Abstract
Cornelia de Lange syndrome (CdLS) is a clinically and genetically heterogeneous developmental disorder. Clinical features include growth retardation, intellectual disability, limb defects, typical facial dysmorphism, and other systemic involvement. The increased understanding of the genetic basis of CdLS has led to diagnostic improvement and expansion of the phenotype. Mutations in five genes (NIPBL, SMC1A, SMC3, RAD21, and HDAC8), all regulators or structural components of cohesin, have been identified. Approximately 60% of CdLS cases are due to NIPBL mutations, 5% caused by mutations in SMC1A, RAD21, and HDAC8 and one proband was found to carry a mutation in SMC3. To date, 311 CdLS-causing mutations are known including missense, nonsense, small deletions and insertions, splice site mutations, and genomic rearrangements. Phenotypic variability is seen both intra- and intergenically. This article reviews the spectrum of CdLS mutations with a particular emphasis on their correlation to the clinical phenotype.
Keywords: Cornelia de Lange syndrome, NIPBL, SMC1A, SMC3, RAD21, HDAC8
Introduction
Cornelia de Lange syndrome (CdLS; MIM #122470, 300590, 610759, 300882, 614701) is a genetically heterogeneous congenital multisystemic disorder with an incidence of between 1:10,000 and 1:30,000 live births. Common characteristics of CdLS include pre- and postnatal growth retardation, microcephaly, developmental delay, cognitive impairment with behavior and neurological problems, facial dysmorphia, hirsutism, and upper extremity defects ranging from small hands to severe reduction defects of the forearms. Additional systemic involvement includes cardiac, gastrointestinal, and musculoskeletal as well as hearing loss and genitourinary anomalies. The typical facial features include fine arched eyebrows, synophrys, long eyelashes, low-set posteriorly rotated ears, long philtrum, thin upper lip, depressed nasal bridge with anteverted nares (Fig. 1) [see specific review for a detailed description Kline et al., 2007; Liu and Krantz, 2008]. Since the first case reported by the Dutch anatomists Gerardus and Willem Vrolik in 1849 and subsequently by the German physician Brachmann in 1916, and two unrelated cases reported by the Dutch pediatrician, Cornelia de Lange, in 1933, who formally described the constellation of clinical findings as a recognizable entity and after whom the disorder has been named [Brachmann, 1916; de Lange, 1933; Oostra et al., 1994; Vrolick, 1849] great progress has been made in our ability to recognize and diagnose what we understand today to be the full clinical spectrum of CdLS. It is now known that there is broad variability in phenotypic expression of CdLS. In addition to the classical more severe clinical phenotype, milder probands have been consistently reported. While diagnostic criteria have been proposed by Kline et al. (2007), our understanding of what “CdLS” is clinically is evolving and being driven in part by the development of diagnostic molecular markers. CdLS is a genetically heterogeneous diagnosis with mutation in NIPBL (MIM #608667), localized to chromosome 5p13, being the first, and most common, genetic cause of CdLS [Krantz et al., 2004; Tonkin et al., 2004]. Heterozygous mutations in NIPBL are found in at least 60% of CdLS probands. In addition, 5% of individuals with CdLS have missense mutations and small in frame deletions in the X-linked SMC1A gene (MIM #300040) and one proband has been identified with an in frame 3 bp deletion in the SMC3 gene (MIM #606062), located on chromosome 10q25 [Deardorff et al., 2007; Musio et al., 2006]. SMC mutations, which maintain the frame of their encoded proteins and do not alter protein expression, are generally associated with milder CdLS phenotypes with moderate neurocognitive impairment and a paucity of major structural defects. Recently, mutations have been found in the HDAC8 gene (MIM #300269) that is also X-linked and encodes a histone, and co-hesin, deacetylase. Mutations in HDAC8 are found in approximately 5% of individuals with CdLS [Deardorff et al., 2012a]. The identified mutations, four missense and one nonsense leading to protein instability, are present in probands with typical clinical features of CdLS but without significant limb involvement and with some additional clinical characteristics that may help in differentiating this group from individuals with CdLS caused by mutations in other genes [Deardorff et al., 2012a]. Finally, mutations in RAD21 (MIM #606462) cause a human cohesinopathy characterized by growth retardation, minor skeletal anomalies, and facial features that overlap findings of individuals with CdLS but can have milder cognitive involvement [Deardorff et al., 2012b].
Figure 1.
Phenotypic characteristics of CdLS caused by mutations in different cohesin regulatory and structural components. A–D: 28-year-old girl with truncating mutation in NIPBL. E–H: 7-year-old boy with missense mutations in NIPBL; I–L: 3-year-old girl with in frame insertion/deletion mutation of HDAC8. M and N: 15-year-old girl with missense mutation in SMC1A. O and P: 3-year-old boy with deletion of RAD21. Q–U: 57-year-old man with in frame deletion of SMC3 (shown as a teenager in “Q”).
The purpose of this review is to review the spectrum of mutations that occur in the five genes that underlie CdLS with emphasis on genotype–phenotype correlations.
NIPBL Overview
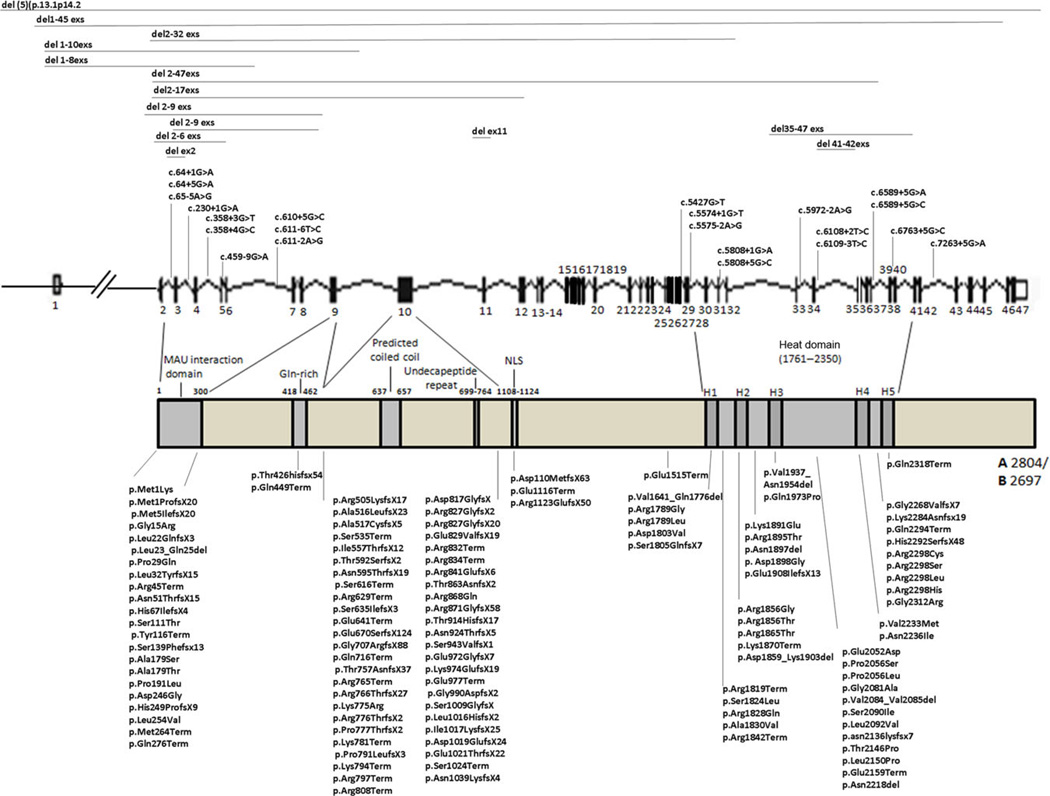
NIPBL consists of 47 exons that encodes two isoforms of delangin, A and B with 2,804 and 2,697 amino acids, respectively [Krantz et al., 2004; Tonkin et al., 2004]. NIPBL contains specific domains including a N-terminal MAU interaction domain, a glutamine-rich domain, a predicted nuclear-localization signal (NLS), an undecapeptide repeat, and a conserved domain with five HEAT repeats that are important for its interaction with other proteins [Neuwald and Hirano, 2000; Yan et al., 2006] (Fig. 2).
Figure 2.
Overview of NIPBL mutations. The 47 exons of NIPBL are indicated with black bars. The different domains of NIPBL are indicated: MAU interaction domain (1–300), glutamine rich domain (Gln-rich, 418–462, predicted coiled coil (637–657), undecapeptide repeat PETPKQK(G/S)(E/D)(G/S)R (699–764), nuclear localization signal (NLS, 1,108–1,124), HEAT domain (1,767–2,350) consisting of five repeats (H1: 1,767–1,805, H2: 1,843–1,881, H3: 1,945–1,984, H4: 2,227–2,267, H5: 2,313–2,351).
NIPBL and its orthologues, Nipped-B in Drosophila, Scc2 in Sac-charomyces cerevisae and Xscc2 in Xenopus, act as regulators of the cohesin complex, composed of SMC1A, SMC3, RAD21, and STAG protein subunits and mediates the sister chromatid cohesion during mitosis. NIPBL is required for loading of cohesin on chromatin, a function that is conserved across evolution, as demonstrated from experimental evidence obtained from model organisms [Ciosk et al., 2000; Gillespie and Hirano, 2004; Rollins et al., 2004; Takahashi et al., 2004]. NIPBL depletion in both Xenopus and Hela cells reduce cohesin association with chromatin and cause defects in sister chromatid pairing, arguing that its function is essential to sister chromatid cohesion [Gillespie and Hirano, 2004; Watrin et al., 2006]. However, analysis performed in lymphoblastoid cell lines derived from CdLS patients with NIPBL mutations revealed that the frequency of sister chromatid defects were not significantly different from healthy controls [Castronovo et al., 2009; Revenkova et al., 2009].
NIPBL is able to interact with the cohesin complex in different ways. It cooperates with MAU2, the human orthologue of S. cerevisae Scc4, through its N-terminal region to form a heterodimeric complex essential to cohesin binding to chromatin [Bermudez et al., 2012; Braunholz et al., 2012; Seitan et al., 2006; Watrin et al., 2006]. In addition, it has been demonstrated that NIPBL interacts directly with cohesin, specifically with the SMC1A-SMC3heterodimer [Bermudez et al., 2012].
NIPBL Mutations in CdLS
Information on NIPBL, SMC1A, SMC3, RAD21, and HDAC8 mutations has been derived from public databases (dbSNP; www.ncbi.nlm.nih.gov and LOVD; www.LOVD.nl) and published reports. To date, 278 NIPBL heterozygous mutations have been identified in CdLS probands: 216 falling in coding sequences, 45 in noncoding regions, and 17 involving gross genomic alterations (Table 1). GenBank NM_133433.3 was used as NIPBL sequence reference. Among coding sequence mutations, 67 are missense, 43 nonsense, 71 small deletions, 33 small insertions, and two small indels. It is worthy to note that exon 10, encoding for the coiled-coil region and for the undecapeptide repeat, has the greatest cluster of mutations with 49 including two missense, 11 nonsense, 24 deletions, and 12 insertions (Fig. 2). Most of the mutations are unique, but recurrent mutations have also been identified in unrelated probands: six missense (c.535G>T in two unrelated probands, c.4013A>G in three unrelated probands, c.6697G>Aintwo unrelated probands, c.6892C>T in two unrelated probands, c.6893G>A, in two unrelated probands, c.7168G>A in two unrelated probands), four nonsense (c.2389C>T in four unrelated probands, c.2494C>T, in two unrelated probands, c.4606C>T in three unrelated probands, c.5167C>T in one unrelated proband and in two siblings), six deletions (c.1445_1448del GAGA in two unrelated probands, c.2479delAG in seven unrelated probands, c.3049delATTA in two unrelated probands, c.3060_3063delAGAG in three unrelated probands, c.6653_6655delATA in two unrelated probands, c.7438_7439delAG, in two unrelated probands) (Supp. Table S1).
Table 1.
Type and Number of NIPBL Mutations Identified in CdLS Probands
| Mutation type | Number of mutations |
|---|---|
| Coding sequence mutations | |
| Missense | 67 |
| Non sense | 43 |
| Deletions | 71 |
| Insertions | 33 |
| Indels | 2 |
| Noncoding sequence mutations | |
| Splicing | 43 |
| Regulatory | 2 |
| Genomic rearrangements | |
| Gross deletions | 16 |
| Translocation | 1 |
Mutations in noncoding regions of NIPBL include: 43 splicing mutations involving both donor and acceptor splice sites. Four recurrent splice site changes have been reported (c.65–5A>G, in three unrelated probands, c.611–6T>C in two unrelated patients, c.358+3G>T in two unrelated patients, c.4321G>T in four unrelated patients, c.6109–3T>C in three unrelated probands), and two different splice changes produced a variant in frame deletion (p.V1414_A1440del) with skipping of exon 19. Alterations in the 5′ untranslated region of the gene have been detected. In particular, the nucleotide change (c.−94C>T) close to the transcription start site that presumably results in an alternative transcript [Selicorni et al., 2007] and the indel, c.−321–320delCCinsA resulting in a reduction of mRNA level [Borck et al., 2006;].
Larger scale genomic alterations have also been reported, including 16 deletions ranging from single to multiple exons, including portions of the nontranslated regulatory regions, or the entire gene as occurs for the del(5)(p13.1p14.2). The de novo balanced translocation t(5;13)(p13.1;q12.1) was the rearrangement which facilitated the identification of NIPBL as a CdLS disease gene [Krantz et al., 2004; Tonkin et al., 2004].
Most of mutations are nonsense, splice site, or frame shifts that result in a predicted truncated protein that presumably leads to the partial reduction in NIPBL production resulting in haploinsufficiency. This notion is supported by the identification of gross deletions leading to the loss of the entire NIPBL region [Hulinsky et al., 2005; Russo et al., 2012].
Though most NIPBL mutations are de novo, 13 familial cases are reported as demonstrated by the observation of two cases where the identified mutations were also seen in mildly affected parents [Borck et al., 2006; Gillis et al., 2004]. Furthermore, in several cases where no mutation was identified in the blood of both parents with multiple affected children carrying the same NIPBL mutation, germline mosaicism was proposed as the pathogenic mechanism [Slavin et al., 2012]. This hypothesis is further supported by the observation that the heterozygous pathogenic NIPBL missense mutation c.7298A>G was found in the sperm of a father with multiple affected offspring and not in his peripheral blood [Niu et al., 2006]. In addition, recently it has been shown that 23% of CdLS probands without detectable mutation in lymphocytes a NIPBL mutation could be detected in buccal cells [Huisman et al., 2013].
SMC1A–SMC3 Mutations in CdLS
To date, mutations in the SMC1A and SMC3 genes that encode core cohesin complex structural components have been identified in about 5% of CdLS probands though only a single proband harbors an SMC3 mutation. The twenty-four SMC1A mutations, all falling within the coding region of the gene, consist of nineteen missense and five small-in frame deletions. GenBank NM_006306.2 was used as SMC1A sequence reference. Common missense mutations among unrelated patients have been found, namely: c.587G>A in the three unrelated probands, c.1193G>A, in three unrelated probands, c.1487G>A, in two unrelated probands and in two different familial cases (Supp. Table S2). The observation that frameshift and nonsense mutations have not been reported suggests that they are likely not tolerated or lead to a different phenotype.
SMC1A mutations do not affect its expression as probands show equals levels of mRNA and proteins when compared with controls [Liu and Krantz, 2009; Revenkova et al., 2009]. In addition, SMC mutations do not prevent the incorporation of mutated proteins into the cohesin complex [Gimigliano et al., 2012], though SMC-mutated hinge dimers appear to have increased affinity for chromatin than the normal proteins [Revenkova et al., 2009]. These findings argue for a dominant negative effect as the pathogenetic mechanism for SMC mutations. SMC1A maps to the X chromosome, in a region which escapes X inactivation [Brown et al., 1995], both hemizygous male and heterozygous female individuals have been identified [Mannini et al., 2010] suggesting that mutated proteins are functional and dominantly interfere with biological processes. The presence of a single-mutated protein would be able to affect ATPase activity or cohesin loading/unloading. This notion is supported by several observations. In fact, data obtained in Bacillus subtilis showed that a mutation in the Walker B domain allowed ATP binding but blocked head–head engagement and ATP hydrolysis, whereas a mutation in the Walker A domain abolished ATP binding [Hirano et al., 2001].
The only mutation identified in SMC3 is a small in-frame deletion of 3 bp in a male proband (Supp. Table S2). GenBank NM_005445.3 was used as SMC3 sequence reference. The low frequency of mutation identification in SMC3 in CdLS suggests that this gene may play important roles beyond cohesion. This is supported by the finding that SMC3 acetylation controls fork processivity in human cells and slow moving forks were found in cells lacking acetyltransferase activity [Terret et al., 2009; Zhang et al., 2008]. In addition, SMC3 deacetylation is involved in cohesin recycling during cell cycle as increased SMC3 acetylation leads to inefficient dissolution of cohesin from chromatin in both prophase and anaphase [Deardorff et al., 2012a]. As SMC3 is a central determinant of these processes, it is likely that its mutations are negatively selected and that only a subset of mutations in specific regions will result in a CdLS phenotype.
SMC proteins contain five domains:P-loopNTPase binding globular domains at both N and C-terminal ends, two coiled coil domains separated by a hinge domain positioned in the middle of the protein. Most of the mutations (12 missense and five in-frame deletions) fall in the coiled-coil domain, whereas no mutations have been identified in the functional hinge domain, with the exception of codon 496 that is located in the hinge-coiled coil transition region [Mannini et al., 2012] suggesting that mutations therein are not tolerated in male cells that have only a single copy and are negatively selected against because no cohesin complex could be formed. However, as SMC1A is in a region that escapes X-inactivation, a hinge mutation that blocks dimerization would likely be equivalent to a null mutation, which is not necessarily lethal in females.
RAD21 Mutations in CdLS
A genome-wide-array-based copy number analysis of 290 probands with typical CdLS phenotype or overlapping features, negative for mutations in NIPBL, SMC1A, and SMC3, led to the identification of a boy with a 8q24.1 microdeletion which included RAD21 gene. The cell line carrying the microdeletion expressed approximately half the normal level of RAD21 RNA suggesting that RAD21 deletion results in haploinsufficiency. In addition, mutational screening of RAD21 exons allowed for the identification of two heterozygous de novo missense mutations, c.1127C>G (leading to p.Pro376Arg) and c.1753T>C (leading to p.Cys585Arg; Supp. Table S3). GenBank NM 006265.2 was used as RAD21 sequence reference. RAD21 interacts with the other cohesin subunits, SMC1A, SMC3, and STAG, to maintain the ring-like structure of the cohesin complex. It has been suggested that p.Pro376Arg mutation might interfere with cohesin activity by increasing the binding of STAG to RAD21, whereas the p.Cys585Arg mutation may alter the interactions at the RAD21–SMC1A interface [Deardorff et al., 2012b].
HDCA8 Mutations in CdLS
Finally, mutations in the HDAC8 gene have recently been identified in six CdLS probands [Deardorff et al., 2012a]. GenBank NM 018486.2 was used as HDAC8 sequence reference. HDAC8, located on chromosome Xq13.1, encodes for a histone deacetylase that deacetylates SMC3 during S-phase to establish cohesiveness of chromatin-loaded cohesin. Four missense and one nonsense, all de novo, have been identified (Supp. Table S4). In addition, the mutation c.1001A>G has been identified in a familial case with an affected boy, his mildly affected sister and his unaffected mother, in which the mutant allele was inactivated in her blood. Functional studies showed the complete skewing toward the normal allele in the blood of affected females arguing a strong selection against HDAC8 mutation. Furthermore, both missense mutations c.539A>G and c.958G>A led to reduced level of HDCA8 protein in fibroblasts and lymphoblastoid cells suggesting that HDAC8 mutations can cause protein instability.
Genotype–Phenotype Correlation
Mutational data analysis demonstrates an NIPBL genotype– phenotype correlation. Mutations in NIPBL are found along the entire gene, with the exceptions of exons 13 and 16 within which no point mutation have been found to our knowledge. This finding may suggest that mutations within these regions are not tolerated indicating that these exons, and the protein domains they code for, could have an important functional role that has yet to be determined. As they are the smallest exons in the NIPBL gene, it is also possible that their mutation rate is very low, if any. Nonsense, splice site and frame shift mutations leading to a truncated and presumably nonfunctional NIPBL protein are associated with a more severe phenotype (Fig. 3) characterized by typical facial features, severe-to-profound developmental and cognitive delay with lack of meaningful communication, severe growth retardation, and structural abnormalities of the limbs and other organs (Supp. Table S1). Missense mutations are, in general, associated with a milder phenotype characterized by absent limb abnormalities and with less severe developmental and growth involvement. Furthermore, some splice site mutations have been identified in probands with a moderate phenotype with typical characteristics but with reduced developmental and cognitive abilities. However, there are a few exceptions of note. Two splice site mutations (c.4320+5G>A, c.4320+2T>A) predicted to yield p.Val1414 Ala1440del with the loss of exon 19 have been identified in probands with a severe phenotype characterized by severe limb anomalies and growth retardation [Pie et al., 2010; Schoumans et al., 2007].
Figure 3.
Genotype–phenotype correlations in CdLS due to NIPBL mutations.
The clinical phenotype of probands with gross genomic rearrangements correlates to the size of the rearrangements and the number of exons involved. In general, probands carrying a deletion of one or a few exons show a mild phenotype with the exception of exon 32. The deletion of exon 32 results in a more severe phenotype presumably because the loss of this exon, encoding a portion of the H3 repeat of HEAT domain, affects NIPBL ability to interact with other proteins. Probands harboring deletions involving large portions of the NIPBL coding sequence or spanning beyond NIPBL sequence show severe cognitive and growth delay and upper limb reduction defects. However, the deletion of the last exons (from 35 to 47), results in a mild phenotype, presumably because the truncated protein maintains some residual function. Together, these data suggest that the loss of specific and larger coding sequences in NIPBL, could have a specific and additive effect on phenotypic severity.
Among unrelated probands sharing the same mutation, differences in clinical phenotype have been observed. For example, only two probands out of three carrying the p.Arg1536Term mutation showed severe limb reduction defects. Similarly probands with the deletion c.2479delAG showed limb defects ranging from small hands to limb reduction. Again, mutations affecting p.Arg2298 codon, namely Arg2298Cys, Arg2298Ser, and Arg2298His were associated with a mild phenotype, with the exception of Arg2298Leu that was identified in a proband with a more severe phenotype. These observations argue that other genetic, or environmental, factors are likely involved in modifying the CdLS phenotype, beyond the effects of the NIPBL mutations themselves.
Experimental evidences in heterozygous Drosophila Nipped-B mutants, Nipbl +/− mouse and CdLS cell lines indicate that NIPBL is tightly regulated. Although NIPBL mutations result in haploinsufficiency, there still remains approximately 70% residual expression and protein levels [Dorsett and Krantz, 2009; Kawauchi et al., 2009; Liu et al., 2009] suggesting that a further reduction is likely lethal to the organism. This notion is supported by the observation that the reduction to 50% Nipped-B mRNA levels by RNAi experiments in Drosophila was lethal to the organism [Rollins et al., 2004]. Therefore, any small environmental or genetic modifying influence that either reduces this expression level (or increases it) will potentially have a profound effect on the phenotypic outcome.
Missense mutations and in-frame deletions involving the HEAT domain, in particular H2–H4 repeats, have been identified in probands with phenotypes ranging from moderate to severe, with limb reduction, severe cognitive impairment and growth retardation. This suggests that mutations affecting the HEAT domains are critical for protein function, likely affecting NIPBL’s interaction with other proteins and/or chromatin. This notion is supported by work demonstrating that HEAT domains play a role in the interaction with histone deacetylases 1 and 3 and mutations involving the HEAT domain lead to a reduced functional interaction between NIPBL and the histone deacetylases [Jahnke et al., 2008].
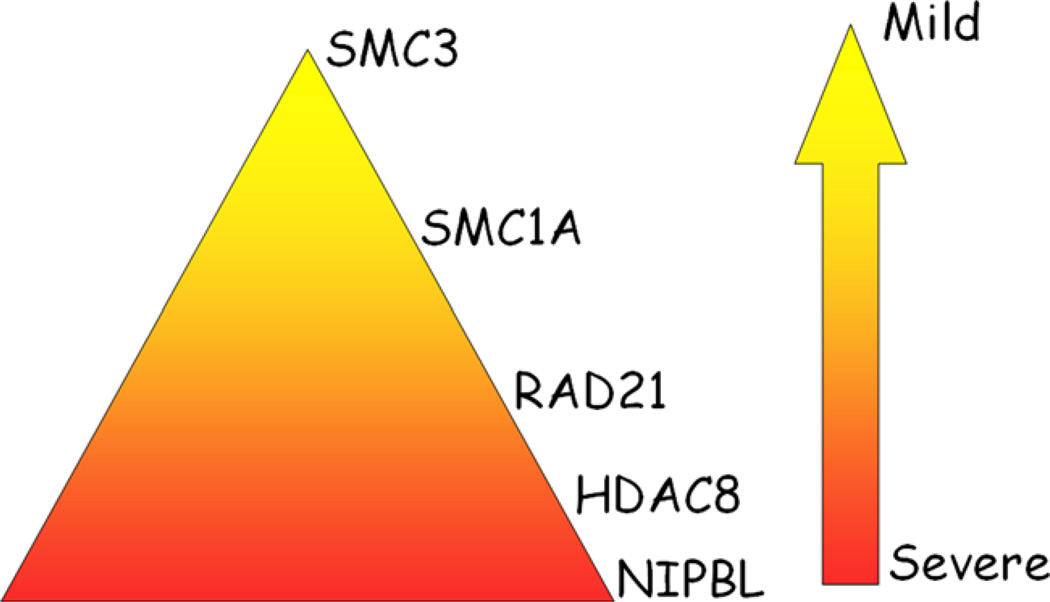
The clinical picture of CdLS probands harboring SMC mutations is more homogenous and is characterized by a mild to moderate phenotype more similar to NIPBL-mutated probands who carry missense changes (Fig. 4). In fact, probands show mild to moderate cognitive impairment, facial features with differences from classical forms of CdLS, with a tendency toward normal birth weight and head circumference and a lack of gross structural anomalies of the limbs.
Figure 4.
Diagram representing the correlation genotype–phenotype of the five CdLS causative genes.
Mutations in RAD21 cause a human cohesinopathy overlapping with the CdLS phenotype. Common features include short stature, synophrys, micrognathia, brachydactyly. The with facial features resemble CdLS but with some divergence and there is remarkably mild cognitive involvement. The finding that SMC1A and RAD21 mutation positive probands display a mild phenotype may be consistent with the observation that all identified mutations do not result in reduced mRNA expression (with the exception of the proband carrying RAD21 deletion) and preserve the protein product, ensuring the formation of the cohesin complex likely with a slightly altered function.
HDAC8 plays a critical role in deacetylating SMC3 that has been removed from chromatin during both prophase and anaphase. This removal allows cohesin to be refreshed and recycled for subsequent cell cycles where it acts in gene transcription and to establish cohesiveness. Failure ofdeacetylation does not cause a defect in unloading of cohesin but appears to cause cohesin to be less stably attached to chromatin. Although the amount of cohesin binding to chromosomes is reduced, the chromosomal residence time has not been measured. Therefore, it cannot be excluded that there is less cohesin available that is fully competent to be loaded onto chromosomes. Similarly to NIPBL haploinsufficiency, HDAC8 mutations result in a modest reduction of transcription [Deardorff et al., 2012a]. Mutations in HDAC8 result in individuals who display clinical features that overlap to some extent with classical forms of CdLS characterized by severe cognitive and growth delay and typical facial dysmorphia, however, like their SMC mutant counterparts they do not display gross limb anomalies, but may have other skeletal findings. The severity of the phenotypes seem to be correlated with the effects of the mutations (missense and nonsense), as they lead to protein instability and a reduced deacetylase activity.
Cohesin Mutations and Gene Expression
The cohesin complex, known to be the key player in sister chromatid cohesion, also plays an evolutionary conserved critical role in gene regulation. Genome-wide data show that cohesin and NIPBL (or Nipped-B in Drosophila) colocalize and bind preferentially to active genes with peaks near the transcription start sites in mammalian cells and Drosophila [Liu et al., 2009; Misulovin et al., 2008]. In addition, cohesin colocalizes with CCCTC-binding factor (CTCF) binding sites [Parelho et al., 2008; Rubio et al., 2008; Stedman et al., 2008; Wendt et al., 2008], which has multiple functions in gene regulation including its insulator role in blocking enhancer–promoter interaction [Ohlsson et al., 2010]. NIPBL is widely expressed with significant accumulations in limb bud, branchial arch and craniofacial mesenchyme [Krantz et al., 2004]. NIPBL heterozygous mutations result in the partial reduction of its expression as is recapitulated in NIPBL animal and cellular models [Kawauchi et al., 2009; Rollins et al., 2004]. A 30% reduction in expression is sufficient to cause strong effects gene expression and resultant developmental anomalies of organs and tissue where NIPBL accumulates. Increasing experimental evidence suggests that the developmental deficits seen in CdLS are the result of alterations in gene expression. It has been shown that cohesin and NIPBL cooperate in transcriptional regulation. In particular, Nipped-B facilitates the activation of both cut and Ultrabithorax homebox genes by interaction with distant enhancers [Rollins et al., 2004]. Conversely, cohesin subunit reduction increased cut expression, suggesting that cohesin has an inhibitory effect on enhancer–promoter communication and Nipped-B reduces these negative effects by cohesin removal or relocation [Dorsett, 2007]. Subsequently, it has been proposed that the opposing dominant effects of heterozygous Nipped-B and cohesin mutations on gene expression reflect a complex overlay of multiple positive and negative roles of cohesin in transcription [Schaaf et al., 2009]. It is worthy to note that cohesin binds and regulates the expression of the c-Myc gene, a key regulator of cell proliferation, in human, mouse, Drosophila and zebrafish [Kawauchi et al., 2009; Liu et al., 2009; Rhodes et al., 2010; Schaaf et al., 2009; Stedman et al., 2008]. Recently, we showed that c-Myc is the hub of dysregulated pathways and is downregulated in SMC-mutated probands suggesting that its dysregulation might the first event during CdLS phenotype development [Gimigliano et al., 2012]. NIPBL, SMC1A, and HDAC8 mutant CdLS cell lines showed an extremely conserved pattern of gene dysregulation (both up and down-regulated genes) [Deardorff et al., 2012a; Liu and Krantz, 2009]. Similarly, the Nipbl +/− mouse showed dysregulation of multiple genes with changes of twofold or less [Kawauchi et al., 2009]. Altogether these findings suggest that modest but cumulative changes in gene expression could be the underlying pathogenetic mechanism of CdLS. Partial reduction of NIPBL expression levels in CdLS could interfere with cohesin loading at regulatory binding sites affecting cohesin distribution and long range enhancer–promoter interactions.
Recent data from model organisms and human cells suggest that the manner in which cohesin regulates gene expression is more complex. In fact, it has been proposed that cohesin directly regulates transcription by multiple mechanisms, including supporting enhancer–promoter looping [Chien et al., 2011; Kagey et al., 2010; Seitan et al., 2011], and both negative and positive direct effects on transition of paused RNA polymerase to elongation [Fay et al., 2011; Schaaf et al., 2013a; Schaaf et al., 2013b]. In addition, cohesin seems to have a direct physical and functional interactions with Polycomb gene silencing complexes in Drosophila cells [Hallson et al., 2008; Schaaf et al., 2013b; Schaaf et al., 2009; Strubbe et al., 2011], which could be highly relevant to the regulation of homeotic genes that control development. Although these interactions remain to be confirmed in humans, it is possible to speculate that cohesin mutations could directly affect these interactions without affect cohesin chromosome binding and sister chromatid cohesion.
Conclusions
To date, mutations in three core cohesin subunits (SMC1A,SMC3, RAD21) and in two cohesin-regulatory proteins (NIPBL, HDAC8) have been identified in CdLS probands. Analysis of the inter- and intragenic mutational spectrum seen in CdLS probands reveals an evolving picture of genotype–phenotype correlations. NIPBL truncating mutations result in a more severe phenotype. On the contrary, NIPBL and SMC1A/SMC3 missense mutations and in-frame deletions are associated with a milder phenotype, with the exception of mutations mapped at the HEAT domain of NIPBL that have been identified in probands with a severe phenotype. Therefore, both the type of mutation and protein domain are important factors to define the clinical picture of CdLS. Although the reduction in the level of both NIPBL and HDAC8 transcripts and proteins are modest, it is sufficient to cause the developmental deficits present in CdLS. NIPBL mutant cell lines show dysregulated gene transcription and a significant reduction in cohesin binding to regulatory regions of expressed genes. This argues that mutated NIPBL affects cohesin distribution at specific loci causing their dysregulation. NIPBL acts together with cohesin. Experimental evidence shows that the downregulation of Nipped-B and cohesin alters expression of the same several hundred genes in Drosophila [Schaaf et al., 2009]. The finding that 30% reduction in NIPBL transcription and dysregulation in cohesin activity affects gene expression and leads to human disease without chromosome missegregation and defects in sister chromatid cohesion indicates that cohesin plays different roles. It is possible that low cohesin binding is necessary for correct segregation, whereas higher level for transcription regulation [Dorsett, 2011]. In this context, the acetylation/deacetylation cycle of cohesin could mark cohesin involved only in gene regulation. Further studies will be necessary to elucidate the molecular mechanisms of gene expression mediated by cohesin and associated regulatory factors and how alterations in their functions contribute to CdLS pathogenesis.
Supplementary Material
Acknowledgment
Contract grant sponsors: Region of Tuscany (AM), NIH/NICHD PO1 HD 052860 (IDK) and the Center for Cornelia de Lange Syndrome and Related Diagnoses at the Children’s Hospital of Philadelphia (IDK).
Footnotes
Additional Supporting Information may be found in the online version of this article.
Disclosure statement: The authors declare no conflict of interest.
References
- Bermudez VP, Farina A, Higashi TL, Du F, Tappin I, Takahashi TS, Hurwitz J. In vitro loading of human cohesin on DNA by the human Scc2-Scc4 loader complex. Proc Natl Acad Sci USA. 2012;109:9366–9371. doi: 10.1073/pnas.1206840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borck G, Zarhrate M, Cluzeau C, Bal E, Bonnefont JP, Munnich A, Cormier-Daire V, Colleaux L. Father-to-daughter transmission of Cornelia de Lange syndrome caused by a mutation in the 5 ’ untranslated region of the NIPBL gene. Hum Mutat. 2006;27:731–735. doi: 10.1002/humu.20380. [DOI] [PubMed] [Google Scholar]
- Brachmann W. Ein fall von symmetrischer monodaktylie durch Ulnadefekt, mit symmetrischer flughautbildung in den ellenbeugen, sowie anderen abnoanderen (zwerghaftogkeit, halsrippen, behaarung) Jarb Kinder Phys Erzie. 1916;84:225–235. [Google Scholar]
- Braunholz D, Hullings M, Gil-Rodriguez MC, Fincher CT, Mallozzi MB, Loy E, Albrecht M, Kaur M, Limon J, Rampuria A, Clark D, Kline A, et al. Isolated NIBPL missense mutations that cause Cornelia de Lange syndrome alter MAU2 interaction. Eur J Hum Genet. 2012;20:271–276. doi: 10.1038/ejhg.2011.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Miller AP, Carrel L, Rupert JL, Davies KE, Willard HF. The DXS423E gene inXp11.21 escapes Xchromosome inactivation. Hum Mol Genet. 1995;4:251–255. doi: 10.1093/hmg/4.2.251. [DOI] [PubMed] [Google Scholar]
- Castronovo P, Gervasini C, Cereda A, Masciadri M, Milani D, Russo S, Selicorni A, Larizza L. Premature chromatid separation is not a useful diagnostic marker for Cornelia de Lange syndrome. Chromosome Res. 2009;17:763–771. doi: 10.1007/s10577-009-9066-6. [DOI] [PubMed] [Google Scholar]
- Chien R, Zeng W, Kawauchi S, Bender MA, Santos R, Gregson HC, Schmiesing JA, Newkirk DA, Kong X, Ball AR, Jr, Calof AL, Lander AD, et al. Cohesin mediates chromatin interactions that regulate mammalian beta-globin expression. J Biol Chem. 2011;286:17870–17878. doi: 10.1074/jbc.M110.207365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Nasmyth K. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- de Lange C. Sur un type nouveau de dégénération (typus Amstelodamensis) Arch Méd Enfants. 1933;36:713–719. [Google Scholar]
- Deardorff MA, Bando M, Nakato R, Watrin E, Itoh T, Minamino M, Saitoh K, Komata M, Katou Y, Clark D, Cole KE, De Baere E, et al. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature. 2012a;489:313–317. doi: 10.1038/nature11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff MA, Kaur M, Yaeger D, Rampuria A, Korolev S, Pie J, Gil-Rodriguez C, Arnedo M, Loeys B, Kline AD, Wilson M, Lillquist K, et al. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of Cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet. 2007;80:485–494. doi: 10.1086/511888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff MA, Wilde JJ, Albrecht M, Dickinson E, Tennstedt S, Braunholz D, Monnich M, Yan Y, Xu W, Gil-Rodriguez MC, Clark D, Hakonarson H, et al. RAD21 mutations cause a human cohesinopathy. Am J Hum Genet. 2012b;90:1014–1027. doi: 10.1016/j.ajhg.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D. Roles of the sister chromatid cohesion apparatus in gene expression, development, and human syndromes. Chromosoma. 2007;116:1–13. doi: 10.1007/s00412-006-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D. Cohesin: genomic insights into controlling gene transcription and development. Curr Opin Genet Dev. 2011;21:199–206. doi: 10.1016/j.gde.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D, Krantz ID. On the molecular etiology of Cornelia de Lange syndrome. Ann N Y Acad Sci. 2009;1151:22–37. doi: 10.1111/j.1749-6632.2008.03450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay A, Misulovin Z, Li J, Schaaf CA, Gause M, Gilmour DS, Dorsett D. Cohesin selectively binds and regulates genes with paused RNA polymerase. Curr Biol. 2011;21:1624–1634. doi: 10.1016/j.cub.2011.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie PJ, Hirano T. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr Biol. 2004;14:1598–1603. doi: 10.1016/j.cub.2004.07.053. [DOI] [PubMed] [Google Scholar]
- Gillis LA, McCallum J, Kaur M, DeScipio C, Yaeger D, Mariani A, Kline AD, Li HH, Devoto M, Jackson LG, Krantz ID. NIPBL mutational analysis in 120 individuals with Cornelia de Lange syndrome and evaluation of genotype-phenotype correlations. Am J Hum Genet. 2004;75:610–623. doi: 10.1086/424698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimigliano A, Mannini L, Bianchi L, Puglia M, Deardorff MA, Menga S, Krantz ID, Musio A, Bini L. Proteomic profile identifies dysregulated pathways in Cornelia de Lange syndrome cells with distinct mutations in SMC1A and SMC3 genes. J Proteome Res. 2012;11:6111–6123. doi: 10.1021/pr300760p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallson G, Syrzycka M, Beck SA, Kennison JA, Dorsett D, Page SL, Hunter SM, Keall R, Warren WD, Brock HW, Sinclair DA, Honda BM. The Drosophila cohesin subunit Rad21 is a trithorax group (trxG) protein. Proc Natl Acad Sci USA. 2008;105:12405–12410. doi: 10.1073/pnas.0801698105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Anderson DE, Erickson HP, Hirano T. Bimodal activation of SMC ATPase by intra- and inter-molecular interactions. EMBO J. 2001;20:3238–3250. doi: 10.1093/emboj/20.12.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman SA, Redeker EJ, Maas SM, Mannens MM, Hennekam RC. High rate of mosaicism in individuals with Cornelia de Lange syndrome. J Med Genet. 2013;50:339–344. doi: 10.1136/jmedgenet-2012-101477. [DOI] [PubMed] [Google Scholar]
- Hulinsky R, Byrne JL, Lowichik A, Viskochil DH. Fetus with interstitial del(5)(p13.1p14.2) diagnosed postnatally with Cornelia de Lange syndrome. Am J Med Genet A. 2005;137A:336–338. doi: 10.1002/ajmg.a.30856. [DOI] [PubMed] [Google Scholar]
- Jahnke P, Xu W, Wulling M, Albrecht M, Gabriel H, Gillessen-Kaesbach G, Kaiser FJ. The Cohesin loading factor NIPBL recruits histone deacetylases to mediate local chromatin modifications. Nucleic Acids Res. 2008;36:6450–6458. doi: 10.1093/nar/gkn688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi S, Calof AL, Santos R, Lopez-Burks ME, Young CM, Hoang MP, Chua A, Lao T, Lechner MS, Daniel JA, Nussenzweig A, Kitzes L, et al. Multiple organ system defects and transcriptional dysregulation in the Nipbl(+/−) mouse, a model of Cornelia de Lange Syndrome. PLoS Genet. 2009;5:e1000650. doi: 10.1371/journal.pgen.1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline AD, Krantz ID, Sommer A, Kliewer M, Jackson LG, FitzPatrick DR, Levin AV, Selicorni A. Cornelia de Lange syndrome: clinical review, diagnostic and scoring systems, and anticipatory guidance. Am J Med Genet A. 2007;143A:1287–1296. doi: 10.1002/ajmg.a.31757. [DOI] [PubMed] [Google Scholar]
- Krantz ID, McCallum J, De Scipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, Nowaczyk MJ, Toriello H, et al. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet. 2004;36:631–635. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Krantz ID. Cohesin and human disease. Annu Rev Genomics Hum Genet. 2008;9:303–320. doi: 10.1146/annurev.genom.9.081307.164211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Krantz ID. Cornelia de Lange syndrome, cohesin, and beyond. Clin Genet. 2009;76:303–314. doi: 10.1111/j.1399-0004.2009.01271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhang Z, Bando M, Itoh T, Deardorff MA, Clark D, Kaur M, Tandy S, Kondoh T, Rappaport E, Spinner NB, Vega H, et al. Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 2009;7:e1000119. doi: 10.1371/journal.pbio.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannini L, Liu J, Krantz ID, Musio A. Spectrum and consequences of SMC1A mutations: the unexpected involvement of a core component of cohesin in human disease. Hum Mutat. 2010;31:5–10. doi: 10.1002/humu.21129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannini L, Menga S, Tonelli A, Zanotti S, Bassi MT, Magnani C, Musio A. SMC1A codon 496 mutations affect the cellular response to genotoxic treatments. Am J Med Genet A. 2012;158A:224–228. doi: 10.1002/ajmg.a.34384. [DOI] [PubMed] [Google Scholar]
- Misulovin Z, Schwartz YB, Li XY, Kahn TG, Gause M, MacArthur S, Fay JC, Eisen MB, Pirrotta V, Biggin MD, Dorsett D. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma. 2008;117:89–102. doi: 10.1007/s00412-007-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musio A, Selicorni A, Focarelli ML, Gervasini C, Milani D, Russo S, Vezzoni P, Larizza L. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet. 2006;38:528–530. doi: 10.1038/ng1779. [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Hirano T. HEAT repeats associated with condensins, cohesins, and other complexes involved in chromosome-related functions. Genome Res. 2000;10:1445–1452. doi: 10.1101/gr.147400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu DM, Huang JY, Li HY, Liu KM, Wang ST, Chen YJ, Udaka T, Izumi K, Kosaki K. Paternal gonadal mosaicism of NIPBL mutation in a father of siblings with Cornelia de Lange syndrome. Prenat Diagn. 2006;26:1054–1057. doi: 10.1002/pd.1554. [DOI] [PubMed] [Google Scholar]
- Ohlsson R, Bartkuhn M, Renkawitz R. CTCF shapes chromatin by multiple mechanisms: the impact of 20 years of CTCF research on understanding the workings of chromatin. Chromosoma. 2010;119:351–360. doi: 10.1007/s00412-010-0262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostra RJ, Baljet B, Hennekam RC. Brachmann-de Lange syndrome “avant la lettre”. Am J Med Genet. 1994;52:267–268. doi: 10.1002/ajmg.1320520303. [DOI] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Pie J, Gil-Rodriguez MC, Ciero M, Lopez-Vinas E, Ribate MP, Arnedo M, Deardorff MA, Puisac B, Legarreta J, de Karam JC, Rubio E, Bueno I, et al. Mutations and variants in the cohesion factor genes NIPBL, SMC1A, and SMC3 in a cohort of 30 unrelated patients with Cornelia de Lange syndrome. Am J Med Genet A. 2010;152A:924–929. doi: 10.1002/ajmg.a.33348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenkova E, Focarelli ML, Susani L, Paulis M, Bassi MT, Mannini L, Frattini A, Delia D, Krantz I, Vezzoni P, Jessberger R, Musio A. Cornelia de Lange syndrome mutations in SMC1A or SMC3 affect binding to DNA. Hum Mol Genet. 2009;18:418–427. doi: 10.1093/hmg/ddn369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes JM, Bentley FK, Print CG, Dorsett D, Misulovin Z, Dickinson EJ, Crosier KE, Crosier PS, Horsfield JA. Positive regulation of c-Myc by cohesin is direct, and evolutionarily conserved. Dev Biol. 2010;344:637–649. doi: 10.1016/j.ydbio.2010.05.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins RA, Korom M, Aulner N, Martens A, Dorsett D. Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol Cell Biol. 2004;24:3100–3111. doi: 10.1128/MCB.24.8.3100-3111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci USA. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo S, Masciadri M, Gervasini C, Azzollini J, Cereda A, Zampino G, Haas O, Scarano G, Di Rocco M, Finelli P, Tenconi R, Selicorni A, et al. Intragenic and large NIPBL rearrangements revealed by MLPA in Cornelia de Lange patients. Eur J Hum Genet. 2012;20:734–741. doi: 10.1038/ejhg.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CA, Kwak H, Koenig A, Misulovin Z, Gohara DW, Watson A, Zhou Y, Lis JT, Dorsett D. Genome-wide control of RNA polymerase II activity by cohesin. PLoS Genet. 2013a;9:e1003382. doi: 10.1371/journal.pgen.1003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CA, Misulovin Z, Gause M, Koenig A, Gohara DW, Watson A, Dorsett D. Cohesin and polycomb proteins functionally interact to control transcription at silenced and active genes. PLoS Genet. 2013b;9:e1003560. doi: 10.1371/journal.pgen.1003560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CA, Misulovin Z, Sahota G, Siddiqui AM, Schwartz YB, Kahn TG, Pirrotta V, Gause M, Dorsett D. Regulation of the Drosophila Enhancer of split and invected-engrailed gene complexes by sister chromatid cohesion proteins. PLoS One. 2009;4:e6202. doi: 10.1371/journal.pone.0006202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoumans J, Wincent J, Barbaro M, Djureinovic T, Maguire P, Forsberg L, Staaf J, Thuresson AC, Borg A, Nordgren A, Malm G, Anderlid BM. Comprehensive mutational analysis of a cohort of Swedish Cornelia de Lange syndrome patients. Eur J Hum Genet. 2007;15:143–149. doi: 10.1038/sj.ejhg.5201737. [DOI] [PubMed] [Google Scholar]
- Seitan VC, Banks P, Laval S, Majid NA, Dorsett D, Rana A, Smith J, Bateman A, Krpic S, Hostert A, Rollins RA, Erdjument-Bromage H, et al. Metazoan Scc4 homologs link sister chromatid cohesion to cell and axon migration guidance. PLoS Biol. 2006;4:e242. doi: 10.1371/journal.pbio.0040242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, Marks H, Adams DJ, et al. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 2011;476:467–471. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selicorni A, Russo S, Gervasini C, Castronovo P, Milani D, Cavalleri F, Bentivegna A, Masciadri M, Domi A, Divizia MT, Sforzini C, Tarantino E, et al. Clinical score of 62 Italian patients with Cornelia de Lange syndrome and correlations with the presence and type of NIPBL mutation. Clin Genet. 2007;72:98–108. doi: 10.1111/j.1399-0004.2007.00832.x. [DOI] [PubMed] [Google Scholar]
- Slavin TP, Lazebnik N, Clark DM, Vengoechea J, Cohen L, Kaur M, Konczal L, Crowe CA, Corteville JE, Nowaczyk MJ, Byrne JL, Jackson LG, et al. Germline mosaicism in Cornelia de Lange syndrome. Am J Med Genet A. 2012;158A:1481–1485. doi: 10.1002/ajmg.a.35381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strubbe G, Popp C, Schmidt A, Pauli A, Ringrose L, Beisel C, Paro R. Polycomb purification by in vivo biotinylation tagging reveals cohesin and Trithorax group proteins as interaction partners. Proc Natl Acad Sci USA. 2011;108:5572–5577. doi: 10.1073/pnas.1007916108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat Cell Biol. 2004;6:991–996. doi: 10.1038/ncb1177. [DOI] [PubMed] [Google Scholar]
- Terret ME, Sherwood R, Rahman S, Qin J, Jallepalli PV. Cohesin acetylation speeds the replication fork. Nature. 2009;462:231–234. doi: 10.1038/nature08550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet. 2004;36:636–641. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- Vrolick W. Tabulae ad illustrandam embryogenesin hominis et mammalium tam naturalem quasm abnormem. Amsterdam: Londonck; 1849. [Google Scholar]
- Watrin E, Schleiffer A, Tanaka K, Eisenhaber F, Nasmyth K, Peters JM. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr Biol. 2006;16:863–874. doi: 10.1016/j.cub.2006.03.049. [DOI] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Yan J, Saifi GM, Wierzba TH, Withers M, Bien-Willner GA, Limon J, Stankiewicz P, Lupski JR, Wierzba J. Mutational and genotype–phenotype correlation analyses in 28 Polish patients with Cornelia de Lange syndrome. Am J Med Genet A. 2006;140:1531–1541. doi: 10.1002/ajmg.a.31305. [DOI] [PubMed] [Google Scholar]
- Zhang J, Shi X, Li Y, Kim BJ, Jia J, Huang Z, Yang T, Fu X, Jung SY, Wang Y, Zhang P, Kim ST, et al. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol Cell. 2008;31:143–151. doi: 10.1016/j.molcel.2008.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.