Abstract
Background and Objectives
Tumor necrosis factor-alpha (TNF-a) was related to inflammation and involved in the development of colorectal cancer. Polymorphisms located in TNF-a promoter region, such as 308G/A and 238G/A, could affect the risk of various types of cancer by regulating TNF-a production. In this study, a meta-analysis was performed to investigate the association between common polymorphisms of TNF-a promoter region and colorectal cancer susceptibility.
Methods
Searching of several databases was performed for all publications on the association between TNF-a polymorphisms and colorectal cancer. Summary odds ratios (ORs) with their 95% confidence intervals (95% CIs) were calculated using random-effects models. Stratified analyses based on ethnicity and control population source were also conducted.
Results
Overall, TNF-a 308A polymorphism showed a significant association with increased risk of colorectal cancer in worldwide populations under homozygote comparison [AA vs. GG, OR (95% CI) = 1.46 (1.07–1.97)] other than heterozygote comparison [AG vs. GG, OR (95% CI) = 1.05 (0.93–1.19)]. TNF-a 238A was not associated with colorectal cancer risk under homozygote or heterozygote comparisons. In stratified analysis, significant association was observed only in Western populations [AA vs. GG, OR (95% CI) = 1.39 (1.01–1.91)] other than in Eastern populations under homozygote comparison. No significant difference was observed between population-based subgroup and hospital-based subgroup.
Conclusions
TNF-a 308A was moderately associated with an increased risk of colorectal cancer in Western populations, and TNF-a 238A polymorphism was not significantly associated with colorectal cancer risk.
Introduction
Colorectal cancer is the third most commonly diagnosed cancer and the fourth leading cause of cancer-related death worldwide [1]. More than 1 million new cases of colorectal cancer were diagnosed globally every year [2], and more than 715,000 deaths were resulted from colorectal cancer in 2010, nearly twice of 490,000 in 1990 [3]. Colorectal cancer is becoming an important public health problem, especially in developed countries. In United States, both incidence rate and mortality rate of colorectal cancer rank third among all cancers in both men and women, and the lifetime risk of developing colorectal cancer is about 1 in 20 (5%) [4].
Recently, a number of risk factors for colorectal cancer were identified [5]. Among them, markers of systemic inflammation, obesity, and diabetes were found to be associated with colorectal cancer risk in prospective epidemiologic studies [6].
Biological and epidemiological studies indicated a clear association between chronic inflammation and colorectal cancer [6]–[8]. Patients with inflammatory bowel disease (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC), have an increased risk of colorectal cancer [7]. Epidemiological studies also strongly suggested the involvement of genetic factors in IBD development, especially those associated with inflammation [9], [10].
Tumor necrosis factor-alpha (TNF-a) is the most important proinflammatory cytokine involved in cell growth, differentiation, and apoptosis [11], [12], which has also been reported to play a critical role in the carcinogenesis [12]. Consistent with this, a large amount of studies indicated that chronic inflammation and pro-inflammatory mediators including TNF-a may increase the risk of malignancy [9], [13]–[15]. As transcription of TNF-a is regulated by the promoter region of TNF-a gene, many studies have shown that polymorphisms located in TNF-a promoter region [such as 238 (rs361525), 308 (rs1800629), 857 (rs1799724), and 1031 (rs1799964)] could regulate TNF-a production, thus affecting the risk of cancers [16]–[18].
Recently, TNF-a 308 polymorphism was confirmed as a risk factor for a range of cancers by meta-analysis, such as gastric, breast and hepatocellular cancers [19]–[21]. However, both of the two previous meta-analyses focused on TNF-a 308 polymorphism claimed that it was not significantly associated with risk of colorectal cancer [22], [23], which appears in contradiction with the results of other cancers [19]–[21]. Considering the small sample size of both the two meta-analyses (1,708 and 1,742, respectively), whether TNF-a 308 polymorphism is associated with colorectal cancer remains inconclusive. Therefore another well-conducted, independent review of this issue was required to get a definitive conclusion.
For the association between colorectal cancer risk and other SNPs such as 238, 857, 863 and 1031, no integrated analysis has been made. In light of the heterogeneity of each published study, none of the work could achieve a reliable and consistent conclusion.
In this article, we gathered data from all previous studies of TNF-α polymorphisms and colorectal cancer, and a meta-analysis containing 3372 cases and 4523 controls was conducted to investigate whether TNF-α 308, TNF-α 238 promoter polymorphisms were associated with the risk of colorectal cancer.
Materials and Methods
Data Sources
This meta-analysis was performed according to the PRISMA guideline (Supporting information: Table S1). A comprehensive searching for all articles that had been published on the association between TNF-a polymorphisms and the risk of colorectal cancer was performed, using the following terms in the MEDLINE, PubMed, EMBASE: (‘Tumor Necrosis Factor-alpha’ [MeSH] OR ‘Tumor Necrosis’ OR TNF) AND (‘Polymorphism, Genetic’ [MeSH] OR polymorphism OR polymorphisms OR risk) AND (colorectal cancer). All articles were updated on Oct 25, 2013. References of all research and review articles were reviewed for additional references. Searching was conducted by two independent researchers to make sure that no published papers were missed.
Inclusion and Exclusion Criteria
Included studies met the following criteria: (i) case–control studies on the association between TNF-a polymorphisms and colorectal cancer; (ii) usable genotype frequencies in cases and controls provided; and (iii) reporting outcomes and risk estimates and/or presentation of data necessary for calculating ORs with 95% CIs. We excluded studies overlapping with other studies or overlapping with data from the same authors.
Data Extraction
Two researchers extracted the data independently. Items of the author’s last name, year of publication, country of origin, source of the study population, genotypes and numbers of cases and controls were extracted. The numbers of studies on the association between TNF-a 308, TNF-a 238 polymorphisms and the risk of colorectal cancer were 14, 4, respectively. Most of the studies used frequency-matched controls to cases by age and sex.
Statistical Analysis
The Hardy–Weinberg equilibrium (HWE) was tested to compare the observed genotype frequencies with expected genotype frequencies in controls of all studies. ORs and 95% CIs were calculated to assess the strength of association between TNF-a polymorphisms and risk of colorectal cancer under homozygote comparison and heterozygote comparison. Random-effects models were used to calculate overall summary ORs and 95% CIs. Study populations were classified as Western (Europe and America) or Eastern (China, Korea, India and Iran).
The significance of the summary ORs was determined by the Z-test, in which two-sided p<0.05 was considered as statistically significant. The Q-statistic was calculated to examine result heterogeneity among studies, and p<0.1 was considered significant. The I2-statistic was also calculated to efficiently test heterogeneity [24], with I2<25%, 25–75% and >75% considered to represent low, moderate and high degree of inconsistency, respectively. Begg’s funnel plot was plotted to examine the underlying publication bias [25]. For sensitivity analysis, relatively smaller studies were excluded and the summary ORs (95% CIs) were recalculated. All analyses were done using STATA 12.0 (STATA Corporation, College Station, TX, USA).
Results
Characteristics of Studies
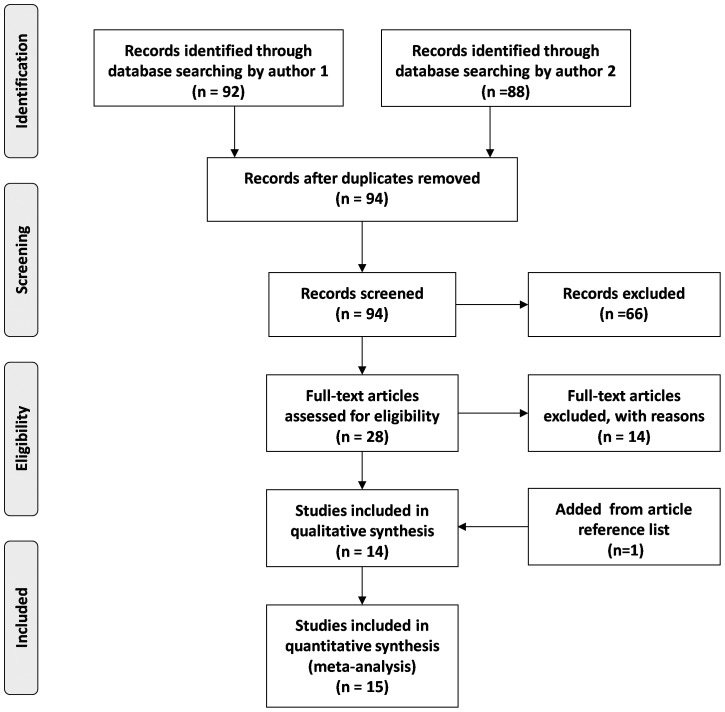
In this article, 15 studies were identified to evaluate the relationship between TNF-a polymorphisms and risk of colorectal cancer, and a total number of 3372 cases and 4523 controls were included. Detailed screening process was shown in Figure 1. The most commonly investigated polymorphisms were TNF-a 308 and TNF-a 238, which were reported in 14 and 4 studies, respectively [2], [26]–[39]. Other SNPs, such as TNF-a 857, TNF-a 863, and TNF-a 1031 were also investigated in this comprehensive searching, but the corresponding sample sizes were too small to perform quantitative data synthesis. Genotype and allele distributions of TNF-a 308 and TNF-a 238 were shown in Table 1. Median frequencies of TNF-a 308A allele were 15.0% in Western populations and 7.1% in Eastern populations. Corresponding frequencies for TNF-a 238A allele were 3.5% and 7.1%, respectively.
Figure 1. Flow diagram of included and excluded studies in meta-analysis.
Table 1. Study characteristics.
| First author | year | Loc. | case | control | controlsource | 308-Afrequency | p HWE | 238-Afrequency | p HWE |
| Madani | 2008 | W | 51 | 46 | PB | – | – | 1.09% | 0.94 |
| Park | 1998 | E | 139 | 324 | PB | 7.07% | 0.79 | – | – |
| Jang | 2001 | E | 27 | 92 | HB | 3.80% | 0.85 | 7.07% | 0.39 |
| Landi | 2003 | W | 363 | 320 | HB | 15.00% | 0.70 | – | – |
| Macarthur | 2005 | W | 246 | 389 | PB | 23.78% | 0.63 | – | – |
| Fei | 2005 | E | 92 | 132 | PB | 7.95% | 0.78 | – | – |
| Theodoropoulos | 2006 | W | 222 | 200 | PB | 16.00% | 0.69 | – | – |
| Gunter | 2006 | W | 217 | 202 | PB | 17.08% | 0.68 | – | – |
| Toth | 2007 | W | 183 | 141 | PB | 10.64% | 0.74 | – | – |
| Wu | 2008 | W | 156 | 116 | PB | 14.66% | 0.70 | 3.73% | 0.32 |
| Wang | 2008 | E | 344 | 669 | PB | 8.89% | 0.77 | – | – |
| Garrity-Park | 2008 | W | 114 | 114 | HB | 10.53% | 0.75 | 3.51% | 0.02 |
| Tsilidis | 2009 | W | 204 | 372 | PB | 13.98% | 0.71 | – | – |
| Suchy | 2008 | W | 350 | 350 | PB | 15.57% | 0.69 | – | – |
| Li | 2011 | E | 180 | 180 | HB | 5.83% | 0.60 | – | – |
Year: Publication year; Loc.: Location of the population; E: Eastern country; W: Western country; p HWE: p value of Hardy–Weinberg equilibrium, chi-square test; 308-A%: Percentage of 308-A allele frequency among controls; 238-A%: Percentage of 238-A allele frequency among controls.
Quantitative Data Synthesis
For TNF-a 308G/A polymorphism, 14 studies with a total number of 2837 cases and 3601 controls were included in this analysis. Figure 2A showed the random-effects overall OR (95% CIs) of TNF-a 308 polymorphism under homozygote comparison [AA vs. GG, OR (95% CI) = 1.46 (1.07–1.97)]. Figure 2B showed the random-effects overall OR (95% CIs) of TNF-a 308 polymorphism under heterozygote comparison [AG vs. GG, OR (95% CI) = 1.05 (0.93–1.19)].
Figure 2. The association between TNF-a 308 polymorphism and risk of colorectal cancer.
A: AA vs. GG, Forest plot; B: AG vs. GG, Forest plot; ORs, odds ratios.
When stratified by ethnicity, for Western populations, a significant association between TNF-a 308 and colorectal cancer risk was observed under homozygote comparison [AA vs. GG, OR (95% CI) = 1.39 (1.01–1.91)], but not heterozygote comparison [AG vs. GG, OR (95% CI) = 1.04 (0.94–1.15)]. For Eastern populations, no significant association was found under homozygote or heterozygote comparison. Subgroups characterized by control population source (hospital-based, HB or population-based, PB), and neoplasm location (colon or rectum) were also analyzed with the same method as above (Table 2). Both homozygote and heterozygote comparisons displayed significant different risk of colorectal cancer in HB subgroup [AA vs. GG, OR (95% CI) = 2.16 (1.17–4.00)] but not in PB subgroup [AA vs. GG, OR (95% CI) = 1.28 (0.90–1.82)].
Table 2. Overall and group-specific summary statistics for TNF-a 308, TNF-a 238 in colorectal cancer.
| SNPs | Number of studies | Comparison | Test of association[OR (95% CI)] | Test of heterogeneity | ||
| P-value (Z test) | I2(%) | P-value | ||||
| 308 | ||||||
| Hospital based | 3 | AA vs. GG | 2.16 (1.17–4.00) | 0.01 | 86% | <0.001 |
| 4 | AG vs. GG | 1.19 (0.97–1.47) | 0.10 | 84% | <0.001 | |
| Population based | 9 | AA vs. GG | 1.28 (0.90–1.82) | 0.18 | 0% | 0.89 |
| 9 | AG vs. GG | 1.00 (0.90–1.10) | 0.95 | 23% | 0.23 | |
| Colon cancer | 3 | AA vs. GG | 1.22 (0.71–2.08) | 0.48 | 64% | 0.06 |
| 3 | AG vs. GG | 0.85 (0.70–1.02) | 0.09 | 0% | 0.66 | |
| Rectal cancer | 2 | AA vs. GG | 0.66 (0.27–1.60) | 0.35 | 46% | 0.17 |
| 2 | AG vs. GG | 0.88 (0.68–1.14) | 0.33 | 0% | 0.86 | |
| 238 | ||||||
| Hospital based | 1 | AA vs. GG | 0.33 (0.01–8.02) | 0.50 | 0% | 0.10 |
| 1 | AG vs. GG | 0.83 (0.26–2.63) | 0.75 | 0% | <0.001 | |
| Population based | 3 | AA vs. GG | 2.46 (0.32–19.11) | 0.39 | 12% | 0.44 |
| 3 | AG vs. GG | 0.75 (0.47–1.19) | 0.22 | 0% | 0.37 | |
CI: confidence interval; OR: odds ratio; TNF-a: tumor-necrosis factor-a.
For TNF-a 238 G/A polymorphism, we included 4 studies with a total number of 535 cases and 922 controls. Overall, no significant association was found between TNF-a 238G/A polymorphism and the risk of colorectal cancer under homozygote comparison [AA vs. GG, OR (95% CI) = 1.23 (0.24–6.23); Figure 3A] or heterozygote comparison [AG vs. GG, OR (95% CI) = 0.74 (0.47–1.17), Figure 3B]. Since there are only 4 studies recruited in this study, no stratified analysis was illustrated in the form of figures. Those results were displayed in the table along with results of TNF-a 308 instead (Table 2).
Figure 3. The association between TNF-a 238 polymorphism and risk of colorectal cancer.
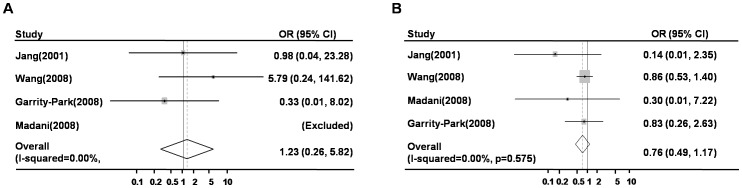
A: AA vs. GG, Forest plot; B: AG vs. GG, Forest plot; ORs, odds ratios.
Test of Heterogeneity
For the overall meta-analysis, the Q-statistic was significant and I2 showed a moderate variation for TNF-a 308 (AA vs. GG, I2 = 40.9%, p = 0.07; AG vs. GG, I2 = 60.3%, p = 0.01), and no variation for TNF-a 238 (AA vs. GG, I2 = 0.0%, p = 0.46; AG vs. GG, I2 = 0.0%, p = 0.58).
In the stratified analyses of TNF-a 308, I2 showed moderate variations in both Eastern (AA vs. GG, I2 = 51.7%, p = 0.13) and Western populations (AA vs. GG, I2 = 43.8%, p = 0.08). For PB and HB subgroups, I2 showed no variation (AA vs. GG, I2 = 0.0%, p = 0.884) and a strong variation (AA vs. GG, I2 = 86.9%, p<0.001), respectively, which indicated that hospital-based studies might be important source of heterogeneity. Moreover, variation in the frequency of this TNF-a 308 polymorphism among the populations or some unidentified factors may also be the source of heterogeneity.
Sensitivity Analysis and Publication Bias
To further strengthen our conclusions, the sensitivity analysis was performed. A single relatively small study involved in the meta-analysis was excluded each time. For TNF-a 308, the corresponding pooled ORs were not changed significantly (ORs ranged from 1.20 to 1.54 under homozygote comparison, from 1.00 to 1.12 under heterozygote comparison, detailed data not shown), indicating that our results were statistically robust. Sensitivity analysis was not performed for TNF-a 238 because there were only 4 studies included.
For publication bias investigation, Begg’s funnel plot and Egger’s weighted regression were performed for the association between TNF-a polymorphisms and colorectal cancer. No evidence for bias was detected in TNF-a 308 polymorphism under homozygote comparison (Figure 4A. Begg’s test p = 0.325, Egger’s test p = 0.180). However, evidence of publication bias was found in the heterozygote comparison (Figure 4B. Begg’s test p = 0.024, Egger’s test p = 0.063). Publication bias investigation was not performed for TNF-a 238 because there were only 4 studies included.
Figure 4. Publication bias tests for the association between TNF-a 308 polymorphism and colorectal cancer.
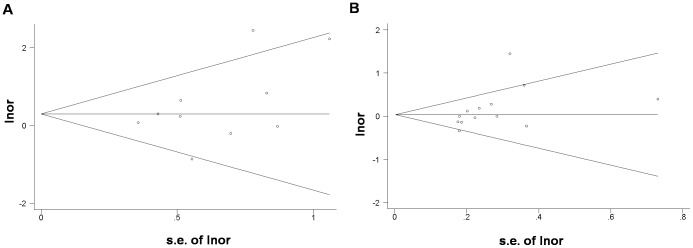
A: AA vs. GG, Begg’s funnel plots; B: AG vs. GG, Begg’s funnel plots.
Discussion
Inflammation is one of key factors involved in carcinogenesis, and people with inflammatory bowel disease are at high risk of colorectal cancer [7]. Therefore polymorphisms of inflammation-related genes have been regarded as potential sources of cancer risk biomarkers [5]. TNF-a, the most extensively studied inflammatory factor in cancer, was proved to affect colorectal tumorigenesis through different pathways [11], [12]. Previous studies indicated that TNF-a-related cellular functions were greatly influenced by polymorphisms in the promoter region of TNF-a gene [16]–[18].
308G/A is the most commonly studied SNP site of TNF-a promoter region. Various meta-analyses indicated that TNF-a 308A polymorphism was associated with an elevated risk of gastric cancer [19], breast cancer [20], cervical cancer [40] and lung cancer [41]. However, for colorectal cancer, two previous meta-analyses reported that TNF-a 308 polymorphism was not associated with colorectal cancer risk [22], [23], and one of these two meta-analyses also demonstrated that TNF-a 308A polymorphism was associated with reduced risk of colorectal cancer in Western population [22]. Additionally, another systematic review focused on field synopsis of genetic association also included a meta-analysis reporting TNF-a 308 polymorphism was not associated with colorectal cancer risk [42]. These results were distinct from nearly all other types of cancer, and were hardly explained by the mechanism of TNF-a. In our study, 2725 cases and 3369 controls from 14 independent studies were included, and the previous conclusion was not supported by our study. According to our study, TNF-a 308A polymorphism was associated with an elevated risk of colorectal cancer under homozygote comparison. This result was in accordance with the meta-analysis of other cancers, and also could be explained by the previous mechanistic study of TNF-a [43], which reported that in humans, the presence of a promoter polymorphism of TNF-a 308A is associated with increased plasma TNF-a concentration, which may lead to an increased risk of cancers.
However, the association between TNF-a 308 polymorphism and colorectal cancer risk became less significant under heterozygote comparison. Compared with AA genotype, the lack of association between TNF-a 308 AG genotype and increased risk of colorectal cancer could be explained by haploinsufficiency effect, that is, the elevated plasma TNF-a concentrations resulted from a single TNF-a 308 A allele might be not high enough to strongly affect cancer risk. Sensitivity analysis indicated that when excluded each study involved in the meta-analysis, ORs under heterozygote comparison stayed over 1.00, suggesting that if more studies were included in the future analysis, the association between TNF-a 308 AG genotype and increased risk of colorectal cancer might also become statistically significant.
Comparable ORs were observed when analyses were stratified to Eastern and Western populations, indicating that there was no obvious race-specific effect in this regard. However, since the sample size of Eastern populations is relatively small, the association between TNF-a 308A and risk of colorectal cancer was not significant. Strictly, we can only conclude that TNF-a 308A was moderately associated with an increased risk of colorectal cancer in Western populations.
As it is shown in Table 2, AA vs. GG comparison results were significant in HB subgroup but not in PB subgroup, which suggested that control source might affect the association between TNF-a 308 polymorphism and colorectal risk in Hospital-Based study. Together, we concluded that TNF-a 308A polymorphism was a general risk factor of colorectal cancer.
TNF-a 238G/A is the second commonly studied SNP site following 308 among all polymorphisms located in the promoter region of TNF-a gene. However, recently no integrated analysis has been made to get a definitive conclusion of whether TNF-a 238G/A is associated with colorectal cancer. Up to now, among all publications, 4 studies investigated the association between TNF-a 238 polymorphism and risk of colorectal cancer, but yielded contradictory results. In our meta-analysis, 535 cases and 922 controls were enrolled and we found that TNF-a 238 polymorphism was not significantly associated with colorectal cancer risk Previously, a meta-analysis including 34 studies with 34,679 cases and 41,186 controls reported that no significant association was found between the TNF-a 238 polymorphism and the overall cancer risk [44], which is in conformity with our present results. However, the number of current studies on TNF-a 238 polymorphism and risk of colorectal cancer is relatively small, thus investigations involving more cases are needed in the future.
Genome-Wide Association Study (GWAS) is also a powerful methodology in genetic studies. As for GWAS studies of colorectal cancer [45]–[48], there is no evidence of association with the TNF-a region. Compared with GWAS studies, meta-analyses are easy to be influenced by sample heterogeneity and inaccuracies in a single study. However, GWAS studies also have demerits. To control the cost, at the first step of GWAS studies, researchers always screen for significant SNP in the whole genome including more than 500,000 SNPs in a small population, and it couldn’t have enough power to identify all SNPs associated with a disease. For example, one GWAS study only identified six possible risk alleles for colorectal cancer [45], which seems too few for such a complex disease. So no evidence of association with the TNF-a region derived from GWAS does not necessarily mean that all SNPs in this region have no association with risk of colorectal cancer. We speculated that, similar to Barton et al. reported on Rheumatoid Arthritis [49], expanding the screen criteria at the first step of GWAS may improve the testing power and generate more SNPs associated with colorectal cancer, and TNF-a 308 polymorphism might be among them. However, for the original GWAS data are not published, we couldn’t verify such speculation. On the other hand, a well-conducted meta-analysis is equal to a multi-centered, randomized, and controlled study, which could also generate a credible conclusion. Even in the GWAS era, combining available information to generate an integrated result is still reasonable and can save a considerable amount of resources.
Several limitations of this study should also be addressed. Firstly, the sample size was relatively small for stratified analyses, which weakened our conclusions, especially under homozygote comparison. Therefore more studies were needed to be included to obtain a more reliable result. Secondly, detailed information of individual level was lacking in this analysis, for which many stratified analyses were not able to be performed. If individual raw data were available, effect induced by age, gender, drug use and other environmental factors could also be investigated. Thirdly, cytokines, such as interleukins, transform growth factor, tumor necrosis factor, may exert interacting functions with each other, so SNPs of other cytokines should also be taken into account to conclude a true effect if possible. Fourthly, results of GWAS studies were not included because the raw data had not been published. Additionally, gene-gene, gene-phenotype, and gene-environment interactions should be considered.
In summary, this meta-analysis had pooled all the available data related to TNF-a polymorphisms and colorectal cancer, and indicated that TNF-a 308 was moderately associated with an increased risk of colorectal cancer in Western populations. TNF-a 238 polymorphism was not significantly associated with colorectal cancer risk. Additionally, large well-designed cohort studies would warrant to confirm this conclusion, and to fully understand the molecular mechanism of colorectal cancer. Further prospective studies in combination with analyses of other cytokines and environmental factors are also required.
Supporting Information
PRISMA Checklist of items to include when reporting a systematic review or meta-analysis.
(DOC)
Acknowledgments
We deeply appreciate Dr. Zhi-Rong Yang (Peking University Center for Evidence-Based Medicine and Clinical Research) and Dr. Qi-Yan Wang (Departments of Genetics, Peking University Cancer Hospital & Institute) for providing revision suggestions.
Funding Statement
This work was supported by the Doctoral Program of Higher Education of China (20110001110050) and National 973 Program of China (2009CB521805). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jemal A, Bray F (2011) Center MM, Ferlay J, Ward E, et al (2011) Global cancer statistics. CA: A Cancer Journal for Clinicians 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 2. Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, et al. (2010) Colorectal cancer. Lancet 375: 1030–1047. [DOI] [PubMed] [Google Scholar]
- 3. Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, et al. (2012) Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380: 2095–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Siegel R, Naishadham D, Jemal A (2012) Cancer statistics, 2012. CA: A Cancer Journal for Clinicians 62: 10–29. [DOI] [PubMed] [Google Scholar]
- 5.Arem H, Mayne ST, Sampson J, Risch H, Stolzenberg-Solomon RZ (2013) Dietary fat intake and risk of pancreatic cancer in the Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial. Ann Epidemiol. [DOI] [PMC free article] [PubMed]
- 6. Yehuda-Shnaidman E, Schwartz B (2012) Mechanisms linking obesity, inflammation and altered metabolism to colon carcinogenesis. Obes Rev 13: 1083–1095. [DOI] [PubMed] [Google Scholar]
- 7.Nieminen U, Jussila A, Nordling S, Mustonen H, Farkkila MA (2013) Inflammation and disease duration have a cumulative effect on the risk of dysplasia and carcinoma in IBD: A case-control observational study based on registry data. Int J Cancer. [DOI] [PubMed]
- 8. Wang D, DuBois RN (2013) The role of anti-inflammatory drugs in colorectal cancer. Annu Rev Med 64: 131–144. [DOI] [PubMed] [Google Scholar]
- 9. Li M, You Q, Wang X (2011) Association between polymorphism of the tumor necrosis factor alpha-308 gene promoter and colon cancer in the Chinese population. Genet Test Mol Biomarkers 15: 743–747. [DOI] [PubMed] [Google Scholar]
- 10. Garrity-Park M, Loftus EV Jr, Sandborn WJ, Smyrk TC (2012) Myeloperoxidase immunohistochemistry as a measure of disease activity in ulcerative colitis: association with ulcerative colitis-colorectal cancer, tumor necrosis factor polymorphism and RUNX3 methylation. Inflamm Bowel Dis 18: 275–283. [DOI] [PubMed] [Google Scholar]
- 11. Waters JP, Pober JS, Bradley JR (2013) Tumour necrosis factor in infectious disease. J Pathol 230: 132–147. [DOI] [PubMed] [Google Scholar]
- 12. Aggarwal BB, Gupta SC, Kim JH (2012) Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey. Blood 119: 651–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gupta R, Sharma SC, Das SN (2008) Association of TNF-alpha and TNFR1 promoters and 3′ UTR region of TNFR2 gene polymorphisms with genetic susceptibility to tobacco-related oral carcinoma in Asian Indians. Oral Oncol 44: 455–463. [DOI] [PubMed] [Google Scholar]
- 14. Guo W, Wang N, Li Y, Zhang JH (2005) Polymorphisms in tumor necrosis factor genes and susceptibility to esophageal squamous cell carcinoma and gastric cardiac adenocarcinoma in a population of high incidence region of North China. Chin Med J (Engl) 118: 1870–1878. [PubMed] [Google Scholar]
- 15. Hong Y, Ge Z, Jing C, Shi J, Dong X, et al. (2013) Functional Promoter -308G>A Variant in Tumor Necrosis Factor alpha Gene Is Associated with Risk and Progression of Gastric Cancer in a Chinese Population. PLoS One 8: e50856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huizinga TW, Westendorp RG, Bollen EL, Keijsers V, Brinkman BM, et al. (1997) TNF-alpha promoter polymorphisms, production and susceptibility to multiple sclerosis in different groups of patients. J Neuroimmunol 72: 149–153. [DOI] [PubMed] [Google Scholar]
- 17. Hellmig S, Fischbach W, Goebeler-Kolve ME, Folsch UR, Hampe J, et al. (2005) A functional promoter polymorphism of TNF-alpha is associated with primary gastric B-Cell lymphoma. Am J Gastroenterol 100: 2644–2649. [DOI] [PubMed] [Google Scholar]
- 18. Lindholm E, Bakhtadze E, Cilio C, Agardh E, Groop L, et al. (2008) Association between LTA, TNF and AGER polymorphisms and late diabetic complications. PLoS One 3: e2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gorouhi F, Islami F, Bahrami H, Kamangar F (2008) Tumour-necrosis factor-A polymorphisms and gastric cancer risk: a meta-analysis. British Journal of Cancer 98: 1443–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang Y, Feng R, Bi S, Xu Y (2011) TNF-alpha polymorphisms and breast cancer. Breast Cancer Res Treat 129: 513–519. [DOI] [PubMed] [Google Scholar]
- 21. Yang Y, Luo C, Feng R, Bi S (2011) The TNF-alpha, IL-1B and IL-10 polymorphisms and risk for hepatocellular carcinoma: a meta-analysis. J Cancer Res Clin Oncol 137: 947–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang J, Cao C, Luo H, Xiong S, Xu Y, et al. (2011) Tumour necrosis factor alpha -308G/A polymorphism and risk of the four most frequent cancers: a meta-analysis. International Journal of Immunogenetics 38: 311–320. [DOI] [PubMed] [Google Scholar]
- 23. Chen Z, Zhu L, Zhang J, Xu H, Chen X, et al. (2012) TNF-α-308 G>A polymorphism and colorectal cancer risk: a meta-analysis. International Journal of Colorectal Disease 28: 431–432. [DOI] [PubMed] [Google Scholar]
- 24. Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sterne JA, Egger M, Smith GD (2001) Systematic reviews in health care: Investigating and dealing with publication and other biases in meta-analysis. BMJ 323: 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Park KS, Mok JW, Rho SA, Kim JC (1998) Analysis of TNFB and TNFA NcoI RFLP in colorectal cancer. Mol Cells 8: 246–249. [PubMed] [Google Scholar]
- 27. Jang WH, Yang YI, Yea SS, Lee YJ, Chun JH, et al. (2001) The -238 tumor necrosis factor-alpha promoter polymorphism is associated with decreased susceptibility to cancers. Cancer Lett 166: 41–46. [DOI] [PubMed] [Google Scholar]
- 28. Landi S, Moreno V, Gioia-Patricola L, Guino E, Navarro M, et al. (2003) Association of common polymorphisms in inflammatory genes interleukin (IL)6, IL8, tumor necrosis factor alpha, NFKB1, and peroxisome proliferator-activated receptor gamma with colorectal cancer. Cancer Res 63: 3560–3566. [PubMed] [Google Scholar]
- 29. Macarthur M, Sharp L, Hold GL, Little J, El-Omar EM (2005) The role of cytokine gene polymorphisms in colorectal cancer and their interaction with aspirin use in the northeast of Scotland. Cancer Epidemiol Biomarkers Prev 14: 1613–1618. [DOI] [PubMed] [Google Scholar]
- 30. Gunter MJ (2006) Inflammation-Related Gene Polymorphisms and Colorectal Adenoma. Cancer Epidemiology Biomarkers & Prevention 15: 1126–1131. [DOI] [PubMed] [Google Scholar]
- 31. Theodoropoulos G, Papaconstantinou I, Felekouras E, Nikiteas N, Karakitsos P, et al. (2006) Relation between common polymorphisms in genes related to inflammatory response and colorectal cancer. World J Gastroenterol 12: 5037–5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Toth EK, Kocsis J, Madaras B, Biro A, Pocsai Z, et al. (2007) The 8.1 ancestral MHC haplotype is strongly associated with colorectal cancer risk. Int J Cancer 121: 1744–1748. [DOI] [PubMed] [Google Scholar]
- 33. Garrity-Park MM, Loftus EV Jr, Bryant SC, Sandborn WJ, Smyrk TC (2008) Tumor necrosis factor-alpha polymorphisms in ulcerative colitis-associated colorectal cancer. Am J Gastroenterol 103: 407–415. [DOI] [PubMed] [Google Scholar]
- 34. Madani S, Noorinayer B, Madani H, Sadrolhefazi B, Molanayee S, et al. (2008) No association between TNF-alpha-238 polymorphism and colorectal cancer in Iranian patients. Acta Oncol 47: 473–474. [DOI] [PubMed] [Google Scholar]
- 35. Suchy J, Klujszo-Grabowska E, Kladny J, Cybulski C, Wokolorczyk D, et al. (2008) Inflammatory response gene polymorphisms and their relationship with colorectal cancer risk. BMC Cancer 8: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu GY, Wang XM, Keese M, Hasenberg T, Sturm JW (2008) [Association between tumor necrosis factor alpha gene polymorphism and colorectal cancer]. Zhonghua Wei Chang Wai Ke Za Zhi 11: 569–571. [PubMed] [Google Scholar]
- 37. Tsilidis KK, Helzlsouer KJ, Smith MW, Grinberg V, Hoffman-Bolton J, et al. (2009) Association of common polymorphisms in IL10, and in other genes related to inflammatory response and obesity with colorectal cancer. Cancer Causes & Control 20: 1739–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fei BY, Lu HX, Chen YW, Yang JM (2005) TNF-alpha and IL-1 genetic polymorphisms in colorectal cancer. Chin J Dig 25: 661–662. [Google Scholar]
- 39. Wang AZ, Zhu ZZ (2008) Association of TNF gene polymorphism wuth colorectal cancer risk. Journal of Tongji University 29: 24–27. [Google Scholar]
- 40. Zhang HL, Zhang YJ (2013) A systemic assessment of the association between tumor necrosis factor alpha 308 G/A polymorphism and risk of cervical cancer. Tumour Biol 34: 1659–1665. [DOI] [PubMed] [Google Scholar]
- 41. Peng WJ, He Q, Yang JX, Wang BX, Lu MM, et al. (2012) Meta-analysis of association between cytokine gene polymorphisms and lung cancer risk. Mol Biol Rep 39: 5187–5194. [DOI] [PubMed] [Google Scholar]
- 42. Theodoratou E, Montazeri Z, Hawken S, et al. (2012) Systematic meta-analyses and field synopsis of genetic association studies in colorectal cancer. J Natl Cancer Inst 104: 1433–1457. [DOI] [PubMed] [Google Scholar]
- 43. Popa C, Netea MG, van Riel PLCM, van der Meer JWM, Stalenhoef AFH (2007) The role of TNF- in chronic inflammatory conditions, intermediary metabolism, and cardiovascular risk. J Lipid Res 48: 751–762. [DOI] [PubMed] [Google Scholar]
- 44. Zhou P, Lv GQ, Li CW, Du LF, et al. (2011) The TNF-Alpha-238 Polymorphism and Cancer Risk: A Meta-Analysis. PLoS One 6: e22092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tenesa A, Farrington SM, Prendergast JGD, et al. (2008) Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet 40: 631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tomlinson IPM, Webb E, Carvajal-Carmona L, et al. (2008) A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet 40: 623–630. [DOI] [PubMed] [Google Scholar]
- 47. Figueiredo JC, Lewinger JP, Song C, et al. (2011) Genotype–environment interactions in microsatellite stable/microsatellite instability-low colorectal cancer: results from a genome-wide association study. Cancer Epidem Biomark Prev 20: 758–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zanke BW, Greenwood CMT, Rangrej J, et al. (2007) Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet 39: 989–994. [DOI] [PubMed] [Google Scholar]
- 49. Barton A, Thomson W, Ke X, et al. (2008) Re-evaluation of putative rheumatoid arthritis susceptibility genes in the post-genome wide association study era and hypothesis of a key pathway underlying susceptibility. Human molecular genetics. Hum Mol Genet 17: 2274–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA Checklist of items to include when reporting a systematic review or meta-analysis.
(DOC)