Abstract
To gain insight into oral squamous cell carcinogenesis, we performed deep sequencing (RNAseq) of non-tumorigenic human OKF6-TERT1R and tumorigenic SCC-9 cells. Numerous homeobox genes are differentially expressed between OKF6-TERT1R and SCC-9 cells. Data from Oncomine, a cancer microarray database, also show that homeobox (HOX) genes are dysregulated in oral SCC patients. The activity of Polycomb repressive complexes (PRC), which causes epigenetic modifications, and retinoic acid (RA) signaling can control HOX gene transcription. HOXB7, HOXC10, HOXC13, and HOXD8 transcripts are higher in SCC-9 than in OKF6-TERT1R cells; using ChIP (chromatin immunoprecipitation) we detected PRC2 protein SUZ12 and the epigenetic H3K27me3 mark on histone H3 at these genes in OKF6-TERT1R, but not in SCC-9 cells. In contrast, IRX1, IRX4, SIX2 and TSHZ3 transcripts are lower in SCC-9 than in OKF6-TERT1R cells. We detected SUZ12 and the H3K27me3 mark at these genes in SCC-9, but not in OKF6-TERT1R cells. SUZ12 depletion increased HOXB7, HOXC10, HOXC13, and HOXD8 transcript levels and decreased the proliferation of OKF6-TERT1R cells. Transcriptional responses to RA are attenuated in SCC-9 versus OKF6-TERT1R cells. SUZ12 and H3K27me3 levels were not altered by RA at these HOX genes in SCC-9 and OKF6-TERT1R cells. We conclude that altered activity of PRC2 is associated with dysregulation of homeobox gene expression in human SCC cells, and that this dysregulation potentially plays a role in the neoplastic transformation of oral keratinocytes.
Keywords: Epigenetic silencing, Polycomb, SUZ12, Chromatin, H3K27me3, Oral squamous cell carcinoma, Head and neck squamous cell, carcinoma, Tumorigenesis, RNA-seq, Homeobox, Retinoic acid
Introduction
Head and neck cancers are very common worldwide [1]. It is estimated that 41,380 new patients will be diagnosed with cancer of the oral cavity and the pharynx and 7890 will die of these diseases in 2013 in the United States alone [2]. More than 90% of all oral cavity cancers are of the squamous cell carcinoma (SCC) type [1]. Many risk factors are known for oral SCC, including tobacco and alcohol abuse and genetic susceptibility [1].
Vitamin A (retinol), its natural metabolites, and its synthetic analogs constitute a class of chemicals often referred to as retinoids. Retinoids play an important role as regulators of cell proliferation and differentiation in embryonic development [3] and organ homeostasis. All-trans-retinoic acid (RA) acts as a ligand for a group of nuclear receptors known as retinoic acid receptors α, β or γ (RARs) [4]. RARs function as heterodimers with members of the retinoid X receptor family (RXR α, β or γ) [4]. RAR/ RXR heterodimers bind to specific DNA sequences, known as retinoic acid response elements (RAREs), and regulate the transcription of downstream genes [4,5]. Normal epithelial cells require retinoid signaling to differentiate and function properly [6]. The expression of RARs and the metabolism of retinoids are aberrant in oral cancer [7–9]. Retinoids in combination with other drugs have been used to treat oral leukoplakia and squamous cell carcinoma [10].
Homeodomain proteins are transcription factors [11]. In vertebrates, groups of homeodomain genes known as Hox genes are located on chromosomes in clusters [11]. Recent reports suggest that HOX genes are often dysregulated in human oral SCC (OSCC). HOXA1, HOXA10, and HOXB7 are expressed at higher levels in OSCC as compared to healthy mucosas and may be prognostic markers for OSCC [12–14]. Expression of HOX genes during embryogenesis is regulated by RA [15]. RA induces expression of genes in the 3′ ends of Hox clusters, resulting in an increase in these transcripts in the anterior part of the body, while transcripts of 5′ Hox cluster genes that are not induced by RA are enriched posteriorly [15]. We and others have identified retinoic acid response elements (RAREs) in the enhancers of Hoxa1, Hoxd1, Hoxa4, and Hoxb1 [5,16–18]. Silencing of Hox gene expression during development and cell differentiation is mediated by the activity of Polycomb complexes [15]. Core components of the human Polycomb repressive complex 2 (PRC2) include: E(Z) homolog 2 (EZH2), SUZ12, embryonic ectoderm development (EED) and retinoblastoma-binding protein p48 (RBAP48, or RBBP4) [19]. EZH2 is enzymatically active, catalyzing the mono-, di- and tri- methylation of lysines 9 and 27 of histone 3; this results in transcriptional silencing of the underlying genes. In addition to EZH2, both SUZ12 and EED are indispensable for PRC2 enzymatic activity. EZH2 can also act independently of other PRC2 proteins [20]. Several recent reports suggest that high expression and activity of EZH2 might be associated with tumor proliferation and poor prognosis in oral SCC patients [21–23].
Our hypothesis is that neoplastic transformation of oral keratinocytes involves changes in the epigenetic regulation of transcription. We have tested this hypothesis by assessing the differences in RNA transcripts between cultured immortalized, non-tumorigenic and neoplastically transformed human oral keratinocytes by RNAseq and qRT-PCR. We have also determined the roles of SUZ12 and RA in the regulation of differentially expressed genes by chromatin immunoprecipitation and by shRNA mediated depletion of SUZ12 in immortalized non-tumorigenic oral keratinocytes and in SCC lines.
Materials and methods
Cell culture and chemicals
Immortalized non-transformed human oral keratinocytes cell lines OKF4-TERT1, OKF6-TERT1 and OKF6-TERT1R [24], were kindly provided by Dr. James G. Rheinwald, Harvard Medical School. These cells were cultured in Keratinocyte-SFM medium (#10744019, Gibco, CA, USA), supplemented with 0.3 mM CaCl2, 0.2 ng/ml EGF, penicillin/streptomycin/L-glutamine (#10378-016, Gibco, CA, USA) and bovine pituitary extract (BPE), as previously described [24]. Human OSCC lines SCC-9, SCC-15 and SCC-25 were cultured in Dulbecco’s Modified Eagle’s Medium: Nutrient Mixture F-12 (DMEM/F-12; Gibco, CA, USA), supplemented with 10% fetal calf serum and 400 ng/ml hydrocortisone. HEK293 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Gibco, CA, USA), supplemented with 10% fetal calf serum. Stock solutions of all-trans retinoic acid (RA; Sigma-Aldrich, St. Louis, MO) were prepared in 100% ethanol at the beginning of each experiment and diluted in growth medium to a concentration of 1 µM. For mRNA analyses, cells were plated in 10 cm2 tissue culture plates at the density of 2 × 106 cells/plate and treated with 1 µM RA or vehicle (0.1% ethanol) for 48 h. For chromatin immunoprecipitation, the cells were plated in 15 cm2 tissue culture plates at a density of 3 × 106 cells/ plate and treated with 1 µM RA or vehicle. For cell proliferation assays, cells were plated in 12-well plates at a density of 2 × 104 cells per well, 3 wells for each cell line in each of three biological repeats of the experiment. The cells were treated with 1 µM RA or vehicle in the growth medium for 6 days and counted using a cell and particle counter (Coulter Z; Beckman Coulter, Inc., Fullerton, CA). Stable Suz12 knockdown (shRNA sequence TRCN0000038728) and control cell populations were established as described [25].
Deep sequencing and data analysis
For deep sequencing, total RNA was isolated from cells using an RNeasy mini kit (Quiagen Inc. Valencia, CA, USA), including an in-column DNAse treatment. RNA quality check, cDNA library preparation, and deep sequencing were performed by the Weill Cornell Medical College Genomics Resources Core Facility. Data analysis was performed with guidance and help from Dr. Fabienne Campagne (Institute for Computational Biomedicine, Weill Cornell Medical College (WCMC)). Sequencing data were uploaded into the GobyWeb application (Campagne lab) for alignment to the genome. Differential expression analysis was performed initially using GobyWeb [26]. Differential expression analysis was repeated using EdgeR, an RNAseq analysis package already cited in published reports [27–29]. To focus on potentially the most relevant genes, we generated lists of transcripts with at least a threefold difference between the groups. Further analysis was performed using software available publicly online from DAVID (The Database for Annotation, Visualization and Integrated Discovery [30]).
cDNA generation and real time polymerase chain reaction (PCR) analysis
Total RNA was isolated from cells with Trizol reagent (#15596, Invitrogen, CA, USA), following the manufacturer’s protocol. Reverse transcription was performed using 1 µg of total RNA and qScript cDNA Synthesis Kit (#95048, Quanta Biosciences, Inc. Gaithersburg, MD, USA). First strand cDNA was diluted 20 × and 2 µl were used as a template in PCR Primers specific for hypoxanthine phosphoribosyltransferase 1 (HPRT1) were described [31].
Chromatin immunoprecipitation (ChIP)
ChIP experiments were performed as described [32], with two modifications: (1) chromatin was sheared using a Branson 150 Sonifier (setting 3) for 3 20 s intervals, and (2) 20 µl of soluble chromatin was removed, diluted to a final volume of 100 µl with ChIP elution buffer (50 mM Tris-HCl (pH 8.0), 1% SDS, 1mM EDTA), 4 µl of 5 M NaCl was added, and samples were incubated at 65 °C overnight to reverse cross-links in the chromatin. The DNA was purified using a PCR purification kit (# 28106 Qiagen, Gaithersburg, MD, USA) according to the manufacturer’s protocol, eluted using 20 µl of the elution buffer, and the DNA concentration was calculated based on 260 nm absorbance measurements. The remaining chromatin sample was stored at −80 °C. For each immunoprecipitation (IP), a volume of chromatin corresponding to 20 µg of DNA was used, and diluted to a total volume of 500 µl with lysis buffer. Antibodies for ChIP analysis were: rabbit monoclonal anti SUZ12 (D39F6) XP® (#3737, Cell Signaling, Danvers, MA, USA), anti trimethyl-histone H3 lysine 27 (H-3K27me3 #07–449 Millipore, Billerica, MA, USA) and normal rabbit IgG (sc-2027, Santa Cruz CA, USA). We designed our primers to amplify sequences near selected homeobox genes that have been previously found to bind SUZ12 in human cells by ChIP-seq, based on data available from ENCODE (The Encyclopedia of DNA Elements Consortium), visualized in the University of California, Santa Cruz (UCSC) Genome Browser.
Western blot analysis
Protein extracts in final sample buffer (0.5 M Tris-HCl, pH 6.8,10% glycerol and 1% sodium dodecyl sulfate) were sonicated, separated by SDS-PAGE, and transferred onto nitrocellulose membranes. Membranes were blocked in 5% non-fat dry milk, 0.1% Tween 20 in PBS, for 1 h. at room temperature and incubated with primary antibodies, diluted in blocking solution, overnight at 4 °C. After washing with 0.1% Tween 20-PBS, the membranes were incubated with appropriate secondary antibodies (horseradish peroxidase conjugated anti-mouse or anti-rabbit IgG), washed again, and developed using a Western Pierce ECL Plus Substrate kit (# 32132; Thermo Scientific, Waltham, MA, USA). Antibodies used were: rabbit monoclonal anti Suz12 (D39F6) XP®, (# 3737; Cell Signaling, Danvers, MA, USA), used at 1:2000 dilution; mouse monoclonal anti actin (Clone C4 #MAB1501; Millipore, Billerica, MA, USA), used at 1:80,000; and horseradish peroxidase- conjugated secondary antibodies (#715-035-150 and # 711-035-152; Jackson ImmunoResearch Laboratories, Inc. West Grove, PA, USA), used at 1:5000 dilution.
Statistical analysis
All experiments include at least 3 independent biological replicates (n ≥ 3). Statistical analyses of the RNAseq results are discussed above. Quantitative PCR experiments were analyzed using the GraphPad Prism program. One-way ANOVA followed by Dunnett’s post-test, setting the result obtained for the OKF6-TERT1cells as the control value to which all other samples were compared; a two-way ANOVA followed by Bonferroni post-test correction; or an unpaired t test were applied, as indicated in the figure legends.
Results
RNA-seq analyses reveal differential expression of large numbers of homeobox genes in non-tumorigenic vs. tumorigenic oral keratinocytes
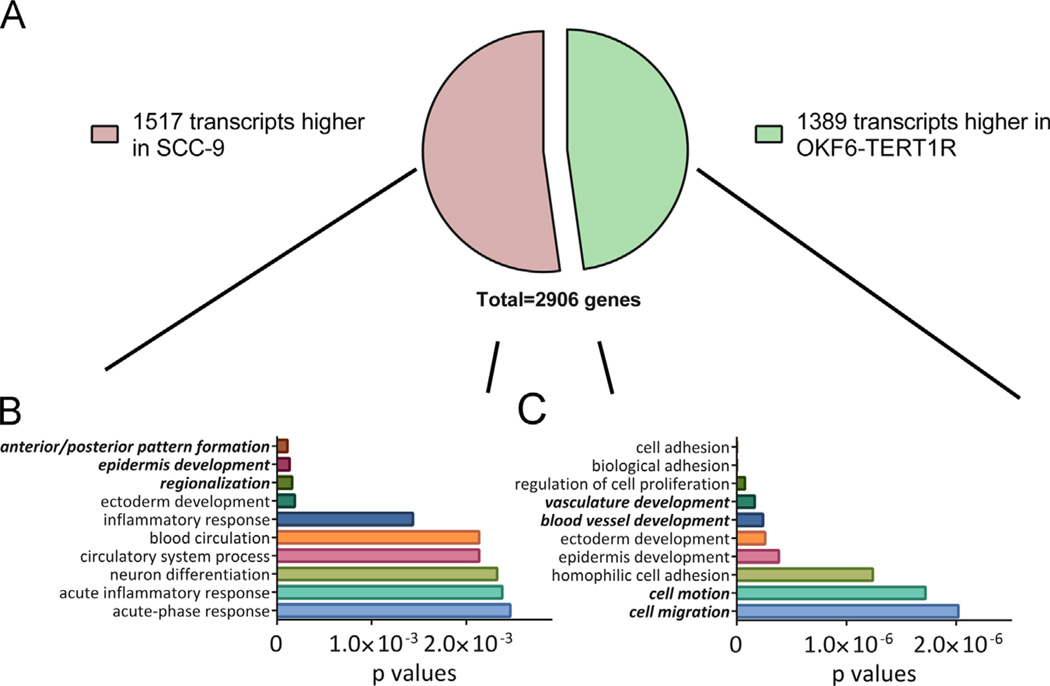
To gain insight into the molecular changes during OSCC carcino-genesis, we performed unbiased, whole genome deep sequencing (RNA-seq) using RNA isolated from cultured, human TERT-immor-talized, non-tumorigenic OKF6-TERT1R and OSCC SCC-9 cells. Since OKF6-TERT1R cells are non-tumorigenic, while SCC-9 cells form rapidly growing tumors when transplanted into nude mice [33], the genes differentially expressed between these two cell types should elucidate the differences between non-tumorigenic and tumorigenic cells. We identified 2906 genes that met our inclusion criteria (see Materials and methods section): 1517 genes exhibited increased mRNA levels in SCC-9 cells compared to OKF6-TERT1R cells and 1389 showed reduced levels in SCC-9 compared to OKF6-TERT1R cells (Fig. 1A). The 50 mRNAs with the highest fold differences between OKF6-TERT1R and SCC-9 cells are shown (Supplementary Tables 1 and 2).
Fig. 1.
RNAseq analyses reveal differential expression of large numbers of genes in non- tumorigenic vs. tumorigenic oral keratinocytes. (A) Pie chart showing the distribution of genes with at least a 3 fold difference in transcript levels betweeen OKF6-TERT1R and SCC-9 cells. Lines indicate the part of gene list used in gene ontology (GO) analysis in (B) and (C). (B) and (C) Results of GO analysis for (B) the genes with transcript levels at least 3 fold higher in SCC-9 than OKF6-TERT1R cells and (C) the genes with transcript levels at least 3 fold higher in OKF6-TERT1R than SCC-9 cells. Ten GO terms with the lowest p values are shown. GO terms associated with homeobox genes are in highlighted in bold italics.
We found that many homeobox genes are highly represented among the genes with large fold differences between the two cell types. Twenty nine homeobox genes exhibited transcript levels higher in SCC-9 than the OKF6-TERT1R cells, and 19 homeobox genes displayed transcript levels that were lower in the SCC-9 than the OKF6-TERT1R cells (see Table 1 for the homeobox genes differentially expressed between OKF6-TERT1R and SCC-9). HOXB3 and HOXB6 ranked among the top 1% of differentially expressed genes, with transcript levels 2519 and 1799 fold higher in SCC-9 than in OKF6-TERT1R cells; in addition, 14 homeobox genes ranked among the top 10% of mRNAs higher in SCC-9 than in OKF6-TERT1R (Table 1). In contrast, IRX1, SIX2, MEIS3, TSHZ2, and PBX1 ranked among the top 10% of mRNAs lower in SCC-9 than in OKF6-TERT1R (Table 1). Dysregulation of homeobox gene expression might constitute a selective advantage for transformed oral keratinocytes.
Table 1.
Homeobox genes with transcript levels higher (top) or lower (bottom) in vehicle treated SCC-9 than in vehicle treated OKF6-TERT1R cell lines (RNAseq).
| No | Ensemble gene ID | Gene symbol |
Gene name | Fold change SCC-9/ OKF6- TERT1R |
t-Test (BUQ)-BH-FDR-q- value |
|---|---|---|---|---|---|
| 1 | ENSG00000120093 | HOXB3 | homeobox B3 | 2519 | 2.46E–34 |
| 2 | ENSG00000108511 | HOXB6 | homeobox B6 | 1799 | 5.63E–36 |
| 3 | ENSG00000123364 | HOXC13 | homeobox C13 | 910 | 1.45E–64 |
| 4 | ENSG00000123388 | HOXC11 | homeobox C11 | 608 | 1.69E–42 |
| 5 | ENSG00000180818 | HOXC10 | homeobox C10 | 493 | 1.15E–58 |
| 6 | ENSG00000180806 | HOXC9 | homeobox C9 | 473 | 1.16E–38 |
| 7 | ENSG00000120087 | HOXB7 | homeobox B7 | 373 | 1.23E–64 |
| 8 | ENSG00000253293 | HOXA10 | homeobox A10 | 287 | 6.24E–30 |
| 9 | ENSG00000172789 | HOXC5 | homeobox C5 | 239 | 1.38E–27 |
| 10 | ENSG00000170166 | HOXD4 | homeobox D4 | 229 | 1.32E–45 |
| 11 | ENSG00000198353 | HOXC4 | homeobox C4 | 218 | 2.02E–26 |
| 12 | ENSG00000128652 | HOXD3 | homeobox D3 | 218 | 2.87E–41 |
| 13 | ENSG00000152192 | POU4F1 | POU class 4 homeobox 1 | 212 | 5.68E–37 |
| 14 | ENSG00000258518 | SHOX2 | short stature homeobox protein 2 isoform c |
166 | 5.81E–23 |
| 15 | ENSG00000132130 | LHX1 | LIM homeobox 1 | 159 | 1.88E–35 |
| 16 | ENSG00000175879 | HOXD8 | homeobox D8 | 155 | 1.86E–52 |
| 17 | ENSG00000170689 | HOXB9 | homeobox B9 | 107 | 2.11E–27 |
| 18 | ENSG00000168779 | SHOX2 | short stature homeobox 2 | 100 | 6.47E–33 |
| 19 | ENSG00000128710 | HOXD10 | homeobox D10 | 94 | 8.40E–34 |
| 20 | ENSG00000163623 | NKX6-1 | NK6 homeobox 1 | 94 | 2.99E–21 |
| 21 | ENSG00000106536 | POU6F2 | POU class 6 homeobox 2 | 94 | 3.15E–22 |
| 22 | ENSG00000134138 | MEIS2 | Meis homeobox 2 | 54 | 9.08E–34 |
| 23 | ENSG00000128713 | HOXD11 | homeobox D11 | 38 | 1.54E–19 |
| 24 | ENSG00000144355 | DLX1 | distal-less homeobox 1 | 35 | 1.31E–24 |
| 25 | ENSG00000119042 | SATB2 | SATB homeobox 2 | 10 | 1.47E–23 |
| 26 | ENSG00000180438 | TPRXL | tetra-peptide repeat homeobox-like | 5 | 2.05E–07 |
| 27 | ENSG00000119547 | ONECUT2 | one cut homeobox 2 | 4 | 1.40E–06 |
| 28 | ENSG00000106852 | LHX6 | LIM homeobox 6 | 3 | 3.59E–05 |
| 29 | ENSG00000105717 | PBX4 | pre-B-cell leukemia homeobox 4 | 3 | 0.001775 |
| No | Ensemble gene ID |
Gene symbol |
Gene name |
Fold change OKF6-TERT1R /SCC-9 |
t-Test (BUQ)-BH-FDR-q- value |
| 1 | ENSG00000170549 | IRX1 | iroquois homeobox 1 | 338 | 4.00E–76 |
| 2 | ENSG00000105419 | MEIS3 | Meis homeobox 3 | 111 | 6.92E–75 |
| 3 | ENSG00000170577 | SIX2 | SIX homeobox 2 | 105 | 5.70E–72 |
| 4 | ENSG00000182463 | TSHZ2 | teashirt zinc finger homeobox 2 | 91 | 3.06E–46 |
| 5 | ENSG00000185630 | PBX1 | pre-B-cell leukemia homeobox 1 | 46 | 2.34E–30 |
| 6 | ENSG00000121297 | TSHZ3 | teashirt zinc finger homeobox 3 | 22 | 6.44E–50 |
| 7 | ENSG00000113430 | IRX4 | iroquois homeobox 4 | 20 | 3.67E–48 |
| 8 | ENSG00000028277 | POU2F2 | POU class 2 homeobox 2 | 18 | 2.16E–29 |
| 9 | ENSG00000126778 | SIX1 | SIX homeobox 1 | 16 | 7.47E–28 |
| 10 | ENSG00000148516 | ZEB1 | zinc finger E-box binding homeobox 1 | 9 | 2.49E–17 |
| 11 | ENSG00000184271 | POU6F1 | POU class 6 homeobox 1 | 7 | 1.49E–13 |
| 12 | ENSG00000143995 | MEIS1 | Meis homeobox 1 | 6 | 5.45E–20 |
| 13 | ENSG00000159556 | ISL2 | ISL LIM homeobox 2 | 6 | 4.10E–10 |
| 14 | ENSG00000171476 | HOPX | HOP homeobox | 5 | 3.90E–11 |
| 15 | ENSG00000169554 | ZEB2 | zinc finger E-box binding homeobox 2 | 4 | 1.40E–06 |
| 16 | ENSG00000182568 | SATB1 | SATB homeobox 1 | 3 | 1.93E–06 |
| 17 | ENSG00000016082 | ISL1 | ISL LIM homeobox 1 | 3 | 6.91E–06 |
| 18 | ENSG00000064195 | DLX3 | distal-less homeobox 3 | 3 | 0.005921 |
| 19 | ENSG00000167081 | PBX3 | pre-B-cell leukemia homeobox 3 | 3 | 2.55E–10 |
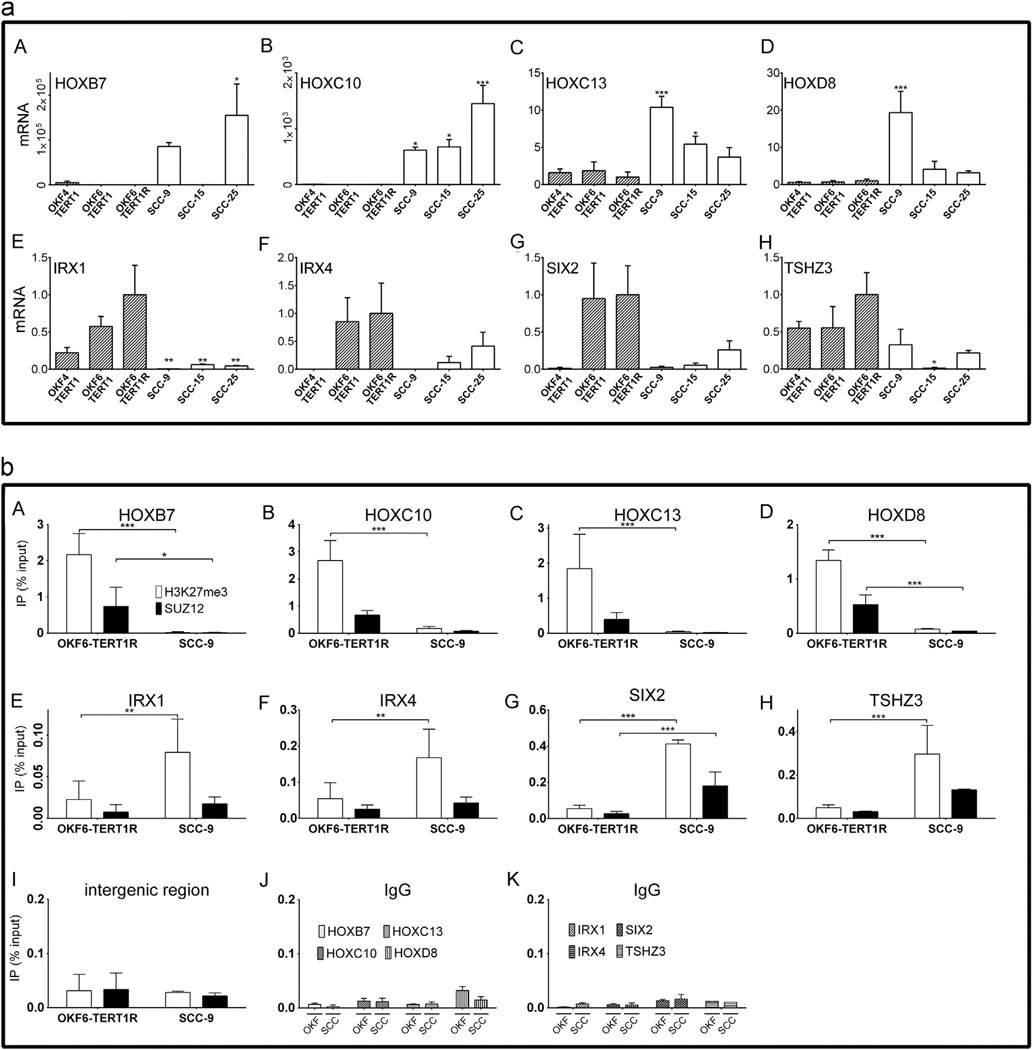
To evaluate whether a similar pattern of homeobox gene expression is characteristic of other non-tumorigenic vs. tumorigenic oral keratinocyte cell lines we assessed transcript levels of selected homeobox genes differentially expressed between OKF6-TERT1R and SCC-9 cells in the TERT-immortalized, non-tumorigenic cell lines OKF4-TERT1 and OKF6-TERT1 and the tumorigenic SCC-15 and SCC-25 lines. For these experiments, we selected homeobox genes with the greatest fold differences between SCC-9 and OKF6-TERT1R cells based on the initial analysis performed using Goby (see: Materials and methods section). HOXB7, HOXC10, HOXC13 and HOXD8 transcripts (Fig. 2(a) A–D) were detected at very low levels in all non-tumorigenic cell lines and at markedly higher levels in at least one tumorigenic SCC cell line. HOXB7 transcripts were 17 and 31 fold higher in SCC-9 and SCC-25 cells, respectively, than in OKF4-TERT1 cells, which showed the highest HOXB7 transcript levels among the non-tumorigenic OKF cell lines (Fig. 2(a) A). HOXC10 transcripts were 106, 111 and 409 fold higher in SCC-9, SCC-15 and SCC-25 cells, respectively, than in OKF4-TERT1 cells (Fig. 2(a) B). HOXC13 transcripts were 10, 5 and 4 fold higher in SCC-9, SCC-15 and SCC-25, respectively, than in OKF6-TERT1cells (Fig. 2(a) C). HOXD8 transcripts were 19, 4 and 3 fold higher in SCC-9, SCC-15 and SCC-25 cells, respectively, than in OKF6-TERT1R cells (Fig. 2(a) D).
Fig. 2.
(a) mRNA levels of homeobox genes expressed differentially in non-tumorigenic and tumorigenic oral keratinocyte cell lines and (b) levels of SUZ12 and the H3K27me3 epigenetic mark proximal to homeodomain genes expressed differentially between OKF6-TERT1R and SCC-9 cells. n=3 independent biological repeats; * indicates p<0.05, ** indicates p<0.01 and *** indicates p<0.001; note differences in scales of y-axes in different panels (a) mRNA levels are normalized to HPRT1 expression and represented relative to transcript levels in control treated OKF6-TERT1R cells (set as 1); differences in transcript levels between the cell lines were analyzed by one-way ANOVA followed by Dunnett’s posttest, setting the result obtained for the OKF6-TERT1cells as control value to which all other samples were comapred: A–D mRNA levels of HOX genes expressed at higher levels in SCC-9 vs. OKF6-TERT1R cells. E–H mRNA levels of genes expressed at lower levels in SCC-9 vs. OKF6-TERT1R cells. (b) Data are represented as percent of chromatin used as input in corresponding IPs; differences in immunoprecipitated chromatin levels between the cell lines were analyzed two-way ANOVA followed by Bonferroni post-test: A–D Levels of the H3K27me3 mark (clear bars) and SUZ12 (black bars) at HOX genes expressed at higher levels in SCC-9 vs. OKF6-TERT1R cells, as assessed by ChIP; E to H: Levels of H3K27me3 and SUZ12 at HOX genes expressed at lower levels in SCC-9 vs. OKF6-TERT1R cells, as assessed by ChIP; I: Negative control: association of H3K27me3 mark and SUZ12 with with an intergenic region, as assessed by ChIP J and K: Negative control: chromatin immunoprecipitated using normal rabbit IgG along with antibodies specific to SUZ12 and H3K27me3; sequences amplified are indicated by the graph legend.
In contrast, IRX1, IRX4, SIX2 and TSHZ3 transcripts were higher in non-tumorigenic than in tumorigenic cells (Fig. 2(a) E–H). Transcript levels of IRX1 were 203, 16 and 23 fold lower in SCC-9, SCC-15 and SCC-25, respectively, than in OKF6-TETRT1R cells (Fig. 2(a) E). IRX4 transcripts were 1015 and 8 fold lower in SCC-9 and SCC-15, respectively, than in OKF6-TERT1R (Fig. 2(a) F), and SIX2 mRNA was 43, 18, and 4 fold less abundant in SCC-9, SCC-15 and SCC-25, respectively, than in OKF6-TERT1R cells (Fig. 2 (a) G). We detected TSHZ3 mRNA at 3, 76 and 5 fold lower levels in SCC-9, SCC-15, and SCC-25, respectively, than in OKF6-TERT1R cells (Fig. 2(a) H). We conclude that differences in homeobox transcripts are a characteristic of several non-tumorigenic and tumorigenic oral keratinocytes.
Homeobox genes differentially expressed between OKF6-TERT1R and SCC-9 cells are also dysregulated in samples directly from HNSCC patients
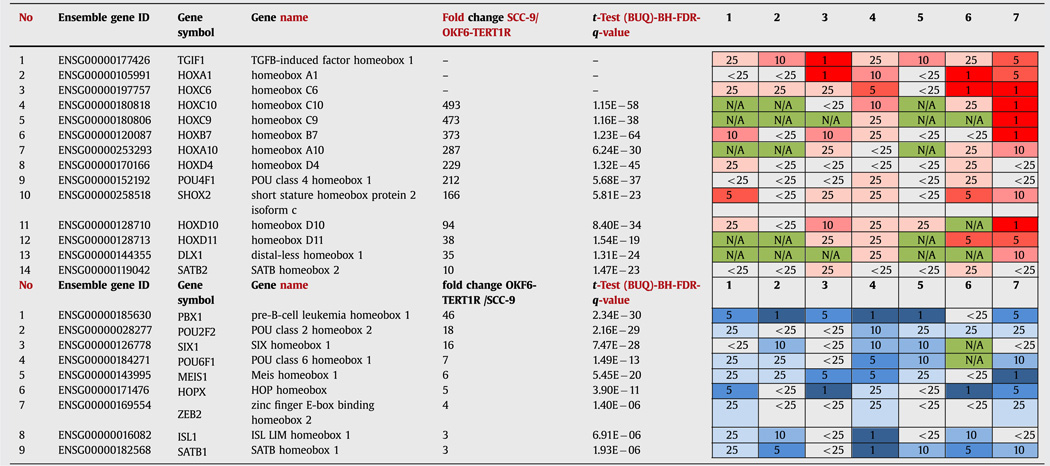
In order to relate our data from this human cell culture model to changes observed in human HNSCC patient tissues, we utilized ONCOMINE, an online high-throughput cancer biology database and analysis platform [34]. For our comparisons, we selected ONCOMINE datasets that compare transcript levels in normal tissues [35–41] with those in HNSCC samples; four out of seven data sets contain tongue carcinoma samples only [36,38,40,41]; one contains oral cavity SCC samples only [39]; and the remaining two include samples from OSCC patients as well as samples from HNSCC patients with tumors arising in oropharynx, hypopharynx, larynx or sinus [35,37]). Analysis of the ONCOMINE data revealed that genes from the homeodomain protein family are often differentially expressed between human HNSCC patient tumor and normal tissues. HOXC6, HOXA1, HOXB7, HOXC10 and HOXD10 rank among the top genes with transcript levels elevated in tumor vs. normal tissue in ONCOMINE datasets (Table 2). PBX1, HOPX, MEIS1 and ISL1 rank within the top genes with transcript levels lower in tumor than in normal tissue in ONCOMINE datasets (Table 2). Thus, homeobox genes are often dysregulated in tumor samples from SCC patients, and as such could be essential in the tumorigenic transformation of oral keratinocytes.
Table 2.
Homeobox genes with transcript levels higher (top) or lower (bottom) in vehicle treated SCC-9 than in vehicle treated OKF6-TERT1R cells and in human HNSCC samples than in control normal tissue (ONCOMINE data sets). Oncomine datasets referred in the table are: (1) Cromer Head–Neck, [35]; (2) Estilo Head–Neck, [36]; (3) Ginos Head-Neck, [37]; (4) Pyeon Multi-cancer, [41]; (5) Talbot Lung, [38]; (6) Toruner Head–Neck, [39]; and (7) Ye Head-Neck, [40]; Top: Homeobox gene transcripts in OKF6-TERT1R and SCC-9 cells (RNAseq data), concomitantly ranking among the top 25% of genes with transcript levels higher in tumor vs. normal tissue in at least two Oncomine datasets are shown. Transcripts are ordered according to fold change between the OKF6-TERT1R and SCC-9 cells. Three top homeobox gene transcripts HOXA1, HOXC6, and TGIF1, that rank high among Oncomine datasets, but not in our RNA-Seq experiments, are also shown; red color intensity /number in the table denote transcript’s ranking among the top 1, 5,10 or 25% of genes with transcript levels elevated in tumor vs. normal tissue in the indicated dataset; Bottom: Homeobox gene transcripts in OKF6-TERT1R and SCC-9 cells (RNAseq data), concomitantly ranking among the top 25% of genes with transcript levels lower in tumor vs. normal tissue in at least two Oncomine datasets are shown. Transcripts are ordered according to fold change between the OKF6-TERT1R and SCC-9 cells; blue color intensity /number in the table denote transcript’s ranking among the top 1, 5, 10 or 25% of genes with transcript levels reduced in tumor vs. normal tissue in the indicated dataset; green color and letters N/A denote transcript levels not assessed by the indicated dataset.
 |
Additional genes differentially expressed between OKF6-TERT1R and SCC-9 cells are also dysregulated in samples directly from HNSCC patients
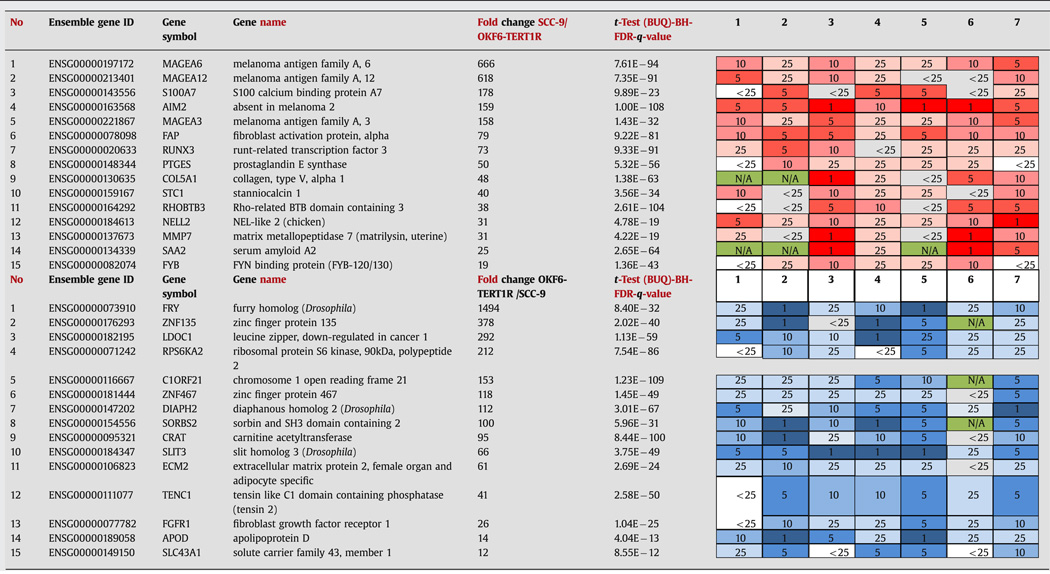
Fifteen of the top 25% of the transcripts higher in SCC-9 than in the OKF6-TERT1R cells in our screen that also ranked within the top 25% of genes expressed at higher levels in tumor compared to normal tissue samples in 7 ONCOMINE datasets [35–41] are shown (Table 3). AIM2, FAP, MAGEA3, MAGEA6, and NELL2 transcripts were significantly higher in tumor than in normal tissue samples in all of the ONCOMINE datasets (Table 3). A comparison of transcripts expressed at lower levels in SCC-9 than in OKF6-TERT1R cells and ranked within the top 25% of genes with mRNA levels lower in tumor than in normal tissue samples by selected ONCOMINE datasets is shown (Table 3). Five transcripts, APOD, DIAPH2, FRY, LDOC1, and SLIT3, were expressed at significantly lower levels in tumor than in normal tissue samples in all of the compared datasets (Table 3). Thus, our data and data obtained from human tissue samples highlight many of the same genes, suggesting that OKF6-TERT1R and SCC-9 can serve as a reasonable model to investigate aspects of neoplastic transformation in human oral SCC.
Table 3.
Genes with transcript levels higher (top) or lower (bottom) in vehicle treated SCC-9 than in vehicle treated OKF6-TERT1R cells compared to human HNSCC samples vs. control normal human tissue (ONCOMINE datasets). Oncomine datasets referred in the table are: (1) Cromer Head-Neck, [35]; (2) Estilo Head-Neck, [36]; (3) Ginos Head-Neck, [37]; (4) Pyeon Multi-cancer, [41]; (5) Talbot Lung, [38]; (6) Toruner Head-Neck, [39]; and (7) Ye Head-Neck, [40]; Top: The top 25% of genes with transcript levels higher in SCC-9 than in OKF6-TERT1R cells (RNA-seq data) concomitantly ranking among the top 25% of genes with transcript levels higher in tumor vs. normal tissue in the highest number of relevant Oncomine datasets- the top 15 are shown. Transcripts are ordered according to fold change between the SCC-9 and OKF6-TERT1R cells; red color intensity /number in the table denote transcript’s ranking among the top 1, 5, 10 or 25% of genes with transcript levels elevated in tumor vs. normal tissue in the indicated dataset; Bottom: The top 25% of genes with transcript levels lower in SCC-9 than in OKF6-TERT1R cells (RNAseq data) concomitantly ranking among the top 25% of genes with transcript levels lower in tumor vs. normal tissue in the highest number of relevant Oncomine datasets- the top 15 are shown. Transcripts are ordered according to fold change between the OKF6-TERT1R and SCC-9 cells; blue color intensity /number in the table denote transcript’s ranking among the top 1, 5, 10 or 25% of genes with transcript levels reduced in tumor vs. normal tissue in the indicated dataset; green color and letters N/A denote transcript levels not assessed by the indicated dataset.
 |
Gene ontology annotation of genes differentially expressed between OKF6-TERT1R and SCC-9 cells highlights homeobox genes
We utilized DAVID (The Database for Annotation, Visualization and Integrated Discovery, an online open access resource) [30] to annotate differentially expressed genes. We identified gene ontology (GO) terms associated with genes differentially expressed between OKF6-TERT1R and SCC-9 cells and enriched as compared to the human genome; ten terms with the highest enrichment significance (p values) are shown (Fig. 1B and C). We also employed DAVID’s gene ontology clustering module, which groups functionally related annotations into clusters, each assigned an enrichment score (a statistical parameter). An overview of clusters with enrichment scores higher or equal to 1.3 (for more details, see: Supplementary methods) is shown (Supplementary Tables 3 and 4).
The gene ontology terms associated with the transcripts expressed differentially between SCC-9 and OKF6-TERT1R cells cover cellular components, biological processes, and molecular functions associated with carcinogenesis and tumor invasion and/or metastasis. For example, SCC-9 cells express higher levels of transcripts associated with the inflammatory response, but lower levels of transcripts associated with cell adhesion or regulation of cell proliferation than OKF6-TERT1R cells. Interestingly- as we did not anticipate a similar result in untreated cells—a subset of transcripts present at lower levels in SCC-9 than in OKF6-TERT1R cells forms a cluster associated with response to vitamin A (cluster 33, enrichment score 1.9, containing genes: KLF4, PDGFA, SOX2, MICB, FADS1, MAP1B; Supplementary Table 4). This last finding suggests that the ability to respond to vitamin A might be compromised in tumorigenic as compared to non-tumorigenic cells.
We also found that homeobox genes are associated with the most enriched GO terms (Fig. 1B and C) and are highly represented among the clusters with the top enrichment scores. This provides further in silico support for our hypothesis that changes in homeo-box gene expression promote tumorigenic transformation.
Homeobox genes with higher transcript levels in SCC-9 than in OKF6-TERT1R cells associate with SUZ12 and possess H3K27me3 chromatin marks in OKF6-TERT1R cells
Silencing of HOX gene expression is often mediated by the activity of Polycomb complexes [15]. We hypothesized that the changes in homeodomain transcript levels in the SCC-9 vs. OKF6-TERT1R cells are a consequence of changes in the epigenetic regulation of their expression. Thus, we used chromatin immunoprecipitation (ChIP) to evaluate the association of the PRC2 core protein SUZ12 with chromatin near homeodomain genes expressed differentially between the OKF6-TERT1R and SCC-9 cell lines, as well as to measure the levels of the repressive histone 3 lysine 27 trimethyl (H3K27me3) epigenetic mark present at the same genes. Homeo-box transcripts expressed at higher levels in SCC-9 than in OKF6-TERT1R cells, specifically HOXB7, HOXC10, HOXC13 and HOXD8 exhibited between 8 (SUZ12) and 124-fold (H3K27me3) lower levels of SUZ12 and the H3K27me3 epigenetic mark in the SCC-9 compared to the OKF6-TERT1 cells (Fig. 2(b) A–D). A summary of these results, including the fold changes in SUZ12 binding and H3K27me3 levels between OKF6-TERT1R and SCC-9 cells at HOXB7, HOXC10, HOXC13, and HOXD4, is shown (Table 4).
Table 4.
Descriptive summary of ChIP results shown in Fig. 2(b) and Fig. 4(b). Note: p values shown in table refer to Fig. 2(b) and were calculated using two-way ANOVA followed by Bonferroni post-test.
| Cell treatment | Gene name |
H3K27me3 |
SUZ12 |
||
|---|---|---|---|---|---|
| Fold increase in OKF6-TERT1R /SCC-9 | p Value | Fold increase in OKF6-TERT1R /SCC-9 | p Value | ||
| Vehicle (0.1% ethanol) | HOXB7 | 124 | < 0.001 | 56 | < 0.01 |
| HOXC10 | 14.8 | < 0.001 | 8 | n.s. | |
| HOXC13 | 47 | < 0.001 | 18 | n.s. | |
| HOXD8 | 17 | < 0.001 | 13 | < 0.001 | |
|
Gene name |
Fold increase in SCC-9/OKF6-TERT1R | p Value | Fold increase in SCC-9/OKF6-TERT1R | p Value | |
| IRX1 | 3.5 | < 0.05 | 2.2 | n.s. | |
| IRX4 | 3.0 | < 0.001 | 1.7 | n.s. | |
| SIX2 | 7.6 | < 0.001 | 6.6 | < 0.001 | |
| TSHZ3 | 6.0 | < 0.05 | 4.3 | n.s. | |
| Cell treatment |
Gene name |
H3K27me3 Fold increase in OKF6-TERT1R /SCC-9 |
p Value |
SUZ12 Fold increase in OKF6-TERT1R /SCC-9 |
p Value |
| 1 µM RA | HOXB7 | 51 | < 0.001 | 45 | < 0.001 |
| HOXC10 | 16 | < 0.001 | 21 | < 0.001 | |
| HOXC13 | 31 | < 0.001 | 22 | n.s. | |
| HOXD8 | 11 | < 0.001 | 12 | < 0.001 | |
|
Gene name |
Fold increase in SCC-9/OKF6-TERT1R | p Value | Fold increase in SCC-9/OKF6-TERT1R | p Value | |
| IRX1 | 3.7 | < 0.01 | 1.6 | n.s. | |
| IRX4 | 5.3 | < 0.001 | 2.3 | n.s. | |
| SIX2 | 7.4 | < 0.001 | 3.6 | < 0.05 | |
| TSHZ3 | 10 | < 0.01 | 2.3 | n.s. | |
Thus, SUZ12 is present and the PRC2 complex is active, as measured by the levels of the H3K27me3 mark, at DNA regions near the assessed HOX genes in OKF6-TERT1R cells. We speculate that the presence of PRC2 contributes to the transcriptional silencing of these genes, or alternatively, that these marks are a consequence of the long term, epigenetic gene silencing in OKF6-TERT1R cells [42]. In contrast, both the SUZ12 and H3K27me3 marks are not enriched above the negative IgG controls at the same genes in SCC-9 cells, in which these HOX genes are transcriptionally active, suggesting that the absence of the PRC2 and H3K27me3 marks is associated with the transcriptional activation of the HOXB7, HOXC10, HOXC13, and HOXD8 genes in the SCC-9 cells.
Homeobox genes with lower transcript levels in SCC-9 than in OKF6-TERT1R cells associate with SUZ12 and possess the H3K27me3 chromatin mark in SCC-9 cells
Conversely, the genes IRX1, IRX4, SIX2, and TSHZ3, expressed at lower levels in SCC-9 than in OKF6-TERT1R cells, exhibited higher levels of SUZ12 and the H3K27me3 mark in SCC-9 than in OKF6-TERT1R cells (Fig. 2(b) E–H). A summary of these results, including the fold changes in SUZ12 association and H3K27me3 deposition between OKF6-TERT1R and SCC-9 cells at IRX1, IRX4, SIX2 and TSHZ3 is shown (Table 4). The presence of SUZ12 and the H3K27me3 mark at the IRX1, IRX4, SIX2 and TSHZ3 genes in the SCC-9 cells suggests that the PRC2 complex is active in SCC-9 cells, so the absence of these marks in SCC-9 cells in the vicinity of assessed HOX genes (Fig. 2(b) A–D) is likely the result of PRC2 displacement rather than of a deficiency in PRC2 expression or enzymatic activity. We also conclude that the transcriptional silencing of homeobox genes in SCC-9 cells (namely: IRX1, IRX4, SIX2 and TSHZ3) is associated with the presence of the H3K27me3 mark nearby.
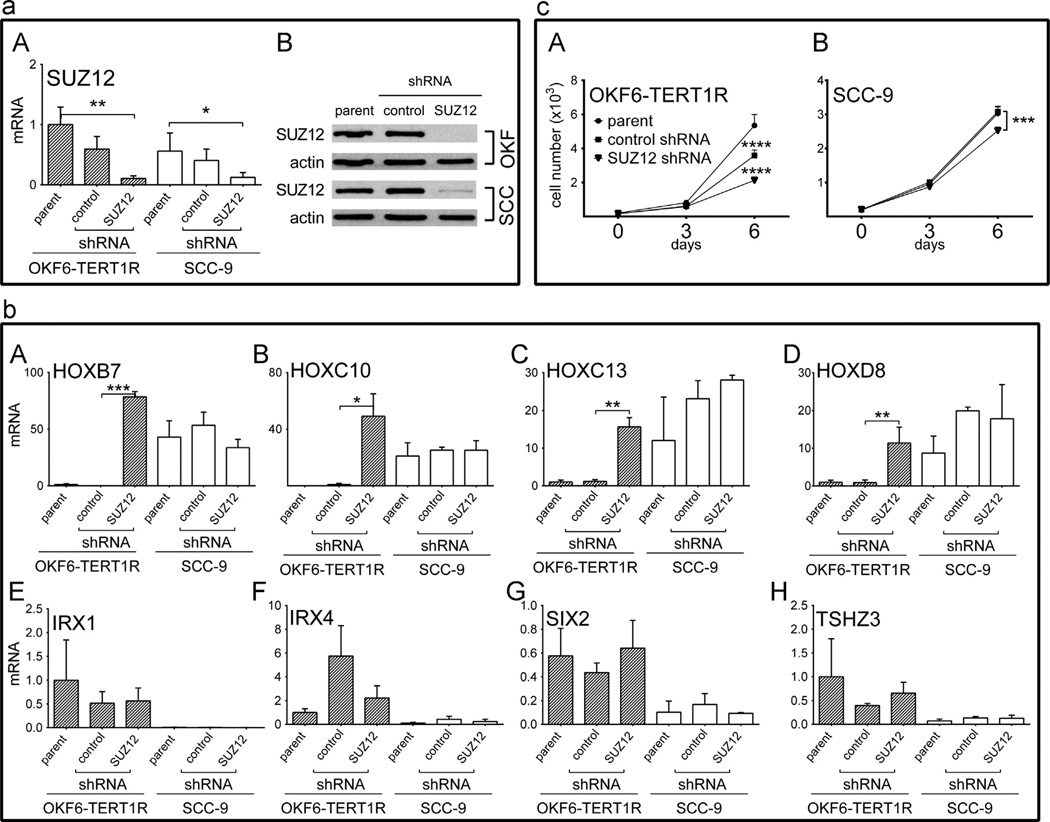
SUZ12 knock down (kd) increases mRNA levels of HOX genes in OKF6-TERT1R cells and reduces the proliferation rate of OKF6-TERT1R
In order to test whether PRC2 activity contributes to the transcriptional silencing of homeobox genes in oral keratinocytes, we knocked down expression of SUZ12 in OKF6-TERT1R and SCC-9 cells. Hairpin sequences targeting SUZ12 were previously validated [25,43] and were introduced into cells through lentiviral transduction. SUZ12 mRNA and protein levels in parental, control and SUZ12 kd cell populations were determined by RT-PCR and Western blot, respectively (Fig. 3(a) A and B). We found that in OKF6-TERT1R cells that express SUZ12 targeting hairpin SUZ12 mRNA levels are only 11% of the SUZ12 mRNA levels in the parental cell line, and that SUZ12 protein levels in OKF6-TERT1R SUZ12 kd cells are below the detection threshold. Similarly, in SUZ12 kd SCC-9 cells mRNA levels were reduced to 22% of the SUZ12 transcript levels in the parental SCC-9 cells, and SUZ12 protein levels in the SUZ12 kd SCC-9 cells were reduced to 9% of SUZ12 protein detected in parental SCC-9 cells.
Fig. 3.
(a) SUZ12 mRNA and protein levels in SUZ12 depleted OKF6-TERT1R and SCC-9 cell populations; (b) mRNA levels of homeobox genes expressed differentially in SUZ12 depleted OKF6-TERT1R and SCC-9 cell populations; and (c) proliferation of SUZ12 depleted OKF6-TERT1R and SCC-9 cell populations. (a) (A) mRNA levels are normalized to GAPDH mRNA levels and are represented relative to transcript levels in OKF6-TERT1R cells (set as 1); y-axis, arbitrary units. Differences in transcript levels between the cell populations were analyzed by unpaired t test, significant differences are indicated. (B) Western blot analysis of SUZ12 protein levels in cells expressing control “scrambled” or SUZ12 targeting shRNA sequence and parental cells. A representative result is shown. (b) PCR results presented and analyzed as in panel (a)(A); A–D mRNA levels of HOX genes expressed at higher levels in parental SCC-9 vs. OKF6-TERT1R cells. E–H mRNA levels of genes expressed at lower levels in parental SCC-9 vs. OKF6-TERT1R cells; (c) cell proliferation assays; data analyzed using two- way ANOVA followed by Bonferroni’s post test, equal numbers of parental and shRNA sequence expressing cell populations were seeded, allowed to attach for 24 h and counted 1, 4 and 7 days following plating (corresponding to 0, 3 and 6 days time points on the graph). n=3 independent biological repeats; note differences in scales of y-axes in different panels, * indicates p<0.05, * indicates p<0.01, *** indicates p<0.001 and **** indicates p<0.0001.
Next, we assessed the transcript levels of selected homeobox genes in OKF6-TERT1R and SCC-9 cells that express either a SUZ12 targeting hairpin or a control “scrambled” sequence and in the parental cell lines not exposed to lentiviral transduction. HOXB7, HOXC10, HOXC13, and HOXD8 transcripts were detected at 78; 49; 15; and 11 fold higher levels, respectively, in the SUZ12 kd OKF6-TERT1R cells as compared to the parental OKF6-TERT1R cells (Fig. 3(b) A–D).
In contrast, transcript levels of the genes IRX1, IRX4, SIX2, and TSHZ3, which are lower in the parental SCC-9 than in parental OKF6-TERT1R cells, did not change in response to SUZ12 depletion (Fig. 3(b) E–H).
These results suggest that removal of the PRC2 complex from HOX genes in the non-tumorigenic oral keratinocytes is sufficient to induce expression of HOX genes in these cells, and thus PRC2 displacement is the most likely mechanism leading to high HOXB7, HOXC10, HOXC13 and HOXD8 transcript levels in the tumorigenic SCC-9 cells. We also conclude that the transcriptional silencing of homeobox genes in SCC-9 cells (namely: IRX1, IRX4, SIX2 and TSHZ3), while correlated with SUZ12 association and H3K27me3 deposition nearby, is likely controlled by an additional mechanism, as SUZ12 knock down/ PRC2 disruption did not suffice to increase the transcript levels of IRX1, IRX4, SIX2 and TSHZ3 in our experiments (Fig. 3(b) E–H).
Importantly, depletion of SUZ12 reduced the proliferation rates of both the non-tumorigenic OKF6-TERT1R and the tumorigenic SCC-9 cells (Fig. 3(c) A and B), albeit to different extents. The number of SUZ12 depleted SCC-9 cells was 83% of parental SCC-9 cells (p ≤ 0.001), while the number of SUZ12 depleted OKF6-TERT1R cells was only 40% of parental OKF6-TERT1R cells (p ≤ 0.0001), at 6 days of culture. This result indicates that non-tumorigenic oral keratinocytes require the functions of SUZ 12 and PRC2 to maintain their full proliferative potential. Tumorigenic SCC-9 cells were less sensitive to the proliferation-inhibitory effects of SUZ12 depletion, suggesting that the process of neoplastic transformation renders some of the SUZ12 functions redundant. This is in agreement with our observation that depletion of SUZ12 was not sufficient to increase mRNA levels of the homeobox genes IRX1, IRX4, SIX2 and TSHZ3 in the SCC-9 cells (Fig. 3(b) E–H).
RNAseq data show that retinoic acid (RA) influences gene expression in both OKF6-TERT1R and SCC-9 cells
So far, we focused on two aspects, HOX gene expression and the activity of PRC2, which were shown to be controlled by the signaling molecule retinoic acid (RA). Expression of HOX genes during embryogenesis is regulated by RA [15]. We and others identified retinoic acid response elements (RAREs) in enhancers of Hoxa1, Hoxd1, Hoxa4, and Hoxb1 [5,16–18]. In murine embryonic stem cells and F9 stem cells, RA induces the removal of Suz12 from the proximal promoter and RARE of Hoxa1 (and other primary RA target genes); this is associated with transcriptional activation [32,44]. Our gene ontology analysis (Supplementary Table 4) indicated that levels of transcripts associated with response to vitamin A are lower in the SCC-9 than in OKF6-TERT1R cells, suggesting that retinoid signaling might be impaired in tumorigenic as compared to non-tumorigenic cells. Finally, in human oral SCC carcinogenesis the silencing of RARβ2, an RA inducible isoform of RARβ, is one of the earliest epigenetic events [9]. Thus, we assessed the effects of RA on gene expression in OKF6-TERT1R and SCC-9 cells.
We performed whole genome deep sequencing (RNAseq) using RNA isolated from RA-treated OKF6-TERT1R and SCC-9 cells. We identified 2424 genes that met our inclusion criteria (see: Materials and methods section): 1224 exhibited increased expression in SCC-9 cells compared to OKF6-TERT1R cells and 1200 showed increased levels in OKF6-TERT1R compared to SCC-9 cells. The 50 genes with the highest fold differences in mRNA levels between RA- treated OKF6-TERT1R and RA-treated SCC-9 cells are shown (Supplementary Tables 5 and 6). Among these 2424 genes with different transcript levels between RA-treated SCC-9 and OKF6-TERT1R cells were 569 genes that were detected only in RA-treated cells. These included HOXA5, HOXD1, PRRX2 (paired related homeobox 2), HDX (highly divergent homeobox) and HOXA2, which showed 930, 53, 46, 8 and 4 fold higher transcript levels in RA-treated SCC-9 than in OKF6-TERT1R cells, respectively.
RA signaling related genes were among those differentially expressed between RA-treated SCC-9 cells and RA-treated OKF6-TERT1R cells. CYP26B1 (cytochrome P450, family 26, subfamily B, polypeptide 1; ENSG00000003137; an enzyme involved in RA metabolism) transcript levels were 6.9 fold (p = 5.89 × 10−23) higher in RA-treated SCC-9 than OKF6TERT1R cells, while LRAT (lecithin: retinol acyltransferase; ENSG00000121207; an enzyme involved in retinol storage within cells), ALDH1A3 (aldehyde dehydrogenase 1 family, member A3; ENSG00000184254; an enzyme closely related to the main retinaldehyde dehydrogenase ALDH1A2, one of the enzymes that increase intercellular RA levels) and RARγ (ENSG00000172819) transcript levels were 191 (p = 3.73 × 10−68), 7.4 (p = 8.00 × 10−32) and 3.5 (p = 5.87 × 10−17) fold higher in RA-treated OKF6-TERT1R than in SCC-9 cells. RARα (ENSG00000131759) transcript levels were 1.5 (p=0.003) fold higher in RA-treated OKF6-TERT1R than in SCC-9 cells, while RARβ (ENSG00000077092) was not differentially expressed between these two cell types. Thus, our data agree with earlier reports of abnormal RA signaling and metabolism in OSCC [7–9] and further validate the use of OKF6-TERT1R and SCC-9 as models to investigate differences in gene expression between non-tumorigenic and tumorigenic oral keratinocytes
Changes in mRNA levels in RA treated vs. control (vehicle treated) SCC-9 cells are less extensive than in OKF6-TERT1R cells
We discovered that 260 transcripts were higher and 165 transcripts were lower in RA-treated compared to control (vehicle treated) OKF6-TERT1R, while only 25 transcripts were higher and 31 transcripts were lower in RA-treated vs. control SCC-9 cells. The log-fold changes (log FC) between control and RA-treated cells, plotted against the log-counts per million (log CPM) for each transcript, are shown (Supplementary Fig. 1). Genes with a greater than 3 fold difference in transcript levels between control and RA-treated cells are highlighted in black. The top 50 genes with transcript levels increased by at least 3 fold by RA treatment of OKF6-TERT1R cells are shown (Supplementary Table 7). The list of all genes with transcript levels increased by at least 3 fold by RA in SCC-9 cells is shown (Supplementary Table 8).
The fold increases in transcript levels induced by RA in the two cell lines were also markedly different. The transcript most highly induced by RA treatment of OKF6-TERT1R cells was CFI (complement factor I), with levels 1093 (p=2.46 × 10−27) fold higher in RA-treated than in control (Supplementary Table 7). In contrast, CD22 (CD22 molecule) transcript was 13.8 (p=1.25 × 10−14) fold higher in RA-treated than in control SCC-9 cells (Supplementary Table 8). In both cell lines STRA6 (stimulated by retinoic acid gene 6) and LOXL4 (lysyl oxidase-like 4) transcripts were highly induced by RA (Supplementary Tables 7 and 8). STRA6 transcript levels were 288 (p=5.99 × 10−120) fold higher in RA-treated than in control OKF6-TERT1R cells (Supplementary Table 7), but only 5.6 (p = 1.40 × 10−18) fold higher in RA-treated than in control SCC-9 cells (Supplementary Table 8). LOXL4 transcript levels were 258 (p = 1.79 × 10−56) fold higher in RA-treated than in control OKF6-TERT1R cells, and only 7.7 (p=3.12 × 10−14) fold higher in RA-treated than in control SCC-9 cells (Supplementary Tables 7 and 8). These data suggest that the transcriptional response to RA is attenuated in SCC-9 as compared to OKF6-TERT1R cells.
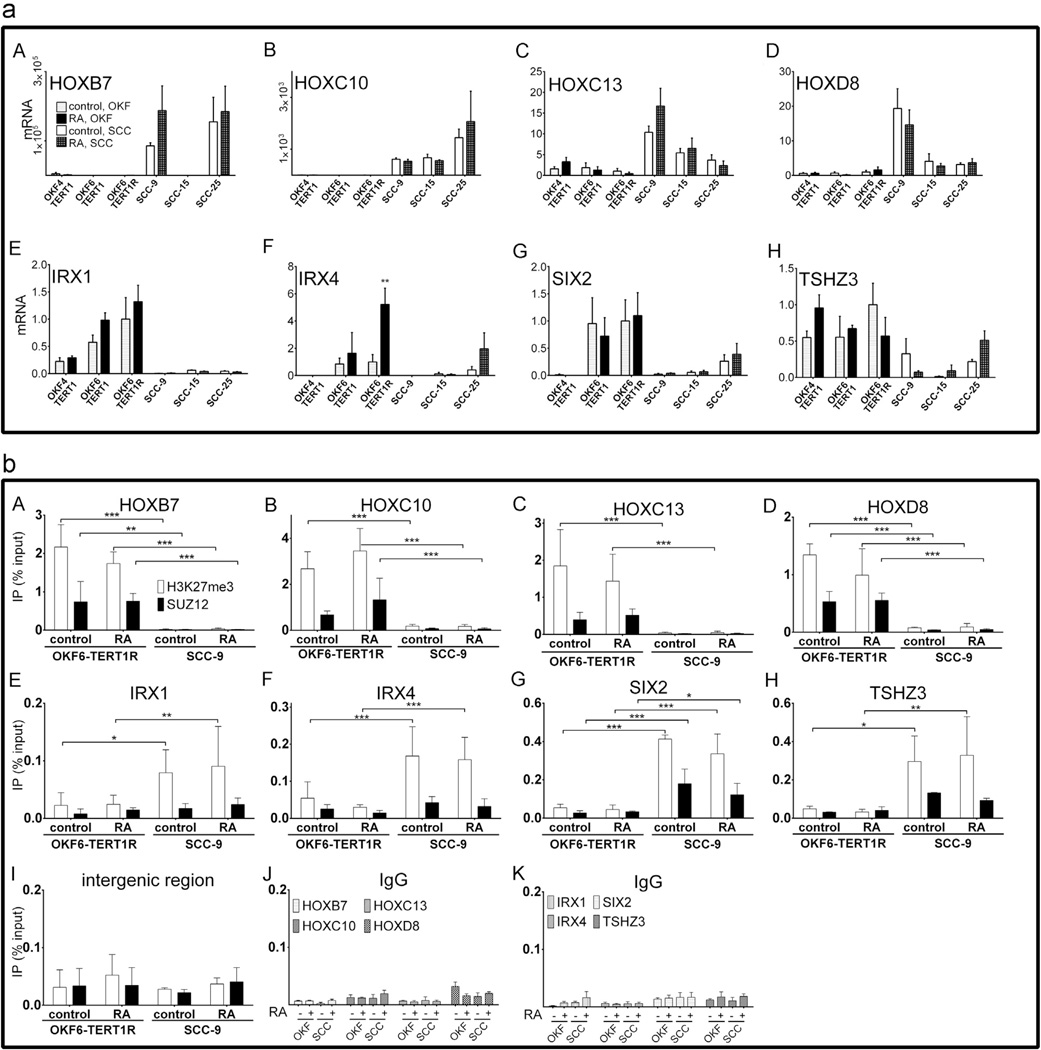
RA had effects on very few homeobox gene transcript levels in both cell lines. Zinc finger E-box binding homeobox 2 (ZEB2) transcript levels were 2.3 (p = 7.79 × 10−5) fold higher (RNAseq data not shown) in RA-treated than in control OKF6-TERT1R cells; HOP homeobox (HOPX) and distal-less homeobox (DLX3) transcript levels were 14 (p=2.64 × 10−20) and 5.5 (p = 6.27 × 10−8) fold lower (RNAseq data not shown) in RA-treated than in control OKF6-TERT1R cells. In SCC-9 cells RA treatment increased paired related homeobox 2 (PRRX2) and homeobox A5 (HOXA5) transcripts by 4.4 (p = 3.31 × 10−7) and 3.7 (p=4.59 × 10−6) fold, respectively (Supplementary Table 8). We also assessed the effects of RA by qRT-PCR on transcript levels of homeobox genes discussed earlier (Fig. 2(a)). We observed no statistically significant RA associated changes in the levels of HOXB7, HOXC10, HOXC13, HOXD4, IRX1, SIX2 or TSHZ3 (Fig. 4(a) A–E, G and H). IRX4 transcript levels, as assessed by qRT-PCR, were ~5 fold higher in RA-treated than in control OKF6-TERT1R cells (p<0.01, Fig. 4(a) F). We conclude that most homeobox genes differentially expressed between OKF6-TERT1R and SCC-9 cells are not regulated by RA signaling.
Fig. 4.
(a) mRNA levels of homeobox genes expressed differentially in vehicle and RA treated, non-tumorigenic and tumorigenic oral keratinocyte cell lines and (b) levels of SUZ12 and H3K27me3 epigenetic marks proximal to homeodomain genes expressed differentially between non-tumorigenic and tumorigenic oral keratinocyte cells in vehicle and RA treated OKF6-TERT1R and SCC-9 cells. n=3 independent biological repeats; * indicates p < 0.05, ** indicates p < 0.01 and *** indicates p < 0.001; note differences in scales of y-axes in different panels (a) mRNA levels are normalized to HPRT1 expression and represented relative to transcript levels in control, untreated OKF6-TERT1R cells (set as 1); differences in transcript levels between the vehicle and RA-treated samples for each cell line were analyzed by two-way ANOVA followed by Bonferroni post-test A–D mRNA levels of HOX genes expressed at higher levels in SCC-9 vs. OKF6-TERT1R cells after 48 h of treatment with 1 µM RA. The cells were treated for 48 h with 1 µM RA or 0.1% ethanol (vehicle control). E–H mRNA levels of HOX genes expressed at lower levels in SCC-9 vs. OKF6-TERT1R cell line after 48 h of treatment with 1 µM RA or 0.1% ethanol (vehicle control). (b) Data are represented as percent of chromatin used as input in corresponding IPs; differences in immunoprecipitated chromatin levels between the cell lines were analyzed by two-way ANOVA followed by Bonferroni post-test; A–D Levels of the H3K27me3 mark and SUZ12 at HOX genes expressed at higher levels in SCC-9 vs. OKF6-TERT1R cell line after 48 h of treatment with 1 µM RA, as assessed by ChIP; E–H Levels of the H3K27me3 mark and SUZ12 at HOX genes expressed at lower levels in SCC-9 vs. OKF6-TERT1R cell line after 48 h of treatment with 1 µM RA, as assessed by ChIP; I Association of H3K27me3 mark and SUZ12 with with an intergenic region after 48 h of treatment with 1 µM RA., as assessed by ChIP. J and K Chromatin immunoprecipitated using normal rabbit IgG along with antibodies specific to SUZ12 and H3K27me3; sequences amplified are indicated by the graph legend.
RA treatment does not alter SUZ12 association or H3K27 trimethylation at homeobox genes that show different transcript levels in SCC-9 and OKF6-TERT1R cells
We have shown that one of the effects elicited by RA at the proximal promoter of Hoxa1 (and other primary RA target genes) in murine embryonic stem and F9 stem cells is the dissociation of SUZ12 and the removal of H3K27me3 marks [32,44]. We performed chromatin immunoprecipitation on RA-treated OKF6-TERT1R and SCC-9 cells. Homeobox genes expressed at higher levels in SCC-9 than in the OKF6-TERT1R cells were associated with SUZ12 and enriched for the H3K27me3 mark in RA-treated OKF6-TERT1R, but not in RA-treated SCC-9 cells (Fig. 4(b) A–D). Homeobox genes expressed at lower levels in SCC-9 than in the OKF6-TERT1R cells were associated with SUZ12 and enriched for the H3K27me3 mark in RA-treated SCC-9, but not in RA-treated OKF6-TERT1R cells (Fig. 4(b) E–H). A summary of these results, including the fold changes in SUZ12 association and H3K27me3 levels between RA-treated OKF6-TERT1R and RA-treated SCC-9 cells at HOXB7, HOXC10, HOXC13, HOXD4, IRX1, IRX4, SIX2 and TSHZ3 is shown (Table 4). From these data we conclude that RA is not a major regulator of PRC2 association and activity at the assessed homeobox genes in OKF6-TERT1R and SCC-9 cells. We also assessed the effects of RA treatment on homeobox mRNA levels in SUZ12 depleted OKF6-TERT1R and SCC-9 cells. We observed no significant differences in the RA responses of SUZ12 depleted cells compared to parental populations (Supplementary Fig. 2). Thus, the interplay between RA signaling and SUZ12/ PRC2 functions is more limited in OKF6-TERT1R and SCC-9 cells, and these cells behave differently from ES cells [32,44].
Discussion
OKF6-TERT1R and SCC-9 cells can serve as a model to investigate aspects of neoplastic transformation of human oral keratinocytes
Whole genome transcriptome sequencing (RNAseq) is increasingly being used for gene expression profiling. This technique has also been employed to study frequently occurring mutations in HNSCC [45,46]. We performed RNA-seq to investigate differences in transcript levels between non-tumorigenic and tumorigenic human cultured oral keratinocytes, and utilized ONCOMINE to relate our data from a human cell culture model to changes observed in oral SCC and other HNSCC patient tissues [34]. Our data and data obtained from human tissue samples include many of the same genes [35–41] (Table 3). Among these genes, AIM2 (Table 3) has been proposed as a putative oncogene [47] and MAGEA12 and S100A7 (Table 3) as putative diagnostic markers for early detection of OSCC [48,49]. Thus, OKF6-TERT1R and SCC-9 cells can serve as a reasonable model to investigate differences (such as differences in gene expression) between non-tumorigenic and neoplastically transformed cells because the cultured cells reflect many of the gene expression differences seen in OSCC patients vs. normal controls.
Homeobox genes are often dysregulated in HNSCC
We focused on the homeobox gene family. ONCOMINE HNSCC datasets rank many homeobox genes high (Table 2). HOXA1 is overexpressed in human OSCC tumor samples as compared to healthy mucosa and a high immunohistological HOXA1 signal is associated with a poor prognosis [12]. HOXA10 transcripts were 287 fold higher in tumorigenic than in non-tumorigenic oral keratinocytes (Table 1); HOXA10 mRNA levels are also higher in primary HNSCC samples than in corresponding normal oral epithelium tissues and HOXA10 protein is significantly associated with higher TNM stages (TNM Classification of Malignant Tumors is a cancer staging system; acronym letters refer to Tumor size, involved regional lymph Nodes and presence of distant Metas-tases) [14]. HOXB7 transcripts are 373 fold higher in tumorigenic than in non-tumorigenic cells (Table 1), and HOXB7 expression is higher in HNSCC compared to normal oral mucosa (Table 2); higher HOXB7 levels are also correlated with higher TNM stage, shorter overall survival, and disease-free survival after treatment [13]. Furthermore, downregulation of HOXB7 in SCC-9 cells decreased the proliferation rate [13], suggesting that HOXB7 may contribute to the abnormal proliferation in oral carcinogenesis. HOXC6 overexpression in HNSCC tissue and cell lines is linked to the activation of anti-apoptotic pathways in tumors through regulation of Bcl-2 expression [50]. IRX1 showed the greatest transcript level difference (338 fold lower in SCC-9) between OKF6-TERT1R and SCC-9 cells (Table 1). IRX1 is also described as a frequently methylated tumor suppressor in HNSCC, with decreased expression in 80% of the tumors assessed [51]. Thus, our data support the idea that homeobox genes are often dysregulated in OSCC tumors. Homeobox genes regulate normal keratinocyte differentiation [52,53]. The roles that homeobox genes play in oral SCC development remain unclear. Based on the aforementioned results [13,50] and data from other malignancies we speculate that dysregulation of homeobox genes can affect cellular processes such as transcription, receptor signaling, proliferation, differentiation, epithelial to mesenchymal transition, and apoptosis (for a review, see: [54]).
Homeobox genes differentially expressed between OKF6-TERT1R and SCC-9 cells are not regulated by RA
Although bona fide RAREs have been identified within a few of Hox gene enhancers [5,16–18], changes in Hox transcript levels upon induction of stem cell differentiation, most often using RA, are well documented [11,15,55]. It is therefore worth noting that RA has only a very limited effect on HOX gene transcript levels (Fig. 4(a), Supplementary Table 8 and data not shown). Most HOX genes in differentiated cells are transcriptionally silenced by histone and/or promoter DNA methylation [15], and it is plausible that the homeobox genes expressed in OKF6-TERT1R and SCC-9 cells are transcriptionally activated as a result of abnormal (and hence RA-independent) mechanisms occurring during cell immortalization or transformation. Alternatively, compromised response mechanisms to RA in SCC-9 cells could underlie the limited changes in HOX gene transcript levels (Fig. 4(a), Supplementary Table 8 and data not shown). In accord with earlier reports [7,8,56], SCC-9 responses to RA treatment, as assessed by RNAseq, were limited to a small number of transcripts (Supplementary Table 8). PRRX2 and HOXA5 were the only two homeobox transcripts that changed upon RA treatment of SCC-9 cells in our RNAseq experiment (Supplementary Table 8). Transcripts of homeobox genes in control and RA-treated SCC-9 cells were evaluated by qRT-PCR by an earlier report; among 84 genes included in the assessed panel only two, HOXD12 and MEOX1, exhibited transcript changes of greater than 3-fold upon RA treatment [56].
SUZ12 binding and H3K27me3 marks correlate inversely with the transcript levels of HOX genes in human oral keratinocytes
In some instances, Polycomb complexes mediate silencing of HOX gene expression (40, 41). A number of publications report Poly-comb protein expression in oral SCC and other types of HNSCC, and several showed that EZH2 overexpression correlated with tumor proliferation and poor prognosis [21–23].
We hypothesized that the changes in homeodomain transcript levels in SCC-9 vs. OKF6-TERT1R cells are a consequence of changes in the epigenetic regulation of their expression and we evaluated the association of SUZ12 [19] and the presence of the H3K27me3 mark at homeodomain genes expressed differentially between the OKF6-TERT1R and SCC-9 cells. In F9 stem cells, RA reduced Suz12 and the associated H3K27me3 mark at the RAREs of Hoxa1, RARβ2, and Cyp26a1 [32]. Thus, we evaluated the effects of RA on SUZ12 binding and H3K27me3 levels in our SCC model. SUZ12 binding and H3K27me3 marks correlated with the tran-scriptional status of HOX genes in OKF6-TERT1R and SCC-9 cells, but were not affected by RA treatment (Fig. 2 and 4). Thus, we conclude that transcription of the assessed HOX genes, SUZ12 dissociation, and H3K27me3 mark removal from these genes are not regulated by RA in this model. Dissecting the mechanisms involved in regulating HOX gene expression will extend our understanding of the processes driving neoplastic transformation of these cells.
Importantly, whether the removal of Suz12/PRC2 and the H3K27me3 mark from transcriptionally active genes is a cause or a consequence of transcription activation remains unclear. A recent report suggests that PRC2 acts as a sensor for dense, transcriptionally inactive chromatin, and that the H3K27me3 repressive epigenetic mark is deposited to maintain the silenced state rather than then to actively inhibit genes transcription [42]. The presence of the H3K27me3 mark in HNSCC has been recently assessed by Gannon et al. [23]. The authors showed elevated EZH2 protein levels in half of HNSCC samples in a tissue microarray and in 70% of assessed HNSCC cell lines (as compared to control normal oral epithelium or normal keratinocytes, respectively). No changes in global levels of H3K27me3 were observed; however, a differentiation related gene, involucrin, exhibited elevated levels of the H3K27me3 mark bound to promoter in SCC lesions relative to normal tissue samples [23]. Gannon et al. [23] additionally established that the EZH2 inhibitor DZNep had cancer cell selective toxicity and reduced initial HNSCC tumor growth in a xenograft model, further suggesting that modulation of Polycomb activity in HNSCC has a therapeutic potential. Here we show that another differentiation related gene family, namely the homeobox gene family, exhibits different levels of SUZ12 and the H3K27me3 mark bound in tumorigenic SCC cells as compared to non-tumorigenic immortalized oral keratinocytes. In our experiments, shRNA mediated depletion of SUZ12 led to increased transcript levels of HOX genes in the non-tumorigenic OKF6-TERT1R cells. We conclude that Polycomb positioning is altered in OSCC as compared to normal epithelial cells, leading to altered gene expression patterns in the malignant cells (Fig. 2). We speculate that PRC2 functions become non-essential in cells as they acquire tumorigenic potential, as illustrated by the fact that SUZ12 depletion was not sufficient to increase transcript levels of assessed homeobox genes in SCC-9 cells (Fig. 3(b) E–H). Interestingly, shRNA mediated EZH2 depletion did not induce apoptosis in HNSCC cells, as assessed by Gannon et al. [23]. This result is similar to ours in that SUZ12 depletion in SCC-9 cells decreased the proliferation rate of the SUZ12 kd population to a lesser extent as compared to OKF6-TERT1R cells (Fig. 3(c)). One exciting further direction would be elucidation of the molecular mechanism driving aberrant homeobox gene expression in transformed oral keratinocytes by (1) assessing additional epigenetic histone and DNA modifications deposited at these genes or (2) investigating the composition of the PRC2 complexes in OKF6-TERT1R and SCC-9 cells by mass spectrometry to explain differential chromatin association within these cells. Such experiments are currently underway.
Supplementary Material
Acknowledgments
This research was supported by NIH grant RO1DE010389 to LJG and by Weill Cornell funds. KM is supported by a Kirschstein-NRSA predoctoral fellowship F31-DE021316, and was supported for a portion of this project on training grant 5T32GM073546-04. We thank Dr. Fabienne Campagne for guidance and help with RNAseq experiment design and analysis. We thank Dr. Alison Urvalek and Dr. Kristian Laursen for critically reading the manuscript and for their insightful suggestions.
Abbreviations
- ChIP
chromatin immunoprecipitation
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GO
gene ontology
- H3K27me3
histone 3 lysine 27 trimethyl
- HNSCC
head and neck squamous cell carcinoma
- HOX
homeobox
- HPRT1
hypoxanthine phosphoribosyltransferase 1
- OSCC
oral squamous cell carcinoma
- PRC
polycomb repressive complexes
- qRT-PCR
quantitative real time polymerase chain reaction
- RA
retinoic acid
- RAR
retinoic acid receptor
- RARE
retinoic acid response element
- RNAseq
RNA sequencing
- RXR
retinoid X receptor
- shRNA
short hairpin RNA
- SCC
squamous cell carcinoma
- TNM
TNM classification of malignant tumours
- WCMC
Weill Cornell Medical College
Footnotes
Conflict of interest
The authors disclose no potential conflicts of interest.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.yexcr.2013.09.011.
REFERENCES
- 1.Curado MP, Hashibe M. Recent changes in the epidemiology of head and neck cancer. Curr. Opin. Oncol. 2009;21:194–200. doi: 10.1097/CCO.0b013e32832a68ca. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA: Cancer J. Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 3.Means AL, Gudas LJ. The roles of retinoids in vertebrate development. Annu. Rev. Biochem. 1995;64:201–233. doi: 10.1146/annurev.bi.64.070195.001221. [DOI] [PubMed] [Google Scholar]
- 4.Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- 5.Langston AW, Gudas LJ. Identification of a retinoic acid responsive enhancer 3′ of the murine homeobox gene Hox-1.6. Mech. Dev. 1992;38:217–227. doi: 10.1016/0925-4773(92)90055-o. [DOI] [PubMed] [Google Scholar]
- 6.Reichrath J, Lehmann B, Carlberg C, Varani J, Zouboulis CC. Vitamins as hormones. Horm. Metab. Res. 2007;39:71–84. doi: 10.1055/s-2007-958715. [DOI] [PubMed] [Google Scholar]
- 7.Guo X, Gudas LJ. Metabolism of all-trans-retinol in normal human cell strains and squamous cell carcinoma (SCC) lines from the oral cavity and skin: reduced esterification of retinol in SCC lines. Cancer Res. 1998;58:166–176. [PubMed] [Google Scholar]
- 8.Hu L, Crowe DL, Rheinwald JG, Chambon P, Gudas LJ. Abnormal expression of retinoic acid receptors and keratin 19 by human oral and epidermal squamous cell carcinoma cell lines. Cancer Res. 1991;51:3972–3981. [PubMed] [Google Scholar]
- 9.Youssef EM, Lotan D, Issa JP, Wakasa K, Fan YH, Mao L, Hassan K, Feng L, Lee JJ, Lippman SM, et al. Hypermethylation of the retinoic acid receptor-beta(2) gene in head and neck carcinogenesis. Clin. Cancer Res. 2004;10:1733–1742. doi: 10.1158/1078-0432.ccr-0989-3. [DOI] [PubMed] [Google Scholar]
- 10.Feng L, Wang Z. Clinical trials in chemoprevention of head and neck cancers. Rev. Recent Clin. Trials. 2012;7:249–254. doi: 10.2174/157488712802281349. [DOI] [PubMed] [Google Scholar]
- 11.Pick L, Heffer A. Hox gene evolution: multiple mechanisms contributing to evolutionary novelties. Ann. N. Y. Acad. Sci. 2012;1256:15–32. doi: 10.1111/j.1749-6632.2011.06385.x. [DOI] [PubMed] [Google Scholar]
- 12.Bitu CC, Destro MF, Carrera M, da Silva SD, Graner E, Kowalski LP, Soares FA, Coletta RD. HOXA1 is overexpressed in oral squamous cell carcinomas and its expression is correlated with poor prognosis. BMC Cancer. 2012;12:146. doi: 10.1186/1471-2407-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Destro MF, De Souza Setubal, Bitu CC, Zecchin KG, Graner E, Lopes MA, Kowalski LP, Coletta RD. Overexpression of HOXB7 homeobox gene in oral cancer induces cellular proliferation and is associated with poor prognosis. Int. J. Oncol. 2010;36:141–149. [PubMed] [Google Scholar]
- 14.Yamatoji M, Kasamatsu A, Yamano Y, Sakuma K, Ogoshi K, Iyoda M, Shinozuka K, Ogawara K, Takiguchi Y, Shiiba M, et al. State of homeobox A10 expression as a putative prognostic marker for oral squamous cell carcinoma. Oncol. Rep. 2010;23:61–67. [PubMed] [Google Scholar]
- 15.Soshnikova N, Duboule D. Epigenetic regulation of vertebrate Hox genes: a dynamic equilibrium. Epigenetics. 2009;4:537–540. doi: 10.4161/epi.4.8.10132. [DOI] [PubMed] [Google Scholar]
- 16.Doerksen LF, Bhattacharya A, Kannan P, Pratt D, Tainsky MA. Functional interaction between a RARE and an AP-2 binding site in the regulation of the human HOX A4 gene promoter. Nucleic Acids Res. 1996;24:2849–2856. doi: 10.1093/nar/24.14.2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langston AW, Thompson JR, Gudas LJ. Retinoic acid-responsive enhancers located 3′ of the Hox A and Hox B homeobox gene clusters. Functional analysis. J. Biol. Chem. 1997;272:2167–2175. doi: 10.1074/jbc.272.4.2167. [DOI] [PubMed] [Google Scholar]
- 18.Pöpperl H, Featherstone MS. Identification of a retinoic acid response element upstream of the murine Hox-4.2 gene. Mol. Cell Biol. 1993;13:257–265. doi: 10.1128/mcb.13.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Meara MM, Simon JA. Inner workings and regulatory inputs that control polycomb repressive complex 2. Chromosoma. 2012;121:221–234. doi: 10.1007/s00412-012-0361-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, Wu X, Stack EC, Loda M, Liu T, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is polycomb-independent. Science. 2012;338:1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kidani K, Osaki M, Tamura T, Yamaga K, Shomori K, Ryoke K, Ito H. High expression of EZH2 is associated with tumor proliferation and prognosis in human oral squamous cell carcinomas. Oral Oncol. 2009;45:39–46. doi: 10.1016/j.oraloncology.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 22.Cao W, Feng Z, Cui Z, Zhang C, Sun Z, Mao L, Chen W. Up-regulation of enhancer of zeste homolog 2 is associated positively with cyclin D1 overexpression and poor clinical outcome in head and neck squamous cell carcinoma. Cancer. 2012;118:2858–2871. doi: 10.1002/cncr.26575. [DOI] [PubMed] [Google Scholar]
- 23.Gannon OM, Merida de Long L, Endo-Munoz L, Hazar-Rethinam M, Saunders NA. Dysregulation of the repressive H3K27 trimethylation mark in head and neck squamous cell carcinoma contributes to dysregulated squamous differentiation. Clin. Cancer Res. 2013;19:428–441. doi: 10.1158/1078-0432.CCR-12-2505. [DOI] [PubMed] [Google Scholar]
- 24.Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg RA, Louis DN, Li FP, G J. Rheinwald, Human kerati-nocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol. Cell Biol. 2000;20:1436–1447. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benoit YD, Lepage MB, Khalfaoui T, Tremblay E, Basora N, Carrier JC, Gudas LJ, Beaulieu JF. Polycomb repressive complex 2 impedes intestinal cell terminal differentiation. J. Cell Sci. 2012;125:3454–3463. doi: 10.1242/jcs.102061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorff KC, Chambwe N, Zeno Z, Simi M, Shaknovich R, Campagne F. GobyWeb: simplified management and analysis of gene expression and DNA methylation sequencing data. PLoS One. 2013;8:e69666. doi: 10.1371/journal.pone.0069666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ozer HG, Parvin JD, Huang K. DFI: gene feature discovery in RNA-seq experiments from multiple sources. BMC Genomics 13. 2012;(Suppl. 8):S11. doi: 10.1186/1471-2164-13-S8-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metpally RP, Nasser S, Malenica I, Courtright A, Carlson E, Ghaffari L, Villa S, Tembe W, Van Keuren-Jensen K. Comparison of analysis tools for miRNA high throughput sequencing using nerve crush as a model. Front. Genet. 2013;4:20. doi: 10.3389/fgene.2013.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang dW, Sherman BT, Lempicki RA. Systematic and inte-grative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Raman JD, Mongan NP, Liu L, Tickoo SK, Nanus DM, Scherr DS, Gudas LJ. Decreased expression of the human stem cell marker, Rex-1 (zfp-42), in renal cell carcinoma. Carcinogenesis. 2006;27:499–507. doi: 10.1093/carcin/bgi299. [DOI] [PubMed] [Google Scholar]
- 32.Gillespie RF, Gudas LJ. Retinoid regulated association of transcriptional co-regulators the polycomb group protein SUZ12 with the retinoic acid response elements of Hoxa1, RARbeta(2), and Cyp26A1 in F9 embryonal carcinoma cells. J. Mol. Biol. 2007;372:298–316. doi: 10.1016/j.jmb.2007.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rheinwald JG, Beckett MA. Tumorigenic keratinocyte lines requiring anchorage and fibroblast support cultures from human squamous cell carcinomas. Cancer Res. 1981;41:1657–1663. [PubMed] [Google Scholar]
- 34.ONCOMINE. City: Editor (ed)\widehat(eds) [Google Scholar]
- 35.Cromer A, Carles A, Millon R, Ganguli G, Chalmel F, Lemaire F, Young J, Dembélé D, Thibault C, Muller D, et al. Identification of genes associated with tumorigenesis and metastatic potential of hypopharyngeal cancer by microarray analysis. Oncogene. 2004;23:2484–2498. doi: 10.1038/sj.onc.1207345. [DOI] [PubMed] [Google Scholar]
- 36.Estilo CL, O-charoenrat P, Talbot S, Socci ND, Carlson DL, Ghossein R, Williams T, Yonekawa Y, Ramanathan Y, Boyle JO, et al. Oral tongue cancer gene expression profiling: identification of novel potential prognosticators by oligonucleotide microarray analysis. BMC Cancer. 2009;9:11. doi: 10.1186/1471-2407-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ginos MA, Page GP, Michalowicz BS, Patel KJ, Volker SE, Pambuccian SE, Ondrey FG, Adams GL, Gaffney PM. Identification of a gene expression signature associated with recurrent disease in squamous cell carcinoma of the head and neck. Cancer Res. 2004;64:55–63. doi: 10.1158/0008-5472.can-03-2144. [DOI] [PubMed] [Google Scholar]
- 38.Talbot SG, Estilo C, Maghami E, Sarkaria IS, Pham DK, O-charoenrat P, Socci ND, Ngai I, Carlson D, Ghossein R, et al. Gene expression profiling allows distinction between primary and metastatic squamous cell carcinomas in the lung. Cancer Res. 2005;65:3063–3071. doi: 10.1158/0008-5472.CAN-04-1985. [DOI] [PubMed] [Google Scholar]
- 39.Toruner GA, Ulger C, Alkan M, Galante AT, Rinaggio J, Wilk R, Tian B, Soteropoulos P, Hameed MR, Schwalb MN, et al. Association between gene expression profile and tumor invasion in oral squamous cell carcinoma. Cancer Genet. Cytogenet. 2004;154:27–35. doi: 10.1016/j.cancergencyto.2004.01.026. [DOI] [PubMed] [Google Scholar]
- 40.Ye H, Yu T, Temam S, Ziober BL, Wang J, Schwartz JL, Mao L, Wong DT, Zhou X. Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics. 2008;9:69. doi: 10.1186/1471-2164-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pyeon D, Newton MA, Lambert PF, den Boon JA, Sengupta S, Marsit CJ, Woodworth CD, Connor JP, Haugen TH, Smith EM, et al. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007;67:4605–4619. doi: 10.1158/0008-5472.CAN-06-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan W, Wu T, Fu H, Dai C, Wu H, Liu N, Li X, Xu M, Zhang Z, Niu T, et al. Dense chromatin activates polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science. 2012;337:971–975. doi: 10.1126/science.1225237. [DOI] [PubMed] [Google Scholar]
- 43.Benoit YD, Laursen KB, Witherspoon MS, Lipkin SM, Gudas LJ. Inhibition of PRC2 histone methyltransferase activity increases TRAIL-mediated apoptosis sensitivity in human colon cancer cells. J. Cell Physiol. 2012 doi: 10.1002/jcp.24224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kashyap V, Gudas LJ. Epigenetic regulatory mechanisms distinguish retinoic acid-mediated transcriptional responses in stem cells and fibroblasts. J. Biol. Chem. 2010;285:14534–14548. doi: 10.1074/jbc.M110.115345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kondo Y, Nagai K, Nakahata S, Saito Y, Ichikawa T, Suekane A, Taki T, Iwakawa R, Enari M, Taniwaki M, et al. Overexpression of the DNA sensor proteins absent in melanoma 2 interferon-inducible 16, contributes to tumorigenesis of oral squamous cell carcinoma with p53 inactivation. Cancer Sci. 2012;103:782–790. doi: 10.1111/j.1349-7006.2012.02211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mollaoglu N, Vairaktaris E, Nkenke E, Neukam FW, Ries J. Expression of MAGE-A12 in oral squamous cell carcinoma. Dis. Markers. 2008;24:27–32. doi: 10.1155/2008/359840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kesting MR, Sudhoff H, Hasler RJ, Nieberler M, Pautke C, Wolff KD, Wagenpfeil S, Al-Benna S, Jacobsen F, Steinstraesser L. Psoriasin (S100A7) up-regulation in oral squamous cell carcinoma and its relation to clinicopathologic features. Oral Oncol. 2009;45:731–736. doi: 10.1016/j.oraloncology.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 50.Moon SM, Kim SA, Yoon JH, Ahn SG. HOXC6 is deregulated in human head and neck squamous cell carcinoma and modulates Bcl-2 expression. J. Biol. Chem. 2012;287:35678–35688. doi: 10.1074/jbc.M112.361675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bennett KL, Karpenko M, Lin MT, Claus R, Arab K, Dyckhoff G, Plinkert P, Herpel E, Smiraglia D, Plass C. Frequently methylated tumor suppressor genes in head and neck squamous cell carcinoma. Cancer Res. 2008;68:4494–4499. doi: 10.1158/0008-5472.CAN-07-6509. [DOI] [PubMed] [Google Scholar]
- 52.Liang Y, Xia L, Du Z, Sheng L, Chen H, Chen G, Li Q. HOXA5 inhibits keratinocytes growth and epidermal formation in organotypic cultures in vitro and in vivo. J. Dermatol. Sci. 2012;66:197–206. doi: 10.1016/j.jdermsci.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 53.Yang JM, Sim SM, Kim HY, Park GT. Expression of the homeobox gene HOPX is modulated by cell differentiation in human keratinocytes and is involved in the expression of differentiation markers. Eur. J. Cell Biol. 2010;89:537–546. doi: 10.1016/j.ejcb.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 54.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat. Rev. Cancer. 2010;10:361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 55.Schughart K, Kappen C, Ruddle FH. Mammalian homeobox-containing genes: genome organization, structure, expression and evolution. Br. J. Cancer Suppl. 1988;9:9–13. [PMC free article] [PubMed] [Google Scholar]
- 56.Acquafreda T, Nunes FD, Soprano DR, Soprano KJ. Expression of homeobox genes in oral squamous cell carcinoma cell lines treated with all-trans retinoic acid. J. Cell Biochem. 2010;111:1437–1444. doi: 10.1002/jcb.22871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.