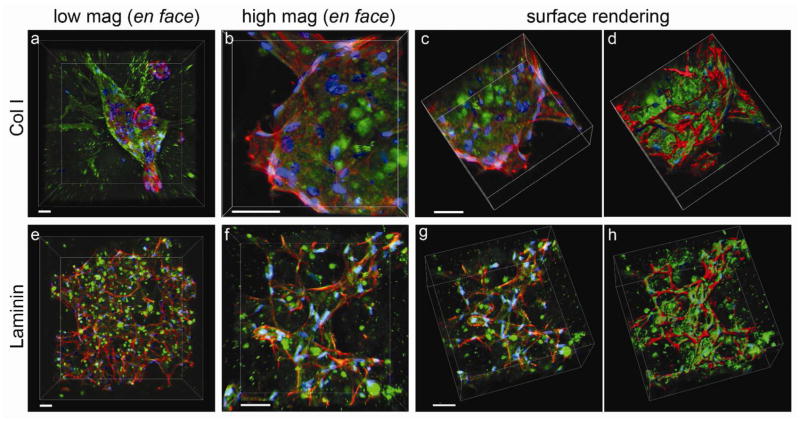
Similarly, the number of cells per unit area in the ECM composites (containing either Col I or Laminin) was maintained over time, but decreased significantly (after 14 days) in PEG hydrogels (no ECM control, Figure 4b). That cell number does not increase substantially in composites containing ECM appears to reflect a decrease in proliferation (Figure S5). Interestingly, live/dead staining revealed that the morphology of hMSCs in each ECM composite was distinctive as shown in Figure 5a. In Col I composites, hMSCs were localized to Col I fibers and maintained elongated phenotypes (Figure 5a, Col I), whereas hMSCs in Laminin composites exhibit more reticulate structures and associated morphology (Figure 5a, Laminin). Since live/dead staining does not allow detailed assessment of the intricate interplay between cell and ECM, we examined this interaction more closely by immunofluorescence staining of ECM and labeling F-actin with fluorescently-tagged phalloidin. The labeled composites were optically sectioned using Multiphoton Laser Scanning Microscopy (MPLSM, Figure 6a–c, e–g; both day 1 and 21 in Figure S6 in Supporting Information). Digital image stacks were processed using Imaris software to generate 3D reconstructions to illustrate the arrangement and interplay between hMSCs and ECM within the composites. In Col I composites, hMSCs appeared to attach to and extend along Col I fibrils that often surrounded individual cell bodies or cell aggregates, confirming the 3-dimensionality of cell-ECM interactions within the ECM composite (Supplementary video S1 and 2). In Laminin composites, hMSCs were less likely to form cell aggregates and instead individually adhered to the reticulate architecture of Laminin. In PEG hydrogels without ECM (Figure 5a, no ECM), the 3D context supported survival of hMSCs up to 7 days after which necrotic areas emerged at the center of hMSC aggregates.
Figure 5.
Representative images showed distinct morphologies of hMSCs (a) and miPSCs (b) encapsulated in ECM composites. Live (green)/dead (red) staining (nuclear counterstain with Hoechst 33342, blue).
Figure 6.
Interaction of hMSCs with ECM of composites. MPLSM was used to optically section (at 1 μm intervals) microenvironments of ECM composites containing hMSCs to reconstruct the three-dimensionality of the sample and the unique interactions of cell with ECM in each case. (a–d) hMSCs in Col I at low magnification (a, 217 planes) and high magnification (b–d, 117 planes). (e–h) hMSCs in Laminin at low magnification (e, 416 planes) and high magnification (f–h, 163 planes). All images were taken after 21 days in culture. Z stacks of images were reconstructed from raw images (c and g) to solid objects using Imaris software for Col I (d) and Laminin (h) composites respectively to best view cell-matrix interactions. ECM (green), phalloidin (red), and DAPI (blue). Scale bar, 50 μm.
Viability and morphology of encapsulated miPSCs in 3D ECM composites
The viability of encapsulated miPSC in composites containing ECM and in the PEG hydrogel alone was greater than 93% at 14 days of culture (Figure 4c). The culture of miPSCs was terminated at 14 days due to rapid proliferation (Figure S5) of miPSCs in ECM composites. Cells quickly over-populated the capacity of the well causing the medium to become acidic within 24 h, even after only 5 or 6 days of culture. The number of miPSCs counted increased from day 1 to 14 (Figure 4d), indicating that miPSCs proliferated extensively in the ECM composites and PEG hydrogels (no ECM control) during the 14 days culture period. The morphology of miPSCs in ECM composites was quite different from that of hMSCs. After 2 or 3 days of encapsulation, miPSCs began forming colonies that eventually became large aggregates (irregular shapes) or spheroids (regular) over 14 days (Figure 5b). This considerable growth proceeded in the presence or absence of ECM, suggesting that cell-cell interactions play an important role in the survival of miPSCs, possibly overshadowing the role of ECM in miPSC survival. Interestingly, a fraction of miPSC aggregates exhibited beating areas (Figure S7) in both ECM composites and PEG hydrogels (no ECM control) but with differential kinetics (see also Supplementary videos S3–5), suggesting ECM and/or three dimensionality may dictate the emergence and function of certain stem cell-derived mature cell types (i.e., cardiomyocytes). Collectively, these viability and morphology data show that ECM composites supported long-term viability of hMSCs and miPSCs, where hMSC viability and morphology were highly dependent on exogenous ECM.
DISCUSSION
Here we employed a mild and chemoselective crosslinking chemistry to polymerize PEG and simultaneously entrap whole molecule ECM and multipotent or pluripotent stem cells. This PEG-NCL approach was previously used to encapsulate insulin secreting MIN6 β cells in PEG hydrogels26 and to achieve surface PEGylation on titanium surfaces.27 We built upon these previously applications, using this unique chemistry to create 3D, ECM-based microenvironments that can be produced with any ECM protein and combinations of such proteins.
The PEG-NCL approach is quite different from other methods used to generate 3D microenvironments such as radical polymerization employing a photoinitiator (e.g. Irgacure 2959) with ultraviolet (UV) light12,52 or eosin Y with visible light.53 Photopolymerization has been an attractive technique because the conversion of liquid photosensitive polymer solution to a hydrogel occurs rapidly and can be controlled in time and space under UV or visible light. During photopolymerization, UV light homolytically splits photoinitiator molecules into radicals, initiating the formation of crosslinked networks from acrylated PEG macromonomers. However, free radicals can directly react with proteins and DNA, thereby directly inducing cellular damage. This process has been shown to negatively affect the viability, proliferation, and differentiation of bone marrow-derived MSC monolayers.54 Further, it is difficult to measure the extent to which polymerization is complete and therefore how many residual functional groups remain. This deserves further attention in light of recent work, showing that low concentrations of small functional groups can affect the lineage commitment of encapsulated MSCs.55 In an attempt to reduce the toxicity of free radicals and the heterogeneity of crosslinking, thiol-ene photopolymerization has been developed. This method of photopolymerization substantially decreased the concentration and lifetime of water-soluble photoinitiators (1 mM of lithium phenyl-2,4,6-trimethylbenzoylphosphinate) and reduced the period of exposure of encapsulated cells to UV light (less than 10 s),56–58 however the complete removal of cytotoxic compounds was not possible. Therefore, our group became interested in developing a crosslinking strategy that does not require catalysts or radicals as well as a chemoselective chemistry whose reaction is orthogonal to the presence of exogenous ECM in the designated synthetic scaffolds. Of note, the system we have developed is best suited to discern this impact of cell exposure to exogenously provided ECM at early time points on stem cell behavior at later time points. It is not the purpose of the system to control downstream events associated with exposure to exogenous ECM, which could include for example, production of endogenous ECM and associated changes in stiffness or sequestration of soluble signaling molecules.
Despite the advantages of NCL, minor limitations of the PEG-NCL approach include the release of potentially harmful thiol by-product (Figure S1) and relatively slow crosslinking kinetics (approximately 15 min). In the future a variation of NCL involving an oxo-ester rather than a thioesters may be implemented. This approach eliminates the release of thiol by-products by covalent capture of the thiol side chain of N-terminal cysteine that initially formed a thioester bond, resulting in rapid crosslinking of PEG hydrogels (from several seconds to less than a minute) with high viability (87 ± 7%) of entrapped 3T3 fibroblasts in 24 h.59 Be that as it may, we also observed high viability of multipotent and pluripotent stem cells in ECM composites (from 78 ± 5% to 84 ± 3% in 24 h) and high retention of both cells and ECM with the thioester approach. The true test of stem cell health will be the effect of both chemistries on lineage commitment and function of mature progeny. Another factor that is likely to impact on cell health in the long term is the inability of the polymer to be remodeled by the cells and therefore limited space for cell expansion or endogenously produced ECM. Studies that require extended time frames may therefore require incorporation of binding sites for matrix metalloproteinases or other moiety that is subject to degradation. This can be accomplished with modification of the system described here by conjugating Cys-functionalized MMP-sensitive sequence60 to PEG-thioester and mix with PEG-precursors.
Here we showed that Col I and Laminin could be effectively entrapped in PEG hydrogels using NCL. We sought to utilize two different families of ECM proteins (fibrillar Col I and reticular Laminin) to establish proof of concept. In addition, we selected these two molecules as they have both been included in protocols designated either to enhance stem cell growth61,62 or to encourage stem cell differentiation to cardiomyocytes,63–66 which is of particular interest for our laboratory. Previously, we reported that Col I and Laminin are two of the primary constituents of the developing myocardium.67,68 Indeed, we found that miPS cells entrapped in PEG hydrogels and to a lesser extent in composites containing either Col I or Laminin can beat spontaneously (Figure S7), indicative of cardiomyocyte differentiation. Differentiation of cardiomyocytes from pluripotent or multipotent cell types has been accomplished, but the process remains inefficient and often expensive.69–72 Progress in this area has been catalyzed by improved understanding of the transcriptional events directing cardiac specification and the soluble factors stimulating these signaling events. However, minimal effort has been devoted to improve understanding of the impact of insoluble factors, including ECM, on cardiomyocyte differentiation. We have shown that individual, whole molecule ECM proteins coated on 2D tissue culture plastics are capable of inducing multilineage differentiation, including cardiomyocyte differentiation, of human MSCs at levels comparable to chemical induction.28 In addition, others have shown that exogenous ECM proteins are required both to promote survival of cardiomyocytes ex vivo and are essential to implement all cardiomyocyte differentiation protocols employed to date.69,72 With the technology described here, it is possible to study the biochemical impact of individual ECM or multi-ECM, 3D composites on the behavior of stem cells and other cell types. The approach is scalable and conducive to systematic multifactorial optimization73 that would maximize the discovery of ECM composites instructive for specific cell behaviors, including differentiation, in vitro. In addition, in vitro results could be used to predict how cells might behave when delivered to particular tissues or organs for regenerative purposes.
CONCLUSIONS
PEG-NCL chemoselective chemistry was utilized to entrap whole molecule ECM proteins in PEG hydrogels, generating ECM composites for 3D culture of multipotent and pluripotent stem cells. The viscoelasticity and pore size of ECM composites were held constant such that the biochemical impact of ECM alone on cell behavior could be studied. Cell viability was maintained in the composites and indeed hMSC viability was enhanced by the addition of exogenous ECM. A brief glimpse of cell behavioral changes was observed with changes in composite composition as hMSCs contributed to reticulate structures in the presence of exogenous Laminin as opposed to fibrillar masses in the case of Col I. In addition, miPSCs were able to form beating areas in composites containing Laminin, but to a lesser extent in composites containing Col I. Thus, this PEG-NCL approach provides an opportunity to independently examine the biochemical impact of ECM on stem cell differentiation in 3D environments. Further understanding of stem cell behaviors and differentiation in potentially multi-ECM composites in vitro would advance the field of regenerative medicine towards the goals of replacing lost or damaged tissues or organs.
Supplementary Material
Acknowledgments
We thank Peiman Hematti and Deepak Srivastava for kindly providing hMSCs and miPSCs, respectively. This research was in part supported by the American Heart Association (11IRG5570039 to BMO and 13POST14080011 to JPJ) and the National Institutes of Health (R01 DE021215 and R01 DE021104 to PBM). Oscillating rheometry and TGA was performed at the University of Wisconsin-Madison Soft Materials Laboratory.
Footnotes
Supporting Information Available
The synthesis of EMPSA, the reaction scheme of PEG-NCL, the initial viability of hMSC and miPSC immediately before encapsulation, details of pore size calculation, the viability of hMSC in PEG10k, the cytotoxicity of PEG macromonomers, proliferation of encapsulated stem cells, the distribution of ECM and hMSCs at day 1 and 21 in ECM composites, the quantification of beating areas in ECM composites, and supplementary videos of 3D reconstructions and beating miPSC aggregates included. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Huxley-Jones J, Robertson DL, Boot-Handford RP. Matrix Biol. 2007;26:2–11. doi: 10.1016/j.matbio.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 2.Özbek S, Balasubramanian PG, Chiquet-Ehrismann R, Tucker RP, Adams JC. Mol Biol Cell. 2010;21:4300–4305. doi: 10.1091/mbc.E10-03-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levental I, Georges PC, Janmey PA. Soft Matter. 2007;3:299–306. doi: 10.1039/b610522j. [DOI] [PubMed] [Google Scholar]
- 4.Tse JR, Engler AJ. Curr Protoc Cell Biol. 2010;10.16:1–16. doi: 10.1002/0471143030.cb1016s47. [DOI] [PubMed] [Google Scholar]
- 5.Reilly GC, Engler AJ. J Biomech. 2010;43:55–62. doi: 10.1016/j.jbiomech.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Krishna OD, Kiick KL. Biopolymers. 2010;94:32–48. doi: 10.1002/bip.21333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collier JH, Segura T. Biomaterials. 2011;32:4198–4204. doi: 10.1016/j.biomaterials.2011.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jung JP, Gasiorowski JZ, Collier JH. Biopolymers. 2010;94:49–59. doi: 10.1002/bip.21326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koepsel JT, Nguyen EH, Murphy WL. Integr Biol. 2012;4:914–924. doi: 10.1039/c2ib20055d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hudalla GA, Kouris NA, Koepsel JT, Ogle BM, Murphy WL. Integr Biol. 2011;3:832–842. doi: 10.1039/c1ib00021g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saik JE, Gould DJ, Keswani AH, Dickinson ME, West JL. Biomacromolecules. 2011;12:2715–2722. doi: 10.1021/bm200492h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin CC, Anseth KS. Proc Natl Acad Sci USA. 2011;108:6380–6385. doi: 10.1073/pnas.1014026108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Badylak SF, Freytes DO, Gilbert TW. Acta Biomater. 2009;5:1–13. doi: 10.1016/j.actbio.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 14.Kleinman HK, Martin GR. Semin Cancer Biol. 2005;15:378–386. doi: 10.1016/j.semcancer.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Badylak SF. Biomaterials. 2007;28:3587–3593. doi: 10.1016/j.biomaterials.2007.04.043. [DOI] [PubMed] [Google Scholar]
- 16.Friess W. Eur J Pharm Biopharm. 1998;45:113–136. doi: 10.1016/s0939-6411(98)00017-4. [DOI] [PubMed] [Google Scholar]
- 17.Blombäck B, Bark N. Biophys Chem. 2004;112:147–151. doi: 10.1016/j.bpc.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 18.Serban MA, Prestwich GD. Methods. 2008;45:93–98. doi: 10.1016/j.ymeth.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia X, Kiick KL. Macromol Biosci. 2009;9:140–156. doi: 10.1002/mabi.200800284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burdick JA, Prestwich GD. Adv Mater. 2011;23:H41–H56. doi: 10.1002/adma.201003963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Battista S, Guarnieri D, Borselli C, Zeppetelli S, Borzacchiello A, Mayol L, Gerbasio D, Keene D, Ambrosio L, Netti P. Biomaterials. 2005;26:6194–6207. doi: 10.1016/j.biomaterials.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 22.Yang F, Cho SW, Son SM, Hudson SP, Bogatyrev S, Keung L, Kohane DS, Langer R, Anderson DG. Biomacromolecules. 2010;11:1909–1914. doi: 10.1021/bm100357t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duan Y, Liu Z, O’Neill J, Wan L, Freytes D, Vunjak-Novakovic G. J Cardiovasc Trans Res. 2011;4:605–615. doi: 10.1007/s12265-011-9304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dawson PE, Muir TW, Clarklewis I, Kent SBH. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 25.Jung JP, Jones JL, Cronier SA, Collier JH. Biomaterials. 2008;29:2143–2151. doi: 10.1016/j.biomaterials.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su J, Hu B-H, Lowe WL, Jr, Kaufman DB, Messersmith PB. Biomaterials. 2010;31:308–314. doi: 10.1016/j.biomaterials.2009.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Byun E, Kim J, Kang SM, Lee H, Bang D, Lee H. Bioconjug Chem. 2010;22:4–8. doi: 10.1021/bc100285p. [DOI] [PubMed] [Google Scholar]
- 28.Santiago JA, Pogemiller R, Ogle BM. Tissue Eng Part A. 2009;15:3911–3922. doi: 10.1089/ten.tea.2008.0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martino MM, Mochizuki M, Rothenfluh DA, Rempel SA, Hubbell JA, Barker TH. Biomaterials. 2009;30:1089–1097. doi: 10.1016/j.biomaterials.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baharvand H, Azarnia M, Parivar K, Ashtiani SK. J Mol Cell Cardiol. 2005;38:495–503. doi: 10.1016/j.yjmcc.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 31.Khatiwala CB, Kim PD, Peyton SR, Putnam AJ. J Bone Miner Res. 2009;24:886–898. doi: 10.1359/JBMR.081240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu BH, Su J, Messersmith PB. Biomacromolecules. 2009;10:2194–2200. doi: 10.1021/bm900366e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trivedi P, Hematti P. Exp Hematol. 2008;36:350–359. doi: 10.1016/j.exphem.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Laake LW, Qian L, Cheng P, Huang Y, Hsiao EC, Conklin BR, Srivastava D. Circ Res. 2010;107:340–347. doi: 10.1161/CIRCRESAHA.109.215434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moreland RB, Smith PK, Fujimoto EK, Dockter ME. Anal Biochem. 1982;121:321–326. doi: 10.1016/0003-2697(82)90487-0. [DOI] [PubMed] [Google Scholar]
- 36.Zustiak SP, Leach JB. Biomacromolecules. 2010;11:1348–1357. doi: 10.1021/bm100137q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raeber GP, Lutolf MP, Hubbell JA. Biophy J. 2005;89:1374–1388. doi: 10.1529/biophysj.104.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leach JB, Bivens KA, Patrick CW, Jr, Schmidt CE. Biotechnol Bioeng. 2003;82:578–589. doi: 10.1002/bit.10605. [DOI] [PubMed] [Google Scholar]
- 39.Lu S, Anseth KS. Macromolecules. 2000;33:2509–2515. [Google Scholar]
- 40.Mellott MB, Searcy K, Pishko MV. Biomaterials. 2001;22:929–941. doi: 10.1016/s0142-9612(00)00258-1. [DOI] [PubMed] [Google Scholar]
- 41.Bott K, Upton Z, Schrobback K, Ehrbar M, Hubbell JA, Lutolf MP, Rizzi SC. Biomaterials. 2010;31:8454–8464. doi: 10.1016/j.biomaterials.2010.07.046. [DOI] [PubMed] [Google Scholar]
- 42.Browning MB, Wilems T, Hahn M, Cosgriff-Hernandez E. J Biomed Mater Res A. 2011;98:268–273. doi: 10.1002/jbm.a.33109. [DOI] [PubMed] [Google Scholar]
- 43.DeForest CA, Sims EA, Anseth KS. Chem Mater. 2010;22:4783–4790. doi: 10.1021/cm101391y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munoz-Pinto DJ, Bulick AS, Hahn MS. J Biomed Mater Res A. 2009;90A:303–316. doi: 10.1002/jbm.a.32492. [DOI] [PubMed] [Google Scholar]
- 45.Tan H, DeFail AJ, Rubin JP, Chu CR, Marra KG. J Biomed Mater Res A. 2010;92:979–987. doi: 10.1002/jbm.a.32438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Dijk M, van Nostrum CF, Hennink WE, Rijkers DTS, Liskamp RMJ. Biomacromolecules. 2010;11:1608–1614. doi: 10.1021/bm1002637. [DOI] [PubMed] [Google Scholar]
- 47.Anderson SB, Lin CC, Kuntzler DV, Anseth KS. Biomaterials. 2011;32:3564–3574. doi: 10.1016/j.biomaterials.2011.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischer RS, Myers KA, Gardel ML, Waterman CM. Nat Protoc. 2012;7:2056–2066. doi: 10.1038/nprot.2012.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guilak F, Cohen DM, Estes BT, Gimble JM, Liedtke W, Chen CS. Cell Stem Cell. 2009;5:17–26. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Higuchi A, Ling QD, Chang Y, Hsu ST, Umezawa A. Chem Rev. 2013;113:3297–3328. doi: 10.1021/cr300426x. [DOI] [PubMed] [Google Scholar]
- 51.Köllmer M, Keskar V, Hauk TG, Collins JM, Russell B, Gemeinhart RA. Biomacromolecules. 2012;13:963–973. doi: 10.1021/bm300332w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hwang N, Varghese S, Li H, Elisseeff J. Cell Tissue Ress. 2011;344:499–509. doi: 10.1007/s00441-011-1153-2. [DOI] [PubMed] [Google Scholar]
- 53.Hillel AT, Unterman S, Nahas Z, Reid B, Coburn JM, Axelman J, Chae JJ, Guo Q, Trow R, Thomas A, Hou Z, Lichtsteiner S, Sutton D, Matheson C, Walker P, David N, Mori S, Taube JM, Elisseeff JH. Sci Transl Med. 2011;3:93ra67. doi: 10.1126/scitranslmed.3002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fedorovich NE, Oudshoorn MH, van Geemen D, Hennink WE, Alblas J, Dhert WJA. Biomaterials. 2009;30:344–353. doi: 10.1016/j.biomaterials.2008.09.037. [DOI] [PubMed] [Google Scholar]
- 55.Benoit DSW, Schwartz MP, Durney AR, Anseth KS. Nat Mater Ure. 2008;7:816–823. doi: 10.1038/nmat2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fairbanks BD, Schwartz MP, Halevi AE, Nuttelman CR, Bowman CN, Anseth KS. Adv Mater. 2009;21:5005–5010. doi: 10.1002/adma.200901808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCall JD, Anseth KS. Biomacromolecules. 2012;13:2410–2417. doi: 10.1021/bm300671s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shih H, Lin CC. Biomacromolecules. 2012;13:2003–2012. doi: 10.1021/bm300752j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strehin I, Gourevitch D, Zhang Y, Heber-Katz E, Messersmith PB. Biomater Sci. 2013;1:603–613. doi: 10.1039/C3BM00201B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lutolf MP, Hubbell JA. Biomacromolecules. 2003;4:713–722. doi: 10.1021/bm025744e. [DOI] [PubMed] [Google Scholar]
- 61.Domogatskaya A, Rodin S, Tryggvason K. Annu Rev Cell Dev Biol. 2012;28:523–553. doi: 10.1146/annurev-cellbio-101011-155750. [DOI] [PubMed] [Google Scholar]
- 62.Jones MB, Chu CH, Pendleton JC, Betenbaugh MJ, Shiloach J, Baljinnyam B, Rubin JS, Shamblott MJ. Stem Cells Dev. 2010;19:1923–1935. doi: 10.1089/scd.2009.0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dijk A, Niessen HWM, Zandieh Doulabi B, Visser FC, Milligen FJ. Cell Tissue Res. 2008;334:457–467. doi: 10.1007/s00441-008-0713-6. [DOI] [PubMed] [Google Scholar]
- 64.Malan D, Reppel M, Dobrowolski R, Roell W, Smyth N, Hescheler J, Paulsson M, Bloch W, Fleischmann BK. Stem Cells. 2009;27:88–99. doi: 10.1634/stemcells.2008-0335. [DOI] [PubMed] [Google Scholar]
- 65.Wan CR, Frohlich EM, Charest JL, Kamm RD. Cel Mol Bioeng. 2011;4:56–66. [Google Scholar]
- 66.Zeng D, Ou DB, Wei T, Ding L, Liu XT, Hu XL, Li X, Zheng QS. BMC Cell Biology. 2013;14:5. doi: 10.1186/1471-2121-14-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hanson KP, Jung JP, Tran QA, Hsu SPP, Iida R, Ajeti V, Campagnola PJ, Eliceiri KW, Squirrell JM, Lyons GE, Ogle BM. Tissue Eng Part A. 2013;19:1132–1143. doi: 10.1089/ten.tea.2012.0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jung JP, Squirrell JM, Lyons GE, Eliceiri KW, Ogle BM. Trends Biotechnol. 2012;30:233–240. doi: 10.1016/j.tibtech.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, Palecek SP. Proc Natl Acad Sci USA. 2012;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lian XJ, Zhang JH, Azarin SM, Zhu KX, Hazeltine LB, Bao XP, Hsiao C, Kamp TJ, Palecek SP. Nat Protoc. 2013;8:162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mummery C, Zhang J, Ng E, Elliott D, Elefanty A, Kamp T. Circ Res. 2012;111:344–358. doi: 10.1161/CIRCRESAHA.110.227512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang J, Klos M, Wilson GF, Herman AM, Lian X, Raval KK, Barron MR, Hou L, Soerens AG, Yu J, Palecek SP, Lyons GE, Thomson JA, Herron TJ, Jalife J, Kamp TJ. Circ Res. 2012;111:1125–1136. doi: 10.1161/CIRCRESAHA.112.273144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jung JP, Moyano JV, Collier JH. Integr Biol. 2011;3:185–196. doi: 10.1039/c0ib00112k. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.